Abstract
STUDY QUESTION
Are uterine fluid-derived extracellular vesicles (UF-EVs) a ‘liquid biopsy’ reservoir of biomarkers for real-time monitoring of endometrial status?
SUMMARY ANSWER
The transcriptomic cargo of UF-EVs reflects the RNA profile of the endometrial tissue as well as changes between the non-receptive and the receptive phase, possibly supporting its use for a novel endometrial receptivity test.
WHAT IS KNOWN ALREADY
EVs have been previously isolated from uterine fluid, where they likely contribute to the embryo-endometrium crosstalk during implantation. Based on a meta-analysis of studies on endometrial tissue implantation-associated genes and the human exosomes database, 28 of the 57 transcripts considered as receptivity markers refer to proteins present in human exosomes. However, the specific transcriptomic content of receptive phase UF-EVs has yet to be defined.
STUDY DESIGN, SIZE, DURATION
Two experimental series were set up. First, we simultaneously sequenced RNA species derived from paired UF-EVs and endometrial tissue samples collected from physiologically cycling women. Second, we analyzed RNA species of UF-EVs collected during the non-receptive (LH + 2) and receptive (LH + 7) phase of proven fertile women and from the receptive (LH + 7) phase of a population of women undergoing ART and transfer of euploid blastocysts.
PARTICIPANTS/MATERIALS, SETTING, METHODS
For paired UF—endometrial tissue sampling, endometrial tissue biopsies were obtained with the use of a Pipelle immediately after UF collection performed by lavage of the endometrial cavity. Overall, n = 87 UF samples were collected and fresh-processed for EV isolation and total RNA extraction, while western blotting was used to confirm the expression of EV protein markers of the isolated vesicles. Physical characterization of UF-EVs was performed by Nanoparticle Tracking Analysis. To define the transcriptomic cargo of UF-EV samples, RNA-seq libraries were successfully prepared from n = 83 UF-EVs samples and analyzed by RNA-seq analysis. Differential gene expression (DGE) analysis was used to compare RNA-seq results between different groups of samples. Functional enrichment analysis was performed by gene set enrichment analysis with g:Profiler. Pre-ranked gene set enrichment analysis (GSEA) with WebGestalt was used to compare RNA-seq results with the gene-set evaluated in a commercially available endometrial receptivity array.
MAIN RESULTS AND THE ROLE OF CHANCE
A highly significant correlation was found between transcriptional profiles of endometrial biopsies and pairwise UF-EV samples (Pearson’s r = 0.70 P < 0.0001; Spearman’s ρ = 0.65 P < 0.0001). In UF-EVs from fertile controls, 942 gene transcripts were more abundant and 1305 transcripts less abundant in the LH + 7 receptive versus the LH + 2 non-receptive phase. GSEA performed to evaluate concordance in transcriptional profile between the n = 238 genes included in the commercially available endometrial receptivity array and the LH + 7 versus LH + 2 UF-EV comparison demonstrated an extremely significant and consistent enrichment, with a normalized enrichment score (NES)=9.38 (P < 0.001) for transcripts up-regulated in LH + 7 in the commercial array and enriched in LH + 7 UF-EVs, and a NES = −5.40 (P < 0.001) for transcripts down-regulated in LH + 7 in the commercial array and depleted in LH + 7 UF-EVs. When analyzing LH + 7 UF-EVs of patients with successful versus failed implantation after transfer of one euploid blastocyst in the following cycle, we found 97 genes whose transcript levels were increased and 64 genes whose transcript levels were decreased in the group of women who achieved a pregnancy. GSEA performed to evaluate concordance in transcriptional profile between the commercially available endometrial receptivity array genes and the comparison of LH + 7 UF-EVs of women with successful versus failed implantation, demonstrated a significant enrichment with a NES = 2.14 (P = 0.001) for transcripts up-regulated in the commercial array in the receptive phase and enriched in UF-EVs of women who conceived, and a not significant NES = −1.18 (P = 0.3) for transcripts down-regulated in the commercial array and depleted in UF-EVs. In terms of physical features, UF-EVs showed a homogeneity among the different groups analyzed except for a slight but significant difference in EV size, being smaller in women with a successful implantation compared to patients who failed to conceive after euploid blastocyst transfer (mean diameter ± SD 205.5± 22.97 nm vs 221.5 ± 20.57 nm, respectively, P = 0.014).
LARGE SCALE DATA
Transcriptomic data were deposited in NCBI Gene Expression Omnibus (GEO) and can be retrieved using GEO series accession number: GSE158958.
LIMITATIONS, REASONS FOR CAUTION
Separation of RNA species associated with EV membranes might have been incomplete, and membrane-bound RNA species—rather than the internal RNA content of EVs—might have contributed to our RNA-seq results. Also, we cannot definitely distinguish the relative contribution of exosomes, microvesicles and apoptotic bodies to our findings. When considering patients undergoing ART, we did not collect UFs in the same cycle of the euploid embryo transfer but in the one immediately preceding. We considered this approach as the most appropriate in relation to the novel, explorative nature of our study. Based on our results, a validation of UF-EV RNA-seq analyses in the same cycle in which embryo transfer is performed could be hypothesized.
WIDER IMPLICATIONS OF THE FINDINGS
On the largest sample size of human EVs ever analyzed with RNA-seq, this study establishes a gene signature to use for less-invasive endometrial receptivity tests. This report is indeed the first to show that the transcriptome of UF-EVs correlates with the endometrial tissue transcriptome, that RNA signatures in UF-EVs change with endometrial status, and that UF-EVs could serve as a reservoir for potential less-invasive collection of receptivity markers. This article thus represents a step forward in the design of less-invasive approaches for real-time monitoring of endometrial status, necessary for advancing the field of reproductive medicine.
STUDY FUNDING/COMPETING INTEREST(S)
The study was funded by a competitive grant from European Society of Human Reproduction and Embryology (ESHRE Research Grant 2016-1). The authors have no financial or non-financial competing interests to disclose.
TRIAL REGISTRATION NUMBER
NA.
Keywords: implantation, receptivity, exosomes, microvescicles, extracellular vesicles, RNA-seq, transcriptome, endometrium, assisted reproduction
Introduction
The endometrium is unique among adult tissues in the extent of remodeling that it undergoes during each menstrual cycle, primarily under the orchestration of ovarian-derived hormones. The cyclical changes include the transition between the proliferative (nonreceptive, oestrogen dependent) and secretory (progesterone-dependent) phase, in preparation for embryo implantation. In women, this ‘receptive phase’ occurs at about 6–10 days after the LH surge that initiates ovulation (Mackens et al., 2017; Evans et al., 2019). This so-called ‘window of implantation’ is a limited timeframe in which the endometrium becomes a fertile soil by providing a nutritive and immune privileged environment for the embryo. A strictly synchronized embryonic-endometrial dialogue, important for the establishment of a successful pregnancy, occurs in this phase (Prapas et al., 1998; Wilcox et al., 1999; Shapiro et al., 2008, 2014, 2016; Healy et al., 2017; Franasiak et al., 2018; Evans et al., 2019; Messaoudi et al., 2019).
Both embryonic and endometrial competence have been investigated extensively over the last decades and advances in comprehensive molecular approaches have allowed the development of tests for diagnostic use in the clinical practice of ART. As far as embryonic competence is concerned, current state-of-the-art diagnostics are based on whole-genome next-generation sequencing (NGS) of blastocyst biopsies (preimplantation genetic testing for aneuploidies, PGT-A), allowing the selection of euploid blastocysts for transfer.
For endometrial competence, some tests using the transcriptome of endometrial biopsies have been introduced into clinical practice for the diagnosis of endometrial receptivity status during the putative window of implantation [endometrial receptivity array (ERA®) by Igenomix, Endometrial receptivity Map (ER-Map®) by iGLS and endometrial receptivity peak (ERPeak) by CooperGenomics™]. Nonetheless, embryo implantation continues to represent a major limiting step in the current understanding and treatment of human fertility. In fact, transfer of euploid blastocysts to a receptive endometrium still fails to achieve successful pregnancy in over 30% of cases (Neves et al., 2019). A substantial drawback of current receptivity diagnostics is that the endometrial tissue needs to be obtained through an invasive endometrial biopsy that hampers the possibility to perform the procedure during the same cycle as embryo transfer. Due to extensive inter-cycle variations in reproductive physiology, the receptive status of the menstrual cycle when the test is performed might thus differ from that of the actual embryo transfer cycle.
Interestingly, based on a recent meta-analysis of nine studies on endometrial tissue implantation-associated genes, 28 of the 57 transcripts currently considered as receptivity markers were also detected in extracellular vesicles (EVs), i.e. secreted lipid bilayer-enclosed lumens (Altmäe et al., 2017). EVs are heterogeneous in size, which roughly distinguishes them as exosomes (50–150 nm), microvesicles (100–1000 nm) and apoptotic bodies (50–5000 nm) (Hauser et al., 2017; Mentkowski et al., 2018). EVs are even more heterogeneous in molecular composition (encompassing functional proteins, DNA, mRNA, non-coding RNA and lipids). Because EVs derive their cargo from the contents of the cells producing them, they are intensively studied as mediators for intercellular communication and novel microenvironment modulators (O’Brien et al., 2020). While thought to be present in uterine fluid (Ng et al., 2013), EVs are however not included in current diagnostics for endometrial receptivity. To some extent, this might even contribute to the limitations of current receptivity tests, and a role for EVs as ‘liquid biopsy’ reservoirs of biomarkers suitable for real-time monitoring of endometrial status deserves consideration.
To explore this possibility, two experimental approaches have been set up. First, in order to assess the concordance between endometrial tissue and the uterine fluid-derived EVs (UF-EVs) transcriptome, we simultaneously sequenced RNA species derived from pairwise samples collected from physiologically cycling women (Fig. 1A). Second, to unravel the transcriptomic profile of EVs released by the endometrium during the window of implantation, we analyzed RNA species of UF-EVs collected from: proven fertile women (Fig. 1B) and a larger population of women undergoing ART and transfer of euploid blastocysts, as a function of pregnancy outcomes (Fig. 1C). Collectively, this unprecedented study includes the largest sample size of human EVs ever analyzed with RNA-seq in order to comprehensively investigate their transcriptomics.
Figure 1.
Workflow summarizing the experimental design. (A) Transcriptomic profile evaluation of UF-EVs and concomitant endometrial biopsies (E-Tissue) collected in the proliferative and secretory phase of regularly cycling volunteer women. A sample of uterine fluid and a concomitant endometrial biopsy were collected from each woman. RNA-seq analysis on UF-EV samples and endometrial biopsies were performed and RNA profiles were compared. A small aliquot of each resuspended UF-EV samples were used for NTA. (B) Transcriptomic profile evaluation in UF-EVs collected before and during the window of implantation from provenly fertile women. Each of these n = 14 fertile (f) women underwent UF sampling twice, 2 and 7 days after LH peak (fUF-EVs LH + 2 and fUF-EVs LH + 7) as detected with an ovulation test. A total of n = 4 samples collected from n = 3 women dropped-out for unsuccessful library preparation. RNA-seq analysis on samples from the remaining n = 11 women were performed to explore the physiological endometrial EVs transcriptomic profile of the receptive phase (DGE of fUF-EVs LH + 7 vs fUF-EVs LH + 2). A small aliquot of each resuspended UF-EV samples (except for one sample due to technical issues) was used for NTA. (C) Transcriptomic profile evaluation of UF-EVs collected during the window of implantation from n = 49 women undergoing ART and PGT-A. Subjects were sampled in the cycle before that of embryo transfer, 7 days after the LH peak as detected with an ovulation test. A small aliquot of each resuspended UF-EV samples (except for n = 1 sample due to technical issues) was used for NTA. Samples collected from n = 7 women dropped-out for low number of read counts. RNA-seq analysis on n = 42 UF-EV samples was performed to investigate whether EVs content was associated with a successful receptivity (DGE of UF-EVs from women who achieved pregnancy vs UF-EVs of women who failed to achieve pregnancy). DGE, differential gene expression analysis; NTA, nanoparticle tracking analysis; PGT-A, preimplantation genetic testing for aneuploidies; UF-EVs, uterine fluid-derived extracellular vesicles.
Materials and methods
Subjects and experimental design
This project received the approval of the local human ethics committee of the IRCCS San Raffaele Scientific Institute, Milan, Italy (#ESHRE2016 approval date 9 March 2017). For all participants, written informed consent was obtained and sampling performed at the ART clinic within the Obstetrics and Gynecology Unit of the Institution. For the first experimental approach to compare the transcriptional profile of UF-EVs and of the corresponding endometrial tissues, tissues and UF samples were obtained simultaneously from n = 10 volunteer women (Fig. 1A). All women selected were of reproductive age (≤38 years), had normal BMI (17–28 kg/m2), reported regular menstrual cycles (25–35 days) and were clinically examined for the absence of hormonal aberrations, uterine pathologies or polycystic ovary syndrome. All women were non-smokers, not using any hormonal treatment and had no previous infertility records (Table I). Following the criteria for dating of the endometrial tissues (Noyes et al., 1950), the first day of last menstrual period was recorded and menstrual cycle phase was accordingly defined as proliferative for n = 5 subjects and secretory for the other n = 5. For the second experimental approach aimed at evaluating the transcriptomic profile of UF-EVs associated with implantation, two groups of subjects were included: a proven fertile women group (n = 14) (Fig. 1B), and a population of n = 49 women undergoing ART with PGT-A, of whom the outcomes of successful versus failed clinical pregnancy in the following euploid blastocyst transfer attempt were recorded (Fig. 1C). For the purposes of brevity, the acronyms UF-EVs ‘pregnant’ and ‘not pregnant’ were used in figures throughout this article, to refer to samples collected from women who achieved or failed implantation, respectively, in the following embryo transfer. This category of women undergoing PGT-A was chosen because—transferring an euploid embryo—their ART outcome is considered dependent on endometrial characteristics (Vaiarelli et al., 2016).
Table I.
Descriptive characteristics of subjects who participated in the study.
| Characteristics | Mean ± SD or n | Range or % |
|---|---|---|
| Descriptive characteristics of normally cycling volunteer women (n = 10) | ||
| Age (years) | 31.4 ± 4.8 | 24–38 |
| BMI (kg/m2) | 21.3 ± 3.9 | 17.5–28.1 |
| Cycle length (days) | 29.1 ± 2.7 | 27–35 |
| Parity | ||
| Nulliparous | 9 | 90% |
| Primiparous | 1 | 10% |
| Previous spontaneous abortions | ||
| None | 10 | 100% |
|
| ||
| Descriptive characteristics of fertile controls (n = 14) | ||
| Age (years) | 36.2 ± 5.5 | 23–42 |
| BMI (kg/m2) | 21.9 ± 1.6 | 19.8–25.7 |
| Parity | ||
| Primiparous | 9 | 64.3 |
| Multiparous | 5 | 35.7 |
| Previous spontaneous abortions | ||
| None | 14 | 100% |
|
| ||
| Descriptive characteristics of patients undergoing PGT-A (n = 49) | ||
| Age (years) | 35.6 ± 3.6 | 25–43 |
| BMI (kg/m2) | 21.6 ± 2.5 | 16.9–28 |
| Basal FSH (IU/l) | 7.0 ± 2.3 | 1.3–14.0 |
| Basal AMH (ng/ml) | 2.9 ± 1.9 | 0.7–8.6 |
| AFC (n) | 13.1 ± 5.2 | 5.0–35.0 |
| Previous failed embryo transfers (n) | 1.9 ± 2.8 | 0–12 |
| Parity | ||
| Nulliparous | 44 | 89.8 |
| Primiparous | 4 | 8.2 |
| Multiparous | 1 | 2.0 |
| Previous spontaneous abortions | ||
| None | 27 | 55.1 |
| One | 10 | 20.4 |
| Two or more | 12 | 24.5 |
| Indication to PGT-A | ||
| Monogenic disease | 15 | 30.6 |
| Recurrent implantation failure | 14 | 28.6 |
| Chromosomal abnormality | 10 | 20.4 |
| Idiopathic recurrent abortion | 5 | 10.2 |
| Other | 5 | 10.2 |
| Not pregnant after IVF cycle | 27 | 55.1 |
| Pregnant after IVF cycle | 22 | 44.9 |
Data are expressed as mean ± SD or number (%).
AFC, antral follicle count; AMH, anti-Müllerian hormone; rFSH, recombinant FSH; PGT, preimplantation genetic testing.
Women with proven fertility were aged 20–42 years, non-smokers, taking no medication, with a BMI ≤ 26 kg/m2 and regular menstrual cycles (25–35 days). Proven fertility was defined as having had at least one previous live birth resulting from a natural conception and no previous history of miscarriage (Table I). To compare UF-EVs between the non-receptive and the receptive phase of the menstrual cycle, ovulation was timed by measuring the urinary LH surge (Clearblue Advanced Ovulation, Swiss Precision Diagnostics, Gmbh, Geneva, Switzerland) and UF samples were collected from each of the n = 14 fertile women twice, on day LH + 2 for the nonreceptive and day LH + 7 for the receptive phase, respectively. Inclusion criteria for the population of women undergoing ART comprised clinical indication to ART and PGT-A treatments (see Tables I and II), age ≤43 years, regular menstrual cycles (25–35 days) and a BMI ≤26 kg/m2.
Table II.
Characteristics of PGT-A patients divided by implantation outcome.
| Characteristics | Successful implantation | Failed implantation | P |
|---|---|---|---|
| (n = 19) | (n = 23) | ||
| Age (years) | 35.6 ± 3.2 | 35.6 ± 4.2 | 1 |
| BMI (kg/m2) | 21.7 ± 2.4 | 21.6 ± 2.8 | 0.98 |
| Basal FSH (IU/l) | 7.4 ± 2.0 | 6.2 ± 2.2 | 0.09 |
| Basal AMH (ng/ml) | 2.8 ± 1.7 | 3.1 ± 2.2 | 0.99 |
| AFC (n) | 12.3 ± 3.8 | 13.8 ± 6.4 | 0.48 |
| Previous failed embryo transfers (n) | 1.4 ± 2.0 | 2.3 ± 2.8 | 0.35 |
| Stimulation protocol data | |||
| Gn-RH antagonist protocol | 19 (100) | 23 (100) | 1.00* |
| Total dose of rFSH/hMG | 2386 ± 1163 | 2809 ± 1235 | 0.48 |
| Number of blastocysts transferred (n) | 1 (100) | 1 (100) | 1.00* |
| Indication to PGT | |||
| Monogenic disease | 7 | 5 | 0.66* |
| Recurrent implantation failure | 5 | 8 | |
| Chromosomal abnormality | 4 | 6 | |
| Other | 2 | 1 | |
| Idiopathic recurrent abortion | 1 | 3 |
Data are expressed as mean ± SD or number (%), P values are reported for Mann–Whitney U test or *Pearson chi square.
ART procedures
All included women underwent routine fertility investigations and treatments were performed according to standard protocols. If indication to ART and PGT was represented by infertility, treatments were started after at least 1 year of unsuccessful attempts to conceive spontaneously. As per standard PGT, women underwent controlled ovarian hyperstimulation with a GnRH antagonist protocol (Vanni et al., 2017). Oocytes were injected with a single spermatozoon 2–4 h after follicle aspiration and embryos were cultured to the blastocyst stage. Blastocysts obtained were biopsied and frozen until confirmation of ploidy status at NGS PGT. When diagnosis of blastocyst euploidy was confirmed, frozen-thawed embryo transfer was scheduled. In these patients, UF samples were collected once, on day 7 after detection of a urinary LH surge (Clearblue Advanced Ovulation, Swiss Precision Diagnostics, Gmbh, Geneva, Switzerland) in the menstrual cycle preceding that of the scheduled euploid embryo transfer. Therefore, on day LH + 7 of the following monitored natural cycle, ultrasound-guided euploid blastocyst transfer was performed. Patients were followed up and their clinical outcomes were recorded. Clinical pregnancy (i.e. successful implantation) was defined as the presence of an intrauterine gestational sac with a fetal beat on ultrasound examination performed at week 4–5 after embryo transfer (week 6–7 of gestation). According to the clinical outcomes of the n = 49 women included, n = 22 UF-EV samples from women who achieved a clinical pregnancy in the immediately following cycle could thus be compared to n = 27 UF-EV samples from women failing to achieve a pregnancy.
Endometrial tissue/uterine fluid collection and isolation of UF-EVs
Endometrial tissue biopsies were obtained with the use of a Pipelle, immediately after UF sampling. UF samples were obtained by lavage of the endometrial cavity with 2.5 ml of sterile saline solution using a balloon hysterosonography catheter to avoid vaginal contamination. As previously reported, recovered volume may vary (from 0.8 to 1.8 ml in our series) (Luddi et al., 2019). For EVs isolation, UFs were freshly processed to avoid freezing/thawing cycles. The samples were immediately recovered from the catheter and centrifuged at 1200 × g for 10 min at 4°C in order to separate the mucus from the liquid fraction and to remove cells and cell debris. The supernatant was held at 4°C while mucus within the sample was retrieved and suspended in PBS (pH 7.6, 1:1) (Euroclone S.p.a, Pero, MI, Italy). To release EVs, the mucus was physically dissociated by vortexing for 3–5 min and then centrifuged at 800 ×g for 5 min. The supernatant was harvested, and the procedure repeated several times combining the supernatants until the mucus was fully dissociated (i.e. no longer evident upon observation) (Ng et al., 2013). The supernatant from mucus dissociation was then added to the liquid fraction of UF and subjected to sequential centrifugations to isolate EVs, following a modification of the protocol described by Théry et al. (2018). Briefly, supernatants were subjected to a 300 ×g centrifugation for 10 min to remove any remaining cells, and then to a second centrifugation at 2000 ×g for 20 min to remove dead cells and debris. The supernatants were diluted in at least an equal amount of PBS and transferred to sterilized 1.5-ml polypropylene tubes (Beckman Coulter Inc., Brea, CA, USA, item no. 357448). Sample was ultra-centrifuged at 110 000 × g for 2 h at 4°C in a TLA-55 rotor (Beckman Coulter Inc.) using an Optima TLX centrifuge (Beckman Coulter Inc.) to pellet EVs. The pellets were resuspended in PBS, again centrifuged at 110 000 × g for 90 min and finally resuspended in a total volume of 110 µl of PBS (filtered three times with 0.1 µm filter). The EV resuspension was finally split in two aliquots, 100 µl for RNA NGS and 10 µl for nanoparticle tracking analysis (NTA). Aliquots of UF-EVs were preserved at −80°C until use.
Nanoparticle tracking analysis
The NanoSight NS300 system (NanoSight Ltd., Amesbury, UK) was used to visualize UF-EVs by laser light scattering in order to assess the concentration/size of UF-EVs. UF-EVs (3 µl) were diluted 1–300 with PBS, filtered three times with a 0.1 µm filter and concentrations adjusted, if necessary, in order to specifically fit the optimal working range (20–120 particles\per frame) of the instrument. Five 30-s videos were recorded under the flow mode for each sample with camera level set at 13 and detection threshold set at 5. At least 700 completed tracks per video were collected and the data were analyzed with NTA software (NTA 3.2 Dev Build 3.2.16, NanoSight Ltd., Amesbury, UK), which provided high-resolution particle size distribution profiles and EV concentration measurements (n° of particles/ml).
Transmission electron microscopy
Preparation for transmission electron microscopy (TEM) analysis was done using the method described by Théry et al. (2018). Briefly, freshly purified UF-EVs were absorbed on glow discharged carbon-coated formvar copper grids, washed with water, contrasted with 2% uranyl acetate and air-dried. Grids were observed with a Zeiss LEO 512 (Zeiss, Oberkochen, Germany) transmission electron microscope. Images were acquired by a 2k × 2k bottom-mounted slow-scan ProScan camera (ProScan, Lagerlechfeld, Germany) controlled by EsivisionPro 3.2 software (Soft Imaging System, Münster, Germany).
Western blotting
For western blotting analysis, endometrial tissues were lysed in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% Tween20, 10 mM β-glycerophosphate) with 1% Protease inhibitor cocktail (Merck KGaA, Darmstadt, Germany), while isolated EVs (20 µg as evaluated by Bradford assay and by NanoDrop8000 measurement) were lysed directly in reducing Laemmli buffer [0.25 M Tris–HCl (pH 6.8), 40% glycerol, 8% sodium dodecyl sulphate (SDS), 5% 2-mercaptoethanol and 0.04% bromophenol blue] and boiled for 5 min at 95°C. For tetraspanins detection, 10 µg of isolated EVs were lysed in non-reducing sample buffer (without 2-mercaptoethanol). For the detection of EV markers, total proteins were resolved by SDS-polyacrylamide gel electrophoresis, electrophoretically transferred to polyvinylidene fluoride membranes, blocked in 5% non-fat powdered milk in TBS-T (0.5% Tween-20) and the membranes were incubated with the following antibodies: anti-CD63 (1:1000; BD Pharmingen, #556019, San Jose, CA, USA), anti-CD9 (1:1000, BD Pharmingen, #555370, San Jose, CA, USA), anti-ALIX (1:500, Santa Cruz, #sc-271975, Santa Cruz, CA, USA), anti-TSG101 (1:500, Novus Bio, #NB200-112, Littleton, CO, USA), anti-calnexin (1:1000, Sigma, #C7617, Sigma/Aldrich, St. Louis, MO, USA), anti-TOM20 (1:1000; Abcam, #ab78547; Cambridge, MA, USA) and anti-TIM44 (1:1000; BD Transduction Laboratories, Franklin Lakes, NJ, USA). Protein bands were detected using X-ray film and enhanced chemiluminescence reagent (ECL, Amersham, Buckinghamshire, UK).
RNA isolation and sequencing library construction
Total RNA from endometrial tissues and EVs was isolated using RNeasy Mini kit and Micro Kit (Qiagen) respectively, following the manufacturer’s protocol (Qiagen, Hilden, Germania) with the optional step of DNase in column. Purified RNA quality/quantity was determined with a Bioanalyzer (Agilent Technologies, Waldbronn, Germany). In order to compare the transcriptomic profile of tissues and UF-EVs, cDNA libraries were generated from total RNA using the QuantSeq 3' mRNA-Seq Library Prep Kit (Lexogen GmbH, Vienna, Austria) according to the manufacturer's instructions. For RNA-seq analysis, on average, 10 M reads of 75 nt length were produced by Illumina NextSeq 500 via SBS technology (Illumina, San Diego, CA, USA). In order to perform transcriptome sequencing on UF-EVs collected from fertile women and ART patients during the window of implantation, cDNA libraries were prepared using the SMART-Seq® v4 Ultra® Low Input RNA Kit (Takara Bio Inc., Otsu, Shiga, Japan) according to the manufacturer’s instructions. Sequencing was performed on both Illumina NextSeq 500 and Illumina NovaSeq 6000 to obtain 15 million single end reads per sample.
RNA sequencing analysis
The raw reads produced from sequencing were trimmed using Trimmomatic, version 0.32, to remove adapters and to exclude low-quality reads from the analysis. The remaining reads were then aligned to the human genome GRCh38, annotated according to Gencode basic annotations version 27, using STAR, version 2.5.3a. Reads were assigned to the corresponding genomic features using featureCounts via Rsubread package. Quality of sequencing and alignment was assessed using FastQC and MultiQC tools. Differences in quantity and quality of the genomic starting material resulted in different depths of sequencing, thus producing RNA libraries of different sizes. For this reason, RNA-seq data were normalized for different library sizes by calculating the Counts Per Million (CPM) values, and also for different gene lengths by calculating the Read Per Kilobase Million (RPKM) values. CPM was used to define the ‘expressed’ genes (Chen et al., 2016), while RPKM were considered for exploratory analysis. ‘Expressed’ genes were defined as those genes showing at least 1 CPM read in a selected number of samples based on the size of the compared groups (Chen et al., 2016). Low expressed genes that did not match this criterion were excluded from the corresponding dataset. CPM values were also used for comparisons and correlation analysis among groups of samples.
For the first experimental approach, in order to explore the RNA content of EVs, two parallel differential gene expression (DGE) analyses were performed: comparing RNA profile of UF-EVs with RNA profile of tissues (paired model), and comparing RNA profile in UF-EVs collected in the secretory phase of the menstrual cycle (sUF-EVs) with RNA profile of EVs collected in the proliferative phase (pUF-EVs). The first comparison was tested using the limma-block model after voom normalization, to consider at the same time the origin of the sample (endometrial (E)-tissues or UF-EVs), the phase of the menstrual cycle and the pairing given by the specific woman, while the second comparison was performed using the edgeR models. For explorative analysis, RNAs in EVs were ranked according to the normalized CPM counts. In each comparison, only genes identified as ‘expressed’ in UF-EV samples were considered.
In the second experimental approach, to identify a specific transcriptomic profile of UF-EVs associated with receptivity, DGE analysis was performed using the R package DESeq2, version 1.6.3, with no assumption about prior distribution (option betaPrior = FALSE) and applying independent filtering to the results. Two comparisons were performed, the first within the fertile women group (fUF-EVs LH + 7 vs fUF-EVs LH + 2) and the second within ART patients grouped by pregnancy outcomes (UF-EVs ‘pregnant’ vs UF-EVs ‘not pregnant’). In each comparison, only genes identified as ‘expressed’ in the corresponding group of samples were considered. In all DGE comparisons, significance was defined using the cut-off suggested by SEQC consortium, that selects genes showing at the same time a raw P-value < 0.01 and a logFC greater than 1 or lower than −1 (SEQC-MAQC-III Consortium, https://doi.org/10.1038/nbt.2957). DGE analysis was performed starting from raw counts, since DESeq2, edgeR and Limma models apply an embedded normalization step.
Analysis of biotype composition of each sample was performed according to Gencode biotype definition (https://www.gencodegenes.org/pages/biotypes.html).
Validation of RNA-seq analysis
For validation of RNA-seq analyses, ANXA2, ITGB8, ALDH1A3, DCDC2 and TMEM37 mRNA levels were validated in a new cohort of UF-EVs samples from volunteers of whom n = 8 were in the proliferative phase and n = 8 in the secretory phase. Since the RNA content of EVs was expected to be limited, UF-EVs were subjected to RNA extraction, reverse transcription and whole transcriptome amplification using a REPLI-g Cell whole genome amplification (WGA) and whole transcriptome amplification (WTA) Kit (Qiagen), according to the manufacturer’s instructions. Briefly, UF-EVs were resuspended in 13 µl PBS as recommended; lysis buffer was added and the solution heated at 95°C for 5 min, to lyse and release the UF-EV contents. Lysed UF-EVs were used for WTA of total RNA. Genomic DNA was removed, cDNA was synthesized and subjected to ligation and amplification steps. The RT-reaction was included in the kit, which used T-Script reverse transcriptase combined with random and oligo-dT primers. REPLI-g SensiPhi DNA Polymerase with high proofreading was used for isothermal amplification of cDNA. The DNA derived from amplification of cDNA was used to perform the PCR protocol. In order to quantify and perform the PCR amplification using the same amount of cDNAs from each sample, purification of amplified DNA by LiCl/EtOH precipitation was performed according to supplementary protocol instructions in the REPLI-g kit. Purified amplified ds-cDNA has been quantified using Fluorimeter QFX with DeNovix high sensitivity Assay (DeNovix Inc., Wilmington, DE, USA). In order to ensure optimal normalization of results on total amplified cDNA quantity, 10 ng of the amplified cDNA of UF-EVs were subjected to PCR using SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). The PCR amplification was carried out for 40 cycles with an annealing temperature of 60°C. The following primers were used: ANXA2 (Fwd 5′-GGACGCGAGATAAGGTCCTG-3′, Rev 5′-ACCATTTCTGGACGCTCAGG-3′); ITGB8 (Fwd 5′-TGGCACTTCAGATGCAGTGT-3′, Rev 5′- GCTCCATGTTGAGTTGTGCG-3′); ALDH1A3 (Fwd 5′-CCTGAGTATTTCACTGGCAGGT-3′, Rev 5′-TCTGTTACGGGCCCTCATTT-3′) DCDC2 (Fwd 5′-AAGACAGAGCAAGGCGTTCA-3′, Rev 5′-AAGCATGCAAGCCTGAAAGC-3′); ACTB (Fwd 5′-GGCACCCAGCACAATGAAG, Rev 5′-CCGATCCACACGGAGTACTTG-3′) except for TMEM37 (Hs_TMEM37_2_SG QuantiTect Primer Assay #QT01530361), purchased from QIAGEN. Ten microliters of PCR products were loaded on a 2% agarose gel and stained with EtBr. The positive control was cDNA from endometrial cells.
Gene set enrichment analysis and over-representation analysis
Over-representation analyses for Gene Ontology (GO) terms and biological pathways (KEGG, Reactome and WikiPathway) were carried out by using the g:Profiler web tool (https://biit.cs.ut.ee/gprofiler). This software was chosen over other functional enrichment analysis tools as it is up-to-date and provides a compact graphical output. The obtained results were corrected for multiple testing by using the g:Profiler tailor-made algorithm g:SCS (Raudvere et al., 2019). The Webgestalt (http://www.webgestalt.org) platform was used for gene set enrichment analysis (GSEA). To compare RNA-seq results with the transcriptional profile of the gene-set evaluated in the ERA® test (Díaz-Gimeno et al., 2011), results from the fUF-EVs LH + 7 versus fUF-EVs LH + 2 comparison and from the UF-EVs ‘pregnant’ versus UF-EVs ‘not pregnant’ comparison were subjected to a Preranked GSEA using GSEA software; Normalized Enrichment Score (NES) was used to indicate the strength of the enrichment in GSEA. With NES being normalized for the number of genes in the considered pathway, the greater the NES absolute value, the stronger the enrichment. The GSEA leading edge, defined as the subset of genes that contributed most to the enrichment score, is also presented (Mootha, et al. 2003; Subramanian et al. 2005).
Statistical analyses
Normality was assessed with the Shapiro–Wilk test. Paired/unpaired Student’s t-test, non-parametric Wilcoxon and Kruskal–Wallis tests followed by the Dunn’s Multiple comparison’s tests were used as appropriate. The Chi-square test was used for the evaluation of gene transcripts detected only in UF-EVs of women who achieved a successful versus a failed pregnancy outcome. All results were expressed as mean ± SD or as median (interquartile range: IQR range). All the analyses and relative graphs were made in Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA) or JASP 0.11.1.0 (JASP Team, T. 2017 JASP, https://jasp-stats.org/).
Results
Characterization of UF-EVs according to the menstrual phase of the cycle
The size (Fig. 2A, left panel), concentration (Fig. 2A, right panel) and morphology (Fig. 2B) of EVs isolated from UF samples of regularly cycling women (n = 10) were characterized.
Figure 2.
Characterization of UF-EVs. (A) Particle size distributions by NTA in proliferative (pUF-EVs; n = 5) and secretory phase samples (sUF-EVs; n = 5) (left panel). Data are displayed as dot-plots showing mean and SD. Particle concentrations/ml by NTA in pUF-EVs and sUF-EV samples (right panel). Data are displayed as boxplots from the minimum to the maximum concentration value, showing median (line) concentration value. Underneath the graph, descriptive table reporting size distribution of UF-EVs collected in proliferative (pUF-EVs) and secretory (sUF-EVs) phase of the cycle. Data represent the median value of mode, D10 (10th percentile), D50 (50th percentile) and D90 (90th percentile) with interquartile range (IQR). (B) Transmission electron microscopy (TEM) representative illustrations of pUF-EVs (left image) and sUF-EV samples (right image) with 100–200 nm size. Scale bar column: 200 nm. The inset represents an area with clear extracellular vesicles structures (C) Western blot showing the presence of different canonic EV markers (CD63, CD9, ALIX, TSG101) in both pUF-EVs and sUF-EV. Two different samples/phase were included. NTA, nanoparticle tracking analysis; UF-EVs, uterine fluid-derived extracellular vesicles.
The evaluation of the 90% of nanoparticles distribution (D90) allowed us to confirm that the absolute majority of the UF-EV population laid in the <350 nm range. pUF-EVs showed a mean diameter ± SD of 185.02 ± 31.16 nm, a median value of D10 (10th percentile) below 110 nm (IQR: 77.0–117.6 nm) and of D90 (90th percentile) below 289.7 nm (IQR: 240.3–341.7 nm). sUF-EVs showed a mean diameter (± SD) of 213.05 ± 30.66 nm, a median value of D10 of 111.7 nm (IQR: 96.1–125.3 nm) and of D90 of 358.4 nm (IQR: 273.3–380 nm). No significant differences in size were thus encountered between pUF-EVs and sUF-EVs. Similarly, no significant changes could be detected in concentrations of EVs from UF samples collected in different phases of the cycle (median concentrations 2.6E + 11, [IQR = 1.09E + 11–2.50E + 12] particles/ml in UFs from proliferative phase women and 5.7E + 11 [IQR = 4.15E + 11–1.86E + 12] particles/ml in UFs from secretory phase women). TEM confirmed detection of EV-like particles in samples collected from both proliferative and secretory phase, with diameters ranging from less than 100 nm to bigger than 200 nm (Fig. 2B). Presence of the commonly used EV positive protein markers ALIX, TSG101, CD63 and CD9 was assessed and successfully confirmed by immunoblotting in UF-EVs samples collected during both phases of the menstrual cycle (Fig. 2C). Using endometrial tissue samples as controls, the western blot of negative EV markers was also performed: lack of calnexin (CNX), TOM20 and TIM44 expression in EVs samples suggests the absence of cellular debris by exclusion of endoplasmic reticulum (CNX) and mitochondrial (TOM 20, TIM44) contamination, respectively (Supplementary Fig. S1).
UF-EVs as a proxy of endometrial tissue throughout the menstrual cycle
As shown in Fig. 1A, to evaluate the correlation between the transcriptional profile of UF-EVs and endometrial tissues, UF-EVs and corresponding tissue biopsies collected from regularly cycling women (Table I) were subjected to RNA-seq analysis. The overall sequencing and alignment passed the quality check (FastQC and MultiQC tools), even if quality statistics varied widely among EV samples, whereas they were more stable and indicated a generally higher library complexity in the corresponding tissue samples. The total number of sequenced reads ranged from 10 to 17 million (M) except for one UF-EV sample (8.5 M). The alignment score was high for most of the samples, with 60–80% of uniquely mapped reads. High levels of duplication were observed (>100), especially in UF-EV samples. Overrepresented sequences (sequences which make up more than 0.1% of the total) partly consisted of poly-A sequences—therefore probably represented technical residuals of libraries preparation, and partly derived from Homo sapiens, as recognized by BLAST (Basic Local Alignment Search Tool from NCBI).
Considering as ‘expressed’ those genes with at least one CPM reads in at least n = 5 samples (since each group of UF-EVs or endometrial tissues in different cycling phases contains five biological replicates) (Chen et al, 2016), the number of detected gene transcripts was 15 268 in tissues and 4132 in UF-EVs (Fig. 3A). Based on this approach, in total 11 175 genes expressed in tissues were undetectable in UF-EVs. On the other hand, out of 4132 gene transcripts detected in EVs samples, n = 39 genes have reached the threshold of one CPM in at least n = 5 samples in EVs but not in endometrial tissues.
Figure 3.
RNA-seq comparisons of paired samples of E-tissues and UF-EVs. (A) Considering as ‘expressed’ those genes with at least 1 count per million (CPM) in at least n = 5 biological replicates (n = 5 E-Tissues or n = 5 EVs), Venn graph shows the number of gene transcripts detected in E-Tissues and in UF-EVs. Note that, based on this approach, out of 4132 gene transcripts detected in EVs samples, n = 39 genes have reached the threshold of 1 CPM in at least n = 5 samples of EVs but not of endometrial tissue. (B) Distribution of the average expression (log-transformed CPM values) of the 4093 gene transcripts detected in in both E-Tissues (upper panel) and UF-EVs (lower panel) (C) Correlation analysis of the gene transcripts detected in both sample types. The Pearson’s r is 0.70 (P<0.0001) and the Spearman’s ñ is 0.65 (P<0.0001). Each dot represents a gene, and values on x and y axes are the average gene expression values in tissue and UF-EVs samples, respectively. The color code represents the number of overlapping dots in a specific position of the graph. This number is quantified by the n-neighbors statistic that is the number of dots in the neighborhood of the considered dot. (D) Volcano plot of RNAs differentially detected in UF-EVs compared to E-Tissues (DGE analysis). DGE, differential gene expression analysis; E-tissues, endometrial tissues; UF-EVs, uterine fluid-derived extracellular vesicles.
Different cut-offs were tested for the definition of the expressed genes, but none of these alternative thresholds in any case resulted in a substantial increase in the number of ‘expressed’ genes, indicating that the great majority of reads in UF samples were distributed on the same ‘few’ genes. Mean gene ‘expression’ was higher in tissues than in UF-EVs but the correlation analysis for transcripts detected in both sample types showed a pattern of gene abundance strongly conserved between the two transcriptional profiles (Pearson’s r = 0.70, P < 0.0001; Spearman’s ρ = 0.65, P < 0.0001) (Fig. 3B and C). Correlation between the endometrial tissue and UF-EV transcriptomes remained significant also when grouping by phase-collection, specifically proliferative phase endometrial tissues versus proliferative phase UF-EVs (r = 0.64 P < 0.001; ρ = 0.60 P < 0.00001) and secretory phase endometrial tissues versus secretory phase UF-EVs (r = 0.64 P < 0.001; ρ = 0.61 P < 0.00001) (Supplementary Fig. S2).
DGE between tissues and UF-EVs could identify n = 908 genes as differentially expressed (Fig. 3D). As expected, the great majority of genes had a lower ‘expression’ in EV samples related to those of tissues, except for 38 genes (Fig. 3D, Supplementary Table SI and Supplementary Data File S1).
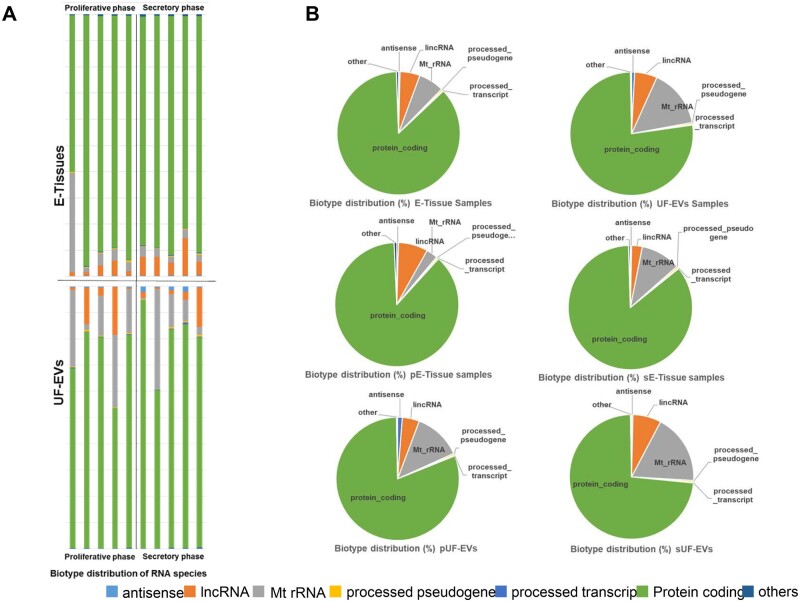
To assess the transcriptomic complexity of EV samples, we evaluated the different classes of transcripts present. Figure 4 shows the composition of each sample library in terms of different biotypes, as defined by Gencode (Fig. 4A) and the average biotype fractions in the different groups of samples (Fig. 4B). Most transcripts (86.45% in endometrial tissues and 76.96% in UF-EVs) derived from protein-coding genes, but fractions of ribosomal Mt-rRNAs were also present, more in UF-EVs than in tissue samples (15.36% and 6.66%, respectively). Fractions of long non-coding RNA (lncRNA) were represented at 5.24% in tissues and 5.93% in UF-EVs; antisense and processed pseudogenes categories accounted for <1% in both groups. The only statistically significant difference was found when comparing the percentage of CPM for protein-coding transcripts between of UF-EVs and endometrial tissues, with a higher abundance in the latter (P = 0.02). Biotype composition of samples sub-grouped based on proliferative versus secretory phase of the cycle of the collected samples was also evaluated but no statistically significant differences for the various classes of transcripts were observed (Fig. 4B).
Figure 4.
Characterization of UF-EV transcriptome. (A) Quantification of different gene biotypes detectable in paired samples of E-tissues and UF-EVs. Bar-plot of the RNA classes detected in the total transcriptome of each E-tissue and UF-EV sample. Biotypes were defined according to Gencode annotations. (B) Pie charts showing the average fraction of each biotype in different groups of samples. Pie charts were designed according to the type of sample (E-Tissue or UF-EV samples, first row) or according to the type of sample and phase of menstrual cycle: pE-Tissues, sE-Tissues (second row), pUF-EVs and sUF-EVs (third row). UF-EV, uterine fluid-derived extracellular vesicles; E-tissues, endometrial tissue samples; sE-Tissues, endometrial tissue samples collected in secretory phase of the menstrual cycle; pE-tissues, endometrial tissue samples collected in proliferative phase of the menstrual cycle; pUF-EVs, uterine fluid-derived extracellular vesicles collected in proliferative phase of the menstrual cycle; sUF-EV, uterine fluid-derived extracellular vesicles collected in secretory phase of the menstrual cycle.
In order to proceed with the characterization of the UF-EV transcriptome as a proxy of that of pairwise tissues, we studied the functional enrichment of the n = 200 most abundant transcripts detected in UF-EVs, excluding rRNAs. The GO analysis highlighted genes related to biological processes such as—among others—endogenous stimuli, responses to hormones, mRNA processing and regulation of cell cycle (orange dots GO:BP; Padj < 0.05, Fig. 5) and to the cellular components extracellular vesicles, focal adhesion and extracellular region (green dots; GO:CC; Padj < 0.05). The vesicles-mediated-transport Reactome pathway and the VEGFA-VEGF2 Signaling WikiPathway term were among the most significantly enriched pathways (dark and light blue dots, Fig. 5). A significant number of genes were also connected with the protein binding and cell adhesion binding (red dots, Gene ontology analysis, Molecular Function-GO: MF).
Figure 5.
Enrichment overrepresentation analysis results of UF-EVs transcriptomes: GOSt multiquery plot of Manhattan. The top n = 200 most abundant transcripts excluding rRNA were selected and tested against BP, MF, CC, Reactome and Wikipathway collections, among others. The most significant results (FDR < 0.05) are highlighted. Y-axis represents Benjamini corrected P-values (−log10) and X-axis represents the functional terms grouped and color coded by data source. The position of the terms in the frames is fixed and the terms of the same branch (Gene Ontology-GO) are close to each other (short link to full g:Profiler results: https://biit.cs.ut.ee/gplink/l/opagQqFsQz). BP, biological process; CC, cellular component; FDR, false discovery rate; G:OSt, Gene ontology Statistics; MF, molecular function; UF-EVs, uterine fluid-derived extracellular vesicles.
An additional DGE analysis was performed comparing gene ‘expression’ profiles in UF-EVs derived from samples collected during the secretory versus the proliferative phase of the cycle (sUF-EVs vs pUF-EVs) (Supplementary Fig. S3). Due to the high variability in RNA concentration among the UF-EV samples, we adjusted the DGE model by adding the original RNA concentration as covariate. The adjusted model detected 63 differentially ‘expressed’ genes based on SEquencing Quality Control (SEQC) cut-off between the two menstrual cycle phases (Supplementary Fig. S3). Among the 63 differentially expressed genes, a total of five (TMEM37, ITGB8, DCDC2, ANXA2andALDH1A3) genes were selected for a validation of the RNA-Seq analysis on UF-EVs, chosen taking into account their biological significance in endometrial activity and receptivity, and their differential expression also within tissues samples (sE-Tissues vs pE-Tissues). The PCR followed by electrophoresis showed a stable presence of TMEM37, ANXA2, ALDH1A3 and ITGB8 transcripts in EVs derived from women in secretory phase while DCDC2 transcript was detected mostly in pUF-EVs (Supplementary Fig. S4), confirming RNA-seq results.
Characterization of UF-EVs collected during the window of implantation
The size (Fig. 6A, left panel) and concentration (Fig. 6A, right panel) of EVs isolated from UF samples of fertile women (n = 13) during both receptive (7 days after the LH surge) and non-receptive endometrial phase (2 days after the LH surge) were characterized by NTA. A total of n = 26 samples (fUF-EVs LH + 2, n = 13; fUF-EVs LH + 7, n = 13) were analyzed. Table I shows the descriptive characteristics of women with proven fertility enrolled in the study. Particles detected in fUF-EVs LH + 7 samples were not statistically different from particles detected in fUF-EVs LH + 2 samples in terms of size. The evaluation of the 90% of nanoparticles distribution (D90) allowed us to confirm that the absolute majority of UF-EV population laid in the <350 nm range. fUF-EVs LH + 2 showed a mean diameter ± SD of 206.21 ± 14.26 nm, a median value of D10 below 120 nm (IQR: 99.8–126.6 nm) and of D90 below 322.8 nm (IQR: 303.9–340.8 nm). fUF-EVs LH + 7 showed a mean diameter (± SD) of 219.5 ± 18.71 nm, a median value of D10 of 114.8 nm (IQR: 101.4–131.9 nm) and D90 of 336.6 nm (IQR: 312.9–371.4 nm). Particle concentrations were also similar in UF samples collected on day LH + 2 (median concentrations 5.09E + 11 particles/ml; IQR = 2.88E + 11–9.04E + 11 particles/ml) and in UF samples collected on day LH + 7 (5.28E + 11 particles/ml; IQR= 3.41E + 11–7.45E + 11 particles/ml).
Figure 6.
NTA characterization of UF-EVs collected during the window of implantation. (A) Particle size distributions (left panel) and concentration (right panel) in samples from fertile women collected 2 (pre-receptive phase) or 7 days (receptive phase) after the LH peak (fUF-EVs LH + 2 n = 13; fUF-EVs LH + 7 n = 13). Data for size are displayed as dot-plot showing mean diameter and SD and as boxplots from the minimum to the maximum concentration (n° particles/ml) value, showing median (line) concentration value. (B) Particle size distributions (left panel) and concentration (right panel) in LH + 7 samples from ART patients who succeeded or not in obtaining a pregnancy (UF-EVs not pregnant n = 26; UF-EVs pregnant n = 22). Data for size are displayed as dot-plot showing mean diameter and standard deviation (SD) and as boxplots from the minimum to the maximum concentration (n° particles/ml) value, showing median (line) concentration value (*P = 0.014). (C) Table reporting size (nm) distribution of EVs isolated from UF collected from fertile women (fUF-EVs LH + 2/LH + 7) and from ART patients with successful (UF-EVs pregnant) or negative pregnancy outcome (UF-EVs not pregnant). Data represent the median value of mode, D10 (10th percentile), D50 (50th percentile) and D90 (90th percentile) with interquartile range (IQR). NTA, nanoparticle tracking analysis; fUF-EVs, uterine fluid-derived extracellular vesicles collected from fertile women; UF-EVs, uterine fluid-derived extracellular vesicles.
To investigate the potential of UF-derived EVs in monitoring the endometrial status, UF samples were also collected from ART patients (n = 48) in whom the outcomes of successful versus failed implantation after the following embryo transfer attempt were recorded. Table I shows the descriptive characteristics of ART women enrolled in the study. This category of women undergoing PGT-A was chosen because—given the diagnosis of embryo competence based on state-of-the-art NGS PGT-A—their implantation potential was considered dependent on endometrial characteristics (Vaiarelli et al., 2016).
NTA on UF-EVs from ART women samples (not pregnant n = 26; pregnant n = 22) revealed that UF-EVs from women who achieved successful pregnancy were smaller (mean diameters ± SD 205.5 ± 22.97 nm) compared with those collected from women who did not get pregnant (mean diameters ± SD 221.5 ± 20.57 nm, P = 0.014) (Fig. 6B, left panel). More specifically, UF-EVs from women who achieved successful pregnancy showed a median value of D10 below 113.3 nm (IQR: 104.9–126.9 nm) and of D90 below 325.5 nm (IQR: 295.5–352.4 nm). Conversely, UF-EVs from women who did not get pregnant showed a median value of D10 of 121.3 nm (IQR: 114.1–131.5 nm) and D90 of 351.4 nm (IQR: 322.5–388.7 nm). Particle concentration was, conversely, similar in UFs from women who achieved a pregnancy and in UFs of women who failed to conceive (median concentrations 3.58E + 11 particles/ml [IQR= 2.29E + 11–6.33E + 11] and 3.84E + 11 particles/ml [IQR = 2.97E + 11–6.31E + 11] particles/ml, respectively) (Fig. 6B, right panel).
Changes in global transcriptome in UF-EVs during the window of implantation
To unravel the transcriptomic profile of EVs released by the endometrium during the receptive phase of the cycle, we analyzed the RNA content derived from pairwise UF samples of n = 14 women with proven fertility 2 and 7 days after the LH surge, as well as of n = 49 ART patients with successful (n = 22) versus failed (n = 27) implantation after the transfer of one euploid embryo. In total, n = 73 samples of RNA extracted from UF-EVs were sequenced and analyzed in three batches. Each batch contained at least n = 6 samples derived from n = 3 fertile women (n = 3 fUF-EVS LH + 2 and n = 3 fUF-EVs LH + 7) and at least n = 12 samples derived from ART women (UF-EVs not pregnant and UF-EVs pregnant). This has been done to allow proper removal of the ‘batch effect’ that may arise when samples are processed in different runs, with inherent bias (Leek et al., 2010). The average quality of sequencing (FastQC statistic) was >28 (mean sequence quality value Phred score) for all the three batches, while the percentage of uniquely mapped reads varied among batches: samples in the first and in the third batches presented percentages of uniquely mapped reads between 40% and 70%, while in the second batch, these percentages were lower than 25% with only few exceptions. In all the three batches, the majority of aligned reads originated from exonic regions (Supplementary Fig. S5A). Read coverage over gene bodies was uniform, with only a slight 3’ bias in n = 5 samples (Supplementary Fig. S5B).
Considering all sequencing data, we defined ‘expressed’ those genes with at least one CPM in at least n = 12 different samples, where n = 12 was the size of the smallest group of samples (fertile women samples). In the original group of n = 14 LH + 2 and n = 14 LH + 7 fUF-EV paired samples from fertile women, n = 2 samples/group (belonging to a total of n = 3 different women) were excluded for unsuccessful library preparation, leaving n = 12 fUF-EV samples per group available. According to this definition, 16 777 transcripts present in UF-EVs of both fertile women and in patients undergoing ART were identified. The great majority of the mapped transcripts originated from protein-coding genes in almost all the samples, except for n = 7 samples belonging to the second batch that contain a high fraction of Mt-rRNAs. This is likely due to the overall lower quality observed for the second batch of libraries (Supplementary Fig. S5C).
Transcriptional profile of UF-EVs from fertile women before and during the window of implantation
DGE analysis on fUF-EV samples from fertile women to explore the physiological endometrial EV transcriptomic profile of the receptive phase was performed using a paired model (DGE of fUF-EVs LH + 7 vs fUF-EVs LH + 2) (Fig. 1B). The comparison within paired samples referred to UF-EVs collected during the window of implantation (LH + 7) versus those collected in the non-receptive phase (LH + 2) from the same women. We did not consider the effect of batch in this model since each pair of samples was sequenced in the same batch. Considering as ‘expressed’ those genes with at least one CPM in at least n = 11 samples (the size of paired samples), we identified 14 228 detected genes in fUF-EVs. Genes that were differentially ‘expressed’ between the two groups were 2247 (942 more abundant gene transcripts and 1305 less abundant in the fUF-EVs LH + 7 group; Fig. 7A and Supplementary Data File S1). The heatmap representing the ‘expression’ of the top 50 most significant genes in the comparison is reported in Fig. 7B, where a clear distinction between the two groups of samples (LH + 7 and LH + 2) is evident. The top 50 genes differentially represented in the fUF-EVs LH + 7 versus fUF-EVs LH + 2 paired sample analysis are shown in Supplementary Table SII.
Figure 7.
DGE of transcripts from EVs isolated from UF samples of fertile women (fUF-EVs) before (LH + 2) and during (LH + 7) the window of implantation. (A) Volcano plots of the differentially detected RNAs in fUF-EVs LH + 7 compared to fUF-EVs LH + 2 samples. Results were obtained using DESeq2 and significance was defined according to SEquencing Quality Control (SEQC) cutoff, that selects genes showing at the same time a raw P-value < 0.01 and a |log2FC| >1. (B) Heatmap representing the ‘expression’ of the top 50 most significant genes in the corresponding comparisons. Expression values have been scaled so that their sum is the same for each gene. Log2FC: log 2 fold change. DGE, differential gene expression analysis; EVs, extracellular vesicles; UF, uterine fluid; fUF-EVs, uterine fluid-derived extracellular vesicles collected from fertile women.
We next focused on the whole gene ‘expression’ profile as a rank gene expression list based on log2 fold change (log2FC). All detected gene transcripts (14 228) were sorted based on their log2FC value between fUF-EVs LH + 7 versus fUF-EVs LH + 2, resulting in a pre-ranked gene list that was further used in a GSEA using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt, http://www.webgestalt.org/). As such, GSEA was performed to uncover the biological relevance of distinct transcriptome profiles of EVs collected during the receptive phase (fUF-EVs LH + 7) and of EVs collected 2 days after LH surge (fUF-EVs LH + 2). A false discovery rate (FDR) < 0.05 with a minimum of 20 genes per category served as the threshold. Analysis of GO revealed that, compared to fUF-EVs LH + 2 samples from fertile women, the transcriptional profile of fUF-EVs LH + 7 samples was highly enriched with genes associated with immune response processes including neutrophil mediated immunity (NES) = 2.91, adaptive immune response (NES = 2.88), humoral immune response (NES = 2.80) and regulation cell-to-cell adhesion (NES = 2.5) (Fig. 8A and Supplementary Table SIII). Moreover, we observed a significant depletion of processes involved in cellular division, such as DNA replication (NES = −2.23) and cell cycle G2/M transition (NES = −1.96) in LH + 7 samples compared to LH + 2 samples (Fig. 8A and Supplementary Table SIII). Cellular component analysis revealed that the majority of transcripts enriched in UF-EVs LH + 7 samples were associated with vesicles and immune system components: tertiary granule (NES = 2.92), vesicles lumen (NES = 37) and endocytic vesicles (NES = 2.17), while a depletion of transcripts associated with DNA replication components such as replication fork (NES = −2.01) and DNA packaging complex (NES = −1.94) was observed (Fig. 8B and Supplementary Table SIII). Among different molecular functions, we found antigen binding (NES = 2.69) and MHC protein binding (NES = 2.01) to be enriched and helicase activity (NES = −1.82) and cyclin dependent protein kinase activity (NES = −1.75) to be depleted among transcripts detected during the implantation window compared to LH + 2 samples (Fig. 8C and Supplementary Table SIII).
Figure 8.
Gene set enrichment analysis using WEB-based GEne SeT AnaLysis Toolkit (WebGestalt, http://www.webgestalt.org/ ) of transcripts from EVs isolated from UF samples of fertile women (fUF-EVs) before (LH + 2) and during (LH + 7) the window of implantation. All the 14 228 ‘expressed’ genes in fertile women UF-EVs were sorted based on their log2FC value for comparison between fUF-EVs LH + 7 and fUF-EVs LH + 2 samples resulting in a pre-ranked gene list that was further used in a gene set enrichment analysis (GSEA) GO analysis. Only categories with an FDR < 0.05 and a minimum of 20 genes are presented. The NES of top enriched (blue bars) and top depleted (orange bars) pathways in fUF-EVs LH + 7 compared to fUF-EVs LH + 2 are listed, categorized by GO annotations. (A) Bar graphs of biological process GO analysis. (B) Bar graph of cellular component GO analysis. (C) Bar graph of molecular function GO analysis. EV, extracellular vesicles; NES, normalized enrichment score; fUF-EVs, uterine fluid-derived extracellular vesicles collected from fertile women; FDR, false discovery rate; log2FC, log 2 fold change.
Transcriptional profile of UF-EVs from women with successful versus failed implantation following ART procedures
To further evaluate the EV transcriptional profile of the receptive endometrium, DGE analysis was performed in UF-EV samples collected from ART patients on day LH + 7 in the month preceding that of the scheduled transfer of one euploid blastocyst diagnosed by NGS PGT-A (Fig. 1C). The comparisons between samples referred to UF-EVs collected from women who achieved a successful implantation versus those collected from women who failed to become pregnant. The choice of women undergoing PGT-A as study population allowed us to exclude the possible bias owing to the genetic contribution of the embryo in the implantation failure. Out of n = 49 UF-EV samples collected from ART patients, n = 7 should not be included in the analysis because of the low number of their total read counts (<2 million reads per sample). The model was then performed adding the sequencing batch as a covariate. Of the remaining n = 42 samples, n = 23 samples derived from women who did not achieve pregnancy and n = 19 from women in which ART procedure was successful. Descriptive characteristics of the two groups are presented in Table II.
Considering as ‘expressed’ those genes with at least 1 CPM in at least n = 19 UF-EV samples where n = 19 was the size of the smallest group (pregnant women), we detected 14 593 transcripts in LH + 7 UF-EVs of ART patients. Genes that were differentially ‘expressed’ between the two groups were 161 considering the SEQC cut-off (97 more abundant gene transcripts and 64 less abundant gene transcripts in the group with successful implantation) (Fig. 9A and Supplementary Table SIV). Figure 9B shows the eight genes with the most significantly different transcript levels between the two groups.
Figure 9.
DGE of transcripts from EVs isolated from LH + 17 UF of women who achieved pregnancy versus those from LH + 17 samples of women who failed to conceive after an ART cycle with euploid blastocyst transfer. (A) Volcano plot of the significantly differentially detected RNAs in UF-EVs from pregnant compared to UF-EVs from not pregnant women. Results were obtained using DESeq2 and significance was defined according to SEQC cutoff, that selects genes showing at the same time a raw P-value < 0.01 and a |log2FC| >1. (B) Table representing genes with highest statistically significant differences between the two sample groups. (C–F) GSEA using WEB-based GEne SeT AnaLysis Toolkit (WebGestalt, http://www.webgestalt.org). All the 14 593 ‘expressed’ genes in ART women UF-EVs were sorted based on their log2FC value for comparison between LH + 7 UF-EV samples derived from women who achieved pregnancy versus LH + 7 UF-EV samples from women who failed to conceive, resulting in a pre-ranked gene list that was further used in a GSEA ontology analysis. Only terms with a minimum of 20 genes are presented. The NES of top enriched (blue bars) and top depleted (orange bars) pathways in LH + 7 UF-EV samples from women achieving pregnancy compared to those from women with failed implantation are listed, categorized by GO annotations. Terms with an FDR < 0.05 are represented with dark blue or orange bars; terms with an FDR ≥ 0.05 are represented with light blue or orange bars. Bar graphs of GO analysis, biological process (C) and cellular component (D) with a minimum of 20 genes per category are presented. Bar graphs of pathway analysis performed using Reactome (E) or KEGG (F) databases. DGE, differential gene expression analysis; EV, extracellular vesicles; GSEA, gene set enrichment analysis; log2FC, log 2 fold change; NES, normalized enrichment score; SEQC, SEquencing Quality Control; UF-EVs, uterine fluid-derived extracellular vesicles.
All detected gene transcripts (14 593) were sorted based on their log2FC value between UF-EVs from women achieving pregnancy versus UF-EVs from women not achieving pregnancy, resulting in a pre-ranked gene list that was further used in a GSEA (WebGestalt). The top 10 most significant categories with a minimum of 20 genes per category served as threshold. Analysis of GO considering an FDR < 0.1 revealed that, compared to samples from women with failed implantation, the transcriptional profile of UF-EVs from women who achieved pregnancy was highly enriched with genes significantly associated with immune response processes, including tumor necrosis factor superfamily cytokine production (NES = 2.02), natural killer cell activation (NES = 1.96), cell killing (NES = 1.83) and response to type I interferon (NES = 1.80) (Fig. 9C and Supplementary Table SV). Moreover, we observed significant depletion of biological processes including epidermis development (NES =−1.88) (Fig. 9C and Supplementary Table SV). Cellular component analysis identified only extracellular matrix (NES = 1.60) as significantly enriched and cornified envelope (NES = −2.67) as significantly depleted in UF-EVs of women with subsequent successful implantation (Fig. 9D and Supplementary Table SV). Among different molecular KEGG and Reactome pathways enriched in EVs from women who achieved a pregnancy, the most significantly enriched were antigen processing and presentation (NES = 1.82), allograft rejection (NES = 1.76), MHC protein binding (NES = 2.01), cell adhesion molecules (CAMs) (NES = 1.66), whereas NF-kappa B signaling pathway (NES = 1.81), Interleukin-10 signaling (NES = −2.06) and IL-17 signaling pathway (NES = −2.12) were depleted (Fig. 9E and F and Supplementary Table SV).
In addition, we aimed at identifying genes ‘selectively expressed’ either by women achieving a pregnancy or failing to conceive. Genes were defined as ‘selectively expressed’ in LH + 7 UF-EV samples from women who achieved pregnancy when they showed ≥1 CPM in at least n = 10 out of n = 19 of these samples and with ≥1 CPM in less than one-fifth of the LH + 7 UF-EV samples from women not achieving pregnancy (4/23 samples). We thus identified 14 transcripts selectively detected in UF-EVs of women with a successful implantation: AC114491.1, AC008608.2, PMS2P5, C10orf99, NPTN-IT1, AC012358.3, ANKRD18A, GLIS2-AS1, AC011447.7, AL009174.1, C1QTNF2, TMED6, AC016355.1 and AL021392.1 (Supplementary Fig. S6, upper panel). Similarly, genes were defined as ‘selectively expressed’ in LH + 7 UF-EV samples from women not achieving pregnancy when they showed ≥1 CPM in at least n = 10 out of n = 23 of these samples and with ≥1 CPM in less than one-fifth of the LH + 7 UF-EV samples from women who achieved pregnancy (3/19 samples). The five genes detected only in UF-EVs of patients who failed implantation were CD200R1, FAM66B, AL391834.1, WNT9B and CECR7 (Supplementary Fig. S6). Notably, 6/19 of the transcripts ‘selectively expressed’ only in one of the two groups of samples analyzed are novel lncRNAs with unknown functions. In Supplementary Fig. S6 lower panel, we report levels of the genes with the most statistically significant ‘selective expression’ in UF-EV samples collected from women who achieved a pregnancy versus samples collected from women who failed to conceive (PMS2P5, ANKRD18A, GLIS2-AS1, NPTN-IT1 and CERC7).
Intersection between genes differentially ‘expressed’ in both comparisons: LH+7 UF-EVs versus LH+2 UF-EVs of fertile women and in LH+7 UF-EVs of women achieving or not an ART pregnancy
Intersection of DGE analysis within fertile women and DGE analysis within women undergoing ART was performed in order to identify genes differentially ‘expressed’ both in LH + 7 UF-EVs of fertile women and in LH + 7 UF-EVs of women achieving an ART pregnancy. To achieve this, we have extracted the significant genes from each DGE model using the SEQC cut-off, and calculated the intersection between these gene lists: 41 genes had a statistically significant different ‘expression’ in both comparisons (Fig. 10A and B). Out of 41 genes included in the intersection, 24 showed the same positive or negative ‘expression’ trend whereas 17 had opposite ‘expression’ trend between the two comparisons (Supplementary Data File S1).
Figure 10.
Analysis of the genes differentially ‘expressed’ both in fertile women UF-EVs (fUF-EVs LH + 7 vs fUF-EVs LH + 2) and in ART patients UF-EVs (LH + 7 UF-EVs of women who achieved pregnancy vs LH + 7 UF-EVs of women who failed to conceive). (A) Venn graph of the significant genes identified by each of the two DGE analyses. (B) Table with Gene symbols, Gene names and Entrez IDs of genes included in the intersection. (C) Gene ontologies and pathways most strongly enriched among endometrial receptivity-associated genes. Genes are presented on the right side on the circle and the correlating GO processes, cellular compartments and pathways are on the left side. DGE, differential gene expression analysis; UF-EVs, uterine fluid-derived extracellular vesicles.
Enrichment analysis for identification of biological processes and pathways connected to these genes (performed with g:Profiler software) revealed them to be mostly involved in biological processes such as cell-cell adhesion, immune system process, chemotaxis, response to external stimuli. Figure 10C shows the connections of the 41 intersection genes with their respective GO biological processes. Three significantly enriched pathways were identified: KEGG pathway of chemokine signaling, Reactome pathway of immune System and a WikiPathway term of type II interferon signaling. A significant number of the genes were also connected with the extracellular region and secretory cell compartments (Fig. 10C and Supplementary Table SVI).
UF-EVs as a potential liquid biopsy for endometrial receptivity status
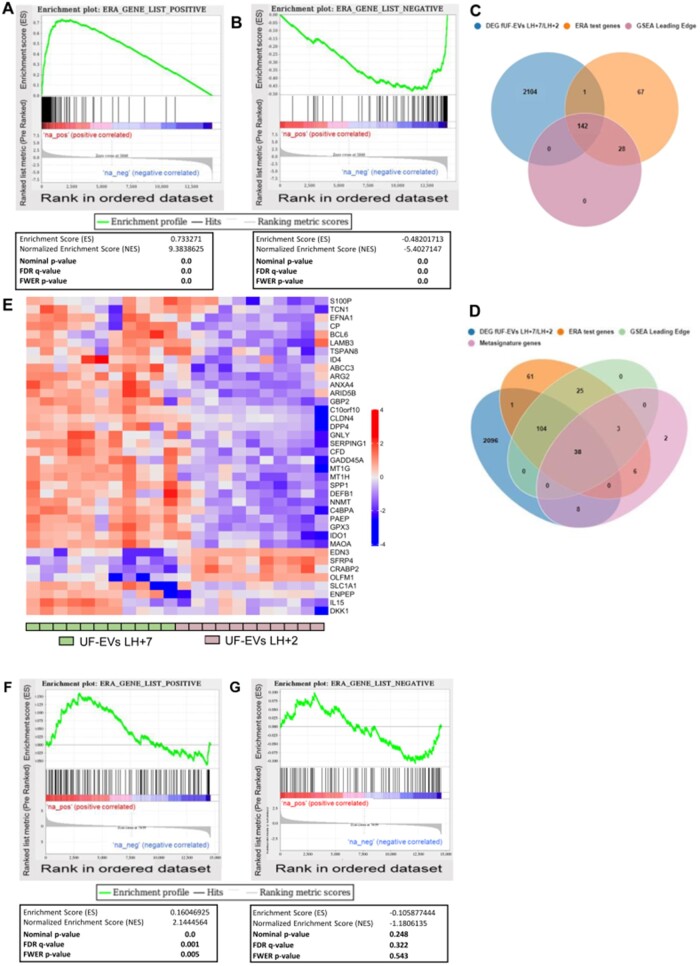
To confirm the possibility that the UF-EV-transcriptome may recognize the receptive endometrium during the implantation window in physiologic conditions, we have performed a Preranked Gene Set Enrichment Analysis (Preranked-GSEA, GSEA software, a joint project of UC San Diego and Broad Institute; Mootha et al. 2003; Subramanian et al., 2005) on the log2FC data resulting from comparison within samples from fertile women (fUF-EVS LH + 7 vs fUF-EVs LH + 2). In particular, we have evaluated whether the gene-set reported in the ERA® (Díaz-Gimeno et al., 2011) was significantly enriched among the more abundant or less abundant transcripts in our ‘LH + 7 versus LH + 2’ fUF-EV pairwise model comparison.
Enrichments were extremely significant and consistent. Using the 238 ERA® gene list, (of which n = 143 were described to be up-regulated and n = 95 to be down-regulated during the implantation window), GSEA highlighted that fUF-EVs have a similar transcriptional profile to that reported in the receptive endometrial tissue. A significant NES = 9.38 (P < 0.001) was found for transcripts up-regulated in LH + 7 in ERA® and more abundant in fUF-EVs LH7, and a similarly significant NES = −5.40 (P < 0.001) was found for transcripts down-regulated in LH + 7 in ERA® and less abundant in fUF-EVs LH + 7. The GSEA leading edge, i.e. the subset of our gene set that contributed most to the enrichment score, counted 106 more abundant gene transcripts and 64 less abundant gene transcripts in EVs derived from day LH + 7 compared to LH + 2 (Fig. 11A and B). Comparing the lists of leading-edge genes with ERA® genes and with differentially ‘expressed’ genes (DEG of fUF-EVs LH + 7/fUF-EVs LH + 2), we found 142 genes in common among these three lists (Fig. 11C). Looking at the Venn diagram (Fig. 11C), it can be observed that only one gene appeared both on the list of ERA® genes and on the list of DEGs (fUF-EVs LH + 7/LH + 2) but not in the list of leading-edge genes. This gene is BIRC3, showing more abundance during the implantation window in LH + 7 UF-EVs and a down-regulation in tissues during the receptive phase according to the ERA® results. A further comparison was performed adding a fourth list of genes, consisting of the ‘metasignature’ transcriptome derived from the published meta-analysis of endometrial receptivity-associated transcripts, comprising 57 genes (Altmäe et al., 2017). This intersection identified 38 genes ‘expressed’ in UF-EVs: showing differential ‘expression’ with the same trends between LH+ 7 vs LH + 2 in both EVs and ERA®; belonging to the GSEA leading edge of genes with the most significant enrichment score; and identified as receptivity-associated genes by the current meta-analysis on endometrial transcriptome (Fig. 11D and Supplementary Table SVII). The Heatmap in Fig. 11E shows the transcriptional profile of these 38 intersection genes in our LH +2 and LH + 7 fUF-EV samples collected from fertile women. Preranked GSEA was also performed to determine whether the ERA® genes are also present in UF-EVs of ART patients and whether they have a similar transcriptional profile in relation to the achievement or not of pregnancy. For the n = 143 transcripts that are up-regulated in the receptive phase according to the ERA® test, a significant enrichment with a NES = 2.14 (P = 0.001) for transcripts more abundant in UF-EVs of women who conceived was found, with 48 genes belonging to the leading edge of the GSEA (Fig. 11E). For the 95 transcripts that are down-regulated in the receptive phase according to the ERA® test, a not significant but concordant trend with a NES = −1.18 (P = 0.3) for transcripts less abundant in UF-EVs of women who conceived was found, with n = 25 genes belonging to the leading edge of the GSEA (Fig. 11F and G).
Figure 11.
Comparison of fUF-EVs RNA-seq results with transcriptional profile of ERA® test genes on endometrial tissue. Preranked Gene Set Enrichment Analysis (Preranked-GSEA, GSEA software https://www.gsea-msigdb.org/gsea/index.jsp) was performed to test the enrichment of the ERA® gene-sets with the transcriptomes of samples from fertile women [Genes were preliminary ranked according to the Log2FC of fUF-EVs LH + 7 (n = 11) vs fUF-EVs LH + 2 (n = 11)]. GSEA curves are obtained for the n = 143 and n = 95 ERA® gene-sets expected to be respectively up-regulated (A) and down-regulated (B) during the implantation window. ERA® genes expected to be up-regulated during the implantation window, were significantly enriched in fUF-EVs LH + 7 (NES = 9.38; P < 0.001) and ERA® genes expected to be down-regulated during the implantation window, were significantly depleted in fUF-EVs LH + 7 (NES = −5.40; P < 0.001) (C) Venn graph showing the overlap between genes in the ERA® test, genes in the GSEA Leading Edge and differentially ‘expressed’ genes in samples from fertile women (fUF-EVs LH + 7/LH + 2, raw P-value <0.01 and a |Log2FC|>1). (D) Venn graph showing the overlap between genes in the ERA® test, genes in the GSEA Leading Edge, Metasignature (Altmäe et al., 2017) and differentially ‘expressed’ genes in samples from provenly fertile women (fUF-EVs LH + 7/LH + 2, raw P-value <0.01 and a |Log2FC|>1). (E) Heatmap representing the ‘expression’ of 38 genes in the LH + 2 (n = 11) and LH + 7 (n = 11) fUF-EV samples of fertile women. Such 38 genes represent the intersection group of genes that (i) are differentially ‘expressed’ with the same trends between LH + 7 versus LH + 2 in both EVs and ERA® (ii) belong to the GSEA leading edge of genes with the most significant enrichment score and (iii) are identified as receptivity-associated genes by the current meta-analysis on endometrial transcriptome (Altmäe et al., 2017). Preranked-GSEA was performed to test the enrichment of the ERA® gene-sets with the transcriptomes of UF-EV samples from women undergoing ART [Genes were preliminary ranked according to the Log2FC of UF-EVs ‘pregnant’ (n = 23) vs UF-EVs ‘not pregnant’ (n = 19)]. GSEA curves are obtained for the n = 143 and n = 95 ERA® gene-sets expected to be respectively up-regulated (F) and down-regulated (G) during the implantation window. ERA® genes expected to be up-regulated during the implantation window were significantly enriched in LH + 7 UF-EVs of women achieving a pregnancy (NES = 2.14; P = 0.001). For ERA® genes expected to be down-regulated during the implantation window, a not significant but concordant trend for depletion with a NES = −1.18 (P = 0.3) for transcripts less abundant in UF-EVs of women who conceived was found. fUF-EVs, uterine fluid-derived extracellular vesicles collected from fertile women; GSEA, gene set enrichment analysis; NES, normalized enrichment score; log2FC, log 2 fold change.
Discussion
In this study, we sought to determine the RNA content of EVs released in the uterine microenvironment in order to understand its potential value as a proxy of endometrial receptivity status. Purified EV populations containing exosomes have been previously isolated and characterized in uterine fluid (Ng et al., 2013; Vilella et al., 2015; Luddi et al., 2019). Different protocols for their isolation have been assessed (Vilella et al., 2015; Campoy et al., 2016) and the standard procedure by differential centrifugation was found as the best approach in terms of providing sufficient materials for proteomic and transcriptomic analyses. We have thus confirmed that UF-EVs can be efficiently isolated. However, the UF-EV transcriptomic profile characterizing the implantation window and the receptive endometrial status had not been investigated yet. The first major finding of our study was the highly significant correlation between the RNA signature of EVs obtained from UF collected throughout the menstrual cycle with the transcriptome profile of pairwise endometrial tissue biopsies. Biotype distribution analysis confirmed the overall similarity between the two types of samples as the protein coding transcripts were in both cases detected as the major class represented. As expected based on the limited material enclosed in EVs, DGE analysis of the 908 genes differentially ‘expressed’ between EVs and endometrial tissues showed that, except for 38 genes, transcripts had a lower ‘expression’ in EV samples related to tissue samples, confirming that the transcriptional profile in EVs reflected the endometrial expression profile. Moreover, among the UF-EV samples, the great majority of reads were distributed on the same consistent genes, as demonstrated by the limited changes in the number of ‘expressed’ genes when different cut-offs in CPM of reads mapped to define ‘expression’ were applied. On one hand, the very high degree of similarity that we observed between the gene expression of the endometrial tissue and of EVs might to some extent suggest that these are more likely to be microvesicles or apoptotic bodies, both including portions of the cytoplasm of the origin cell. However, we cannot definitely distinguish the relative contribution of exosomes, microvesicles and apoptotic bodies to our findings. As a matter of fact, assigning an EV to a particular biogenesis pathway remains extraordinarily difficult and current guidelines of the International Society for Extracellular Vesicles suggest not to operate such a distinction ‘unless, for example, the EV is caught in the act of release by live imaging techniques or labelled with specific markers’ (Théry et al., 2018). On the other hand, the fact that 38 gene transcripts were found to be more abundant in UF-EVs than in endometrial tissue by DGE analysis supports the idea of some selective RNA incorporation in UF-derived vesicles. However, mechanisms that influence packaging of RNAs into EVs, the extent to which these are specific to selected RNA species, and how RNA cargo loading is regulated by human cells remain elusive (O'Brien et al., 2020). Nonetheless, our results overall identify UF-EVs as sources of biomarkers for endometrial receptivity. Most abundant transcripts in EVs revealed their role in cell adhesion, response to hormone and stimulus processes and confirmed both the endometrial origin (from stromal and glandular cells) and their connection with the extracellular region and vesicles-mediated transport. Transcripts enriched in EVs from the secretory phase of the cycle comprised genes involved in focal adhesion processes, such as RAB14, ITGB8, PPP2R5C, RPS6KB2 and genes that are known to be specifically up-regulated in the tissue during this phase, such as TMEM37 (Tseng et al., 2010). In mice, ITGB8, activates the VAV-RAC1 signaling axis via FAK, facilitating endometrial epithelial cell receptivity towards blastocyst attachment (Kumar et al., 2015).
In light of these findings, confirming the hypothesis that EV cargo may represent a comprehensive picture of the tissue transcriptomic profile, we proceed with our wider experiments aimed at exploring the transcriptomic and biophysical UF-EV profile characterizing the implantation window. DGE analysis comparing the RNA content of UF-EVs collected from women with proven fertility at LH + 7 versus RNA content of non- receptive LH + 2 EV samples identified 942 transcripts enriched in LH + 7 samples. These transcripts are involved in the same processes that are up-regulated in endometrial tissue in the same phase of the cycle. Indeed, the proteins encoded by these RNAs are mainly related to cell adhesion, immune responses, cell communication and negative regulation of proliferation and development (Talbi et al., 2006; Díaz-Gimeno et al., 2011, 2014). As significant examples, among the most significantly different transcripts in the LH + 7 versus LH + 2 comparison are PAEP (progestagen associated endometrial protein) and GPX3 (glutathione peroxidase 3), recently reported by single-cell level transcriptomic studies to be upregulated first in the luminal and then in the glandular endometrial epithelium during the transition to the window of implantation (Wang et al., 2020). Similarly, among enriched genes in our fUF-EVs LH + 7 versus LH + 2 comparison are metallothionein I genes (MT1E, MT1F, MT1G, MT1X), also described at single-cell level to be a key regulatory module associated with the window of implantation in endometrial unciliated epithelium (Wang et al., 2020). However, we draw particular attention to our results comparing transcripts enriched in EVs of the mid-secretory phase with the ERA® genes. The ERA®, together with a computational algorithm, detects the unique transcriptomic signature of endometrial receptivity by analyzing 238 DEGs in an endometrial biopsy and predicting the window of implantation (Díaz-Gimeno et al., 2013). The second major finding of this study was that the gene-set reported in the ERA® (Díaz-Gimeno et al., 2011) was significantly enriched among the mRNA species representatives of our ‘fUF-EVs LH + 7 versus fUF-EVs LH + 2’ pairwise mode comparison. Specifically, the GSEA analysis revealed a significant concordance in transcriptional profile between ERA® genes and the transcripts more or less abundant in the LH + 7 versus LH + 2 fUF-EV comparison and that the leading edge of this enrichment counted 170 genes. Out of 170 gene transcripts detected in EVs with the same profile of those expressed in a receptive endometrial tissue, 142 were also significantly differentially represented between UF-EVs of the pre-receptive versus UF-EVs of the receptive phase (DGE fUF-EVs LH + 7 vs fUF-EVs LH + 2). Further comparison of this 142-gene list (intersection of LH + 7 vs LH + 2 in EVs and receptive vs non-receptive phase in ERA®) with the endometrial receptivity ‘metasignature’ published genes list (Altmäe et al., 2017) highlighted changes in 38 gene transcripts in both UF-EVs and endometrial tissue during the window of implantation. Among them is DPP4 (dipeptidyl peptidase 4). DPP4 (CD26) is a membrane-binding extracellular glycoprotein that acts as a positive regulator of TCR-mediated T-cell coactivation. DPP4 is known to be expressed on the surface of endometrial cells during the window of implantation, when its enzymatic activation facilitates both blastocyst adhesion via fibronectin binding and trophoblast invasion through pericellular proteolysis of the extracellular matrix (Shimomura et al., 2006; Dolanbay et al., 2016). At single-cell level, DPP4 transcription has recently been shown to be activated in both glandular and luminal endometrial epithelium (Wang et al., 2020), possibly suggesting an endometrial epithelial contribution to UF-EVs release. Overall, these results, thus support the reliability of EV isolation from UF samples as a ‘proxy’ of the endometrium and also hint at an EV-mediated RNA transport related to the endometrial timing at UF collection. This RNA cargo may thus be potentially useful to set up a less-invasive endometrial receptivity assay.
By including a population of women undergoing ART and performing a DGE analysis of transcriptomes from UF-EVs collected from women who achieved a pregnancy versus those from women who failed to conceive, we further established a signature of 161 genes as putative receptivity biomarkers. UF-EVs collected from women who achieved pregnancy were enriched in genes mostly related to the defence response, including allograph rejection and chemotaxis process, while UF-EVs from women who failed to conceive were enriched in transcripts involved in epidermal cell differentiation. These results, which represent a third major finding of our study, are in line with the consistent body of literature showing a balance between protective anti-infectious mechanisms and immune tolerance toward a semi-allogeneic embryo in the establishment of a hospitable endometrial environment (Kao et al., 2002; Altmäe et al., 2010; Robertson et al., 2018; Diao et al., 2020). The enrichment analysis also confirmed the involvement of these differentially ‘expressed’ genes in intricate cytokine pathways (IL-17, IL-10 and TNF) that govern selective immune regulation and control the adhesion and vascularization processes during the embryo-endometrium dialogue. A dysregulation of these pathways has been associated with implantation failure (Chaouat et al., 2002; Winger et al., 2011; Wang et al., 2019). In addition, we identified some lncRNAs selectively present in UF-EVs from women achieving successful or failed implantation, suggesting an additional regulatory function of RNA transported by EVs. For example, CERC7, detected only in UF-EVs of women failing to achieve pregnancy, regulates immune cell differentiation through the modulation of CTLA4 expression by targeting miR-429 (Yao et al., 2017). CTLA4, expressed by activated Foxp3+ TREG cells, participates in the balance between effectors (i.e. innate immunity and NK cells, B cells, T helper [Th] 1 and Th17 immunity) and regulators (Th2 cells, regulatory T cells) (Vignali et al., 2008), which is essential for establishment of a pregnancy. Interestingly, the list of differentially ‘expressed’ genes deriving from the comparison of EV samples collected from ART women achieving successful versus failed implantation shared 41 genes in common with the differentially ‘expressed’ genes in the LH + 7 EVs versus LH + 2 EVs comparison performed on samples from fertile women. This gene list highlights the importance of defence responses, specifically modulation of the immune system, chemotaxis, the response to external stimuli and stress, and of cell adhesion pathways in the correct timing of endometrial receptivity. Even though for 24 of these 41 genes the negative or positive trend of ‘expression’ was concordant between the two sets of samples while it was discordant for 17 genes, this result overall supports the idea that these specific genes belong to pathways that undergo fine regulation at endometrial level during the window of implantation. As an example, one such gene is CLU (clusterin), which is over-represented in both DGE analyses. Clusterin is a secreted glycoprotein that has been previously suggested to play an immunosuppressive role during the receptive period, by inhibiting membrane attack of activated complement proteins and interacting with IgG at endometrial epithelial level (Tapia et al., 2008). Another over-represented gene is CYBB. Recently described as a discriminatory gene that is over-expressed by endometrial macrophages, CYBB over-representation in UF-EVs might thus suggest a leukocyte contribution to their release (Wang et al., 2020). Interestingly, these processes are similar to those detected by Altmäe et al. (2017) in their meta-signature study based on endometrial tissue. Finally, from the genes excluded from the intersection, it can deduce that other transcripts might be finely tuned to ensure correct embryo implantation in the context of ART. As an exploratory analysis, we also tested whether ERA® genes used for timing the endometrial transfers have the same transcriptional profile of the mRNA species characterizing our UF-EVs ‘pregnant’ versus UF-EVs ‘not pregnant’ comparison. Results derived from enrichments were not as significant and consistent as for those derived from the comparison of LH + 7 fUF-EVs versus LH + 2 fUF-EVs, as expected based on the fact that the ERA® gene list was obtained based on the evaluation of pre-receptive versus LH + 7 endometrial biopsies and not based on successful versus failed implantation. Altogether, gene transcripts with the most robust rationale for being used as predictive UF-EV markers of endometrial receptivity in ART might currently be considered those (n = 41) derived from the intersection between the two DGE comparisons, fUF-EVs versus LH + 2 fUF-EVs and LH + 7 UF-EVs ‘pregnant’ versus LH + 7 UF-EVs ‘not pregnant’.
In terms of physical features, UF-EVs revealed a homogeneity among the different groups analyzed, except for a slight but significant difference in EV size, being smaller in women with a successful implantation compared to patients who failed to conceive. The presence of a higher fraction of apoptotic bodies (the largest type of EVs, with a diameter of 50–5000 nm) in EV preparation from UF of patients with failed implantation compared to patients with successful pregnancy could thus be hypothesized.
Some limitations of the present study deserve consideration. To enhance the consistency and clinical feasibility of our methods (O’Brien et al., 2020), we did not treat pelleted EVs with RNase. Consequently, separation of RNAs associated with EV membranes might have been incomplete, and membrane-bound RNAs—rather than the internal RNA content of EVs—might have to some extent contributed to our RNA-seq results. In addition, when considering patients undergoing ART, we did not collect UFs in the same cycle of the embryo transfer but rather in the cycle immediately preceding the transfer attempt. We considered this approach as the most appropriate in relation to the novel, explorative nature of our study. Based on our results, a validation of UF-EV RNA-seq analyses in the same cycle in which embryo transfer is performed should be provided. Finally, stronger significant differences in DGE analyses when we considered EVs from ART patients could have been hidden by the great inter-sample variability characterizing this group of patients who proceed to embryo PGT owing to various clinical indications.
Among the strengths of the study is the homogeneous collection of UF samples on day 2 and day 7 after the urinary LH surge of a monitored natural cycle throughout an ovulation test. This test has an over 99% accuracy at detecting the LH surge (Clearblue Advanced Ovulation, Swiss Precision Diagnostics, Gmbh, Geneva, Switzerland). This allows us to reliability predict the implantation window, eliminating the women’s cycle variability. More importantly, to the best of our knowledge, this study includes the highest number of RNA-seq datasets from human EVs particles reported to date. The data presented do not only pave the way to understanding the molecular mechanisms that underlie intercellular communication during embryo implantation but also could be useful for those approaching the RNA-seq based method on EVs.
In conclusion, we report the transcriptional signature of EVs released in the uterine microenvironment during the window of implantation. This report is the first demonstrating that EVs from UF could be a good proxy of endometrial tissue, by showing that: the transcriptome of UF-EVs correlates with the endometrial tissue transcriptome and includes genes known to regulate cell adhesion and implantation; RNA signatures in UF-EVs change with endometrial phase and status; UF-EVs could serve as a reservoir for potential less-invasive collection of receptivity markers. Currently, biopsy-based invasive methods are the only available approaches to evaluate endometrial receptivity. Therefore, UF collection may be proposed to become a less-invasive routine practice compared to endometrial biopsy, avoiding the risk of endometrial damage (Luddi et al., 2019; Grasso et al., 2020). This paper thus represents a step forward in the design of less-invasive, personalized approaches for real-time monitoring of endometrial status, necessary for advancing the field of reproductive medicine. In fact, our results set the basis for further clinical studies based on UF-EVs sampling and aimed at identifying cut-offs of expression levels of gene sets or pathways for the prediction of implantation in ART.
Data availability
The data underlying this article are available in NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and can be accessed with GSE158958 deposition number.
Authors’ roles
E.G., S.M., E.P., G.T., M.C. and P.V. contributed to conceiving and designing the study and to data acquisition. S.S. contributed to sample acquisition and article revision. P.V. also contributed to article drafting and revision. E.G., S.G.M., V.M., D.L., L.C., V.B. and E.G. contributed to data analysis, data interpretation, article drafting and revision. V.S.V. and L.P. contributed to data acquisition, data interpretation and article drafting and revision. All authors approved the final version of the manuscript.
Funding
This research was possible thanks to the funding provided by the European Society of Human Reproduction and Embryology (ESHRE Research Grant 2016-1).
Conflict of interest
The authors have no financial and non-financial competing interests to disclose.
Supplementary Material
References
- Altmäe S, Martínez-Conejero JA, Salumets A, Simón C, Horcajadas JA, Stavreus-Evers A.. Endometrial gene expression analysis at the time of embryo implantation in women with unexplained infertility. Mol Hum Reprod 2010;16:178–187. [DOI] [PubMed] [Google Scholar]
- Altmäe S, Koel M, Võsa U, Adler P, Suhorutšenko M, Laisk-Podar T, Kukushkina V, Saare M, Velthut-Meikas A, Krjutškov K. et al. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomic biomarkers. Sci Rep 2017;7:10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy I, Lanau L, Altadill T, Sequeiros T, Cabrera S, Cubo-Abert M, Pérez-Benavente A, Garcia A, Borrós S, Santamaria A. et al. Exosome-like vesicles in uterine aspirates: a comparison of ultracentrifugation-based isolation protocols. J Transl Med 2016;14:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouat G, Zourbas S, Ostojic S, Lappree-Delage G, Dubanchet S, Ledee N, Martal J.. A brief review of recent data on some cytokine expressions at the materno-foetal interface which might challenge the classical Th1/Th2 dichotomy. J Reprod Immunol 2002;53:241–256. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lun AT, Smyth GK.. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 2016;5: 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao L, Cai S, Huang C, Li L, Yu S, Wang L, Liu S, Li Y, Zeng Y.. New endometrial immune cell-based score (EI-score) for the prediction of implantation success for patients undergoing IVF/ICSI. Placenta 2020;99:180–188. [DOI] [PubMed] [Google Scholar]
- Díaz-Gimeno P, Horcajadas JA, Martínez-Conejero JA, Esteban FJ, Alamá P, Pellicer A, Simón C.. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril 2011;95:50–60. [DOI] [PubMed] [Google Scholar]
- Díaz-Gimeno P, Ruiz-Alonso M, Blesa D, Bosch N, Martínez-Conejero JA, Alamá P, Garrido N, Pellicer A, Simón C.. The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil Steril 2013;99:508–517. [DOI] [PubMed] [Google Scholar]
- Díaz-Gimeno P, Ruíz-Alonso M, Blesa D, Simón C.. Transcriptomics of the human endometrium. Int J Dev Biol 2014;58:127–137. [DOI] [PubMed] [Google Scholar]
- Dolanbay EG, Yardimoglu M, Yalcinkaya E, Yazir Y, Aksoy A, Karaoz E, Caliskan E.. Expression of trophinin and dipeptidyl peptidase IV in endometrial co-culture in the presence of an embryo: A comparative immunocytochemical study. Mol Med Rep 2016;13:3961–3968. [DOI] [PubMed] [Google Scholar]
- Evans J, Rai A, Nguyen HPT, Poh QH, Elglass K, Simpson RJ, Salamonsen LA, Greening DW.. Human endometrial extracellular vesicles functionally prepare human trophectoderm model for implantation: understanding bidirectional maternal-embryo communication. Proteomics 2019;19:e1800423. [DOI] [PubMed] [Google Scholar]
- Franasiak JM, Forman EJ, Patounakis G, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT. Jr.,. Investigating the impact of the timing of blastulation on implantation: management of embryo-endometrial synchrony improves outcomes. Hum Reprod Open 2018;4:hoy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso A, Navarro R, Balaguer N, Moreno I, Alama P, Jimenez J, Simón C, Vilella F.. Endometrial liquid biopsy provides a miRNA roadmap of the secretory phase of the human endometrium. J Clin Endocrinol Metab 2020;105:dgz146. [DOI] [PubMed] [Google Scholar]
- Hauser P, Wang S, Didenko VV.. Apoptotic bodies: selective detection in extracellular vesicles. Methods Mol Biol 2017;1554:193–200. [DOI] [PubMed] [Google Scholar]
- Healy MW, Yamasaki M, Patounakis G, Richter KS, Devine K, DeCherney AH, Hill MJ.. The slow growing embryo and premature progesterone elevation: compounding factors for embryo-endometrial asynchrony. Hum Reprod 2017;32:362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC.. Global gene profiling in human endometrium during the window of implantation. Endocrinology 2002;143:2119–2138. [DOI] [PubMed] [Google Scholar]
- Kumar V, Maurya VK, Joshi A, Meeran SM, Jha RK.. Integrin beta 8 (ITGB8) regulates embryo implantation potentially via controlling the activity of TGF-B1 in mice. Biol Reprod 2015;92:109. [DOI] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA.. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010;11:733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luddi A, Zarovni N, Maltinti E, Governini L, Leo V, Cappelli V, Quintero L, Paccagnini E, Loria F, Piomboni P.. Clues to non-invasive implantation window monitoring: isolation and characterisation of endometrial exosomes. Cells 2019;8:811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackens S, Santos-Ribeiro S, van de Vijver A, Racca A, Van Landuyt L, Tournaye H, Blockeel C.. Frozen embryo transfer: a review on the optimal endometrial preparation and timing. Hum Reprod 2017;32:2234–2242. [DOI] [PubMed] [Google Scholar]
- Mentkowski KI, Snitzer JD, Rusnak S, Lang JK.. Therapeutic potential of engineered extracellular vesicles. AAPS J 2018;20:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi S, El Kasmi I, Bourdiec A, Crespo K, Bissonnette L, Le SC, Bissonnette F, Kadoch IJ.. 15 years of transcriptomic analysis on endometrial receptivity: what have we learnt? Fertil Res Pract 2019;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–273. [DOI] [PubMed] [Google Scholar]
- Neves AR, Devesa M, Martínez F, Garcia-Martinez S, Rodriguez I, Polyzos NP, Coroleu B.. What is the clinical impact of the endometrial receptivity array in PGT-A and oocyte donation cycles? J Assist Reprod Genet 2019;36:1901–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YH, Rome S, Jalabert A, Forterre A, Singh H, Hincks CL, Salamonsen LA.. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One 2013;8:e58502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J.. Dating the endometrial biopsy. Fertil Steril 1950;1:3–25. [DOI] [PubMed] [Google Scholar]
- O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO.. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol 2020;21:585–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prapas Y, Prapas N, Jones EE, Duleba AJ, Olive DL, Chatziparasidou A, Vlassis G.. The window for embryo transfer in oocyte donation cycles depends on the duration of progesterone therapy. Hum Rep 1998;13:720–723. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J.. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SA, Care AS, Moldenhauer LM.. Regulatory T cells in embryo implantation and the immune response to pregnancy. J Clin Invest 2018;128:4224–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BS, Daneshmand ST, Garner FC, Aguirre M, Ross R.. Contrasting patterns in in vitro fertilization pregnancy rates among fresh autologous, fresh oocyte donor, and cryopreserved cycles with the use of day 5 or day 6 blastocysts may reflect differences in embryo-endometrium synchrony. Fertil Steril 2008;89:20–26. [DOI] [PubMed] [Google Scholar]
- Shapiro D, Boostanfar R, Silverberg K, Yanushpolsky EH.. Examining the evidence: progesterone supplementation during fresh and frozen embryo transfer. Reprod Biomed Online 2014;29:S1–14. [DOI] [PubMed] [Google Scholar]
- Shapiro BS, Daneshmand ST, Desai J, Garner FC, Aguirre M, Hudson C.. The risk of embryo-endometrium asynchrony increases with maternal age after ovarian stimulation and IVF. Reprod Biomed Online 2016;33:50–55. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Ando H, Furugori K, Kajiyama H, Suzuki M, Iwase A, Mizutani S, Kikkawa F.. Possible involvement of crosstalk cell-adhesion mechanism by endometrial CD26/dipeptidyl peptidase IV and embryonal fibronectin in human blastocyst implantation. Mol Hum Reprod 2006;12:491–495. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA. et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006;147:1097–1121. [DOI] [PubMed] [Google Scholar]
- Tapia A, Gangi LM, Zegers-Hochschild F, Balmaceda J, Pommer R, Trejo L, Pacheco IM, Salvatierra AM, Henríquez S, Quezada M. et al. Differences in the endometrial transcript profile during the receptive period between women who were refractory to implantation and those who achieved pregnancy. Hum Reprod 2008;23:340–351. [DOI] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng LH, Chen I, Chen MY, Yan H, Wang CN, Lee CL.. Genome-based expression profiling as a single standardized microarray platform for the diagnosis of endometrial disorder: an array of 126-gene model. Fertil Steril 2010;94:114–119. [DOI] [PubMed] [Google Scholar]
- Vaiarelli A, Cimadomo D, Capalbo A, Orlando G, Sapienza F, Colamaria S, Palagiano A, Bulletti C, Rienzi L, Ubaldi FM.. Pre-implantation genetic testing in ART: who will benefit and what is the evidence? J Assist Reprod Genet 2016;33:1273–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni VS, Somigliana E, Reschini M, Pagliardini L, Marotta E, Faulisi S, Paffoni A, Vigano’ P, Vegetti W, Candiani M. et al. Top quality blastocyst formation rates in relation to progesterone levels on the day of oocyte maturation in GnRH antagonist IVF/ICSI cycles. PLoS One 2017;12:e0176482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali DA, Collison LW, Workman CJ.. How regulatory T cells work. Nat Rev Immunol 2008;8:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martínez S, Marcilla A, Simón C.. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 2015;142:3210–3221. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Zhang H, Chen ZQ, Zhang W, Liu XM, Fang JY, Liu FJ, Kwak-Kim J.. Endometrial TGF-β, IL-10, IL-17 and autophagy are dysregulated in women with recurrent implantation failure with chronic endometritis. Reprod Biol Endocrinol 2019;17:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, Pan W, Simon C, Quake SR.. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 2020;26:1644–1653. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Baird DD, Clarice R, Weinberg CR.. Time of Implantation of the Conceptus and Loss of Pregnancy. N Engl J Med 1999;340:1796–1799. [DOI] [PubMed] [Google Scholar]
- Winger EE, Reed JL, Ashoush S, El-Toukhy T, Ahuja S, Taranissi M.. Degree of TNF-α/IL-10 cytokine elevation correlates with IVF success rates in women undergoing treatment with Adalimumab (Humira) and IVIG. Am J Reprod Immunol 2011;65:610–618. [DOI] [PubMed] [Google Scholar]
- Yao K, Wang Q, Jia J, Zhao H.. A competing endogenous RNA network identifies novel mRNA, miRNA and lncRNA markers for the prognosis of diabetic pancreatic cancer. Tumour Biol 2017;39:1010428317707882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and can be accessed with GSE158958 deposition number.