Abstract
Metastasis is a major contributor to cancer-associated deaths. It is characterized by a multistep process that occurs through the acquisition of molecular and phenotypic changes enabling cancer cells from a primary tumour to disseminate and colonize at distant organ sites. Over the past decade, the discovery and characterization of long noncoding RNAs (lncRNAs) have revealed the diversity of their regulatory roles, including key contributions throughout the metastatic cascade. Here, we review how lncRNAs promote metastasis by functioning in discrete pro-metastatic steps including the epithelial–mesenchymal transition, invasion and migration and organotrophic colonization, and by influencing the metastatic tumour microenvironment, often by interacting within ribonucleoprotein complexes or directly with other nucleic acid entities. We discuss well-characterized lncRNAs with in vivo phenotypes and highlight mechanistic commonalities such as convergence with the TGFβ–ZEB1/ZEB2 axis or the nuclear factor-κB pathway, in addition to lncRNAs with controversial mechanisms and the influence of methodologies on mechanistic interpretation. Furthermore, some lncRNAs can help identify tumours with increased metastatic risk and spur novel therapeutic strategies, with several lncRNAs having shown potential as novel targets for antisense oligonucleotide therapy in animal models. In addition to well-characterized examples of lncRNAs functioning in metastasis, we discuss controversies and ongoing challenges in lncRNA biology. Finally, we present areas for future study for this rapidly evolving field.
Metastatic disease accounts for the vast majority of cancer-associated deaths. Despite advances in the diagnosis and treatment of cancer, the prognosis for patients with metastatic cancer remains poor, with median survival time measured in months in certain cancers1. The dissemination of cancer cells from primary tumour to distant organs involves an orchestrated, multistep process known as the invasion–metastasis cascade2-4. First, cells of the primary tumour must locally invade surrounding normal tissue. Then, these cells intravasate into the systemic circulation and subsequently extravasate at a distant site, where the metastatic cells are required to proliferate and colonize an often-foreign tissue environment.
Long noncoding RNAs (lncRNAs) are operationally defined as RNA transcripts longer than 200 nucleotides, and there is no evidence that they encode peptides5,6 (FIG. 1). While initially described only as intergenic transcripts7, lncRNAs now encompass natural antisense transcripts, overlapping transcripts and intronic transcripts, among others, depending on the genomic arrangement of the lncRNA with respect to nearby protein-coding genes8. Tens of thousands of lncRNAs have been identified by high-throughput RNA sequencing9-12, but only a small percentage of these have been functionally characterized. Through differential expression analysis and comparative transcriptomic studies of cancer specimens, various lncRNAs have been prioritized for functional studies, revealing a diversity of phenotypes and mechanisms (FIG. 1).
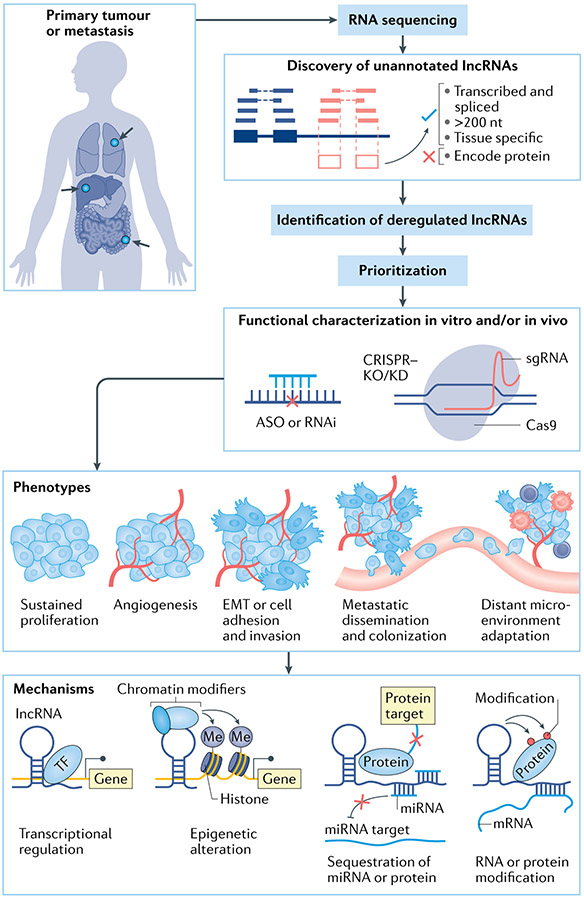
Fig. 1 ∣. Long noncoding RNAs in cancer.
Starting from discovery to characterization of their functions and mechanisms. First, tumour specimens and normal tissues sampled from patients with cancer are subject to RNA sequencing. Sequencing reads are then aligned to the reference genome, which allows quantification of RNA abundance and discovery of unannotated long noncoding RNAs (lncRNAs). Comparisons of transcript abundance between tumour and normal and/or between metastasis and primary tumour allow identification of deregulated lncRNAs. Subsequent omics data integration, computational analysis and high-throughput functional screening using techniques such as CRISPRi allow prioritization of lncRNA candidates for further in-depth functional and mechanistic characterization. Perturbation and manipulation of the lncRNAs can be performed via antisense oligonucleotide (ASO), small interfering RNA (siRNA), CRISPR-based methods in vitro and in vivo to generate phenotypic changes that can be observed and evaluated. LncRNAs are known to have both oncogene and tumour suppressor roles and contribute to tumour progression and metastasis. Mechanistically, lncRNAs can regulate the expression of other genes by interacting with other molecules resulting in transcriptional regulation, epigenetic alteration, sequestration of RNA or protein and enhancer regulation. EMT, epithelial–mesenchymal transition; KO/KD, knockout/knock-down; Me, methyl; miRNA, microRNA; RNAi, RNA interference; sgRNA, single guide RNA; TF, transcription factor.
LncRNAs serve important roles in gene regulation, including modulating gene activation and silencing13,14, X chromosome inactivation15,16, alternative splicing17 and post-translational regulation18-20. LncRNAs perform these functions through a variety of different mechanisms, including acting as molecular scaffolds that ‘guide’ chromatin-modifying enzymes as in the case of HOTAIR21-23 or DLX6AS24,25, acting as competing endogenous RNAs (ceRNAs) that ‘sponge’ microRNAs or proteins26, facilitating or inhibiting long-range chromatin interactions (for example, LUNAR1 or CCAT1)27-29, or even functioning through the act of transcription itself14,30-33 (FIG. 1). Additional mechanisms are also emerging, such as orchestration of nuclear architecture, formation of circular lncRNAs and destabilization of interacting mRNAs. These and other mechanisms of lncRNA function have been reviewed elsewhere5,6,13,34, and additional mechanisms are expected to be unearthed. Of note, particular mechanisms such as the ceRNA model have attracted scepticism (BOX 1), primarily owing to stoichiometric imbalances between target mRNAs and their putative binding sites on ceRNAs, and some lncRNAs have multiple mechanisms described35-37, contributing to the complexity of this rapidly evolving field.
Box 1 ∣. The controversy of the competing endogenous RNA hypothesis.
The competing endogenous RNA (ceRNA) hypothesis proposes that some RNA molecules with shared microRNA (miRNA) binding sites compete for post-transcriptional control, leading to diminished target gene repression. Despite the growing number of studies focused on long noncoding RNAs (lncRNAs) acting as ceRNAs in metastasis, scepticism remains about whether physiological expression levels of a single lncRNA, which can represent a small fraction of the total miRNA targets, is sufficient to alter miRNA regulation156,157. Others suggest that a ceRNA would need to introduce a similar quantity of miRNA binding sites to the existing pool of targets to alter their repression, or that a ceRNA would need to be at equimolar concentration to all miRNA targets158-160; however, this could be compensated through multiple tandem miRNA binding sites. This highlights the importance of establishing standardized strategies to demonstrate the physiological relevance of ceRNA candidates.
The miRNA prediction methods for establishing a ceRNA interaction can vary significantly between algorithms161. Moreover, sequence content alone does not take into account the physiological expression levels of the ceRNA, miRNA and target genes. Gene expression data across patient cohorts can help to prioritize ceRNAs162 that have a positive expression correlation with target genes163, although it does not discriminate between correlated genes owing to alternative regulatory mechanisms. Another strategy is to prioritize ceRNAs displaying biochemical enrichment with RNA-induced silencing complex (RISC) components164. Numerous databases leverage various miRNA prediction algorithms, biochemical data and gene expression data to facilitate research on ceRNAs165-167. A caveat of existing databases is that they lack a global perspective on the miRNA target pool size and the fidelity of the miRNA interactions within the target pool that may affect the regulatory ability of the ceRNA. Nonetheless, they provide an opportunity for hypothesis generation.
The most common experimental validation strategies use models of ceRNA overexpression or silencing. Consequently, altered miRNA target gene regulation is assessed by manipulating levels of miRNA or RISC components followed by gene expression analysis. However, it is experimentally challenging to overexpress miRNAs or ceRNAs at physiological levels. Furthermore, direct evidence is necessary to show that the ceRNA–miRNA relationship is dependent on miRNA biogenesis168. In addition, it is crucial to demonstrate that the miRNA-binding sites within the target genes are dependent on the ceRNA. This can be achieved in an endogenous system via CRISPR genome editing of the miRNA binding site within the target gene and appropriate rescue experiments. Also, although cell lines offer practical advantages, they cannot mimic patient tissues. However, the use of mouse models169 (if an orthologue exists) or patient-derived xenografts and organoids could serve as more relevant physiological models. Last, future studies will also have to consider the possibility that multiple genes are altered, which collectively increases the abundance of multiple miRNA-binding sites.
Collectively, the ease of identifying ceRNA interactions coupled with a common experimental validation framework may have led to a bias of metastasis-associated lncRNAs being mechanistically characterized as ceRNAs while under-representing alternative regulatory mechanisms.
LncRNAs have emerged as key regulators of cancer pathways and also as biomarkers of disease38,39. Initially a peculiarity of molecular biology, lncRNAs have now been described in the context of most if not all of the classic hallmarks of cancer — sustained proliferation, replicative immortality, evasion of growth suppressors, induction of angiogenesis, resistance to cell death and metastasis40 (FIG. 1).
Importantly, much like protein-coding genes, in which genetic alterations are foundational aspects of oncogenesis, lncRNAs are amplified, deleted or mutated in malignancies38,40. A broad set of lncRNAs is located in recurrent copy number-altered regions in the genomes of tumours, including FAL1, whose genomic amplification represses the growth suppressor gene CDKN1A. PVT1 is frequently amplified in many cancers as well36,41, with recurrent mutations found in its promoter region in breast and other tumours36. Although these lncRNAs are appealing to study owing to a potential genetic mechanism of activation or deletion, it is important to evaluate whether the lncRNA gene resides within the minimal common deleted or amplified region, suggesting that its genetic aberration is the result of selective pressure. Epigenetic alteration of lncRNAs also occurs in cancer, as is the case with CCAT1 activation in the CpG island methylator phenotype28,42. Thus, genetic and epigenetic alterations in lncRNAs might account for the apparent rarity of driver mutations in early cancer genome studies that focused on protein-coding genes43.
In contrast to protein-coding genes in which their putative domains provide insight into their function (and can be disrupted), the regulatory mechanisms of lncRNAs are diverse and still emerging. Of note, PVT1 has been shown to have important functions as both a DNA regulatory element (that is, cis mechanism of action) and a RNA transcript (that is, trans mechanism of action)36,41. This concept is not unique to PVT1, and underscores the need for appropriate models to dissect how the DNA or RNA may be affected and its ultimate contribution to tumorigenesis and metastasis44-48. As the lncRNA tumour biology field has evolved, independent groups investigating the few lncRNAs that have been studied have often used different methods (Supplementary table 1) leading to potentially contradictory findings. Although these contradictory findings may cast some doubt on their importance, it also highlights the complexity of lncRNAs and ultimately led to the development of improved strategies for deciphering their functions and potentially resolving existing discrepancies. An experimental framework for studying cis- and trans-acting lncRNAs has been detailed in a previous review5,6.
Our understanding of how certain lncRNAs regulate the invasion–metastasis cascade and related processes such as organ-specific tropism and how they affect the tumour microenvironment (TME) is steadily increasing (FIGS 2,3). Although lncRNAs in cancer metastasis have been reviewed before49-51, several recent studies have been published, challenging previously held tenets about lncRNA function and mechanism35,36,52. Importantly, new tools such as modified antisense oligonucleotides (ASOs) and CRISPR–Cas9 and its derivatives have transformed the way lncRNAs are studied53-57, elucidating novel revelations about their function14,36,44, as well as lending insight into new therapeutic targets58. In this Review, we discuss recent work that has demonstrated in vivo evidence of lncRNA function in cancer metastases, along with controversies pertaining to some of these lncRNAs.
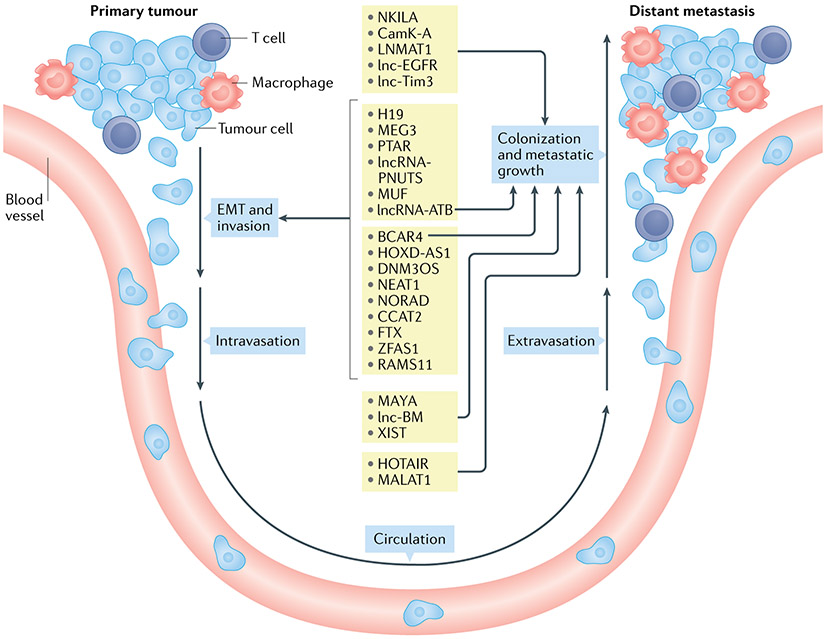
Fig. 2 ∣. Long noncoding RNAs involved in the multiple processes of the invasion–metastasis cascade.
Metastasis is a result of a series of orchestrated cellular processes (invasion–metastasis cascade). First, changes in morphology and cellular adhesion are facilitated through epithelial–mesenchymal transition (EMT). Cancer cells then invade the surrounding normal tissue (local invasion) and make way into (intravasation) and out of (extravasation) the systemic circulation to land at a distant site. There, the metastatic cells proliferate and colonize an often-foreign tissue environment. Numerous long noncoding RNAs (lncRNAs) have been reported to regulate one or more of these processes (shown in the centre of the figure with arrows indicating the involvement in the corresponding processes).
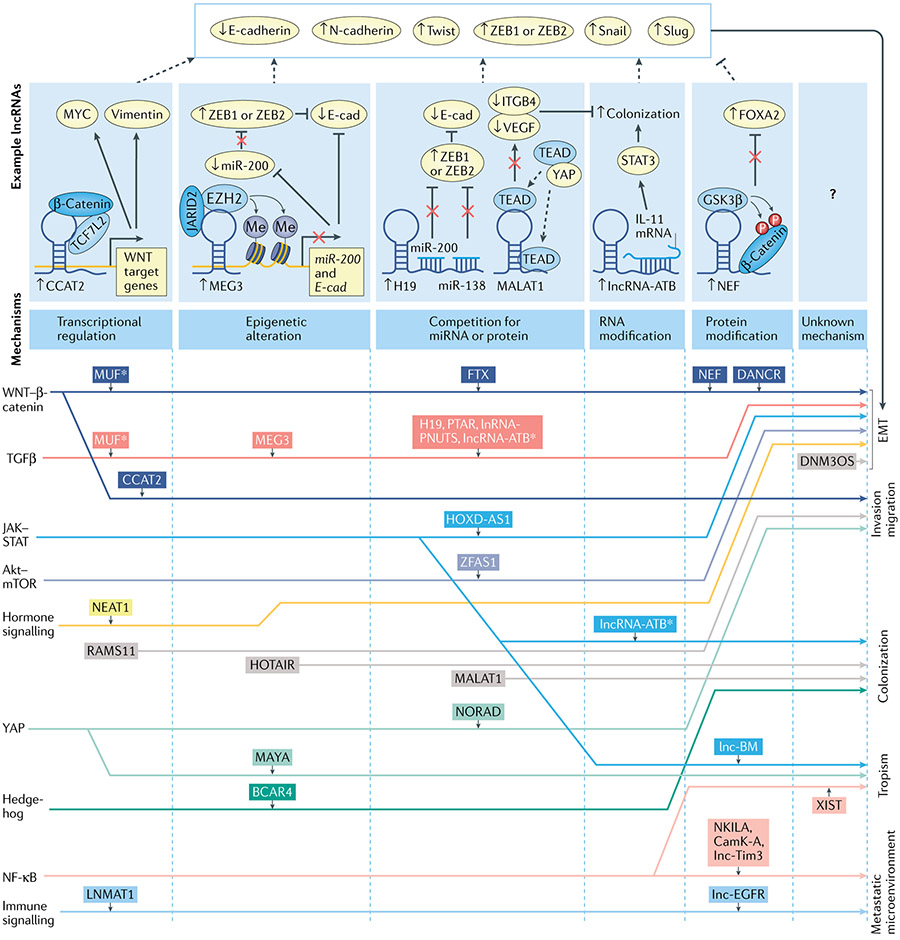
Fig. 3 ∣. Long noncoding RNAs regulate metastasis via various pathways using diverse mechanisms.
Shown are long noncoding RNAs (lncRNAs) with their associated pathways (left) and the biological processes in metastasis that they regulate (right) linked by lines to show lncRNAs interacting with specific pathways to regulate certain biological processes. LncRNAs are grouped from left to right by their downstream mechanism shown on the top panel. For each downstream mechanism, an example detailed mechanism of a lncRNA is presented. LncRNAs in grey boxes are those for which consensus pathway involvement has not been characterized. An asterisk (*) indicates lncRNAs that are involved in multiple pathways or that regulate multiple biological processes in metastasis. In the top panel downstream mechanisms are shown by which representative lncRNAs regulate epithelial–mesenchymal transition (EMT). NF-κB, nuclear factor-κB.
Local invasion and EMT
Local invasion of surrounding tissue is a necessary initial step for tumour metastasis. The epithelial–mesenchymal transition (EMT) and its reverse counterpart the mesenchymal–epithelial transition, which are processes normally activated in embryonic development and tissue homeostasis to ensure proper morphogenesis of tissues and organs, are crucial for the execution of the invasion–metastasis cascade59-61. Through activation of master regulators such as SNAI1, TWIST1 and ZEB1, carcinomas with epithelial phenotypes are conferred traits characteristic of mesenchymal cells that allow invasion, intravasation and dissemination to distant sites62. Although many lncRNAs have been described to modify growth and invasion, we focus our discussion on lncRNAs with roles in invasion and EMT that are supported by in vivo experiments in the context of metastases (FIG. 2).
H19 in the EMT.
H19, one of the earliest described lncRNAs63, is overexpressed in several cancers64 and involved in tumour cell invasion (FIG. 3). In bladder cancer cells, it was shown that H19 overexpression drives migration of malignant cells in vitro, by associating with the polycomb repressive complex 2 (PRC2) component EZH2, leading to increased β-catenin activity and decreased E-cadherin, supporting a transition towards a mesenchymal state65. A challenge in the interpretation of these results is the tendency of PRC2 to undergo promiscuous RNA interactions66, which may obfuscate the primary molecular interactions responsible for H19 function. Nonetheless, H19-mediated transformation towards a mesenchymal identity also occurs in colorectal cancer (CRC) cells, where H19 was shown to function as a ceRNA that sequesters microRNAs miR-138 and miR-200a, leading to the derepression of vimentin, ZEB1 and ZEB2 protein expression67. Consistently, H19 expression was higher in mesenchymal subtypes of CRC cells than in epithelial subtypes. The role of H19 in promoting invasion ultimately leading to metastases was further supported in spontaneous mammary tumour mouse models. Here, H19 was specifically overexpressed in clones that are capable of seeding metastases (that is, 4T1, 4T07 and 168FARN lines), and short hairpin RNA (shRNA)-mediated knock-down of H19 in metastasis-capable clones abrogated metastasis from the primary site to distal organs (for example, lung, kidney and liver)68. In this study, H19 acted as a ceRNA against miR-200b and miR-200c, leading to derepression of ZEB1 and GIT2 in primary tumour and distal metastases. GIT2 was shown to promote colonization in distal metastases. However, in circulating tumour cells (CTCs), H19 differentially sponged the miRNA let-7b, leading to derepression of CYTH3, which promoted a mesenchymal phenotype conducive to extravasation. This highlights a striking influence of environmental context on lncRNA function, a theme that has been observed as a general property of lncRNA biology54. One strength of this study was the use of endogenous Argonaut pulldown, adding biochemical support instead of relying on nucleic acid sequence analysis or indirect measurements of ceRNA activity, and targeted mutagenesis to support the ceRNA role of H19, despite the controversy surrounding ceRNAs in general (BOX 1). Therefore, H19 coordinates a stepwise cascade of metastasis initiation and colonization with potentially different mechanisms of action based on the cellular and environmental context.
These above-mentioned investigations of H19 function, however, do not separate the microRNA sponging capacity of the H19 transcript from its own ability to give rise to miR-675, which is encoded in the first exon of the H19 gene. In prostate cancer cells, high levels of H19 are unexpectedly associated with reduced metastatic potential69, and H19-encoded miR-675 expression led to downregulation of transforming growth factor β-induced protein (TGFBI), which is involved in progression of the EMT70. Therefore, transcripts derived from the H19 gene locus may have separable mechanisms for the regulation of the invasion–metastasis cascade. Although it is simpler to ascribe a single regulatory function to a lncRNA, it is plausible that a lncRNA (such as H19) that is altered in multiple cancer types may have a more crucial role and multiple regulatory mechanisms. As such, reconciling these differences requires the use of models that can either separate out each mechanism (specifically manipulating the PRC2 interaction site or miRNA product) or minimally take into account how each model used could impact each regulatory mechanism (Supplementary table 1). Collectively, this highlights both the importance of H19 and the complexity of studying this and other lncRNAs.
Other competitive endogenous lncRNAs in the EMT.
Several additional lncRNAs have been implicated in the EMT. Among these, pro-transition associated RNA (PTAR) has been shown to be associated with the mesenchymal subtype of ovarian cancer and act as a ceRNA against miR-101, resulting in derepression of ZEB1 expression71. Indeed, shRNA-mediated knock-down of PTAR increased E-cadherin expression and reduced FN1, ZEB1 and vimentin expression in ovarian cancer xenografts. Also, intraperitoneal injection of ovarian cancer cells with stable PTAR knock-down resulted in fewer tumour nodules in infected mice compared with controls, overall indicative of the role of PTAR in promoting EMT71.
Another intriguing lncRNA, lncRNA-PNUTS, is transcribed from a locus that produces both a lncRNA variant (lncRNA-PNUTS) and a mRNA variant (PNUTS)72. While the lncRNA acts as a ceRNA, the mRNA produces a regulator of protein phosphatase 1. Treatment of cells with TGFβ led to PI3K–Akt-dependent phosphorylation and subsequent derepression of lncRNA-PNUTS transcription. LncRNA-PNUTS expression correlated with ZEB1 and/or ZEB2 mesenchymal marker expression and was shown to be a ceRNA that binds to miR-205, leading to derepression of ZEB1/ZEB2 (FIG. 4). Overexpression of lncRNA-PNUTS induced EMT in both lung carcinoma and murine mammary gland cell lines, as evidenced by altered morphology and increased expression of ZEB1/ZEB2 and vimentin, and this effect is abrogated by co-transfection with miR-205, supporting the ceRNA function of lncRNA-PNUTS72. ShRNA-mediated knock-down of lncRNA-PNUTS in MDA-231-LM2 breast cancer cells resulted in decreased burden of lung metastases when injected into mammary fat pads of mice.
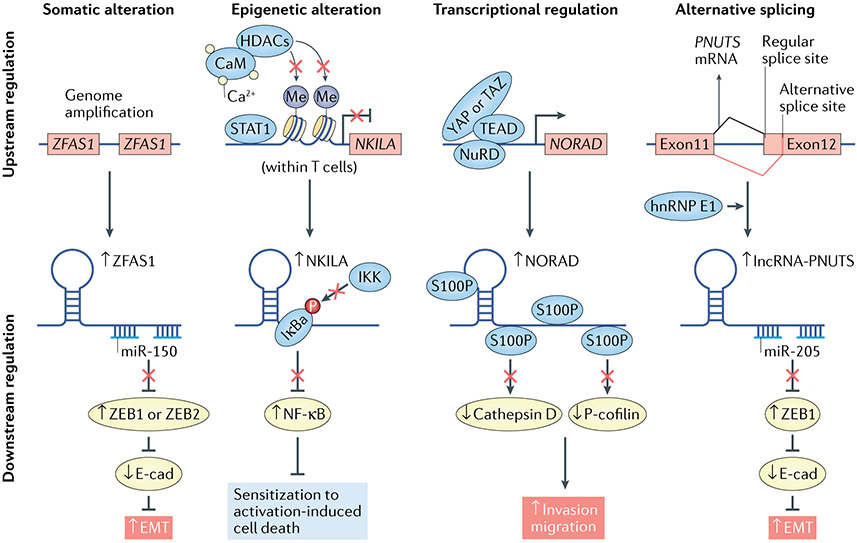
Fig. 4 ∣. Metastasis-associated long noncoding RNAs are regulated by various upstream mechanisms.
Shown are examples of metastatic long noncoding RNAs (lncRNAs) activated by representative mechanisms (from left to right). For each mechanism, the mechanistic model of an example lncRNA is presented (top: upstream regulation of the lncRNA; bottom: regulation of the downstream target and metastatic phenotype). EMT, epithelial–mesenchymal transition; hnRNP, heterogeneous nuclear ribonucleoprotein; NF-κB, nuclear factor-κB; miR, microRNA.
LncRNA-activated by TGFβ (lncRNA-ATB) is another ceRNA that acts as a regulator of the TGFβ–ZEB1/ZEB2 axis. As a transcriptional target of TGFβ, lncRNA-ATB competitively binds to the miR-200 family, leading to upregulation of ZEB1/ZEB2 (REFS73,74). Consistent with this mechanism, overexpression of miR-200a in hepatoma cells abolished EMT. As a result, lncRNA-ATB induced EMT, invasion, increased levels of CTCs and distant organ colonization in an in vivo orthotopic xenograft model of hepatocellular carcinoma (HCC)75. Also in HCC cells, lncRNA ZFAS1 is amplified at the level of genomic DNA (FIG. 4), competitively binds miR-150, derepressing ZEB1, MMP14 and MMP16 gene expression to promote metastasis76. Another ceRNA, FTX, acts by competitively sponging miR-374a and inhibiting HCC cell EMT and invasion, in addition to binding DNA replication licensing factor MCM2, thereby impeding DNA replication and inhibiting proliferation in HCC cells77.
As discussed above, models of competitive binding between lncRNAs and miRNA are frequently described, and caution should be exercised when interpreting experiments claiming such mechanisms of action. This is because of the potential presence of unbalanced physiological abundances of these transcripts and other concerns (BOX 1). Nonetheless, PTAR, lncRNA-PNUTS and lncRNA-ATB have each been shown to function in multiple cancer types, lending credence to their importance. However, it is possible that as these lncRNAs continue to be mechanistically interrogated additional mechanisms will be revealed that are consistent across cancer types.
EMT regulation by lncRNAs via alternative pathways.
In contrast to lncRNAs interfering with TGFβ signalling and transcriptional EMT pathways (FIG. 3), STAT3-activated lncRNA HOXD-AS1 promoted lymph node metastases in HCC by sequestering miR-130a-3p to derepress SOX4 mRNA expression78, ultimately resulting in enhanced migration and invasion via upregulation of MMP2, EZH2 and other SOX4 signalling targets79. Moreover, shRNA-mediated suppression of HOXD-AS1 in multiple HCC cell lines reduced migration and invasion in vitro (independently of proliferation and apoptosis). In line with this, stable HOXD-AS1 knock-down in HCC cells led to reduced numbers of lung metastases in a tail vein injection mouse model.
TGFβ-responsive lncRNA MEG3 promotes EMT through interacting with EZH2 and indirectly upregulating ZEB1 (REFS80,81). LncRNA-MUF promotes EMT in CRC cells through activation of WNT–β-catenin signalling82, while it leads to activation of the TGFβ–SMAD2/SMAD3 signalling pathway through inhibition of SMAD4 protein degradation75. Several additional lncRNAs have also been demonstrated to regulate the EMT via alternative pathways75,82-86 and are summarized in FIG. 3.
Promotion of invasion and migration.
LncRNA NORAD, initially discovered for its role in maintaining genomic stability by sequestering cytoplasmic Pumilio proteins and thus maintaining expression of mitotic and DNA repair transcripts20,87, also functions in invasion and metastases in cancer (FIG. 4). NORAD deficiency is associated with lymph node metastasis in samples from patients with lung and breast cancer88. In breast and lung cancer cells in vitro, it has been shown that ectopic NORAD expression inhibited invasion and migration. In the same study, when intravenously injected into immunodeficient mice, breast cancer cells harbouring NORAD knock-down exhibited greater seeding of lung metastases. Mechanistically, NORAD acted as a molecular decoy, binding to and inhibiting the action of S100P, thereby suppressing S100P-mediated pro-metastatic signalling, which includes invasion activated by cathepsin D and cofilin88.
In patients with colon cancer, CCAT2 has been shown to be overexpressed in microsatellite-stable CRC and metastatic CRC89. It has been shown that CCAT2 can activate MYC transcription and WNT signalling through its interaction with TCF7L2 (observed using RNA immunoprecipitation in CRC cells). Consistent with these results, retroviral vector-mediated overexpression of CCAT2 increased migration of HCT116 cells in vitro and increased the frequency of liver metastasis when cells were injected into the spleen. Interestingly, other studies described CCAT2 facilitating metastases through metabolism, by promoting a pro-glycolysis isoform of the enzyme glutaminase through regulation of alternative splicing90.
More recently, using transcriptome sequencing, the lncRNA RAMS11 was found to be overexpressed in liver metastasis compared with primary tumour from patients with metastatic CRC91. Genomic deletion of RAMS11 using CRISPR–Cas9 decreased cellular invasion and migration of colon cancer cells in vitro. Further, RAMS11 knockout in both tail vein and hemi-splenectomy orthotopic models reduced liver metastasis and lung metastasis, respectively. These metastatic phenotypes were associated with RAMS11-dependent chromobox homologue 4 transcriptional activation of DNA topoisomerase II-α (TOP2A).
Unanswered questions about lncRNA regulation of invasion, migration and the EMT.
An emerging theme is that lncRNAs interface with known master regulators of the EMT, such as TGFβ, ZEB1, SNAI1 and the WNT–β-catenin pathway to enhance or repress metastatic potential. These interactions may be direct through ribonucleoprotein complexes, or indirect through epigenetic regulators or miRNA sequestration (FIGS 1,3). LncRNAs such as NORAD, CCAT2 and RAMS11 can induce invasion and migration phenotypes independently of these pathways. Although these and several other lncRNAs have been reported to contribute to these processes, they do not function in isolation but instead via well-established protein-coding regulators, supporting the notion that lncRNAs tend to function as ‘fine tuners’ rather than master regulators of metastases. Future work will be aimed at corroborating the mechanism of these lncRNAs in contributing to EMT, invasion and migration, and elucidating the mechanisms of action of lncRNAs that use various other pathways. Furthermore, whether these above-described lncRNAs function during other steps of the invasion–metastasis cascade remains to be explored.
Regulation of metastatic colonization
Malignant cells that escape the primary tumour through invasive and mesenchymal phenotypic transformations must intravasate into the bloodstream, overcome anoikis during haematogenous spread, extravasate at a distal site and colonize distant organ sites3. Although little is known about the role of lncRNAs in the first three of these cellular processes in vivo, several lncRNAs have been implicated in promoting the overall ability of metastatic cells to colonize and flourish at distant organ sites.
MALAT1 in metastatic colonization.
MALAT1, one of the first lncRNAs to be described in the process of cancer metastasis, is associated with lung cancer metastases and is also prognostic for survival in patients with early-stage non-small cell lung cancer92. Promoter deletion or ASO-mediated knock-down of MALAT1 in a mouse mammary tumour virus (MMTV)–polyomavirus middle T antigen (PyMT) model of human luminal B breast cancer results in decreased formation of lung metastases, greater tumour differentiation (as evidenced by cystic and encapsulated tumour appearance) and elevated E-cadherin (supporting an epithelial phenotype)93. MALAT1 can regulate these functions by localizing to nuclear speckles and altering co-transcriptional alternative splicing based on these initial descriptions.
Furthermore, when MALAT1 expression was inhibited in A549 lung carcinoma cells (via insertion of a premature polyadenylation signal into the MALAT1 locus), this led to decreased expression of various metastasis-associated genes (for example, ROBO1, MIA2, GPC6, LPHN2 and ABCA1) and reduced migration in vitro compared with control cells94. Also, MALAT1 knock-down led to fewer lung metastases upon tail vein injections in mice, and cells were more likely to remain within blood vessels compared with control cells, suggesting their inability to intravasate into tissue. Cells that did invade distal lung sites formed micrometastases, implying ineffective colonization following MALAT1 knockout. Suggesting therapeutic potential for MALAT1 targeting in metastatic lung cancer, subcutaneous administration of MALAT1 ASO decreased the burden of lung nodules in pulmonary mouse models of metastatic non-small cell lung cancer94. Therefore, perturbation of both the genetic locus and the RNA transcript resulted in concordant phenotypes in this individual study94, an agreement not universally observed in lncRNA studies, some reasons for which are dependent on the methodologies used to study lncRNAs (Supplementary table 1).
Although MALAT1 is generally implicated as a pro-tumour metastasis lncRNA, some evidence has suggested that MALAT1 can contribute to suppressing metastasis. Transcriptional inactivation of MALAT1 in MMTV-PyMT mouse models using insertion of premature polyadenylation sequences promoted lung metastasis35, in stark contrast to earlier experiments that also used premature termination (in addition to ASO targeting) as methods of lncRNA inactivation94. Furthermore, previous experiments that used promoter deletion or transcript knock-down of MALAT1 implicate the lncRNA in regulation of mRNA splicing and neighbouring gene expression93,95, adding to the range of differing mechanisms proposed for this gene. The pro-metastasis effects of MALAT1 depletion observed by Kim and colleagues35 were rescued by transgenic overexpression of MALAT1 (REF.35). In this study, it was identified that MALAT1 can bind to and inactivate transcriptional activity of the pro-metastatic TEAD transcription factor family, when analysed using modern techniques such as comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS). This can result in the abolition of ITGB4 and VEGFA expression, which may be required for migration and invasion35. These disparate results suggest that the phenotype of lncRNA function may depend not only on the cellular context54, as is the case when comparing MALAT1 function in breast cancer35 versus lung cancer94, but also on the dominant mechanism of action (for example, regulation of gene expression and splicing93,95, or inactivation of transcription factors35). MALAT1 also illustrates a major challenge in lncRNA research. As it is not a priori established whether the genomic locus or the RNA gene product (or both) is responsible for lncRNA function, the lncRNA mechanism must be interpreted within the context of the methods used to disrupt varying degrees of the genomic locus or RNA transcript (Supplementary table 1), as different methods may lead to conflicting conclusions if taken at face value.
Pro-colonization pathways connecting lncRNA and cytokines.
Metastatic colonization also exploits non-canonical hedgehog signalling, whereby GLI transcriptional networks are activated independently of the PTCH1-SMO transmembrane receptors96. In breast cancer, GLI2 activation of pro-metastatic transcriptional programmes is dependent on BCAR4 (REF.97), a lncRNA initially discovered for its role in anti-oestrogen resistance98. In response to CCL21 chemokine signalling, BCAR4 interacts with SNIP1 and PPP1R10 to epigenetically derepress GLI2 transcriptional targets. In vivo knock-down of Bcar4 using either shRNA or ASOs suppresses lung metastases derived from orthotopically implanted MDA-MB-231 breast cancer cells in mice97. Clinically, BCAR4 expression is correlated with metastatic burden of breast cancer and is prognostic of patient survival, highlighting its biological role in metastatic dissemination and its potential as a disease marker.
LncRNA-ATB, which in addition to its role in promoting the EMT through upregulation of ZEB1 and/or ZEB2 (REF.74), induces distant organ colonization of subcutaneously transplanted HCC cells in mice73. This pro-colonization function is attributed to lncRNA-ATB binding and stabilizing IL11 mRNA, leading to STAT3 signalling, which inhibits apoptosis99. Therefore, this lncRNA may represent an important mediator of multiple steps in the invasion–metastasis cascade, despite the challenges of resolving ceRNA function (BOX 1).
HOTAIR promotes colonization.
The HOX loci lncRNA HOTAIR is a relatively well-established regulator of metastasis38. Initially discovered as a trans repressor of certain HOXD cluster genes in embryonic development22, HOTAIR is overexpressed in metastatic breast cancer and is also prognostic of patient survival21. In patients with CRC, high HOTAIR expression correlates with the presence of liver metastases100. Ectopic expression of HOTAIR in non-metastatic SK-BR3 breast cancer cells and metastasis-competent MDA-MB-231 cells results in lung colonization following tail vein injection into mice, with MDA-MB-231 cells exhibiting persistent metastatic colonization and expansion21. By acting as a molecular scaffold for the PRC2 and histone H3K4 demethylase LSD1 (REF.23), HOTAIR reprogrammes the epigenome of breast cancer cells to resemble those of embryonic fibroblasts21. Interestingly, the metastatic potentiation of HOTAIR in breast cancer cells can be enhanced in a paracrine mechanism101. TGFβ1 secreted by carcinoma-associated fibroblasts (CAFs) in vitro is capable of upregulating HOTAIR expression levels through transcriptional activation of HOTAIR by SMAD2, SMAD3 or SMAD4, thereby inducing EMT in breast cancer cells (MDA-MB-231 and MCF-7). Consistent with this observation and prior studies, HOTAIR knock-down decreases lung metastases101. Although HOTAIR was one of the first lncRNAs studied in metastasis, and therefore more groups have been able to continue dissecting its role, it has nonetheless been broadly found to promote metastasis across cancer types.
As tumour cells disseminate towards distant organs, the lncRNAs described in this can promote colony expansion in what are often foreign microenvironments. Further investigation is warranted to determine whether this effect is a generalized consequence of enhanced growth and proliferation, or whether these and other lncRNAs strengthen the relative fitness of metastatic cells within specific niches.
Regulation of site-specific tropism
Metastatic cells exhibit site-specific tropism, as various organ environments require specific adaptations for the survival of disseminated tumour cells102-105. In other words, specific organ environments are more hospitable for the survival of disseminated tumour cells. This has been supported by expression of specific protein-coding genes that facilitate organ-specific metastatic tropism (such as connective tissue growth factor (CTGF) in bone metastases106), which allow disseminated tumour cells to overcome the demands of the tissue microenvironment at distant sites. An outstanding question in such organotropic metastasis is how breast cancer, the most common malignancy in women, gives rise to metastases that are distributed to distant organs (for example, bones, lungs, central nervous system (CNS), lymph nodes and gastrointestinal tract) in non-random ways. For instance, up to 70% of breast cancer metastases are disseminated to the bone, while 10–30% of metastases localize to the brain107. As such a pervasive disease, breast cancer has motivated the development of several well-characterized in vivo mouse genetic models108-110, enabling dissection of this crucial decision-making process. Certain lncRNAs have been identified to direct these organ preferences, the principles of which may be applicable to lncRNAs in other primary malignancies (FIG. 5).
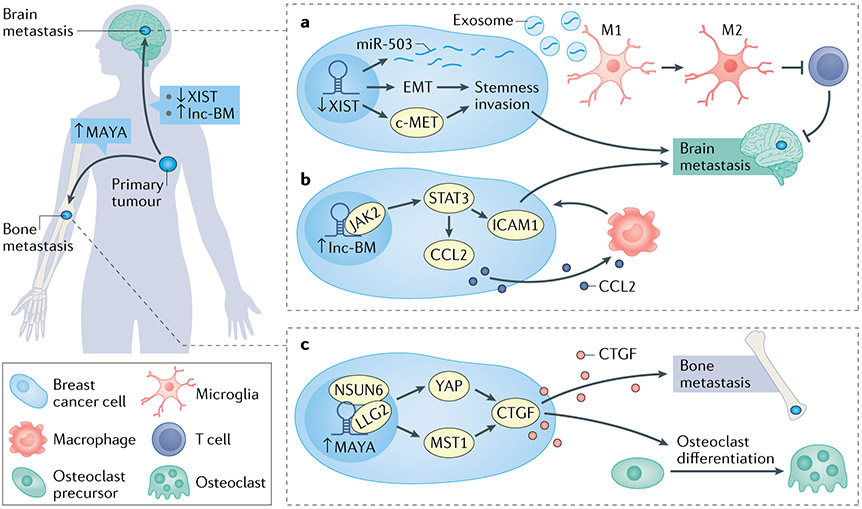
Fig. 5 ∣. Long noncoding RNAs in metastasis site-specific tropism.
a ∣ Downregulation of XIST expression levels activates epithelial–mesenchymal transition (EMT) and MET, leading to stemness, invasion, increased production and release of miR-503, which subsequently induces M2 microglia polarization and T cell responses in breast cancer brain metastases. b ∣ lnc-BM interacts with JAK2 to activate STAT1/STAT3 leading to activation of ICAM1, thereby permitting malignant cell co-option of brain endothelial cells. This also activates CCL2 and induces CCL2-dependent macrophage recruitment to brain metastases. c ∣ MAYA promotes bone metastases via activation of YAP1 and methylation of the Hippo pathway gene MST1, leading to increased connective tissue growth factor (CTGF) and osteoblast differentiation.
Bone metastases.
Within bone metastases derived from primary breast cancer, paracrine release of CTGF from malignant cells activates the YAP pathway in osteoclasts to induce differentiation and osteolysis, thereby promoting bone metastasis colonization106. Using a high-throughput RNA interference (RNAi) screen, it was shown that the lncRNA MAYA was required for YAP1 activation in MCF-7 breast cancer cells111 (FIG. 5). Knock-down of MAYA in cells derived from human breast cancer bone metastases (BoM-1833) led to abrogation of CTGF secretion, which impaired cancer cell-induced osteoclast differentiation and bone resorption. Mechanistically, it was shown that MAYA functions in a molecular complex with LLG2 and the methyltransferase NSUN6, which methylates the Hippo pathway component MST1. Methylation of MST1 then leads to activation of YAP1-regulated genes111. Tumours in mice derived from BoM-1833 cells with MAYA knock-down showed a reduced burden of bone metastases compared with controls. Furthermore, intravenous injection of locked nucleic acids (LNAs) against MAYA reduced the bone metastasis burden in mice that had previously been inoculated with breast cancer cells or A549 lung cancer cells, suggesting therapeutic targeting of MAYA as a precise way of treating bone metastases111.
Brain metastases.
By contrast with bone metastases, breast cancer cells that metastasize to the CNS exploit different pathways to achieve site-specific colonization. Brain metastases derived from primary breast cancer express L1 cell adhesion molecule (L1CAM) to co-opt vascular endothelia, endowing metastatic outgrowth throughout the brain112. Lnc-BM is a lncRNA enriched in brain metastases compared with lung or bone metastases derived from orthotopically introduced breast cancer cells113 (FIG. 5). Lnc-BM expression was inversely associated with survival in patients, and positively correlated with CNS recurrence. Intracardiac injection of mice with breast cancer cells that were depleted of lnc-BM (via RNAi or CRISPR–Cas9) reduced the burden of brain metastases, along with reducing co-option of blood vessels within the brain by cancer cells. Mechanistically, lnc-BM activates expression of ICAM1 through phosphorylation of STAT1/3, thereby permitting malignant cell co-option of brain endothelial cells in an analogous manner to L1CAM112,113. Furthermore, once cells colonized within the brain, lnc-BM mediated JAK2 activation to recruit macrophages via CCL2, thereby regulating multiple pathways conducive to brain metastases.
In a departure from its role in X chromosome inactivation16,114, lncRNA XIST was shown to be repressed in breast cancer brain metastases in mice, when compared with metastases in bone, liver or lung115. In line with this, silencing of XIST in MCF-7 and SK-BR3 breast cancer cells that were intracardiacally injected into mice resulted in increased burden of brain metastases. Further, genomic knockout of Xist enhanced brain metastases in MMTV-PyMT mouse models of spontaneous breast cancer metastasis, through induction of EMT and activation of MET, promoting a stem cell phenotype115. XIST knock-down can also lead to release of exosomal miR-503, which induces polarization of M2 microglia, thereby altering the metastasis microenvironment115.
As the expression and function of lncRNAs are generally cell type specific (with notable exceptions such as MALAT1 and XIST)9,54, it is perhaps unsurprising that lncRNAs play important roles in the organ-specific tropism of cancer metastases. However, it is unclear whether site-specific lncRNA regulatory programmes are triggered before metastatic colonization, thereby acting as directors of cell fate. Alternatively, lncRNAs may exert their influence on the maintenance of metastatic clones through the microenvironment.
lncRNAs in the metastatic microenvironment
On arrival at distant organ sites, metastatic cells form and interact with the TME, which includes diverse processes such as angiogenesis116, suppression and/or co-option of the innate and adaptive immune system117-119, and the reprogramming of stromal populations to promote metastatic outgrowth120,121. The establishment and maintenance of a supportive microenvironment, including resident stromal cells and innate and adaptive immune cells, is required for the outgrowth of metastatic colonies2,122. Through suppression of the immune system, recruitment of angiogenesis, paracrine signalling and deposition of pro-metastatic extracellular matrices, the TME contributes to cancer metastases in diverse ways. The roles of lncRNAs in the tumour and metastatic microenvironment are emerging (FIG. 6).
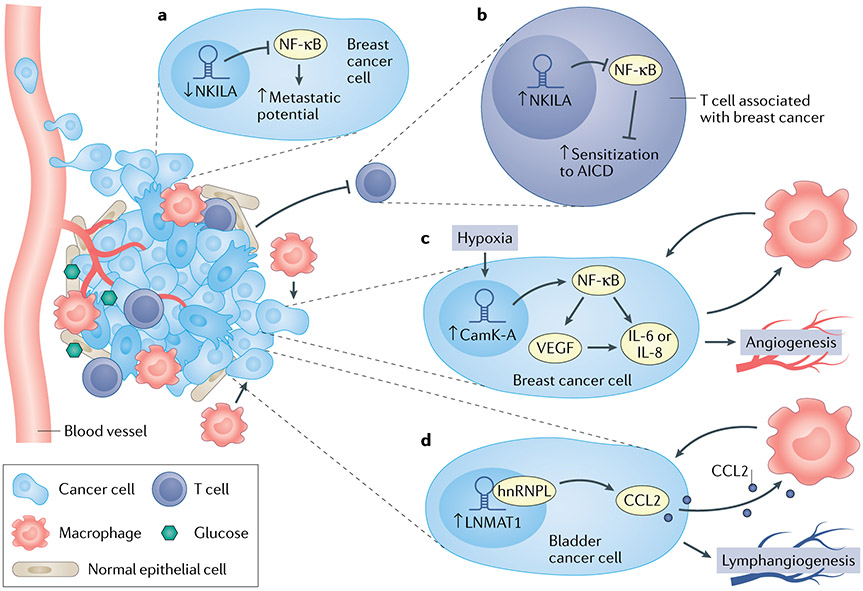
Fig. 6 ∣. Long noncoding RNAs and tumour microenvironment.
a ∣ NKILA inactivates the nuclear factor-κB (NF-κB) pathway in breast cancer cells, and NKILA downregulation leads to decreased metastatic potential. In tumour-infiltrating T lymphocytes, NKILA leads to sensitization to activation-induced ceLL death. b ∣ CamK-A activates NF-κB in response to microenvironmental hypoxia, leading to activation of IL-6/IL-8 and VEGF, which increase macrophage recruitment and angiogenesis. c ∣ LNMAT1 activates CCL2 and CCL2-dependent macrophage recruitment in bladder cancer metastasis. hnRNPL, heterogeneous nuclear ribonucleoprotein L.
NF-κB-associated lncRNAs in the microenvironment.
Initially discovered as a cytoplasmic lncRNA that directly inhibits the NF-κB complex within breast cancer cells, leading to increased metastatic potential when downregulated123 (FIG. 6a), the NF-κB-interacting lncRNA (NKILA) also regulates intrinsic antitumour activity in T lymphocytes in a STAT1–histone acetylation-dependent mechanism124 (FIGS 4,6b). In breast tumour-infiltrating T cells, NKILA suppresses NF-κB signalling, leading to activation-induced cell death of those T cells (FIG. 6b). Knock-down of NKILA using shRNA in CD8+ T cells increased their penetration into lungs when adoptively transferred into breast cancer xenograft mouse models, and those T cells upregulated perforin and CD107a expression and exhibited increased antitumour activity124. CD8+ T cell exhaustion can also be induced through lnc-Tim3, which was shown to promote activation of p53 and RelA transcriptional targets125.
Whereas NKILA negatively regulates the NF-κB pathway in both tumour cells and immune cells, lncRNA CamK-A was shown to activate the NF-κB pathway in breast cancer cells through degradation of IκB in response to hypoxia-induced calcium influx126 (FIG. 6b). This led to activation of gene transcription of IL6, IL8 and VEGF (among other genes), which ultimately promoted macrophage recruitment and angiogenesis in patient-derived breast cancer flank xenograft models. Consistently, conditioned medium from CamK-A-deficient breast cancer cells failed to induce angiogenesis in HUVEC endothelial cell culture, whereas angiogenesis was induced in conditioned medium from CamK-A-proficient cells126.
Immune cell regulation by lncRNAs.
Regulatory T cells, which contribute to avoidance of immune surveillance in tumours, are also influenced by lncRNA activity. In CD4+ T cells, lnc-EGFR was shown to bind to cytoplasmic EGFR, thereby stabilizing it for downstream signal transduction127. This led to induction of FOXP3, signifying differentiation towards the regulatory T cell lineage. Consistent with the expansion of this suppressive immune population, lnc-EGFR overexpression in T cells led to increased tumour growth compared with controls when these cells were implanted in xenograft models of HCC127.
In bladder cancer, LNMAT1 is overexpressed in lymph node-positive bladder cancer and is prognostic of overall survival in patients with bladder cancer128. ShRNA-mediated knock-down of LNMAT1 reduced lymph node metastases of bladder cancer cells injected into footpads of nude mice. However, the function of LNMAT1 was not intrinsic to the tumour cell, as conditioned medium from LNMAT1-transduced bladder cancer cells was capable of stimulating macrophage activation, and this effect could be abrogated by CCL2-neutralizing antibodies (FIG. 6c). Interestingly, LNMAT1 activated the expression of CCL2 by forming a DNA–RNA triplex with the CCL2 promoter and facilitated the deposition of the transcriptionally active histone modification, H3K4 trimethylation. Furthermore, LNMAT1 overexpression increased lymphangiogenesis in vitro through macrophage-dependent upregulation of VEGF-C128. Therefore, lncRNAs can have potent non-cell-autonomous effects on the microenvironment.
LncRNAs are crucial regulators of differentiation and function in immune cells belonging to the progenitor, innate and adaptive lineages129. Furthermore, lncRNAs are necessary for the development and physiology of neural, pulmonary, cardiac and embryonic tissues130,131. As all these cell types can contribute to the metastatic microenvironment or premetastatic niche, it is conceivable that additional lncRNAs will be implicated in these processes as well.
Translational potential of lncRNAs in metastases
Survival of patients with metastatic cancer is poor, and resistance to therapy remains a major barrier to effective treatment. One intriguing approach to improving the treatment of cancer is to leverage tissue- and cancer-specific expression profiles to develop prognostic markers for progression from primary to metastatic disease.
LncRNAs as novel diagnostic and prognostic tools.
While lncRNA PCA3 has been identified as a non-invasive upfront diagnostic marker of prostate cancer with reliable test characteristics and is used in clinical practice132, overexpression of lncRNA SChLAP1 is independently prognostic of prostate cancer metastasis within 10 years following prostatectomy based on a microarray analysis of 1,008 patients133. Application of SChLAP1 measurement to risk stratification of patients at time of surgery may enable precision medicine strategies for proactive medical treatment of aggressive tumours. Indeed, SChLAP1 expression is a critical component of a genomic classifier used in clinical practice for prostate cancer133. Mechanistically, SChLAP1 binds to and antagonizes the SWI/SNF complex, leading to decreased genome-wide occupancy of SNF5 and decreased expression of its target genes134. Consequently, SChLAP1 knock-down in 22Rv1 prostate cancer cells reduced metastatic seeding when these cells were intracardiacally injected into SCID mouse models. However, this proposed mechanism of SChLAP1 has been challenged, as subsequent investigation demonstrated that overexpression of SChLAP1 did not evict the SWI/SNF complex from chromatin, suggesting a SWI/SNF-independent mechanism of action for SChLAP1 in prostate cancer metastases52. It is possible that the mechanism of SChLAP1 exhibits context or cell-type specificity while still maintaining utility as a clinical prognosticator.
Clinical outcomes of patients with metastatic cancer vary dramatically, especially across patients with different histologies of metastatic cancer135,136. In patients with metastatic renal cell carcinoma (RCC), high lncARSR expression was prognostic for shorter overall survival compared with tumours with low lncARSR levels137. lncARSR can confer resistance to sunitinib by sequestering miR-34 and miR-449, resulting in derepression of AXL and MET expression. By shuttling across cells via exosomes, lncARSR is capable of promoting drug resistance in a paracrine fashion. In line with lncARSR conferring sunitinib resistance, intravenous administration of LNA ASOs against lncARSR in an orthotopic xenograft model of RCC conferred sensitivity to concurrent sunitinib therapy137.
RAMS11 expression was associated with poor prognosis in patients from two independent cohorts of patients with colon cancer91. Interestingly, a drug screen of FDA-approved agents revealed that increased RAMS11 expression promoted resistance to floxirudine (FUDR), a chemotherapy that is commonly used to treat metastatic CRC, and topoisomerase inhibitors. Indeed, RAMS11 CRISPR knockout cells showed increased sensitivity to FUDR and topoisomerase inhibitors91. Further work is necessary to translate the prognostic and predictive utility of RAMS11 to clinical trials.
As a caveat, the biomarker discoveries discussed thus far may still be at an early stage. Whereas SChLAP1 ranked highest for elevated expression in patients with metastatic progression compared with patients without metastatic progression133, many lncRNAs with cancer- or tissue-specific expression profiles may not be highly expressed enough to be used as a reliable biomarker. Moving forwards, the increasing availability of transcriptome sequencing data will greatly facilitate lncRNA biomarker discovery. However, independent validation is crucial and could be more challenging for lncRNAs. Importantly, only a subset of microarray platforms contained probes monitoring RAMS11 and SChLAP1 expression that enabled independent validation across cohorts of patients with long-term clinical outcome91,133. However, many lncRNAs will not be captured by any existing microarray platform. Although some studies may use gene-specific validation in small independent cohorts, more widespread adoption of a biomarker requires a systematic comparison of the diagnostic or prognostic significance of a lncRNA relative to existing protein-coding gene biomarkers, when available. Given the longer follow-up time associated with specific cancer types (for example, prostate cancer) coupled with the need for well-annotated clinical follow-up associated with metastatic end points, this may require leveraging older retrospective cohorts. Further, owing to complex mechanisms of patient progression and response to treatments, a single lncRNA may not be the most clinically useful biomarker, but instead potential lncRNA biomarkers need to be evaluated in conjunction with protein-coding genes. Moving forwards, it will also be important to consider the ability to reliably detect putative lncRNA biomarkers non-invasively to minimize the need for biopsies. This will be particularly important for using lncRNA biomarkers for serial monitoring of patients for whom repeat tumour biopsy is impractical. Overall, SChLAP1, lncARSR and RAMS11 highlight the contributions of lncRNAs in cancer treatment response and their potential uses as prognostic and predictive biomarkers.
Therapeutic targeting of lncRNAs.
Through in vivo preclinical models of metastatic disease, lncRNAs have shown promise as therapeutic targets for cancer treatment. For instance, the lncRNAs MAYA, MALAT1 and lncARSR, which we discuss above, have each been targeted for in vivo silencing using ASOs to ameliorate the burden of metastatic disease in mouse models93,94,111,137. However, it will be crucial to ensure minimal off-target effects when targeting lncRNAs using ASO therapeutics138,139, which could pose further issues given the generally lower abundance of lncRNA transcripts in vivo9. Emerging evidence demonstrates that ASOs disrupt target RNAs through premature transcriptional termination140,141, in addition to RNase H-mediated degradation of mature RNA, which should be considered when predicting the activity of ASO therapeutics. Hepatotoxicity has also been observed with certain ASOs142, and chemical modification of the ASO has been shown to mitigate this and off-target effects as well143,144. A larger practical issue will be the optimization of ASO delivery, which is a general area of development to increase the impact of ASOs targeting lncRNAs or protein-coding genes. Despite these challenges, it is reassuring that protein-coding genes have been targeted using ASOs in both mouse models of cancer145,146 and clinical trials, as in the case of SMN2 in patients with spinal muscular atrophy147, STAT3 in treatment-refractory lymphoma148 and SMAD7 in Crohn’s disease149. Perhaps the once-lofty vision of targeting lncRNAs for cancer metastasis treatment appears hopeful.
Current limitations
Although many seminal studies demonstrate the contributions of lncRNAs throughout metastasis (FIG. 2), gaps of knowledge remain. In particular, the steps in the invasion–metastasis cascade between cellular transformation and distant colonization, which would include extravasation, haematogenous (or leptomeningeal as may be the case in CNS malignancies) spread, intravasation and formation of micrometastases, have less well-established contributions from lncRNAs. Overcoming these limitations would require additional patient-matched molecular profiling data from granular stages of metastasis (for example, primary tumour, CTCs, micrometastases and well-defined metastases).
In addition, the majority of the lncRNAs discussed in this Review act to enhance metastatic potential. Whether these observations present inherent attributes of lncRNA biology or are a consequence of methodological bias in respective studies remains to be determined (Supplementary table 1). Nonetheless, additional studies of lncRNAs that potentially suppress metastasis are warranted. Generalizing the roles of lncRNAs in metastasis across cancer types has also been elusive, and perhaps this should be expected based on the cell type-specific function of most lncRNAs. Improved methodology to dissect the mechanisms of lncRNAs in experimental models that faithfully recapitulate human disease are also required, as characterization of lncRNAs in cancer still lags behind that of their protein-coding counterparts.
Another conundrum in the field of lncRNA biology is why seemingly disparate lncRNAs tend to converge upon a few firmly established protein-coding pathways, instead of functioning as independent regulators of metastasis. It is possible that decades of research on protein-coding genes have enabled the discovery and convergence of key metastasis regulator genes whereas the comparative infancy of research into many lncRNAs has not yet enabled similar lncRNA discoveries and validation across independent laboratories. However, emerging data suggest that ‘functional conservation’ in lieu of primary sequence or secondary structure conservation may be a salient feature of lncRNAs150,151. Furthermore, it is possible that multiple RNA species fulfill similar roles interacting with established protein-coding genes, a concept highlighted by the requirement of PRC2 for RNA interaction to properly function152, despite exhibiting promiscuous protein–RNA interactions66. Taken together, it is conceivable that even a menagerie of lncRNAs generated from various locations in the genome with divergent sequences can influence a similar set of potent metastatic pathways.
Conclusions
Metastases are an often-deadly consequence of cancer. LncRNAs are not only important regulators of cancer hallmarks in general38,39, but also key players in the pathogenesis of metastases49. As more lncRNAs are rapidly characterized, as has been the case in recent years, it is evident that lncRNAs function at critical junctures in the invasion–metastasis cascade, organotropic colonization of distant organs and TME. By functioning together with pro- or anti-metastatic protein complexes, or by interacting directly with other RNA species or DNA (as in the case of RNA–DNA triple helices), lncRNAs exert their influence on cancer pathways via diverse mechanisms5,6,13. Outstanding questions remain, such as the role of lncRNAs in CTCs, formation of the premetastatic niche and stromal cell-specific functions120,153. Future studies necessitate investigation into these additional processes and also further delineation of the mechanisms of lncRNAs involved in metastasis. Moving forwards, in addition to selecting appropriate models for inactivating a lncRNA, it will be crucial for labs to perform appropriate genetic rescue experiments. Independent validation of results between different investigators will be necessary as well. While undertaking these endeavours, it is also crucial to understand the impact of various methodologies on interpreting lncRNA function, and to be open minded to different possibilities for how lncRNAs function.
LncRNAs have also emerged as an underappreciated cache of novel therapeutic targets. Their diverse roles in cancer progression provide new opportunities for undermining metastases in the clinical setting. The maturation of technologies that enable in vivo targeting of lncRNAs53,154, such as antisense oligonucleotides that are analogous to those currently used to treat human disease155, will greatly foster the realization of lncRNA therapeutics against metastases.
Supplementary Material
Invasion–metastasis cascade.
A stepwise succession of events whereby primary tumour cells adopt a phenotype that promotes local invasion, intravasation into the bloodstream, extravasation and migration to distant organ sites, and colonization of those sites, which often involves physiological adaptation.
Long noncoding RNAs.
(lncRNAs). Noncoding RNA transcripts longer than 200 nucleotides, which are frequently polyadenylated, and that show no evidence that they encode proteins.
Competing endogenous RNAs.
(ceRNAs). RNA transcripts (which may be lncRNAs, mRNAs or pseudogenes) that are capable of binding and influencing the activity of microRNAs through complementary base pairing.
Organ-specific tropism.
The propensity for disseminated cancer cells to colonize and proliferate at specific organ sites due to diverse physiological and molecular factors.
Antisense oligonucleotides.
(ASOs). Exogenous oligonucleotides, often chemically modified to resist degradation, that alter the amount, stability or activity of complementary RNA transcripts, usually through RNase-based mechanisms.
Epithelial–mesenchymal transition.
(EMT). A set of molecular processes that convert cancer cells of polarized epithelial phenotypes into more invasive mesenchymal phenotypes.
Anoikis.
A subtype of programmed cell death triggered by inappropriate cellular or extracellular matrix interactions, often in the setting of tumour cells escaping their primary environment.
Micrometastases.
Collections of metastasized tumour cells that are clinically detectable but typically under 2 mm in diameter in human patients.
Comprehensive identification of RNA-binding proteins by mass spectrometry.
(ChiRP-MS). A method of identifying endogenous RNA–protein interactions with high specificity, using in vivo crosslinking, RNA pulldown and mass spectrometry.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41568-021-00353-1.
References
- 1.Amin MB AJCC Cancer Staging Manual. 8th Edn. (Springer, 2017). [Google Scholar]
- 2.Lambert AW, Pattabiraman DR & Weinberg RA Emerging biological principles of metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valastyan S & Weinberg RA Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292 (2011).Seminal primer on invasion–metastasis cascade and pathogenesis of tumour metastasis.
- 4.Gupta GP & Massagué J Cancer metastasis: building a framework. Cell 127, 679–695 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ulitsky I & Bartel DP lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013).Comprehensive review of lncRNA concepts.
- 6.Kopp F & Mendell JT Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St Laurent G, Wahlestedt C & Kapranov P The landscape of long noncoding RNA classification. Trends Genet. 31, 239–251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabili MN et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).Establishes the widespread and cell type-specific nature of lncRNA expression.
- 10.Djebali S et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer MK et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet 47, 199–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL & Chang HY Genome regulation by long noncoding RNAs. Annu. Rev. Biochem 81, 145–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engreitz JM et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr ML & Bertram EG A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163, 676–677 (1949). [DOI] [PubMed] [Google Scholar]
- 16.Chen C-K et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Tripathi V et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willingham AT et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Carrieri C et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Lee S et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta RA et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).Establishes HOTAIR as an important epigenetic regulator in breast cancer metastasis.
- 22.Rinn JL et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai M-C et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bond AM et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci 12, 1020–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berghoff EG et al. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140, 4407–4416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tay Y, Rinn J & Pandolfi PP The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trimarchi T et al. Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang J-F et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma W et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods 12, 71–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latos PA et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Kornienko AE, Guenzl PM, Barlow DP & Pauler FM Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11, 59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mele M & Rinn JL ‘Cat’s cradling’ the 3D genome by the act of LncRNA transcription. Mol. Cell 62, 657–664 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Yao R-W, Wang Y & Chen L-L Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Kim J et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet 50, 1705–1715 (2018).Challenges the concept of MALAT1 as a pro-metastasis lncRNA by revealing an alternative molecular mechanism.
- 36.Cho SW et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keniry A et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol 14, 659–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt AM & Chang HY Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slack FJ & Chinnaiyan AM The role of non-coding RNAs in oncology. Cell 179, 1033–1055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu X et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng Y-Y et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86 (2014).Establishes genomic alterations of lncRNA loci as important features in cancer.
- 42.McCleland ML et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Invest 126, 639–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin Y et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16, 504–516 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Paralkar VR et al. Unlinking an lncRNA from its associated cis element. Mol. Cell 62, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groff AF et al. In vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 16, 2178–2186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimitrova N et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 54, 777–790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huarte M et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidle UH, Birzele F, Kollmorgen G & Rüger R Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteom. 14, 143–160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Q, Yan J & Agami R Long non-coding RNAs in metastasis. Cancer Metastasis Rev. 37, 75–81 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Li J, Meng H, Bai Y & Wang K Regulation of lncRNA and its role in cancer metastasis. Oncol. Res 23, 205–217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raab JR et al. SWI/SNF remains localized to chromatin in the presence of SCHLAP1. Nat. Genet 51, 26–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu SJ & Lim DA Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 19, e46955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu SJ et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, eaah7111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y et al. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat. Biotechnol 1656, 175–1210 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Zhu S et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat. Biotechnol 34, 1279–1286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boettcher M et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat. Biotechnol 36, 170–178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu SJ et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 21, 83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiery JP, Acloque H, Huang RYJ & Nieto MA Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Nieto MA, Huang RYJ, Jackson RA & Thiery JP EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Yang J et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Craene B & Berx G Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13, 97–110 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Brannan CI, Dees EC & Ingram RS The product of the H19 gene may function as an RNA. Mol. Cell 10, 28–36 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rainier S et al. Relaxation of imprinted genes in human cancer. Nature 362, 747–749 (1993).One of the earliest reports of a noncoding RNA involved in cancer.
- 65.Luo M et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333, 213–221 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Davidovich C, Zheng L, Goodrich KJ & Cech TR Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol 20, 1250–1257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang W-C et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou W et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal 10, eaak9557 (2017).Demonstrates the role of H19 in a stepwise cascade of metastasis initiation and colonization, with alternative molecular mechanisms based on the cellular context.
- 69.Zhu M et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 281, 3766–3775 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Chen W-Y et al. Loss of SPDEF and gain of TGFBI activity after androgen deprivation therapy promote EMT and bone metastasis of prostate cancer. Sci. Signal 10, eaam6826 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Liang H et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer 17, 119–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grelet S et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol 19, 1105–1115 (2017).A well dissected example of a lncRNA acting as a ceRNA to regulate the EMT.
- 73.Yuan J-H et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Shi S-J et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget 6, 11652–11663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu N et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. 65, 87–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li T et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 75, 3181–3191 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Liu F et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 35, 5422–5434 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Wang H et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 16, 136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y et al. Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J. Cell. Biochem 120, 10633–10642 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Terashima M, Tange S, Ishimura A & Suzuki T MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J. Biol. Chem 292, 82–99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mondal T et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun 6, 7743–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yan X et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Cancer Res. 77, 6704–6716 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Mitra R et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat. Commun 8, 1604–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Z et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 24, 59–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li W et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Invest 127, 3421–3440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liang W-C et al. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene 37, 1445–1456 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Tichon A et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun 7, 12209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan B-S et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 38, 5612–5626 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Ling H et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Redis RS et al. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol. Cell 61, 520–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Silva-Fisher JM et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat. Commun 11, 2156–13 (2020).A lncRNA that is both prognostic of survival in metastatic colon cancer and predictive of response to chemotherapy and targeted molecular therapy.
- 92.Ji P et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Arun G et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gutschner T et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189 (2013).Seminal loss of function study of MALAT1 as a lncRNA regulator of cancer metastasis.
- 95.Zhang B et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sterling JA et al. The hedgehog signaling molecule Gli2 induces parathyroid hormone-related peptide expression and osteolysis in metastatic human breast cancer cells. Cancer Res. 66, 7548–7553 (2006). [DOI] [PubMed] [Google Scholar]
- 97.Xing Z et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meijer D, van Agthoven T, Bosma PT, Nooter K & Dorssers LCJ Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol. Cancer Res 4, 379–386 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Calon A et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kogo R et al. Long noncoding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 (2011). [DOI] [PubMed] [Google Scholar]
- 101.Ren Y et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-β 1 secretion. Mol. Cancer 17, 5–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gupta GP et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb. Symposia Quant. Biol 70, 149–158 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Minn AJ et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuzhalin AE & Yu D Brain metastasis organotropism. Cold Spring Harb. Perspect. Med 10, a037242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weidle UH, Birzele F, Kollmorgen G & Rüger R Molecular basis of lung tropism of metastasis. Cancer Genomics Proteom. 13, 129–139 (2016). [PubMed] [Google Scholar]
- 106.Shimo T et al. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J. Bone Miner. Res 21, 1045–1059 (2006). [DOI] [PubMed] [Google Scholar]
- 107.Chen W, Hoffmann AD, Liu H & Liu X Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol 2, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guy CT, Cardiff RD & Muller WJ Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Muller WJ, Sinn E, Pattengale PK, Wallace R & Leder P Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54, 105–115 (1988). [DOI] [PubMed] [Google Scholar]
- 110.Annunziato S, Barazas M, Rottenberg S & Jonkers J Genetic dissection of cancer development, therapy response, and resistance in mouse models of breast cancer. Cold Spring Harb. Symposia Quant. Biol 81, 141–150 (2016). [DOI] [PubMed] [Google Scholar]
- 111.Li C et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol 19, 106–119 (2017).Studies the role of MAYA in directing organ-specific tropism towards bone metastases and demonstrates therapeutic in vivo targeting of this lncRNA in mice.
- 112.Valiente M et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang S et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Invest 127, 4498–4515 (2017).Demonstrates how lnc-BM promotes brain metastases through activation of cell adhesion molecules and cytokine release.
- 114.Lee JT Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol 12, 815–826 (2011). [DOI] [PubMed] [Google Scholar]
- 115.Xing F et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 78, 4316–4330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.De Palma M, Biziato D & Petrova TV Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 17, 457–474 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Kitamura T, Qian B-Z & Pollard JW Immune cell promotion of metastasis. Nat. Rev. Immunol 15, 73–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karki R & Kanneganti T-D Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 19, 197–214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huntington ND, Cursons J & Rautela J The cancer-natural killer cell immunity cycle. Nat. Rev. Cancer 7, 703–718 (2020). [DOI] [PubMed] [Google Scholar]
- 120.Peinado H et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017). [DOI] [PubMed] [Google Scholar]
- 121.Zanconato F, Cordenonsi M & Piccolo S YAP and TAZ: a signalling hub of the tumour microenvironment. Nat. Rev. Cancer 19, 454–464 (2019). [DOI] [PubMed] [Google Scholar]
- 122.Quail DF & Joyce JA Microenvironmental regulation of tumor progression and metastasis. Nat. Med 19, 1423–1437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu B et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Huang D et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat. Immunol 19, 1112–1125 (2018).Broadens the role of NKILA toinclude both tumour-intrinsic and tumour microenvironment factors.
- 125.Ji J et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 9, 478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sang L-J et al. LncRNA CamK-A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Mol. Cell 72, 71–83.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Jiang R et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun 8, 15129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen C et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun 9, 3826–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen YG, Satpathy AT & Chang HY Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol 18, 962–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fatica A & Bozzoni I Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet 15, 7–21 (2014). [DOI] [PubMed] [Google Scholar]
- 131.Sauvageau M et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2, e01749 (2013).Genetic deletion of multiple lncRNAs in mouse models demonstrating essential phenotypes.
- 132.Roobol MJ et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur. Urol 58, 475–481 (2010). [DOI] [PubMed] [Google Scholar]
- 133.Prensner JR et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15, 1469–1480 (2014).Large-scale translational study establishing SChLAP1 as a prognostic marker for metastatic prostate cancer risk.
- 134.Prensner JR et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet 45, 1392–1398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reyes DK & Pienta KJ The biology and treatment of oligometastatic cancer. Oncotarget 6, 8491–8524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Palma DA et al. The oligometastatic state - separating truth from wishful thinking. Nat. Publ. Group 11, 549–557 (2014). [DOI] [PubMed] [Google Scholar]
- 137.Qu L et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).Demonstrates paracrine transfer of lncRNAs through exosomes leading to drug resistance.
- 138.Yoshida T et al. Estimated number of off-target candidate sites for antisense oligonucleotides in human mRNA sequences. Genes Cell 23, 448–455 (2018). [DOI] [PubMed] [Google Scholar]
- 139.Yoshida T et al. Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes Cell 24, 827–835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee J-S & Mendell JT Antisense-mediated transcript knockdown triggers premature transcription termination. Mol. Cell 77, 1044–1054.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lai F, Damle SS, Ling KK & Rigo F Directed RNase H cleavage of nascent transcripts causes transcription termination. Mol. Cell 77, 1032–1043. e4 (2020). [DOI] [PubMed] [Google Scholar]
- 142.Swayze EE et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Meng L et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Scharner J et al. Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic Acids Res. 48, 802–816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zielinski R & Chi KN Custirsen (OGX-011): a second-generation antisense inhibitor of clusterin in development for the treatment of prostate cancer. Future Oncol. 8, 1239–1251 (2012). [DOI] [PubMed] [Google Scholar]
- 146.Yamamoto Y et al. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin. Cancer Res 21, 1675–1687 (2015). [DOI] [PubMed] [Google Scholar]
- 147.Finkel RS et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med 377, 1723–1732 (2017).Randomized clinical trial of an antisense oligonucleotide-based therapeutic in human patients demonstrating successful clinical response.
- 148.Hong D et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med 7, 314ra185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Monteleone G et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn’s disease. N. Engl. J. Med 372, 1104–1113 (2015). [DOI] [PubMed] [Google Scholar]
- 150.Cornelis G, Souquere S, Vernochet C, Heidmann T & Pierron G Functional conservation of the lncRNA NEAT1 in the ancestrally diverged marsupial lineage: evidence for NEAT1 expression and associated paraspeckle assembly during late gestation in the opossum Monodelphis domestica. RNA Biol. 13, 826–836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Karner H et al. Functional conservation of LncRNA JPX despite sequence and structural divergence. J. Mol. Biol 432, 283–300 (2020). [DOI] [PubMed] [Google Scholar]
- 152.Long Y et al. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet 52, 931–938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fares J, Fares MY, Khachfe HH, Salhab HA & Fares Y Molecular principles of metastasis: a hallmark of cancer revisited. Signal. Transduct. Target. Ther 5, 28–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bassett AR et al. Considerations when investigating lncRNA function in vivo. eLife 3, e03058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wurster CD & Ludolph AC Antisense oligonucleotides in neurological disorders. Ther. Adv. Neurol. Disord 11, 1756286418776932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Thomson DW & Dinger ME Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet 17, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- 157.Denzler R, Agarwal V, Stefano J, Bartel DP & Stoffel M Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ala U et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl Acad. Sci. USA 110, 7154–7159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Figliuzzi M, Marinari E & De Martino A MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys. J 104, 1203–1213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hausser J & Zavolan M Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat. Rev. Genet 15, 599–612 (2014). [DOI] [PubMed] [Google Scholar]
- 161.Thomson DW, Bracken CP & Goodall GJ Experimental strategies for microRNA target identification. Nucleic Acids Res. 39, 6845–6853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gennarino VA et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 22, 1163–1172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Du Z et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun 7, 10982–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Paraskevopoulou MD et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 41, D239–D245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang P et al. miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database 2015, bav098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hoffmann M et al. SPONGEdb: a pan-cancer resource for competing endogenous RNA interactions. NAR Cancer 10.1093/narcan/zcaa042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li J-H, Liu S, Zhou H, Qu L-H & Yang J-H starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Karreth FA et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Piwecka M et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.