ABSTRACT
Colorectal cancer (CRC) is the third most commonly diagnosed cancer, the third leading cause of cancer-related deaths, and has been on the rise among young adults in the United States. Research has established that the colonic microbiome is different in patients with CRC compared to healthy controls, but few studies have investigated if and how the microbiome may relate to CRC progression through the serrated pathway versus the adenoma-carcinoma sequence.
Our view is that progress in CRC microbiome research requires consideration of how the microbiome may contribute to CRC carcinogenesis through the distinct pathways that lead to CRC, which could enable the creation of novel and tailored prevention, screening, and therapeutic interventions. We first highlight the limitations in existing CRC microbiome research and offer corresponding solutions for investigating the microbiome’s role in the adenoma-carcinoma sequence and serrated pathway. We then summarize the findings in the select human studies that included data points related to the two major carcinogenic pathways. These studies investigate the microbiome in CRC carcinogenesis and 1) utilize mucosal samples and 2) compare polyps or tumors by histopathologic type, molecular/genetic type, or location in the colon.
Key findings from these studies include: 1) Fusobacterium is associated with right-sided, more advanced, and serrated lesions; 2) the colons of people with CRC have bacteria typically associated with normal oral flora; and 3) colons from people with CRC have more biofilms, and these biofilms are predominantly located in the proximal colon (single study).
KEYWORDS: Colorectal cancer, microbiome, serrated, adenoma, carcinogenesis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer, the third leading cause of cancer-related deaths1, and is increasing in young adults in the United States.2 It is estimated that over 50% of the screening-age population (≥50 years) have one or more precancerous adenomas or polyps.
The Cancer Genome Atlas project, utilizing extensive genomic and transcriptomic characterization of colorectal cancer, has proposed the classification of CRC into two major groups: tumors with microsatellite instability (MSI) and tumors with chromosomal instability.3 Cancers with microsatellite instability largely result from defective DNA mismatch repair caused by inactivating mutations or epigenetic silencing of mismatch repair genes such as the MLH1 tumor suppressor gene. Epigenetic silencing frequently occurs due to CpG island promoter methylation of MLH1. A majority of tumors arising from this pathway, termed the serrated pathway, also have BRAF V600E mutations, and a minority have DNA polymerase Epsilon or Delta 1 mutations. In contrast, cancers with chromosomal instability have alterations in chromosome number and activating mutations in oncogenes K-ras, PIK3CA or inactivating mutations in tumor suppressor genes Apc, p53 and SMAD4. These are the hallmark alterations seen in tumors that arise by the adenoma-carcinoma sequence, first characterized by Fearon and Vogelstein.4
While the majority of CRC develops through the adenoma-carcinoma sequence, up to a third of CRC develops via the serrated pathway.5 Precursor lesions of the serrated pathway or an alternate pathway include a broad group of serrated polyps, including benign hyperplastic polyps (HPs), precancerous traditional serrated adenomas (TSAs) and sessile serrated polyps (SSPs). TSAs may exhibit elements of the adenoma-carcinoma sequence such as K-ras mutation, or may have BRAF mutation6 and CpG island hypermethylation (a signature of the sessile serrated pathway7). TSAs are typically found in the left (distal) colon.8 In contrast, SSPs rarely demonstrate elements of the chromosomal instability pathway but frequently have BRAF mutations and appear to progress toward dysplasia and carcinoma as a result of microsatellite instability due to MLH1 promoter CpG island hypermethylation. SSPs are found in the right (proximal) side of the colon 80% of the time,9 consistent with findings that tumors characterized by BRAF mutations, microsatellite instability and CpG island hypermethylation phenotype (CIMP) have shown a linear increase in frequency from distal to proximal colon.10 A summary of the major pathways of carcinogenesis can be seen in Figure 2.
Figure 2.
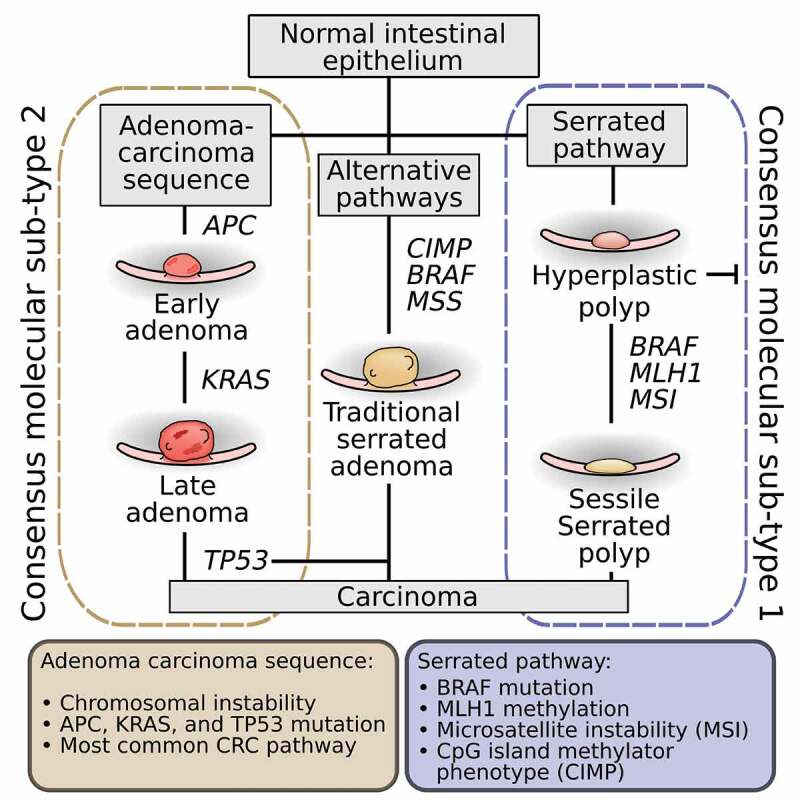
It is not yet known what factors influence the progression of a CRC precursor through the adenoma-carcinoma sequence, serrated pathway or an alternate pathway. It is tempting to hypothesize that the microbiome may play a role, but most research has focused on the microbiome alterations in patients with CRC or precursor polyps without further specification of the histopathologic, genetic, or epigenetic type. This review will briefly summarize the proposed mechanisms of how the microbiome contributes to CRC from existing research, the most common limitations in this research, and offer corresponding solutions. We then summarize the key findings in the select studies that have added data points related to the major carcinogenic pathways to CRC.
Mechanisms of the microbiome in CRC carcinogenesis
Research has established the importance of diet and lifestyle in CRC carcinogenesis.11 The microbiome, intimately related to the environmental factors linked to CRC, has been postulated to play a role in CRC carcinogenesis since the 1960s. While single bacterial strains have been associated with CRC Table 1, the current belief is that intestinal microbial dysbiosis and a subsequent inappropriate or altered immune response can confer a predisposition to chronic inflammation,17–19 which is known to contribute to the development of disease and cancer. Microbes may contribute to genetic and epigenetic alterations via the production of superoxide radicals and genotoxins and toll-like receptor mediated induction of carcinogenic pathways.15,20–22 Diets rich in fiber have been associated with a decreased risk of CRC,23 which may be due to the production of butyrate by colonic bacteria.24,25 Studies on butyrate have shown that it reduces inflammation and can inhibit growth and induce apoptosis in cancer cells.26 An imbalance of butyrate, folate, and biotin-producing bacteria27 could contribute to carcinogenesis as these molecules are involved in epithelial proliferation either directly or epigenetically.28–30 Further, secondary bile acids may be carcinogenic by acting as mutagens, eliciting reactive oxygen species, and increasing NF-kappa B activation, resulting in inflammation.31 Additionally, a diet low in fiber leaves the colon devoid of Microbiota Accessible Carbohydrates32 and open for bacteria to feed on the protein-rich mucus layer that protects the colon epithelium.33 It is possible that the microbial switch from metabolizing carbohydrates to proteins generates inflammatory side products and loss of the protective mucus layer results in direct contact of bacteria with the epithelium. This direct contact has been proposed as a step in inciting cellular changes or inflammation in the colon epithelium.34 Landmark studies on the microbiome and CRC are summarized in Figure 1.
Table 1.
The proposed mechanisms of individual bacteria implicated in colorectal cancer
| Bacteria | Proposed CRC Contribution Mechanism |
|---|---|
| Enterococcus faecalis | Produces reactive oxygen species that can cause DNA damage, contributing to chromosomal instability.12 |
| Enterotoxigenic Bacteroides fragilis (ETBF) | Produces the B. fragilis toxin, which is directly genotoxic and cleaves the tumor suppressor protein E-cadherin, resulting in enhanced Wnt/beta-catenin and NF-kB signaling leading to increased mucosal permeability and colonocyte proliferation. ETBF may also increase tumorigenesis through its association with upregulated Interleukin-17, which activates STAT3, and T-helper cell 17 inflammatory response.13 |
| Streptococcus gallolyticus | S. gallolyticus in the blood has long been a red flag for carcinoma of the colon. It thrives in environments seen in colonic tumors, is able to translocate through the epithelium, and is associated with enhanced inflammatory signaling.14 |
| Escherichia coli | Particular strains can secrete colibactin toxin and induce double stranded DNA breaks in mice,15 and may also downregulate DNA mismatch repair proteins. |
| Fusobacterium nucleatum | Generates the FadA adhesin protein which allows it to bind cell E-cadherin and activate Wnt signaling.16 It also appears to recruit immune cells to the tumor environment and upregulate inflammatory genes. |
Figure 1.
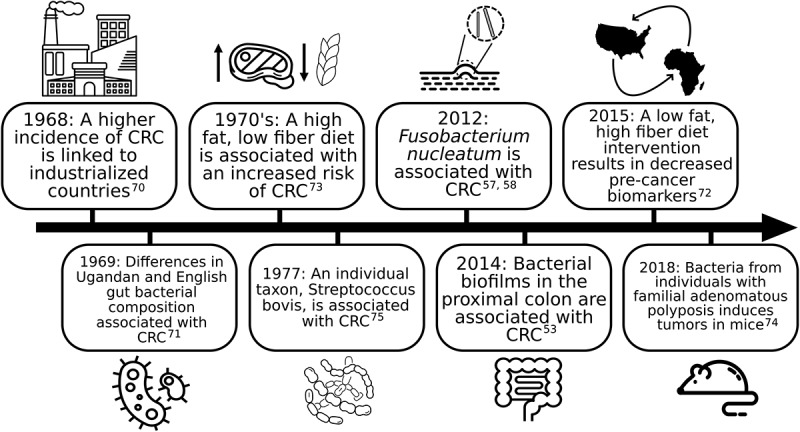
Landmark research on the microbiome and colorectal cancer
Research limitations and solutions
To date, most studies have focused on comparing the intestinal microbiome between healthy individuals and those with CRC tumors or adenomas using stool samples. Research has demonstrated that there are differences in the microbiota of colons with CRC, such as an increase in Prevotella35 and Fusobacteria,36–39 reduction in butyrate-producing bacteria,40–42 Bifidobacteria,43 and overall diversity.41 However, some of these findings are inconsistent across the literature, potentially due to differing study design and a number of common limitations found in CRC microbiome studies. Additionally, these microbial differences may therefore be consequential to the altered biochemical environment of CRC tumor mucosa44 and unrelated to processes that triggered the development of CRC.
We suggest five limitations to existing human studies and present potential corresponding solutions. The first frequent limitation among human studies is the sole use of stool samples. This may be problematic as the composition of mucosa-associated microbes which adhere to the intestinal epithelia has been shown to be different from microbes found in the lumen, which are more likely found in stool.40–43 Stool samples have a blend of the microbial communities present in the colon and are unable to provide the same location information as localized samples from the mucosa. Given that microbes in close contact with the epithelium may have greater potential to influence progression into CRC, studying mucosa-associated microbes by obtaining mucosal samples from precursor lesions and tumors may provide more useful data.
Second, the majority of studies have compared the microbiome of healthy individuals and patients with CRC tumors. The list of microbes that has been found to be enriched or deficient in CRC tumors may be related to the altered environment of the tumor rather than the microbe’s involvement in CRC carcinogenesis. CRC tumors have been found to have decreased glucose, lower pH, and elevated amino acids and fatty acids.44 To determine if and how the microbiome may contribute to CRC carcinogenesis, we suggest that sample collection be broadened to include precursor lesions in the sequential stages of carcinogenesis as well as CRC tumors. Examination of precursor lesions is important for elucidating early changes in the microbiome that may incite or accelerate the progression of CRC carcinogenesis.
A third limitation is that many of the studies that examine precursor lesions do not differentiate based on polyp type (adenoma vs. SSP vs. TSA). Given that CRC carcinogenesis proceeds predominantly through two unique pathways, it is possible that the microbiome may contribute to the incitement and/or progression of each pathway in specific ways. We believe that changes in microbial populations specific to polyp and tumor type should be investigated, which can be done using genetic, molecular, or histopathological characterization of colorectal polyps and tumors.
A fourth limitation is that when mucosa has been sampled, the information regarding the site in the colon where the sample was collected, a vital piece of metadata, is often not reported. Location data are important because while the adenoma-carcinoma pathway is found throughout the colon, the epigenetic sessile serrated pathway is more uniquely right sided Figure 3. For example, several groups have demonstrated that Fusobacteria is associated with right-sided colonic lesions with features of the serrated pathway.36–38 We suggest that additional data points of location in the colon be added during the sample collecting process, such as if the sample was obtained from the proximal or distal colon, or rectum. The differences in terms of anatomical location within the colon and characteristics of each pathway are summarized in Figures 2 and 3.
Figure 3.
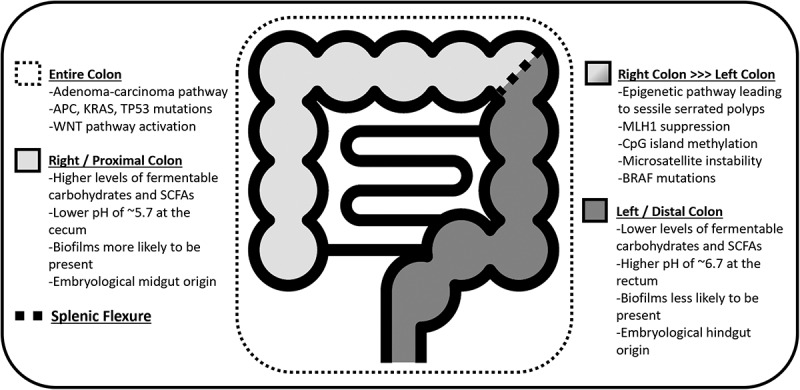
Biogeographical differences and CRC pathways throughout the colon.(53,76,80)
A fifth limitation is that some studies have been limited in the type of sequencing used to characterize the microbiome. The prevailing approach is 16S rRNA amplicon sequencing, which focuses on an individual, universal marker gene. 16S rRNA gene sequencing can elucidate which bacteria are present in a sample but does not include information about specific metabolic capacities of individual strains or species. The increased resolution provided by whole community shotgun metagenomic sequencing may in part explain why a recent meta-analysis of 16S rRNA fecal microbiomes from CRC patients failed to find biomarkers of CRC,45 while subsequent shotgun sequencing meta-analyses detected increased protein and mucin catabolism genes and reduced carbohydrate degradation genes.46 Thus, we recommend that future studies consider using shotgun sequencing for their sample analysis. In addition, it is critical that microbiome researchers use consistent standards, such as having positive and negative controls as part of every study and using spike-in standards to get semi-quantitative data.47 It is also beneficial to use consistent taxonomic resolution. Research data and analyses should be open access to support rigorous and reproducible science.
Microbiome of the serrated pathway vs. the adenoma-carcinoma sequence
It is our belief that human studies that use mucosal samples and differentiate polyp or tumor samples by either location in the colon, histopathologic type, or molecular/genetic type will provide key information on how the microbiome may 1) contribute to CRC carcinogenesis, and 2) play specific roles in the two major pathways of CRC carcinogenesis. We found that the number of studies that investigated the mucosal microbiome and differentiated lesions in any of the aforementioned ways was very limited. Four studies included data on lesions classified by molecular/genetic type,36,48–50 seven included location in the colon,36,38,49–53 and three by histopathologic type.36–38 These 9 studies are summarized in Table 2, with key findings discussed below. There is a notable gap in existing research with regards to differentiating between TSAs and SSPs in addition to adenomas. One of the studies that categorized lesions by genetic mutation revealed some interesting patterns that could be theoretically extrapolated to the different polyp types. Burns et al. found that lesions with Apc mutations (seen in the adenoma-carcinoma sequence) have an increase in Finegoldia, which is an opportunistic pathogen at sites of epithelial disruption. Lesions with mutations in KMTC2, which is commonly mutated with K-ras (part of the adenoma-carcinoma sequence and frequently mutated in TSAs) could be predicted by the abundance of Ruminococcus, which has been linked to IBD and CRC.48 Another study found that SSPs with dysplasia, but not without dysplasia or adenomas, had characteristic Clostridium perfringens infection. They hypothesized that C. perfringens may enhance carcinogenesis via Yes-associated protein activation.54
Table 2.
Microbiome studies differentiating by CRC carcinogenic pathway or location histopathologic type
| Author | Sample type | Location in colon | Histopathologic type | Molecular/Genetic type | Sample source |
|---|---|---|---|---|---|
| Flemer et al.51 2017 | Mucosa, stool | X | On and off tumor | ||
| Gao et al.52 2015 | Mucosa | X | On and off tumor | ||
| Dejea et al.53 2014 | Mucosa | X | Biofilms on and off tumors and precursor lesions | ||
| Burns et al.48 2016 | Mucosa | X | On and off tumor | ||
| Ito et al.36 2015 | Mucosa | X | X | X | Tumors and precursor lesions |
| Purcell et al.49 2017 | Mucosa | X | X | Tumors | |
| Hale et al.50 2018 | Mucosa | X | X | On and off tumor | |
| Park et al.37 2016 | Mucosa | X | Precursor lesions, tumors | ||
| Yu et al.38 2016 | Mucosa | X | X | F. nucleatum in tumors and precursor lesions |
Biofilm presence in left vs. right colon
Studies by Flemer et al., Gao et al., and Dejea et al. compared left versus right CRC microbiota. These studies revealed that there are significantly different bacteria in patients with CRC versus those without, and in proximal versus distal CRC.51,52 More importantly, different clusters of bacteria were associated with different gut mucosa gene expression patterns, which could provide the link between colon microenvironments and their unique microbial inhabitants and ensuing pathogenesis. The most significant finding was in the study by Dejea et al.53 examining the presence of biofilms on adenomas and carcinomas. Biofilms contain an array of microorganisms that are adherent to one another as well as a surface by way of extracellular matrix produced by bacteria. The polysaccharide layer promotes bacterial survival in nutrient poor conditions and allows the inhabitants to exchange metabolites with the environment external to the biofilm. Biofilms may create pro-carcinogenic environments that are as important to the development of cancer as the specific taxa present. Dejea et al. found that biofilms were predominantly found on proximal CRC and adenomas, and tumors with biofilms displayed bacterial invasion. Biofilm-covered epithelial cells were shown to have decreased E-cadherin (a transmembrane inter-cellular binding protein) leading to increased mucosal permeability, increased Interleukin-6 (an inflammatory mediator) and increased STAT-3 activation leading to proliferation. In theory, biofilm formation and bacterial invasion could allow bacteria to persistently interact with the epithelium and lead to chronic inflammation and carcinogenesis. Of note, the aforementioned changes were also seen in a subset of healthy individuals with biofilms, begging the question of whether biofilm presence can contribute to the incitement of CRC carcinogenesis. A subsequent study by Johnson and Dejea found that biofilms contained increased levels of polyamine metabolites (molecules in all eukaryotic cells essential for growth) associated with cellular proliferation and colon cancer.55 Specifically, the level of a polyamine metabolite that has been a proposed marker for early-stage CRC56 was elevated in tumors regardless of biofilm presence and was further elevated in the presence of biofilms on tumor and normal tissue. While biofilms are predominantly present on right-sided colon cancers, the increased acetylated polyamine metabolites on biofilm-positive cancer and paired normal tissues on the left side suggests that biofilm presence, not colon location, is what alters polyamine metabolism leading to metabolites associated with CRC. The association of biofilms with proximal CRC raises the question of whether biofilms may specifically contribute to the sessile serrated pathway. Further research is needed to define the genetic and histopathologic type of polyps and tumors that biofilms are found on.
Fusobacterium and serrated pathway lesions
Many studies have implicated the genus Fusobacterium in CRC, as early as the 2012 studies by Kostic et al.57 and Castellarin et al.58 Some studies, such as the reproducibility study by Repass have not found the same association, but these mixed findings could be related to varying population samples, sample types, and sequencing methods.59 Our analysis of the included studies revealed a clear association between Fusobacterium and lesions that were proximal, of higher histological grade, and with features of the serrated pathway. The study by Yu et al. found that Fusobacterium was found more frequently in SSPs than adenomas, and in proximal more than distal CRC. While Ito et al. did not detect a difference in Fusobacterium between adenomas, TSAs, or SSPs, his group did find that the rate of Fusobacterium positive SSPs increased when moving from distal to proximal colon.36,39 Studies by Park and Ito found Fusobacterium to be more associated with CRC than less advanced lesions, with F. nucleatum’s presence increasing as the histological grade increased, which could be the result of new microenvironments of CRC tumors.36,37 SSPs often have a mucus cap and overexpress mucin forming genes such as MUC6, MUC5aC, MUC17, and MUC2, which has been associated with increased metastasis.60 It is possible that this mucus cap can help Fusobacterium and other bacteria survive. Mucus caps may function similar to biofilms; however, Yu et al.38 found that Fusobacterium presence and its ability to invade the mucosa did not depend on biofilm presence. All of the studies differentiating microbial populations by molecular or genetic mutation found Fusobacterium to be associated with lesions characterized by features of the serrated pathway including mismatch repair (MMR) deficiency, MLH1 methylation, CpG island methylator phenotype, or high microsatellite instability.36,49,50 Interestingly, Hale et al. found that MMR status was a stronger predictor of the microbe community variance than even location in the colon and off versus on tumor. Of note, these findings are associations, and whether Fusobacterium is a “passenger” or a driver of CRC is a current debate.
Oral bacteria in CRC pathogenesis
Bacterial species typically found in the oral cavity have been often found in the colon, though it is unknown whether these microbes originated in the oral cavity or whether particular strains are specialized to live in the colon.61 However, a study by Komiya et al.62 found that 40% of patients had identical strains of F. nucleatum in their CRC tumor tissue and saliva, postulating that F. nucleatum may originate in the oral cavity. A number of studies including a multicenter metagenome sequencing study across five countries have found that a number of oral cavity associated bacteria are over-represented in CRC, including Fusobacterium, Porphyromonas, Parvimonas, and Prevotella.63 Other studies have found that tumors with characteristics of the adenoma-carcinoma sequence, which typically develop from adenomas, were found to have increased abundance of Capnocytophaga,48 Selenomonas, and Prevotella.49 In contrast, the oral microbes Fusobacterium nucleatum, Parvimonas micra, Peptostreptococcus stomatis and Porphyromonas gingivalis were associated with Consensus Molecular Subtype 1 (CMS1) tumors,49 which are closely associated with the serrated pathway.64 This study also noted that these oral microbes are capable of forming biofilms, and that P. gingivalis co-aggregates with Treponema denticola and Tannerella forsythia, which are also enriched in CMS1 tumors.49 A number of previously mentioned studies also associate Fusobacterium with premalignant polyps with features of the serrated pathway. A study by Hale et al. found that MMR deficient CRC tumors, seen in the serrated pathway, were enriched with Fusobacterium nucleatum and periodonticum.50
It remains unknown whether alterations in the colonic mucosa attract oral flora associated microbes or if they become present upon the development of the CRC tumor environments. A 2018 study by Flemer et al. found that networks of typically oral bacteria were more abundant on polyps and CRC tumors than on the mucosa of healthy individuals. The microbial networks from polyps and CRC tumors were similar to those from the patient’s oral swabs as well as their healthy mucosa. They postulated that this may indicate that oral cavity-associated bacterial networks may exist prior to and contribute to the development of CRC.
Our group questions whether dietary habits could lead to changes in the gut epithelium, such as increased permeability or alterations in pH, that allow oral bacteria to survive in the intestines.65 Gliadin intake or a low fiber diet leading to reduced butyrate, crucial for the maintenance of cell adhesion and epithelial barrier defense, could in theory alter barrier function. Further, if colonocytes lack butyrate, they may switch to metabolizing other energy sources, such as carbohydrates. In line with this thinking, Flemer et al. found that when oral pathogens were present in the colonic mucosa, their presence was strongly negatively correlated with the abundance of butyrate-producer Lachnospiraceae. The abundance of Lachnospiraceae was weakly negatively correlated with a Western diet and was postulated to prevent colonic colonization by the typically oral bacteria that are associated with CRC. While there have not been any confirmed mechanisms by which these bacteria contribute to CRC carcinogenesis, some mechanisms have been proposed regarding communities of oral cavity associated bacteria found in biofilms. An example of an oral biofilm is dental plaque, which is colonized by Streptococcus and Actinomyces, creating an anaerobic environment conducive to F. nucleatum and other oral bacteria survival.66 Biofilms in the colon may also harbor oral cavity associated bacteria, including commensal bacteria and the pathogenic periodontal bacteria F. nucleatum and P. gingivalis.67 The new environment of the biofilm has been suggested to allow for opportunistic pathogens associated with the oral cavity, such as Peptostreptococcus and Porphyromonas, to survive and promote CRC tumorigenesis. Proposed mechanisms include bacterial secretion of enzymes that disrupt the protective mucus layer and destruction of defensin peptides and IgA antibodies, allowing bacteria to attach to and invade the mucosa and incite inflammation via IL-6, STAT3,66 and IL-8 pathways.68
Conclusion
Research has established that the intestinal microbiome is different in patients with CRC. Most studies have utilized stool samples, and some have used mucosal samples. Of the studies that use mucosal samples, the comparison of the microbiome is often between an adenoma (without further specification of type) or tumor to normal tissue. However, there are very few studies that have investigated the microbiome of adenomas, SSPs, TSAs, and tumors in the adenoma-carcinoma sequence and serrated pathway specifically. This distinction can be made by specifying each sample’s genetic, epigenetic, or histopathologic type, or location in the colon as a proxy. Analysis of the select studies that have made these distinctions revealed that microbes typically considered normal oral flora can be found in the colons of patients with CRC and in biofilms, and that Fusobacterium is associated with right sided, more advanced, and serrated pathway lesions. One study suggested that the colons from people with CRC have more biofilms, particularly on proximal tumors.53 While these findings cannot yet change clinical practice, they indicate a possible future where microbial signatures may be used to screen for and characterize precursor polyps and be used to characterize which carcinogenic pathway they were derived from or might progress through. This level of detail could open the door for novel prevention, screening, and treatment interventions for CRC based on carcinogenic pathway characterization. Eventually, CRC interventions could expand to include microbial manipulation. For this to become a possibility, more clinical studies are required to investigate the microbiome of precursor polyps as well as tumors in each stage of carcinogenesis in the adenoma-carcinoma sequence and serrated pathways. This will require mucosal samples and characterization by histopathology, genetic and molecular profile, and location in the colon. Longitudinal studies should begin early on, prior to the occurrence of any colon pathology, and extend through CRC treatment and monitoring. If particular microbial populations or metabolites are associated with CRC carcinogenesis, further bench research will be necessary to investigate possible mechanisms of carcinogenesis and interventions. Specifically, experimental studies with model organisms and specific in vitro cell cultures that reflect the distinct pathways to CRC to investigate how oral cavity associated microbes and biofilms may further CRC progression. Finally, it is critical that microbiome researchers use consistent standards and make their data and analyses open access to support rigorous and reproducible science.69,70–80
Funding Statement
American Cancer Society, Institutional Research Grant IRG-16-187-13American Cancer Society [IRG-16-187-13];
References
- 1.Siegel, R.L., Miller, K.D. and Jemal, A . (2019), Cancer statistics, 2019. CA A Cancer J Clin, 69:7-34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Ballester V, Rashtak S, Boardman L.. Clinical and molecular features of young-onset colorectal cancer. World J. Gastroenterol. 2016;22:1736–1744. doi: 10.3748/wjg.v22.i5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller MF, Ibrahim AEK, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125–134. doi: 10.1007/s00428-016-1956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Nakanishi Y, Diaz-Meco MT, Moscat J. Serrated colorectal cancer: the road less travelled? Trends Cancer. 2019;5:742–754. doi: 10.1016/j.trecan.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crockett SD, Nagtegaal ID. Terminology, molecular features, epidemiology, and management of serrated colorectal neoplasia. Gastroenterology. 2019;157:949–966.e4. doi: 10.1053/j.gastro.2019.06.041. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am. J. Surg. Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy AJ, Serra S, Chetty R. Traditional serrated adenoma: an overview of pathology and emphasis on molecular pathogenesis. BMJ Open Gastroenterol. 2019;6:e000317. doi: 10.1136/bmjgast-2019-000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JF, Tang S-J, Lash RH, Wu R, Yang Q. Anatomic distribution of sessile serrated adenoma/polyp with and without cytologic dysplasia. Arch. Pathol. Lab. Med. 2015;139:388–393. doi: 10.5858/arpa.2013-0523-OA. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology. 2015;148:1244–1260.e16. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Rhee K-J, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J. Cell Sci. 2007;120:1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer. 2010;9:249. doi: 10.1186/1476-4598-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede J-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke JD, Serino M, Tilg H, Watson A, Wells JM. Intestinal permeability – a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW, Buurman WA, de Vos WM, Rensen SS. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity. 2013;21:E607–E615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 19.Collins SM, Denou E, Verdu EF, Bercik P. The putative role of the intestinal microbiota in the irritable bowel syndrome. Dig. Liver Dis. 2009;41:850–853. doi: 10.1016/j.dld.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2006;12:782–786. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68:9909–9917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, Hu -L-L, Gonzalez-Navajas J, Seo GS, Shen C, Brick J, Herdman S, Varki N, Corr M, Lee J, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat. Med. 2010;16:665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aune D, Chan DSM, Lau R, Vieira R, Greenwood DC, Kampman E, Norat T. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ. 2011;343:d6617. doi: 10.1136/bmj.d6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 2014;87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 25.Nielson T. Baxter, Alexander W . Schmidt, Arvind Venkataraman, Kwi S. Kim, Clive Waldron, Thomas M. SchmidtmBio Jan 2019, 10 (1) e02566-18; doi: 10.1128/mBio.02566-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hague A, Elder DJ, Hicks DJ, Paraskeva C. Apoptosis in colorectal tumour cells: induction by the short chain fatty acids butyrate, propionate and acetate and by the bile salt deoxycholate. Int. J. Cancer. 1995;60:400–406. doi: 10.1002/ijc.2910600322. [DOI] [PubMed] [Google Scholar]
- 27.Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 2018;57:1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindzon GM, Medline A, Sohn K-J, Depeint F, Croxford R, Kim Y-I. Effect of folic acid supplementation on the progression of colorectal aberrant crypt foci. Carcinogenesis. 2009;30:1536–1543. doi: 10.1093/carcin/bgp152. [DOI] [PubMed] [Google Scholar]
- 29.Cherbonnel‐Lasserre CL, Linares‐Cruz G, Rigaut J-P, Sabatier L, Dutrillaux B. Strong decrease in biotin content may correlate with metabolic alterations in colorectal adenocarcinoma. Int. J. Cancer. 1997;72:768–775. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Said HM. Cellular Uptake of Biotin: mechanisms and Regulation. J. Nutr. 1999;129:490S–493S. doi: 10.1093/jn/129.2.490S. [DOI] [PubMed] [Google Scholar]
- 31.da Silva Lima F, Rogero MM, Ramos MC, Borelli P, Fock RA. Modulation of the nuclear factor-kappa B (NF-κB) signalling pathway by glutamine in peritoneal macrophages of a murine model of protein malnutrition. Eur. J. Nutr. 2013;52:1343–1351. doi: 10.1007/s00394-012-0443-0. [DOI] [PubMed] [Google Scholar]
- 32.Utzschneider KM, Kratz M, Damman CJ, Hullarg M. Mechanisms linking the gut microbiome and glucose metabolism. J. Clin. Endocrinol. Metab. 2016;101:1445–1454. doi: 10.1210/jc.2015-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenburg ED, Sonnenburg JL. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 2019;17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 34.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U. S. A. 2011;108 Suppl 1:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Van Nhieu JT, JP Furet. Microbial dysbiosis in colorectal cancer (CRC) patients. Plos One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 37.Park CH, Han DS, Oh Y-H, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci. Rep. 2016;6:25271. doi: 10.1038/srep25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int. J. Cancer. 2016;139:1318–1326. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 39.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano H-O, Sugai T, An B, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. Isme J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. JNCI J. Natl. Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. Plos One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 45.Sze MA, Schloss PD. Leveraging existing 16S rRNA gene surveys to identify reproducible biomarkers in individuals with colorectal tumors [Internet]. mBio. 2018;9. [cited 2019 Jan 27]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5989068/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. - PubMed - NCBI [Internet] . [cited 2020 Apr 2]. https://www.ncbi.nlm.nih.gov/pubmed/30936547 [DOI] [PMC free article] [PubMed]
- 47.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall L-I, McDonald D, et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 48.Burns MB, Montassier E, Abrahante J, Priya S, Niccum DE, et al . Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PLOS Genetics 2018;14(6): e1007376. doi:10.1371/journal.pgen.1007376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hale VL, Jeraldo P, Chen J, Mundy M, Yao J, Priya S, Keeney G, Lyke K, Ridlon J, White BA, et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers [Internet]. bioRxiv. 2018. http://biorxiv.org/content/early/2018/06/13/346510.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flemer B, Lynch DB, Brown JMR, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, O’Toole PW. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota dysbiosis is associated with colorectal cancer [Internet]. Front. Microbiol. 2015;6. [cited 2018 Aug 30]. https://www.frontiersin.org/articles/10.3389/fmicb.2015.00020/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujiwara-Tani R, Fujii K, Mori S, Kishi S, Sasaki T, Ohmori H, Nakashima C, Kawahara I, Nishiguchi Y, Mori T, et al. Role of clostridium perfringens enterotoxin on YAP activation in colonic sessile serrated adenoma/polyps with dysplasia. Int. J. Mol. Sci. 2020;21:3840. doi: 10.3390/ijms21113840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson CH, Dejea CM, Edler D, Hoang L, Santidrian A, Felding B, Ivanisevic J, Cho K, Wick E, Hechenbleikner E, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hiramatsu K, Takahashi K, Yamaguchi T, Matsumoto H, Miyamoto H, Tanaka S, Tanaka C, Tamamori Y, Imajo M, Kawaguchi M, et al. N(1),N(12)-Diacetylspermine as a sensitive and specific novel marker for early- and late-stage colorectal and breast cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005;11:2986–2990. doi: 10.1158/1078-0432.CCR-04-2275. [DOI] [PubMed] [Google Scholar]
- 57.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Repass J, Iorns E, Denis A, Williams SR, Perfito N, Errington TM. Reproducibility project: cancer biology. replication study: fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. eLife. 2018;7:e25801. doi: 10.7554/eLife.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delker DA, McGettigan BM, Kanth P, Pop S, Neklason DW, Bronner MP, Burt RW, Hagedorn CH. RNA sequencing of sessile serrated colon polyps identifies differentially expressed genes and immunohistochemical markers. Plos One. 2014;9:e88367. doi: 10.1371/journal.pone.0088367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O‘Riordain M, Shanahan F, O‘Toole PW, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68:1335–1337. doi: 10.1136/gutjnl-2018-316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai Z, Coker OO, Nakatsu G, Wu WK, Zhao L, Chen Z, Chan FK, Kristiansen K, Sung JJ, Wong SH, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers [Internet]. Microbiome. 2018;6. [cited 2020 Sep 29]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5896039/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang K, Willis JA, Reumers J, Taggart MW, San Lucas FA, Thirumurthi S, Kanth P, Delker DA, Hagedorn CH, Lynch PM, et al. Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018;29:2061–2067. doi: 10.1093/annonc/mdy337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, et al. Extensive transmission of microbes along the gastrointestinal tract. eLife. 2019; Feb;8. doi: 10.7554/elife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koliarakis I, Messaritakis I, Nikolouzakis TK, Hamilos G, Souglakos J, Tsiaoussis J. Oral bacteria and intestinal dysbiosis in colorectal cancer [Internet]. Int. J. Mol. Sci. 2019;20. [cited 2020 Sep 14]. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6747549/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol. Med. 2017;23:18–30. doi: 10.1016/j.molmed.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Huang GT, Kim D, Lee JK, Kuramitsu HK, Haake SK. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect. Immun. 2001;69:1364–1372. doi: 10.1128/IAI.69.3.1364-1372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R. Establishing microbial composition measurement standards with reference frames. Nat. Commun. 2019;10:2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doll R. The geographical distribution of cancer. Br. J. Cancer. 1969;23:1–8. doi: 10.1038/bjc.1969.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hill MJ, Drasar BS, Hawksworth G, Crowther JS, Hawksworth G, Williams RE. Bacteria and aetiology of cancer of large bowel. Lancet Lond. Engl. 1971;1:95–100. doi: 10.1016/S0140-6736(71)90837-3. [DOI] [PubMed] [Google Scholar]
- 72.O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Health NRC (US) C on D and. Cancer [Internet] . National academies press (US); 1989. [cited 2020 Apr 2]. https://www.ncbi.nlm.nih.gov/books/NBK218760/
- 74.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of streptococcus bovis with carcinoma of the colon. N. Engl. J. Med. 1977;297:800–802. doi: 10.1056/NEJM197710132971503. [DOI] [PubMed] [Google Scholar]
- 76.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 77.Fallingborg J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999;46:183–196. [PubMed] [Google Scholar]
- 78.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, Hardcastle JD. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noun Project [Internet] . Noun Proj. [cited 2020 Apr 2]. https://thenounproject.com
- 80.Macfarlane GT, Gibson GR, Cummings JH. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 1992;72:57–64. [DOI] [PubMed] [Google Scholar]


