Abstract
Salt stress is a major environmental factor limiting plant growth and productivity. We recently discovered an important new salt tolerance pathway, where the cell wall leucine-rich repeat extensins LRX3/4/5, the RAPID ALKALINIZATION FACTOR (RALF) peptides RALF22/23 and receptor-like kinase FERONIA (FER) function as a module to simultaneously regulate plant growth and salt stress tolerance. However, the intracellular signaling pathways that are regulated by the extracellular LRX3/4/5-RALF22/23-FER module to coordinate growth, cell wall integrity and salt stress responses are still unknown. Here, we report that the LRX3/4/5-RALF22/23-FER module negatively regulates the levels of jasmonic acid (JA), salicylic acid (SA) and abscisic acid (ABA). Blocking JA pathway rescues the dwarf phenotype of the lrx345 and fer-4 mutants, while disruption of ABA biosynthesis suppresses the salt-hypersensitivity of these mutants. Many salt stress-responsive genes display abnormal expression patterns in the lrx345 and fer-4 mutants, as well as in the wild type plants treated with epigallocatechin gallate (EGCG), an inhibitor of pectin methylesterases, suggesting cell wall integrity as a critical factor that determines the expression pattern of stress-responsive genes. Production of reactive oxygen species (ROS) is constitutively increased in the lrx345 and fer-4 mutants, and inhibition of ROS accumulation suppresses the salt-hypersensitivity of these mutants. Together, our work provides strong evidence that the LRX3/4/5-RALF22/23-FER module controls plant growth and salt stress responses by regulating hormonal homeostasis and ROS accumulation.
Keywords: salt stress, cell wall integrity, growth, LRX, RALF, FER, hormone
INTRODUCTION
Leucine-rich repeat extensins (LRXs) are cell wall-localized proteins important for the regulation of cell wall integrity. In Arabidopsis, there are 11 LRX proteins, named LRX1-LRX11 [1,2]. Mutations in LRX8, LRX9 and LRX11 lead to altered compositions of extensins, arabinogalactan proteins, rhamnogalacturonan I and xyloglucan in the cell wall of pollen tubes [3,4]. It has also been reported that callose in the apical walls of pollen tubes accumulates in the lrx8 lrx9 lrx11 and lrx9 lrx10 lrx11 mutants [3–5]. In the lrx3 lrx4 lrx5 (hereafter referred to as lrx345) triple mutant, the amounts of rhamnose, galactose, arabinogalactan proteins, extensins and arabinan are reduced, while the levels of mannose and lignin are increased compared with the wild type [6]. The mechanisms by which LRX proteins regulate the deposition of cell wall components remain to be elucidated.
LRX proteins harbor an N-terminal LRR domain and a C-terminal extensin domain, both of which function in the extracellular region [2,7]. The lack of an intracellular domain suggests that LRX proteins need partners to transduce cell wall signals to the cell interior. LRX8-LRX11 physically interact with RALF4/19 in the cell wall of pollen tubes and feed into the ANX1/2-mediated signaling pathway, and thus regulate pollen germination and pollen tube growth [8,9]. The binding of LRXs with RALF4 has been confirmed by a recent structural study, which showed that these two components exhibit a strong affinity and LRX proteins preferentially bind folded RALF peptides [10]. It has also been shown that RALF4/19 are ligands for ANX1/2 and BUPS1/2 [11]. Our previous study showed that LRX3, LRX4 and LRX5 function together with RALF22/23 and FER to regulate plant growth and salt stress responses [12]. FER is a plasma membrane kinase that acts as a receptor of several RALF peptides [13,14]. Recently, it was reported that THE1 is a receptor for RALF34 and the THE1-RALF34 module is required for the regulation of lateral root initiation [15]. Notably, ANX1/2, FER and THE1 all belong to the Catharanthus roseus receptor-like kinase 1-like (CrRLK1L) protein family [9]. Based on these data, we can speculate that LRXs, RALFs and CrRLK1L family proteins function as modules to monitor cell wall status and transduce cell wall signals and regulate cell wall integrity in different tissues or upon exposure to different environmental stresses.
The lrx345 and fer-4 mutants as well as transgenic plants overexpressing RALF22 or RALF23 display similar phenotypes, including dwarfism, salt-hypersensitivity, increased accumulation of anthocyanin and increased susceptibility to bacteria [12,14]. It was proposed that FER regulates cell expansion via phosphorylation of the proton pump AHA2 [13,16], although genetic evidence for this is still lacking. FER positively regulates the phosphatase activity of ABI2, a negative regulator of the ABA core signaling pathway, and thus negatively modulates ABA sensitivity [17]. FER directly binds with pectins and is required for the activation of downstream cell wall repair pathways via Ca2+-mediated signals under salt stress [18]. The reduced root elongation of the fer mutant under salt stress could be caused by impaired cell wall integrity, which results in dramatic root cell burst during growth recovery [18]. FER is also involved in the positive regulation of plant immunity through the regulation of the association of the FLS2/EFR-BAK1 complex and the stability of MYC2 [14,19].
So far, the mechanisms underlying the enhanced cell death of the lrx345 and fer mutants under high salinity remain unknown. Here, we investigated intracellular signaling pathways that are regulated by the LRX3/4/5-RALF22/23-FER module. Our results show that three hormones, JA, SA and ABA, are consitutively increased in the lrx345 and fer mutants, and that elevated levels of these hormones are responsible for the dwarf phenotype and salt-hypersensitivity of the lrx345 and fer-4 mutants. Our data also reveal that LRX3/4/5-RALF22/23-FER module-mediated cell wall integrity signals play a crucial role in determining the expression of stress-responsive genes after both short and prolonged exposure to high salinity.
RESULTS
Jasmonic acid (JA)- and salicylic acid (SA)-responsive genes are constitutively up-regulated in the lrx345 triple mutant
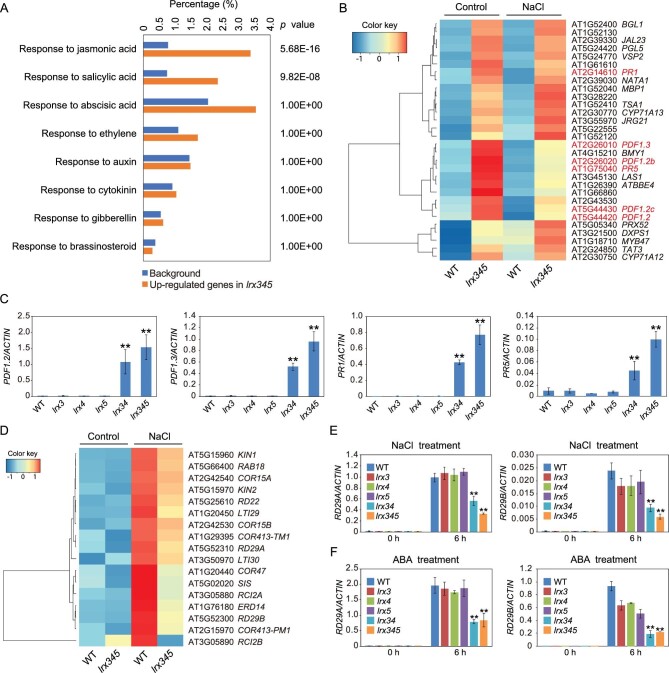
To gain insight into the mechanisms underlying the dwarf phenotype and salt-hypersensitivity of the lrx345 triple mutant [12], we performed RNA-seq analysis for the wild type and lrx345 mutant seedlings before and after NaCl treatment for 6 h. We first compared gene expression between the wild type and lrx345 mutant without NaCl treatment. In total, 1418 genes were significantly up-regulated (fold change >2, P value < 0.05) and 1609 genes were significantly down-regulated (fold change >2, P value < 0.05) in the lrx345 mutant (Table S1). Gene ontology (GO) enrichment analysis showed that genes belonging to categories ‘response to jasmonic acid’ (P value = 5.68E-16) and ‘response to salicylic acid’ (P value = 9.82E-08) were significantly enriched among the up-regulated genes (Fig. 1A). Specifically, 34.2% of JA-responsive genes, including PDF1.2, PDF1.2b, PDF1.2c, PDF1.3 and VSP2, and 30.8% of SA-responsive genes, including PR1 and PR5, were constitutively up-regulated in the lrx345 mutant (Fig. 1B). Quantitative real-time (qRT)-PCR analysis showed that the increased expression of PDF and PR genes occurred only in the lrx34 double and lrx345 triple mutants but not in any of the lrx single mutants, suggesting that the three LRX proteins are redundant in the regulation of JA and SA pathways (Fig. 1C). By contrast, genes responsive to other hormones, including abscisic acid, ethylene, auxin, cytokinin, gibberellin and brassinosteroid, were not significantly enriched (P value < 0.05) among the up-regulated genes (Fig. 1A). These results indicate that LRX3/4/5 are involved in the negative regulation of JA and SA signaling pathways.
Figure 1.
Transcriptome profiles of wild type and lrx345 mutant plants before and after NaCl treatment. (A) GO enrichment analysis of genes that were up-regulated in the lrx345 mutant compared with the wild type under normal conditions. The enrichment of genes responsive to eight plant hormones are shown. (B) Heat map of the 30 genes that were up-regulated with highest fold changes in the lrx345 mutant plants compared with the wild type. PDF and PR genes are indicated in red. (C) qRT-PCR analysis of the transcript levels of PDF1.2, PDF1.3, PR1 and PR5 in wild type and lrx single, double and triple mutants. ACTIN8 was used as an internal control. Values are means of three biological replicates ± SD, **P < 0.01 (Student's t-test). (D) Heat map of the abiotic stress-responsive genes that exhibited lower transcript levels in the lrx345 mutant compared to wild type after NaCl treatment for 6 h. (E, F) qRT-PCR analysis of the gene expression of RD29A and RD29B in the indicated mutant plants before and after NaCl (150 mM) (E) or ABA (50 μM) (F) treatment for 6 h. ACTIN8 was used as an internal control. Values are means of three biological replicates ± SD, **P < 0.01 (Student's t-test).
Genes involved in abiotic stress response and cell wall modification display lower transcript levels in the lrx345 mutant after salt treatment
Comparison of gene expression between the wild type and lrx345 mutant after NaCl treatment showed that 1330 genes were up-regulated and 575 genes were down-regulated in the lrx345 mutant (Table S2). As under normal conditions, up-regulated genes were enriched in the categories ‘response to jasmonic acid’ (P value = 9.81E-21) and ‘response to salicylic acid’ (P value = 7.88E-13). Down-regulated genes were enriched in the categories ‘cell wall organization’ (P value = 9.89E-15), ‘cell wall modification’ (P value = 6.15E-05), ‘xyloglucan metabolic process’ (P value = 2.70E-04) and ‘hemicellulose metabolic process’ (P value = 1.30E-03) (Fig. S1A). The cell wall-related genes that are regulated by the LRX proteins include those encoding pectin lyase-like superfamily proteins, expansins, xyloglucan endotransglucosylase/hydrolases, casparian strip membrane domain proteins and glycosyl hydrolases (Fig. S1B). The down-regulation of these cell wall-related genes in the lrx345 mutant under salt stress was confirmed by qRT-PCR analysis (Fig. S1C). The down-regulated genes were also enriched in the category ‘response to water deprivation’ (P value = 2.80E-04) (Fig. S1A). The genes belonging to this category include many well-known abiotic stress responsive genes, such as RD29A, RD29B, RD22, RAD18, COR15A and KIN1 (Fig. 1D). qRT-PCR analysis confirmed that the expression of RD29A and RD29B genes was substantially down-regulated in the lrx34 double and lrx345 triple mutants, but was not significantly affected in the lrx single mutants after salt treatment compared with the wild type (Fig. 1E). Also ABA-induced up-regulation of RD29A and RD29B was attenuated in the lrx345 mutant (Fig. 1F). Together, these results indicate that LRX3/4/5 proteins are involved in the regulation of abiotic stress-responsive genes and cell wall modification genes under salt stress.
Turgor pressure is important for cell expansion. Under high salinity, maintainance of turgor pressure requires the uptake of K+ and increased water flow to the cell [20]. From our RNA-seq data, we found that the expression of K+ transporters AtHAK5 [21] and KUP11 [22], was significantly decreased in the lrx345 mutant under salt stress (Table S2). Interestingly, among 36 aquaporin genes that were investigated, none of them showed a significantly increased expression but 16 genes displayed significantly reduced expression in the lrx345 mutant compared with the wild type under high salinity (Table S3). The results suggest that the LRX proteins are important for the regulation of turgor pressure and water homeostasis under salt stress.
PDF and PR genes are highly up-regulated in the fer-4 mutant and in transgenic plants overexpressing RALF22/23
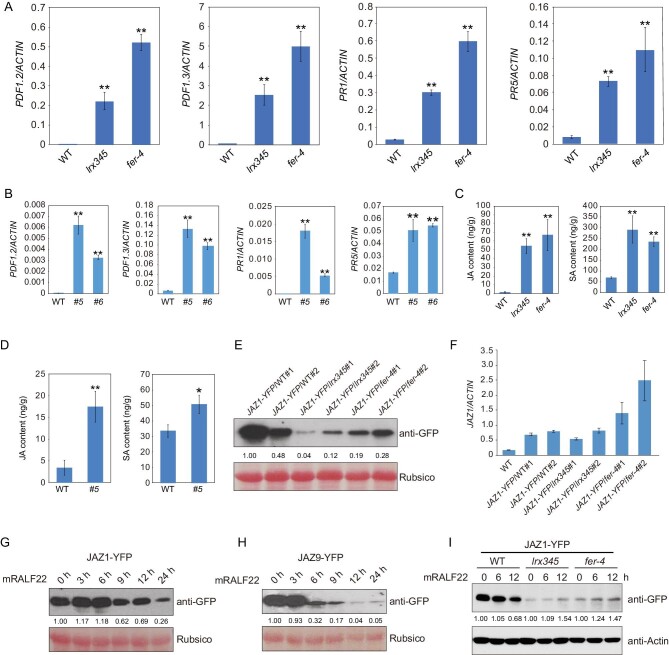
The JA- and SA-responsive genes PDF1.2, PDF1.3, PR1 and PR5 were also constitutively up-regulated in the fer-4 mutant and in transgenic plants overexpressing RALF22 or RALF23 (Fig. 2A and B, and Fig. S2A), consistent with previous evidence that LRX3/4/5, RALF22/23 and FER function in the same pathway [12]. To determine whether increased hormone accumulation underpins the up-regulation of JA- and SA-responsive genes, we measured the levels of JA and SA in the lrx345 and fer-4 mutants. In both mutants, the level of JA was more than 30-fold higher, while the level of SA was more than 3-fold higher, than that in the wild type (Fig. 2C). Transgenic plants overexpressing RALF22 also showed higher levels of JA and SA than the wild type (Fig. 2D).
Figure 2.
LRX3/4/5-RALF22/23-FER module negatively regulates JA signaling pathway. (A) qRT-PCR analysis of the transcript levels of PDF1.2, PDF1.3, PR1 and PR5 in wild type, lrx345 and fer-4 seedlings. ACTIN8 was used as an internal control. (B) qRT-PCR analysis of the transcript levels of PDF1.2, PDF1.3, PR1 and PR5 in wild type and two independent RALF22 overexpressing lines (#5 and #6). ACTIN8 was used as an internal control. (C) Jasmonic acid (JA) and salicylic acid (SA) contents in wild type, lrx345 and fer-4 seedlings. (D) JA and SA contents in transgenic plants overexpressing RALF22. (E) Immunoblotting analysis of JAZ1 protein in wild type, lrx345 and fer-4 seedlings expressing JAZ1-YFP. (F) qRT-PCR analysis of the JAZ1 transcript levels in the transgenic plants. (G, H) Transgenic plants expressing JAZ1-GFP (G) and JAZ9-GFP (H) were treated with mature RALF22 (mRALF22) for 0, 3, 6, 9, 12 and 24 h. Immunoblottings were performed using anti-GFP antibody. (I) Ten-day-old seedlings were treated with mRALF22 (1 μM) for 0, 6 and 12 h. Immunoblotting assays were performed using anti-GFP and anti-Actin antibodies. The band intensity in (E), (G) and (H) was qualified using ImageJ software. Values in (A–D) are means of three biological replicates ± SD, *P < 0.05 and **P < 0.01 (Student's t-test).
To understand the causes of JA and SA over-accumulation in the lrx345 mutant, we analyzed the expression of genes involved in JA and SA biosynthesis in the RNA-seq data. Of the 25 genes involved in JA biosynthesis, 14 were increased by more than 2-fold in the lrx345 mutant (Fig. S2B). qRT-PCR confirmed that JA biosynthesis-related genes were constitutively up-regulated in the lrx345 and fer-4 mutants (Fig. S2C). Both RNA-seq and qRT-PCR assays showed that ICS1, EPS1, CBP60G and PBS3, which are important for SA biosynthesis, were significantly up-regulated in the lrx345 mutant (Fig. S3A and B). These results indicate that the elevated contents of JA and SA in the lrx345 and fer-4 mutants are caused by the up-regulation of JA- and SA-biosynthesis genes.
Accumulation of JA results in the degradation of JASMONATE ZIM-DOMAIN (JAZ) proteins [23]. To further elucidate the role of the LRX3/4/5-RALF22/23-FER module in regulating the JA signaling pathway, we generated transgenic plants expressing JAZ1-YFP in the wild type, lrx345 and fer-4 backgrounds. JAZ1 protein but not its transcript levels were markedly lower in the lrx345 and fer-4 mutants (Fig. 2E and F). Consistent with the result that overexpression of RALF22 triggered the accumulation of JA (Fig. 2D), application of exogenous mature RALF22 (mRALF22) induced the degradation of JAZ1 (Fig. 2G). Similarly, mRALF22 also triggered the degradation of JAZ9 protein (Fig. 2H). To understand whether mRALF22-triggered degradation of JAZs is mediated by LRX3/4/5 and FER, we examined the protein level of JAZ1 in lrx345 and fer-4 mutants after mRALF22 treatment. We found that mRALF22 could not trigger JAZ1 degradation in the lrx345 and fer-4 mutants (Fig. 2I). Together, the above results indicate that the LRX3/4/5-RALF22/23-FER module negatively regulates the JA and SA signaling pathways.
Disruption of the JA pathway suppresses the dwarf phenotype of the lrx345 and fer-4 mutants
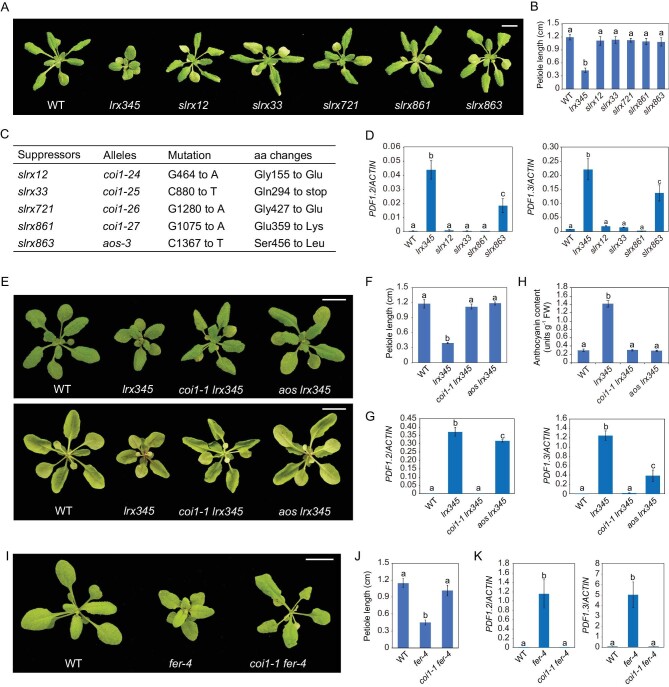
To identify downstream factors of the LRX3/4/5-RALF22/23-FER signaling module, we screened for the suppressors of lrx345 (referred to as slrx) using ethyl methanesulfonate (EMS)-mediated mutagenesis. Approximately 15 000 lrx345 seeds were used for EMS mutagenesis and 18 suppressors that could rescue the dwarf phenotype of the lrx345 mutant were identified. As these suppressors displayed a similar growth phenotype (Fig. 3A and B), five of them were chosen for whole-genome sequencing-based mapping. We identified mutations in the COI1 gene, encoding the JA receptor [24], in four of the five suppressors, while a mutation in the AOS gene, encoding a JA biosynthetic enzyme [25], was found in the remaining suppressor (Fig. 3C). Consistent with these results, we found that the expression of the JA-responsive genes PDF1.2 and PDF.3 was attenuated in the suppressors (Fig. 3D). By crossing lrx345 to previously published coi1-1 and aos mutants [24,25], we confirmed that mutations in these genes fully recovered the growth of the lrx345 mutant (Fig. 3E and F), and also reduced the expression of PDF genes (Fig. 3G). It should be noted that the reduction of PDF genes expression was less pronounced in the aos mutant than that in the coi1 mutant (Fig. 3D and G). The increased accumulation of anthocyanin in the lrx345 mutant was also suppressed by the AOS and COI1 mutations (Fig. 3E and H). In the fer-4 mutant, the dwarf phenotype and the increased expression of PDF genes were similarly suppressed by the coi1 mutation (Fig. 3I–K). These results indicate that constitutive activation of the JA pathway is responsible for the dwarfism and accumulation of anthocyanin in the lrx345 and fer-4 mutants.
Figure 3.
Disruption of the JA pathway suppresses the dwarf phenotype of the lrx345 and fer-4 mutants. (A) Rosette morphology of the wild type, lrx345, and five suppressors of lrx345 (slrx) grown in soil. Scale bar, 1 cm. (B) Petiole length of each genotype grown in soil for 4 weeks. (C) Mutations identified in the lrx suppressors by whole-genome sequencing-based mapping. (D) qRT-PCR analysis of the transcript levels of PDF1.2 and PDF1.3. ACTIN8 was used as an internal control. (E) Morphology of the upper epidermis (upper panel) and lower epidermis (bottom panel) of plants grown in soil. Scale bar, 1 cm. (F) Petiole lengths of the wild type, lrx345, coi1-1 lrx345 and aos lrx345 grown in soil for 4 weeks. (G) qRT-PCR analysis of the transcript levels of PDF1.2 and PDF1.3. ACTIN8 was used as an internal control. (H) Quantification of anthocyanin accumulation in 12-day-old seedlings of the wild type, lrx345, coi1-1 lrx345 and aos lrx345. (I) Rosette morphology of the wild type, fer-4 and coi1-1 fer-4. Scale bar, 1 cm. (J) Petiole lengths of the wild type, fer-4 and coi1-1 fer-4 grown in soil for 4 weeks. (K) qRT-PCR analysis of the transcript levels of PDF1.2 and PDF1.3 in wild type, fer-4 and coi1-1 fer-4. ACTIN8 was used as an internal control. Values in (B), (F) and (J) are means of 10 petioles ± SD. Values in (D), (G), (H) and (K) are means of three biological replicates ± SD. Different letters represent statistically significant differences (P < 0.01, one-way ANOVA).
We then tested whether disrupting the JA pathway can reverse the salt-hypersensitivity of lrx345 and fer-4 mutants. Both coi1-1 and aos mutants are male-sterile [24,25], but the aos mutant can produce seeds when exogenous JA is applied, so we obtained aos lrx345 homozygous plants and tested their response to salt stress. The aos mutation only slightly suppressed the salt-hypersensitivity of lrx345 mutant (Fig. S4A and B). Further, we crossed the coi1-16 mutant allele [26], which can produce seeds at low temperatures (16°C), with the lrx345 mutant, and found that the coi1-16 mutation also only slightly suppressed the salt-hypersensitivity of lrx345 mutant (Fig. S4A and B).
The jazQ mutant, which lacks five JAZ transcriptional repressors, exhibits constitutive JA responses [27]. Similar to the lrx345 mutant, the jazQ mutant displays a dwarf phenotype [27] (Fig. S4C and D). To determine whether constitutive activation of JA pathway results in enhanced cell death under salt stress, we grew the jazQ mutant on medium with a high concentration of salt. The jazQ mutant survived as well as the wild type under salt stress (Fig. S4E). Besides, mutation of MYC2, which encodes a key transcription factor of the JA signaling pathway, partially suppressed the dwarf phenotype, but only slightly suppressed the salt-hypersensitivity of the lrx345 mutant (Fig. S4F–H). Together, these results indicate that activation of the JA pathway alone is not sufficient to cause cell death in plants under high salinity.
To assess the contribution of the increased SA content to the phenotypes of the lrx345 mutant, we crossed lrx345 to the sid2-2 mutant, in which SA biosynthesis is highly impaired [28]. The mutation of SID2 did not reverse the dwarf phenotype of lrx345, but partially suppressed its salt-hypersensitivity (Fig. S3C–E). As expected, the up-regulation of PR1 was completely reversed in the sid2-2 lrx345 quadruple mutant, while the expression of PDF1.2 and PDF1.3 was not significantly changed (Fig. S3F), indicating that the increased expression of PR1 in the lrx345 mutant depends on the SA pathway.
In our suppressor screen, we also identified three genes, TT3, TT4 and TT6, that are required for the anthocyanin accumulation in the lrx345 mutant, but mutations in these genes did not suppress the dwarf phenotype and salt-hypersensitivity of the lrx345 mutant (Fig. S5). These three genes, encoding dihydroflavonol reductase, chalcone synthase and flavanone 3-hydroxylase, respectively, are required for anthocyanin biosynthesis in Arabidopsis [29–31]. Disruption of JA biosynthesis or perception suppresses both the anthocyanin accumulation and plant growth phenotypes of the lrx345 mutant (Fig. 3E and H), but abolishing anthocyanin biosynthesis only affects the accumulation of anthocyanin, suggesting that JA signaling can feed forward into anthocyanin production.
Salt-hypersensitivity of the lrx345 and fer-4 mutants is dependent on ABA
As many salt stress-responsive genes were down-regulated in the lrx345 and fer-4 mutants after NaCl treatment for 6 h, we speculated that the salt-hypersensitivity of lrx345 and fer-4 mutants may be caused by an impaired stress response. ABA is an important hormone for the up-regulation of stress-responsive genes in plants [32]. To determine whether the salt-hypersensitivity of the lrx345 mutant is associated with a defective ABA pathway, we crossed the lrx345 mutant with the aba2-1 mutant, in which ABA biosynthesis is substantially decreased [33] (Fig. S6A). Unexpectedly, the aba2-1 mutation almost fully rescued the cell-death phenotype of the lrx345 mutant under high salinity (Fig. 4A–C). To confirm this result, we crossed the lrx345 mutant with another aba2 mutant allele, aba2-3. Similarly, the aba2-3 mutation almost fully suppressed the salt-hypersensitivity of the lrx345 mutant (Fig. S6B and C). The suppression of the cell-death phenotype of the lrx345 mutant by aba2 mutation under high salinity was alleviated by applying exogenous ABA (Fig. 4D). The salt-hypersensitivity of the fer-4 mutant was also suppressed by the aba2-1 mutation (Fig. S6D and E). We then tested the accumulation of ABA in the lrx345 and fer-4 mutants and found that ABA content was higher in these mutants than in the wild type plants, both in basal conditions and upon salt treatment (Fig. 4E). Gene expression analysis showed that ABA de novo biosynthesis genes were not significantly up-regulated and that ABA catabolism genes were not significantly down-regulated in the lrx345 mutant (Table S4). However, the β-glucosidase gene AtBG1, which is required for conversion of the inactive glucose-conjugated form of ABA (ABA-GE) to the active form of ABA [34], was highly up-regulated in the lrx345 mutant (Table S4), suggesting that a LRX3/4/5-mediated pathway may modulate ABA accumulation via the regulation of AtBG1 expression.
Figure 4.
Suppression of the salt-hypersensitive phenotype of lrx345 mutant plants by a mutation in ABA2. (A) Phenotypes of seedlings of the indicated genotypes grown on MS and MS+NaCl (120 mM) media. (B) Survival rates of the wild type, lrx345 and aba2-1 lrx345 seedlings grown on MS+NaCl (120 mM) medium. (C) Phenotypes of seedlings transferred from MS to MS+NaCl (100 mM) medium. The photograph was taken 5 days after 3-day-old seedlings were transferred. (D) Survival rate of the wild type, lrx345 and aba2-1 lrx345 seedlings grown on MS+NaCl (120 mM) media supplemented with or without ABA (1 μM). (E) ABA contents of 10-day-old seedlings before and after NaCl (150 mM) treatment (1 h). (F) qRT-PCR analysis of the transcript levels of PDF1.2, PDF1.3, PR1 and PR5 in the indicated genotypes. ACTIN8 was used as an internal control. Values in (B–F) are means of three biological replicates ± SD, *P < 0.05 and **P < 0.01 (Student's t-test). (G) Measurement of JA and SA contents in the wild type, lrx345 and aba2-1 lrx345 seedlings. Values are means of three biological replicates ± SD. Different letters represent statistically significant differences (P < 0.01, one-way ANOVA). (H) qRT-PCR analysis of the expression of RD29A and RD29B in the indicated genotypes before and after NaCl treatment. ACTIN8 was used as an internal control. Values are means of three biological replicates ± SD.
We found that the aba2-1 single mutant exhibited a higher survival rate than the wild type when grown on the medium with a high concentration of NaCl (150 mM) (Fig. S6F and G), suggesting that accumulation of ABA promotes salt-induced cell death. Consistent with the previous study showing that fer mutant plants are hypersensitive to ABA [17,35], the lrx345 mutant also exhibited delayed seed germination on the medium supplemented with ABA (Fig. S6H).
Previous study showed that the increased sensitivity of fer mutant plants to ABA may be a result of inactivation of ABI2 [17,35], a phosphatase that negatively regulates ABA signaling. To test whether the hypersensitivity of lrx345 and fer-4 to salt stress involves ABI2, we crossed lrx345 and fer-4 mutants with the abi2-1 mutant. The abi2-1 is a dominant mutation that blocks ABA signaling [36]. We found that the abi2-1 mutation could not suppress the salt-hypersensitive phenotype of the lrx345 and fer-4 mutants (Fig. S7), suggesting that ABI2 alone cannot account for LRX3/4/5- and FER-mediated regulation of salt stress responses.
The aba2-1 mutation also partially suppressed the growth defects and anthocyanin accumulation of the lrx345 mutant (Fig. S6I–K). The transcript levels of PDF1.2 and PDF1.3 were markedly lower, while the transcript levels of PR1 and PR5 were moderately lower in the aba2-1 lrx345 quadruple mutant than that in the lrx345 mutant (Fig. 4F), which is consistent with a drastic reduction in JA content and a slight decrease in SA content in the quadruple mutant (Fig. 4G). These results indicate that ABA accumulation contributes to the activation of JA signaling pathway and partially contributes to the activation of SA signaling pathway in the lrx345 mutant.
LRX345-FER module is required for both activation and attenuation of salt stress-responsive genes
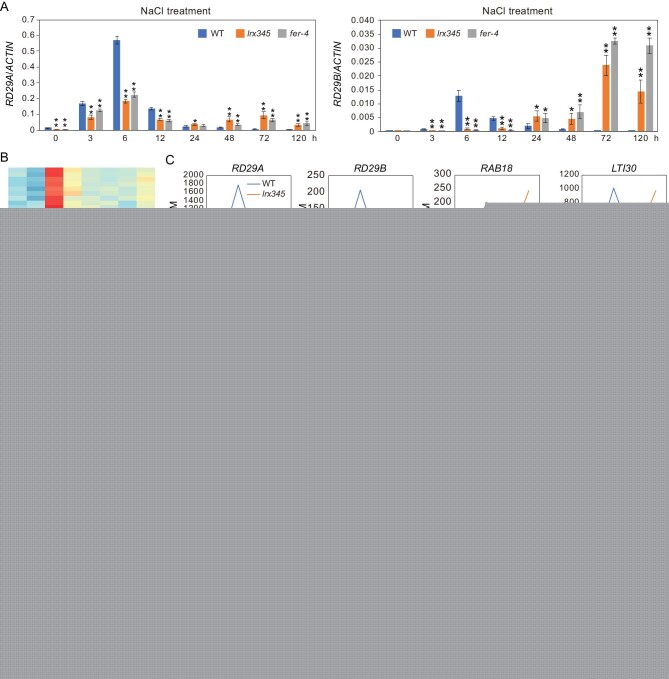
Although the aba2-1 mutation suppressed the cell-death phenotype of the lrx345 mutant under high salinity, the salt-induced expression of RD29A and RD29B was further impaired in the aba2-1 lrx345 quadruple mutant (Fig. 4H), suggesting that the cell-death phenotype of the lrx345 triple mutant under high salinity is not caused by the decreased expression of these salt stress-responsive genes. It has been proposed that the outcome of salt stress responses (adaptation or cell death) depends to some extent on when exactly these responses start and end [37]. Overactivation or prolonged activation of stress responses is apparently detrimental for plants under stress conditions. To test whether LRX3/4/5 and FER may be involved in the attenuation of salt stress-responsive genes, we examined the expression of RD29A and RD29B genes after exposure to high salinity for 0, 3, 6, 12, 24, 48, 72 and 120 h. In the wild type, the expression of both RD29A and RD29B reached peak levels after salt treatment for 6 h, and then went down after longer exposure to high salinity (Fig. 5A). In the lrx345 and fer-4 mutants, the expression of RD29A also reached peak levels after salt treatment for 6 h but showed a second smaller wave of expression after 24 h, while the expression of RD29B continuously increased after exposure to salt stress (Fig. 5A). Similarly, the expression of RD29A and RD29B genes was increased in both RALF22 and RALF23 overexpressing plants compared with the wild type after prolonged exposure to high salinity (Fig. S8A). These results indicate that the LRXs-RALFs-FER module is not only required for the up-regulation of salt stress-responsive genes soon after salt treatment, but is also essential for attenuating the expression of salt stress-responsive genes after longer exposure to salt stress. The increased expression of RD29A and RD29B genes in the lrx345 mutant after salt treatment for 72 h was completely abolished by the aba2 mutation (Fig. S8B), suggesting that the ABA-mediated pathway is required for prolonged expression of salt stress-responsive genes in the lrx345 mutant.
Figure 5.
LRX3/4/5 and FER are required for the dynamic regulation of salt stress-responsive gene expression. (A) Ten-day-old seedlings of the wild type, lrx345 and fer-4 were treated with NaCl for 0, 3, 6, 12, 24, 48, 72 and 120 h. The transcript levels of RD29A and RD29B were analyzed by qRT-PCR. ACTIN8 was used as an internal control. (B) RNA-seq assay was performed for the wild type and lrx345 after NaCl treatment for 0, 6, 24 and 72 h. Heat map shows the genes that exhibited lower transcript levels after NaCl treatment for 6 h, and showed higher transcript levels after treatment for 72 h in the lrx345 mutant compared with that in the wild type. (C) Representative genes that display similar expression patterns as RD29A and RD29B after salt treatment for 0, 6, 24 and 72 h. The transcript profile of each gene in the wild type and lrx345 mutant was generated from RNA-seq data. (D) Wild type seedlings were pretreated with or without EGCG (100 μM) for 2 h before being subjected to NaCl treatment for 0, 6, 24 and 72 h. Transcript levels of RD29A, RD29B and RAB18 were analyzed. ACTIN8 was used as an internal control. Values in (A) and (D) are means of three biological replicates ± SD, *P < 0.05 and **P < 0.01 (Student's t-test).
To investigate how many salt stress-responsive genes display prolonged expression in the lrx345 mutant under salt stress, we performed RNA-seq analysis for the wild type and lrx345 mutant after salt treatment for 0, 6, 24 and 72 h (Table S5). RNA-seq data confirmed that the expression of RD29A and RD29B genes was lower at 6 h but higher at 72 h after NaCl treatment in the lrx345 mutant than in the wild type (Fig. 5B and C, Table S6). We identified 160 salt stress-inducible genes, including RAB18, LTI30, ABR, DAA1, NADP-ME1, TZF4, ABCG6, AT1G16850 and LEA4-5, that displayed similar expression patterns to RD29A and RD29B (Fig. 5B and C, Table S6). The abnormal expression patterns of these genes in the lrx345 mutant were confirmed by qRT-PCR analysis (Fig. S9). Gene ontology (GO) enrichment analysis of these 160 genes showed that they are enriched in the categories ‘phenylpropanoid metabolic process’ (P value = 1.19E-12), ‘response to acid chemical’ (P value = 5.17E-10), and ‘response to oxygen-containing compound’ (P value = 1.54E-08). However, 173 salt stress-inducible genes, including COR15B, COR47, COR413-TM1, KIN2 and RD22, had lower expression in the lrx345 mutant than in the wild type after salt treatment for 6 h but did not show higher expression in the mutant after salt treatment for 72 h (Table S7). These genes are enriched in the categories ‘starch metabolic process’ (P value = 3.35E-06), ‘lipid metabolic process’ (P value = 4.72E-06) and ‘glucan metabolic process’ (P value = 1.28E-05). Together, these results suggest that the LRX3/4/5- and FER-mediated pathway is required for both activation and attenuation of a specific set of stress-responsive genes.
LRXs and FER maintain cell wall integrity likely via a communication with pectin [18,38]. To understand whether the abnormal expression pattern of stress-responsive genes in the lrx345 and fer-4 mutants results from disrupted cell wall properties under high salinity, we pretreated wild type plants with epigallocatechin gallate (EGCG), an inhibitor of pectin methylesterases (PMEs) [39], before subjecting them to salt treatment for 0, 6, 24 and 72 h. PMEs are enzymes that remove methyl ester groups from homogalacturonan (HGA), a major constituent of pectin in the cell wall [40]. Demethylesterified pectins either form Ca2+ bonds and contribute to wall firmness or are more accessible to pectin-degrading enzymes [41]. Gene expression analysis showed that, similar to the lrx345 and fer-4 mutants, the wild type seedlings pretreated with EGCG lost the ability to maintain proper expression patterns of RD29A, RD29B and RAB18 genes after both short and prolonged exposure to high salinity conditions (Fig. 5D). These results suggests that EGCG-triggered modifications of cell wall components likely determine the expression patterns of stress-responsive genes under high salinity. Consistent with a previous study [38], we found that EGCG treatment inhibited the root elongation of the wild type plants, but had a less effect on the root growth inhibition of fer-4 mutant (Fig. S10).
Accumulation of reactive oxygen species (ROS) is responsible for salt-induced cell death of the lrx345 and fer-4 mutants
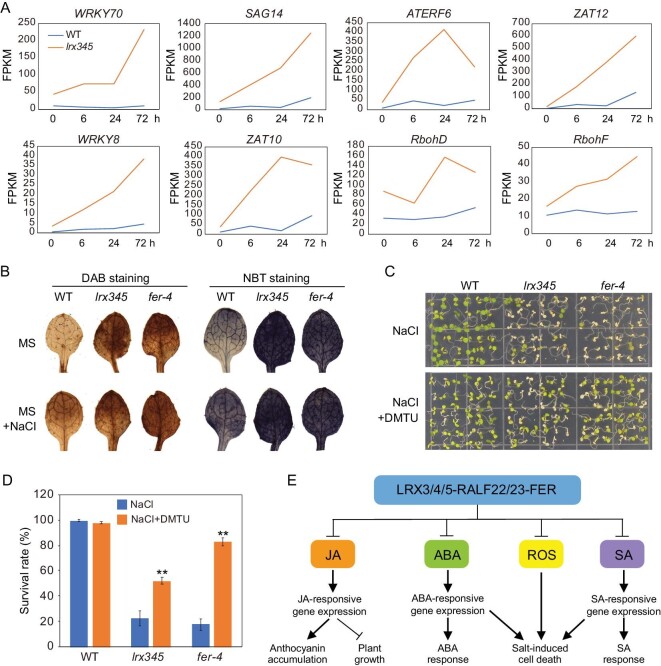
Our RNA-seq data also revealed that genes constitutively up-regulated in the lrx345 mutant were enriched in the category ‘response to oxidative stress’ (P value = 1.97E-07). Specifically, 59 oxidative stress-responsive genes, including WRKY8, WRKY70, SAG14, ATERF6, ZAT10 and ZAT12, were constitutively up-regulated in the lrx345 mutant (Fig. 6A). Under high salinity, the transcripts of these six genes either did not change or slowly increased in the wild type, but they were dramatically up-regulated in the lrx345 mutant (Fig. 6A). Interestingly, RbohD and RbohF, encoding two NADPH oxidative enzymes required for the production of ROS, were up-regulated in the lrx345 mutants both under basal conditions and upon NaCl treatment (Fig. 6A). In addition, the protein level of RbohD was increased in the lrx345 mutant under both normal conditions and salt stress treatment (Fig. S11A).
Figure 6.
Inhibition of ROS production restores the cell-death phenotype of lrx345 and fer-4 mutants under high salinity. (A) Transcript levels of RbohD, RbohF and six oxidative stress-responsive genes before and after NaCl treatment. The expression values of each gene were calculated based on RNA-seq data. (B) Wild type, lrx345 and fer-4 seedlings before and after NaCl treatment were stained with DAB and NBT. (C) Phenotype of seedlings grown on MS+NaCl (120 mM) media supplemented with or without DMTU (5 mM). (D) Survival rate of the wild type, lrx345 and fer-4 seedlings grown on MS+NaCl (120 mM) media supplemented with or without DMTU (5 mM). Values are means of three biological replicates ± SD, **P < 0.01 (Student's t-test). (E) Diagram summarizing the role of LRX3/4/5-RALF22/23-FER module in the regulation of JA, SA, ABA and ROS accumulation.
We measured the content of ROS, including hydrogen peroxide (H2O2) and superoxide radicals (O2·−), in the lrx345 and fer-4 mutants using 3,3′-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining. Levels of both H2O2 and O2·− were higher in the leaves of the lrx345 and fer-4 mutants than in wild type leaves (Fig. 6B). The increased accumulation of ROS in the leaves of the lrx345 and fer-4 mutants is opposite to previous results showing that FER promotes the accumulation of ROS in the root and filiform apparatus and positively regulates flg22-, elf18- and chitin-induced ROS production [14,42,43]. These contradicting results suggest that FER may control ROS production in tissue-specific and environmental stress-dependent manners. To understand whether the salt-induced cell-death phenotype of the lrx345 and fer-4 mutants is caused by the increased production of ROS, we grew seedlings on NaCl medium supplemented with dimethylthiourea (DMTU), a scavenger of H2O2. Application of DMTU largely reversed the cell-death phenotype of fer-4 mutant and to a lesser extent suppressed that of lrx345 mutant under high salinity (Fig. 6C and D). We also found that the wild type plants grown on NaCl media supplemented with H2O2 displayed a lower survival rate than the plants grown on media with NaCl alone (Fig. S11B and C), supporting that ROS over-accumulation enhances salt-induced cell death in plants.
Study has shown that ABA promotes the production of ROS via RbohD and RbohF NADPH oxidases [44]. To investigate whether aba2 mutation-mediated inhibition of the cell-death phenotype of lrx345 and fer-4 mutants is a result of reduced ROS production, we assessed H2O2 and O2·− in the aba2-1 lrx345 and aba2-1 fer-4 mutants using DAB and NBT staining. The aba2 mutation did not obviously reduce the accumulation of H2O2 and O2·− in the lrx345 and fer-4 mutants (Fig. S12A). However, gene expression analysis indicated that RbohF, but not RbohD, was substantially down-regulated in the aba2-1 lrx345 mutant compared with the lrx345 mutant (Fig. S12B). In addition, the expression of ROS-responsive genes, such as ZAT10, SAG1, WRKY8 and AtERF6, was largely attenuated in the aba2-1 lrx345 mutant, especially after NaCl treatment for 24 h or longer (Fig. S12B). These results suggest that the suppression of the salt-induced cell death of the lrx345 and fer-4 mutants by aba2 mutation is at least partially a result of attenuated ROS response under salt stress.
DISCUSSION
Environmental stresses, such as high salinity, not only trigger intracellular stress responses, but also interfere with cell wall integrity and inhibit plant growth [45]. Coordinated regulation of cell wall integrity, growth and stress response is critical for plants to survive under unfavorable environmental conditions. Previous results have indicated that the LRX3/4/5-RALF22/23-FER module is involved in regulating cell wall integrity, growth and salt stress responses in plants. Here, we provide transcriptomic, hormonal and genetic data to show that this regulation is achieved by the modulation of JA, SA and ABA hormones (Fig. 6E). As several genes involved in JA and SA biosynthesis were constitutively up-regulated in the lrx345 and fer-4 mutants, we proposed that LRX3/4/5-RALF22/23-FER may regulate JA and SA pathways by controlling the expression of their biosynthesis genes. A recent study showed that SA- and JA-responsive genes are also highly up-regulated in the fer-4 mutant and that FER interacts with and destabilizes MYC2, a master regulator of JA response [19]. These results suggest that FER may regulate the expression of JA biosynthesis genes via the modulation of MYC2 stability.
Genetic or chemical perturbation of the cell wall was known to activate JA signaling [46,47], but the components linking cell wall stress to JA signaling had been missing. We found that the JA pathway is constitutively activated in the lrx345 and fer-4 mutants and in transgenic plants overexpressing RALF22 or RALF23, suggesting that the LRX3/4/5-RALF22/23-FER module is involved in the relay of cell wall stress signals to the JA pathway. It is likely that stress-induced perturbation of cell wall integrity triggers the dissociation of the RALF22/23 from LRX3/4/5, and the released RALF22/23 peptides promote the internalization of FER [12], which then triggers the activation of the JA pathway. Our results showed that the SA pathway, which is generally considered as antagonistic to the JA signaling pathway [48,49], is also constitutively up-regulated in the lrx345 and fer-4 mutants. Therefore, how these two hormones are simultaneously increased in the lrx345 and fer-4 mutants is of great interest. Although disruption of SA biosynthesis partially suppresses the salt-hypersensitivity of the lrx345 mutant, further work is required to address whether SA is induced by the perturbation of cell wall integrity and the exact roles of SA in the LRX3/4/5-RALF22/23-FER module-mediated pathways.
JA is an important hormone that is required for defense against diverse pathogens and herbivores [50,51]. The JA-mediated defense responses are usually accompanied by growth repression. In recent years, the molecular mechanisms underlying the roles of JA in the regulation of plant growth have been well addressed. JA promotes the accumulation of DELLA proteins that interact with and repress the transcription activity of growth-promoting PIF transcription factors [52–54]. Our genetic analysis showed that the dwarf phenotype of the lrx345 and fer-4 mutants is almost fully suppressed by mutations in COI1 and AOS genes, which shows that JA is involved in the regulation of plant growth in the cell wall integrity pathways. Studies have shown that salt stress can activate the JA pathway [55,56], but the biological significance of JA pathway in salt stress tolerance is still largely unknown. We propose that the JA pathway, which is triggered by salt-induced perturbation of cell wall integrity via the LRX3/4/5-RALF22/23-FER module, is required to inhibit cell expansion under high salinity. The inhibition of cell expansion under high salinity allows priority to cell wall repair before cell growth can resume, thus preventing cell bursting.
The enhanced cell death of the lrx345 and fer-4 mutants under high salinity is similar to that observed in the sos1 mutant [57]. SOS1, a Na+/H+ antiporter, is required for the extrusion of excessive Na+ from the cytosol and maintainance of ion homeostasis under salt stress [57,58]. Unlike the sos1 mutant, Na+ is not significantly accumulated in the fer mutant under salt stress [59], which suggests a different mechanism for the salt-induced cell death in the lrx345 and fer mutants. Salt stress can induce diverse cellular responses, such as Ca2+, ROS and ABA [32]. Our results showed that ABA and ROS contents are consitutively elevated in the lrx345 and fer-4 mutants, and the cell-death phenotype of the lrx345 and fer-4 mutants is almost fully suppressed by the ABA2 mutation that disrupts ABA biosynthesis, and largely suppressed by DMTU, a H2O2 scavenger, suggesting that the abnormal ABA homeostasis and accumulation of ROS are the causes of the cell-death phenotype of the lrx345 and fer-4 mutants under high salinity. The accumulation of ROS in the lrx345 and fer-4 mutants is probably caused by the increased expression of RbohD and RbohF genes. Extensive studies have shown that over-accumulation of ROS results in cell death in plants [60–62], but the mechanisms underlying the roles of ABA in the regulation of cell death are still largely unknown. Based on the knowledge that ABA promotes the production of ROS via RbohD and RbohF NADPH oxidases [44], and that elevated expression of RbohF gene in the lrx345 mutant was attenuated by the aba2 mutation, we speculate that ABA promotes the salt-induced cell death of the lrx345 and fer-4 mutants via RbohF-mediated production of ROS. We found that the abi2-1 mutation did not suppress the salt-hypersensitive phenotype of lrx345 and fer-4 mutants. This result may be explained because the abi2-1 mutation does not fully block ABA signaling. Alternatively, LRX3/4/5- and FER-mediated regulation of salt stress response may depend on ABA pathway component(s) other than ABI2. Studies have shown that FER regulates the GEFs-ROP11 complex [17] and ROP11 protects the activity of ABI1 and ABI2 by alleviating the ABA receptor PYL9-mediated inhibition [63]. It is possible that besides regulating ABA accumulation, FER may control salt stress tolerance by regulating ABA response through several signaling components.
Proper regulation of stress-responsive gene expression is critical for plants to survive under adverse environmental conditions. Although extensive studies have elucidated the genes and signaling pathways that are required for the regulation of the expression of stress-responsive genes [32,64,65], the physical properties of cells that determine the dynamic expression of stress-responsive genes in response to high salinity are still unclear. Our data showed that the lrx345 and fer-4 mutants, and the wild type plants treated with EGCG, an inhibitor of PMEs, lost the ability to activate and attenuate the expression of many stress-responsive genes after short and prolonged exposure to high salinity, suggesting that expression patterns of salt stress-responsive genes are determined by the properties of the cell wall, the signals of which are monitored and transduced by the LRX3/4/5-RALF22/23-FER module. In future, the association between pectin structure and salt stress response needs to be investigated in more detail.
In summary, identification of the downstream signaling pathways that are regulated by the LRXs-RALFs-FER module helps us to understand the intracellular events that plants explore to respond to cell wall perturbation under a variety of environmental stresses. The intermediate steps leading from the LRX3/4/5-RALF22/23-FER module to the induction of downstream responses including the increases in ABA, JA and SA levels remain to be elucidated. Nonetheless, our results provide a mechanistic framework to understand how phytohormone content and signaling are modulated to provide a homeostatic mechanism allowing for maintenance of cell wall integrity and growth regulation as well as for plant survival under salt stress. The simultaneous increase of multiple hormones in the lrx345 and fer-4 mutants provides an invaluable opportunity to investigate the cross-talk among these hormones and how too much of these defense hormones under salt stress can cause plant death. Understanding the biological function of the LRX3/4/5-RALF22/23-FER module may ultimately enable engineering of crop plants that not only can survive salt stress but also can grow well.
METHODS AND MATERIALS
The methods and materials are described in detail in the Supplementary data.
Data resources
RNA-seq data have been deposited in the NCBI GEO under accession numbers GEO: GSE136269.
Supplementary Material
Acknowledgements
We thank Rebecca Stevenson for technical assistance, Prof. Daoxin Xie for providing coi1-1 and aos seeds, Prof. Yiwei Jiang for help with anthocyanin measurement, and Prof. Yan Liang for providing anti-RbohD antibody.
Contributor Information
Chunzhao Zhao, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China; Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA; Key laboratory of Plant Stress Biology, School of Life Sciences, Henan University, Kaifeng 475004, China.
Wei Jiang, Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA; National Center for Soybean Improvement, Key Laboratory of Biology and Genetics and Breeding for Soybean, Ministry of Agriculture, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, China.
Omar Zayed, Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA; Genetics Department, Faculty of Agriculture, Menofia University, Shebeen Elkoum 32511, Egypt.
Xin Liu, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Kai Tang, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China; Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA.
Wenfeng Nie, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China.
Yali Li, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China.
Shaojun Xie, Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA.
Yuan Li, Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA; State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Tiandan Long, Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA; State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu 611130, China.
Linlin Liu, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Yingfang Zhu, Key laboratory of Plant Stress Biology, School of Life Sciences, Henan University, Kaifeng 475004, China.
Yang Zhao, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China; Key laboratory of Plant Stress Biology, School of Life Sciences, Henan University, Kaifeng 475004, China.
Jian-Kang Zhu, Shanghai Center for Plant Stress Biology and Center of Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, China; Department of Horticulture and Landscape Architecture, Purdue University, West Lafayette, IN 47907, USA.
FUNDING
This work was supported by the Strategic Priority Research Program (XDB27040101) of the Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
C.Z. and J.-K.Z. designed the research; C.Z., W.J., O.Z., X.L., W.N., Y.L., Y.L., T.L. and L.L. performed the experiments; C.Z., O.Z., K.T., S.X., Y.Z., Y.Z. and J.-K.Z. analyzed the data; C.Z. and J.-K.Z. wrote the paper.
REFERENCES
- 1. Baumberger N, Doesseger B, Guyot R et al. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol 2003; 131: 1313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borassi C, Sede AR, Mecchia MA et al. An update on cell surface proteins containing extensin-motifs. J Exp Bot 2016; 67: 477–87. [DOI] [PubMed] [Google Scholar]
- 3. Wang X, Wang K, Yin G et al. Pollen-expressed leucine-rich repeat extensins are essential for pollen germination and growth. Plant Physiol 2018; 176: 1993–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fabrice TN, Vogler H, Draeger C et al. LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol 2018; 176: 1981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sede AR, Borassi C, Wengier DL et al. Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett 2018; 592: 233–43. [DOI] [PubMed] [Google Scholar]
- 6. Draeger C, Ndinyanka Fabrice T, Gineau E et al. Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol 2015; 15: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ringli C. The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J 2010; 63: 662–9. [DOI] [PubMed] [Google Scholar]
- 8. Mecchia MA, Santos-Fernandez G, Duss NN et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017; 358: 1600–3. [DOI] [PubMed] [Google Scholar]
- 9. Franck CM, Westermann J, Boisson-Dernier A. Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annu Rev Plant Biol 2018; 69: 301–28. [DOI] [PubMed] [Google Scholar]
- 10. Moussu S, Broyart C, Santos-Fernandez G et al. Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc Natl Acad Sci USA 2020; 117: 7494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ge Z, Bergonci T, Zhao Y et al. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 2017; 358: 1596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao C, Zayed O, Yu Z et al. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc Natl Acad Sci USA 2018; 115: 13123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haruta M, Sabat G, Stecker K et al. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014; 343: 408–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stegmann M, Monaghan J, Smakowska-Luzan E et al. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017; 355: 287–9. [DOI] [PubMed] [Google Scholar]
- 15. Gonneau M, Desprez T, Martin M et al. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr Biol 2018; 28: 2452–8. [DOI] [PubMed] [Google Scholar]
- 16. Masachis S, Segorbe D, Turra D et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 2016; 1: 16043. [DOI] [PubMed] [Google Scholar]
- 17. Yu F, Qian L, Nibau C et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA 2012; 109: 14693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng W, Kita D, Peaucelle A et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 2018; 28: 666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo H, Nolan TM, Song G et al. FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr Biol 2018; 28: 3316–24. [DOI] [PubMed] [Google Scholar]
- 20. Shabala SN, Lew RR. Turgor regulation in osmotically stressed Arabidopsis epidermal root cells. Direct support for the role of cell turgor measurements. Plant Physiol 2014; 129: 290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rubio F, Nieves-Cordones M, Alemán F et al. Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol Plant 2008; 134: 598–608. [DOI] [PubMed] [Google Scholar]
- 22. Ahn SJ, Shin R, Schachtman DP. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 2004; 134: 1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thines B, Katsir L, Melotto M et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007; 448: 661–5. [DOI] [PubMed] [Google Scholar]
- 24. Xie DX, Feys BF, James S et al. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998; 280: 1091–4. [DOI] [PubMed] [Google Scholar]
- 25. Park JH, Halitschke R, Kim HB et al. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 2002; 31: 1–12. [DOI] [PubMed] [Google Scholar]
- 26. Ellis C, Karafyllidis I, Turner JG. Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 2002; 15: 1025–30. [DOI] [PubMed] [Google Scholar]
- 27. Campos ML, Yoshida Y, Major IT et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 2016; 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wildermuth MC, Dewdney J, Wu G et al. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001; 414: 562–5. [DOI] [PubMed] [Google Scholar]
- 29. Ferrer JL, Jez JM, Bowman ME et al. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol 1999; 6: 775–84. [DOI] [PubMed] [Google Scholar]
- 30. Kubasek WL, Shirley BW, McKillop A et al. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis Seedlings. Plant Cell 1992; 4: 1229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wisman E, Hartmann U, Sagasser M et al. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 1998; 95: 12432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu JK. Abiotic Stress Signaling and Responses in Plants. Cell 2016; 167: 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzalez-Guzman M, Apostolova N, Belles JM et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002; 14: 1833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee KH, Piao HL, Kim H-Y et al. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006; 126: 1109–20. [DOI] [PubMed] [Google Scholar]
- 35. Chen J, Yu F, Liu Y et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc Natl Acad Sci USA 2016; 113: E5519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSlTIVE2 (AB12) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 1997; 9: 759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ismail A, Takeda S, Nick P. Life and death under salt stress: same players, different timing? J Exp Bot 2014; 65: 2963–79. [DOI] [PubMed] [Google Scholar]
- 38. Dünser K, Gupta S, Herger A et al. Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J 2019; 38: e100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewis KC, Selzer T, Shahar C et al. Inhibition of pectin methyl esterase activity by green tea catechins. Phytochemistry 2008; 69: 2586–92. [DOI] [PubMed] [Google Scholar]
- 40. Willats WG, McCartney L, Mackie W et al. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 2001; 47: 9–27. [PubMed] [Google Scholar]
- 41. Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci 2007; 12: 267–77. [DOI] [PubMed] [Google Scholar]
- 42. Duan Q, Kita D, Li C et al. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 2010; 107: 17821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duan Q, Kita D, Johnson EA et al. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun 2014; 5: 3129. [DOI] [PubMed] [Google Scholar]
- 44. Kwak JM, Mori IC, Pei Z et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 2003; 22: 2623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voxeur A, Höfte H. Cell wall integrity signaling in plants: “To grow or not to grow that's the question.” Glycobiology 2016; 26: 950–60. [DOI] [PubMed] [Google Scholar]
- 46. Ellis C, Karafyllidis I, Wasternack C et al. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 2002; 14: 1557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Denness L, McKenna JF, Segonzac C et al. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in arabidopsis. Plant Physiol 2011; 156: 1364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 2009; 47: 177–206. [DOI] [PubMed] [Google Scholar]
- 49. Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 2005; 43: 205–27. [DOI] [PubMed] [Google Scholar]
- 50. Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 2013; 111: 1021–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 2009; 60: 183–205. [DOI] [PubMed] [Google Scholar]
- 52. Yang DL, Yao J, Mei CS et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 2012; 109: E1192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feng S, Martinez C, Gusmaroli G et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008; 451: 475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Lucas M, Daviere JM and, Rodriguez-Falcon M et al. A molecular framework for light and gibberellin control of cell elongation. Nature 2008; 451: 480–4. [DOI] [PubMed] [Google Scholar]
- 55. Zhao Y, Dong W, Zhang N et al. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol 2014; 164: 1068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Valenzuela CE, Acevedo-Acevedo O, Miranda GS et al. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J Exp Bot 2016; 67: 4209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi H, Ishitani M, Kim C et al. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 2000; 97: 6896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol 2018; 60: 796–804. [DOI] [PubMed] [Google Scholar]
- 59. Yu Y, Assmann SM. Inter-relationships between the heterotrimeric Gbeta subunit AGB1, the receptor-like kinase FERONIA, and RALF1 in salinity response. Plant Cell Environ 2018; 41: 2475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petrov V, Hille J, Mueller-Roeber B et al. ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci 2015; 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu J, Sun Y, Zhao Y et al. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Res 2015; 25: 621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 2002; 277: 559–65. [DOI] [PubMed] [Google Scholar]
- 63. Li Z, Li Z, Gao X et al. ROP11 GTPase negatively regulates ABA signaling by protecting ABI1 phosphatase activity from inhibition by the ABA receptor RCAR1/PYL9 in Arabidopsis. J Integr Plant Biol 2012; 54: 180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 2009; 149: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Singh D, Laxmi A. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 2015; 6: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.