Abstract
The microRNA miR396 directly represses GROWTH-REGULATING FACTORs (OsGRFs) and has been implicated in regulating rice yield and in nitrogen assimilation. Overexpressing the miR396 targets OsGRF4 and OsGRF6 improves rice yield via increased grain size and panicle branching, respectively. Here, we used CRISPR/Cas9 to assess the function of miR396 genes in rice. Knockout of MIR396ef (MIR396e and MIR396f), but not other isoforms, enhanced both grain size and panicle branching, resulting in increased grain yield. Importantly, under nitrogen-deficient conditions, mir396ef mutants showed an even higher relative increase in grain yield as well as elevated above-ground biomass. Furthermore, we identified OsGRF8 as a new target of miR396, in addition to the known targets OsGRF4 and OsGRF6. Disruption of the miR396-targeting site in OsGRF8 was sufficient to both enlarge grain size and elongate panicles. Our results suggest that rice-seed and panicle development are regulated by miR396ef-GRF4/6/8-GIF1/2/3 modules and that miR396ef are promising targets of genome editing for breeding environmentally friendly rice varieties that require less nitrogen fertilization.
Keywords: microRNA, miR396, OsGRF4/6/8, OsGIF1/2/3, nitrogen deficiency, grain yield
Introduction
MicroRNAs (miRNAs) are single-stranded, non-coding RNAs of 21–24 nucleotides, which typically load into a silencing complex to direct cleavage or translational inhibition, and thus repression, of complementary messenger RNAs (mRNAs) [1–3]. In plants, miRNAs act as key regulators to control development, growth and stress tolerance [4–6].
Several miRNAs have been shown to control rice yield. For example, miR156 and miR397 affect rice yield by regulating their targets, OsSPLs and OsLACs, respectively [5–9]. The microRNA miR396 was recently shown to regulate rice-grain yield by targeting the transcription factors growth-regulating factors (GRFs), which execute their function via GRF-interacting factors (GIFs) [10]. miR396 can target and thus limit the expression of both OsGRF4 and OsGRF6 in rice [11]. OsGRF4 controls grain development by regulating many brassinosteroid-induced genes and disruption of the miR396 target site in OsGRF4 enhances grain size and yield in rice [12,13]. OsGRF4 also balances the inhibitory activities of DELLA to promote and integrate nitrogen assimilation, carbon fixation and growth [14], but whether this is regulated by miR396 is not known. OsGRF6 directly promotes the expression of panicle-branching factor genes OsTAWAWA1 and OsMADS34, and decreased expression of MIR396b or overexpression of OsGRF6 increases the panicle-branching number and, in turn, grain yield [15]. Overexpression of the miR396 target genes AtGRF1, AtGRF2 and AtGRF5 in Arabidopsis leads to the production of larger seeds, indicating that a miR396-GRF regulatory network broadly participates in seed development [16].
miR396 antagonizes the expression of its target GRF2 in the distal part of leaves, restricting cell proliferation in developed tissues [17]. miR396 also negatively regulates cell proliferation by markedly decreasing the expression of cell-cycle-related genes, such as CYCLINB1;1 and TCP4 [17,18]. An miR396-GRF-GIF regulatory network has also been reported to regulate growth, development and abiotic stress tolerance in Arabidopsis [19,20]. Overexpression of miR396 in Arabidopsis reduces the accumulation of GRF7 [19,21]. GRF7 directly represses DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) and other genes that are up-regulated in response to abscisic acid; these genes enhance tolerance of dehydration and high-salinity conditions in Arabidopsis plants [19].
Given that a single miRNA often targets multiple genes to regulate diverse biological processes, identifying and manipulating key miRNAs is considered a potential strategy to improve agronomic traits of crop plants [22]. Gene-editing technology has been applied to improve agronomic traits and we were interested in using this approach to manipulate key miRNAs and generate resources for breeding elite crop varieties. Here, we studied the individual functions of the eight members of the miR396 gene family in rice. We discovered that two members, miR396e and miR396f (miR396ef), regulate seed and panicle development. mir396ef mutants showed increases in grain size and panicle branching, which resulted in increased grain yield. Importantly, mir396ef mutations showed a more substantial increase in grain yield and above-ground biomass under conditions of nitrogen deficiency. We demonstrated that OsGRF4, OsGRF6 and OsGRF8 are the main targets of miR396ef and that miR396-GRF4/6/8-GIF1/2/3 modules regulate rice-grain and panicle development. Our work suggests that miR396ef is an attractive target for generating high-yield rice plants that are less dependent on nitrogen fertilization.
Results
miR396ef isoforms regulate grain development in rice
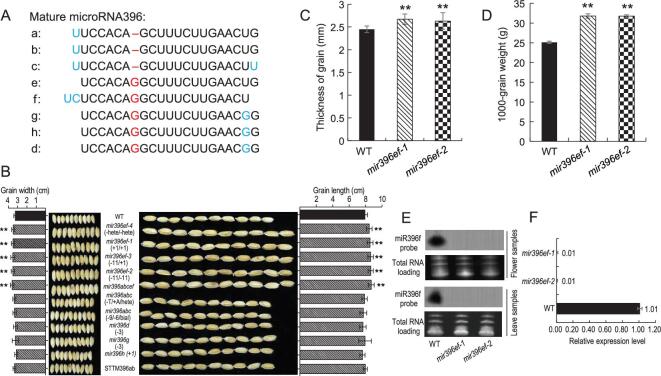
Rice contains five different miR396 isoforms, encoded by eight genes (Fig. 1A). According to the plant miRNA database PmiREN (Plant miRNA ENcyclopedia, http://www.pmiren.com/), the family members MIR396e and f have the highest expression in reproductive tissues, such as inflorescence, florets and spikelets (Supplementary Table 1) [23,24]. MIR396c is also expressed in these reproductive tissues, but at a relatively lower level (Supplementary Table 1) [23,24]. To identify the miR396 isoforms that modulate rice-seed and panicle development, we used CRISPR-Cas9 and five sgRNA constructs to knock out the eight MIR396 members (Supplementary Fig. 1). miR396a–c and miR396e–f have the same mature sequences, respectively, and each group was targeted by one construct. Although miR396 d, g and h also have the same mature sequence, they are located on the complementary strands of different genes, and therefore were targeted by three different constructs to avoid functional disruptions of overlapping genes (Supplementary Fig. 1D).
Figure 1.
miR396ef isoforms regulate rice-grain development (A) miR396 members in rice. Isoform differences are highlighted in blue. Red color indicates the difference at position seven. (B) Comparison of the 10-grain width and 10-grain length between mir396 mutant plants and wild-type (WT). The genotypes are labeled after the name of mutants in the brackets. Plus (+) and minus (−) and subsequent number represent the nucleotides inserted and deleted at the sgRNA target sites, and mutations on different mir396 members are separated by slashes. The ‘hete’ means heterozygous mutation on the corresponding members. The ‘bial’ means biallelic mutation on the corresponding member. (C) The grain thickness of mir396ef and WT plants. (D) The 1000-grain weight of mir396ef and WT plants. (E) Northern blotting detection of miR396ef in flowers and leaf samples of WT and mir396ef plants. Total RNA stained with ethidium bromide was used as a loading control. (F) Stem-loop RT-qPCR of miR396ef in WT and mir396ef leaf samples. U6 was used to normalize samples. Data are presented as means ± SD (n = 100 in (B)–(D); n = 3 in (F)). P-values (versus the WT) were calculated with Student’s t-test, two-tailed. **P < 0.01.
CRISPR lines with mutations in mature miR396 regions, which should disrupt function, were selected for further study. Unexpectedly, only mir396ef mutants exhibited grain-size phenotypes, whereas the remaining mir396 mutants displayed no differences compared with wild-type (WT) (Fig. 1B and Supplementary Figs 2 and 3). In addition, mir396abc triple mutants, mir396d and mir396g single mutants (with non-frameshift mutations in overlapping genes) and mir396h mutants exhibited no differences in morphological phenotypes compared to WT, including plant height (Supplementary Figs 2A and 3A, E and I), grain length, grain width (Fig. 1B and Supplementary Figs 2B and C and 3B, C, F, G, J and K) and panicle architecture (Supplementary Figs 2D and E and 3M and N). We also applied the short tandem target MIMICs (STTM) technology to block miR396ab functions (Supplementary Fig. 4A and E). The STTM-miR396 lines showed similar seed length, seed width and panicle branching compared with WT (Supplementary Fig. 4B–D).
In contrast, compared with the WT, mir396ef mutants showed obvious increases in grain length, width and thickness (increased 8.46%, 7.95% and 8.16%, respectively) (Fig. 1B and C). The 1000-grain weight was also significantly increased in mir396ef (31.78 ± 0.55 g) compared to WT (25.12 ± 0.30 g) (Fig. 1D). We obtained many mir396ef double-mutant CRISPR lines with different genotypes, all of which displayed the same seed phenotypes, confirming the functions of MIR396ef during seed development (Fig. 1B and Supplementary Fig. 5A and B). We generated mir396abcef mutants by crossing mir396abc with mir396ef mutants, which showed similar grain length and width as mir396ef (Fig. 1B). The mature isoform of miR396ef was almost undetectable in mir396ef leaf and flower (Fig. 1E and F), indicating successful elimination of MIR396e and f.
Due to the extremely high genome-editing efficiency at the mi396e and f target sites, we obtained only mir396ef double mutants. To obtain an mir396e single mutant, we designed an sgRNA to specifically disrupt MIR396e (Supplementary Fig. 6A and B). The grain length and width of mir396e mutants are 4.77% and 5.58% larger than the WT, respectively (Supplementary Fig. 6C and D). The seed phenotypes of mir396e are weaker than that of mir396ef double mutants (Supplementary Fig. 6C), suggesting that both MIR396e and MIR396f contribute to the grain-size development. We examined putative off-target loci for miR396abc and miR396ef target sequences and did not observe any mutations (Supplementary Fig. 7). These results indicate that both miR396e and f isoforms are involved in rice-grain development, although mir396f single mutants are needed to confirm the function of miR396f.
mir396ef mutants display simultaneous increases in grain size and panicle branching
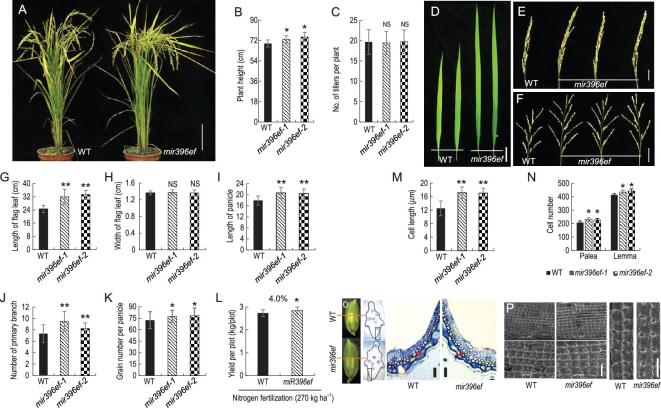
The increased seed size of the mir396ef mutants was associated with a significantly enlarged grain husk, as demonstrated by both histological sectioning and scanning electron microscopy (Fig. 2O and P). The length and number of inner palea and lemma cells were significantly increased in the mir396ef grain husk compared with the WT (Fig. 2M and N). An MIR396e-promoter::GUS assay indicated that MIR396e was expressed in grain hull, spikelet, leaf and root (Supplementary Fig. 8), consistently with its regulation of grain-husk development.
Figure 2.
Morphological and growth phenotypes of mir396ef plants. (A) Gross morphologies of WT and mir396ef plants at maturity. Scale bar, 10 cm. (B) Plant height of WT and mir396ef. (C) Tiller number per plant of WT and mir396ef. Leaf (D), panicle (E) and panicle-branching (F) phenotypes of WT and mir396ef plants. Scale bars: 50 mm in (D); 30 mm in (E) and (F). Flag-leaf blade length (G), flag-leaf blade width (H), panicle length (I), number of primary branchings per panicle (J) and grain number per panicle (K) of WT and mir396ef plants. (L) Grain yield per plot of WT and mir396ef with nitrogen fertilization. One hundred plants (10 × 10) were cultivated in 2.0 × 2.0 (m) plots. (M) Cell length of grain spikelet hulls in WT and mir396ef plants. (N) Cell number of spikelet hull palea and lemma in WT and mir396ef plants. (O) Cross-sections of spikelet hulls of WT and mir396ef plants, and magnified view of the boxed cross-section area. The red line indicates the position of the cross-section. The rectangle indicates the position of the magnification of the hull. The red arrow points to the palea cells. Scale bars, 50 μm. (P) Scanning election microscopy images of the outer surface of glume in WT and mir396ef. Scale bars, 100 μm. Data are presented as means ± SD (n = 20 in (B), (C), (G)–(K) and (M); n = 5 in (L) and (N)). P-values (versus the WT) were calculated with Student’s t-test, two-tailed. *P < 0.05; **P < 0.01. NS, not significant.
We found that mir396ef mutant plants (74.13 ±3.54 cm) were taller than the WT (69.33 ± 3.20 cm), though the tiller number was similar to WT (Fig. 2A–C). In addition, compared to the WT flag leaf, the mir396ef flag leaf was 32.8% longer but had the same width (Fig. 2D, G and H), resulting in an increased leaf area. The photosynthetic rates of the flag leaf showed no difference between the mir396ef and the WT in the paddy field (Supplementary Fig. 9). The unchanged photosynthetic rates with increased leaf area would enhance overall photosynthesis, perhaps leading to the seed increase in mir396ef mutants relative to WT.
Rice grains grow on the panicle with numerous branches. Longer panicles and more panicle branches generally correlate with more grains. Compared to WT, mir396ef lines had longer panicles (20.55 ± 1.83 vs 17.93 ± 1.64 cm), more primary branches (8.8 ± 1.34 vs 7.33 ± 1.56) and higher grain numbers per panicle (77.82 ± 8.89 vs 72.35 ± 10.96) (Fig. 2E, F, I, J and K). However, the mir396ef panicles were partly enwrapped by the flag leaf, which affects the fertility and filling of the enwrapped panicle (Supplementary Fig. 10A–C).
The phenotypes of mir396ef, including a longer flag leaf for enhanced photosynthesis, larger grain size for more grain weight and more grains per panicle, suggest that mir396ef could be valuable targets to increase grain yield. Our paddy-field plot-yield test revealed an approximately 4% increase in grain yield on conventional agricultural 270 kg ha−1 nitrogen fertilizer (Fig. 2L). Like WT mature pollens, mir396ef pollens were deeply stained by iodine–potassium iodide (I2-KI) and their fertility was not changed (Supplementary Fig. 10D–F). The yield increase of 396ef was below what we expected, perhaps due to a higher rate of empty grains in the mir396ef mutants than in WT (Supplementary Fig. 10C).
We also tested grain-quality traits in the mir396ef mutants. The brown and polished grains of mir396ef mutants had a more ‘white splotch’ appearance compared with the WT-grain appearance (Supplementary Fig. 11A and B). The alkali-spreading value (gelatinization temperature) as well as the percentage and degree of chalkiness were increased (Supplementary Fig. 11C, D and F), whereas the gel consistency was decreased in the mir396ef mutants compared with the WT (Supplementary Fig. 11E).
mir396ef mutants exhibit significantly increased grain yield under nitrogen-deficient conditions
The miR396 target OsGRF4 confers increased nitrogen assimilation and biomass accumulation in rice [14]. To investigate the response of miR396 to nitrogen treatments, we examined the expression of miR396 family members in seedlings grown in hydroponic culture either with or without nitrogen supply. We found that the levels of miR396f increased transiently in seedlings grown under nitrogen-deficient conditions, reaching a maximum at about 6 hours after exposure to nitrogen deficiency, then decreasing and returning to normal levels at about 24 hours (Supplementary Fig. 12). In contrast, miR396e expression did not change in response to nitrogen deficiency (Supplementary Fig. 12).
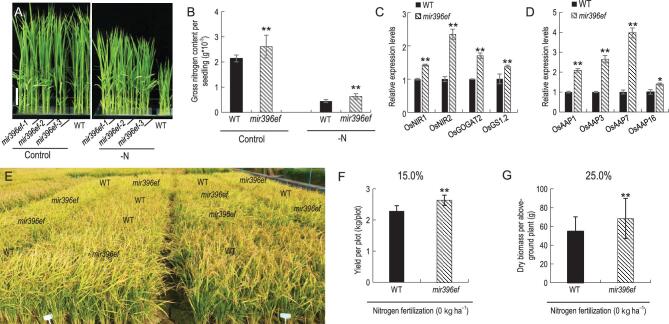
Next, we examined the nitrogen assimilation of the mir396ef mutants at the rice-seedling stage. We found that seedlings from mir396ef plants had a significantly higher nitrogen content than those from WT plants, when grown in hydroponic culture either with or without nitrogen supply (Fig. 3A and B). Recently, Li et al. showed that OsGRF4 recognizes the GCGG core motif by ChIP-Seq; ChIP-PCR and Electrophoretic Mobility Shift Assay (EMSA) confirmed that OsGRF4 associates with promoters that contain this motif and regulate genes involved in ammonia and nitrate metabolism, including GOGAT2, NIR1 and GS1.2, etc. [14]. To determine whether genes involved in nitrogen assimilation and utilization are differentially expressed in mir396ef mutant and WT seedlings, we performed reverse-transcription quantitative PCR (RT-qPCR). Specifically, we examined the expression of NIR1, NIR2, GOGAT2 and GS1.2, which encode enzymes involved in nitrogen assimilation [14]. We also examined the AMINO ACID PERMEASE genes (OsAAPs), which are nitrogen-use-efficiency-related genes that determine amino-acid delivery from source to sink in plants [25,26]. All of these genes we examined showed increased expression in mir396ef mutant seedlings compared to WT seedlings (Fig. 3C and D). These results suggest that MIR396ef is involved in nitrogen assimilation and utilization in rice.
Figure 3.
Grain yield of WT and mir396ef in a nitrogen-deficient environment. (A) Nitrogen-deficiency response assay in WT and mir396ef plants. Scale bar, 5 cm. (B) The gross nitrogen content per seedling with and without nitrogen supplied in WT and mir396ef plants. (C) and (D) The expression level of nitrogen-related genes in WT and mir396ef seedling samples detected by RT-qPCR. ACTIN1 was used to normalize samples; relative expression levels were measured using the 2–ΔΔCt analysis method. (E) Plots yield test of WT and mir396ef in nitrogen-deficient paddy fields. (F) Grain yield per plot of the WT and mir396ef without nitrogen fertilization. One hundred plants (10 × 10) were cultivated in 2.0 × 2.0 (M) plots. (G) Dry biomass of the above-ground per WT and mir396ef plants in nitrogen-deficient paddy fields. Data are presented as means ± SD (n = 3 in (B)–(D); n = 5 in (E)–(G)). P-values (versus the WT) were calculated with Student’s t-test, two-tailed. *P < 0.05; **P < 0.01.
Next, we determined whether the improved nitrogen assimilation and utilization of mir396ef mutant plants affect growth and yield under nitrogen-deficient conditions. We found that mir396ef mutants grew substantially taller than WT plants when grown in hydroponic culture without nitrogen supply (Fig. 3A). Moreover, mir396ef plants showed a substantial yield increase (∼15%) compared with the WT under nitrogen-deficient conditions (0 kg ha−1 nitrogen fertilizer application) in the paddy field (Fig. 3E and F). The yield of mir396ef under low-nitrogen conditions was comparable to the yield of WT under normal nitrogen-fertilization conditions (Supplementary Table 2). Interestingly, mir396ef showed a 25% increase in above-ground dry biomass per plant under nitrogen-deficient conditions compared to WT plants (Fig. 3G).
Regulation of grain size and branch length by an miR396ef-GRFs-GIFs module
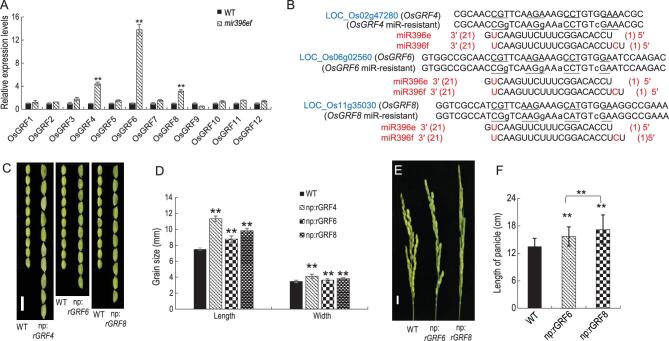
Twelve GRF transcription factors in rice harbor the miR396 target site [27]. We found that only OsGRF4, OsGRF6 and OsGRF8 were up-regulated in mir396ef plants compared to WT (Fig. 4A). RLM-RACE (5′ RNA ligase mediated rapid amplification of cDNA ends) analysis showed that miR396 could direct the cleavage of OsGRF4 and OsGRF6 mRNAs in vivo at a specific site within the miR396 pairing region with leaf samples (Supplementary Fig. 13A). Interestingly, in leaf samples, miR396 caused the cleavage of OsGRF8 mRNA at a site 118 bp downstream of the miR396 pairing region, and results of transient co-expression assays in Nicotiana benthamiana leaves are consistent with such a cleavage (Supplementary Figs 13A and 14). However, in flower samples, miR396 could direct the cleavage of OsGRF8 mRNA at the predicted miR396 pairing region (Supplementary Fig. 13A).
Figure 4.
miR396 regulates grain size and panicle length via OsGRF4, OsGRF6 and OsGRF8. (A) The expression profiles of 12 OsGRFs genes in WT and mir396ef leaf samples. (B) The mutations introduced at the miR396 binding site of OsGRF4, OsGRF6 and OsGRF8 mRNAs to generate miR396-resistant OsGRF4, OsGRF6 and OsGRF8 driven by their respective native promoters (namely np:rOsGRF4, np:rOsGRF6 and np:rOsGRF8). (C) The grain-length phenotypes of np:rOsGRF4, np:rOsGRF6 and np:rOsGRF8. Scale bar, 10 mm. (D) Comparison of the grain width and grain length between np:rOsGRF4, np:rOsGRF6 and np:rOsGRF8 with WT, respectively. (E) Panicle length of np:rOsGRF6 and np:rOsGRF8. Scale bar, 15 mm. (F) Comparison of panicle length between np:rOsGRF6 and np:rOsGRF8 with WT. Data are presented as means ± SD (n = 20 in (D); n = 5 in (F)). P-values (versus the WT) were calculated with Student’s t-test, two-tailed. **P < 0.01.
To investigate whether miR396-mediated regulation of OsGRF4, OsGRF6 and OsGRF8 control seed and panicle development, we constructed miR396 target-resistant versions of OsGRF4, OsGRF6 and OsGRF8 (designated np:rGRF4, np:rGRF6 and np:rGRF8, respectively) (Fig. 4B) and introduced them into rice under the control of their respective native promoters. We found that np:rGRF4, np:rGRF6 and np:rGRF8 lines exhibited larger grain sizes than WT plants (Fig. 4C and Supplementary Fig. 13B–D), with np:rGRF4 showing the largest increase and np:rGRF6 showing the smallest increase (Fig. 4D and Supplementary Fig. 13B–D). Both np:rGRF6 and np:rGRF8 lines showed increased panicle lengths (Fig. 4E and F and Supplementary Fig. 13I), but their branching numbers remained unchanged compared to WT. The np:rGRF4 lines displayed similar panicle length and branching numbers to WT (Supplementary Fig. 13H). These results suggest that the miR396ef members regulate seed and panicle development by regulating their target genes OsGRF4, OsGRF6 and OsGRF8.
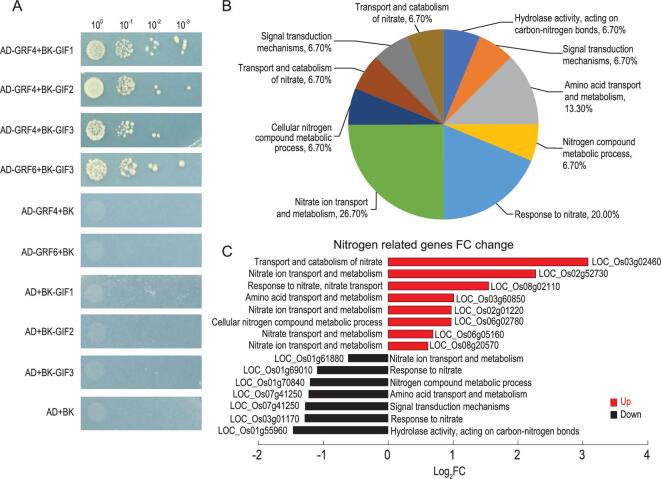
GRFs have been shown to interact with transcription coactivators GIFs [12,20]. To uncover GIFs that interact with OsGRF4 and OsGRF6, we performed a yeast two-hybrid assay. We found that OsGIF1, OsGIF2 and OsGIF3 interacted with OsGRF4, and OsGIF3 also interacted with OsGRF6 (Fig. 5A). Biomolecular fluorescence complementation (BiFC) assays confirmed these interactions in tobacco leaves (Supplementary Fig. 15). The expression of OsGIF1 was significantly increased in mir396ef plants (Supplementary Fig. 16A). We used the CRISPR/Cas9 technology to generate gif1 mutants and observed the expected short-leaf phenotype (Supplementary Fig. 16B). osgif1 mutants also exhibited smaller plant stature and aborted seed phenotypes compared to WT (Supplementary Fig. 16B and C). These findings suggest that rice-seed and panicle development are regulated by the miR396ef-GRF4/6/8-GIF1/2/3 module.
Figure 5.
The interactions between OsGIFs and OsGRFs, and differentially expressed genes in mir396ef plants. (A) OsGRF4 and OsGRF6 interact with OsGIF1/2/3 in a yeast two-hybrid assay. (B) Nitrogen-related genes with differential expression in mir396ef versus WT leaf. (C) The expression of nitrogen-related genes in mir396ef compared with WT leaf. FC, fold change (mir396ef/WT).
To further explore the function of miR396ef in plants, we used RNA-sequencing analysis to examine the transcriptomes of leaves and young spikelet tissues from mir396ef and WT plants. We detected 558 DEGs (differentially expressed genes, ratio > 1.5) in young spikelet tissues, but only 189 DEGs in leaves. We observed 314 and 127 up-regulated genes in mir396ef mutant spikelet and leaves, respectively, compared to WT. In addition, 244 and 62 genes were down-regulated in mir396ef mutant spikelet and leaves compared to WT, respectively (Supplementary Fig. 17A). We observed 15 DEGs encoding ‘nitrogen-related genes’ in leaves and enrichment analysis of the DEGs identified ‘nitrate ion transport and metabolism’ as the top annotated category (Fig. 5B and C). In addition, 26 DEGs were related to brassinonsteroid-induced genes and 21 DEGs were related to auxin signaling, consistently with auxin- and brassinosteroid-mediated regulation of spikelet number and grain size (Supplementary Fig. 18A and B) [12,15]. We confirmed the expression of key genes within brassinosteroid and auxin-signaling pathways by RT-qPCR (Supplementary Fig. 19A and B). We also observed increased expression of the cell-proliferation-related genes, two CYCLINB1;1 homologous genes and one TCP4 homologous gene, in mir396ef mutant, consistently with the increases in cell number (Supplementary Fig. 19C–E).
Discussion
Here, we investigated the role of the miR396 gene family in controlling grain yield in rice, with the goal of identifying targets and strategies for breeding elite crop varieties. The miR396 target gene OsGRF4 increases grain size by activating brassinosteroid responses [12], whereas the miR396 target gene OsGRF6 promotes panicle branching through the activation of several auxin biosynthesis factors [15]. Thus, targeting miR396 itself would be expected to enhance both grain size and panicle branching. We examined the miR396 members by CRISPR/Cas9 editing (Fig. 1A and Supplementary Fig. 1) and found that mir396ef mutants showed the expected traits of increased grain size and panicle branching. It was reported that miR396b negatively regulates panicle branching by targeting OsGRF6, and STTM-miR396b plants showed increased panicle branching and grain yield [15]. However, in our data, STTM-miR396ab lines showed unchanged panicle branching compared with the WT. This discrepancy may be caused by the different genetic backgrounds, since the reported STTM-miR396b lines were in the indica background whereas our STTM-miR396ab lines are in the japonica background. More recently, Miao et al. [28] reported that mir396e single mutant rice had a similar grain size to the WT Xiushui 134. However, in our data, the grain length and width of mir396e mutants are larger than the WT Nipponbare, which may also be explained by the background difference.
We found that mir396ef mutants have several beneficial traits relative to WT plants, including increased plant height (Fig. 2A and B), enhanced length of the flag leaves and panicles (Fig. 2D and E), increased primary branch and grain number in panicle (Fig. 2F, J and K) and increased grain size and weight (Fig. 1B–D). Furthermore, the cell length and cell number in the grain hull of mir396ef mutants are both increased, which enlarged the sink size. Just as in OsGRF4-overexpression grains, the mir396ef mutant showed decreased grain quality because of increased chalkiness (Supplementary Fig. 11), which could be offset by alleles of high-quality genes [29].
miR396 attenuates cell proliferation in developing leaves and negatively regulates leaf growth in Arabidopsis thaliana [17,27]. We found that mir396ef mutant rice plants also showed an obvious increase in leaf length. In addition, the np:rGRF4, np:rGRF6 and np:rGRF8 lines all exhibited increased leaf length and enwrapped panicles (Supplementary Fig. 13E–G). The np:rGRF4 plants are obviously shorter than the WT, perhaps because the np:rGRF4 panicles were severely enwrapped by the flag leaf (Supplementary Fig. 13E). The significant increases in the source leaves of mir396ef mutants could explain the increased grain yield and biomass. Although the grain yield of mir396ef mutants was only 4.0% greater than that of WT in test plots with normal nitrogen fertilization (Fig. 2L), it was 15.0% greater in natural paddy fields without nitrogen fertilization (Fig. 3E and F). Further, the above-ground biomass of mir396ef mutant plants was 25.0% more than that of WT plants under nitrogen-deficient conditions (Fig. 3G). These results suggest that mir396ef mutations cause enhanced nitrogen utilization, consistently with increased OsGRF4 function, which has been shown to balance the inhibitory activities of DELLA to promote and integrate nitrogen assimilation, carbon fixation and growth (Fig. 3C and D) [14]. This notion is supported by our discovery that nitrogen transport and metabolism genes are up-regulated in mir396ef mutants (Fig. 5B and C).
OsGRF4 and OsGRF6 have been reported as targets of miR396ef. Based on expression and mRNA cleavage studies, we uncovered OsGRF4, OsGRF6 and OsGRF8 as major targets of miR396ef (Fig. 4A and Supplementary 13A). Plants expressing miR396ef-resistant versions of target genes, np:rGRF4, np:rGRF6 and np:rGRF8, all exhibited larger grain size to varying degrees, with the largest increase in np:rGRF4. However, the panicle length and branching numbers of np:rGRF4 were unchanged compared to WT (Fig. 4C and D and Supplementary Fig. 13E). Panicle lengths were increased in np:rGRF6 and np:rGRF8 (Fig. 4E and F), but branching numbers were unaffected (Supplementary Fig. 13H and I). In summary, we did not observe synchronous increases in grain size and panicle branching in np:rGRF4, np:rGRF6 and np:rGRF8 plants. These data suggest that the seed-size and panicle-length improvement of mir396ef mutants is due to the combined functions of OsGRF4, OsGRF6 and OsGRF8.
We found that GIFs OsGIF1, OsGIF2 and OsGIF3 interact with OsGRF4, and OsGIF3 also interacts with OsGRF6 (Fig. 5A). Knockout of OsGIF1 led to an expected short-leaf phenotype, suggesting that miR396ef-GRFs-GIFs modules directly control the development of seeds, panicles and leaves in rice. Previous studies demonstrated that miR396 regulates seed and panicle development through brassinosteroid and auxin-signaling pathways [12,15] and our RNA-seq data revealed an altered expression of many brassinosteroid-induced genes and auxin-signaling genes in the mir396ef mutant (Supplementary Fig. 18). We also found that np:rGRF4 and np:rGRF6 have enlarged leaf angles, exhibited as losing plant structure, which are typical phenotypes of brassinosteroid pathway mutants, such as m107 and Gi-2 [12]. These data suggest that miR396ef might influence rice architecture by activating brassinosteroid responses.
Taken together, our findings reveal that manipulation of miR396ef can simultaneously improve grain size and panicle branching. miR396ef thus represents a promising target to increase grain size and grain yield, especially in nitrogen-deficient environments, which might help breeders develop environmentally friendly elite rice varieties.
Materials and methods
Plant materials and growth conditions
The rice variety Nipponbare (Oryza. Sativa L. spp. Japonica, var Nipponbare) was used in this study. Rice plants were cultivated under field conditions at two different experimental stations located in Shanghai (30°N, 121°E) and Lingshui (Hainan Province, 18°N, 110°E). The phytotron, with a 30/24 ± 1°C day/night temperature, 50%–70% relative humidity and a light/dark period of 14 hours/10 hours was used to culture rice seedlings.
Nitrogen-deficiency response assay
Nitrogen deficiency in hydroponic culture conditions was modified from previous work [14]. Seeds were disinfected in 20% sodium hypochlorite solution for 20 min, thoroughly washed with deionized water and then germinated in a dish with sterilized water. The germinated grains were then selected and transplanted to PVC culture pots that contained hydroponic nutrient solution (0.5 mM NaH2PO4, 0.75 mM K2SO4, 1 mM CaCl2, 1.667 mM MgSO4, 40 μM Fe-EDTA (Na), 19 μM H3BO3, 9.1 μM MnSO4, 0.15 μM ZnSO4, 0.16 μM CuSO4 and 0.52 μM (NH4)3Mo7O24, pH 5.5) with or without 1.25 mM NH4NO3. All nutrient solutions were changed every 2 days. The temperature was maintained at 30°C day and 24°C night, and the relative humidity was 70% in phytotron.
Vector construction
The CRISPR-Cas9 vectors targeting MIR396abc, MIR396d, MIR396ef, MIR396g, MIR396h, MIR396e and OsGIF1 were constructed as previously described [30] and the oligos used are listed in Supplementary Table 3. To construct the miR396-resisitant vectors for OsGRF4, OsGRF6 and OsGRF8, the genomic sequences of OsGRF4, OsGRF6 and OsGRF8, fused with the OsGRF4 promoter, OsGRF6 promoter and OsGRF8 promoter, respectively, were amplified and the synonymous mutations were introduced in the miR396 target sequences. All the vectors described above were used to transform rice Nipponbare by Agrobacterium tumefaciens-mediated methods.
Phenotyping and histological experiments
Plant materials were photographed using a Canon EOS7D digital camera and observed using an OLYMPUS BX53 microscope. Grain size was analysed using an SC-A grain analysis system (Wseen Company, China).
Young rice grains were collected and fixed overnight at 4°C in FAA (50% ethanol, 10% formalin and 5% acetic acid) solution and dehydrated in a graded ethanol series. The samples were then embedded in Technovit 7100 resin (Hereaus Kulzer) and made into 2-μm sections using a Leica RM 2265 programmable rotary microtome (Leica Microsystems). After being stained with 0.05% Toluidine Blue, transverse sections were photographed using an OLYMPUS BX53 microscope. Dehydrated grain-husk samples were stocked on the copper stub and sputter-coated with gold-palladium. The seed samples were examined under a scanning electron microscope, JSM-6360LV. Cell number and cell length in the outer parenchyma layer of the spikelet hulls were measured using the OLYMPUS stream software.
Northern blotting
Total RNA was isolated using the Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. About 40 μg of total RNA from leaves and flowers were analysed on a denaturing 19% polyacrylamide gel, transferred to Nytran Super Charge Nylon Membranes (Schleicher & Schuell BioScience) and cross-linked using a Stratagene UV Crosslinker. DNA oligonucleotides complementary to different sequences of miRNAs were synthesized and labeled with [32P]-γ-ATP (PerkinElmer) using T4 polynucleotide kinase (TaKaRa). The membranes were pre-hybridized with PerfectHyb (Sigma) hybridization solution and then hybridized with the labeled probes. After several times of washing, the membranes were autoradiographed using an X-ray film (Carestream, X-OMAT BT Film). For RNA loading, 18S and 5S rRNAs were used as controls. The probe sequences are listed in Supplementary Table 3.
Plot-yield tests
Plants of the WT and mir396ef were grown in Shanghai paddy fields under natural conditions. The area per plot was 2.0 × 2.0 m and 100 plants were cultivated in each plot with planting density of 20 × 20 cm. The nitrogen fertilizer was 270 kg ha−1 in the control field, whereas the nitrogen-deficient fields were without nitrogen fertilizer. Plants were irrigated with river water.
RNA extraction and RT-qPCR and stem-loop PCR
Total RNA was isolated from roots, leaves and flowers from different developmental stages of rice plants. The RNA extraction followed the method mentioned above. After being treated with RNase-free DNase I (Promega), total RNA (1 μg) was reverse transcribed using the TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix kits (TransGen Biotech). The reverse-transcription products were used as templates for RT-qPCR performed on a CFX96 real-time PCR system (Bio-Rad) using SYBR Premix Taq (TransGen Biotech) according to the manufacturer’s protocol. ACTIN1 was used to normalize gene expression and OsU6 was used in microRNA expression. Relative expression levels were measured using the 2–ΔΔCt analysis method. The primers used in RT-qPCR or stem-loop RT-qPCR are listed in Supplementary Table 3.
Transient expression assays in N. benthamiana leaves
The CDS of OSGIF1, OsGIF2 and OsGIF3 were fused in frame with nYFP (yellow fluorescent protein) sequence. The CDS of OsGRF4, OsGRF6 and OsGRF8 were fused in frame with cYFP sequence. nYFP-OsGIF1, nYFP-OsGIF2, nYFP-OsGIF3, cYFP-OsGRF4, cYFP-OsGRF6 and cYFP-OsGRF8 constructs were transformed into Agrobacteria. These stains containing nYFP and cYFP plasmids were co-infiltrated into leaves of N. benthamiana. YFP signals were detected by confocal microscopy LSM800 (ZEISS). The full-length and C-terminal deletion cDNAs of OsGRF8 were fused in frame with eGFP (enhancer Green Fluorescent Protein) sequence and driven by the 35S promoter. The stem-loop sequences of miR396e and miR396f were cloned and inserted after the 35S promoter. nYFP-OsGIF1, nYFP-OsGIF2, nYFP-OsGIF3, cYFP-OsGRF4, cYFP-OsGRF6, cYFP-OsGRF8, 35S::GRF8-eGFP, 35S::GRF8(1–574)-eGFP, 35S::GRF8(1–694)-eGFP, 35S::miR396e and 35S::miR396f constructs were transformed into Agrobacteria. These stains were co-infiltrated into leaves of N. benthamiana. YFP and GFP signals were detected by confocal microscopy LSM800 (ZEISS).
Yeast two-hybrid assay
The prey vector pGADT7 and bait vector pGBKT7 were used in the yeast two-hybrid assay. Full-length cDNAs of OSGIF1, OsGIF2, OsGIF3, OsGRF4, OsGRF6 and OsGRF8 were amplified and separately cloned into pGADT7 and pGBKT7. The prey and bait plasmids were co-transformed into the yeast AH109 strain and grew on SD-Leu-Trp solid media (Clontech). The strains were transferred to the SD-Ade-His-Leu-Trp solid media to test the interactions between prey and bait.
RLM-RACE
RLM-RACE was performed with the FirstChoice™ RLM-RACE Kit (Ambion). In general, total RNA was extracted from samples and the first and second PCRs were performed, with the primers of OsGRF4/6/8-inner and OsGRF4/6/8-outer (Supplementary Table 3), respectively. The products from the second PCR were purified by agarose gel electrophoresis and then cloned for sequencing. For OsGRF4 and OsGRF6, leaf samples were used in these experiments. For OsGRF8, flower samples were also used in these experiments.
Transcriptome analyses
Total RNA was extracted using the RNeasy Plant mini Kit (Qiagen) according to the manufacturer’s instructions. Two independent replicates were used for each sample. The cDNA synthesis, purification and labeling were then performed following the standard protocols. The RNA-seq data were analysed with TopHat and Cufflinks software. Genes with at least 1.5-fold up- and down-regulation in the mir396ef plants compared with those in WT plants were considered as DEGs.
Net photosynthetic rate measurements
The net photosynthetic rate was measured using an LI-6400XT (LICOR Biosciences) at a light intensity of 1000 μmol m−2 s−1 and the reference CO2 concentration was 400 μmol mol−1. The measurement followed the method from the LI-6400XT manual. Two leaves were used in one measurement and placed side by side in order to fill the area of the measure room because the rice leaves were narrower than the diameter of the measure room. The net photosynthetic rate was calculated and recorded by the machine. All experiments were conducted with at least five replicates.
Supplementary Material
FUNDING
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27040101 to J.-K.Z.), Postdoctoral Science Foundation Grant of China (2017 M621553 to J.Z.), National Natural Science Foundation of China (31801016 to J.Z. and 31871223 to H.Z.) and Shanghai Pujiang Program (18PJ1411000 to H.Z.).
AUTHOR CONTRIBUTIONS
J.Z., Z.Z., J.B., X.T. and L.W. performed the experiments and analysed the data. J.-K.Z. and H.Z. supervised the project, designed the research and participated in data analysis. J.Z., H.Z. and J.-K.Z. wrote the paper.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 2006; 57: 19–53. [DOI] [PubMed] [Google Scholar]
- 2. Song X, Li Y, Cao Xet al. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol 2019; 70: 489–525. [DOI] [PubMed] [Google Scholar]
- 3. Yu Y, Jia T, Chen X. The 'how' and 'where' of plant microRNAs. New Phytol 2017; 216: 1002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Secco D, Jabnoune M, Walker Het al. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 2013; 25: 4285–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiao Y, Wang Y, Xue Det al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 2010; 42: 541–4. [DOI] [PubMed] [Google Scholar]
- 6. Zhang YC, Yu Y, Wang CYet al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 2013; 31: 848–52. [DOI] [PubMed] [Google Scholar]
- 7. Miura K, Ikeda M, Matsubara Aet al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 2010; 42: 545–9. [DOI] [PubMed] [Google Scholar]
- 8. Dai Z, Wang J, Yang Xet al. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3.8 pathway in rice. J Exp Bot 2018; 69: 5117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H, Zhang J, Yan Jet al. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc Natl Acad Sci USA 2017; 114: 5277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot 2015; 66: 6093–107. [DOI] [PubMed] [Google Scholar]
- 11. Tsukaya H. Yield increase: GRFs provide the key. Nat plants 2015; 2: 15210. [DOI] [PubMed] [Google Scholar]
- 12. Che R, Tong H, Shi Bet al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat plants 2015; 2: 15195. [DOI] [PubMed] [Google Scholar]
- 13. Duan P, Ni S, Wang Jet al. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat plants 2015; 2: 15203. [DOI] [PubMed] [Google Scholar]
- 14. Li S, Tian Y, Wu Ket al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 2018; 560: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao F, Wang K, Liu Yet al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat plants 2015; 2: 15196. [DOI] [PubMed] [Google Scholar]
- 16. Van Daele I, Gonzalez N, Vercauteren Iet al. A comparative study of seed yield parameters in Arabidopsis thaliana mutants and transgenics. Plant Biotechnol J 2012; 10: 488–500. [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez RE, Mecchia MA, Debernardi JMet al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010; 137: 103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L, Gu X, Xu Det al. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 2011; 62: 761–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim JS, Mizoi J, Kidokoro Set al. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stressresponsive genes including DREB2A. Plant Cell 2012; 24: 3393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Omidbakhshfard MA, Proost S, Fujikura Uet al. Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Mol Plant 2015; 8: 998–1010. [DOI] [PubMed] [Google Scholar]
- 21. Liu D, Song Y, Chen Zet al. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 2009; 136: 223–36. [DOI] [PubMed] [Google Scholar]
- 22. Shriram V, Kumar V, Devarumath RMet al. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci 2016; 7: 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo Z, Kuang Z, Wang Yet al.. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res 2019; doi:10.1093/nar/gkz894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuang Z, Wang Y, Li Let al. miRDeep-P2: accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics (Oxford, England) 2018; 35: 2521–2. [DOI] [PubMed] [Google Scholar]
- 25. Perchlik M, Tegeder M. Improving plant pitrogen use efficiency through alteration of amino acid transport processes. Plant Physiol 2017; 175: 235–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taylor MR, Reinders A, Ward JM. Transport function of rice amino acid permeases (AAPs). Plant Cell Physiol 2015; 56: 1355–63. [DOI] [PubMed] [Google Scholar]
- 27. Debernardi JM, Rodriguez RE, Mecchia MAet al. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 2012; 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao C, Wang D, He Ret al. Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol J 2019; doi:10.1111/pbi.13214. [DOI] [PMC free article] [PubMed]
- 29. Hu J, Wang Y, Fang Yet al. A rare allele of GS2 enhances grain size and grain yield in Rice. Mol Plant 2015; 8: 1455–65. [DOI] [PubMed] [Google Scholar]
- 30. Zhang H, Zhang J, Wei Pet al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 2014; 12: 797–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.