Abstract
Background
Coronavirus disease-2019 (COVID-19) ranges from asymptomatic to severe. Several comorbidities are associated with worse clinical outcomes. Antibiotic use is common in COVID-19 and penicillin (PCN) allergy can affect antibiotic choice and may influence COVID-19 outcomes.
Objective
To investigate the impact of PCN allergy label on COVID-19 outcomes.
Methods
For this retrospective, cohort study, a Web-based tool for population cohort research, TriNetX, was used to identify adult COVID-19 patients with and without PCN allergy label. The two cohorts were matched using 1:1 propensity score matching for baseline demographics and conditions associated with risk for severe COVID-19. The 30-day risks for hospitalization, acute respiratory failure, intensive care unit requirement, mechanical ventilation requirement, and mortality were then compared between groups. Because bacterial infection can drive alternative antibiotic regimens, additional analyses focused on patients without bacterial infection.
Results
After propensity score matching, each cohort consisted of 13,183 patients. COVID-19 patients with PCN allergy had higher risks for hospitalization (risk ratio [RR] = 1.46; 95% confidence interval [CI], 1.41-1.52) acute respiratory failure (RR = 1.25; 95% CI, 1.19-1.31), intensive care unit requirement (RR = 1.20; 95% CI, 1.08-1.34), and mechanical ventilation (RR = 1.17; 95% CI 1.03-1.32) compared with patients without PCN allergy; however, there was no mortality difference (RR = 1.09; 95% CI, 0.96-1.23). Although the bacterial infection risk was higher in PCN allergic COVID-19 patients, exclusion of patients with bacterial infections yielded similar results.
Conclusions
Penicillin allergic patients have higher risk for worse COVID-19 outcomes and should be considered for risk mitigation strategies.
Key words: Drug allergy, Penicillin, Penicillin allergy, COVID-19, Mortality, Hospitalization, Respiratory failure, Severe acute respiratory syndrome coronavirus-2
Abbreviations used: COVID-19, Coronavirus disease-2019; CPT, Current Procedural Terminology; ICD, International Classification of Disease; ICU, Intensive care unit; PCN, Penicillin; PSM, Propensity score matching; SMD, Standardized mean difference
What is already known about this topic? In coronavirus disease-2019 (COVID-19), antibiotic use is common. Penicillin (PCN) allergy is prevalent and affects antibacterial treatment options, risking poorer response to treatment and increased side effects.
What does this article add to our knowledge? In COVID-19, PCN allergy is associated with increased risks for worse outcomes including hospitalization, intensive care requirement, acute respiratory failure, and mechanical ventilation.
How does this study impact current management guidelines? Patients with PCN allergy could be prioritized as a higher-risk group for COVID-19 for the development of risk mitigation strategies, including antibiotic stewardship programs to minimize unnecessary antibiotic use in these patients.
Introduction
A novel coronavirus, severe acute respiratory syndrome coronavirus-2, was identified in late 2019 as the causative agent of coronavirus disease-2019 (COVID-19) and rapidly spread to become a pandemic. Viral pneumonia of unknown etiology was the initial presentation of patients with COVID-19 in Wuhan, China.1 , 2 Common symptoms include fever, cough, myalgia, dyspnea, and fatigue.1 , 3, 4, 5 Although most patients have mild symptoms, a considerable proportion of patients experience more severe disease that requires hospitalization and respiratory support; mortality in COVID-19 is estimated at 1% to 2% in the United States.6 Multiple studies have identified populations at increased risk for severe disease to mitigate risk and stratify those who may benefit from early treatment options, hospitalization, and early vaccination in the setting of limited supplies. Patients with underlying conditions such as diabetes, heart disease, and chronic lung disease are at an especially increased risk for severe disease.7
Upon initial presentation, COVID-19 patients can have overlapping clinical features with acute bacterial infection. Furthermore, a variable proportion of COVID-19 patients (range, 8.2% to 15%) experience secondary bacterial infection, as defined by clinical symptoms and laboratory evidence of bacterial infection after admission.8, 9, 10 Therefore, in addition to the use of supportive therapy and antivirals, antibiotics are commonly used during the treatment course of COVID-19.2 , 8, 9, 10, 11 Multiple studies on the treatment of COVID-19 patients from early in the pandemic documented the use of antibiotics in over 60% of COVID-19 patients (range, 6% to 95%).8 , 10, 11, 12
In hospitalized patients, penicillin (PCN) allergy label is a recognized cause of morbidity. Penicillin allergy label results in the use of second-line antibiotics and broader-spectrum antibiotics that may put the patient at risk for poorer response to treatment, longer hospital stay, increased health care costs, and increased frequency of infection with drug-resistant organisms.13 , 14 Surgical patients with reported PCN allergy have higher probability of a surgical site infection.15 Penicillin allergy label is common; up to 10% of patients report PCN allergy, with symptoms varying from rash and hives to severe drug reactions including anaphylaxis.16 However, most patients with PCN allergy label do not have the allergy after undergoing further evaluation, and less than 10% of patients with documented PCN allergy test positive by PCN skin testing and/or drug challenge.17, 18, 19
Because there is frequent use of empiric antibiotics and increased risk for bacterial infection during the course of COVID-19 treatment, we hypothesized that the presence of PCN allergy label would be associated with worse clinical outcomes in COVID-19 patients. Here, we present a retrospective cohort study of population-based data that examined the 30-day risk for hospitalization, acute respiratory failure, intensive care unit (ICU) requirement, mechanical ventilation, and mortality in COVID-19 patients with PCN allergy label. We also investigated the use of antibiotics in these patients.
Methods
Description of data source
Data were obtained from TriNetX (Cambridge, Mass), a Web-based tool for population cohort research that provides access to electronic medical records including visit information, demographics, diagnoses, procedures, and laboratory test values. We used the TriNetX Research Network, which encompasses electronic medical records from approximately 64 million patients from 48 health care organizations across the United States. The database is deidentified and users do not have access to personal or protected health information.
Study cohorts
We conducted study cohort identification on March 31, 2021. We used a modified algorithm recommended by TriNetX to identify adult patients (aged ≥18 years) with a COVID-19 diagnosis20 , 21 (see Table E1 in this article’s Online Repository at www.jaci-inpractice.org) excluding International Classification of Disease (ICD)-10 code 079.89 (other specified viral infection). Meeting these criteria defined the index event of COVID-19 diagnosis. Patients with a COVID-19 diagnosis were then divided into two study cohorts based on the presence or absence of a PCN allergy label based on an ICD-10 code of allergy status to penicillin (Z88.0). The presence of a PCN allergy label was identified before, on the date of, or after the index event of the COVID-19 diagnosis. Patients were identified from January 20, 2020 to January 20, 2021; analyses were further divided into early (January 20, 2020 to July 20, 2020) and late (July 21, 2020 to January 20, 2021) time frames of the pandemic.
Control variables
Matching of study cohorts was based on demographic and common comorbid conditions that have been reported to be associated with severe COVID-19 or might be associated with severe COVID-19, as listed on the Centers for Disease Control and Prevention website7 (Table I ). These variables were identified using TriNetX-specific term codes as well as ICD-10 codes and included age at COVID-19 diagnosis, female sex, Black race, Hispanic or Latino ethnicity, I50.9 (heart failure), E11 (type 2 diabetes mellitus), Z68.3 (body mass index 30-39, adult), Z68.4 (body mass index 40 or greater, adult), J44 (other chronic obstructive pulmonary disease), J43 (emphysema), N18.3 (chronic kidney disease, stage 3 [moderate]), N18.4 (chronic kidney disease, stage 4 [severe]), N18.5 (chronic kidney disease, stage 5), Z72.0 (tobacco use), F17 (nicotine dependence), Z48 (encounter for aftercare after organ transplant), l20-l25 (ischemic heart diseases), Z33 (pregnant state), D57 (sickle-cell disorders), J45 (asthma), K74 (fibrosis and cirrhosis of liver), and B20 (human immunodeficiency virus disease).
Table I.
Conditions that increase or might increase the risk for severe illness for COVID-19 patients∗
| At increased risk for severe illness |
| Cancer |
| Chronic kidney disease |
| Chronic obstructive pulmonary disease |
| Heart conditions (such as heart failure, coronary artery disease, or cardiomyopathies) |
| Immunocompromised state from solid organ transplant |
| Obesity (BMI of ≥30 kg/m2 but <40 kg/m2) |
| Severe obesity (BMI > 40 kg/m2) |
| Pregnancy |
| Sickle cell disease |
| Smoking |
| Type 2 diabetes mellitus |
| Might be at an increased risk for severe illness |
| Moderate-to-severe asthma |
| Cerebrovascular disease |
| Cystic fibrosis |
| Hypertension |
| Immunocompromised state from blood or bone marrow transplant, immune deficiencies, human immunodeficiency virus disease, use of corticosteroids, or use of other immunosuppressive medications |
| Neurologic conditions (such as dementia) |
| Liver disease |
| Overweight (BMI >25 kg/m2, but <30 kg/m2) |
| Pulmonary fibrosis |
| Thalassemia |
| Type 1 diabetes mellitus |
BMI, body mass index.
Adapted from Centers for Disease Control.7
Study outcomes
Study outcomes were the 30-day risk for hospitalization, acute respiratory failure, ICU requirement, mechanical ventilation requirement, and mortality after COVID-19 diagnosis as representatives of severe outcomes of COVID-19.1, 2, 3 Hospitalization was defined as a record of any of the following Current Procedural Terminology (CPT) codes: 1013699 (initial inpatient consultation services), 1013659 (hospital inpatient services), including visit: short stay and visit: inpatient encounter, or 1013729 (critical care services). Acute respiratory failure was defined as an ICD-10 code of J80 (acute respiratory distress syndrome), J96.0 (acute respiratory failure), J96.2 (acute and chronic respiratory failure), R06.03 (acute respiratory distress), R09.02 (hypoxemia), or J96.9 (respiratory failure, unspecified). Intensive care was defined by CPT code 1013729 (critical care services). Mechanical ventilation was defined as having a CPT code of 31500 (intubation, endotracheal, emergency procedure), 94002 (ventilation assist and management, initiation of pressure or volume preset ventilators for assisted or controlled breathing; hospital inpatient/observation, initial day), 94003 (ventilation assist and management, initiation of pressure or volume preset ventilators for assisted or controlled breathing; hospital inpatient/observation, each subsequent day), 5A1935Z (respiratory ventilation, <24 consecutive hours), 5A1945Z (respiratory ventilation, 24-96 consecutive hours), or 5A1955Z (respiratory ventilation, >96 consecutive hours). Mortality was identified using the TriNetX specific term “deceased.”
Patients with 30-day concurrent bacterial infection after COVID-19 diagnosis in each study cohort were defined as having an ICD-10 code for either bacterial pneumonia, not elsewhere classified (J15) or bacteremia (R78.81). These diagnoses of bacterial pneumonia and bacteremia were chosen as bacterial infections possibly superimposed on the COVID-19 diagnosis.
Antibiotic use
Antibiotic use was identified within 30 days after COVID-19 diagnosis using PCNs (penicillin G related PCNs, PCN amino derivatives, penicillinase-resistant PCNs, extended spectrum PCNs), cephalosporins (first, second, third, and fourth generation), carbapenems (ertapenem, imipenem, and meropenem), monobactams (aztreonam), erythromycins/macrolides, quinolones, tetracyclines, vancomycin, daptomycin, linezolid, sulfamethoxazole or trimethoprim, and aminoglycosides. The use of the macrolide azithromycin was also investigated.
Statistical analyses
Analyses were performed in real time using the TriNetX analytics built-in feature. Continuous variables were described as means with SDs, and categorical variables were described as numbers and proportions. We used a two-tailed t test to compare continuous data and chi-square to compare categorical data. We estimated the absolute risk, risk ratio (RR), and 95% confidence interval (CI) of the various study outcomes across the different study cohorts. We used 1:1 propensity score matching (PSM) to match cohorts for baseline characteristics. A standardized mean difference (SMD) of less than 0.1 was used to indicate adequate balance in PSM. An α level of less than 0.05 was set to indicate statistical significance. Graphs were plotted using GraphPad Prism (version 9.0.0, GraphPad Software, San Diego, Calif).
Results
We identified 13,186 COVID-19 patients with PCN allergy label and 525,562 patients with COVID-19 but without PCN allergy between January 20, 2020 and January 20, 2021. After 1:1 PSM for the baseline variables, each cohort consisted of 13,183 patients; they were well-balanced with SMD less than 0.1. Table II lists baseline characteristics before and after PSM.
Table II.
Baseline characteristics of COVID-19 patients with and without PCN allergy
| Code | Characteristic | Before propensity score matching |
After propensity score matching |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PCN allergy (n = 13,186) | No PCN allergy (n = 525,562) | P | SMD | PCN allergy (n = 13,183) | No PCN allergy (n = 13,183) | P | SMD | ||
| Age at index, y | 56.32 (18.92) | 47.89 (18.73) | <.001 | 0.45 | 56.32 (18.92) | 56.67 (18.89) | .13 | 0.02 | |
| Female | 8967 (68.00) | 290,799 (55.33) | <.001 | 0.26 | 8964 (68.00) | 8942 (67.83) | .77 | <0.01 | |
| 2054-5 | Black | 2752 (20.87) | 91,095 (17.33) | <.001 | 0.09 | 2751 (20.87) | 2746 (20.83) | .94 | <0.01 |
| 2135-2 | Hispanic or Latino | 541 (4.10) | 63,397 (12.06) | <.001 | 0.30 | 541 (4.10) | 500 (3.79) | .19 | 0.02 |
| E11 | Type 2 diabetes mellitus | 4526 (34.32) | 74,445 (14.17) | <.001 | 0.48 | 4523 (34.31) | 4598 (34.88) | .33 | 0.01 |
| I20-I25 | Ischemic heart disease | 3921 (29.74) | 46,355 (8.82) | <.001 | 0.55 | 3918 (29.72) | 3890 (29.51) | .71 | <0.01 |
| I50 | Heart failure | 2567 (19.47) | 27,395 (5.21) | <.001 | 0.44 | 2564 (19.45) | 2493 (18.91) | .27 | 0.01 |
| Z68.3 | Body mass index 30-39 kg/m2 | 3303 (25.05) | 40,242 (7.66) | <.001 | 0.48 | 3300 (25.03) | 3286 (24.93) | .84 | <0.01 |
| Z68.4 | Body mass index ≥40 kg/m2 | 2135 (16.19) | 23,960 (4.56) | <.001 | 0.39 | 2132 (16.17) | 2113 (16.03) | .75 | <0.01 |
| F17 | Nicotine dependence | 2880 (21.84) | 42,449 (8.08) | <.001 | 0.39 | 2877 (21.82) | 2891 (21.93) | .83 | <0.01 |
| Z72.0 | Tobacco use | 975 (7.39) | 11,882 (2.26) | <.001 | 0.24 | 974 (7.39) | 922 (6.99) | .22 | 0.02 |
| J45 | Asthma | 3537 (26.82) | 50,888 (9.68) | <.001 | 0.46 | 3534 (26.81) | 3509 (26.62) | .73 | <0.01 |
| J44 | Other chronic obstructive pulmonary disease | 2482 (18.82) | 23,182 (4.41) | <.001 | 0.46 | 2479 (18.81) | 2407 (18.26) | .25 | 0.01 |
| J43 | Emphysema | 849 (6.44) | 7946 (1.51) | <.001 | 0.25 | 846 (6.42) | 775 (5.88) | .07 | 0.02 |
| N18.3 | Chronic kidney disease, stage 3 | 1336 (10.13) | 17,220 (3.28) | <.001 | 0.28 | 1334 (10.12) | 1241 (9.41) | .05 | 0.02 |
| N18.4 | Chronic kidney disease, stage 4 | 519 (3.94) | 5868 (1.12) | <.001 | 0.18 | 518 (3.93) | 441 (3.35) | .01 | 0.03 |
| N18.5 | Chronic kidney disease, stage 5 | 237 (1.80) | 3076 (0.59) | <.001 | 0.11 | 236 (1.79) | 193 (1.46) | .04 | 0.03 |
| Z33 | Pregnant state | 501 (3.80) | 14,295 (2.72) | <.001 | 0.06 | 501 (3.80) | 533 (4.04) | .31 | 0.01 |
| K74 | Fibrosis and cirrhosis of liver | 373 (2.83) | 4951 (0.94) | <.001 | 0.14 | 372 (2.82) | 323 (2.45) | .06 | 0.02 |
| Z48.2 | Encounter for aftercare following organ transplant | 191 (1.45) | 1672 (0.32) | <.001 | 0.12 | 190 (1.44) | 162 (1.23) | .13 | 0.02 |
| D57 | Sickle-cell disorders | 118 (0.90) | 1781 (0.34) | <.001 | 0.07 | 118 (0.90) | 104 (0.79) | .35 | 0.01 |
| B20 | Human immunodeficiency virus disease | 100 (0.76) | 2329 (0.44) | <.001 | 0.04 | 100 (0.76) | 84 (0.64) | .24 | 0.01 |
PCN, penicillin; SMD, standardized mean difference.
Values are expressed as means (SDs) for continuous variables and n (%) for categorical variables.
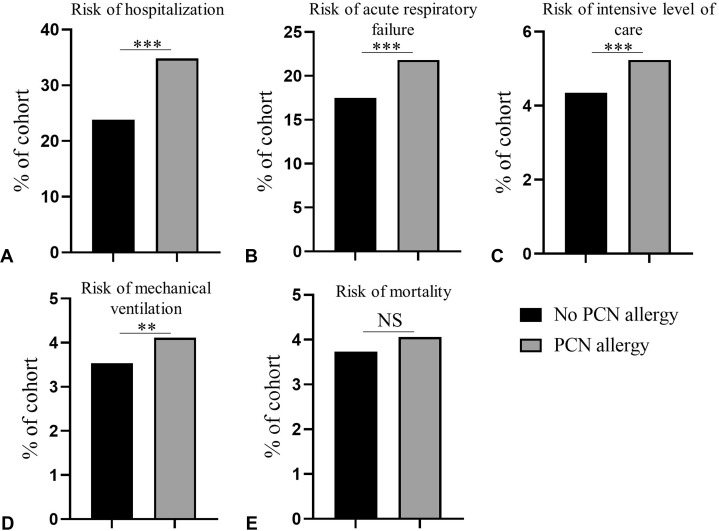
During a 30-day observation period from COVID-19 diagnosis, 4591 of the PCN-allergic patients (34.8%) required hospitalization, compared with 3136 patients without PCN allergy (23.8%; RR = 1.46; 95% CI, 1.41-1.52; P < .001) (Figure 1 , A). Acute respiratory failure occurred in 2877 of the PCN-allergic patients (21.8%) compared with 2305 patients without PCN allergy (17.5%; RR = 1.25; 95% CI, 1.19-1.31; P < .001) (Figure 1, B). A total of 690 PCN-allergic patients (5.23%) required ICU level of care compared with 574 patients without PCN allergy (4.35%; RR = 1.20; 95% CI, 1.08-1.34; P < .001) (Figure 1, C). Moreover, 542 PCN-allergic patients required mechanical ventilation (4.11%) compared with 465 patients without PCN allergy (3.53%; RR = 1.17; 95% CI, 1.03-1.32; P = .01) (Figure 1, D). During a 30-day observation period, 535 patients with PCN allergy died (4.06%) compared with 492 patients without PCN allergy (3.73%) (Figure 1, E). This slightly higher mortality risk in PCN-allergic patients was not statistically significant (RR = 1.09; 95% CI, 0.96-1.23; P = .17).
Figure 1.
Penicillin (PCN) allergy label is associated with worse outcomes in coronavirus disease-2019 (COVID-19). The 30-day risks of (A) hospitalization, (B) acute respiratory failure, (C) intensive level of care, (D) mechanical ventilation, (E) and mortality in COVID-19 patients without PCN allergy label (black bars) (n = 13,183) compared with COVID-19 patients with PCN allergy (gray bars) (n = 13,183). ∗∗P = .01; ∗∗∗P < .001; NS, not statistically significant.
During the 30-day observation from COVID-19 diagnosis, antibiotics were ordered for 4423 patients with PCN allergy (33.6%) compared with 3418 patients without PCN allergy (25.9%; RR = 1.29; 95% CI, 1.25-1.34; P < .001). Examining the specific classes of antibiotics, there was decreased use of PCNs in PCN-allergic patients (n = 458; 3.47%) compared with patients without PCN allergy (n = 989; 7.50%) (RR = 0.46; 95% CI, 0.42-0.52; P < .001). However, there was greater use of other classes of antibiotics in PCN-allergic patients, including cephalosporins (RR = 1.13; 95% CI, 1.07-1.20; P < .001), carbapenems (RR = 2.29; 95% CI, 1.89-2.76; P < .001), quinolones (RR = 2.45; 95% CI, 2.21-2.73; P < .001), macrolides (RR = 1.30; 95% CI, 1.22-1.39; P < .001), tetracyclines (RR = 1.65; 95% CI, 1.49-1.83; P < .001), and vancomycin (RR = 1.62; 95% CI, 1.48-1.77; P < .001) (Figure 2 ). Azithromycin, a macrolide antibiotic, was used in 1803 patients with PCN allergy (13.7%) compared with 1382 patients without PCN allergy (10.5%; RR = 1.31; 95% CI, 1.22-1.39; P < .001).
Figure 2.
Antibiotic use in coronavirus disease-2019 (COVID-19) patients with and without penicillin (PCN) allergy. Rates of use of PCNs, cephalosporins, carbapenems, quinolones, macrolides, tetracyclines, and vancomycin in COVID-19 patients without PCN allergy (black bars) and with PCN allergy (gray bars). ∗∗∗P < .001.
Because this observed higher risk for hospitalization, acute respiratory failure, ICU level of care, and mechanical ventilation in COVID-19 patients with a PCN allergy label could be attributed to alternative antibiotic regimens used to treat concurrent bacterial infection, we determined the proportion of patients who developed bacterial pneumonia or bacteremia within 30 days after COVID-19 diagnosis in each study cohort. Bacterial infection occurred in 515 PCN-allergic COVID-19 patients (3.91%) but in only 347 COVID-19 patients without PCN allergy (2.63%; RR = 1.48; 95% CI, 1.30-1.70; P < .001).
We then conducted exploratory analyses to account for this confounding factor by excluding patients who had bacterial pneumonia or bacteremia within 30 days after COVID-19 diagnosis. After excluding these patients, we identified two new cohorts of patients: PCN-allergic patients with no bacterial infection during the 30 days after COVID-19 diagnosis (n = 12,634) and PCN nonallergic patients with no bacterial infection during the 30 days after COVID-19 diagnosis (n = 518,434). These two cohorts were also successfully matched (SMD < 0.1) for the same baseline variables; 12,630 matched patients in each group were included in the final analysis (Table III ). After excluding patients with bacterial infection, COVID-19 patients with PCN allergy still had an increased risk for hospitalization: 4093 patients hospitalized (32.4%) compared with 2707 PCN nonallergic patients hospitalized (21.4%; RR = 1.51; 95% CI, 1.45-1.58; P < .001) (Figure 3 , A). The increased risk for acute respiratory failure also persisted; 2451 PCN-allergic COVID-19 patients developed acute respiratory failure (19.4%) compared with 1846 COVID-19 patients without PCN allergy (14.6%; RR = 1.33; 95% CI, 1.26-1.49; P < .001) (Figure 3, B). The increased need for intensive care also remained in PCN-allergic patients (n = 508; 4.02%) compared with patients without PCN allergy (n = 421; 3.33%; RR = 1.21; 95% CI, 1.06-1.37; P = .004) (Figure 3, C). More patients with PCN allergy required mechanical ventilation (n = 372; 2.95%) compared to patients without PCN allergy (n = 297; 2.35%; RR = 1.25; 95% CI, 1.08-1.46; P = .003) (Figure 3, D). There was a statistical difference in mortality risk between COVID-19 patients with PCN allergy (n = 447; 3.54%) and COVID-19 patients without PCN allergy (n = 390; 3.09%; RR = 1.15; 95% CI, 1.00-1.31; P = .045) (Figure 3, E). Despite the exclusion of patients with bacteremia and bacterial pneumonia, 3963 patients with PCN allergy received antibiotics (31.4%) and 3005 patients without PCN allergy received antibiotics (23.8%; RR = 1.32; 95% CI, 1.27-1.37; P < .001).
Table III.
Baseline characteristics of COVID-19 patients with and without PCN allergy for cases without bacterial pneumonia or bacteremia
| Code | Characteristic | Before propensity score matching |
After propensity score matching |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PCN allergy (n = 12,634) | No PCN allergy (n = 518,434) | P | SMD | PCN allergy (n = 12,630) | No PCN allergy (n = 12,630) | P | SMD | ||
| Age at index, y | 55.89 (18.95) | 47.68 (18.68) | <.001 | 0.44 | 55.89 (18.96) | 56.17 (18.90) | .23 | 0.01 | |
| Female | 8652 (68.48) | 288,017 (55.56) | <.001 | 0.27 | 8648 (68.47) | 8641 (68.42) | .92 | <0.01 | |
| 2054-5 | Black | 2631 (20.83) | 89,637 (17.29) | <.001 | 0.09 | 2628 (20.81) | 2627 (20.80) | .99 | <0.01 |
| 2135-2 | Hispanic or Latino | 522 (4.13) | 62,307 (12.02) | <.001 | 0.29 | 522 (4.13) | 473 (3.75) | .11 | 0.02 |
| E11 | Type 2 diabetes mellitus | 4216 (33.37) | 71,209 (13.74) | <.001 | 0.48 | 4212 (33.35) | 4205 (33.29) | .93 | <0.01 |
| I20-I25 | Ischemic heart disease | 3617 (28.63) | 43,946 (8.48) | <.001 | 0.54 | 3613 (28.61) | 3545 (28.07) | .34 | 0.01 |
| I50 | Heart failure | 2322 (18.38) | 25,515 (4.92) | <.001 | 0.43 | 2318 (18.35) | 2137 (16.92) | .003 | 0.04 |
| Z68.3 | Body mass index 30-39 kg/m2 | 3119 (24.69) | 39,041 (7.53) | <.001 | 0.48 | 3115 (24.66) | 3150 (24.94) | .61 | <0.01 |
| Z68.4 | Body mass index ≥40 kg/m2 | 2015 (15.95) | 23,238 (4.48) | <.001 | 0.39 | 2011 (15.92) | 1968 (15.58) | .46 | <0.01 |
| F17 | Nicotine dependence | 2759 (21.84) | 41,442 (7.99) | <.001 | 0.40 | 2755 (21.81) | 2827 (22.38) | .27 | 0.01 |
| Z72.0 | Tobacco use | 928 (7.35) | 11,567 (2.23) | <.001 | 0.24 | 927 (7.34) | 868 (6.87) | .15 | 0.02 |
| J45 | Asthma | 3384 (26.79) | 49,947 (9.63) | <.001 | 0.46 | 3380 (26.76) | 3392 (26.86) | .86 | <0.01 |
| J44 | Other chronic obstructive pulmonary disease | 2287 (18.10) | 21,881 (4.22) | <.001 | 0.45 | 2283 (18.08) | 2166 (17.15) | .05 | 0.02 |
| J43 | Emphysema | 760 (6.02) | 7414 (1.43) | <.001 | 0.24 | 757 (5.99) | 655 (5.19) | .005 | 0.04 |
| N18.3 | Chronic kidney disease, stage 3 | 1199 (9.49) | 16,111 (3.11) | <.001 | 0.27 | 1197 (9.48) | 1136 (8.99) | .18 | 0.02 |
| N18.4 | Chronic kidney disease, stage 4 | 460 (3.64) | 5376 (1.04) | <.001 | 0.17 | 458 (3.63) | 376 (2.98) | .004 | 0.04 |
| N18.5 | Chronic kidney disease, stage 5 | 208 (1.65) | 2770 (0.53) | <.001 | 0.11 | 206 (1.63) | 158 (1.25) | .01 | 0.03 |
| Z33 | Pregnant state | 495 (3.92) | 14,238 (2.75) | <.001 | 0.07 | 495 (3.92) | 542 (4.29) | .14 | 0.02 |
| K74 | Fibrosis and cirrhosis of liver | 345 (2.73) | 4626 (0.89) | <.001 | 0.14 | 344 (2.72) | 313 (2.48) | .22 | 0.02 |
| Z48.2 | Encounter for aftercare following organ transplant | 167 (1.32) | 1513 (0.29) | <.001 | 0.12 | 165 (1.31) | 122 (0.97) | .01 | 0.03 |
| D57 | Sickle-cell disorders | 114 (0.90) | 1737 (0.34) | <.001 | 0.07 | 113 (0.90) | 114 (0.90) | .95 | <0.01 |
| B20 | Human immunodeficiency virus disease | 94 (0.74) | 2273 (0.44) | <.001 | 0.04 | 94 (0.74) | 86 (0.68) | .55 | <0.01 |
PCN, penicillin; SMD, standardized mean difference.
Values are expressed as means (SDs) for continuous variables and n (%) for categorical variables.
Figure 3.
Penicillin (PCN) allergy label is associated with worse outcomes in coronavirus disease-2019 (COVID-19) irrespective of concurrent bacterial infection. The 30-day risks for (A) hospitalization, (B) acute respiratory failure, (C) intensive level of care, (D) mechanical ventilation, (E) and mortality in COVID-19 patients without PCN allergy (black bars) (n = 12,630) compared to COVID-19 patients with PCN allergy (gray bars) (n = 12,630) after excluding patients who developed concurrent bacterial infection. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Because management of COVID-19 patients, including use of antibiotics, might have changed from early to later in the pandemic with increased data on rates of concurrent bacterial infections and recommendations for antibiotic use in COVID-19,22, 23, 24 we investigated COVID-19 outcomes early (January 20, 2020 to July 20, 2020) and later in the pandemic (July 21, 2020 to January 20, 2021) between patients with and without PCN allergy after similar PSM methods. The 30-day risks for hospitalization, acute respiratory failure, ICU level of care, and mechanical ventilation for COVID-19 patients were higher both early and late in the pandemic in PCN-allergic patients compared to patients without PCN allergy (see Table E2 in this article’s Online Repository at www.jaci-inpractice.org).
Discussion
To the authors’ knowledge, this is the first large retrospective cohort study that assessed the impact of PCN allergy label on clinical outcomes in COVID-19. Although not clearly defined, the development of symptoms and clinical outcomes in COVID-19 patients has been shown to take days to weeks.25 We showed that PCN allergy is associated with a 1.5-fold increased risk for hospitalization, a 1.3-fold increased risk for acute respiratory failure, a 1.2–fold increase in risk for intensive level of care, and a 1.2-fold increased risk for mechanical ventilation within 30 days of COVID-19 diagnosis. The basis for this negative impact of PCN allergy label on COVID-19 disease outcome is unclear; however, it does not appear to be explained solely by concurrent bacteremia and bacterial pneumonia, because the higher risks for hospitalization, acute respiratory failure, ICU level of care, and mechanical ventilation in our study persisted after we excluded patients with bacterial infection during the 30-day observation period.
Early in the pandemic, the World Health Organization issued guidance for managing COVID-19, which advocated for starting empiric antimicrobials to treat all likely pathogens in severe illness, even if COVID-19 was suspected, and then to deescalate based on testing results.26 In the United States during the pandemic, the Veterans’ Health Administration noticed a spike in antibiotic use with greatest increases in ceftriaxone, cefepime, doxycycline, and azithromycin.27 Indeed, antibiotic use in COVID-19 patients is common; several studies documented antibiotic use in more than 60% of patients (range, 6% to 95%).8 , 10, 11, 12 Frequently used antibiotics included moxifloxacin, azithromycin, cephalosporins, and PCNs.8 , 9 , 11 , 28 With such frequent use of antibiotics in COVID-19 patients,2 , 8, 9, 10, 11 PCN allergy label may result in increased side effects from the use of broader-spectrum antibiotics and alternative antibiotics. The impact of such broader and alternative antibiotic regimens could be mediating the increased risk for worse outcomes in COVID-19 for PCN-allergic patients.
In contrast to the higher rate of antibiotic use among COVID-19 patients, the reported rates of secondary bacterial infection in COVID-19 patients are lower and ranged from 8.2% to 15% of patients.8, 9, 10 Our study demonstrated a lower rate of concurrent bacterial infections; however, it is possible that our composite ICD-10 codes for concurrent bacterial infections of bacterial pneumonia and bacteremia did not account for all such bacterial infections. Nonetheless, this observed discrepancy between the reported rate of bacterial infection and the use of antibiotics indicates that antibiotic use in COVID-19 patients is largely empiric, and PCN allergy label is likely to modify the choice of empiric antibiotic coverage.
Owing to the risks of PCN allergy label, including use of broader antibiotics, longer hospitalization, poorer response to treatment,13 , 14 and concern for antibiotic resistance, the position statement from the American Academy of Allergy, Asthma, and Immunology supports further workup and testing for patients with documented PCN allergy,29 because greater than 90% of patients, when properly evaluated, are able to be cleared of the PCN allergy label.17, 18, 19 The findings that COVID-19 patients with PCN allergy are at increased risk for severe outcomes further supports the evaluation of patients with PCN allergy label, because clearance of the PCN allergy might reduce the risk for poorer outcomes in COVID-19. This is an area for further investigation.
We acknowledge several limitations to our study. (1) The use of ICD-10 codes is subject to reported bias; however, we expect this to affect both study cohorts equally. (2) Although the cohorts were matched for multiple underlying conditions that are associated with increased risk for severe disease in COVID-19, we could not account for other diagnoses such as cancer because of ICD-10 code limitations, and we could not account for possible higher-risk conditions that have not yet been identified, which could represent confounding variables in these cohorts. (3) Not every patient in the no–PCN allergy cohort was tested for an allergy to PCNs, and so some patients in this cohort could have an unrecognized PCN allergy. (4) Finally, because most patients with a PCN allergy label may not prove to have true PCN allergy when properly evaluated, our results should be interpreted with caution.
Based on these findings of higher risk of severe outcomes in COVID-19, patients with PCN allergy could be prioritized to a higher risk group. Strategies for mitigating these increased risks for patients with PCN allergy should be considered, including evaluating PCN allergy for possible de-labeling and early vaccination.
Footnotes
The project was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR002014. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest: The authors declare that they have no relevant conflicts of interest.
Online Repository
Table E1.
International Classification of Disease (ICD)-10 and Current Procedural Terminology (CPT) codes with matching terms used to identify COVID-19 patients
| Code | Term |
|---|---|
| B97.29 | Other coronavirus as cause of diseases classified elsewhere |
| B34.2 | Coronavirus infection, unspecified |
| J12.81 | Pneumonia due to SARS-associated coronavirus |
| U07.1 | COVID-19 |
| U07.2 | COVID-19, virus not identified (World Health Organization) |
| 94307-6 | SARS coronavirus 2 N gene [presence] in unspecified specimen by nucleic acid amplification using primer-probe set N1 |
| 94308-4 | SARS coronavirus 2 N gene [presence] in unspecified specimen by nucleic acid amplification using primer-probe set N2 |
| 94309-2 | SARS coronavirus 2 RNA [presence] in unspecified specimen by NAA with probe detection |
| 94310-0 | SARS-like coronavirus N gene [presence] in unspecified specimen by NAA with probe detection |
| 94314-2 | SARS coronavirus 2 RdRp gene [presence] in unspecified specimen by NAA with probe detection |
| 94315-9 | SARS coronavirus 2 E gene [presence] in unspecified specimen by NAA with probe detection |
| 94316-7 | SARS coronavirus 2 N gene [presence] in unspecified specimen by NAA with probe detection |
| 94500-6 | SARS coronavirus 2 RNA [presence] in respiratory specimen by NAA with probe detection |
| 94502-2 | SARS-related coronavirus RNA [presence] in respiratory specimen by NAA with probe detection |
| 94532-9 | SARS-related coronavirus plus Middle East respiratory syndrome coronavirus RNA [presence] in respiratory specimen by NAA with probe detection |
| 94533-7 | SARS coronavirus 2 N gene [presence] in respiratory specimen by NAA with probe detection |
| 94534-5 | SARS coronavirus 2 RDRP gene [presence] in respiratory specimen by NAA with probe detection |
| 94559-2 | SARS coronavirus 2 orf1ab region [presence] in nasopharynx by NAA with non-probe detection |
| 94565-9 | SARS coronavirus 2 RNA [presence] in nasopharynx by NAA with non-probe detection |
| 94639-2 | SARS coronavirus 2 orf1ab region [presence] in unspecified specimen by NAA with probe detection |
| 94640-0 | SARS coronavirus 2 S gene [presence] in respiratory specimen by NAA with probe detection |
| 94641-8 | SARS coronavirus 2 S gene [presence] in unspecified specimen by NAA with probe detection |
| 94647-5 | SARS-related coronavirus RNA [presence] in unspecified specimen by NAA with probe detection |
| 94660-8 | SARS coronavirus 2 RNA [presence] in serum or plasma by NAA with probe detection |
| 94756-4 | SARS coronavirus 2 N gene [presence] in respiratory specimen by nucleic acid amplification using Centers for Disease Control and Prevention primer-probe set N1 |
| 94757-2 | SARS coronavirus 2 N gene [presence] in respiratory specimen by nucleic acid amplification using Centers for Disease Control and Prevention primer-probe set N2 |
| 94758-0 | SARS coronavirus 2 E gene [presence] in respiratory specimen by NAA with probe detection |
| 94759-8 | SARS coronavirus 2 RNA [presence] in nasopharynx by NAA with probe detection |
| 94765-5 | SARS coronavirus 2 E gene [presence] in serum or plasma by NAA with probe detection |
| 94766-3 | SARS coronavirus 2 N gene [presence] in serum or plasma by NAA with probe detection |
| 94767-1 | SARS coronavirus 2 S gene [presence] in serum or plasma by NAA with probe detection |
| 94508-9 | SARS coronavirus 2 IgM antibody [presence] in serum or plasma by rapid immunoassay |
| 94564-2 | SARS coronavirus 2 IgM antibody [presence] in serum or plasma by immunoassay |
COVID-19, Coronavirus disease-2019; NAA, nucleic acid amplification; SARS, Severe acute respiratory syndrome.
Table E2.
Risk of clinical outcomes, risk of concurrent bacterial infection, and rates of antibiotic use in COVID-19 patients early and late in the pandemic
| Outcome | Early (January 20, 2020 to July 20, 2020) |
Late (July 21, 2020 to January 20, 2021) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No PCN allergy, n (%) (n = 3419) | PCN allergy, n (%) (n = 3419) | Risk ratio | 95% confidence interval | P | No PCN allergy, n (%) (n = 10,220) | PCN allergy, n (%) (n = 10,220) | Risk ratio | 95% confidence interval | P | |
| Hospitalization | 929 (27.2) | 1353 (39.6) | 1.46 | 1.36-1.56 | <.001 | 2312 (22.6) | 3370 (33.0) | 1.46 | 1.39-1.53 | <.001 |
| Acute respiratory failure | 652 (19.1) | 858 (25.1) | 1.32 | 1.20-1.44 | <.001 | 1698 (16.6) | 2103 (20.6) | 1.24 | 1.17-1.31 | <.001 |
| Intensive care unit level of care | 197 (5.76) | 248 (7.25) | 1.26 | 1.05-1.51 | .01 | 380 (3.72) | 463 (4.53) | 1.22 | 1.07-1.39 | .004 |
| Mechanical ventilation | 163 (4.77) | 202 (5.91) | 1.24 | 1.01-1.52 | .04 | 289 (2.83) | 352 (3.44) | 1.22 | 1.05-1.42 | .01 |
| Mortality | 185 (5.41) | 176 (5.15) | 0.95 | 0.78-1.16 | .63 | 325 (3.18) | 372 (3.64) | 1.15 | 0.99-1.33 | .07 |
| Bacterial infection | 119 (3.48) | 181 (5.29) | 1.52 | 1.21-1.91 | <.001 | 248 (2.43) | 357 (3.49) | 1.44 | 1.23-1.69 | <.001 |
| Antibiotic use | 1044 (30.5) | 1277 (37.4) | 1.22 | 1.14-1.31 | <.001 | 2448 (24.0) | 3272 (32.0) | 1.34 | 1.28-1.40 | <.001 |
CI, Confidence interval; COVID-19, coronavirus disease-2019; ICU, intensive care unit; PCN, penicillin.
Results are after 1:1 propensity score matching for baseline variables.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jutzeler C.R., Bourguignon L., Weis C.V., Tong B., Wong C., Rieck B. Comorbidities, clinical signs and symptoms, laboratory findings, imaging features, treatment strategies, and outcomes in adult and pediatric patients with COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020;37:101825. doi: 10.1016/j.tmaid.2020.101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Deng H., Ou C., Liang J., Wang Y., Jiang M. Clinical symptoms, comorbidities and complications in severe and non-severe patients with COVID-19: A systematic review and meta-analysis without cases duplication. Medicine (Baltimore) 2020;99:e23327. doi: 10.1097/MD.0000000000023327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alimohamadi Y., Sepandi M., Taghdir M., Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J Prev Med Hyg. 2020;61 doi: 10.15167/2421-4248/jpmh2020.61.3.1530. E304-E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant M.C., Geoghegan L., Arbyn M., Mohammed Z., McGuinness L., Clarke E.L. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): a systematic review and meta-analysis of 148 studies from 9 countries. PLoS One. 2020;15:e0234765. doi: 10.1371/journal.pone.0234765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention COVID data tracker. https://covid.cdc.gov/covid-data-tracker/#cases_casesper100klast7days Available from:
- 7.Centers for Disease Control and Prevention Coronavirus disease 2019 (COVID-19): people with certain medical conditions. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html?CDC_AA_refValhttps3A2F2Fwww.cdc.gov2Fcoronavirus2F2019-ncov2Fneed-extra-precautions2Fgroups-at-higher-risk.html Available from:
- 8.Feng Y., Ling Y., Bai T., Xie Y., Huang J., Li J. COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201:1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L.S., Chau S.K., Tso E.Y., Tsang S.W., Li I.Y., Wong B.K. Bacterial co-infections and antibiotic prescribing practice in adults with COVID-19: experience from a single hospital cluster. Ther Adv Infect Dis. 2020;7 doi: 10.1177/2049936120978095. 2049936120978095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothe K., Feihl S., Schneider J., Wallnöfer F., Wurst M., Lukas M. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis. 2021;40:859–869. doi: 10.1007/s10096-020-04063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan S.H., Ng T.M., Tay H.L., Yap M.Y., Heng S.T., Loo A.Y.X. A point prevalence survey to assess antibiotic prescribing in patients hospitalized with confirmed and suspected coronavirus disease 2019 (COVID-19) J Glob Antimicrob Resist. 2021;24:45–47. doi: 10.1016/j.jgar.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macy E., Contreras R. Health care use and serious infection prevalence associated with penicillin "allergy" in hospitalized patients: a cohort study. J Allergy Clin Immunol. 2014;133:790–796. doi: 10.1016/j.jaci.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Sousa-Pinto B., Cardoso-Fernandes A., Araújo L., Fonseca J.A., Freitas A., Delgado L. Clinical and economic burden of hospitalizations with registration of penicillin allergy. Ann Allergy Asthma Immunol. 2018;120:190–194.e2. doi: 10.1016/j.anai.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Blumenthal K.G., Ryan E.E., Li Y., Lee H., Kuhlen J.L., Shenoy E.S. The impact of a reported penicillin allergy on surgical site infection risk. Clin Infect Dis. 2018;66:329–336. doi: 10.1093/cid/cix794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joint Task Force on Practice Parameters; American Academy of Allergy, Asthma and Immunology; American College of Allergy, Asthma and Immunology; Joint Council of Allergy, Asthma and Immunology Drug allergy: an updated practice parameter. Ann Allergy Asthma Immunol. 2010;105:259–273. [Google Scholar]
- 17.Macy E., Schatz M., Lin C., Poon K.Y. The falling rate of positive penicillin skin tests from 1995 to 2007. Perm J. 2009;13:12–18. doi: 10.7812/tpp/08-073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacco K.A., Bates A., Brigham T.J., Imam J.S., Burton M.C. Clinical outcomes following inpatient penicillin allergy testing: a systematic review and meta-analysis. Allergy. 2017;72:1288–1296. doi: 10.1111/all.13168. [DOI] [PubMed] [Google Scholar]
- 19.Gadde J., Spence M., Wheeler B., Adkinson N.F. Clinical experience with penicillin skin testing in a large inner-city STD clinic. JAMA. 1993;270:2456–2463. [PubMed] [Google Scholar]
- 20.Harrison S.L., Fazio-Eynullayeva E., Lane D.A., Underhill P., Lip G.Y.H. Comorbidities associated with mortality in 31,461 adults with COVID-19 in the United States: a federated electronic medical record analysis. PLoS Med. 2020;17:e1003321. doi: 10.1371/journal.pmed.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taquet M., Luciano S., Geddes J.R., Harrison P.J. Bidirectional associations between COVID-19 and psychiatric disorder: retrospective cohort studies of 62 354 COVID-19 cases in the USA. Lancet Psychiatry. 2021;8:130–140. doi: 10.1016/S2215-0366(20)30462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieswerda E., de Boer M.G.J., Bonten M.M.J., Boersma W.G., Jonkers R.E., Aleva R.M. Recommendations for antibacterial therapy in adults with COVID-19 - an evidence based guideline. Clin Microbiol Infect. 2021;27:61–66. doi: 10.1016/j.cmi.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staub M.B., Beaulieu R.M., Graves J., Nelson G.E. Changes in antimicrobial utilization during the coronavirus disease 2019 (COVID-19) pandemic after implementation of a multispecialty clinical guidance team. Infect Control Hosp Epidemiol. 2020;42:810–816. doi: 10.1017/ice.2020.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta S.D., Talwar A., Lee J.T. A proposed framework and timeline of the spectrum of disease due to SARS-CoV-2 infection: illness beyond acute infection and public health implications. JAMA. 2020;324:2251–2252. doi: 10.1001/jama.2020.22717. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected, 13 March 2020. https://apps.who.int/iris/handle/10665/331446 Available from:
- 27.Dieringer T.D., Furukawa D., Graber C.J., Stevens V.W., Jones M.M., Rubin M.A. Inpatient antibiotic utilization in the Veterans' Health Administration during the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol. 2021;42:751–753. doi: 10.1017/ice.2020.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abelenda-Alonso G., Padullés A., Rombauts A., Gudiol C., Pujol M., Alvarez-Pouso C. Antibiotic prescription during the COVID-19 pandemic: a biphasic pattern. Infect Control Hosp Epidemiol. 2020;41:1371–1372. doi: 10.1017/ice.2020.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penicillin Allergy in Antibiotic Resistance Workgroup. Penicillin allergy testing should be performed routinely in patients with self-reported penicillin allergy. J Allergy Clin Immunol Pract. 2017;5:333–334. doi: 10.1016/j.jaip.2016.12.010. [DOI] [PubMed] [Google Scholar]