Abstract
Objective:
To test the hypothesis that mutations in the parathyroid hormone 1 receptor (PTH1R) include effects in both primary and permanent teeth.
Materials and Methods:
DNA was extracted from saliva samples of 29 patients (8 familial and 21 sporadic) who presented with clinical evidence of infraoccluded teeth, and their unaffected relatives (N = 22). Sequencing followed by mutational analysis of the coding regions of PTH1R gene was completed for all individuals (N = 29).
Results:
Eight of 29 cases revealed a heterozygous pathogenic variant in the PTH1R gene; five of eight variants represented distinct mutations based on comparison with the dbSNP, HGMD, and ESP databases. One mutation (c.1765 T>C p.Trp89Arg) was found to segregate within a family (n = 3). In silico analyses for all variants revealed a putative pathogenic effect. A genotype-phenotype correlation was reported as defined by a functional mutation in PTH1R and corresponding effects on one or more posterior teeth only; unilateral or bilateral involvement, infraoccluded primary teeth.
Conclusions:
Novel mutations were reported in the PTH1R gene that included PFE-affected primary molars, thus providing the basis for using a genetic diagnostic tool for early diagnosis leading to proper management.
Keywords: Primary failure of eruption (PFE), Dental eruption, PTH1R
INTRODUCTION
Tooth eruption is the developmental process whereby the developing tooth within the bony crypt traverses the bone of the jaws into the oral cavity until it reaches functional occlusion.1 The role of the dentist and orthodontist is to monitor eruption patterns to avoid subsequent occlusal problems secondary to eruption disorders. It is not uncommon for the general dentist or dental specialist to encounter patients with occlusal problems resulting from eruption disorders. Pediatric dental and orthodontic specialists routinely monitor for aberrations in normal dental eruption such as timing, sequence, and extent of eruption.
The normal eruptive process requires the passage of the teeth through the bones and oral epithelium in a precise bilateral temporal sequence that must be coordinated with the growth of the maxilla and mandible in the three spatial planes. Three factors contribute to this complex process: (1) bone resorption,( 2) gingival resorption (both above the dental follicle), and (3) root elongation at the apex of the follicle.2 Bone resorption is the result of osteoclastogenesis, which does not require the force of an erupting tooth but is instead a genetically programmed process.3,4 This alteration in balance between the resorptive and the appositional processes are the putative factors underlying the development of primary failure of eruption (PFE).5 Recent investigations have, however, focused on gene discovery as an important logical first step into uncovering the mechanistic underpinnings of PFE.
The term PFE was coined by Proffit and Vig (1981) to indicate a defect in the eruption mechanism that is not due to an obvious obstruction. In most cases, PFE-affected teeth erupt partially (supracrestally) and then are arrested before they reach functional occlusion.6,7 A diagnostic approach to delineate PFE from other eruption disorders, based on clinical and genetic characterization of PFE-affected cohorts compared with other eruption disorders, provides a diagnostic rubric.8 This rubric defines a critical first step to determine whether the eruption pathway is clear. That is, the presence of a mechanical obstruction should be ruled out using current and historical clinical radiographs and photos. This includes knowledge of whether a past obstruction has led to the current infraoccluded position of the affected teeth. A second step is to determine whether at least one first molar is affected. Other characteristics that occur with varying frequency include predominantly affected posterior teeth, involvement of the primary and permanent teeth, unilateral or bilateral occurrence tendency to develop ankylosis after application of orthodontic force, and isolated localization without systemic involvement.6
Since the discovery that the PTH1R gene was responsible for the development of PFE, the literature continues to amass PFE-associated mutations in PTH1R. The road to unravel the genotype-phenotype correlation is still quite elusive and even more puzzling since the spectrum of disorders arising from PTH1R mutations is so broad. Indeed, mutations in PTH1R are associated with lethal dwarfism, chondrodysplasia, and isolated dental disorders specific to eruption.9 The complete understanding of this wide phenotypic spectrum for similar mutations is enigmatic, requiring a complete understanding of the structure and function of the PTH1R gene.
This gene is located on chromosome 3p21-p22.1, MIM # and contains 16 exons.10 The resultant protein is a transmembrane glycoprotein (593 amino acids) consisting of an extended extracellular ligand-binding amino terminal, an intracellular G-protein–associated carboxyl terminal domain, a signal peptide formed by 28 amino acids, an extracellular domain of 150 amino acids with the hormone binding site, and the transmembrane domain containing 7 alpha-helices and an intercellular domain.11 The extracellular domain binds two different ligands: parathyroid hormone (PTH), which acts principally at the level of the liver and osteocytes, and parathyroid hormone-related peptide (PTHrP). Both these ligands allow regulation of the intracellular calcium levels and favor bone metabolism.12,13 PTH is produced exclusively by the parathyroid gland. However, a number of tissues produce PTHrP and cellular types including endothelium, chondrocytes, bone, liver, smooth muscle cells, and tooth buds.14 Only one normal copy of PTH1R is sufficient—but necessary—to carry out its functions in most of the tissues. In tooth development, significant quantities of PTH1R are necessary.15 For this reason, it was speculated that the PFE phenotype could be the result of a dose-dependent inactivation of PTH1R. Moreover, the observation that some autosomal dominant mutations of the PTH1R gene do not affect the teeth or other skeletal structures except for osteoarthritis has been noted. Recent studies have confirmed the association between osteoarthritis and low levels of PTH1R in chondrocytes.11,16
An overarching goal of genetic studies in this postgenomic era is to provide meaningful clinical applications to improve diagnostic accuracy. Studies of PFE continue to refine the descriptions of the eruption disorder, but a complete understanding of its pathogenesis remains elusive. Further, clinical parameters based on a complete dissection of the PFE phenotype should include a genotype:phenotype correlation of the dentition and craniofacial structures. Previous studies have revealed that PFE occurs more rarely in the primary dentition but always includes permanent teeth. In this report, families and multiple individuals who showed clear involvement of the mixed dentition as well as permanent teeth created an opportunity to expand the window of diagnosis and therefore management at an earlier age. These findings, therefore, mark an opportunity to refine the genotype:phenotype correlation of PFE and PTH1R and improve diagnostic accuracy.
MATERIALS AND METHODS
Subjects
A cohort of 51 patients (ranging in age between 7 and 45 years) with a familial eruption disorder (infraocclusion of at least one tooth in at least one member of the family) was recruited to participate in this study. Twenty-nine of these patients had features of PFE.8 Twenty-two unaffected relatives were recruited and served as control subjects. Diagnosis of infraocclusion was determined through (1) clinical examination consisting of intraoral and extraoral photos, (2) panoramic X-ray, and (3) patient interview. The criteria for eruption disorder diagnosis included the phenotypical characteristics previously described including PFE types.8,17 Clinical examination of patients was conducted at the Dental Institute of the Catholic University of the Sacred Heart in Rome (UCSC), and mutational studies were carried out at the laboratories of the Medical Genetic Institute and the Dental Institute of the UCSC. This study was approved by the Ethics Committee of the UCSC.
Mutational Analysis
The DNA from saliva was extracted using a Wizard Genomic DNA Purification Kit (Promega Corp, Fitchburg, Wisc). Sequencing followed by mutational analysis of PTH1R was carried out for all subjects using primers (Genamics Expression 1.1 Bio-Soft.Net Sequence Analysis and Alignment for Windows; Table 1) designed for all coding regions and the intron-exon junctions of the PTH1R gene. Purification of PCR products with the PCR Product Pre-Sequencing Kit (Amersham Pharmacia Biotech, Piscataway, NJ) containing serum alkaline phosphatase (SAP) and an exonuclease was completed. Sequencing of PCR products was carried out with Big Dye Terminator 3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, Calif), purified with Big Dye X-Terminator Kit (Life Technologies), and analyzed using an automated sequencer AB1 3130 Genetic Analyzer (Life Technologies). Variants were compared with the dbSNP, HGMD, and ESP databases to determine whether the variant was novel or previously reported.
Table 1.
Primer Sets Designed and Used for PCR Amplification of PTH1R Gene (Annealing Temperature, 55°C)
| Gene |
Exon |
Sequence |
| PTH1R | 3 FORWARD | AGCCTGACGCAGCTCTGCA |
| 3 REVERSE | CCCACAGTCCAGACATCCCA | |
| PTH1R | 4 FORWARD | AGAGCAGATTCCCCACATGC |
| 4 REVERSE | TTCACCTGGCTCTGTATCCT | |
| PTH1R | 5 FORWARD | TCCTCACCCATCGTCTCAGAT |
| 5 REVERSE | AAGAGCCAAGAAGCATGAGC | |
| PTH1R | 6 FORWARD | AGATGTATTCATCCTTCTGGG |
| 7 REVERSE | TAAGGTTGCTGGAGGAGTCAAG | |
| PTH1R | 8 FORWARD | AAATTCACTCCCACCCCACG |
| 8 REVERSE | TGGACAGGAAGCTGGGTTGT | |
| PTH1R | 9 FORWARD | ACAACCCAGCTTCCTGTCCA |
| 9 REVERSE | GTTGCGAGGGACCCTATAAG | |
| PTH1R | 10 FORWARD | AAACGAAGCCTGCCCCTTC |
| 10 REVERSE | GCCTGGAATAGGGTCAGGAT | |
| PTH1R | 11 FORWARD | GGAATGACCTTGTGGACAGC |
| 11 REVERSE | TAGCTGTTGAGGACACAGGG | |
| PTH1R | 12 FORWARD | AGGGTCACAGGAGGCTACTT |
| 12 REVERSE | TGTCACTGCATCTCTGGGTG | |
| PTH1R | 13 FORWARD | CCAGCCCAGAAAGGAAAACC |
| 13 REVERSE | TAGTGCAGGGCCTGGTACAA | |
| PTH1R | 14 FORWARD | AGGTGAACTGGGTTGTCCTC |
| 15 REVERSE | GAATGTCCTCAGGGGTGTTC | |
| PTH1R | 16 FORWARD | CACTTGGCCTTGGAGTTTCC |
| 16 REVERSE | CCACCCATCTTTTGGTCC |
In Silico Analysis
Sequence data were analyzed using Mutation Surveyor V3.3. Unknown sequence variants were evaluated by SIFT, PolyPhen2, and AlignGVGD. Variants suspected to affect correct splicing were evaluated for splice-site strength or branch point prediction based on five different algorithms (SpliceSiteFinder, MaxEntScan, NNSPLICE, GeneSplicer, Human Splicing Finder) using the bioinformatics tools of the Alamut software v.1.5 (Interactive Biosoftware, http://www.interactive-biosoftware.com/alamut.html). An in silico analysis was used to evaluate the folding of the protein and the eventual pathogenicity of such a variant by using the dedicated software Sorting Intolerant from Tolerant (SIFT) and Phenotyping (PolyPhen-2).
RESULTS
Mutational and in Silico Analysis
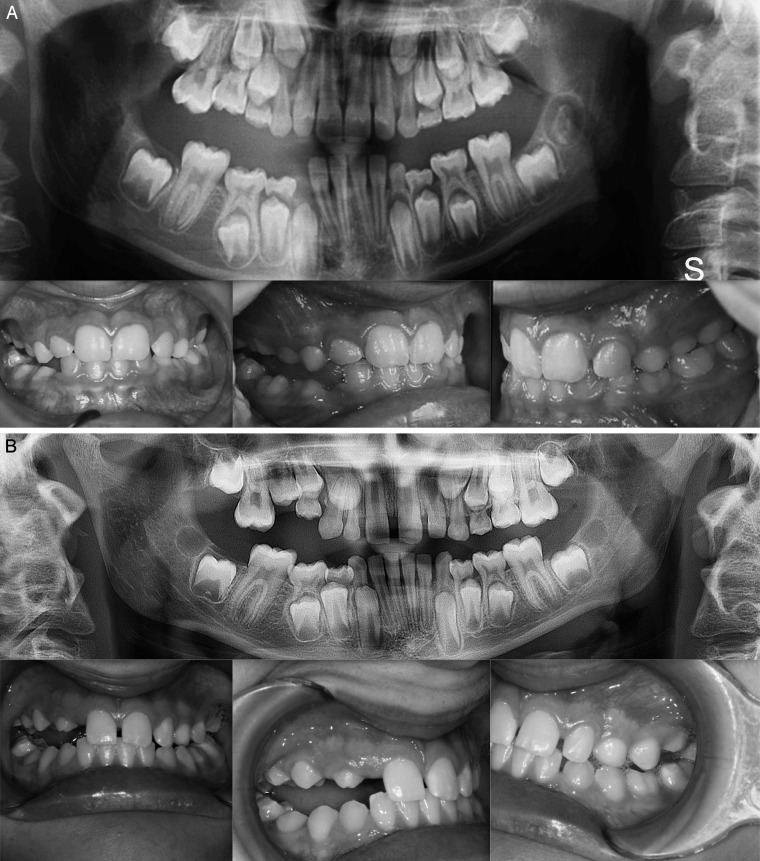
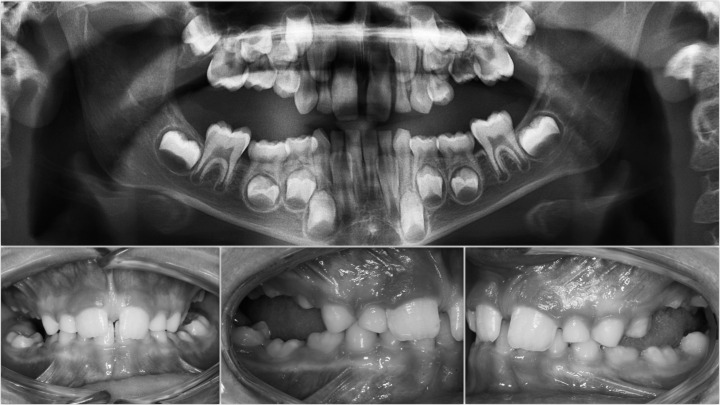
Eight of 29 cases revealed a heterozygous pathogenic variant in the PTH1R gene. A summary of the mutations identified in this study is shown in Table 2. Briefly, three missense variants were identified in five of eight affected subjects that have not been described previously based on comparison with published databases (Table 3). A variant in the CTCF binding site (c.313+32 A>G rs113566258 SNP) was identified in two brothers (Figure 1A,B). The siblings were diagnosed as Type I PFE and showed bilateral presentation. This variant (c.313+32 A>G downstream of exon 5 of the PTH1R gene) occurred in a regulatory region that interacts with transcription factors. In addition to this mutation, the brothers were affected by a second mutation that was unique to each of them: one sibling (II:1) also harbored a frameshift deletion that occurred in the amino terminus of the gene in exon 16, c.1593DelC, p.(Pro532Leufs*) (Figure 1A,B). In the other sibling (II:3), a synonymous variant in exon 10, c.1152 G>A, (rs200475872; Figure 1A,B) was identified that did not cause variation at the level of the protein amino acid sequence, but involved a change in the nitrogenous base. Based on in silico analysis, this change may compromise the folding of mRNA and influence its half-life and protein production. In this sample of patients, the same synonymous variant, c.1152G>A, was found in an isolated case, featuring a severe open bite due to PFE (Figure 2).
Table 2.
Summary of Mutations in PFE Patients
| Nucleotide Change |
Variant |
Reference |
| c.64 C>T | Missense variant | This study |
| p.Ala22Val | Novel Mutation | |
| c.313+32 A>G | (CTCF Binding Site) | rs113566258 |
| c.1152 G>A | Synonymous variant | rs200475872 |
| c.1593 Del C | Frame shift deletion | This study |
| p.(Pro532Leufs*) | Novel mutation | |
| c.1765 T>C | Missense variant | This study |
| p.Trp589Arg | Novel mutation |
Table 3.
Databases
| 1. ENSEMBL: http://www.ensembl.org/index.htm |
| 2. GENBANK: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide&itool=toolbar |
| 3. UCSC: https://genome.ucsc.edu/ |
| 4. 1000 Genomi: http://www.1000genomes.org |
Figure 1.
(A, B) Intraoral photos of two brothers. Panoramic: variant downstream of exon 5 (CTCF Binding site) rs113566258 SNP c.313+32 A>G; variant in exon 16 (position 3:46903467) c.1593-95 Del C, p.531/532; synonymous variant in exon 10 (position 3:46899419) c.1152 G>A.
Figure 2.
Intraoral photos. Panoramic: synonymous variant in exon 10 (position 3:46899419) c.1152 G>A.
In another isolated case of PFE, a missense mutation de novo, c.64 C>T p. Ala22Val, not described previously, was found in exon 3. Such a variant occurred in the extracellular domain of the PTH1R protein at the level of the signal peptide, and it determined the change of an amino acid. However, since the two amino acids have the same charge, the mutation is not considered highly pathogenic.
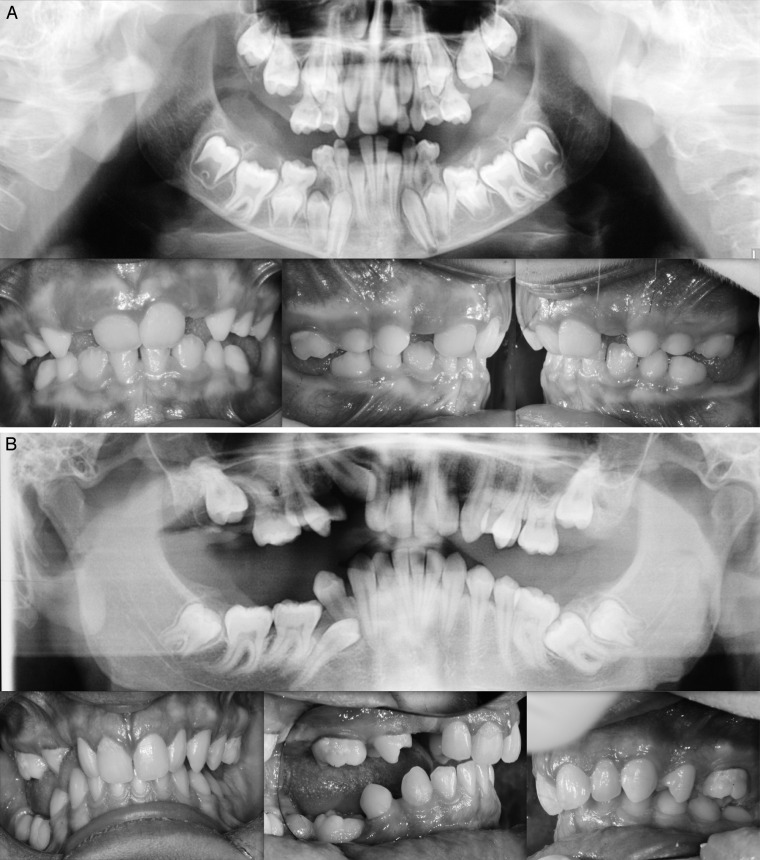
Finally, a missense variant, not previously reported in the data base, was identified in exon 16 c.1765 T>C p.Trp89Arg. The missense variant, found in three members of the same family—mother, daughter, and son (Figure 3A,B)—was responsible for substituting the amino acid tryptophan with arginine p: Trp89Arg, an amino acid with different chemical characteristics. This mutation occurred in the cytoplasmic domain of the PTH1R protein and is involved in the interaction of the receptor with the G proteins that activate the intracellular cascade signal. Further, the in silico results suggest that this amino acid substitution in the PTH1R protein alters the structure and function of protein since it occurred within a catalytic domain of the protein.
Figure 3.
(A, B) Intraoral photos of two brothers. Panoramic: familial missense variant in exon 16 c.1765 T>C p.589 W>R.
The younger daughter (II:2) belonging to family 2 showed a more complex clinical picture with a lack of eruption of the elements from the permanent as well as deciduous series and with cystic formations (Figure 3A,B). A severe, bilateral, posterior open bite due to infraocclusion of numerous permanent teeth was found in the elder brother (II:1; Figure 3A,B). Finally, the mother of patients II:1 and II:2 presented with bilateral infraocclusion of both upper first molars and lower premolars. An absence of occlusal contacts at the level of the premolars was found (Figure 3A,B).
Clinical Features
Clinical characterization of the infraocclusion identified in this study was consistent with PFE based on a genetic diagnosis and application of an eruption disorder diagnostic rubric.7 A careful clinical examination of the specific associated dental features includes the following: at least one tooth with infraocclusion, the infraocclusion is limited to the posterior region, uni- (38%; Figure 1A) and bilateral presentation (35%; Figures 1B and 2), an involvement of at least one deciduous molar (48% deciduous; Figures 1A,B, and 3A; 55% permanent, Figure 3B). Moreover, dentofacial features were observed as follows: vertical skeletal asymmetry due to severity of the lateral open bite on the affected side (35%; Figure 1A,B, and Figure 3A,B); facial asymmetry consisting of lateral deviation of the mandible (Figure 1A,B, and Figure 2; Table 4). This asymmetry was more evident in patients with unilateral open bite. It was further discovered that affected patients cosegregated maxillary constriction and Class III dental/skeletal relationships (28%; Figure 1A,B, and Figure 2) as previously reported.16 Infraocclusion was present in at least one blood relative of nine patients. The phenotypic findings and associated mutational analysis of PTH1R (N = 8) are summarized in Table 4.
Table 4.
Summary of Features in Patient With Mutation in PTH1R
| Mutations: Nucleotide Change |
c.313+32 A>G c.I593 Del C p•(Pro532Leufs9) Fig 1A,B |
c.313+32 A>6 c.1152 GA Fig 1A,B |
c.1152 G>A Fig 2 |
c. 1152 G>A p.(Pro532Leufs*) |
c.64 C>T p. Ala22Val |
c.1765T>C p. Trp58963/ Fig 3A,B |
c. 1765T>C p. Trp589 arl |
c. 1765T>C |
| Permanent molars | x | x | x | x | x | x | x | |
| Deciduous molars | x | x | x | x | x | x | ||
| Dental Class I | x | x | x | x | x | |||
| Dental Class II | ||||||||
| Dental Class III | x | x | x | |||||
| Unilateral PFE | x | x | ||||||
| Bilateral PFE | x | x | x | x | x | x | ||
| Skeletal Class III | x | x | x | |||||
| Deepbite | x | x | x | |||||
| Unilateral open bite | x | x | x | |||||
| Bilateral open bite | x | x | x | |||||
| Facial asymmetry | x | x | ||||||
| Others dental anomalies (agenesis, follicular cysts) | x | x | x |
DISCUSSION
In this study, five mutations were identified: three novel and two reported previously (rs113566258 and rs20047872). The PTH1R alterations included (1) a variant at the level of the gene regulatory region to which transcriptional factors (CTCF binding site) bind, (2) a frameshift deletion, (3) a synonymous variant, and (4) two missense variants. Estimating the pathogenicity of certain variants was difficult, but the cause-and-effect relationship between PTH1R gene mutations and PFE was supported through in silico analysis in those variants that led to the production of a truncated protein. Future functional studies will provide a more detailed analysis of the consequence of the mutations described in this report.
Isolated and familial cases were investigated in this cohort that included five families with more than one member affected by infraocclusion of at least one molar. A key finding of this study was that the variants found in two families and three unrelated patients was associated with infraocclusion in the mixed dentition. A positive correlation with PFE in the mixed dentition was reported and hence the finding supports the development of a protocol for early genetic diagnosis during the mixed dentition stage.
These results mark the most comprehensive documentation of the involvement of the mixed dentition for individuals with functional mutations in the PTH1R gene. The missense variant found in family 2 was similar to those reported by Yamaguchi and also revealed a similar phenotype.16 While the emphasis has been placed on familial inheritance as a compelling sign to detect PFE, the findings of this study support the investigation of infraoccluded primary teeth and family history as a combined approach to improve early detection.8,18,19 Rhoads et al. identified a c.1092delG, a frameshift mutation in a young boy and his parent that showed involvement of the mixed dentition.7 The c.1092delG variant that caused a truncated protein was also associated with a skeletal and dental Class III malocclusion, leading to a moderate open bite on the right side. The current report expands the finding of Rhoads et al. by identifying novel mutations in multiple families (Figure 1A,B).
CONCLUSIONS
The results of this research confirm that mutations of the PTH1R gene are responsible for eruption failure with a varied clinical presentation consistent with previous descriptions of PFE. These phenotypes can be found early at the level of the deciduous and then in the permanent dentition.
A clinical diagnosis combined with genetic analysis of the PTH1R gene will not only ensure an early diagnosis, but also serve as an alert for screening of potentially affected relatives.
Taken together, the frequency of PFE and the number of patients displaying genetic heterogeneity of the PTH1R gene suggest that PFE caused by mutations in PTH1R is underestimated.
Based on these observations, genetically-guided orthodontics can bring clinical, therapeutic, and even economic advantages in managing problems of dental eruption.
The knowledge acquired through a genetic exam could guide the clinician in elaborating a correct orthodontic treatment plan, avoiding unnecessary and long therapy for patients that result in failure and expose them to iatrogenic damage.
REFERENCES
- 1.Ten Cate AR. Oral Histology, Development, Structure and Function. St Louis: Mosby; 1989. [Google Scholar]
- 2.Cahill DR, Marks SC. Tooth eruption: evidence for the central role of the dental follicle. J Oral Path. 1983;9:189–200. doi: 10.1111/j.1600-0714.1980.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 3.Cahill DR. Eruption pathway formation in the presence of experimental tooth impaction in puppies. Anat Rec. 1969;164:67–77. doi: 10.1002/ar.1091640105. [DOI] [PubMed] [Google Scholar]
- 4.Wise GE, Frazier Bowers S, D'Souza RN. Cellular, molecular, and genetic determinants of tooth eruption. Crit Rev Oral Biol Med. 2002;13(4):323–334. doi: 10.1177/154411130201300403. [DOI] [PubMed] [Google Scholar]
- 5.Frazier Bowers S, Puranik CP, Nahaney MC. The etiology of eruption disorders—further evidence of a “genetic paradigm.”. Semin Orthod. 2010;16-3:180–185. doi: 10.1053/j.sodo.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proffit WR, Vig KW. Primary failure of eruption: a possible cause of posterior open bite. Am J Orthod. 1981;80:173–190. doi: 10.1016/0002-9416(81)90217-7. [DOI] [PubMed] [Google Scholar]
- 7.Pilz P, Meyer-Marcotty P, Eigenthaler M, Roth H, Weber BH, Stellzig-Eisenhauer A. Differential diagnosis of primary failure of eruption (PFE) with and without evidence of pathogenic mutations in the PTHR1 gene. J Orofac Orthop. 2014 May;75(3):226–239. doi: 10.1007/s00056-014-0215-y. [DOI] [PubMed] [Google Scholar]
- 8.Rhoads SG, Hendricks HM, Frazier-Bowers S. Establishing the diagnostic criteria for eruption disorders based on genetic and clinical data. Am J Orthod Dentofacial Orthop. 2013;144:194–202. doi: 10.1016/j.ajodo.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Roth H, Fritsche LG, Meier C, Pilz P. Expanding the spectrum of PTH1R mutations in patients with primary failure of tooth eruption. Clin Oral Invest. 2013;18(2):377–384. doi: 10.1007/s00784-013-1014-3. [DOI] [PubMed] [Google Scholar]
- 10.Decker E, Stellzing-Eisenhauer A, Fiebig BS et al. PTH1R loss-of-function mutations in familial, nonsyndromic primary failure of tooth eruption. Am J Hum Genet. 2008;83:781–786. doi: 10.1016/j.ajhg.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frazier Bowers S, Hendricks HM, Wight JL, Long K, Dibble CF, Bencharit S. Novel mutations in PTH1R associated with primary failure of eruption and ostheoarthritis. J Dent Res. 2014;93(2):134–139. doi: 10.1177/0022034513513588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta NS, Abou-Samra AB. PTH and PTHrP signaling in osteoblasts. Cell Signal. 2009;21:1254. doi: 10.1016/j.cellsig.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strewler GJ. The physiology of parathyroid hormone-related protein. N Eng J Med. 2000;342:177–185. doi: 10.1056/NEJM200001203420306. [DOI] [PubMed] [Google Scholar]
- 14.Lavi-Moshayoff V, Wasserman G, Meier T, Silver J, Naveh-Many T. PTH increases FGF23 gene expression and mediates the high FGF23 levels of experimental kidney failure: a bone parathyroid feedback loop. Am J Physiol Renal Physiol. 2010;299:F882–F889. doi: 10.1152/ajprenal.00360.2010. [DOI] [PubMed] [Google Scholar]
- 15.Wise GE, King GJ. Mechanisms of tooth eruption and orthodontic tooth movement. J Dent Res. 2008;87:414–434. doi: 10.1177/154405910808700509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frazier Bowers S, Koeheler K, Ackerman J, Proffit W. Primary Failure of eruption: further characterization of a rare eruption disorder. Am J Orthod Dentofacial Orthop. 2007;131:578e1–578e11. doi: 10.1016/j.ajodo.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi T, Hosomichi K, Narita A, Shirota T, et al. Exon resequensing combined with linkage analysis identifies novel PTH1R variants in primary failure of tooth eruption in Japanese. J Bone Miner Res. 2011;26:1655–1661. doi: 10.1002/jbmr.385. [DOI] [PubMed] [Google Scholar]
- 18.Frazier Bowers S, Simmons D, Wright T, Proffitt WR, Ackerman JL. Primary failure of eruption and PTH1R: the importance of a genetic diagnosis for orthodontic treatment planning. Am J Orthod Dentofacial Orthop. 2010;137:160–161. doi: 10.1016/j.ajodo.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Stellzig-Eisenhauer A, Decker E, Meyer-Marcotty P, et al. Primary failure of eruption (PFE). Clinical and molecular genetics analysis. Orthod Fr. 2013 Sep;84(3):241–250. doi: 10.1051/orthodfr/2013055. [DOI] [PubMed] [Google Scholar]