Abstract
Skeletal muscle is one of the most important organs of the animal body. Long noncoding RNAs play a crucial role in the regulation of skeletal muscle development via several mechanisms. We recently identified obesity-related lncRNA (lnc-ORA) in a search for long noncoding RNAs that influence adipogenesis, finding it impacted adipocyte differentiation by regulating the PI3K/protein kinase B/mammalian target of rapamycin pathway. However, whether lnc-ORA has additional roles, specifically in skeletal muscle myogenesis, is not known. Here, we found that lnc-ORA was significantly differentially expressed with age in mouse skeletal muscle tissue and predominantly located in the cytoplasm. Overexpression of lnc-ORA promoted C2C12 myoblast proliferation and inhibited myoblast differentiation. In contrast, lnc-ORA knockdown repressed myoblast proliferation and facilitated myoblast differentiation. Interestingly, silencing of lnc-ORA rescued dexamethasone-induced muscle atrophy in vitro. Furthermore, adeno-associated virus 9–mediated overexpression of lnc-ORA decreased muscle mass and the cross-sectional area of muscle fiber by upregulating the levels of muscle atrophy–related genes and downregulating the levels of myogenic differentiation–related genes in vivo. Mechanistically, lnc-ORA inhibited skeletal muscle myogenesis by acting as a sponge of miR-532-3p, which targets the phosphatase and tensin homolog gene; the resultant changes in phosphatase and tensin homolog suppressed the PI3K/protein kinase B signaling pathway. In addition, lnc-ORA interacted with insulin-like growth factor 2 mRNA-binding protein 2 and reduced the stability of myogenesis genes, such as myogenic differentiation 1 and myosin heavy chain. Collectively, these findings indicate that lnc-ORA could be a novel underlying regulator of skeletal muscle development.
Keywords: skeletal muscle, myogenesis, lnc-ORA, miR-532-3p, insulin-like growth factor 2 mRNA-binding protein 2, PTEN/PI3K/AKT signaling pathway
Abbreviations: AAV, adeno-associated virus; Dex, dexamethasone; EdU, 5-ethynyl-20-deoxyuridine; GAS, gastrocnemius; GO, Gene Ontology; IGF2BP2, insulin-like growth factor 2 mRNA-binding protein 2; KEGG, Kyoto Encyclopedia of Genes and Genomes; lnc-ORA, obesity-related lncRNA; lncRNAs, long noncoding RNAs; MAFbx, muscle atrophy F-box; MRFs, myogenic regulatory factors; MuRF1, muscle RING finger 1; MyHC, myosin heavy chain; MyoG, myogenin; MyoD, myogenic differentiation 1; PTEN, phosphatase and tensin homolog; RIP, RNA immunoprecipitation; silnc-ORA, siRNA-lnc-ORA
Skeletal muscle development is regulated by a series of myogenic regulatory factors (MRFs) (1). The MRF family plays a crucial positive role in skeletal muscle myogenic determination and differentiation during embryogenesis and postnatal myogenesis and includes myogenic differentiation 1 (MyoD), myogenic factor 5, myogenin (MyoG), and myogenic regulatory factor 4 (2). Once MRFs have been activated, many myogenic transcription factors form obligate heterodimers with their coregulators to activate the myoblast differentiation program by regulating the transcription of many genes, including coding and noncoding genes. In addition, myoblast proliferation leads to an increase in the number of nuclei, contributing to muscle growth in some forms of muscle hypertrophy in adults.
Skeletal muscle atrophy is controlled by the balance between the protein degradation rate and protein synthesis rate and induced by various stressors, including starvation, denervation, mechanical unloading, inflammation, and aging (3). This balance reflects the physiological condition of the muscle fiber, and breakage results in muscular dystrophy (4). Two critical protein degradation pathways, the autophagic–lysosomal and ubiquitin–proteasome systems, are activated during skeletal muscle atrophy (5, 6). These pathways include many atrophy-related genes, which are regulated by specific transcription factors. Muscle RING finger 1 (MuRF1) and muscle atrophy F-box (MAFbx) are atrophy markers, which represent two of many E3 ubiquitin ligases that are mostly expressed in skeletal muscle. Knockdown of MAFbx prevents skeletal muscle loss during fasting, and the absence of MuRF1 alleviates dexamethasone (Dex)-induced muscle atrophy in mice (7). The PI3K/protein kinase B (AKT) signaling pathway is one of the signaling pathways that regulate mammalian skeletal muscle atrophy (8).
The animal genome contains abundant noncoding RNAs, which serve as regulators of gene expression at the transcriptional, translational, and epigenetic levels. Noncoding RNAs regulate various muscle biological processes (9, 10, 11). Recent studies have confirmed that functional long noncoding RNAs (lncRNAs) are involved in skeletal muscle development, including skeletal muscle cell proliferation, differentiation, injury, atrophy, and regeneration by chromatin remodeling, transcription regulation, and microRNA sponge absorption (12, 13, 14, 15, 16, 17, 18). Although the effects of these lncRNAs in skeletal muscle myogenesis have been partially characterized, the function and regulatory mechanism of obesity-related lncRNA (lnc-ORA) in this process remains elusive.
In the present study, we found that lnc-ORA was significantly differentially expressed in skeletal muscle between two important developmental stages. Furthermore, the results indicated that lnc-ORA promoted myoblast proliferation, inhibited myoblast differentiation, and induced muscle atrophy in vitro. Moreover, overexpression of lnc-ORA reduced muscle mass and the cross-sectional area of muscle fibers by upregulating the levels of muscle atrophy–related genes and downregulating the levels of myoblast differentiation–related genes in vivo. Mechanistic investigations showed that lnc-ORA functioned as a sponge for miR-532-3p and insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), which activated phosphatase and tensin homolog (PTEN) and attenuated PI3K/AKT signaling, a critical pathway of myogenesis and muscle atrophy. Based on the aforementioned results, our findings provide a novel strategy for the regulation of skeletal muscle development.
Results
Lnc-ORA is a potential regulator of skeletal muscle development
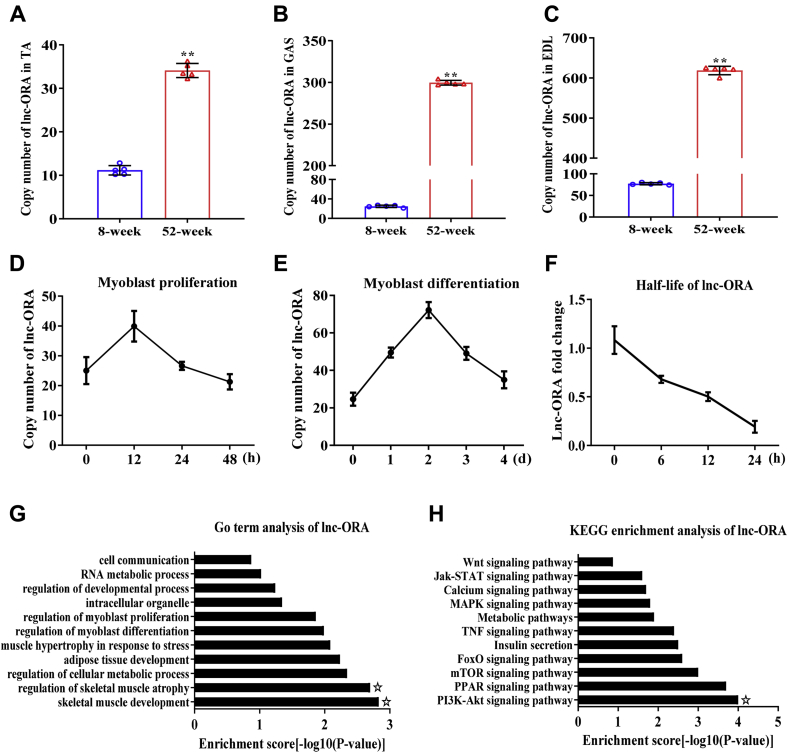
To confirm whether lnc-ORA is associated with skeletal muscle development, the absolute expression of lnc-ORA in the tibialis anterior, gastrocnemius (GAS), and extensor digitorum longus muscles from 8-week-old and 52-week-old mice was examined. The results showed that the levels of lnc-ORA were much higher in 52-week-old mice than in 8-week-old mice (Fig. 1, A–C). Furthermore, the level of lnc-ORA was the highest at 12 h and then gradually decreased from 24 to 48 h during myoblast proliferation (Fig. 1D). The level of lnc-ORA gradually increased during the early stage and then decreased in the late stage of C2C12 cell differentiation (Fig. 1E), indicating that lnc-ORA was involved in myoblast proliferation and differentiation during muscle formation. To confirm the stability of lnc-ORA in C2C12 cells, a half-life experiment was performed. The results showed that the half-life of lnc-ORA was approximately 12 h, indicating that lnc-ORA is stably expressed in C2C12 cells (Fig. 1F). In addition, based on lnc-ORA–targeted genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed. GO term analysis revealed that lnc-ORA participated in the regulation of skeletal muscle development, muscle atrophy, cellular metabolic process, muscle hypertrophy, especially myoblast proliferation and differentiation (Fig. 1G). KEGG analysis indicated that lnc-ORA could regulate biological processes through the PI3K/AKT, peroxisome proliferator–activated receptors, mammalian target of rapamycin, and forkhead box O signaling pathway (Fig. 1H). Collectively, these results imply that lnc-ORA is a potential regulator of skeletal muscle development, partly through the PI3K/AKT signaling pathway.
Figure 1.
Lnc-ORA is a regulator of skeletal muscle development.A, copy number of lnc-ORA in the TA of 8-week-old and 52-week-old mice (n = 5). B, copy number of lnc-ORA in the GAS of 8-week-old and 52-week-old mice (n = 5). C, copy number of lnc-ORA in the EDL of 8-week-old and 52-week-old mice (n = 5). D, copy number of lnc-ORA during myoblast proliferation (n = 5). E, copy number of lnc-ORA during myoblast differentiation (n = 5). F, half-life of lnc-ORA in myoblasts (n = 5). G, GO term analysis of the lnc-ORA. H, KEGG enrichment analysis of lnc-ORA. Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. EDL, extensor digitorum longus; GAS, gastrocnemius; GO, Gene Ontology; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; TA, tibialis anterior.
Overexpression of lnc-ORA promotes myoblast proliferation, whereas knockdown of lnc-ORA inhibits myoblast proliferation
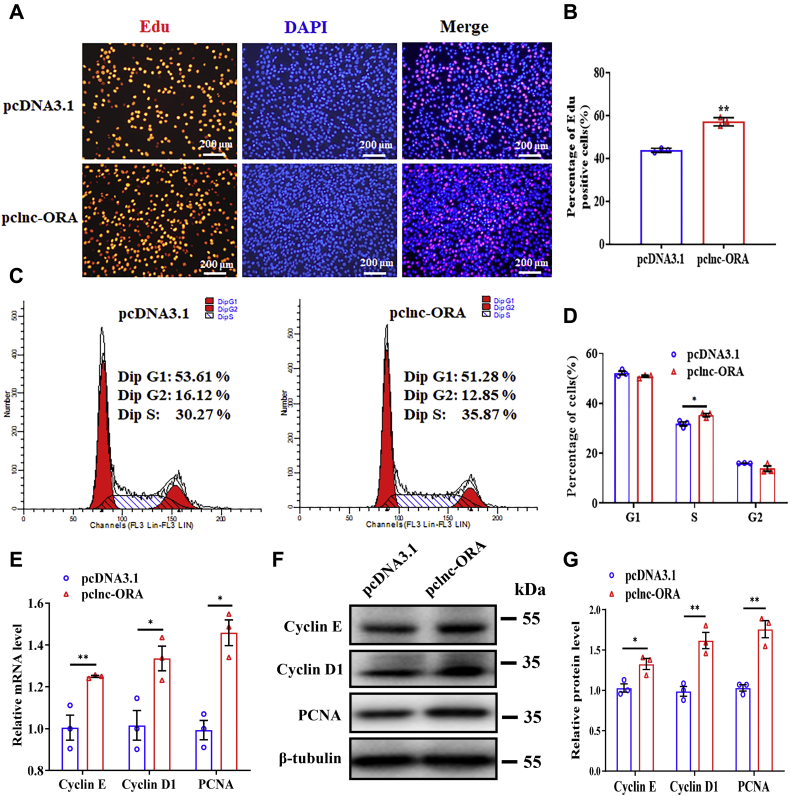
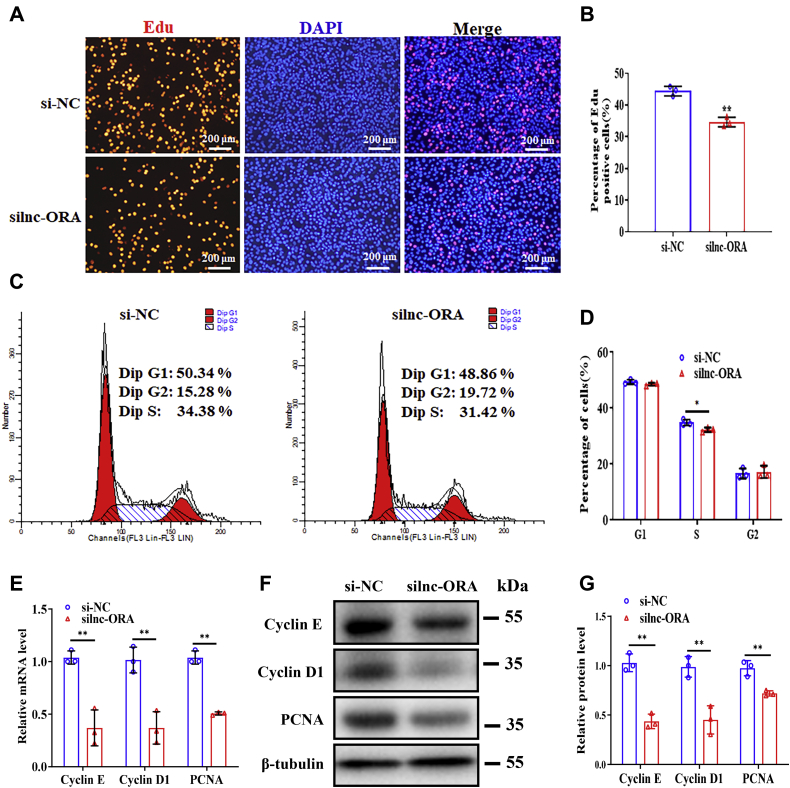
To investigate the role of lnc-ORA in myoblast proliferation, lnc-ORA overexpression and knockdown experiments were carried out in proliferating C2C12 cells. The results showed that overexpression and knockdown of lnc-ORA worked well (Fig. S1, A and B). Overexpression of lnc-ORA increased the number of 5-ethynyl-20-deoxyuridine (EdU)-positive cells (Fig. 2, A and B). A flow cytometry analysis also indicated that overexpression of lnc-ORA increased the number of cells that progressed to S phase (Fig. 2, C and D). In addition, cell count assay showed that overexpression of lnc-ORA increased the total cell number (Fig. S1C). Moreover, overexpression of lnc-ORA increased the mRNA and protein levels of proliferation-related genes, including cyclin E, cyclin D1, and proliferating cell nuclear antigen (Fig. 2, E–G). Furthermore, knockdown of lnc-ORA decreased the number of EdU-positive cells (Fig. 3, A and B), the total cell number (Fig. S1D), the number of cells in S phase (Fig. 3, C and D), and the mRNA and protein levels of proliferation-related genes (Fig. 3, E–G). Taken together, these results indicate that lnc-ORA promotes myoblast proliferation.
Figure 2.
Overexpression of lnc-ORA promotes myoblast proliferation.A, EdU and DAPI (nuclei) staining analysis 24 h after transfection of pclnc-ORA and pcDNA3.1 empty plasmid in proliferating myoblasts (n = 3). The scale bar represents 200 μm. B, percentage of EdU-positive cells/total cells (n = 3). C, flow cytometry analysis 24 h after transfection of pclnc-ORA and pcDNA3.1 empty plasmid in proliferating myoblasts (n = 3). D, statistical results of flow cytometry (n = 3). E, mRNA levels of cyclin E, cyclin D1, and PCNA 24 h after overexpression of lnc-ORA (n = 3). F, Western blot detection of cyclin E, cyclin D1, and PCNA (n = 3). G, quantitation of the protein level in F (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-20-deoxyuridine; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; pclnc-ORA, pcDNA3.1-lnc-ORA; PCNA, proliferating cell nuclear antigen.
Figure 3.
Knockdown of lnc-ORA inhibits myoblast proliferation.A, EdU and DAPI (nuclei) staining analysis 24 h after transfection of silnc-ORA and si-NC in proliferating myoblasts (n = 3). The scale bar represents 200 μm. B, percentage of EdU-positive cells/total cells (n = 3). C, flow cytometry analysis 24 h after transfection of silnc-ORA and si-NC in proliferating myoblasts (n = 3). D, statistical results of flow cytometry (n = 3). E, mRNA levels of cyclin E, cyclin D1, and PCNA 24 h after knockdown of lnc-ORA (n = 3). F, Western blot detection of cyclin E, cyclin D1, and PCNA (n = 3). G, quantitation of protein level in F (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-20-deoxyuridine; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; PCNA, proliferating cell nuclear antigen; silnc-ORA, siRNA-lnc-ORA.
Overexpression of lnc-ORA inhibits myogenic differentiation, whereas knockdown of lnc-ORA promotes myogenic differentiation
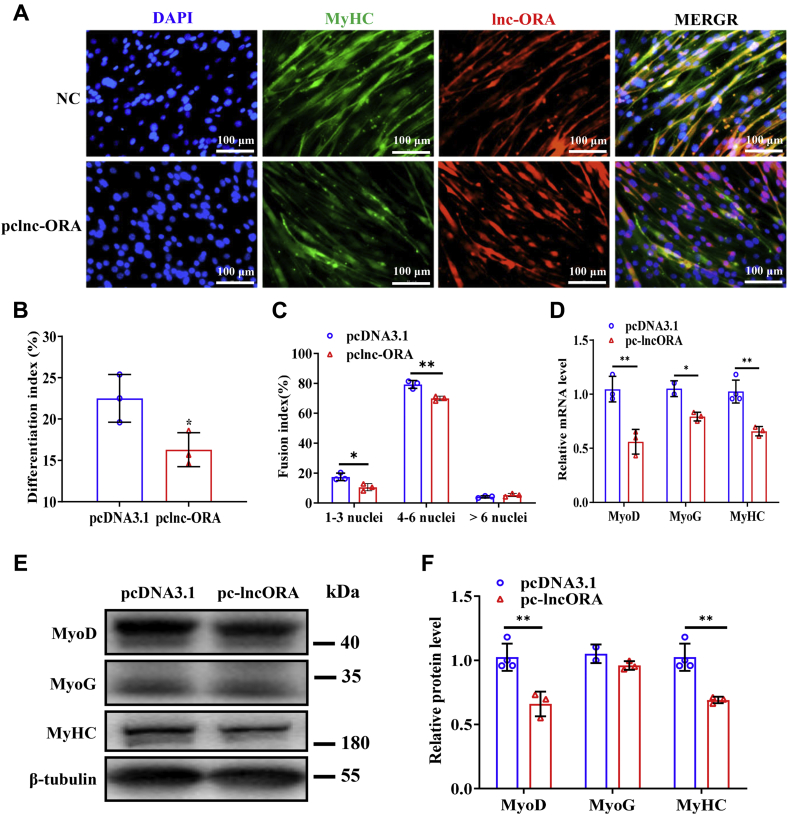
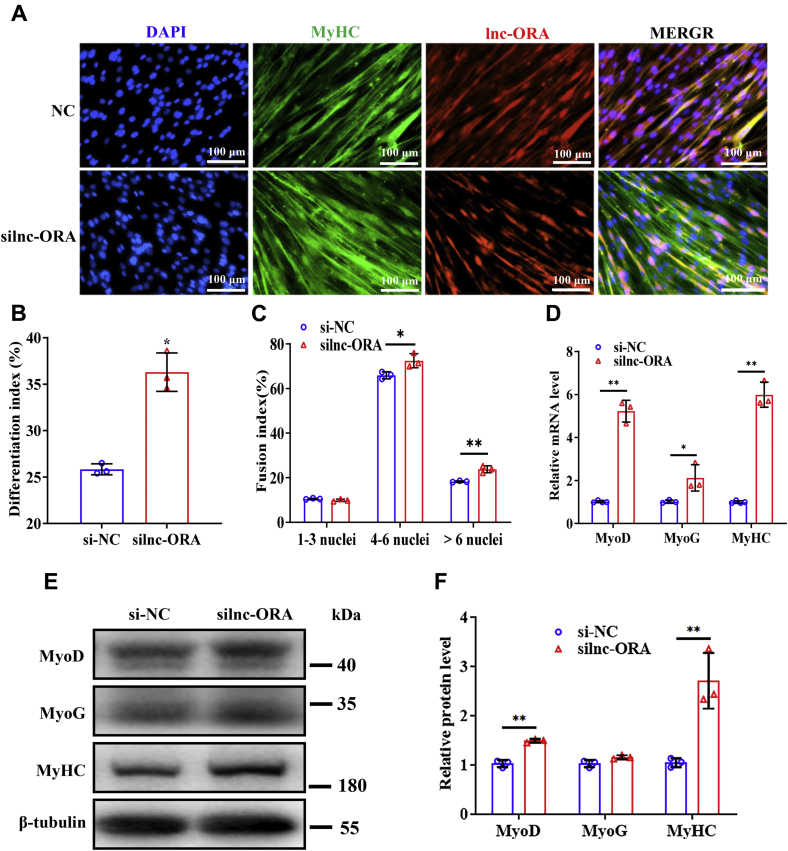
To verify the effect of lnc-ORA on myogenic differentiation, overexpression and knockdown experiments were performed during C2C12 cell differentiation. C2C12 cells were induced by differentiation culture medium (Fig. S2A). After myoblasts were transfected with pcDNA3.1-lnc-ORA or siRNA-lnc-ORA (silnc-ORA) vector, the levels of lnc-ORA markedly changed after myogenic induction (Fig. S2, B and C), showing that overexpression and knockdown of lnc-ORA worked well. Overexpression of lnc-ORA decreased the number of myosin heavy chain (MyHC)-positive cells, the differentiation index, and the multinuclear fusion index (Fig. 4, A–C). Moreover, overexpression of lnc-ORA downregulated the mRNA and protein levels of myogenic markers, including MyoD, MyoG, and MyHC (Fig. 4, D–F). Conversely, knockdown of lnc-ORA increased the number of MyHC-positive cells and the differentiation index, as well as the multinuclear fusion index at day 4 after myogenic induction (Fig. 5, A–C). Knockdown of lnc-ORA upregulated the mRNA and protein levels of these markers (Fig. 5, D–F). Taken together, these results demonstrate that lnc-ORA inhibits myogenic differentiation of myoblasts.
Figure 4.
Overexpression of lnc-ORA inhibits myogenic differentiation.A, immunofluorescent staining of MyHC and DAPI (nuclei) on day 4 of differentiation after overexpression of lnc-ORA (n = 3). The scale bar represents 100 μm. B, statistical analysis of the differentiation index in A (n = 3). C, statistical analysis of the fusion index in A (n = 3). D, mRNA levels of myogenic genes on day 4 of differentiation after overexpression of lnc-ORA (n = 3). E, Western blot detection of MyoD, MyoG, and MyHC (n = 3). F, quantitation of protein levels in E (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; MyHC, myosin heavy chain; MyoD, myogenic differentiation 1; MyoG, myogenin.
Figure 5.
Knockdown of lnc-ORA promotes myogenic differentiation.A, immunofluorescent staining of MyHC and DAPI (nuclei) on day 4 of differentiation after knockdown of lnc-ORA (n = 3). The scale bar represents 100 μm. B, statistical analysis of the differentiation index in A (n = 3). C, statistical analysis of the fusion index in A (n = 3). D, mRNA levels of myogenic genes on day 4 of differentiation after knockdown of lnc-ORA (n = 3). E, Western blot detection of MyoD, MyoG, and MyHC (n = 3). F, quantitation of protein levels in E (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; MyHC, myosin heavy chain; MyoD, myogenic differentiation 1; MyoG, myogenin.
Knockdown of lnc-ORA rescues Dex-induced muscle atrophy in vitro
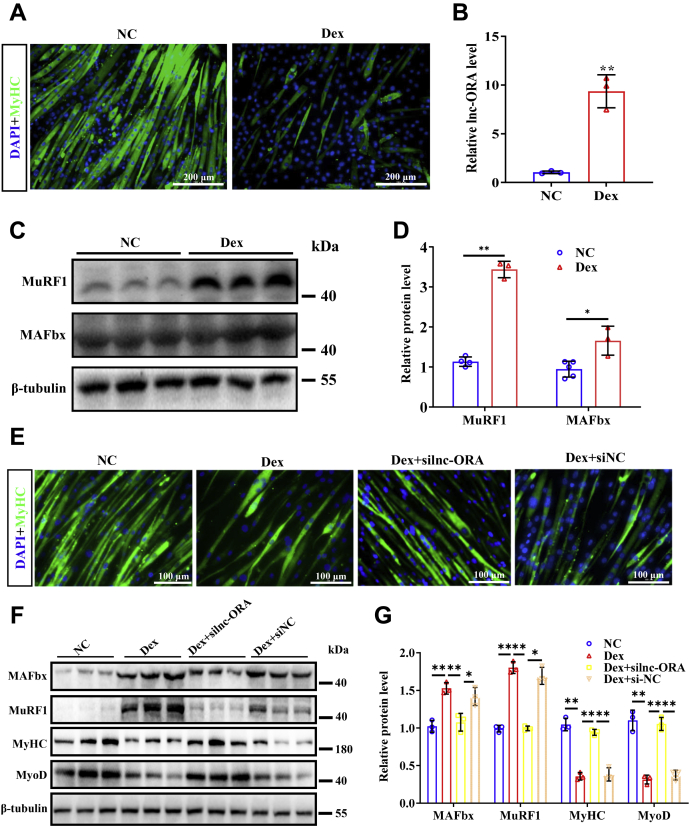
To examine whether lnc-ORA could influence muscle atrophy, a Dex-induced muscle atrophy model was used in C2C12 cells (Fig. 6A). The level of lnc-ORA was significantly upregulated by the Dex treatment (Fig. 6B), and an increase in the protein levels of MAFbx and MuRF1 also occurred (Fig. 6, C and D). Furthermore, knockdown of lnc-ORA markedly rescued Dex-induced muscle atrophy, as shown by MyHC staining (Fig. 6E). Knockdown of lnc-ORA decreased the levels of MAFbx and MuRF1 and increased the levels of myogenic differentiation factors (Fig. 6, F and G). Therefore, knockdown of lnc-ORA rescued Dex-induced muscle atrophy in vitro, suggesting that lnc-ORA could be a potential therapeutic target for treating muscle atrophy.
Figure 6.
Knockdown of lnc-ORA rescues Dex-induced muscle atrophy in vitro.A, morphology of C2C12 cells induced by Dex. The scale bar represents 200 μm. B, level of lnc-ORA in atrophy myotube. C, protein levels of MuRF1 and MAFbx (n = 3). D, quantitation of protein levels in C (n = 3). E, immunofluorescence staining of MyHC and DAPI (nuclei) after cotreatment of silnc-ORA and Dex. The scale bar represents 100 μm. F, Western blot detection of MuRF1, MAFbx, MyHC, and MyoD after cotreatment with silnc-ORA and Dex (n = 3). G, quantitation of protein levels in F (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole; Dex, dexamethasone; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; MAFbx, muscle atrophy F-box; MuRF1, muscle RING finger 1; MyHC, myosin heavy chain; MyoD, myogenic differentiation 1; silnc-ORA, siRNA-lnc-ORA.
Overexpression of lnc-ORA decreases muscle mass and induces muscle atrophy in vivo
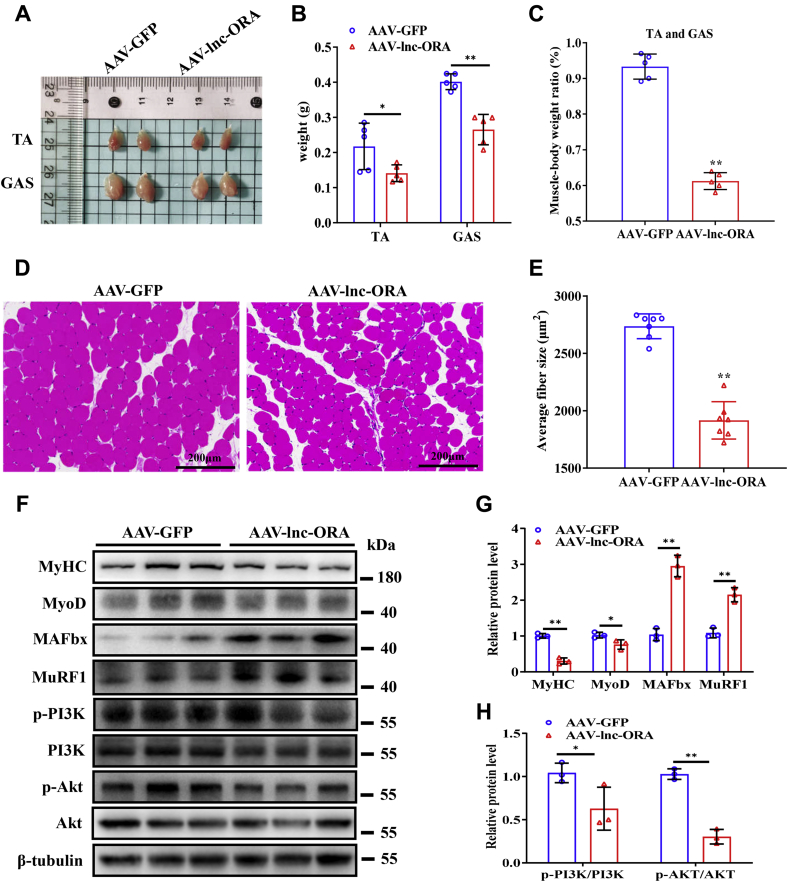
To investigate the role of lnc-ORA in the regulation of muscle development in vivo, 10-week-old mice were injected with adeno-associated virus-GFP (AAV-GFP) or AAV-lnc-ORA overexpression virus and sacrificed after 8 weeks. Compared with the AAV-GFP control group, the AAV treatment group showed decreased size and mass of the tibialis anterior and GAS muscles (Fig. 7, A and B). Meanwhile, the muscle percentage of AAV-lnc-ORA–injected mice was lower (Fig. 7C). H&E staining showed that the cross-sectional areas of GAS fibers were dramatically smaller in the AAV-lnc-ORA group than in the AAV-GFP group (Fig. 7, D and E). Overexpression of lnc-ORA significantly decreased the expression levels of MyHC and MyoD but increased the expression levels of MAFbx and MuRF1 (Fig. 7, F and G). The PI3K/AKT signaling pathway was inhibited in the GAS muscles of AAV-lnc-ORA mice (Fig. 7, F and H). Together, these results suggest that overexpression of lnc-ORA decreases muscle mass and induces muscle atrophy in vivo.
Figure 7.
Overexpression of lnc-ORA decreases muscle mass and induces muscle atrophy in vivo.A, images of mouse skeletal muscles. B, statistical analysis of muscles mass (n = 5). C, percentage of skeletal muscle (TA and GAS) (n = 5). D, H&E staining images of GAS cross section. The scale bar represents 200 μm. E, statistics of fiber area in D (n = 5). F, Western blot detection of MyHC, MyoD, MAFbx, MuRF, p-PI3K, PI3K, p-AKT, and AKT in the GAS (n = 3). G and H, quantitation of protein level in F (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. AKT, protein kinase B; GAS, gastrocnemius; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; MAFbx, muscle atrophy F-box; MuRF, Muscle RING finger; MyHC, myosin heavy chain; MyoD, myogenic differentiation 1; p-AKT, phosphorylated AKT; p-PI3K, phosphorylated PI3K; TA, tibialis anterior.
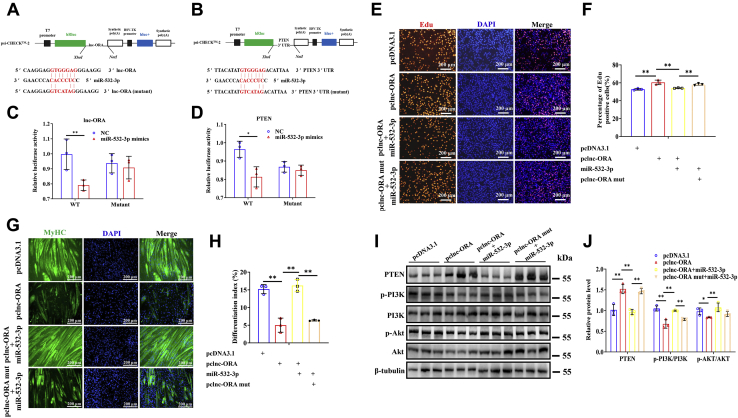
Lnc-ORA sponges miR-532-3p to control myogenesis through the PTEN/PI3K/AKT signaling pathway
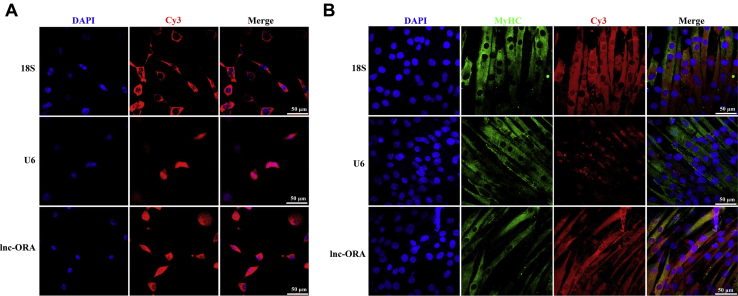
To investigate the molecular mechanism of lnc-ORA in myogenesis, FISH experiments were performed to confirm the subcellular locations of lnc-ORA in myoblasts and myotubes. The results showed that lnc-ORA was predominantly expressed in the cytoplasm of both myoblasts and myotubes (Fig. 8, A and B). This finding indicates that lnc-ORA plays a role in the post-transcriptional regulation mechanism in the cytoplasm. Moreover, we predicted that miR-532-3p with a conserved seed sequence could be adsorbed by lnc-ORA (Fig. S3, A and B) and that PTEN acts as a target of miR-532-3p (Fig. S3C). To confirm this hypothesis, we constructed wildtype and mutant dual luciferase reporters, psi-CHECK 2.0-lnc-ORA or psi-CHECK 2.0-PTEN 3’UTR (Fig. 9, A and B). The dual-luciferase reporter assay revealed that miR-532-3p bound to lnc-ORA transcripts and the PTEN 3′UTR (Fig. 9, C and D). Furthermore, significantly enriched miR-532-3p and lnc-ORA were measured through argonaute 2 RNA immunoprecipitation (RIP) assay (Fig. S3D). Moreover, lnc-ORA was significantly enriched by biotin-labeled miR-532-3p compared with the control or mutated miR-532-3p (Fig. S3E). We have also detected the absolute copy number of miR-532-3p under the different conditions. The results showed that the absolute copy number of miR-532-3p exhibited opposite trends as that of lnc-ORA under different conditions (Fig. S4, A–E). Based on the aforementioned results, we speculated that lnc-ORA acted as a competing endogenous RNA to sponge miR-532-3p. Furthermore, miR-532-3p mimics promoted myogenic differentiation, as shown by immunofluorescence staining of MyHC-positive myotubes (Fig. S5, A and B). Besides, rescue experiments indicated that miR-532-3p attenuated the positive effect of lnc-ORA on myoblast proliferation (Fig. 9, E and F), whereas it rescued the repressive effect of lnc-ORA on myoblast differentiation (Fig. 9, G and H). Moreover, miR-532-3p showed the opposite effect against lnc-ORA on the protein levels of PTEN, phosphorylated PI3K (p-PI3K), and phosphorylated AKT (p-AKT) (Fig. 9, I and J). In contrast, lnc-ORA knockdown reduced the protein level of PTEN and increased the protein levels of p-PI3K and p-AKT (Fig. S6). Therefore, lnc-ORA inhibited myogenic differentiation and promoted muscle atrophy through absorption of miR-532-3p via PTEN/PI3K/AKT signaling pathway.
Figure 8.
Subcellular localization of lnc-ORA in proliferating and differentiating myoblasts.A, subcellular localization of lnc-ORA by FISH in myoblasts. Special FISH probes against lnc-ORA, U6, and 18S were modified by Cy3 in red. 18S is a cytoplasmic marker, and U6 is a nuclear marker. The nucleus was stained by DAPI in blue. The scale bar represents 50 μm. B, subcellular localization of lnc-ORA by FISH in myotubes. Special FISH probes against lnc-ORA, U6, and 18S were modified by Cy3 in red. 18S is a cytoplasmic marker, and U6 is a nuclear marker. The nucleus was stained by DAPI in blue. The scale bar represents 50 μm. DAPI, 4′,6-diamidino-2-phenylindole; lnc, long noncoding; lnc-ORA, obesity-related lncRNA.
Figure 9.
Lnc-ORA functions as a ceRNA for miR-532-3p to control myogenesis.A and B, schematic of the double luciferase assay vector. C and D, analysis of the luciferase reporter assay (n = 3). E, EdU and DAPI (nuclei) staining analysis. The scale bar represents 200 μm. F, statistical analysis in E (n = 3). G, immunofluorescent staining of MyHC analysis. The scale bar represents 200 μm. H, statistical analysis in G (n = 3). I, Western blot detection of PTEN and myogenic factors (n = 3). J, statistical analysis in I (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. ceRNA, competing endogenous RNA; DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-20-deoxyuridine; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; MyHC, myosin heavy chain; PTEN, phosphatase and tensin homolog.
Lnc-ORA contributes to myogenesis by competitively binding IGF2BP2
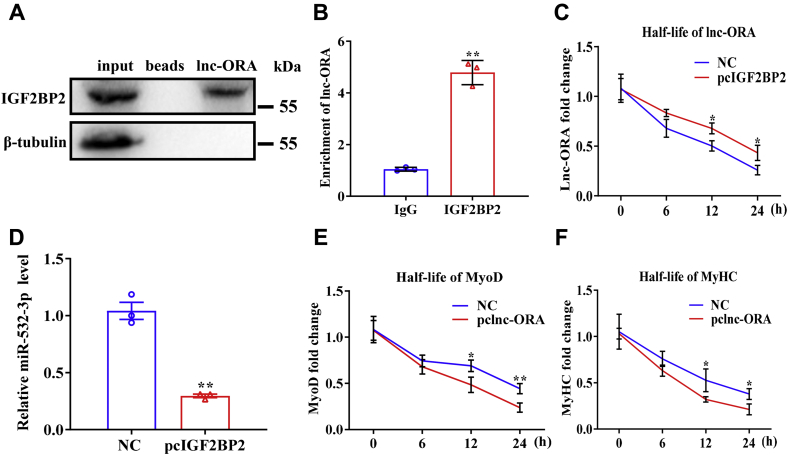
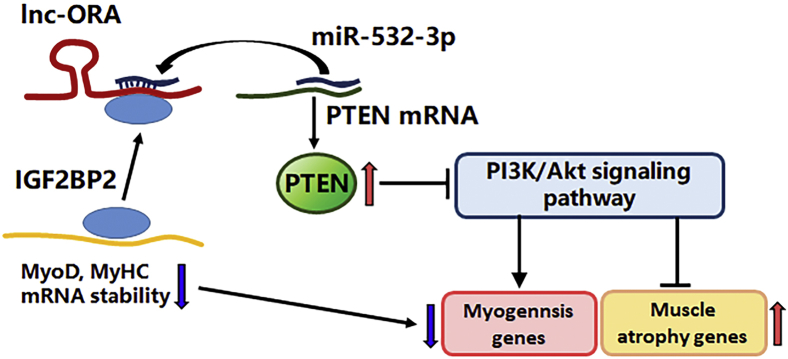
To further explore the mechanism by which lnc-ORA regulates myogenesis, we used a biotinylated lnc-ORA probe to perform an RNA pull-down assay, followed by mass spectrometry. Through this analysis, we identified IGF2BP2, an RNA-binding protein, as being bound to lnc-ORA, and we confirmed this interaction by Western blotting (Fig. 10A). Next, RIP was performed in C2C12 cells using an IGF2BP2 antibody. Lnc-ORA was present in the IGF2BP2 RIP sample at a much higher level than in the control IgG RIP sample (Fig. 10B). It has been demonstrated that IGF2BP2 could promote RNA stability. We then used this half-life assay to explore the RNA stability of lnc-ORA. The results showed that the interaction of lnc-ORA and IGFBP2 increased lnc-ORA stability and the ability to sponge miR-532-3p (Fig. 10, C and D). Moreover, lnc-ORA directly bound to IGF2BP2 and negatively regulated the IGF2BP2-mediated stability of myogenesis genes such as MyoD and MyHC (Fig. 10, E and F). Together, we used RNA pull-down experiments to identify IGF2BP2 as an important binding partner of lnc-ORA in the myogenesis process. Based on the aforementioned results, we suggest that lnc-ORA, acting as a miR-532-3p sponge and interacting with IGF2BP2 to control PTEN-mediated PI3K/AKT signaling, inhibits skeletal muscle myogenesis and induces muscle atrophy (Fig. 11).
Figure 10.
Lnc-ORA contributes to myogenesis by competitively binding IGF2BP2.A, Western blotting identified IGF2BP2 bound with lnc-ORA. B, RIP assay revealed that IGF2BP2 was bound with lnc-ORA (n = 3). C, lnc-ORA RNA stability assays in C2C12 myoblasts (n = 3). D, expression level of miR-532-3p when IGF2BP2 was overexpressed (n = 3). E and F, MyoD and MyHC RNA stability assays in C2C12 cells (n = 3). Data represent the mean ± SD. ∗p < 0.05; ∗∗p < 0.01. IGF2BP2, insulin-like growth factor 2 mRNA-binding protein 2; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; MyHC, myosin heavy chain; MyoD, myogenic differentiation 1; RIP, RNA immunoprecipitation.
Figure 11.
Molecular regulatory mechanism of lnc-ORA that inhibits skeletal muscle myogenesis and induces muscle atrophy. Lnc-ORA controls PTEN protein levels and further activates the PI3K/AKT signaling pathway by functioning as ceRNA to sponge miR-532-3p and interact with IGF2BP2. AKT, protein kinase B; ceRNA, competing endogenous RNA; IGF2BP2, insulin-like growth factor 2 mRNA-binding protein 2; lnc, long noncoding; lnc-ORA, obesity-related lncRNA; PTEN, phosphatase and tensin homolog.
Discussion
Skeletal muscle is closely associated with physiological function, muscle strength, and metabolic performance, which influence human chronic disease, quality of life, and animal meat production. Recently, several studies have indicated that lncRNAs are implicated in skeletal muscle formation. However, the effect and molecular mechanism of lnc-ORA on myoblast proliferation, differentiation, and muscle atrophy are unknown. In the present study, we found that lnc-ORA was differentially expressed in mouse skeletal muscle with age, functioning as a miR-532-3p sponge and interacting with IGF2BP2 to inhibit myogenesis and induce muscle atrophy, and it could be a novel target for the regulation of skeletal muscle development.
LncRNAs have been demonstrated to regulate myoblast proliferation and differentiation, such as Myoparr (19), Linc-smad7 (20), Lnc-31 (21), and Gm26917 (22). Our previous study revealed that knockdown of lnc-ORA inhibited preadipocyte proliferation and differentiation (23). In the current study, we found that overexpression of lnc-ORA promoted myoblast proliferation but inhibited myotube formation. Furthermore, its knockdown had the opposite effect, implying that lnc-ORA is a vital negative regulatory factor of skeletal muscle myogenesis.
An increasing number of studies have also indicated that lncRNAs, including lncIRS1 (24), lncMUMA (25), Atrolnc-1 (26), Pvt1 (27), and SMN-AS1 (28), play a significant role in the regulation of muscle atrophy. In the current study, the level of lnc-ORA markedly increased in a Dex-induced muscle atrophy model, and knockdown of lnc-ORA significantly rescued muscle atrophy by inhibiting MAFbx and MuRF1 and promoting myogenic differentiation factor expression. In addition, skeletal muscle atrophy is defined as a decline in skeletal muscle mass because of muscle cellular shrinkage (29, 30). Interestingly, we found that overexpression of lnc-ORA significantly decreased skeletal muscle mass and the cross-sectional area of muscle fibers in vivo. Therefore, it is suggested that lnc-ORA could be a promising therapeutic target for muscle atrophy.
LncRNAs not only regulate target genes at the transcriptional level in the nucleus but also participate at the post-transcriptional level in the cytoplasm, suggesting that the subcellular localization of lncRNAs is related to their regulatory mechanism (31, 32, 33). Here, we found that lnc-ORA is distributed in both the nucleus and cytoplasm of either myoblasts or myotubes, but the proportion in the cytoplasm is much greater than that in the nucleus, indicating that lnc-ORA may play an important role in post-transcriptional regulation. Generally, lncRNAs function by absorbing miRNAs to regulate the expression of target genes (34, 35, 36). Bioinformatics prediction showed that lnc-ORA acts as a competing endogenous RNA to absorb miRNA-532-3p, which could target PTEN. As an important factor of myogenic genes, PTEN could negatively regulate the PI3K signaling pathway, which has well-known anabolic effects to promote myoblast differentiation in skeletal muscles (37). Furthermore, inhibition of the PI3K signaling pathway could activate the transcription factor forkhead box O family (38, 39), which induces the expression of the MuRF-1 and MAFbx genes implicated in the induction of muscle atrophy (40, 41). Based on our results, we confirmed that lnc-ORA inhibited myogenic differentiation and aggravated muscle atrophy by sequestering miRNA-532-3p from inhibition of PTEN. In general, a single miRNA is able to recognize and inhibits a large number of target genes. Conversely, a single gene may be targeted by numerous miRNAs. Of course, lnc-ORA could be predicted to sequester other miRNAs in some biological processes, such as miR-149-5p, miR-320-5p, and miR-760-5p. Here, we confirmed that lnc-ORA acts as an miR-532-3p sponge in skeletal muscle myogenesis. IGF2BP2 could promote the stability and translation of RNAs (42). Moreover, IGF2BP targets 76 genes that are associated with muscle development (43). Consistent with the molecular mechanism of lncMYOD (44), we found that lnc-ORA competitively bound to IGF2BP2 and negatively regulated the IGF2BP2-mediated stability of myogenic genes such as MyoD and MyHC.
In conclusion, we found that lnc-ORA acts as a miR-532-3p sponge and interacts with IGF2BP2 to suppress the PI3K/AKT signaling pathway, resulting in the inhibition of myogenesis and induction of skeletal muscle atrophy. Based on our findings, we hypothesize that lnc-ORA could be a novel underlying regulator of skeletal muscle development.
Experimental procedures
Animals
C57/BL6J male mice were purchased from the Medical Laboratory Animal Center of Xi'an Jiaotong University (Xi'an, China). All animal experiments were approved by ethics committee of animal welfare and health of Northwest A&F University (NWAFU-314020038).
Cell culture, transfection, and RNA stability assay
The C2C12 mouse myoblast cell line was acquired from American Type Culture Collection. Cells were cultured with growth medium, which was composed of high-glucose Dulbecco's modified Eagle's medium and 10% fetal bovine serum (Gibco). After the cells reached 80 to 90% confluence, C2C12 cell differentiation was induced by differentiation medium consisting of Dulbecco's modified Eagle's medium and 2% horse serum (Gibco). To study myoblast proliferation, transfection experiments were performed when cells reached 40 to 50% confluence. Cells were transfected for studying myogenic differentiation when cells reached 70 to 80% density. All transfection procedures were performed with X-treme-GENE Transfection Reagent (Roche) in accordance with the manufacturer's instructions. The miR-532-3p mimics and inhibitor, silnc-ORA and siNC, were synthesized by RiboBio. The overexpression vectors of lnc-ORA and lnc-ORA mutant were constructed with the pcDNA3.1 plasmid. The empty pcDNA3.1 vector was used as a control plasmid. To construct Dex-induced muscle atrophy in vitro, well-differentiated C2C12 cells were treated with either 50 μM Dex (D4902; Sigma–Aldrich) or PBS (control), and then siRNA targeting lnc-ORA was transfected 36 h after treatment. The half-life of lnc-ORA was detected according to our previous report (45).
RNA-fluorescent in situ hybridization
In situ hybridization of lnc-ORA in myoblasts and myotubes was performed with a FISH kit (RiboBio). After washing three times with PBS, C2C12 cells were fixed with 4% paraformaldehyde for 30 min. Next, the cells were prehybridized for 30 min with prehybridization solution after permeabilization with 0.5% Triton X-100 buffer for 5 min. Then, the specific oligodeoxynucleotide probes of anti-18S RNA, anti-U6, and anti-lnc-ORA were hybridized at 37 °C overnight. Finally, the cells were stained with 4′,6-diamidino-2-phenylindole dye and photographed using a confocal laser-scanning microscope.
RNA isolation and relative quantitative real-time PCR
Total RNA from different cell and tissue samples was extracted by TRIzol reagent (TaKaRa) in accordance with the manufacturer's protocol. Complementary DNA synthesis was carried out by reverse transcription kits (TaKaRa). Quantitative real-time RT-PCR (qRT-PCR) was performed with a Bio-Rad iQTM5 system (Bio-Rad). For the miR-532-3p expression level, the specific primers of miR-532-3p and reference gene U6 (RiboBio) were used for reverse transcription. The 2−ΔΔCt method was used to analyze qRT-PCR data. Mouse GAPDH was used as a reference gene. The primer sequences of genes detected in this study are listed in Table S1.
Western blotting
Total protein of the cell or tissue samples was harvested using radioimmunoprecipitation lysis buffer after washing three times with PBS. Immunoblotting was carried out according to our previous method (10). The primary antibodies targeted the following proteins: MyHC (1:500; no. MAB4470; R and D Systems), MyoD (1:500; no. NB100-56511; Novus Biologicals), AKT (1:1000; no. 9272S; CST), p-AKT (Ser473) (1:2000; no. 4060S; CST), PI3K (p55α) (1:1000; no. 11889S; CST), p-PI3K p55 (Tyr199) (1:1000; no. 4228S; CST), β-tubulin (1:200; no. sc-58880; Santa Cruz Biotechnology), MyoG (1:1000; no. NB100-56510SS; Novus Biologicals), MuRF1 (1:200; no. sc-398608; Santa Cruz Biotechnology), MAFbx (1:200; no. sc-166806, Santa Cruz Biotechnology), cyclin E (1:100; no.sc-377100; Santa Cruz Biotechnology), cyclin D1 (1:100; no. sc-450; Santa Cruz Biotechnology), PTEN (1:500; no. sc-7974; Santa Cruz Biotechnology), IGF2BP2 (1:100; no. sc-377014; Santa Cruz Biotechnology), and proliferating cell nuclear antigen (1:100; no. sc-56; Santa Cruz Biotechnology). The secondary antibodies included goat anti-rabbit IgG (1:3000; no. BA1054; BosterBio) and goat antimouse IgG (1:3000; no. BA1050; BosterBio).
Overexpression of lnc-ORA by AAV infection in vivo
The recombinant AAV viruses (AAV9 serotype) expressing lnc-ORA or GFP were acquired by Hanbio (46). Ten-week-old C57/BL6J male mice with eight mice in each group were injected with 150 μl AAV virus including full-length lnc-ORA (AAV-lnc-ORA) or control (AAV-GFP) at 5 × 1012 vg/ml titers.
Frozen section and H&E staining
Frozen sectioning and H&E staining of GAS samples were performed according to our published method (47).
Flow cytometry, EdU, and cell counting kit-8 assays
Flow cytometry, EdU, and cell counting kit-8 experiments were performed according to our previous report (48). Briefly, after the C2C12 cell density reached 40% confluence, the cells were treated with pcDNA3.1-lnc-ORA or silnc-ORA as well as miR-532-3p mimics or miR-532-3p inhibitor. After treatment for 24 h, the cells were used to detect proliferation-related analysis.
Immunofluorescence assay
The immunofluorescence assay was performed as described in our previous procedure (49). Briefly, cells were fixed with 4% paraformaldehyde and blocked with 5% bovine serum albumin for 30 min. Then, the cells were incubated with MyHC primary antibody overnight at 4 °C. Next, the samples were incubated with secondary antibody for 1 h at 37 °C and 4′,6-diamidino-2-phenylindole for 10 min. The differentiation index was determined as the percentage of MyHC-positive nuclei among the total nuclei, and the myotube fusion index was determined as the distribution of the nucleus number among the total myotubes according to a previous report (50).
Bioinformatics analysis
miRbase (http://www.mirbase.org/) and TargetScan (http://www.targetscan.org/vert_71/) were used to predict target genes of miR-532-3p. GO analysis is a functional analysis for neighboring genes and coexpressed genes of lnc-ORA with GO categories. The GO categories are derived from GO (http://www.geneontology.org), which consists of three structured networks of defined terms that describe gene product attributes. The KEGG pathway enrichment analysis for neighboring genes and coexpressed genes of lnc-ORA is based on the latest KEGG database (https://www.genome.jp/kegg).
Dual-luciferase activity assay
The putative binding sites (wildtype and mutated) of miR-532-p within lnc-ORA and the 3′-UTR of PTEN were synthesized using General Biosystems and inserted into psiCHECK-2 vector (Promega). Dual-luciferase activity was analyzed according to our previous report (48). Briefly, human embryonic kidney 293T cells were transfected with miR-532-3p mimics or negative control mimics as well as psi-CHECK2 reporter wildtype or mutant vector. After transfection for 48 h, the relative luciferase activity was analyzed following the manufacturer's protocol (Promega).
Absolute quantitative real-time PCR
Absolute qRT-PCR was used to analyze copy number of lnc-ORA and miR-532-3p under different conditions. Briefly, T-vectors containing a fragment of lnc-ORA and miR-532-3p were constructed and diluted via titration to generate the standard curve. Total RNA (200 ng) from each sample was reverse transcribed, and complementary DNA was used as a template in qRT-PCR. The Ct values were used to calculate the copy numbers of lnc-ORA and miR-532-3p in each sample, according to the standard curve.
RNA pull-down assay
Linearizing DNA was biotin labeled and transcribed in vitro using Biotin RNA Labeling Mix and T7/SP6 RNA polymerase (Roche) and purified with the RNeasy Mini Kit (QIAGEN). One milligram of protein was incubated with 3 μg of biotinylated RNA for 1 h at room temperature. After that, 40 μl streptavidin-coupled beads were added to each reaction and incubated for 1 h at room temperature. Finally, the beads were washed in RIP buffer for five times, and the pulled-down proteins were used for Western blotting. For mass spectrometry, the pulled-down proteins in C2C12 cells were separated by 10% SDS-PAGE and then subjected to silver staining. The differentially expressed bands were excised and analyzed by mass spectrometry (Novogene).
miRNA pull-down assay
A miRNA pull-down assay was performed as previously reported (51). C2C12 cells were transfected with biotinylated miR-532-3p: wildtype miR-532-3p (miR-532-3p-Bio) or mutated miR-532-3p (G–C mutation in the lnc-ORA–binding sites, miR-532-3p-MutBio) and biotinylated control (negative control-Bio) when 70 to 80% of confluency was reached and induced into myogenic differentiation at full confluence. Six days after induction, cell lysates were collected and incubated with Dyna M-280 streptavidin magnetic beads (#11205D; Thermo Fisher Scientific). The product was then treated with RNase-free DNase I (Roche), and RNA was purified using an RNeasy Mini Kit (QIAGEN). The enrichment of lnc-ORA was detected through qRT-PCR.
RNA immunoprecipitation assay
Well-differentiated C2C12 cells were subjected to an RIP assay after 6 days of induction. Briefly, cells were collected, lysed in complete RIP buffer provided in the EZ-Magna RIP kit (Millipore), and then incubated with RIP buffer containing magnetic beads conjugated to anti-argonaute 2 or anti-IGF2BP2 antibody. Subsequently, the samples were digested with proteinase K, and then the RNA was purified from the precipitates. The concentration of RNA was determined using a NanoDrop system (Thermo Fisher Scientific). Finally, qRT-PCR was performed to detect the existence of miR-532-3p and lnc-ORA.
Statistical analysis
All replicate experiments (including cell-based and mouse-based experiments) were biological replicates that were repeated at least three times. All analyses involved the use of SPSS, version 23 (SPSS, Inc). All data are represented as the mean ± SD. Comparisons of two groups were determined by Student's t test, and comparisons of multiple groups were determined by one-way ANOVA with Tukey's post hoc test. The assumption of normality was tested by the Shapiro−Wilks test. All statistical tests were two tailed, and p < 0.05 was considered statistically significant.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Supporting information
This article contains supporting information.
Author contributions
R. C. and Q. Z. data curation; R. C., Y. W., and W. Y. investigation; R. Z. methodology; R. C. writing original draft; R. C. and W. P. writing - review and editing; W. P. project administration; R. Z. conceptualization; W. P. funding acquisition; and W. P. supervised the project.
Funding and additional information
This work was supported by grants from the National Natural Science Foundation (31872979 and 31572366) and the National Key Research and Development Program of China (2017YFD0502002).
Edited by Qi-Qun Tang
Supporting information
References
- 1.Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell. Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M., Rigby P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Schiaffino S., Dyar K.A., Ciciliot S., Blaauw B., Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 4.Bonaldo P., Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackman R.W., Cornwell E.W., Wu C.L., Kandarian S.C. Nuclear factor-kappaB signalling and transcriptional regulation in skeletal muscle atrophy. Exp. Physiol. 2013;98:19–24. doi: 10.1113/expphysiol.2011.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiao J., Demontis F. Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr. Opin. Pharmacol. 2017;34:1–6. doi: 10.1016/j.coph.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Bodine S.C., Baehr L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 2014;307:E469–484. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wimmer R.J., Russell S.J., Schneider M.F. Green tea component EGCG, insulin and IGF-1 promote nuclear efflux of atrophy-associated transcription factor Foxo1 in skeletal muscle fibers. J. Nutr. Biochem. 2015;26:1559–1567. doi: 10.1016/j.jnutbio.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jandura A., Krause H.M. The new RNA world: Growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33:665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Cai R., Sun Y.M., Qimuge N.R., Wang G.Q., Wang Y., Chu G.Y., Yu T.Y., Yang G.S., Pang W.J. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:420–432. doi: 10.1016/j.bbalip.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Pang W.J., Lin L.G., Xiong Y., Wei N., Wang Y., Shen Q.W., Yang G.S. Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation. J. Cell. Biochem. 2013;114:2500–2512. doi: 10.1002/jcb.24595. [DOI] [PubMed] [Google Scholar]
- 12.Wang S.S., Jin J.J., Xu Z.Y., Zuo B. Functions and regulatory mechanisms of lncRNAs in skeletal myogenesis, muscle disease and meat production. Cells. 2019;8:1107. doi: 10.3390/cells8091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng T., Liu Y., Xu Y., Jiang Y., Zhang N., Wang Z., Carmichael G.G., Taylor H.S., Li D., Huang Y. H19 lncRNA promotes skeletal muscle insulin sensitivity in part by targeting AMPK. Diabetes. 2018;67:2183–2198. doi: 10.2337/db18-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Zhao Y., Bao X., Zhu X., Kwok Y.K., Sun K., Chen X., Huang Y., Jauch R., Esteban M.A. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X., Zhang Y., Li T., Ma Z., Jia H., Chen Q., Zhao Y., Zhai L., Zhong R., Li C. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017;8:14016. doi: 10.1038/ncomms14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L., Sun K., Zhao Y., Zhang S., Wang X., Li Y., Lu L., Chen X., Chen F., Bao X. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 17.Jin J.J., Lv W., Xia P., Xu Z.Y., Zheng A.D., Wang X.J., Wang S.S., Zeng R., Luo H.M., Li G.L., Zuo B. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E9802–9811. doi: 10.1073/pnas.1801471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., He L., Zhao Y., Li Y., Zhang S., Sun K., So K., Chen F., Zhou L., Lu L., Wang L., Zhu X., Bao X., Esteban M.A., Nakagawa S. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017;3:17002. doi: 10.1038/celldisc.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitachi K., Nakatani M., Takasaki A., Ouchi Y., Uezumi A., Ageta H., Inagaki H., Kurahashi H., Tsuchida K. Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO. Rep. 2019;20 doi: 10.15252/embr.201847468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song C.C., Wang J., Ma Y.L., Yang Z.X., Dong D., Li H., Yang J., Huang Y., Plath M., Ma Y., Chen H. Linc-smad7 promotes myoblast differentiation and muscle regeneration via sponging miR-125b. Epigenetics. 2018;13:591–604. doi: 10.1080/15592294.2018.1481705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballarino M., Cazzella V., D'Andrea D., Grassi L., Bisceglie L., Cipriano A., Santini T., Pinnarò C., Morlando M., Tramontano A., Bozzoni I. Novel long noncoding RNAs (lncRNAs) in myogenesis: A miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol. Cell. Biol. 2015;35:728–736. doi: 10.1128/MCB.01394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z., Bu N.P., Qiao X.H., Zuo Z.X., Shu Y.H., Liu Z.L., Qian Z., Chen J., Hou Y. Forkhead Box M1 transcriptionally regulates the expression of long noncoding RNAs Snhg8 and Gm26917 to promote proliferation and survival of muscle satellite cells. Stem Cells. 2018;36:1097–1108. doi: 10.1002/stem.2824. [DOI] [PubMed] [Google Scholar]
- 23.Cai R., Tang G.R., Zhang Q., Yong W.L., Zhang W.R., Xiao J.Y., Wei C.S., He C., Yang G.S., Pang W.J. A novel lnc-RNA, named lnc-ORA, is identified by RNA-Seq analysis, and its knockdown inhibits adipogenesis by regulating the PI3K/AKT/mTOR signaling pathway. Cells. 2019;8:477. doi: 10.3390/cells8050477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z.H., Cai B.L., Abdalla B.A., Zhu X.N., Zheng M., Han P.G., Nie Q.H., Zhang X.Q. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle. 2019;10:391–410. doi: 10.1002/jcsm.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z.K., Li J., Guan D., Liang C., Zhuo Z., Liu J., Lu A., Zhang G., Zhang B.T. Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol. Ther. 2018;26:2669–2680. doi: 10.1016/j.ymthe.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L., Si M., Liu X., Choi J.M., Wang Y., Thomas S.S., Peng H., Hu Z. Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachexia Sarcopenia Muscle. 2018;9:962–974. doi: 10.1002/jcsm.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alessio E., Buson L., Chemello F., Peggion C., Grespi F., Martini P., Massimino M.L., Pacchioni B., Millino C., Romualdi C., Bertoli A., Scorrano L., Lanfranchi G., Cagnin S. Single cell analysis reveals the involvement of the long non-coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Res. 2019;47:1653–1670. doi: 10.1093/nar/gkz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.d'Ydewalle C., Ramos D.M., Pyles N.J., Ng S.Y., Gorz M., Pilato C.M., Ling K., Kong L., Ward A.J., Rubin L.L., Rigo F., Bennett C.F., Sumner C.J. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron. 2017;93:66–79. doi: 10.1016/j.neuron.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumitru A., Radu B.M., Radu M., Cretoiu S.M. Muscle changes during atrophy. Adv. Exp. Med. Biol. 2018;1088:73–92. doi: 10.1007/978-981-13-1435-3_4. [DOI] [PubMed] [Google Scholar]
- 30.Pin F., Novinger L.J., Huot J.R., Harris R.A., Couch M.E., O'Connell T.M., Bonetto A. PDK4 drives metabolic alterations and muscle atrophy in cancer cachexia. FASEB J. 2019;33:7778–7790. doi: 10.1096/fj.201802799R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarroux J., Morillon A., Pinskaya M. History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 32.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang T.T., Zhou B., Shi L., Wang H., Chu Q.P., Xu F.L., Li Y., Chen R.N., Shen C.Y., Schinckel A.P. lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 2018;32:377–389. doi: 10.1096/fj.201700560RR. [DOI] [PubMed] [Google Scholar]
- 35.Sun X.M., Li M.X., Sun Y.J., Cai H.F., Lan X.Y., Huang Y.Z., Bai Y.Y., Qi X.L., Chen H. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim. Biophys. Acta Mol. Cell Res. 2016;1863:2835–2845. doi: 10.1016/j.bbamcr.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Small E.M., O'Rourke J.R., Moresi V., Sutherland L.B., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White J.P., Gao S., Puppa M.J., Sato S., Welle S.L., Carson J.A. Testosterone regulation of Akt/mTORC1/FoxO3a signaling in skeletal muscle. Mol. Cell. Endocrinol. 2013;365:174–186. doi: 10.1016/j.mce.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kline W.O., Panaro F.J., Yang H., Bodine S.C. Rapamycin inhibits the growth and muscle-sparing effects of clenbuterol. J. Appl. Physiol. 2007;102:740–747. doi: 10.1152/japplphysiol.00873.2006. [DOI] [PubMed] [Google Scholar]
- 40.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stitt T.N., Drujan D., Clarke B.A., Panaro F., Timofeyva Y., Kline W.O., Gonzalez M., Yancopoulos G.D., Glass D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 42.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L., Hu Y.C., Hüttelmaier S., Skibbe J.R., Su R., Deng X. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Yao Y., Han J., Yang Y., Chen Y., Tang Z., Gao F. Longitudinal epitranscriptome profiling reveals the crucial role of N6-methyladenosine methylation in porcine prenatal skeletal muscle development. J. Genet. Genomics. 2020;47:466–476. doi: 10.1016/j.jgg.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Gong C., Li Z., Ramanujan K., Clay I., Zhang Y., Lemire-Brachat S., Glass D.J. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Pang W.J., Wei N., Xiong Y., Wu W.J., Zhao C.Z., Shen Q.W., Yang G.S. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene. 2014;539:117–124. doi: 10.1016/j.gene.2014.01.037. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y., Yu L., Pan S., Gao S., Chen W., Zhang X., Dong W., Li J., Zhou R., Huang L., Han Y., Bai L., Zhang L., Zhang L. CRISPR/Cas9-mediated targeting of the Rosa26 locus produces Cre reporter rat strains for monitoring Cre-loxP-mediated lineage tracing. FEBS J. 2017;284:3262–3277. doi: 10.1111/febs.14188. [DOI] [PubMed] [Google Scholar]
- 47.Wang G.Q., Zhu L., Ma M.L., Chen X.C., Gao Y., Yu T.Y., Yang G.S., Pang W.J. Mulberry 1-deoxynojirimycin inhibits adipogenesis by repression of the ERK/PPARgamma signaling pathway in porcine intramuscular adipocytes. J. Agric. Food Chem. 2015;63:6212–6220. doi: 10.1021/acs.jafc.5b01680. [DOI] [PubMed] [Google Scholar]
- 48.Ma M., Wang X., Chen X., Cai R., Chen F., Dong W., Yang G., Pang W. MicroRNA-432 targeting E2F3 and P55PIK inhibits myogenesis through PI3K/AKT/mTOR signaling pathway. RNA Biol. 2017;14:347–360. doi: 10.1080/15476286.2017.1279786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai R., Qimuge N.R., Ma M.L., Wang Y.Q., Tang G.R., Zhang Q., Sun Y.M., Chen C.X., Yu T.Y., Dong W.Z., Yang G.S., Pang W.J. MicroRNA-664-5p promotes myoblast proliferation and inhibits myoblast differentiation by targeting serum response factor and Wnt1. J. Biol. Chem. 2018;293:19177–19190. doi: 10.1074/jbc.RA118.003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millay D.P., O'Rourke J.R., Sutherland L.B., Bezprozvannaya S., Shelton J.M., Bassel-Duby R., Olson E.N. Myomaker: A membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dou M., Yao Y., Ma L., Wang X., Shi X., Yang G., Li X. The long noncoding RNA MyHC IIA/X-AS contributes to skeletal muscle myogenesis and maintains the fast fiber phenotype. J. Biol. Chem. 2020;295:4937–4949. doi: 10.1074/jbc.RA119.010498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.