Abstract
NM23 (NME) is a metastasis suppressor that significantly reduces metastasis without affecting primary tumor size, however, the precise molecular mechanisms are not completely understood. We examined the role of dynamin (DNM2), a GTPase regulating membrane scission of vesicles in endocytosis, in NME1 and NME2 regulation of tumor cell motility and metastasis. Overexpression of NMEs in MDA-MB-231T and MDA-MB-435 cancer cell lines increased endocytosis of transferrin and EGF receptors (TfR and EGFR) concurrent with motility and migration suppression. The internalized vesicles costained with Rab5, had AP2 depleted from the cell surface, and exhibited increased Rab5-GTP levels, consistent with endocytosis. Dynamin inhibitors Iminodyne-22 and Dynole-34-2, or shRNA-mediated downregulation of DNM2, impaired NME’s ability to augment endocytosis or suppress tumor cell motility. In a lung metastasis assay NME1 overexpression failed to significantly suppress metastasis in the DNM2 knockdown MDA-MB-231T cells. Using the EGF-EGFR signaling axis as a model in MDA-MB-231T cells, NME1 decreased pEGFR and pAkt expression in a DNM2-dependent manner, indicating the relevance of this interaction for downstream signaling. NME/DNM2 interaction was confirmed in two-way co-immunoprecipitations. Transfection of a NME1 site directed mutant lacking histidine protein kinase activity but retaining nucleoside diphosphate kinase (NDPK) activity, showed that the NDPK activity of NME was insufficient to promote endocytosis or inhibit EGFR signaling. We show that addition of NME1 or NME2 to DNM2 facilitates DNM2 oligomerization and increases GTPase activity, both required for vesicle scission. NME-DNM2 interaction may contribute to metastasis suppression by altering tumor endocytic and motility phenotypes.
Keywords: NM23, NME, Metastasis, Metastasis Suppressor, Endocytosis, Nucleoside Diphosphate Kinase, Histidine protein kinase, Motility, Epidermal Growth Factor Receptor
Introduction
Metastasis is the leading cause of cancer patient mortality, either through direct organ compromise or the consequences of its treatment. The complexity of the metastatic process has rendered it difficult to therapeutically target (1,2). Metastasis suppressors are genes that, when re-expressed in metastatic tumor cells to levels observed in their nonmetastatic counterparts, exert no inhibitory effect on primary tumor growth but significantly reduce metastasis. The specificity of metastasis suppressors may provide a window into the regulation of the metastatic process (3,4).
The NME (Nm23, awd, Ndpk) gene was the first metastasis suppressor identified. Reduced NME expression was observed in highly metastatic melanoma cell lines as compared to related, tumorigenic but less metastatic lines in the K-1735 model system (5). Overexpression of NME in metastatic tumor cell lines significantly reduced in vivo metastasis with no effect on primary tumor size (6). An in vitro hallmark of NME1 function has been its suppression of tumor cell motility in Boyden chamber assays and migration in wound healing assays; NME1 overexpression significantly reduced tumor cell movement to multiple attractants suggesting a regulatory function downstream of any particular receptor (7–9). NME is a family of ten genes, although the NME1 and NME2 members have been best studied in metastasis. It is likely that NMEs suppresses tumor motility and metastasis through complex mechanisms, which may form the basis for development of metastasis-preventive therapies.
The discovery of the nme homolog, Drosophila abnormal wing discs (awd) provided an intriguing connection between the regulation of tumor metastasis and embryonic development. Loss of awd resulted in phenotypic instability and widespread, lethal defects in epithelial outgrowths from imaginal discs (10). AWD is 78% identical to NME1 (11) and re-expression of NME1 in null awd larvae overcame most developmental defects (12). The Hsu laboratory reported migratory and invasion defects in developing epithelial cells from null awd larvae (13), linking the developmental and migration phenotypes. A recent study has reported a role for awd in chromosomal instability, cellular delamination and apoptosis (14).
Multiple developmental studies in Drosophila point to the interesting hypothesis that the endocytic process is intimately involved in the myriad awd developmental phenotypes. Aberrant endocytosis was associated with mutant awd phenotypes and complemented rab5 or shibire (shi)/dynamin genes (15–17). Studies in other organisms have also focused on dynamin (DNM), a family of three GTPases in the human that oligomerize for scission of membrane vesicles in endocytosis (rev in (18,19)). A role for DNM in NME function in mammalian cells has been proposed (20–22) but remains unproven, and the role of this pathway in the tumor metastatic process is unknown.
Herein, we investigated the role of DNM2 in NME-regulated endocytosis, and suppression of tumor cell motility and metastasis. Overexpression of NMEs in two cell lines increased endocytosis of transferrin receptor (TfR) and EGF receptor (EGFR) concurrent with motility and migration suppression and altered signaling. Importantly, dynamin inhibitors or shRNA-mediated downregulation of DNM2 impaired NME1 and NME2 ability to augment endocytosis, suppress tumor cell motility in vitro, and significantly suppress experimental metastasis in vivo. Mechanism of action studies for NME1 and NME2 revealed that both co-immunoprecipitated DNM2. Finally, we report a novel mechanism of action: NME1 and NME2 bind DNM2 and promote its oligomerization into a functional GTPase.
Materials and methods
Additional materials and methods are in Supplementary Files.
Cell culture conditions
Human triple negative breast cancer cell line MDA-MB-231T (a subline of human MDA-MB-231 cells, generously provided by Dr. Zach Howard, National Cancer Institute, Bethesda, MD) and MDA-MB-435 tumor cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Grand Island, NY) supplemented with 10% FBS in a humidified 5% CO2 incubator maintained at 37°C. MDA-MB-231T cells have been authenticated by our laboratory to the ATCC MDA-MB-231 line by short tandem repeat profiling. Growth conditions for additional breast cancer cell lines are provided in Supplementary File.
Cell transfection
MDA-MB-231T cells overexpressing NME1, NME2 and Nme1 was generated using lentivirus system as described previously (23). Briefly, a viral suspension carrying NME1/NME2 or Nme1 (0.5 ml, 107–109 TU/ml, C-Flag-SV40-eGFP-IRES-Puromycin; GeneCopoea) was added to overnight grown MDA-MB-231T cells (5×104 cells/well in 24-well plate) for 12 hrs in a growth medium containing Polybrene (0.5 μl-5 μg/ml). After 12 hrs regular growth media was added and GFP expression was visualized at 72 hrs. For stable cells generation, Puromycin selection was performed (5–10 μg/ml) and GFP positive cells were sorted using FACS and maintained in Puromycin medium. For experiments reported herein, the expression profiles of each cell line were confirmed by western blot.
Experimental metastasis assays
All the animal experiments were carried out at NCI-Frederick under an approved National Cancer Institute Animal Use Agreement. Six-week-old Balb/c athymic nude female mice were injected with 7.5 × 105 MDA-MB-231T cells (Vector-scr, Vector-shDNM2, NME1-scr, NME1-shDNM2) into their lateral tail vein. Nine weeks post-injection, at necropsy, the lungs were collected and fixed in Bouins’ solution. Lung metastatic lesions were counted on H&E step sections under a microscope and reported as a median for each group.
Mutagenesis
Mutagenesis of Wild type NME1 (EX-M0930-Lv203 and vector control EX-NEG-Lv203, GeneCopoeia) to NME1P96S and NME1H118F was performed as previously described (23). Briefly, QuikChange II Site-Directed Mutagenesis Kit (Cat# 200523, Agilent technologies, Santa Clara, USA) was used for mutating P96S (F-5’-CGGGGAGACCAACTCTGCAGACTCCAAGC-3’) and H118F (F-5’-ACAGAATCACTGCCAAATATAATGTTCCTGCCAACTTGTATGCAGA-3’) amino acid residues. Mutagenesis PCR conditions used was 95°C-1 min; 18 cycles of 95°C-50 sec, 60°C-50 sec, 68°C for 6.5 min; 68°C for 7 min. PCR amplified product was finally transfected and bacterial colonies were screened for positive clones and sequence verified. For mapping DNM2-NME region of interaction, different deletion constructs were made at Creative Biomart (Cat#DNM2–20H, DNM2–21H and DNM2–15H). All the deletion constructs are 6xHis tagged.
Gene silencing
DNM2 targeting shRNAs were purchased from GeneCopoeia (Stable selection marker: Hygromycin and Reporter gene: mcherry). shRNAs were transfected into MDA-MB-231T- vector and NME1 overexpressing cells and were selected using Hygromycin. To optimize knockdown, mCherry positive cells were sorted using FACS. Two shRNAs (HSH004403–31-LVRU6MH- Target sequence- cctcatgatcaacaatacgaa and HSH004403–32-LVRU6MH- target sequence-gggtatatcaacttcccatta, shRNA2) were utilized. A scrambled control (CSHCTR001–1-LVRU6MH) was used.
Lysates preparation, protein quantitation and immunoblotting
Cells were washed twice in 1X Phosphate-buffered saline and were lysed in RIPA buffer (20mM Tris-HCl, pH 8.0; 100mM NaCl; 10% Glycerol; 1% NP-40; 0.5% Sodium deoxycholate; 0.1% SDS; Protease Inhibitor Cocktail) by directly adding to the cells (10 cm2 dish-100 μl), followed by 10 min incubation on ice. Cells were scraped and supernatants were collected by spinning at 10,000 rpm for 10 min at 4°C. Protein estimation on lysates was performed by BCA analysis (Pierce® BCA Protein Assay Reagent A and B- Prod# 23228). Lysates were frozen at −80 °C until use. Equal amount of protein was loaded on Any KD™ or 4–20% Mini-PROTEAN TGX™ Gels (BIO-RAD, Cat# 456–9033) and was transferred to Nitrocellulose membrane using Trans-Blot Turbo Mini Nitrocellulose Transfer Packs (BIO-RAD, Cat #1704158). Membrane was blocked using Odyssey® Blocking Buffer (Part # 927–50000) for 1 hr at room temperature. This was followed by the primary antibody incubation in blocking buffer containing 0.2% Tween 20 (SIGMA, P2287) overnight at 4°C. Membrane was washed thrice in 0.1% TBST (for 10 min each) and was incubated with secondary antibody for 1hr, followed by TBST wash. After washing, blot was visualized in Multi-application gel imaging system PXi (Syngene).
Co-Immunoprecipitation (Co-IP)
Protein-protein interactions between NME1, NME2 or mutant NME1s (P96S and H118F) and DNM2 were studied using Co-Immunoprecipitation Kit (ab206996, Abcam). Confluent cultures of cells were lysed in cold Lysis buffer 1 and 500 μg of protein of different lysates were incubated with 20 μg of NME/ DNM2 primary antibody (NME antibody: sc-343 and DNM2 antibody: sc-6400) overnight at 4°C on a rotary mixer. Protein A/G Sepharose® beads was washed twice with 1X Wash buffer and suspended as 50% slurry. Following antibody binding, Protein A/G Sepharose® was (30 μl/ reaction) added to each tube and incubated for 1 hr at 4°C. The beads were collected by centrifugation (2000 x g for 2 min at 4°C) and washed thrice with wash buffer. Finally, the beads were collected by centrifugation. Protein complexes were eluted by boiling (5 min) the beads in denaturing 2X SDS-PAGE loading buffer. All the samples were resolved on Any KD™ Mini-PROTEAN TGX™ (BIO-RAD, Cat# 456–9033) SDS-PAGE gel and processed as Western blot. As control, 1% of total lysate used in the assay was loaded on a separate gel. For in vitro co-immunoprecipitation assay recombinant NME1 protein (1 μg) was incubated with DNM2 (5 μg) or with its deletion constructs 1/2/3 in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, 0.5% Triton-X100 with protease inhibitors cocktail for 30 min. NME1 was immunoprecipitated using αNME antibody and bound proteins were detected with α-DNM2 antibody/α−6xHis antibody.
Dynamin oligomerization assay (Co-sedimentation assay)
Before co-sedimentation, human recombinant DNM2 (DNM2–18H, MYC/DDK-tagged, Creative BioMart) was centrifuged at 4°C for 15 min at 100,000x g to remove potential aggregates. DNM2 (3.5 μg) was incubated for 15 min at 22°C in the absence or presence of NME1, NME2, NME1P96S or NME1H118F (1.5μg) in HCB75 buffer (20 mM HEPES, 1 mM MgCl2, 100 mM EGTA, 75 mM NaCl and 0.5 mM DTT) in a total volume of 25 μl. The solution was then centrifuged at 214,000 × g for 15 min (TLA-100 rotor, Beckman Coulter, Inc.) at 4°C and the resulting pellets (P) and supernatants (S) fractions were processed for western blotting and detected by NME or DNM2 antibody.
Statistical analyses
All experiments were repeated at least three times unless noted. Statistical significance was calculated by a 1-way ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.001). For metastasis 1-way ANOVA (Nonparametric) test was performed comparing median across all the groups with P < 0.05 considered significant (*).
Results
A role for NME overexpression, and DNM2, in cancer cell endocytosis.
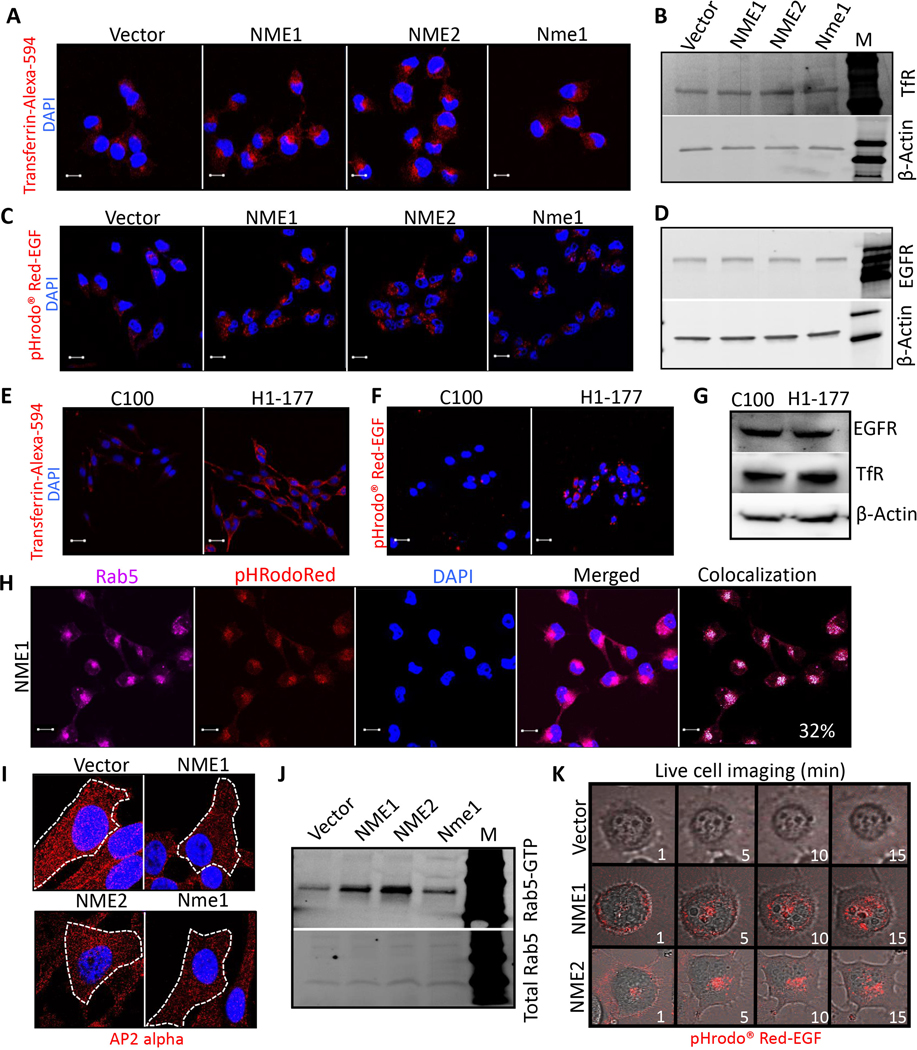
MDA-MB-231T cells, a triple-negative human breast cancer line, were transfected with vector, human NME1 or NME2, or murine Nme1 (Supplementary Fig. S1A, for the passages used herein); all NME transfectants were suppressed in both motility in Boyden chambers and migration in scratch assays in vitro (Supplementary Fig. S1B and C). MDA-MB-435 cells, reported to be either triple-negative breast cancer or melanoma, were transfected with vector (C-100) or NME1 (H1–177) (Supplementary Fig. S1D for the passages used herein); H1–177 cells were both motility and migration suppressed (Supplementary Fig. S1E and F) (23). To examine a potential role of endocytosis in NME function, these cell lines were cultured in serum free culture medium and exposed to labeled ligands for either transferrin receptor (TfR) or epidermal growth factor receptor (EGFR). Overexpression of any form of NME increased the perinuclear internalization of TfR and EGFR (Fig. 1A-D). The pHrodo™ Red-EGF probe fluoresces when exposed to acidic pH, which increases from early- to the late endosomes that fuse with lysosomes, confirming an internal localization of the signal. No nuclear accumulation of EGFR was observed (Fig. 1C). Comparable amounts of total TfR and EGFR were present in each of the transfectants (Fig. 1B and D, respectively). Similar results were observed in MDA-MB-435 model system overexpressing NME (Fig. 1E-G). Increased endocytosis would be expected to reduce surface level of receptors in NME overexpressing cells compared to control. Lower expression of surface Biotin-labeled EGFR and other receptors were confirmed in NME1 overexpressing cells compared to vector control (Supplementary Fig. S2A). To confirm that this was an endocytic process, staining of the NME1 transfectant showed that approximately 32% of pHrodo™ Red-EGF endocytic vesicles costained with Rab5 (early endosomes) (Fig. 1H, Supplementary Fig. S2B and C). Also, decreased cell surface CCP (Clathrin Coated Pits detected by AP2 alpha / alpha Adaptin) staining (Fig. 1I), and elevated Rab5-GTP levels were observed (Fig. 1J), corroborating NME mediated increased endocytosis. Time lapse video microscopy images of EGFR internalization indicated that NME overexpression increased endocytosis rather than just speeding it up over a 15-min time course (Fig. 1K and Supplementary Video S1).
Figure 1: NME overexpression increased tumor cell endocytosis.
Previously reported transfectants of the MDA-MB-231T breast cancer cell line expressed a vector construct or NME genes (human NME1, NME2 or mouse ortholog Nme1), while transfectants of the MDA-MB-435 cell line expressed a vector (C-100) or NME1 (H1–177). A-D MDA-MB-231T transfectants were incubated with Transferrin-Alexa-594 (A) or pHrodo™ Red-EGF (C) (both red) for 15 min, whereupon cells were fixed in PFA and nuclei stained with DAPI (blue). Representative merged images are shown at 63x magnification. Scale bar-10 μm. Lysates of the transfectants were probed for the cognate receptors on western blots, Transferrin receptor (B) and EGFR (D). β-actin was used as normalizing control. E, F MDA-MB-435 transfectants similarly incubated with Transferrin-Alexa-594 (E) or pHrodo™ Red-EGF (F). G Western blot of MDA-MB-435 cells for the cognate receptors. H Vector and NME1 overexpressing MDA-MB-231T cells were incubated with pHrodo™ Red-EGF dye (red) for 15 min followed by immunostaining with Rab5 antibody (far-red). Approximately 32% of pHrodo™ Red-EGF co-localized with Rab5 in NME1 overexpressing cells (white). Scale bar-10 μm. I Immunofluorescence staining of vector and NME overexpressing MDA-MB-231T cells for AP2 alpha to detect clathrin coated pits (red). Cells are outlined in dashed lines with nuclei stained with DAPI (magnification-100x). J Rab5-GTP levels determined by active Rab5 Pull-down assay from whole cell lysates of vector and NME overexpressing MDA-MB-231T cells. Upper panel: Rab5-GTP, Lower panel, Total Rab 5. K Time lapse real time imaging was performed on vector and NME overexpressing (NME1 and NME2) MDA-MB-231T cells incubated with pHrodo™ Red-EGF dye (Supplementary Video S1). Frames at 1, 5, 10 and 15 min are shown. Data are representative of three independent experiments.
NME1 and NME2 interact with the endocytic protein DNM2.
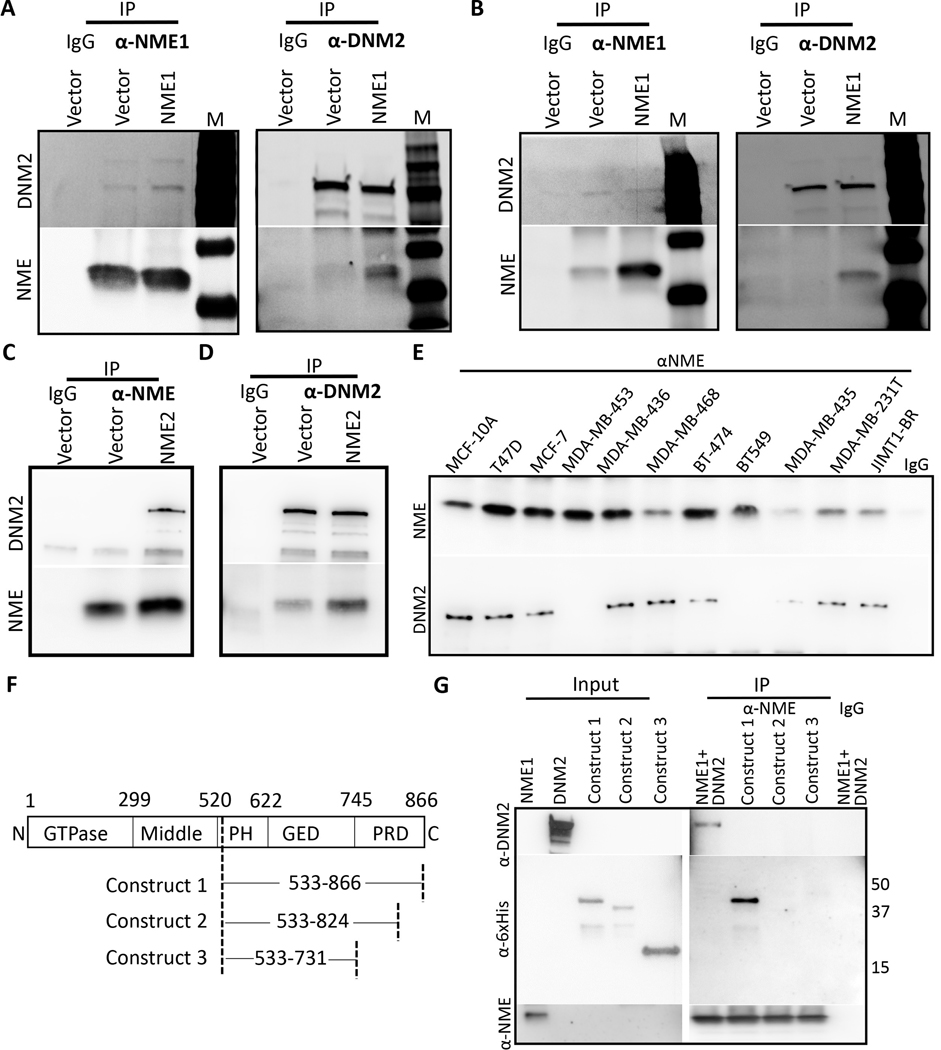
The DNM2 GTPase effects the scission of endocytic vesicles from the plasma membrane as a rate limiting step in endocytosis. Previous reports have shown a genetic interaction of NME (awd) and DNM (shi) in Drosophila development (24), suggesting the hypothesis that a similar interaction may be germane to NME-regulated tumor progression. Co-immunoprecipitations were performed with MDA-MB-231T and MDA-MB-435 transfectants (Fig. 2A, B and Supplementary Fig. S2D, E). Immunoprecipitation with anti-DNM2 pulled down comparable levels of DNM2 in each pair of cell lysates but increased co-immunoprecipitation with NME1 was apparent in the NME1 transfectants. Conversely, immunoprecipitation with anti-NME resulted in greater pulldown in the NME1 transfectants; a proportion of total DNM2 was bound to NME1, higher in the NME1 transfectants. Similar results were observed for NME2 overexpressing cells (Figure 2C, D and Supplementary Fig. S2F). The data confirm a NME-DNM2 interaction and demonstrate that NME protein level was limiting for the extent of NME:DNM interaction.
Figure 2: NME is a binding partner of DNM2.
A Two-way co-immunoprecipitation of NME1 and DNM2 was performed on vector and NME1 overexpressing MDA-MB-231T cells using rabbit anti-NME1 and goat anti-DNM2 antibody. B Two-way co-immunoprecipitation in vector and NME1 overexpressing MDA-MB-435 cells was performed with the above antibodies. C& D Two-way co-immunoprecipitation of NME2 and DNM2 was performed on vector and NME2 overexpressing MDA-MB-231T cells using rabbit anti-NME and mouse anti-DNM2 antibodies. E Co-immunoprecipitation of NME1 and DNM2 was performed on a panel of breast cancer cell lines with varying metastatic status (low to high) using anti-NME antibody for immunoprecipitation. F DNM2 construct map showing deletion in PRD domain [Construct 1: Contains full PRD domain, Construct 2: truncated PRD domain (Δ42 amino acid), Construct 3: deleted PRD domain]. All the constructs contain PH and GED domain. G In vitro co-immunoprecipitation of NME1 was performed with DNM2 or with deletion constructs using anti-NME antibody. Data are representative of three independent experiments.
To further explore the association of NME1-DNM2 interaction and tumor aggressiveness, co-immunoprecipitations were performed in a panel of breast cancer cell lines. Lysates of cells reported as immortalized (MCF10A), tumorigenic but low or non-metastatic (T47D, MCF-7, MDA-MB-453, MDA-MB-436, MDA-MB-468, BT-474) or tumorigenic and metastatic (BT549, MDA-MB-435, MDA-MB-231T, JIMT-1-BR) were immunoprecipitated with anti-NME1 and blotted for DNM2 (Figure 2E). NME1 levels were strong in the immortal line, 4/6 tumorigenic lines (1/6 moderate, 1/6 weak), and only 1/4 metastatic lines (2/4 moderate, 1/ 4 weak). DNM2 binding to NME1 was detectable in the immortal line, 5/6 tumorigenic lines and only 2/4 metastatic lines. The data suggest that the NME1-DNM2 interaction occurs generally and is associated with low metastatic potential. Mapping of DNM2 for its interaction domain with NME1 was performed (Figure 2F). Other DNM2-interacting proteins such as Grb2, Amphiphysin and Endophilin bind the C-terminal domain of DNM2, specifically the PRD domain (25). In a previous report, the PRD domain has been shown to interact with NME1, however a detailed mapping of the C-terminal region is missing (21). For more detailed mapping of the C-terminal region of DNM2, three domains were expressed (construct 1: PH-GED-PRD domain). A series of deletion constructs contained PH-GED-PRDΔ42 (construct 2: 42 amino acid deletion in PRD domain), PH-GED-ΔPRD (construct 3: Full PRD domain deleted) (Figure 2F). The PRD domain contains several motifs characteristic of a type II polyproline helix and deletion of 42 amino acids (construct 2) was performed to delete 4 class-II (PxxPxR) and 2 class-I (RxxPxxP) motifs which are reported to mediate binding of DNM2 to other proteins (26). Binding of these constructs to NME1 was assessed using in vitro immunoprecipitation. Deletion of part of PRD domain (42 amino acid) or full PRD domain in constructs 2–3 abrogated NME1 binding (Figure 2G).
NME-mediated increased endocytosis is DNM2 dependent.
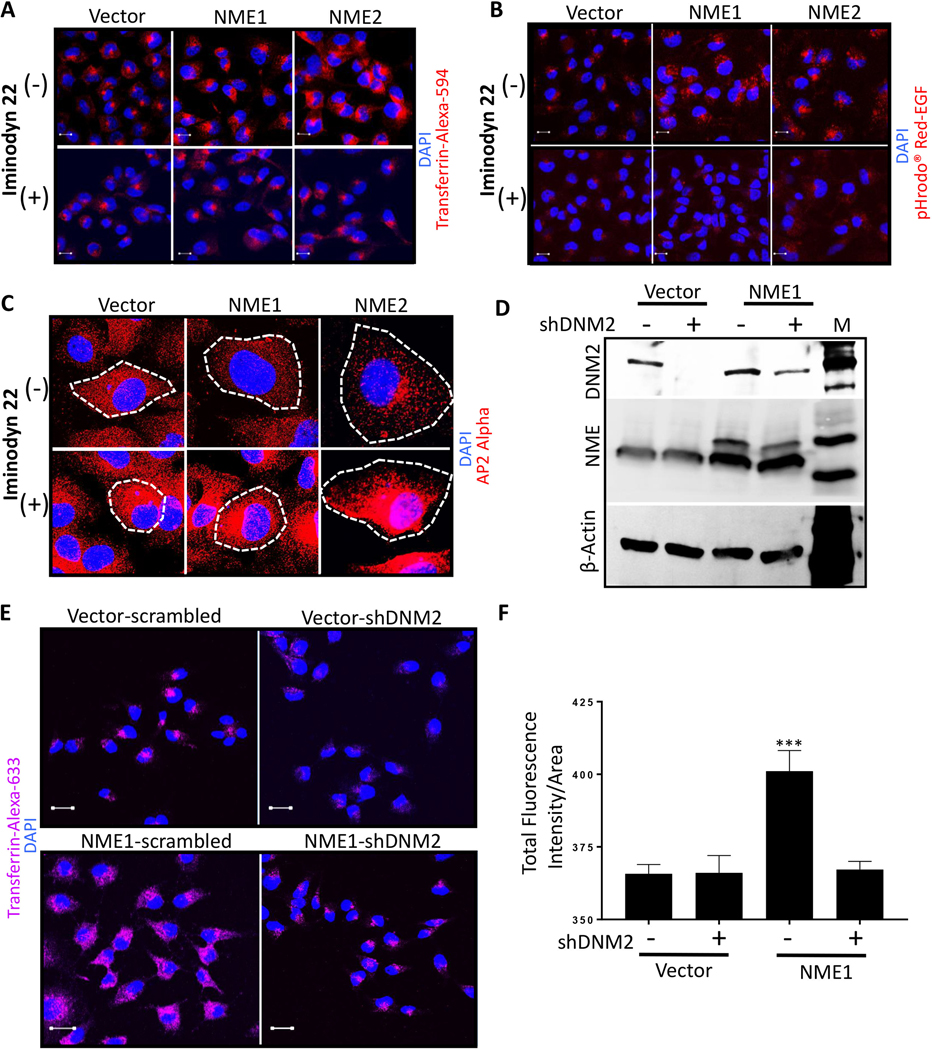
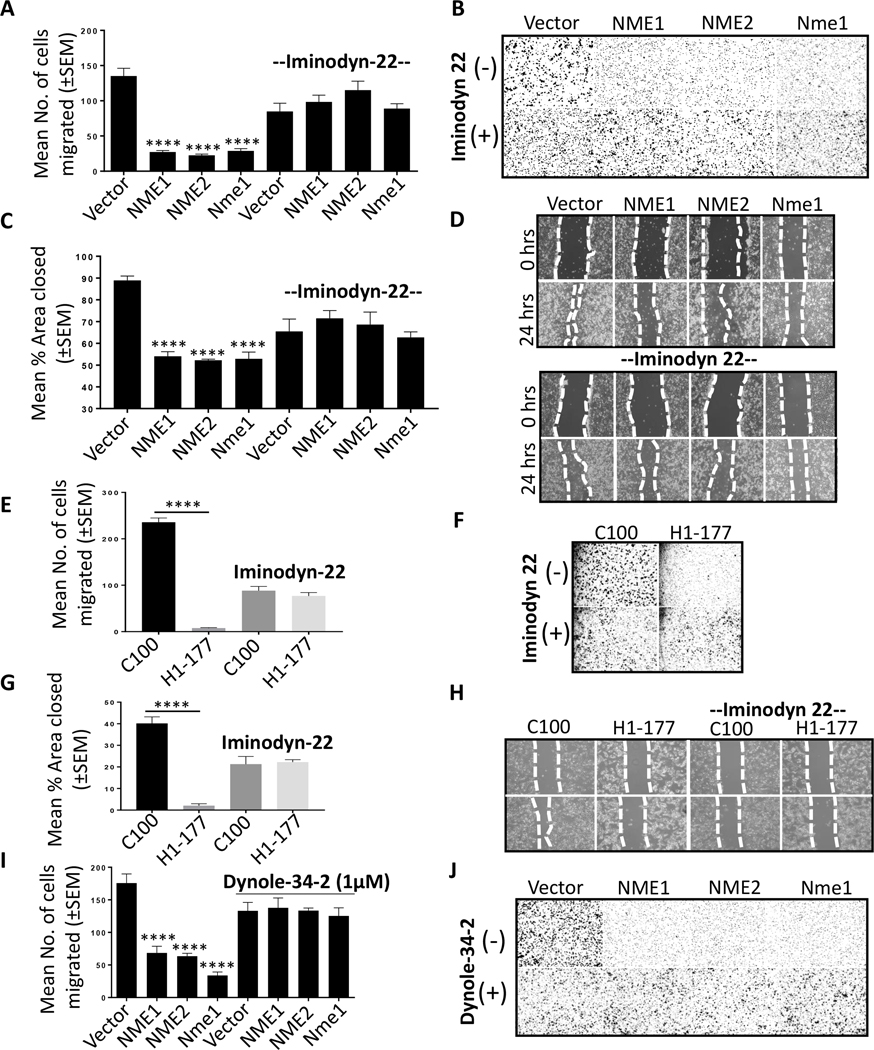
We hypothesized that NME1 function in increasing endocytosis in cancer cells was DNM-dependent. Initial experiments used well characterized dynamin inhibitors (27). Preliminary experiments established noncytotoxic and endocytosis inhibiting dose for Iminodyn-22 (Supplementary Fig. S3A, B). Iminodyn-22, an inhibitor of the GTPase Allosteric Site (GAS) domain of DNM2, abrogated the increased endocytic phenotype of the NME transfectants using Transferrin-Alexa-594 and pHRodo™ Red-EGF probe (Fig. 3A and B, respectively). Similarly, NME1 failed to reduce AP2 staining (CCPs) in presence of Iminodyn-22 (Fig. 3C).
Figure 3: NME-mediated increased tumor cell endocytosis is DNM2 dependent.
A, B Vector and NME overexpressing (NME1 and NME2) MDA-MB-231T cells were plated in chamber slides and treated with a non-cytotoxic dose (2 μM) of dynamin inhibitor (Iminodyn-22) or vehicle for 24 hrs. Cells were then incubated with Transferrin-Alexa-594 (A) or pHrodo™ Red-EGF dye (B) for 15 min., fixed in PFA and nuclei stained with DAPI (blue). C Vector and NME overexpressing MDA-MB-231T cells were plated in chamber slides and cultured with and without 2 μM Iminodyn-22 for 24 hrs. Cells were then fixed in PFA and immunofluorescence was performed with AP2 alpha to detect clathrin coated pits (red). Cells are outlined by dotted lines and nuclei counterstained with DAPI. D-F Effect of DNM2 shRNA. D Western blot of MDA-MB-231T vector and NME1 transfectants, each transfected with a scrambled shRNA (−) or DNM2 shRNA (+). β-actin was used as loading control. E Vector and NME overexpressing MDA-MB-231T cells stably transfected with scrambled or shDNM2 were incubated with Transferrin-Alexa-633 for 15 min. Following acid washes, cells were fixed in PFA and nuclei were visualized by DAPI (blue); staining was visualized using confocal microscopy with representative images shown. F Quantification of Transferrin-Alexa-633 uptake in the transfectants described in (E) using Zeiss software (ZEN). For all images, merged views are shown. Scale bar-10 μm. All experiments shown are representative of 3 replicates and statistical significance was calculated by a 1-way ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.001).
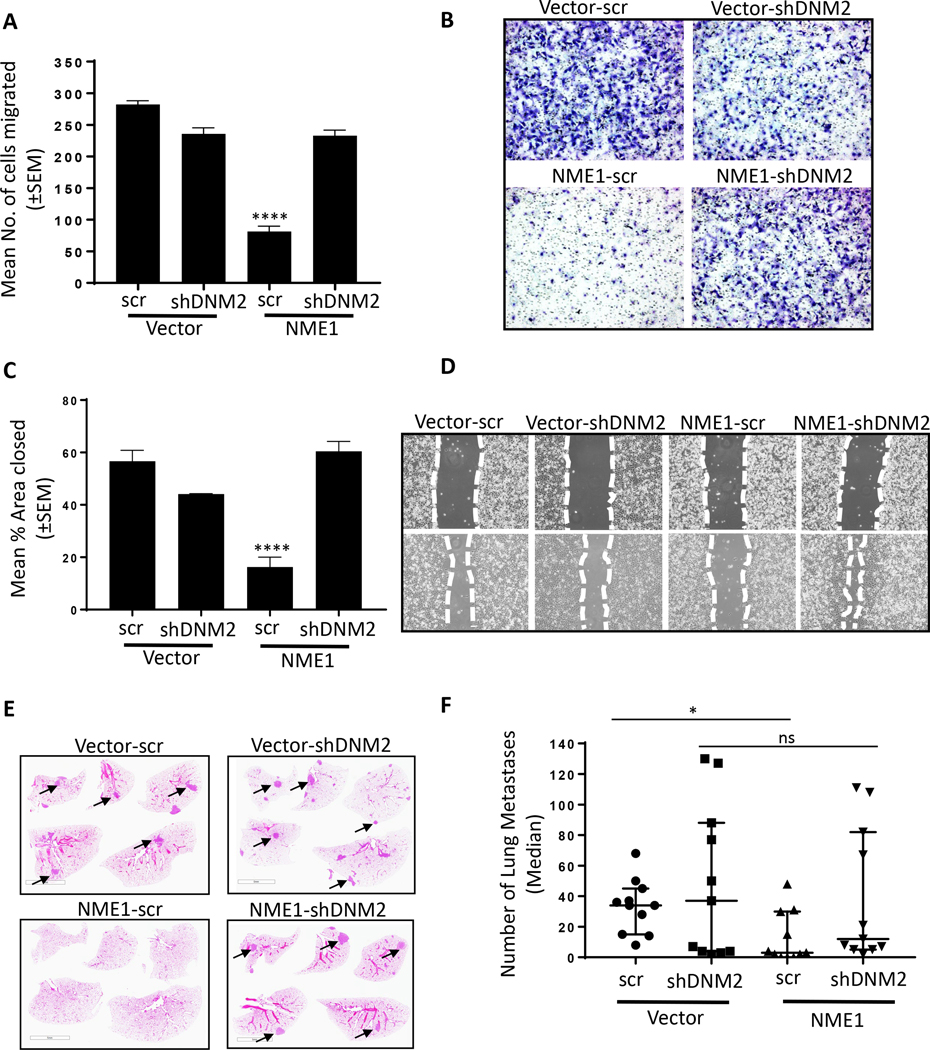
Since inhibitors can be nonspecific, DNM2 was knocked down by shRNAs in vector and NME1 transfectants of MDA-MB-231T cells, with a scrambled shRNA knockdown as a control (Fig. 3D). Endocytosis of TfR was not increased above background levels in cells overexpressing NME1 and shDNM2 (Fig. 3E and F). Endocytosis of EGFR could not be performed due to the overlapping fluorescent tags on the pHrodo™ Red-EGF and shDNM2-mCherry. The data demonstrate that NME modulation of tumor cell endocytosis required DNM2 function.
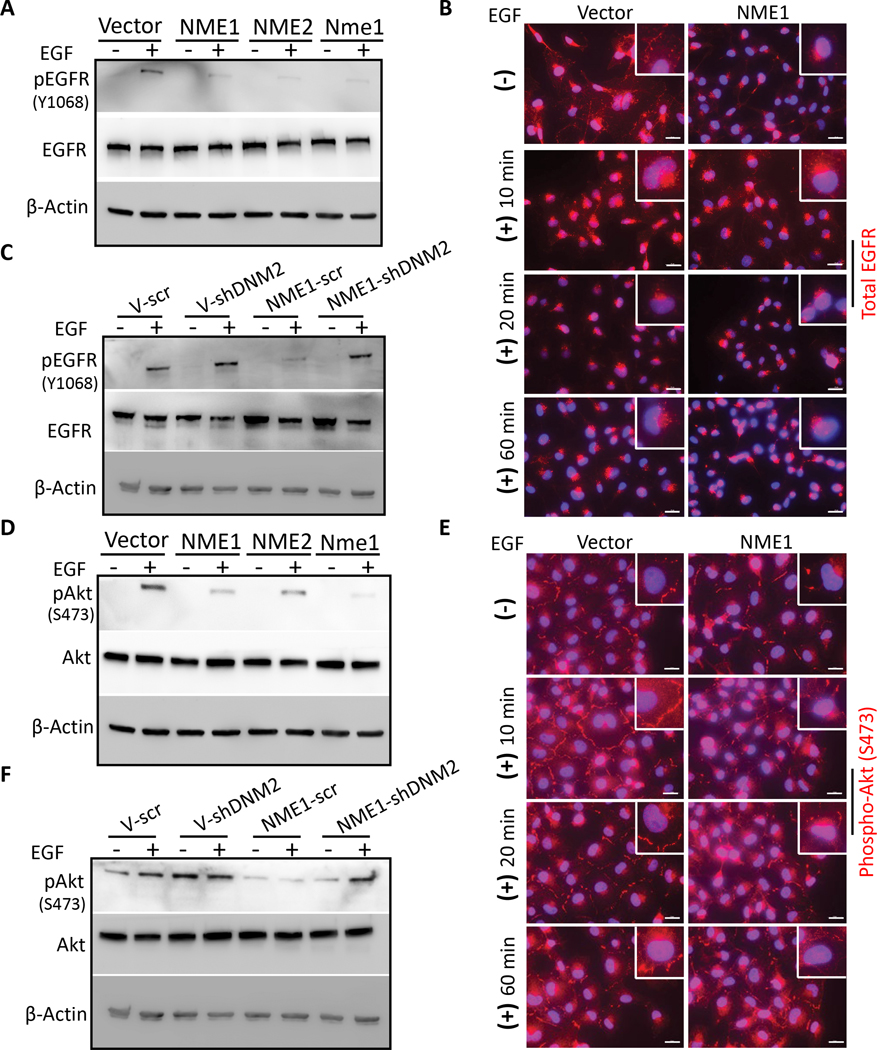
Endocytosis is known to affect signal transduction patterns in tumor cells (28,29). Western blots from lysates of confluent cultures and IF on tumor cells migrating in scratch assays were used to ascertain if EGFR signaling in MDA-MB-231T cells was affected by the NME1-DNM2 axis upon EGF stimulation. Total EGFR levels were comparable between control and NME1 transfectants, but NME overexpression resulted in decreased activated EGFR (pEGFR Y1068) (Fig. 4A), and redistribution to a perinuclear location (Fig. 4B), consistent with endocytosis. The decreased pEGFR (Y1068) observed upon NME1 overexpression was not seen with concomitant DNM2 shRNA (Fig. 4C). Of the targets downstream of EGFR, phosphorylated Akt (S473) was reduced by NME overexpression (Fig. 4D) and lost its membranous localization (Fig. 4E). Knockdown of DNM2 in the NME1 overexpressing cells restored EGF stimulation of pAkt (Fig. 4F). The data demonstrate that the NME1-DNM2 pathway affects specific signaling pathways that may be functional to tumor cell motility and migration.
Figure 4: NME1 overexpression alters EGFR activation in MDA-MB-231T breast cancer cells.
A MDA-MB-231T cells transfected with vector and NME were cultured, and then washed in serum-free medium; cells were incubated with or without EGF (50 ng/ml) in serum-free medium for 1 hr. A western blot of cell lysates is shown quantifying total EGFR and Y1068 pEGFR, with β-actin as a loading control. B Immunofluorescence of EGFR in vector and NME1 transfected MDA-MB-231T cells in a scratch assay were treated as described in panel (A). Before addition of EGF, and at various timepoints through the 1 hr EGF treatment, cells were washed, fixed with PFA and stained for EGFR (red). C Similar experiment performed using MDA-MB-231T cells expressing vector or NME1, and either scrambled shRNA or shDNM2 (described in Figure 3D), to detect EGFR and Y1068 pEGFR. D For detection of Akt activation in MDA-MB-231T tumor cells transfected with vector or NMEs, cells were cultured and treated with 50 ng/ml EGF as described above (A). Lysates were probed for total Akt and S473 pAkt on western blots, with β-actin as a loading control. E Immunofluorescence of S473 pAkt in vector and NME1 transfected MDA-MB-231T cells at various timepoints through the 1 hr of EGF treatment was performed as described above (B). Nuclei were visualized using DAPI (blue). Representative merged images are shown at 63x magnification. Scale bar-20 μm. F MDA-MB-231T cells expressing vector or NME1, and either scrambled shRNA or shDNM2, described above (C) were assessed by western blot for total and pAkt. Data are representative of three independent experiments.
DNM2 impacts NME-inhibition of in vitro tumor cell motility and migration.
DNM2 bound NME and was functionally involved in its acceleration of endocytosis, but was this process critical to tumor cell motility and metastasis? Initial experiments used the Iminodyn-22 dynamin inhibitor in Boyden chamber motility assays to the combination of factors present in 1% fetal bovine serum (FBS) as an attractant. Overexpression of NME1, NME2 or murine Nme1 in MDA-MB-231T cells resulted in ~70–80% reduction in motility as previously reported (Supplementary Fig. S1B) (23). Addition of 2uM Iminodyn-22 resulted in lower overall motility in the vector transfectants, approximately ~40% of the vehicle treated cells (Fig. 5A). However, the motility of the NME transfectants was not suppressed beyond this point, and never reached the levels of motility suppression observed in the vehicle treated NME transfectants (Fig. 5A, B). Similar trends were observed in migration assays, in which confluent cultures were disrupted with a scratch and tumor cell migration into the wound was quantified. Overexpression of any NME suppressed migration by 50–60%. Addition of Iminodyn-22 lowered overall migration in the control transfectant, but the NME transfectants were unable to reduce migration below this point (Fig. 5C, D). The same trends were observed for control and NME1 transfectants of the MDA-MB-435 cell line for motility and migration (Fig. 5E-H). NME mediated motility suppression reversal was also observed with other dynamin inhibitors (Supplementary Fig. S3C-G). Dynole-34–2 (1 μM) was not cytotoxic to MDA-MB-231T tumor cells (Supplementary Fig. S3C). Addition of Dynole-34–2 to MDA-MB-231T cells in motility assays exerted a minimal effect on the motility of vector transfectants compared to vehicle treated; no suppression of motility was observed in the NME transfectants compared to vector control (Fig. 5I, J). Other dynamin inhibitors are shown on Supplementary Fig. S3, including MiTMAB and OcTMAB, resulting in the same trends but with more pronounced inhibition of vector motility. As a control for these inhibitors Pro-myristic acid, which is incapable of dynamin inhibition (30), was without significant effect (Supplementary Fig. S3H, I).
Figure 5: NME-mediated suppression of tumor cell motility and migration are DNM2 dependent.
A Boyden chamber motility assays using vector and NME transfectants of the MDA-MB-231T cell line, in the presence of 2 μM Iminodyn-22 or vehicle control. Motility to 1% FBS over four hr was quantified. B Representative photographs of the undersides of Boyden chamber membrane showing migrated tumor cells. C, D Similar experiments conducted using a scratch migration assay. Cells were pretreated with 2 μM Iminodyn-22 for 24 hrs in 6-well plates. A line was scratched with a P200 pipette tip and migration of cells into the scratch was photographed after 24 hrs. Percent area closed was quantitated using ImageJ. E, F Boyden chamber assays using vector (C-100) and NME1 (H1–177) transfected MDA-MB-435 cells in the presence of 2 μM Iminodyn-22 or vehicle, with representative membrane shown. G, H Similar experiment conducted with a scratch assay on MDA-MB-435 cells as described in panel (D). I, J Boyden chamber motility assays (see panel A) using vector and NME transfectants of the MDA-MB-231T cell line, in the presence of 1 μM Dynole-34–2 or vehicle control. All experiments shown are representative of 4 replicates and statistical significance was calculated by a 1-way ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.001).
Knockdown of DNM2 was used to confirm the specificity of the above results. Vector and NME1 transfectants of MDA-MB-231T were transfected with a scrambled or DNM2 shRNA (Fig. 3D). The motility of the vector and NME1 overexpressing/scrambled shRNA tumor cells mirrored that of NME1 overexpressing line, showing ~70% reduced motility in Boyden chamber assays. DNM2 knockdown exerted a less suppressive effect on motility than the DNM inhibitors, ~17% as compared to vehicle control. Interestingly, the NME1 overexpressing cells with DNM2 knockdown failed to demonstrate any additional motility suppression (Fig. 6A, B). Similar trends were observed in migration assays (Fig. 6C, D). We assessed NME functionality in DNM2 knockdown cells and observed that cells with depleted DNM2 had functional NME1 in terms of its nucleoside diphosphate kinase (NDPK) and 1-phosphohistidine phosphorylation status (Supplementary Fig. S4A, B). A second shRNA to DNM2 showed similar trends (Supplementary Fig. S4C-E). The data indicate that NME1 suppression of motility and migration in vitro was dependent on DNM2.
Figure 6: shRNA-mediated knockdown of DNM2 abrogates NME1 suppression of tumor cell motility and migration, and contributes to metastasis suppression.
A Vector and NME transfected MDA-MB-231T cells stably transfected with scrambled or shDNM2, described in the legend to Figure 3, were assessed for motility in Boyden chambers as described in the legend to Figure 4. B Representative photographs of the Boyden chamber membrane underside showing migrated tumor cells. C Migration in scratch assays of the same cells, as described in the legend to Figure 4. D Representative photographs of the scratch invaded. E 7.5 × 105 MDA-MB-231T cells expressing vector and NME1 overexpression with or without DNM2 knockdown (Vector-scr, Vector-shDNM2, NME1-scr, NME1-shDNM2) were injected into the lateral tail vein of athymic nude mice (each group, n=11). At 9 weeks post-injection, the mice were sacrificed, and the lungs were fixed in Bouin’s solution followed by H&E staining. A representative image of each group is presented with arrows pointing to metastases. F All metastases in the lung sections were counted and presented as scatter plot showing median with interquartile range. Each dot represents a single mouse. All in vitro experiments shown are representative of (motility n=4 and migration n=3) replicates and statistical significance was calculated by a 1-way ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.001). For metastasis 1-way ANOVA (nonparametric) test was performed comparing median across all the groups with P < 0.05 considered significant (*).
DNM2 contributes to NME1 suppression of experimental metastasis.
An experimental metastasis assay was performed on a set of DNM2 knockdown cells (MDA-MB-231T cells overexpressing Vector or NME1, each transfected with a scrambled or DNM2 shRNA) (Fig. 6E and F). Briefly, tumor cells were injected into the tail veins of immunocompromised mice and metastases enumerated 65 days later in lung step sections as a model of metastatic colonization. NME1 overexpression/scrambled shRNA were metastasis suppressed as compared to vector/scrambled shRNA (median of 3 versus 34 metastases/section, respectively, 91.1% reduction, P= 0.04). Inhibition of DNM2 in the vector transfectant resulted in a comparable median number of lung metastases as the vector transfectant/scrambled shRNA (median of 37 versus 34 metastases/section, respectively). Interestingly, the NME1 overexpression/DNM2 shRNA transfectant was only partially suppressed (median of 12 metastases/section, 67.5% reduction compared to vector, P-value >0.99) (Fig. 6E and F). Thus, DNM2 expression is contributory to NME1 suppression of metastasis.
The NDPK activity of NME1 is insufficient for increased endocytosis.
Enzymatically, NME1 has two unusual activities dependent on autophosphorylation of its histidine 118: a NDPK activity reversibly removes a phosphate from nucleoside triphosphates and transfers it to NME1 and subsequently to nucleoside diphosphates, and a HPK activity in which autophosphorylated NME1 transfers the phosphate to protein substrates (31–33). NME has been previously hypothesized to stimulate dynamin function by local provision of GTP via its NDPK activity (21). We and others have previously characterized site directed mutants of NME1 that retain specific enzymatic activities and their associations with in vitro motility suppression (23,31,34,35). MDA-MB-231T cells expressing a vector, NME1, NMEH118F or NMEP96S were previously described (23). Briefly, NMEH118F abolishes the phosphohistidine and both the NDPK and HPK activities, and is not motility suppressive (31,32,35). NMEP96S separates the two enzymatic activities as transfectants have significant NDPK but not HPK activity; that this transfectant, generated independently several times is not motility suppressive has been used to conclude that the NDPK activity of NME1 is insufficient and that the HPK activity may be contributory to NME’s phenotypic effects (23,36). We asked if either of these two mutant NMEs would augment endocytosis, bind DNM2 and downregulate signaling similarly to the wild type protein.
Increased endocytosis of TfR and EGFR were observed in this independent wild type NME1 transfectant as compared to its vector transfectant. Overexpression of NME1H118F or NME1P96S failed to increase endocytosis to the levels of the wild type protein (Supplementary Fig. S5A-D and Supplementary Video S2). Diminution of pEGFR and pAkt upon EGF stimulation of NME overexpressing cells was not observed in the NME1H118F or NME1P96S transfectants (Supplementary Fig. S6A). The data suggest that NME1 histidine phosphorylation was necessary for increased endocytosis and altered signaling, but suggest that the NDPK activity of NME1 was dispensable (the P96S mutant transfectant retains NDPK activity). In vitro experiments showed that DNM2 bound wild type and mutant forms of NME1 (Supplementary Fig. S6B-D). No evidence was observed suggesting that DNM2 is an HPK substrate of NME1 (Supplementary Fig. S7A-E).
A new role for NME1 and NME2 in DNM2 oligomerization.
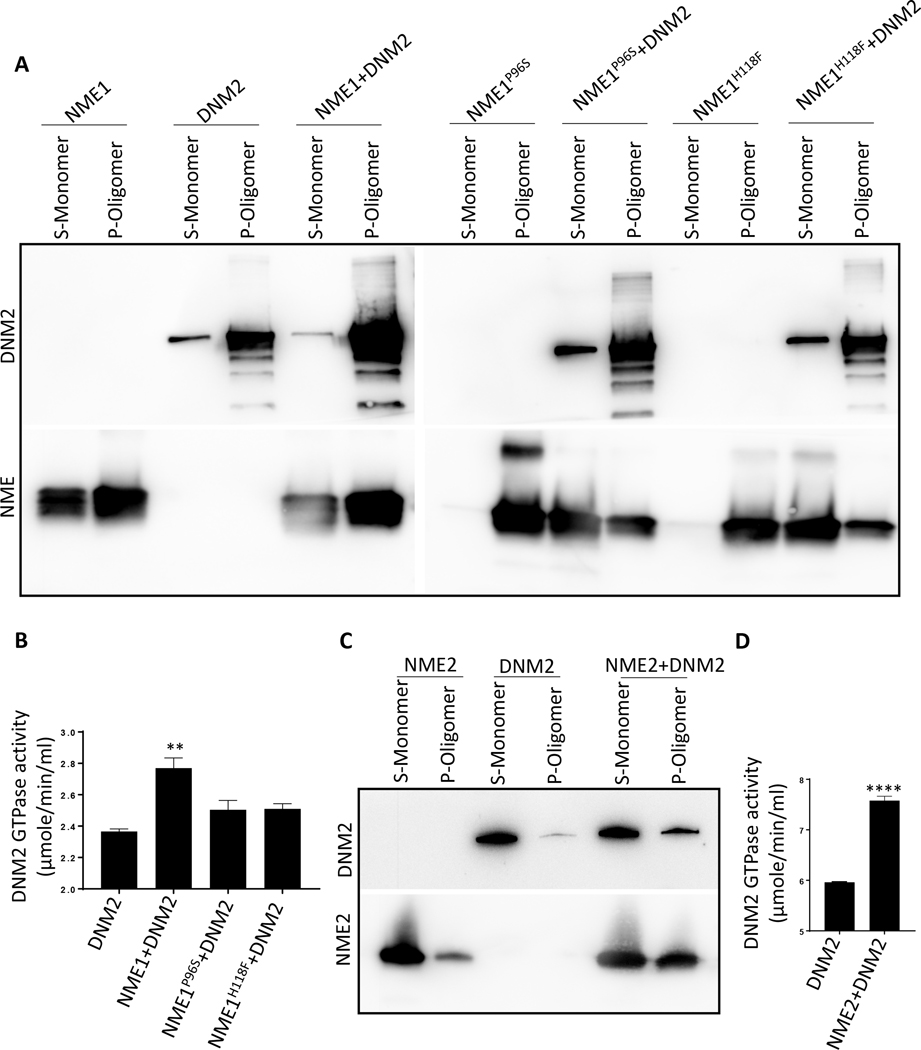
Knowing that DNM2 interacted with NMEs and that DNM2 function was essential for NME1 increased endocytosis, inhibition of motility and migration, and contributed to metastasis, we hypothesized that NME1 regulated the active, oligomeric form of DNM2. DNM2 exists predominantly as monomers, dimers, tetramers and octamers, as shown by crosslinking experiments and these can assemble into higher order ring and helical structures (reviewed in (25)). DNM2 assembly into higher order oligomers increases its GTPase activity leading to endocytosis (scission) of newly formed vesicles from the plasma membrane (37). To ask whether the association of NME1 with DNM2 altered its oligomerization state, partially purified recombinant NME1 was incubated with commercially available DNM2 in HCB75 buffer; the mixtures were subjected to ultracentrifugation, which has previously been reported to separate DNM2 monomers in the supernatant (S) from oligomers in the pellet (P) (38). Western blots of DNM2 and NME were performed (Fig. 7A). DNM2 alone was a mixture of monomers and oligomers, the latter in the pellet. Addition of NME1 to DNM2 greatly increased the oligomer proportion of DNM2 (pellet), with a corresponding decrease in monomeric DNM2 (supernatant). The proportion of NME1 in the supernatant was also decreased, in keeping with an admixture of NME1 and DNM2 forming oligomeric structures. Addition of partially purified recombinant NME1H118F or NME1P96S failed to increase the oligomerization of DNM2 to the level of the recombinant wild type protein.
Figure 7: NME1 and NME2 increased DNM2 oligomerization and GTPase activity in vitro.
A Western blot of DNM2 and NME. Recombinant DNM2 (3.5 μg) was incubated with wild type or mutant recombinant, partially purified NME1s (1.5 μg) in HCB75 buffer for 15 min at 22°C. Each mixture was ultracentrifuged at 214,000 × g for 15 min at 4°C and the resulting pellets (P) and supernatants (S) fractions were processed on western blots. In this assay, DNM2 monomers remained in the supernatant (S) while oligomers precipitated into the pellet (P). B A GTPase assay was performed on DNM2 (3 μg), alone or incubated with wild type NME1, NME1P96S or NME1H118F (1μg) in the presence of liposomes for 15 min, followed by addition of 4 mM GTP and color reagent. OD at 620 nm was measured after 30 min incubation. C Recombinant DNM2 (3.5 μg) was incubated with recombinant NME2 (1.5 μg) as per the oligomerization assay protocol described above and was processed on western blot. D GTPase assay was performed on DNM2 (3 μg), alone or incubated with NME2 (1μg) as per the GTPase protocol described above. All experiments shown are representative of three (n=3) replicates and statistical significance was calculated by a 1-way ANOVA (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.001).
To assess the functionality of DNM2 oligomerization induced by NME1, a DNM2 GTPase assay was performed. It was observed that wild type NME1 increased DNM2 GTPase activity while both the mutants failed to so (Fig. 7B). Similar results were observed with NME2 and it was also able to oligomerize DNM2 leading to increased GTPase activity (Fig. 7C, D). Taken together, NME promoted the GTPase activity of DNM2 by increasing its oligomerization.
Discussion:
Genetic and limited biochemical evidence has suggested an interaction of NME/AWD with an endocytic pathway and identified potential biological roles in development, mitosis and endocytosis. In Drosophila, follicle cells lacking awd had abnormal adherens junctions, with epithelial sheets breaking and piling up in oogenesis. Rab5, a mediator of vesicle trafficking in endocytosis, mediated the awd phenotype (15). Follicle cells lacking awd also showed abnormal Notch signaling due to accumulation in early endosomes (16). In tracheal development, an abnormal epithelial motility response to FGF disrupted pattern formation. A similar endocytic phenotype was observed in shibire (shi)/dynamin mutants, and shi mutation exacerbated the awd phenotype (17). In the nervous system, null awd resulted in a failure of synaptic membrane internalization resulting in behavioral paralysis, similar to shi mutants (24,39). Data from other organisms have also been reported, including one way co-immunoprecipitations of NME and DNM2 (20). It was suggested that NME, via its NDPK activity, provided GTP to facilitate DNM2 scission of endocytic vesicles (21). A similar mechanism may regulate cytokinesis (20). Other potential mechanisms included NME2 modulation of endocytosis via a Rac1-dependent mechanism (40). Potential additional interacting partners include Arf6 and Phoecin (22,41).
Herein we have examined the potential contribution of endocytosis and DNM2 to NME suppression of tumor cell motility, migration, and metastasis suppression. Using two human tumor cell lines transfected with a vector or NME(s), increased perinuclear internalization of two receptors was observed with increased NME expression. For EGFR, the probe was only detectable in acid conditions typical of internalized vesicles. Internalization of receptors showed costaining with Rab5, depleted AP2 from the cell surface, exhibited increased Rab5-GTP levels, and showed low surface levels of receptors, all consistent with an endocytic process. It is likely from the literature that the endocytosis of multiple membrane proteins and receptors is altered by NME expression levels (16,17,21,22,40).
MDA-MB-231 breast cancer cells express relatively high levels of wild type EGFR and were used for more detailed analyses of this pathway. The endocytosis of EGFR influences recycling versus degradation fate decisions, alters signaling via complex pathways, and affects cancer progression (rev. in (28,29,42)). By IF, tumor cells in serum-stripped culture medium (lacking exogenous growth factor ligands) showed increased perinuclear EGFR endocytosis and decreased EGFR at the membrane with NME overexpression. Time course analysis under these conditions showed that the endocytic process for EGFR was qualitatively altered in the NME1 overexpressing tumor cells and not just temporally accelerated. EGFR autophosphorylation at Y1068 was also attenuated by NME1 overexpression. Using combinations of NME overexpression and shDNM2 knockdown, the interaction of these two proteins altered downstream signaling. NME1-mediated reductions in pEGFR and pAkt expression were not observed in the presence of shDNM2. In addition to overall levels on western blots, IF showed differences in subcellular localization through a time course. Future experiments will investigate other potential signaling events downstream of EGFR and other receptors, including the actin cytoskeleton and chemotherapeutic sensitivity (43). The data to date reveals that increased endocytosis is readily observable upon NME overexpression and is associated with altered signaling.
NME augmentation of endocytosis was sensitive to DNM2 inhibition using multiple chemical inhibitors and shRNA mediated knockdown. These same approaches demonstrated a profound effect on tumor cell motility in Boyden chamber assays and migration in scratch assays. Here, NME overexpression was unable to significantly reduce tumor cell motility/migration in the presence of DNM2 inhibition (by both inhibitors and shRNAs). Each of these approaches has weaknesses. The chemical inhibitors each exerted some effect on tumor cell motility per se, with relatively minor inhibition of motility using Iminodyn-22 and Dynole-34–2, and more potent anti-motility activity with MiTMAB and OcTMAB. Both shRNA knockdowns of DNM2 were partial. The fact that similar trends were obtained with each of the approaches lends strength to the conclusions drawn. Thus, DNM2 is required for NME inhibition of tumor cell motility/migration, in keeping with its requirement for AWD restoration of normal Drosophila development (12).
An experimental metastasis assay using MDA-MB-231 breast cancer cells expressing either a vector or NME1, and either a scrambled or DNM2 shRNA, was used to ask if the in vitro trends in motility and migration were applicable to the metastatic process. While an experimental metastasis assay is an incomplete representation of the full metastatic process, owing to injection of tumor cells directly into the circulation, it models the metastatic colonization of a distant site, a translationally important, difficult step in the metastatic process (1). Downregulation of DNM2 accounted for a portion of the NME1 metastasis suppressive effect, resulting in a loss of statistical significance in comparison to vector controls. These data are understandable in that other, independent pathways for NME suppression of metastasis have also been reported (9,44–47) or proposed (48).
Given the known role of NME as a nucleoside diphosphate kinase (NDPK), it has been tempting to conclude that its role in DNM GTPase function is the provision of GTP (20,21,24). For Drosophila neuronal synaptic signaling, a mutation enhancing SHI paralysis was in AWD; a mutant MSM95/MSM95 strain showed reduced NDPK activity but also a reduction in total AWD protein (24). The Boissan lab quantified DNM1/2 GTPase activity in the presence of 1mM ATP and 10 μM GDP, and demonstrated only a 30% increase in DNM1 GTPase activity upon addition of NME1/2 (21). Another report identified an ARF6/DNM2/NME complex and hypothesized that NME provided GTP to ARF6 (22). These reports do not account for the fact that the NDPK activity of NME is comparable for all nucleotides, i.e., that NME can make GTP, ATP, CTP or TTP from GDP + ATP, or that the NDPK reaction is reversible. Provision of spatially high GTP levels in microdomains around G-proteins, where a local excess of GDP could exist, is a plausible mechanism of action. Using a site directed mutation approach, our data indicate that the NDPK activity could be contributory but is insufficient for the endocytic phenotypes observed: NME1P96S, which lacks HPK activity but retains significant NDPK activity in comparison to the wild type protein, was unable to increase endocytosis.
Our data provide the first demonstration of two-way co-immunoprecipitations of DNM2 and NME, initially conducted in transfectants of two cancer cell lines. The amount of NME expression was limiting for the formation of the complex. NME-DNM2 binding was mapped to 42 amino acid in the DNM2-PRD domain. This domain is the site of DNM2 binding to multiple other proteins and the impact of NME binding on those proteins and associated phenotypes should be determined. Among a panel of breast cancer cell lines, NME1-DNM2 co-immunoprecipitation was also observed in non-modified cells; a trend was observed of greater NME1-DNM2 co-immunoprecipitation and less aggressive behavior.
Herein, we report a unique mechanism of action, the promotion of DNM2 oligomerization by NME. Using partially purified proteins and a sedimentation assay, the incubation of NME1 with DNM2 decreased DNM2 monomer concentrations with a corresponding increase in the oligomers, its active form. These results were confirmed by the increased DNM2 GTPase activity in the mixture. NME1 is not the only protein reported to regulate DNM2 oligomerization; others include lipids, microtubules and SH3-domain containing proteins (25). Further work is needed to understand how NME1 participates with other proteins to promote DNM2 oligomerization. NME1 site directed mutants bound DNM2 in co-immunoprecipitation assays but failed to increase its oligomerization, supporting the hypothesis that the simple NDP kinase function of NME1 may be insufficient. We hypothesize that the mutant NMEs possessed an altered conformation which was not permissive for DNM2 oligomerization.
The biochemical assays and several cell biology assays were also conducted using NME2 (Nm23-H2) with similar results. NME2 has been reported to have both similarities and differences with NME1 (49,50), but in this case the data are generalizable. NME1 and NME2 are also known to form heterooligomers which could influence their interactome, sub-cellular localization and possibly their function.
Taken together, NME suppression of tumor cell motility, migration and metastasis has been linked to its interaction with, and promotion of DNM2 oligomerization. This interaction facilitates the endocytosis of receptors and possibly other proteins, likely altering their availability for motility signaling and/or downstream signaling patterns which in turn affect motility, migration and metastasis. The data provide support to the endocytic pathway as a common contributing pathway to NME/AWD regulation of differentiation and cancer progression. These data, if confirmed and extended, could provide a translational lead for drug screening using a NME:DNM2 interaction as a target.
Supplementary Material
Statement of significance.
NME1 suppresses metastasis via changes in tumor endocytosis and motility, mediated by dynamin (DNM2) GTPase activity
Acknowledgments
The authors thank the NCI confocal and FACS facilities. Patricia Steeg is supported by the Intramural Program of the National Cancer Institute, National Institutes of Health. (Investigator-Initiated Intramural Research Projects (ZIA). Project #1ZIASC000892-33, Application #9344091).
Financial support: This work is supported by an NIH intramural grant.
Footnotes
Conflict of interest:
The authors declare no potential conflicts of interest.
References:
- 1.Steeg PS. Targeting metastasis. Nature Reviews Cancer 2016;16:201–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan LL, Pantel K, Kang YB. Tumor metastasis: moving new biological insights into the clinic. Nature Medicine 2013;19:1450–64 [DOI] [PubMed] [Google Scholar]
- 3.Bohl CR, Harihar S, Denning WL, Sharma R, Welch DR. Metastasis suppressors in breast cancers: mechanistic insights and clinical potential. Journal of Molecular Medicine 2014;92:13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nature Reviews Cancer 2009;9:253–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Nat’l Cancer Inst 1988;80:200–4 [DOI] [PubMed] [Google Scholar]
- 6.Salerno M, Ouatas T, Palmieri D, Steeg PS. Inhibition of signal transduction by the nm23 metastasis suppressor: possible mechanisms. Clin Exp Metastasis 2003;20:3–10 [DOI] [PubMed] [Google Scholar]
- 7.Kantor JD, McCormick B, Steeg PS, Zetter BR. Inhibition of cell motility after nm23 transfection of human and murine tumor cells. Cancer Res 1993;53:1971–3 [PubMed] [Google Scholar]
- 8.Marino N, Marshall JC, Collins JW, Zhou M, Qian YZ, Veenstra T, et al. Nm23-H1 Binds to Gelsolin and Inactivates Its Actin-Severing Capacity to Promote Tumor Cell Motility and Metastasis. Cancer Res 2013;73:5949–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, Maga TK, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res 2007;67:7238–46 [DOI] [PubMed] [Google Scholar]
- 10.Dearolf C, Hersperger E, Shearn A. Developmental consequences of awdb3, a cell autonomous lethal mutation of Drosophila induced by hybrid dysgenesis. Developmental Biology 1988;129:159–68 [DOI] [PubMed] [Google Scholar]
- 11.Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IMK, et al. Reduced Nm23/Awd protein in tumor metastasis and aberrant Drosophila development. Nature 1989;342:177–80 [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Liu LZ, Deng XF, Timmons L, Hersperger E, Steeg PS, et al. The enzymatic activity of Drosophila AWD/NDP kinase is necessary but not sufficient for its biological function. Developmental Biology 1996;177:544–57 [PubMed] [Google Scholar]
- 13.Nallamothu G, Woolworth JA, Dammai V, Hsu T. awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Molecular and Cellular Biology 2008;28:1964–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romani P, Duchi S, Gargiulo G, Cavaliere V. Evidence for a novel function of Awd in maintenance of genomic stability. Scientific Reports 2018;7:16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolworth JA, Nallamothu G, Hsu T. The Drosophila Metastasis Suppressor Gene Nm23 Homolog, awd, Regulates Epithelial Integrity during Oogenesis. Molecular and Cellular Biology 2009;29:4679–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ignesti M, Barraco M, Nallamothu G, Woolworth JA, Duchi S, Gargiulo G, et al. Notch signaling during development requires the function of awd, the Drosophila homolog of human metastasis suppressor gene Nm23. Bmc Biology 2014;12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dammai V, Adryan B, Lavenburg KR, Hsu T. Drosophila awd, the homolog of human nm23, regulates FGF receptor levels and functions synergistically with shi/dynamin during tracheal development. Genes & Development 2003;17:2812–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocucci E, Gaudin R, Kirchhausen T. Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Molecular Biology of the Cell 2014;25:3595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sever S Dynamin and endocytosis. Current Opinion in Cell Biology 2002;14:463–7 [DOI] [PubMed] [Google Scholar]
- 20.Conery AR, Sever S, Harlow E. Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal stability by activating the GTPase dynamin during cytokinesis. Proceedings of the National Academy of Sciences of the United States of America 2010;107:15461–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boissan M, Montagnac G, Shen QF, Griparic L, Guitton J, Romao M, et al. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 2014;344:1510–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios F, Schweitzer J, Boshans R, D’Souza-Schorey C. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nature Cell Biol 2002;4:929–36 [DOI] [PubMed] [Google Scholar]
- 23.Khan I, Steeg PS. The relationship of NM23 (NME) metastasis suppressor histidine phosphorylation to its nucleoside diphosphate kinase, histidine protein kinase and motility suppression activities. Oncotarget 2018;9:10185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan KS, Rikhy R, Rao S, Shivalkar M, Mosko M, Narayanan R, et al. Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron 2001;30:197–210 [DOI] [PubMed] [Google Scholar]
- 25.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 2004;5:133–47 [DOI] [PubMed] [Google Scholar]
- 26.Solomaha E, Szeto FL, Yousef MA, Palfrey HC. Kinetics of Src homology 3 domain association with the proline-rich domain of dynamins: specificity, occlusion, and the effects of phosphorylation. J Biol Chem 2005;280:23147–56 [DOI] [PubMed] [Google Scholar]
- 27.Hill TA, Mariana A, Gordon CP, Odell LR, Robertson MJ, McGeachie AB, et al. Iminochromene inhibitors of dynamins I and II GTPase activity and endocytosis. J Med Chem 2010;53:4094–102 [DOI] [PubMed] [Google Scholar]
- 28.Zhou XP, Xie S, Wu SS, Qi YH, Wang ZH, Zhang H, et al. Golgi phosphoprotein 3 promotes glioma progression via inhibiting Rab5-mediated endocytosis and degradation of epidermal growth factor receptor. Neuro-Oncology 2017;19:1628–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakker J, Spits M, Neefjes J, Berlin I. The EGFR odyssey - from activation to destruction in space and time. Journal of Cell Science 2017;130:4087–96 [DOI] [PubMed] [Google Scholar]
- 30.Quan A, McGeachie AB, Keating DJ, van Dam EM, Rusak J, Chau N, et al. Myristyl trimethyl ammonium bromide and octadecyl trimethyl ammonium bromide are surface-active small molecule dynamin inhibitors that block endocytosis mediated by dynamin I or dynamin II. Mol Pharmacol 2007;72:1425–39 [DOI] [PubMed] [Google Scholar]
- 31.Biggs J, Hersperger E, Steeg PS, Liotta LA, Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell 1990;63:933–40 [DOI] [PubMed] [Google Scholar]
- 32.Wallet V, Mutzel R, Troll H, Barzu O, Wurster B, Veron M, et al. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. J Natl Cancer Inst 1990;82:1199–202 [DOI] [PubMed] [Google Scholar]
- 33.Wagner PD, Vu ND. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J Biol Chem 1995;270:21758–64 [DOI] [PubMed] [Google Scholar]
- 34.Munoz-Dorado J, Inouye S, Inouye M. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J Biol Chem 1990;265:2707–12 [PubMed] [Google Scholar]
- 35.Tepper AD, Dammann H, Bominaar AA, Veron M. Investigation of the active site and the conformational stability of nucleoside diphosphate kinase by site-directed mutagenesis. J Biol Chem 1994;269:32175–80 [PubMed] [Google Scholar]
- 36.Wagner PD, Steeg PS, Vu ND. Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci U S A 1997;94:9000–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J Biol Chem 1996;271:22310–4 [DOI] [PubMed] [Google Scholar]
- 38.Byers CE, Barylko B, Ross JA, Southworth DR, James NG, Taylor CAt, et al. Enhancement of dynamin polymerization and GTPase activity by Arc/Arg3.1. Biochim Biophys Acta 2015;1850:1310–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rikhy R, Ramaswami M, Krishnan KS. A temperature-sensitive allele of Drosophila sesB reveals acute functions for the mitochondrial adenine nucleotide translocase in synaptic transmission and dynamin regulation. Genetics 2003;165:1243–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rochdi MD, Laroche G, Dupre E, Giguere P, Lebel A, Watier V, et al. Nm23-H2 interacts with a G protein-coupled receptor to regulate its endocytosis through an Rac1-dependent mechanism. J Biol Chem 2004;279:18981–9 [DOI] [PubMed] [Google Scholar]
- 41.Baillat G, Gaillard S, Castets F, Monneron A. Interactions of Phocein with Nucleoside diphosphate kinase, Eps15, and Dynamin I. J Biol Chem 2002;277:18961–6 [DOI] [PubMed] [Google Scholar]
- 42.Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Molecular Oncology 2018;12:3–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura Y, Bereczky B, Ono M. The EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis of EGFR via the early/late endocytic pathway in non-small cell lung cancer cell lines. Histochem Cell Biol 2007;127:541–53 [DOI] [PubMed] [Google Scholar]
- 44.D’Angelo A, Garzia L, Andre A, Carotenuto P, Aglio V, Guardiola O, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell 2004;5:137–49 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki E, Ota T, Tsukuda K, Okita A, Matsuoka K, Murakami M, et al. nm23-H1 reduces in vitro cell migration and the liver metastatic potential of colon cancer cells by regulating myosin light chain phosphorylation. Int J Cancer 2004;108:207–11 [DOI] [PubMed] [Google Scholar]
- 46.Marino N, Marshall JC, Collins JW, Zhou M, Qian Y, Veenstra T, et al. Nm23-h1 binds to gelsolin and inactivates its actin-severing capacity to promote tumor cell motility and metastasis. Cancer Res 2013;73:5949–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boissan M, Wever O, Lizarraga F, Wendum D, Poincloux R, Chignard N, et al. Implication of Metastasis Suppressor NM23-H1 in Maintaining Adherens Junctions and Limiting the Invasive Potential of Human Cancer Cells. Cancer Res 2010;70:7710–22 [DOI] [PubMed] [Google Scholar]
- 48.Shevde LA, Welch DR. Metastasis suppressor pathways--an evolving paradigm. Cancer Lett 2003;198:1–20 [DOI] [PubMed] [Google Scholar]
- 49.Lee JJ, Kim HS, Lee JS, Park J, Shin SC, Song S, et al. Small molecule activator of Nm23/NDPK as an inhibitor of metastasis. Sci Rep 2018;8:10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potel CM, Fasci D, Heck AJR. Mix and match of the tumor metastasis suppressor Nm23 protein isoforms in vitro and in vivo. FEBS J 2018;285:2856–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.