Abstract
Introduction
Macrophages polarization is essential in infection control. Llipopolysaccharide (LPS) plays an essential role in host innate immune system–pathogen interaction. The LPS structure of Pseudomonas aeruginosa modifies in the adaptation of this pathogen to biofilm-related chronic infection.
Gap statement
There have been several studies on LPS induced polarization of human and mouse macrophages with different results. And it was reported that the lipid A structure of the LPS derived from biofilm-forming Pseudomonas aeruginosa strain PAO1 was modified.
Aim
This study aimed to investigate the effect and the involved pathway of LPS from biofilm-forming PAO1 on human and murine macrophage polarization.
Methodology
LPS was isolated from biofilm-forming and planktonic PAO1 and quantified. Then the LPS was added to PMA-differentiated human macrophage THP-1 cells and Raw264.7 murine macrophage cells. The expression of iNOS, Arg-1, IL4, TNF-α, CCL3, and CCL22 was analysed in the different cell lines. The expression of TICAM-1 and MyD88 in human THP-1 macrophages was quantified by Western blot. PAO1 infected macrophages at different polarization states, and the intracellular bacterial growth in macrophages was evaluated.
Results
LPS from biofilm-forming PAO1 induced more marked hyperinflammatory responses in THP-1 and Raw264.7 macrophages than LPS derived from planktonic PAO1, and these responses were related to the up-regulation of MyD88. Intracellular growth of PAO1 was significantly increased in THP-1 macrophages polarized by LPS from biofilm-forming PAO1, but decreased both in THP-1 and Raw264.7 macrophages polarized by LPS from planktonic PAO1.
Conclusion
The presented in vitro study indicates that LPS derived from biofilm-forming PAO1 induces enhanced M1 polarization in human and murine macrophage cell lines than LPS from planktonic PAO1.
Keywords: lipopolysaccharide, biofilm, Pseudomonas aeruginosa, macrophages, polarization
Introduction
Pseudomonas aeruginosa is one of the most common opportunistic Gram-negative bacteria, and is responsible for a wide range of infections, especially in immunocompromised hosts [1]. Pseudomonas aeruginosa can cause not only acute nfections, but also chronic infections in humans. In chronic infections, P. aeruginosa behaves as biofilm-forming communities [2]. Biofilms are communities of microorganisms that are typically embedded in a matrix and often attached to a surface [3]. Bacterial biofilms are recalcitrant to antibiotics and responsible for the most persistent human bacterial infections [4]. Two explanations have been proposed for the increased resistance of biofilm-forming bacteria: (1) the interaction between bacterial cells and antibiotics or immune cells is reduced because of the biofilm matrix; (2) infiltrating phagocytes are less effective in killing bacterial cells that are enclosed in biofilms [5].
As antigen-presenting cells, macrophages play an important role in innate immune response and adaptive immune response [6]. Owing to their differentiation mechanisms, tissue distribution, and response to stimuli [7, 8], macrophages have been broadly classified into two groups: M1 and M2 macrophages. Classically activated macrophages (M1) are characterized by the secretion of proinflammatory cytokines such as tumor necrosis factor-α (TNF-α), cytokine receptors such as IL-7R, chemokines such as CCL2, CCL3, and CXCL8, and the chemokine receptor CCR7. Other M1-associated upregulated genes encode the enzymes indoleamine-pyrrole 2,3 dioxygenase and NO synthase (iNOS) [9]. By contrast, the M2 macrophages express anti-inflammatory cytokines, such as Arginase-1 (Arg1) and IL10, as well as increasing the expression of cell surface receptor CD206, and chemokines such as CCL22 [10]. M1 macrophages are microbicidal and inflammatory, whereas M2 macrophages limit excessive inflammatory responses, and promote tissue repair [11]. In infectious diseases such as sepsis, the imbalance between M1 and M2 macrophages often leads to serious complications [12, 13].
Macrophages perform essential roles in the defence against pathogens through pattern recognition receptors (PRRs). PRRs bind to conserved molecular patterns on microorganisms and initiate appropriate responses. Toll-like receptors (TLRs) represent a highly conserved class of PRRs, which are glycoprotein receptors consisting of an extracellular ligand-binding domain, a transmembrane domain, and an intracellular signalling portion [14, 15]. Lipopolysaccharide (LPS) is a major component of the outer membrane in Gram-negative bacteria and plays an essential role in host innate immune system–pathogen interaction [16]. TLR2 and TLR4 present on macrophage cell surfaces bind to LPS from P. aeruginosa using MyD88 and TICAM-1 as an adaptor protein [17, 18] to induce an inflammatory response. The classical LPS molecule is composed of the core region, O-antigen and lipid A [19, 20]. A basic lipid A structure containing an N- and O-acylated diglucosamine bisphosphate backbone [4-P-β-d-GlcpNII - (1→6)-α-d-GlcpNI -(1→P)] with chemical variation in the number of primary acyl groups and the types of fatty acids substituting the primary and secondary acyl groups. Most of the laboratory-adapated strains of P. aeruginosa synthesize a penta-acylated (75 % of the molecules) LPS, with some proportion made as a hexa-acylated LPS (25 % of the molecules). The difference between the two isoforms is the lack of an O-linked 3-hydroxy decanoic acid (10 : 0 (3-OH)) group at position 3 of the first glucosamine in the penta-acylated isoform [21]. Compared with that of planktonic bacteria, the LPS profile of a biofilm-forming clinical P. aeruginosa strain (KK11) showed an almost complete loss of O-antigen, and the penta-acyl lipid A was characterized by a lower 12 : 0 (2-OH) content. The structurally modified LPS of the biofilm-forming bacteria enhanced inflammatory cytokine responses in human monocytes, but not in murine macrophages [22]. Although the biofilm-forming PAO1 strain presented the same smooth-type LPS pattern as that of the planktonic culture, the hexa- and hepta-acyl lipid A of PAO1 contained 10 : 0 (3-OH) at both the C3 and C3′ positions [22]. The binding of P. aeruginosa LPS to TLRs depends on the structure of the lipid A of particular strain, as well as the species-specific variation in the LPS-binding domain of the TLR. In the present study, we investigated the effect and the involved pathway of LPS derived from the biofilm-forming P. aeruginosa PAO1 strain on human and murine macrophage polarization.
Methods
Bacterial strains and medium
The wild-type, non-mucoid P. aeruginosa PAO1 strain was stored in our laboratory. Luria broth (LB) was used as the growth medium for both biofilm-forming and planktonic bacteria. The strain was stored at −80 °C in 50 % (v/v) glycerol and subcultured from storage onto LB medium.
Static biofilm formation and microscopy
PAO1 was inoculated into 3 ml of LB and grown with shaking at 37 °C overnight and standardized to an optical density of 1 at 600 nm (OD600=1). This culture was diluted 1 : 100 in LB and grown in triplicate in polystyrene 6-well plates with one glass coverslip in each well at 37 °C. After 2, 4, 6 or 8 days, the coverslips were stained using the BacLight LIVE/DEAD viability kit reagent according to the manufacturer’s instructions. Images were obtained by fluorescence microscopy (Leica, DMI3000 B, Germany).
LPS isolation and quantification
Based on the LIVE/DEAD staining results, 6 day PAO1 biofilms were selected for LPS isolation. Coverslips were rinsed five times with PBS to remove planktonic bacteria. The biofilm was scraped off using a sterilized blade. The biofilm-producing PAO1 and overnight cultured planktonic PAO1 densities were adjusted to OD600=1. The bacterial concentration was confirmed to be 109 c.f.u. ml−1 by plating on agar Petri dishes. LPS was isolated using an LPS extraction kit (iNtRON) following the manufacturer’s instructions. Endpoint Chromogenic LAL Assays (Shanghai Yeasen BioTechnologies, China) were used for LPS quantification.
Cell culture and treatment
The human THP-1 monocyte cell line was kindly provided by Professor Xiongwen Wu from the Department of Immunology, Tongji Medical College, Huazhong University of Science and Technology. The mouse-derived Raw264.7 macrophage cell line was preserved in our department. Cells were seeded in 24-well flat-bottom cell culture plates at 8×105 cells per well in RPMI-1640 medium (the THP-1 cell line) and DMEM (the Raw264.7 cell line) supplemented with 10 % foetal bovine serum (Thermo Fisher Scientific). To determine the appropriate concentration of phorbol-12-myristate-13-acetate (PMA) to differentiate THP-1 monocytes into macrophages, THP-1 cells were treated with different concentrations (0, 12.5, 25, 50, 75 and 100 ng ml−1) of PMA (Solebao Technology Co., Ltd) for 48 h. The supernatants were collected for cell counting (to evaluate cell adherence) and analysis of IL-10 expression (enzyme-linked immunosorbent assay (ELISA), MultiSciences (Lianke) Biotechnology, China). After washing with PBS, adherent cells were harvested. The expression of CD14 was detected by Real-time Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) (primers were synthesized by Wuhan Qingke Biological Co., Ltd; Table 1). All experiments were repeated three times to determine the appropriate PMA concentration. LPS derived from biofilm-forming or planktonic PAO1 was added to Raw264.7 cells and PMA-differentiated THP-1 cells respectively at a concentration of 100 ng ml−1. After 5 h, the supernatants were collected and the cells harvested.
Table 1.
The sequence of primers used in experiments
|
Primers |
Sequence |
|---|---|
|
Human-GAPDH-F |
TGATGACATCAAGAAGGTGGTGAAG |
|
Human-GAPDH-R |
TCCTTGGAGGCCATGTAGGCCAT |
|
CD14-F |
TAGACCTCAGCCACAACTCG |
|
CD14-R |
GTCTGTTGCAGCTGAGATCG |
|
TNFα-F |
CCTGTGAGGAGGACGAACAT |
|
TNFα-R |
GGTTGAGGGTGTCTGAAGGA |
|
CCL3-F |
AGTTCTCTGCATCACTTGCTG |
|
CCL3-R |
CGGCTTCGCTTGGTTAGGAA |
|
CCL22-F |
GCCTACTCTGATGACCGTGG |
|
CCL22-R |
AGAGAGTTGGCACAGGCTTC |
|
CD206-F |
TGGACTGGGCTGAATGATGT |
|
CD206-R |
TAGCCTCGTTTACTGTCGCA |
|
Mice-GAPDH-F |
GTGTTTCCTCGTCCCGTAG |
|
Mice-GAPDH-R |
ATGGCAACAATCTCCACTTT |
|
iNOS-F |
GAGCGAGTTGTGGATTGTC |
|
iNOS-R |
AGGGCTTGGCTGAGTGA |
|
IL4-F |
TCGTCTGTAGGGCTTCC |
|
IL4-R |
TGCTCTTTAGGCTTTCC |
Measurement of cytokine levels
Cytokine (TNF-α, IL-10) expression levels were measured by ELISA kit (MultiSciences (Lianke) Biotechnology, China) according to the manufacturer’s instructions. All experiments were repeated three times.
Quantitative RT-PCR
The total RNA was extracted by acid-guanidinium-phenol method (Trizol LS Reagent, Invitrogen, USA). RNA was dissolved in 50 μl of RNase-free water. For cDNA synthesis, each 25 µl reaction mixture contained 2 µg of RNA, 0.5 µg of random hexamer primers, 5 µl of 5×RT buffer, 1.25 µl of 10 mml−1 dNTPs, 25 U of RNase inhibitor (Promega), and 200 U of Murine Leukaemia Virus (MLV) Reverse Transcriptase (Promega). cDNA synthesis was performed in a PCR Thermal Cycler (Eppendorf) according to the following procedure: an annealing step for 5 min at 70 °C, followed by reverse transcription for 60 min at 37 °C, and reverse transcriptase inactivation for 10 min at 95 °C. A Light Cycler (Roche) was used for qPCR. The primer sequences are described in Table 1. Human and mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control for THP-1 and Raw264.7 cells, respectively. The Ct was acquired using the CFX Connect RT-PCR system (Bio-Rad, Hercules, CA, USA). All experiments were repeated three times.
Immunofluorescence staining
LPS obtained from biofilm-forming PAO1, planktonic PAO1, or Escherichia coli (Sigma–Aldrich) was added to PMA-differentiated THP-1 cells at a concentration of 100 ng ml−1. After 5 h, the cells were fixed in 4 % paraformaldehyde for 15 min at room temperature. The cells were then treated with 3 % bovine serum albumin (BSA) for 30 min at 37 °C to block nonspecific staining, and incubated overnight at 4 °C with an anti-iNOS antibody (Abcam). The next day, the cells were rinsed with PBS, and then incubated for 1 h at room temperature with a horseradish peroxidase (HRP)-conjugated IgG antibody (Abcam). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole, Abcam). The cells were evaluated under an Olympus microscope (CellSens system).
Western blot
PMA-differentiated THP-1 macrophages were treated with LPS as mentioned above and harvested after 5 h. Protein samples were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE), transferred onto nitrocellulose membranes, and probed with anti-Arg-1, anti-iNOS, anti-TICAM-1 (TIR-containing adapter molecule-1), and anti-MyD88 (myeloid differentiation factor 88) primary antibodies (Abcam), followed by incubation with a secondary HRP-conjugated IgG antibody (Abcam). Protein bands were visualized using the Fujifilm LAS-4000 luminescent image analyser.
PAO1 infection of RAW264.7 macrophages and PMA-differentiated THP-1 macrophages
Different macrophages were seeded into 48-well plates at a density of 8×105 ml−1 in complete culture medium (RPMI-1640 or DMEM supplemented with 10 % foetal bovine serum). LPS derived from biofilm-forming or planktonic PAO1 was added to Raw264.7 cells and PMA-differentiated THP-1 cells respectively at a concentration of 100 ng ml−1. The cells were washed twice with PBS 5 h later. Overnight cultured planktonic PAO1 were adjusted to OD600=1. Macrophages were infected with PAO1 at an MOI of 10 and incubated at 37 °C under an atmosphere of 5 % CO2 for 2 h. The cells were washed twice with PBS, gentamicin (200 µg ml−1) was added to kill non-phagocytosed bacteria and macrophages were incubated at 37 °C for another 2 h. The cells were washed twice with PBS. Intracellular bacteria development was estimated by lysing the infected macrophages with 0.1 % Triton X-100. Serial dilutions of lysates were plated on Luria-Bertani (LB) agar (Difco, Detroit, Mich.) plates and CFUs were counted.
Statistical analysis
Data shown are the means±SD of triplicates and are representative of three experiments. Unpaired Student’s t-tests were applied to determine statistical differences. In some experiments, one-way ANOVA was applied to determine statistical differences among groups, followed, if significant, by a Student-Newman-Keuls test for post-hoc comparison. Differences were considered significant when P<0.05.
Results
PMA-induced differentiation of human THP-1 monocytes into macrophages
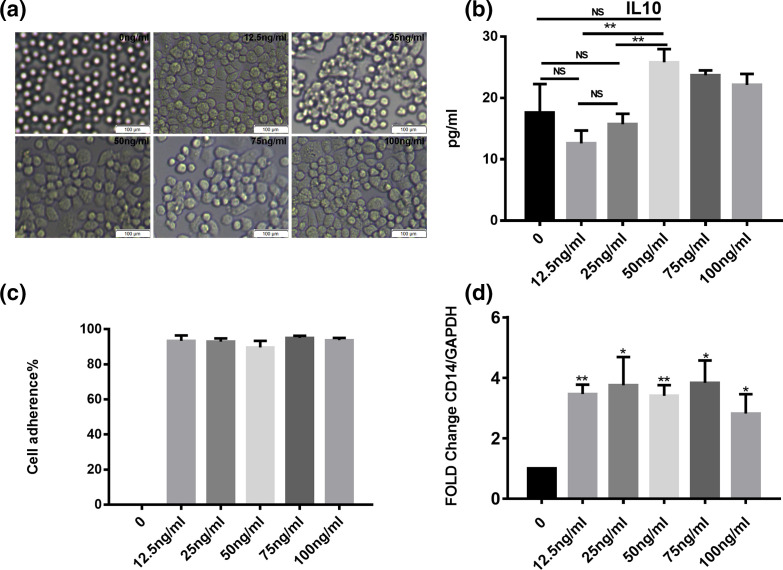
Although human monocytes are generally considered to represent macrophages, PMA was used to differentiate THP-1 cells into macrophages to better compare their polarization with that of murine macrophages. To find appropriate PMA concentration to induce THP-1 macrophage differentiation, cell adherence and CD14 expression was evaluated. To avoid the effects of PMA on macrophage polarization, IL-10 production was compared (Fig. 1). After PMA treatment, THP-1 cells began to grow adherently and the expression of CD14 increased. However, with increasing PMA concentration, especially at 50 ng ml−1, the production of IL-10 by THP-1 cells increased significantly (P<0.01, Fig. 1b). Based on the above results, the PMA concentration of 12.5 ng ml−1 was selected as it could induce macrophage differentiation while only minimally affecting the polarization of THP-1 cells.
Fig. 1.
(a) Cytoscopic images of human THP-1 cells differentiated using different concentrations of PMA (0, 12.5, 25, 50, 75 and 100 ng ml−1). (b) IL-10 production by THP-1 cells after treatment with PMA at different concentrations. (c) Cell adherence rate after washing with PBS. (d) CD14 expression in THP-1 cells treated with different concentrations of PMA. Significant differences among THP-1 cells treated with different PMA concentrations were calculated using the Student’s t-test; *P<0.05, **P<0.01.
PAO1 biofilm formation
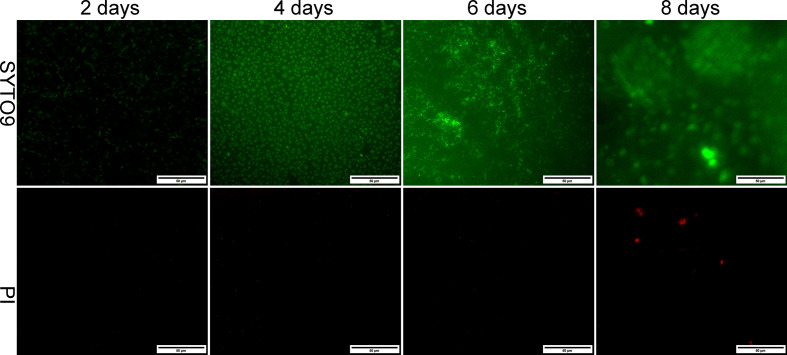
Formation of non-mucoid PAO1 biofilms was evaluated at 2, 4, 6 and 8 days by fluorescence microscopy. After 48 h, PAO1 formed small microcolonies. Four days post-inoculation, a layer of PAO1 cells covered the entire surface. After 6 days, a flat biofilm was formed covering the entire glass. By day 8, dead cells could be observed in the mature cluster of PAO1 biofilm (Fig. 2). This process was similar to that observed by confocal microscopy in our previous study [23]. Six-day PAO1 biofilms were selected for LPS isolation.
Fig. 2.
Biofilm formation by PAO1. Fluorescence micrographs of PAO1 biofilms developed on glass coverslips visualized using BacLight LIVE/DEAD viability staining. Cells with green fluorescence are viable, whereas those with red fluorescence are dead. a–d represent 2, 4, 6 and 8 day P . aeruginosa biofilms, respectively (×40).
LPS from biofilm-forming PAO1 induces hyperinflammatory responses in THP-1 cells to a greater extent than LPS from planktonic PAO1
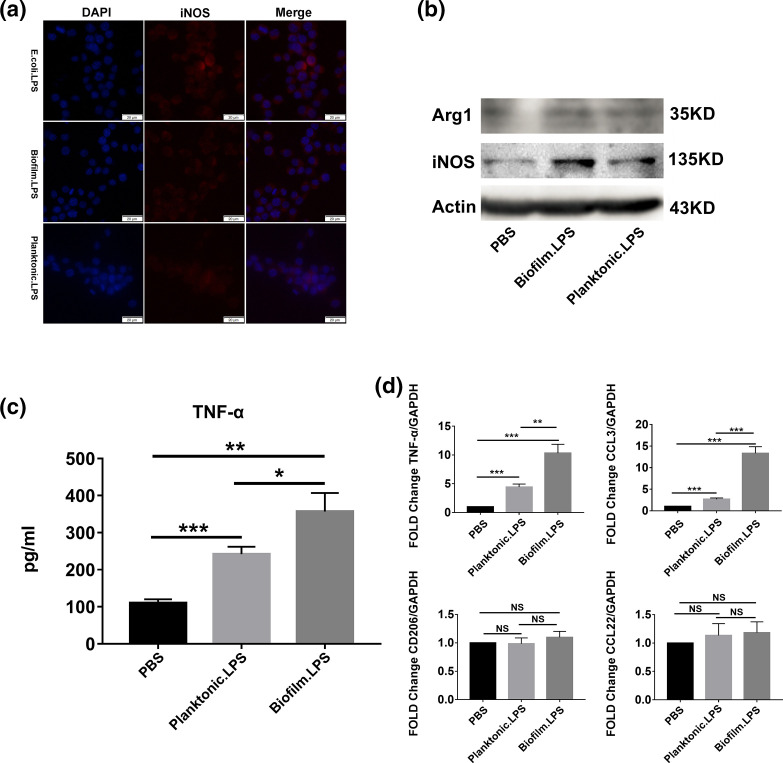
To investigate the effect of LPS from biofilm-forming and planktonic PAO1 on human macrophage polarization, some polarization markers were analysed. The expression of iNOS was estimated by immunofluorescence and Western blot (Fig. 3a, b). When THP-1 cells were treated with LPS, it was higher compared to the control. The expression of iNOS was higher in THP-1 cells treated with LPS from biofilm-forming PAO1 than in those treated with LPS from planktonic PAO1. The production of TNF-α was quantified by ELISA and qRT-PCR (Fig. 3c, d), and the expression of CCL3 was estimated by qRT-PCR (Fig. 3d). Compared with the blank control and THP-1 cells treated with LPS from planktonic PAO1, the expression of TNF-α and CCL3 were increased significantly when THP-1 cells were treated with LPS from biofilm-forming PAO1. However, the expression of Arg-1, CD206 and CCL22 did not change when treated with LPS from PAO1 (Fig. 3b, d). These results suggested that LPS from biofilm-forming PAO1 bacteria promoted M1 polarization in human THP-1 macrophages to a greater extent than LPS from planktonic PAO1; the results also indicated that LPS from biofilm-forming PAO1 induced a more marked inflammatory response in THP-1 cells than LPS from planktonic PAO1.
Fig. 3.
Comparison of the effects of LPS from biofilm-forming and planktonic PAO1 on human THP-1 macrophage polarization. The expression of iNOS (a: immunofluorescence staining; b: Western blot), Arg-1 (b: Western blot), TNF-α (c: ELISA; D: qRT-PCR), CCL3, CD206 and CCL22 (d: qRT-PCR) in THP-1 macrophages after treatment with LPS from biofilm-forming or planktonic PAO1. Significant differences were calculated by the Student’s t-test; *P<0.05, **P<0.01, ***P<0.001.
LPS from biofilm-forming PAO1 induces hyperinflammatory responses in Raw264.7 cells to a greater extent than LPS from planktonic PAO1
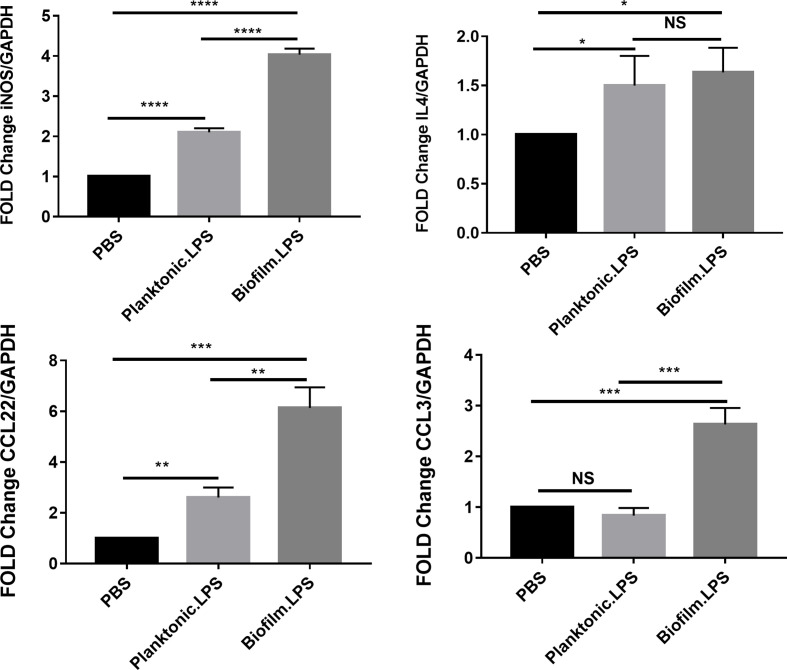
To evaluate the effects of LPS from biofilm-forming PAO1 on murine macrophage polarization, the expression of iNOS, IL4, CCL3, and CCL22 in Raw264.7 was quantified by qRT-PCR (Fig. 4). As observed in human THP-1 macrophages, the expression of M1 polarization-related genes, iNOS and CCL3, was significantly (P<0.05) increased in Raw264.7 cells treated with LPS from biofilm-forming PAO1 when compared with that in cells treated with LPS from planktonic PAO1 and blank control. The inflammatory responses of Raw264.7 cells were enhanced by treatment with LPS from biofilm-forming PAO1 when compared with those of cells treated with LPS from planktonic PAO1. However, the expression of IL4 and CCL22, which is representative of M2 polarization, showed a significant increase in murine macrophages when treated with LPS from PAO1. When comparing the result between LPS from biofilm-forming PAO1 and LPS from planktonic PAO1, the expression of CCL22 was higher significantly in cells treated with LPS from biofilm-forming PAO1. Yet there was no difference was observed in the expression of IL4. This result was not in agreement with that observed in human macrophages.
Fig. 4.
The effects of LPS from biofilm-forming and planktonic PAO1 on murine Raw264.7 macrophage polarization. The expression of iNOS, IL4, and CCL22 and CCL3 in Raw264.7 macrophages after treatment with LPS from biofilm-forming or planktonic PAO1. Significant differences were calculated using the Student’s t-test; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001,.
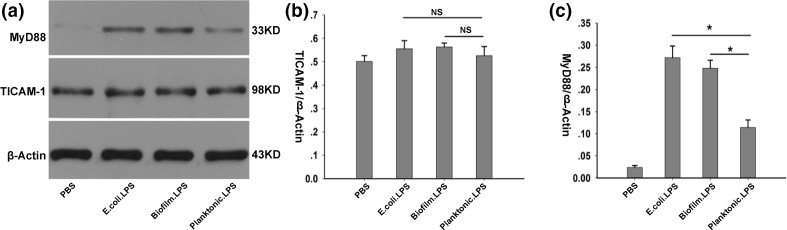
The MyD88 pathway was involved in macrophages polarization induced by LPS from biofilm-forming PAO1
MyD88- and TICAM-1-dependent pathways play an important role in host defenses against P. aeruginosa infection [24]. The expression of TICAM-1 and MyD88 in human THP-1 macrophages was quantified by Western blot. No significant change in the expression of TICAM-1 was recorded. When the cells were treated with LPS from biofilm-forming PAO1 and E. coli , the expression of MyD88 showed a significant increase, compared to treating with LPS from planktonic PAO1 (Fig. 5).
Fig. 5.
The expression of TICAM-1 and MyD88 in human THP-1 macrophages treated with LPS from E.coli, biofilm-forming PAO1 and planktonic PAO1. The expressions of TICAM1 and MyD88 in human THP-1 macrophages were quantified 5 h after the cells treated with LPS from E.coli, biofilm-forming PAO1 and planktonic PAO1. One-way ANOVA was applied to determine statistical differences among groups, followed, if significant, by a Student-Newman-Keuls test for post-hoc comparison.; *P<0.05.
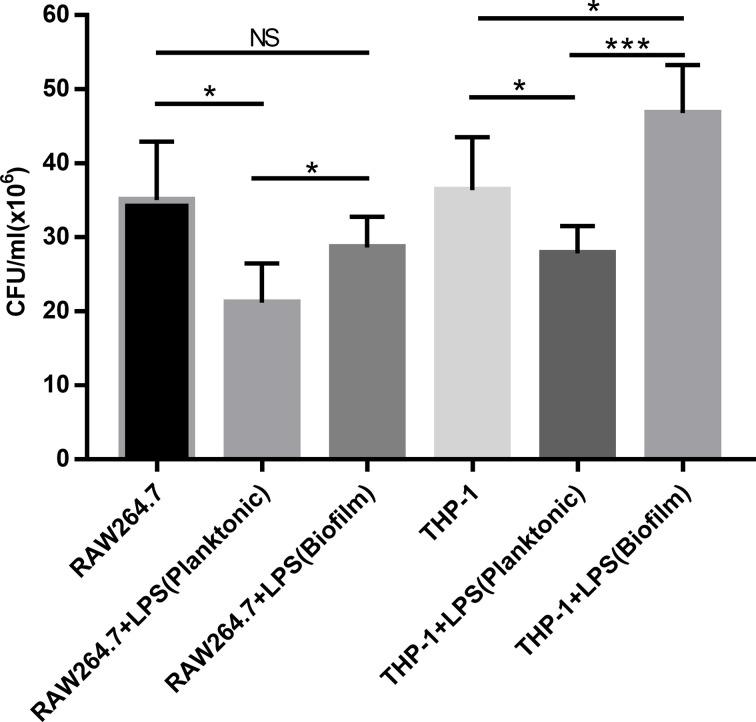
Intracellular bacterial growth in polarized macrophages induced by different LPS
To investigate the phagocytic activity of polarized macrophages, the growth of PAO1 in polarized RAW264.7 and THP-1 cells were shown in Fig. 6. In macrophages polarized by LPS from planktonic PAO1, intracellular growth of PAO1 was reduced (P<0.05) in comparison to the control (without LPS). However, when macrophages polarized by LPS from biofilm PAO1, we observed a significant increase of intracellular PAO1 growth in THP-1 macrophages.
Fig. 6.
Intracellular bacterial growth in polarized macrophages induced by different LPS. LPS derived from biofilm-forming or planktonic PAO1 was added to Raw264.7 cells and PMA-differentiated THP-1 cells respectively at a concentration of 100 ng ml−1. The cells were washed twice with PBS 5 h later. Polarized macrophages were infected with PAO1 at an MOI of 10. Two hours later, the cells were washed twice with PBS, gentamicin (200 µg ml−1) was added to kill non-phagocytosed bacteria. Intracellular bacteria were determined 2 h later. Data are shown as the mean of triplicate wells±SD. Significant differences were calculated using the Student’s t-test; *P<0.05, **P<0.01, ***P<0.001.
Discussion
Biofilm infections are among the greatest challenges facing the modern medical world. It is possible that all non-obligate intracellular bacteria and fungi can adopt a biofilm lifestyle [25]. There are numerous infections related to biofilms. Lipopolysaccharide (LPS) modification seems to play an important role in the adaptation of P. aeruginosa to biofilm-related chronic infection [22, 26].
LPS is a main component of the outer membrane in Gram-negative bacteria. The classical LPS molecule has a tripartite structure consisting of lipid A, core oligosaccharide, and O antigen [19, 20]. Binding of P. aeruginosa LPS to TLRs depends on the structure of the lipid A and the species-specific variation in the LPS-binding domain of TLRs. Murine TLR4 appears to react with penta- and hexa-acylated forms lipid A, however, human TLR4 signalling complexes react strongly with the hexa-acylated form [11, 27]. Pseudomonas aeruginosa may produce several varieties of lipid A during chronic infections or under different growth conditions [21]. The typically hexa-acylated lipid A induces robust inflammatory responses when recognised by TLR4 of macrophages, monocytes, and dendritic cells [27–31]. Modification of the lipid A acylation pattern confers protection against host innate defenses through dampening of the host’s inflammatory responses [32]. The hexa- and hepta-acyl lipid A of biofilm-forming PAO1 contained 10 : 0 (3-OH) at both the C3 and C3′ positions [22]. We analysed the effect of LPS of biofilm-forming PAO1 on macrophage polarization. The results showed that LPS from biofilm-forming PAO1 induced M1 polarization in both human and murine macrophages. The inflammatory response was markedly enhanced when compared with LPS from planktonic PAO1. These data are consistent with the reported effects of LPS from biofilm-forming clinical P. aeruginosa strains on primary human monocytes [22, 33], as well as that observed in cystic fibrosis (CF) patients and in a CF mouse model, in other words, P. aeruginosa biofilm infection can induce a more pronounced inflammatory response [34–37]. Because biofilm-forming PAO1 bacteria still exhibit complete LPS capped with O antigen [22], these results also convinced us that modification of the lipid A structure is key to inducing macrophage hyperinflammatory responses. It was reported that LPS from E. coli mediates time-dependent polarization of murine bone marrow-derived macrophages, whereby short-term LPS stimulation inhibits M1 polarization, while long-term stimulation promotes M1 polarization [38]. We found that the expression of M1 markers (iNOS, CCL3) increased significantly when Raw264.7 murine macrophage cell line was short-term stimulated with LPS from PAO1, but some M2 markers (CCL22, IL4) also increased at the same time. These varied results that have been obtained for murine macrophages may be related to the type of cells used. In the presented study, we selected the Raw264.7 murine macrophage cell line, while murine peritoneal macrophages [22] or bone marrow-derived macrophages [38] were used in other studies. Recent studies suggested that M1 macrophages could also express M2 markers, but M2 macrophages expressed higher M2 markers [39]. Macrophages activation has been shown to be plastic, rapid, and fully reversible, suggesting that macrophage populations are dynamic, heterogeneous and may first take part in inflammation and then participate in its resolution [40–44]. Briefly, because of the modification of the lipid A structure, short-term stimulation with LPS from biofilm-forming PAO1 promote human THP-1 and murine Raw264.7 cells’ M1 polarization. And LPS from biofilm-forming PAO1 induced more marked inflammatory responses than LPS from planktonic PAO1.
After the binding of P. aeruginosa LPS, TLR4 has been reported to activate MyD88- and TICAM-1-dependent pathways for host defenses against P. aeruginosa infection [45]. TLR2 also use MyD88 as the adaptor protein to activate the inflammatory response. MyD88 acts as a central role in inflammatory responses and can induce signalling from several receptors [46, 47]. MyD88-dependent signalling induces the sequential phosphorylation of IRAK-4, IRAK-1, TRAF-6 and TAK-1. The latter activates a cascade of inhibitor of kappa B kinases (IKKs), resulting in the eventual ubiquitination and degradation of IκB and the release of NF-κB, which translocates into the nucleus and regulates the transcription of numerous genes [17], including cytokines, chemotactic factors, and adhesion molecules, all of which are important for initiating inflammatory responses [17, 48, 49]. In MyD88-deficient mice, forty-six pathogens displaying higher growth rates and thirty-three pathogens for which the mortality was greater in vivo, both including P. aeruginosa [50]. Moreover, MyD88-deficiency is vital in infancy and early childhood. The MyD88-deficient patients are susceptible to invasive pyogenic infections, such as severe tonsillitis due to P. aeruginosa in particular [50]. TLR4 can also recruit TICAM-1 (TRIF) when bind to TRAM priorly [44]. The complex activates the kinase TBK1, which phosphorylates IRF3 and −7, mediating the expression of IFN-α and -β. In our study, the MyD88- but not TICAM-1-pathway was involved in the M1 polarization of human THP-1 cells induced by LPS of PAO1.
In our study, both LPS derived from planktonic and biofilm-forming PAO1 induced M1 polarization of macrophages. The M1 polarized macrophages induced by the latter LPS produced more inflammatory factors compared to the macrophages induced by LPS from planktonic PAO1, but there were more bacteria that grew intracellularly in polarized THP-1 macrophages induced by LPS derived from biofilm-forming PAO1, suggesting enhanced M1 polarized THP-1 macrophages were inefficient in killing ingested bacteria. The limitation of this experiment should also be acknowledged. When we evaluated intracellular PAO1 growth in macrophages, although obvious death of macrophages was not observed, we could not rule out the effect of the number of living macrophages on the results since LPS treatment could activate cell death pathways in macrophages. It was reported that macrophages respond to antigenic stimuli and secrete a range of over 100 substances, which vary in their biological activities and metabolic functions [51]. The macrophages secretory products (MSPs) include peptide hormones, components of complement, enzymes, lipids, reactive oxygen and other biological substances [52]. The enhancement in growth of intracellular P. aeruginosa was observed in presence of MSPs [52–54]. It was indicated that there was a threshold of cellular activation at which phagocytic cells effectively killed ingested bacteria. Above this threshold of cellular activation, however, the intracellular micromilieu become favourable to the survival and replication of the ingested bacteria [54]. The interplay between biofilm infection and the host response has been best characterized for CF patients suffering from chronic P. aeruginosa lung infections [55]. Several studies have indicated that LPS induces hyperinflammatory responses by CF macrophages [34–37]. However, in CF patients, hyperinflammatory responses result in severe tissue damage but not pathogen clearance and infection control [56, 57]. The mechanisms for the impaired phagocytic activity of enhanced M1 polarized macrophages needs further study.
Taken together, the results of our in vitro study show that LPS derived from biofilm-forming PAO1 induces enhanced M1 polarization more in human and murine macrophage cell lines than LPS from planktonic PAO1. The MyD88- but not TICAM-1-pathway was involved in this process. Additional research is required to evaluate the phagocytic activity of the enhanced M1 polarized macrophages and clarify the mechanisms during the process.
Funding information
This work was supported by a grant from the Health Commission of Hubei Province (NO. WJ2017M063) and the Wuhan Science and Technology Bureau (NO. 2017060201010176).
Acknowledgement
We would like to thank Professor Xiongwen Wu from the Department of Immunology, Tongji Medical College, Huazhong University of Science and Technology, for providing the human THP-1 monocyte cell line.
Author contributions
S. W., Performed the experiments and manuscript preparation; D. X., contributed significantly to analyse the data; F. T., helped to perform the analysis with constructive discussions; M. N., conceived the experiments and wrote the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: Arg-1, Arginase-1; BSA, bovine serum albumin; CF, cystic fibrosis; DAPI, 4′,6-diamidino-2-phenylindole; E.coli, Escherichia coli; ELISA, enzyme-linked immunosorbent assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IKKs, inhibitor of kappa B kinases; iNOS, indoleamine-pyrrole 2,3 dioxygenase and NO synthase; IRAK, IL‐1R‐associated kinase; IRF, IFN regulatory factor; LPS, Llipopolysaccharide; MLV, murine leukemia virus; MSPs, macrophages secretory products; MyD88, myeloid differentiation factor 88; P. aeruginosa, Pseudomonas aeruginosa; PMA, phorbol-12-myristate-13-ace-tate; PRRs, pattern recognition receptors; qRT-PCR, real-time quantitative reverse transcription-polymerase chain reaction; SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TAK-1, TGF‐β‐activated protein kinase 1; TBK-1, TRAF family member‐associated NF‐κB activator‐binding kinase 1; TICAM-1, TIR-containing adapter molecule-1; TLRs, toll-like receptors; TNF-α, tumor necrosis factor-α; TRAF-6, TNF receptor associated factor 6; TRAM, Toll/IL‐1R domain‐containing adapter‐inducing IFN‐β‐related adaptor molecule.
References
- 1.Gellatly SL, Hancock REW. Pseudomonas aeruginosa : new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 2.Rybtke M, Hultqvist LD, Givskov M, Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J Mol Biol. 2015;427:3628–3645. doi: 10.1016/j.jmb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 3.O’Toole GA. Microbiology: a resistance switch. Nature. 2002;46:695–696. doi: 10.1038/416695a. [DOI] [PubMed] [Google Scholar]
- 4.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 5.Hirschfeld J. Dynamic interactions of neutrophils and biofilms. J Oral Microbiol. 2014;6:26102. doi: 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 7.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 8.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 9.Benoit M, Desnues B, Mege J-L. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 10.Makita N, Hizukuri Y, Yamashiro K, Murakawa M, Hayashi Y. Il-10 enhances the phenotype of M2 macrophages induced by IL-4 and confers the ability to increase eosinophil migration. Int Immunol. 2015;27:131–141. doi: 10.1093/intimm/dxu090. [DOI] [PubMed] [Google Scholar]
- 11.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 12.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 14.Akira S, Takeda K. Toll-Like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 15.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 17.McIsaac SM, Stadnyk AW, Lin T-J. Toll-like receptors in the host defense against Pseudomonas aeruginosa respiratory infection and cystic fibrosis. J Leukoc Biol. 2012;92:977–985. doi: 10.1189/jlb.0811410. [DOI] [PubMed] [Google Scholar]
- 18.Cui J, Chen Y, Wang HY, Wang R-F. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother. 2014;10:3270–3285. doi: 10.4161/21645515.2014.979640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 20.Maldonado RF, Sá-Correia I, Valvano MA. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev. 2016;40:480–493. doi: 10.1093/femsre/fuw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pier G. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciornei CD, Novikov A, Beloin C, Fitting C, Caroff M, et al. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immun. 2010;16:288–301. doi: 10.1177/1753425909341807. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Yu B, Tian D, Ni M. Rhamnolipid but not motility is associated with the initiation of biofilm seeding dispersal of Pseudomonas aeruginosa strain PA17. J Biosci. 2013;38:149–156. doi: 10.1007/s12038-012-9297-0. [DOI] [PubMed] [Google Scholar]
- 24.Lagoumintzis G, Xaplanteri P, Dimitracopoulos G, Paliogianni F. TNF-alpha induction by Pseudomonas aeruginosa lipopolysaccharide or slime-glycolipoprotein in human monocytes is regulated at the level of mitogen-activated protein kinase activity: a distinct role of Toll-like receptor 2 and 4. Scand J Immunol. 2008;67:193–203. doi: 10.1111/j.1365-3083.2007.02053.x. [DOI] [PubMed] [Google Scholar]
- 25.Moser C, Pedersen HT, Lerche CJ, Kolpen M, Line L, et al. Biofilms and host response - helpful or harmful. APMIS. 2017;125:320–338. doi: 10.1111/apm.12674. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy RR, Mazon-Moya MJ, Moscoso JA, Hao Y, Lam JS, et al. Cyclic-di-GMP regulates lipopolysaccharide modification and contributes to Pseudomonas aeruginosa immune evasion. Nat Microbiol. 2017;2:17027. doi: 10.1038/nmicrobiol.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst RK, Hajjar AM, Tsai JH, Moskowitz SM, Wilson CB, et al. Pseudomonas aeruginosa lipid a diversity and its recognition by Toll-like receptor 4. J Endotoxin Res. 2003;9:395–400. doi: 10.1177/09680519030090060201. [DOI] [PubMed] [Google Scholar]
- 28.Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, et al. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa . Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 29.Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3:354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 30.Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res. 2001;7:167–202. doi: 10.1179/096805101101532675. [DOI] [PubMed] [Google Scholar]
- 31.Park BS, Lee J-O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45:e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid a modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciszek-Lenda M, Strus M, Walczewska M, Majka G, Machul-Żwirbla A, et al. Pseudomonas aeruginosa biofilm is a potent inducer of phagocyte hyperinflammation. Inflamm Res. 2019;68:397–413. doi: 10.1007/s00011-019-01227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonfield TL, Konstan MW, Burfeind P, Panuska JR, Hilliard JB, et al. Normal bronchial epithelial cells constitutively produce the anti-inflammatory cytokine interleukin-10, which is downregulated in cystic fibrosis. Am J Respir Cell Mol Biol. 1995;13:257–261. doi: 10.1165/ajrcmb.13.3.7544594. [DOI] [PubMed] [Google Scholar]
- 35.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med. 1995;152:2111–2118. doi: 10.1164/ajrccm.152.6.8520783. [DOI] [PubMed] [Google Scholar]
- 36.Bruscia EM, Zhang P-X, Ferreira E, Caputo C, Emerson JW, et al. Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator −/−mice. Am J Respir Cell Mol Biol. 2009;40:295–304. doi: 10.1165/rcmb.2008-0170OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaman MM, Gelrud A, Junaidi O, Regan MM, Warny M, et al. Interleukin 8 secretion from monocytes of subjects heterozygous for the ΔF508 cystic fibrosis transmembrane conductance regulator gene mutation is altered. CVI. 2004;11:819–824. doi: 10.1128/CDLI.11.5.819-824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Zhang Y, He Y, Wang X, Fang Q. Lipopolysaccharide mediates time-dependent macrophage M1/M2 polarization through the Tim-3/Galectin-9 signalling pathway. Exp Cell Res. 2019;376:124–132. doi: 10.1016/j.yexcr.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 40.Porcheray F, Viaud S, Rimaniol AC, Leone C, Samah B, et al. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17:34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 43.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porcheray F, Viaud S, Rimaniol AC, Léone C, Samah B, et al. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostuni R, Zanoni I, Granucci F. Deciphering the complexity of Toll-like receptor signaling. Cell Mol Life Sci. 2010;67:4109–4134. doi: 10.1007/s00018-010-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oda K, Kitano H. A comprehensive map of the toll‐like receptor signaling network. Mol Syst Biol. 2006;2:2006.0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deguine J, Barton GM. Myd88: a central player in innate immune signaling. F1000Prime Rep. 2014;6:97. doi: 10.12703/P6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor B. Proc Natl Acad Sci U S A. 2009;106:14978–14983. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Wang Z, Zou Y, Lu E, Duan J, et al. Pretreatment with lipopolysaccharide attenuates diethylnitrosamine-caused liver injury in mice via TLR4-dependent induction of Kupffer cell M2 polarization. Immunol Res. 2015;62:137–145. doi: 10.1007/s12026-015-8644-2. [DOI] [PubMed] [Google Scholar]
- 50.von Bernuth H, Picard C, Puel A, Casanova J-L. Experimental and natural infections in MyD88- and IRAK-4-deficient mice and humans. Eur J Immunol. 2012;42:3126–3135. doi: 10.1002/eji.201242683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nathan CF. Secretory products of macrophages. J Clin Invest. 1987;79:319–326. doi: 10.1172/JCI112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mittal R, Sharma S, Chhibber S, Harjai K. Effect of macrophage secretory products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comp Immunol Microbiol Infect Dis. 2006;29:12–26. doi: 10.1016/j.cimid.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Meduri GU, Kanangat S, Stefan J, Tolley E, Schaberg D. Cytokines IL-1β, IL-6, and TNF- α enhance in vitro growth of bacteria. Am J Respir Crit Care Med. 1999;160:961–967. doi: 10.1164/ajrccm.160.3.9807080. [DOI] [PubMed] [Google Scholar]
- 54.Kanangat S, Meduri GU, Tolley EA, Patterson DR, Meduri CU, et al. Effects of cytokines and endotoxin on the intracellular growth of bacteria. Infect Immun. 1999;67:2834–2840. doi: 10.1128/IAI.67.6.2834-2840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen Peter Østrup, Givskov M, Bjarnsholt T, Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2010;59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 56.SenGupta S, Hittle LE, Ernst RK, Uriarte SM, Mitchell TC. A Pseudomonas aeruginosa hepta-acylated lipid A variant associated with cystic fibrosis selectively activates human neutrophils. J Leukoc Biol. 2016;100:1047–1059. doi: 10.1189/jlb.4VMA0316-101R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shih C-C, Liu P-Y, Chen J-H, Liao M-H, Hsieh C-M, et al. Macrophage expression of E3 ubiquitin ligase Grail protects mice from lipopolysaccharide-induced hyperinflammation and organ injury. PLoS One. 2018;13:e0208279. doi: 10.1371/journal.pone.0208279. [DOI] [PMC free article] [PubMed] [Google Scholar]