Abstract
Human coronaviruses present a substantial global disease burden, causing damage to populations’ health, economy, and social well-being. Glycans are one of the main structural components of all microbes and organismic structures, including viruses—playing multiple essential roles in virus infection and immunity. Studying and understanding virus glycans at the nanoscale provide new insights into the diagnosis and treatment of viruses. Glycan nanostructures are considered potential targets for molecular diagnosis, antiviral therapeutics, and the development of vaccines. This review article describes glycan nanostructures (eg, glycoproteins and glycolipids) that exist in cells, subcellular structures, and microbes. We detail the structure, characterization, synthesis, and functions of virus glycans. Furthermore, we describe the glycan nanostructures of different human coronaviruses, such as human coronavirus 229E (HCoV-229E), human coronavirus OC43 (HCoV-OC43), severe acute respiratory syndrome-associated coronavirus (SARS-CoV), human coronavirus NL63 (HCoV-NL63), human coronavirus HKU1 (HCoV-HKU1), the Middle East respiratory syndrome-associated coronavirus (MERS-CoV), and how glycan nanotechnology can be useful to prevent and combat human coronaviruses infections, along with possibilities that are not yet explored.
Keywords: glycan, nanotechnology, glycoproteins, glycolipids, coronaviruses, SARS-CoV-2, diagnostics, vaccine
Introduction
Glycan nanotechnology is an integrated approach that combines nanotechnology with glycan biology and chemistry.1 Glycans are present in mammalian cells and their subcellular structures and in most microbes, in the form of glycoproteins and glycolipids. Owing to glycans’ structural diversity, they can store and transmit biological information and have essential functions in intercellular recognition, signal transduction, infection, immune response, and many other physiological and pathological processes. Virus glycans (ie, structural or functional glycans in virus particles) play crucial roles in the process of viral infection of host cells, including viral attachment, entry into the cell, cleavage of viral proteins, recognition and neutralization of virus by host immune system. Therefore, studying glycans at the nanoscale will enable biomedical applications such as pathogen detection, drug delivery carriers, vaccine development, and biomolecular therapeutics for viruses and infectious diseases.2–4
Human coronaviruses cause respiratory tract infection, mainly leading to mild symptoms, severe pneumonia, dyspnea, and even death.5 So far, seven human coronaviruses (HCoVs) have spread in different regions of the world, posing a significant threat to human health. The outbreak of SARS, MERS, and the very recent emergence of SARS-CoV-2 show that coronaviruses can emerge unexpectedly, causing life-threatening infections that spread rapidly and impose a challenging threat to public health and economic development. Although various approaches have been studied and tested for HCoVs treatment, no specific antiviral is available to date, and patients mainly receive symptomatic treatment and supportive care. In order to better prevent and control HCoVs infections, developing new strategies and technologies to diagnose and treat HCoVs is indispensable.
Glycan Nanostructures in Cells, Subcellular Structures, and Microbes
Glycans—simple or complex saccharides—together with nucleic acid, protein, and lipid constitute all the known cellular (eg, mammalian cells, bacteria, algae, and fungi) non-cellular (eg, viruses and organelles) life forms. Glycans perform various structural and modulatory functions in cells and sub-cellular particles. On the cell surface, glycans are widely distributed, allowing highly selective and responsive interaction with the surrounding environment (uptake nutrients and release of metabolites). Lipid- and protein-bound glycans (ie, glycolipids and glycoproteins) are also present in the nucleus, mitochondria, ribosomes, and cytoplasm and modulate their functions in the synthesis and transfer of nucleic acids proteins, and energy production.
Glycoproteins
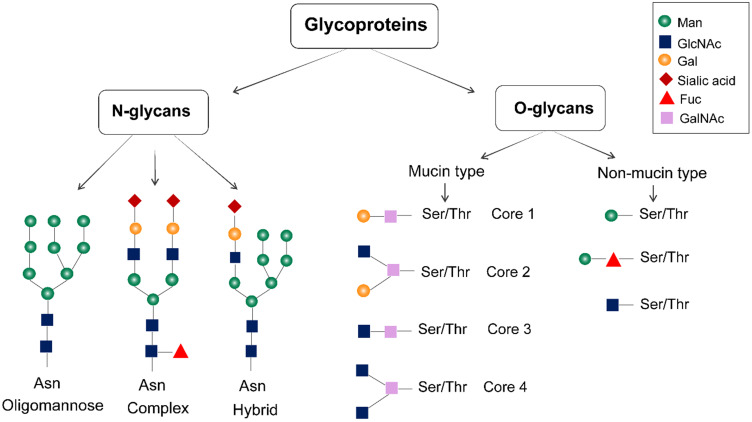
Proteins (on the cell surface, cellular organelles, and in the cytoplasm) are often attached to sugar chains. The carbohydrate moieties in cell surface-associated glycoproteins include both large hetero- and homo-polysaccharides and disaccharides.6 Sialic acid, D-glucose, xylose, glucuronic acid, D-mannose, N-acetyl-D-galactosamine (GalNAc), N-acetylneuraminic acid, L-fucose, N-acetyl-D-glucosamine (GlcNAc), and D-galactose are some of the common types of carbohydrate units found in cell surface-associated glycoproteins.6 Glycans can be divided into N-glycans and O-glycans according to the different connection modes between glycans and proteins. N-glycans are connected to the asparagine (Asn) residues, while O-glycans are attached to serine, threonine, or tyrosine residues (Figure 1). The nascent carbohydrate-protein conjugate is modified by removal or re-addition of glucose as part of a quality-control process in the endoplasmic reticulum (ER); then, the structure moves to the Golgi apparatus for further cell-type-specific glycosylation.7 O-glycans are further categorized into mucin-type O-glycans and non-mucin-type O-glycans based on the monosaccharide residue attached to the amino acid. The former is often found in extracellular glycoproteins, and the latter is usually in the intracellular glycoproteins of the nucleus and mitochondria.8 The glycosylation site of glycoprotein is determined by the secondary and tertiary structures of the polypeptide chain. Glycoproteins can vary in the number of the constituent units of N- and O-linked glycans and the glycosylation sites, where glycans unites are linked to the polypeptide chain.8 Analytical techniques such as capillary electrophoresis (CE), high-performance liquid chromatography (HPLC), electrospray ionization (ESI), mass spectroscopy (MS), liquid chromatography-mass spectrometry (LC-MS), matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS), computational biology, and next-generation sequencing technologies can be used to isolate and analyze the structure of glycan to determine the glycan composition, glycan sequence and interglycosidic linkages in glycoproteins.9
Figure 1.
Structure of glycoproteins. Glycoproteins are mainly glycosylated with N-linked and O-linked glycans. N-glycans are attached to the asparagine residues, and they are processed in the Golgi apparatus to yield oligomannose, hybrid, and complex-type N-glycan structures. The mucin-type O-linked glycosylation mainly contains four common O-linked glycan cores. Non-mucin-type O-glycosylations are produced by attaching GlcNAc, mannose, fucose, glucose, galactose, or xylose to the amino acid.
It is well established that glycans not only affect protein conformation but also participate in glycoprotein recognition and binding.10,11 N-glycans participate in glycoprotein folding by mediating ER chaperone interactions during biosynthesis. If the nascent carbohydrate–protein conjugate is correctly folded, it is processed further in the ER and Golgi apparatus, if the glycoprotein is incorrectly assembled, it is re-glycosylated.10,11 O-glycans are often distributed in clusters on the highly glycosylated segments of proteins, helping to stabilize the structure of the polypeptide chain. Functionally, the N-glycans located in the Fc segment of immunoglobulin G (IgG) on B-cells can bind to Fc receptors on natural killer (NK) cells, monocytes, macrophages or neutrophils and trigger subsequent cytotoxic reactions leading to the release of lytic compounds and apoptosis of the target cell. NK cells can release perforin and granzymes, pro-inflammatory cytokine like IFN-γ to cause target cell apoptosis. Besides, NK cell induces apoptosis through signaling of TNF family death receptors and mitochondrial pathway. These functional interactions of glycans indicate that the structure and expression of glycans are related to the occurrence of diseases and changes in physiological state. Thus, glycans can be used to aid disease biomarker discovery.12
Glycolipids
Glycolipids are widely found in various organisms like bacteria, fungi, plants and animals and can be classified into glyceroglycolipids, glycosphingolipids, in addition to other types with various lipid structures.13 Glycolipids are synthesized on the ER by the binding of a monosaccharide (glucose or galactose) to the lipid, following which linear or branched complex carbohydrate chains are added via glycosyltransferase. The carbohydrate structure of glycolipids is variable, and the formed glycolipids can be neutral (with one or more uncharged sugars), acidic, basic, or amphoteric to play different functions in the cell.14
Glycolipids constitute the backbone of cell membranes, providing ion impermeability and sidedness. Glycosphingolipids distribute in the outer layer of membrane lipids, playing a critical role in cellular homeostasis and necessary cellular activities.15 Lipopolysaccharide (LPS) is the most studied glycolipid on the bacterial surface, which creates innate resistance against many antimicrobials.16 The composition of glycolipids on cell membranes is specific, and they can be used as disease biomarkers. For instance, glycosphingolipids are the antigenic determinants (ie, A, B, and H determinants) of the ABO blood group system. Glycolipids can also act as receptors involved in cell recognition and information transfer.17 In addition, glycolipids are involved in the pathogenesis, development, differentiation, and metastasis of tumors.18
Glycan in Cell Organelles
In the ER, N-glycans, called high-mannose glycans, play a crucial role in protein quality control mediated by lectins and glycan-related enzymes. The trimming status of mannose moieties in high-mannose glycans is a signal to degrade proteins that fail to correct folding.19 During glycoprotein synthesis, the N-glycans are also indispensable in protein folding, glycoprotein transport, and targeting.20 For instance, when the mannose on the lysosomal enzyme is phosphorylated as mannose 6-phosphate, it can recognize and bind to the mannose receptor on the lysosomal membrane so that the lysosomal enzyme can be directed to the lysosome.
Glycans of Viruses
Structure
Viruses can be classified into two groups of enveloped and non-enveloped viruses. A virus envelope is a lipid bilayer membrane, usually modified with virus-encoded glycoproteins. Glycoproteins in the envelope of virus particles are necessary to initiate a viral infection. N-linked glycans have a core formed by three mannose (Man) and two N-acetylglucosamine (GlcNAc) and can be classified into high mannose, hybrid, and complex-type. Unlike N-linked glycans, O-linked glycans do not have a common core structure. However, four common O-linked glycan cores have been found in the mucin-type O-linked glycosylation.21 In addition to envelope glycoproteins, glycans are also present in many secreted viral proteins, such as the secreted glycoprotein G of herpes simplex virus22 and the secreted GP of Ebola.23
Synthesis
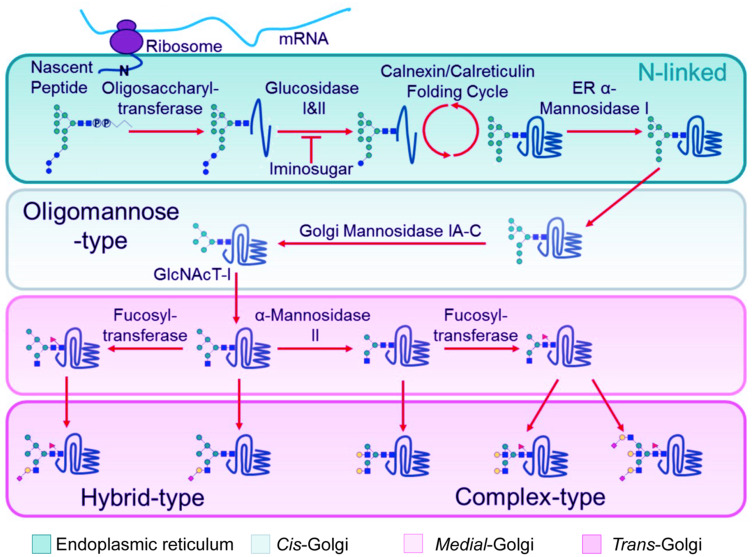
As viruses are obligate parasites, protein glycosylation is jointly determined by the viral genome and the host cell. N-linked glycans are synthesized in the ER, Golgi apparatus and mitochondria, which can be performed simultaneously with the synthesis of peptide chains (Figure 2). The initial glycan moiety begins with nine mannose (Man), three glucose (Glc), and two N-acetylglucosamine (GlcNAc) residues. N-acetylglucosamine binds to dolichol phosphate (Dol-P) on the bilipid membrane of the ER. N-glycan biosynthesis via dolichol intermediates also occurs in the outer and inner membrane of mitochondria. Once the formation of Dol-P–carbohydrate structure is completed, the carbohydrate chain is transferred to the nascent polypeptide chain at an Asn-X-Ser/Thr site by oligosaccharyltransferase.24 Subsequently, the N-glycan is extensively modified and trimmed by glycosidases and glycosyltransferases; then, the structure is transported to the Golgi apparatus for further cell-type-specific glycosylation.25 However, not all viral glycoproteins are synthesized in this way, eg, hepatitis C virus traffic directly from the ER to the plasma membrane.26
Figure 2.
N-linked glycosylation of proteins in the ER and Golgi apparatus.
Notes: After mRNA synthesis, the mature tri-glucosylated N-linked glycan precursor of dolichol-P-Pglycan is transferred en bloc by oligosaccharyltransferase to the asparagine residue of an Asn-X-Ser/Thr sequon on anascent polypeptide chain. Following transfer of the precursor glycan to the protein, glucosidases in the endoplasmic reticulum (ER) remove the three glucose residues while the protein folds in the Calnexin/Calreticulin cycle. Then, aseries of ER and Golgi mannosidases subsequently cleave mannose residues down to the Man5GlcNAc2 glycan. The action of GlcNAc transferase-I (GlcNAcT-I) initiates the first branch of the N-glycan. Once α-Mannosidase II removes the two remaining outer mannose residues, other glycan processing enzymes such as galactosyl-, fucosyl- and sialyl-transferases, can act to construct ahuge assortment of complex-type glycans. Reprinted with permission from Watanabe Y, Bowden TA, Wilson IA, Crispin M.Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863(10):1480–1497.21 Creative Commons (https://creativecommons.org/licenses/by/4.0/legalcode).
The synthesis of O-linked glycans is performed after the synthesis of the polypeptide chain and does not require a glycan carrier. The O-linked GlcNAc is formed by a monosaccharide GlcNAc linked to nucleus and cytoplasm proteins. The GalNAc monosaccharide residue is attached to a serine, threonine, or tyrosine residue of a nascent polypeptide chain by GalNAc-transferases. Then O-linked glycans can be further elongated by competing glycosyltransferases.
Assembly and Characterization
The maturation step of virus glycoproteins takes place in the ER and starts with the removal of glucose residues from the N-linked glycan precursor by ER α-glucosidases I and II; then the glycoprotein bearing monoglucosylated glycans are retained in the ER bound to ER chaperones, calnexin and calreticulin.21 If the glycoprotein is appropriately folded, the final glucose residue is cleaved off by α-glucosidase II, and the glycoprotein is transplanted to the Golgi compartments to acquire modifications. However, the improperly folded glycoprotein is re-glycosylated by glycoprotein glucosyltransferase (UGGT) located in the lumen of the ER.27 Re-glycosylated glycoproteins are re-folded into the correct formulation order-glycosylated and degraded. During this process, glycan contributes a lot to glycoprotein folding and assembly.
Advances in structural biology techniques, particularly mass spectrometry, have enabled us to study and analyze glycoproteins at different levels.28 In addition to confirming the glycan sequences and precise structures it is also possible to confirm the positions of glycans on viral envelope glycoproteins.29 This information uncovered various details on the differences of glycosylation in different viruses, which paved the way for studies on the functions of glycosylation on the virus.
Function
Viral glycans are not only required for structural and stability but also play functional roles in signaling, attachment, and shielding. During the process of protein folding, the molecular chaperone, calnexin, and calreticulin, recognize and bind the glycan to help the protein fold correctly, allowing the misfolded protein to enter the degradation system.20 The importance of N-linked glycosylation has been studied in HIV-1, which suggested integral glycans necessary for correct gp160 folding and incorporation into virions.30 Glycans often protrude on the surface of virus particles and may play a structural role by restricting the conformational freedom of the polypeptide chains connected to them. O-linked glycans often distribute in clusters on highly glycosylated segments of proteins, helping to stabilize the structure of the polypeptide chain. Also, glycans can help glycoprotein to sort, traffic, and release. Another significant role of enveloping glycans lies in virus entry into its susceptible target cells and efficiently transmits the pathogen. For example, Hemagglutinin (HA) is an N-glycoprotein on the surface of the influenza virus, playing a role in the early infection of host cells by the virus. Glycosylation not only regulates the binding ability and specificity between influenza virus and host cell surface receptors but also has an effect on virulence and evolution. The virulence and pathogenicity of rabies virus are also closely related to glycosylation of glycoproteins, which can be applied to the prevention and treatment of rabies.31 Many other pathogens, including noroviruses,32 polyomaviruses,33 and coronaviruses,34 also possess sialic acid and glycan recognition. Sialyllactose-containing glycolipids on the HIV-1 viral membrane have been reported to bind siglec-1 on dendritic cells, thereby mediating the trans-infection of T cells.35 Moreover, it has been shown that the high levels of glycosylation on the envelope of viruses is served as a protective shield from the immunological surveillance of the host.36 Viruses use glycan shielding to limit their recognition by the immune system. Strong evidence for which has been reported for simian immunodeficiency virus (SIV) mutants lacking glycans in the V1 region of Env.37 Other trimeric class I fusion proteins from Influenza,38 Coronavirus39 have also been shown to be quite useful in allowing the virus to thwart the immune system.
Glycans of Coronaviruses
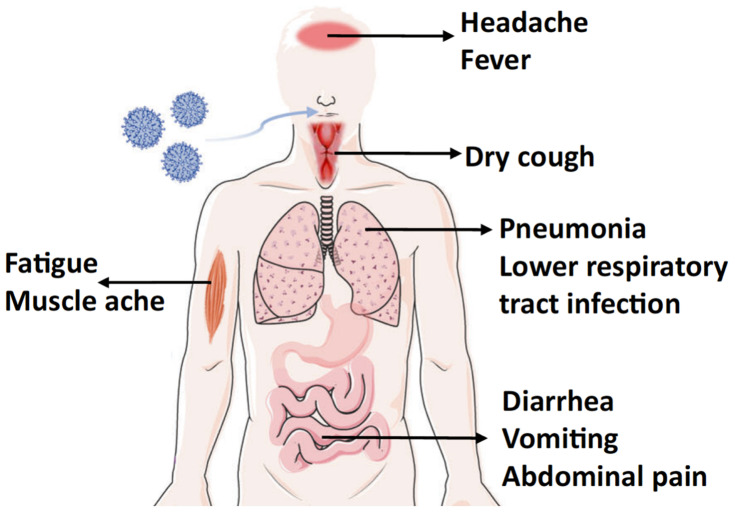
Human coronaviruses (CoVs) are the largest single-stranded RNA viruses. Currently, seven human CoVs have been reported, including HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS, MERS, and SARS-CoV-2. According to the structural characteristics of serology and genome, coronaviruses are usually divided into four groups: alphacoronaviruses, betacoronaviruses, gammacoronaviruses, and deltacoronaviruses. HCoV-229E, HCoV-NL63 belong to alpha-CoV, and HCoV-OC43, SARS-CoV, HCoV-HKU1, MERS-CoV, SARS-CoV-2 belong to beta-CoV. All HCoVs are associated with respiratory and gastrointestinal infections, leading to mild, self-limiting symptoms or severe respiratory disease (Figure 3).40 Early diagnosis of a coronavirus infection requires not only clinical manifestations but also specific testing. The current diagnostic algorithm for coronaviruses combines various diagnostic criteria that expand from the patient’s history and the possibility of exposure to the virus to precise laboratory-based nucleic acid testing (Table 1).
Figure 3.
The symptoms of coronaviruses infection.
Notes: COVID-19 symptoms are similar to other respiratory diseases, such as influenza, SARS, and MERS. However, influenza initiates with cough, whereas COVID-19, like other coronavirus-related diseases initiates with fever.86 Also, COVID-19 differs from SARS and MERS in the order of gastrointestinal symptoms, where upper gastrointestinal symptoms are seen last after fever and respiratory symptoms. Results supported the use of fever as screen for entry into facilities; however rapid highly sensitive methods for viral confirmation are needed. Illustration reprinted with permission from Chen B, Tian EK, He Betal Overview of lethal human coronaviruses. Signal Transduction and Targeted Therapy. 2020;5(1):1–16.113 Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
Table 1.
The Current Diagnostic Algorithm for Coronavirus Infection
| Diagnostics of the Coronaviruses | Manifestations |
|---|---|
| Epidemiological exposure | History of contact with probable or confirmed cases; a history of travel or residence within the high-risk areas |
| Symptoms | Fever, cough, chills, malaise, and myalgia |
| Chest imaging | Radiographic features of pneumonia |
| Precise laboratory tests | Positive tests for by at least one assay, such as positive tests by a PCR-based detection method using respiratory samples or by serological tests |
| Other relevant clinical examinations | Complete blood counts (CBC), C-reactive protein (CRP), inflammatory markers |
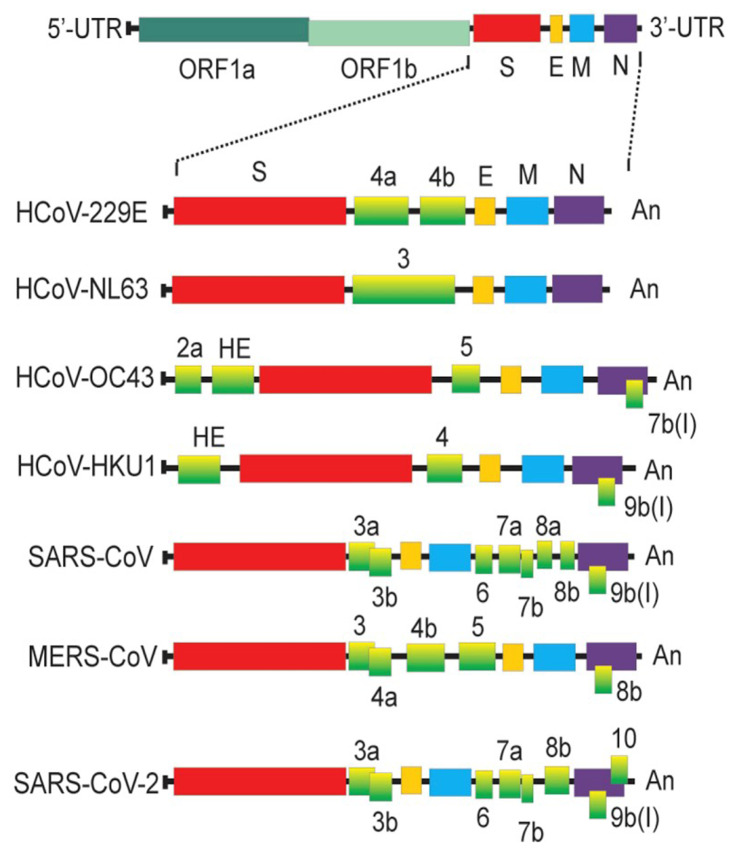
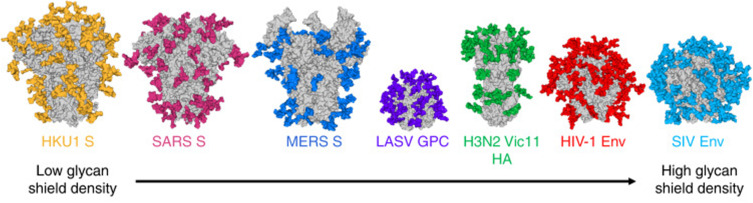
The coronaviruses have four structural proteins E (envelope protein), M (membrane protein), N (nucleocapsid protein), and S (spike protein) (Figure 4). The S glycoprotein of CoV is a highly glycosylated trimeric class I viral fusion protein, composed of three parts: extracellular domain, a transmembrane domain, and cytoplasmic domain.39 The extracellular domain can be cleaved by enzymes into two subunits of S1 and S2. S1 subunit contains four domains: A, B, C, and D, of which domain A or B often serves as the receptor-binding domain (RBD), and S2 is responsible for the fusion of viral envelope with the host cell membrane. During entry, S protein is further cleaved by the host protease at the S2′ site which activates the S protein through extensive irreversible conformational changes for membrane fusion. Three-dimensional (3D) classification of the cryo-electron microscopy (EM) data revealed that there are multiple, distinct conformational states of S trimer resulting from RBD structure changes in SARS-CoV,41 MERS-CoV42 and SARS-CoV-2.43 RBD can transit between “down” and “up” conformations. The “up” conformation is the domain accessible to the host receptor and is in an unstable and active state. However, when RBD is in the “down” conformation, the S protein enters a static and stable state. Glycans on the trimer can affect the conformational changes of RBD, thereby affecting the exposure of receptor-binding sites.44 N-terminal domain (NTD) glycans near the RBD can influence these RBD states’ conformational equilibrium, which affects the viral escape and vaccine design.45 However, only closed S trimers have been observed for the four other human-infecting coronaviruses: HCoV-NL63,39 HCoV-229E,46 HCoV-HKU1,47 and HCoV-OC43.48 Studies have shown that the overall density of glycan shielding in seven coronaviruses was significantly different (Figure 5).49 In addition, viruses belonging to lineage A of the betacoronaviruses also encode a protein called hemagglutinin esterase (HE). The HE protein is also a type I transmembrane glycoprotein that mediates the binding of virus particles to O-acetylated sialic acid through the functions of lectins and receptor destroying enzymes.50 The M protein is the most widespread structural protein in coronaviruses, which is often glycosylated on its amino-terminal domain. Different coronaviruses have different types of glycosylation of M protein; some M proteins are modified with O-linked sugars, whereas some M proteins are N-glycosylated. The M protein determines the budding site of the coronavirus, and binding with the S protein can trigger the assembly of virus particles.51 Although all human coronaviruses share many common features, there exist differences among them in the target cell receptor and the involved glycans in the virus attachment and entry (Table 2).
Figure 4.
Comparison of different genome structures of coronaviruses.
Notes: Reprinted from:Artika IM, Dewantari AK, Wiyatno A. Molecular biology of coronaviruses: current knowledge. Heliyon. 2020;6(8):e04743.118 Creative Commons CC BY-NC-ND (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abbreviations: UTR, untranslated region; ORF, open reading frame; S, spike protein; E, envelope protein; M, membrane protein; N, nucleocapsid protein.
Figure 5.
The difference in the glycans on the virus particles of coronaviruses.
Notes: Reprinted with permission from Watanabe Y, Berndsen ZT, Raghwani Jet al Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):2688.49 Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
Abbreviations: HKU1 S, spike protein of human coronavirus HKU1; SARS S, spike protein of severe acute respiratory syndrome-associated coronavirus; MERS S, sike protein of the Middle East respiratory syndrome-associated coronavirus; LASV GPC, glycoprotein complex of Lassa virus; H3N2 Vic11 HA, haemagglutinin of Victoria-2011 (Vic-11) strain of H3N2 subtype influenza Avirus; HIV-1 Env, envelope protein of human immunodeficiency virus 1; SIV Env, envelope protein of Simian immunodeficiency virus.
Table 2.
Comparison of the Surface Receptors Involved in Human Coronavirus Infection
| Name | Family | Receptora | Receptor-Binding Glycoproteinb | Glycosite Reported |
|---|---|---|---|---|
| HCoV-229E | Alphacoronavirus | APN | S protein | |
| HCoV-OC43 | Betacoronavirus | 9-O-acetylated sialic acid | S protein, HE protein | N133 |
| SARS-CoV | Betacoronavirus | ACE2, Heparan sulfate proteoglycans | S protein, DC-SIGN | 330NAT, 357NST, 783NFS, N158, N227 |
| HCoV-NL63 | Alphacoronavirus | ACE2, Heparan sulfate proteoglycans | S protein | Asn 358 |
| HCoV-HKU1 | Betacoronavirus | 9-O-acetylated sialic acid | S protein, HE protein | Asn171, N132, N19 |
| MERS-CoV | Betacoronavirus | DPP4, sialic acid | S protein | N155, N166, N236 |
| SARS-CoV-2 | Betacoronavirus | ACE2 | S protein | N343, N234, N165, 801NFS, 331NIT |
Notes: aAPN, aminopeptidase N; ACE2, angiotensin-converting enzyme 2; DPP4, dipeptidyl-peptidase 4. bS protein, spike protein; DC-SIGN, dendritic cell-specific intracellular adhesion molecule-3-grabbing nonintegrin; HE, hemagglutinin-esterase.
Human Coronavirus 229E (Alphacoronavirus)
HCoV-229E was first identified in 1966, causing the common cold, but the elderly and immune-compromised people can suffer from a severe respiratory health condition. HCoV-229E uses human aminopeptidase N (hAPN/CD13) as a receptor, which is different from other coronaviruses.52 Immune pressure caused by glycans can affect the APN recognition mode or the conformation of RBD, thus affecting the virus entry into the cell.53 The HCoV-229E S-protein contains as many as 30 predicted N-glycosylation sites. The ~60 residue HR2 region is the most highly glycosylated region, which may play a role in the transition from the pre-fusion to the post-fusion conformation.54
Human Coronavirus OC43 (Betacoronavirus, Lineage A)
HCoV-OC43 emerged as a zoonotic disease. In addition to causing the common cold in adults, it can also cause asymptomatic infections and occasionally serious lower respiratory tract infections. The receptor binding is related to S protein and hemagglutinin esterase (HE). HCoV-OC43 uses 9-O-acetylated sialic acid as a receptor, and the sialic acid glycan-binding site is located in the groove on the surface of S protein domain A. The HE protein can mediate the binding of S protein to the receptor, but this ability will disappear after the mutation of the HE protein domain.55 The cryo-EM reconstruction resolves 14 N-linked glycans extending from the surface of each protomer. It reveals that the HCoV-OC43 S oligosaccharide density is comparable to SARS-CoV S and MERS-CoV S.48
SARS-CoV (Betacoronavirus, Lineage B)
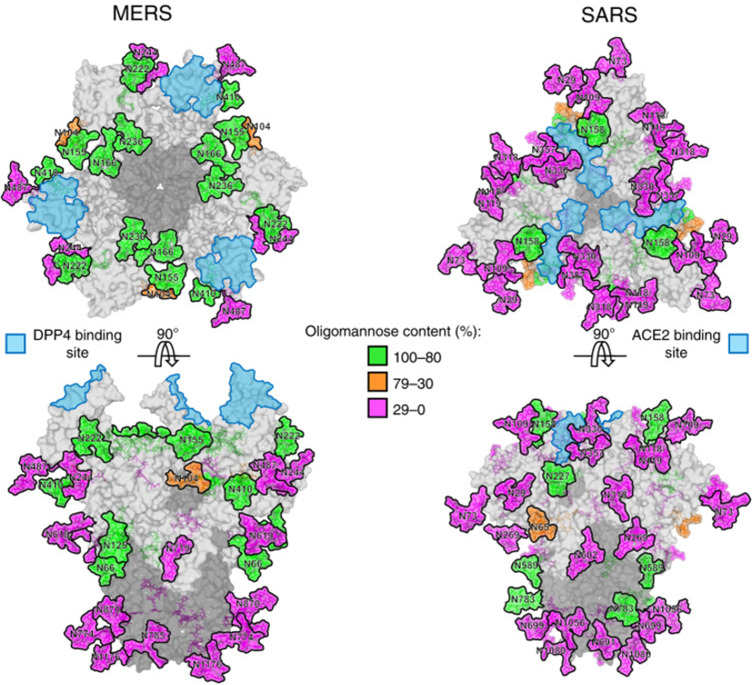
SARS-CoV began in November 2002 in Guangdong Province, China, and lasted for about seven months. The main clinical manifestations of SARS are fever, cough, progressive dyspnea and hypoxemia, and severe cases develop into acute respiratory distress syndrome. SARS-S uses ACE2 as the infection receptor, and C-type lectin DC-SIGN has also been found to be a selective receptor for SARS-CoV.56 The SARS-CoV S glycoprotein has 23 potential N-linked glycosylation sites, and 12 have been considered to be effectively glycosylated (Figure 6).57 Major glycans consist of high-mannose (Man5–9GlcNAc2), hybrid and bi-, tri-, and tetra-antennary complex with and without both bisecting GlcNAc and core fucose.58 The antibody 5H10 targets the S protein of SARS-CoV and has been shown to neutralize SARS-CoV and may be used for prevention and treatment.59 Besides, a glycosylated hydrophobic protein with a single potential N-glycosylation site and three potential O-glycosylation sites were found in SARS-M.
Figure 6.
N-linked glycans on the middle east respiratory syndrome-associated coronavirus (MERS) and severe acute respiratory syndrome (SARS)-associated coronavirus S proteins. Reprinted with permission from Watanabe, Y et al Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1): 2688.49 Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
Abbreviations: DPP4, dipeptidyl-peptidase 4; ACE2, angiotensin-converting enzyme 2.
Human Coronavirus NL63 (Alphacoronavirus)
Human coronavirus NL63 was first discovered in 2004 in a child with bronchiolitis in the Netherlands.60 It can lead to respiratory and digestive diseases in children and adults, and in severe cases, pneumonia and acute gastroenteritis. HCoV-NL63 also uses angiotensin-converting enzyme 2 (ACE2) as a receptor for infecting target cells. It has been found that heparan sulfate proteoglycan acts as the adhesion receptor of HCoV-NL63, enhancing the affinity of S protein and receptor ACE2. The adhesion of HCoV-NL63 and heparan sulfate proteoglycan enhances the ability of virus infection.61 The HCoV-NL63 S trimer is covered by 102 N-linked oligosaccharides, and the S2 subunit has a higher glycan density than the S1 subunit. Alexandra et al reported that a glycan linked to Asn358 could mask the receptor-binding loops and prevent neutralizing antibodies from functioning.39 The N-terminal part of the S protein of HCoV-NL63 contains a unique 179-amino-acid domain where several potential glycosylation sites exist.62 It has high variability and may play a role in immune evasion.
Human Coronavirus HKU1 (Betacoronavirus, Lineage A)
The human coronavirus HKU1 was first discovered in 2005 in a 71-year-old pneumonia patient in Hong Kong.63 HCoV-HKU1 mainly causes mild upper respiratory tract infections, with symptoms such as fever, cough, runny nose, and wheezing. It can lead to severe pneumonia and even death in the elderly and patients with underlying diseases. The S protein of HCoV-HKU1 uses 9-O-Ac-Sia as the attachment receptor, and HE is used as the receptor destroying enzyme.64 The S protein has 28 potential N-linked glycosylation sites (18 in S1 ectodomain and 10 in S2 endodomain). RBD is the domain A of the S protein, and the S2 domain is a hybrid typed N-linked glycan containing minor terminal mannose residues.65 Removal of the glycan attached to Asn171 has been reported to lower the binding affinity of HKU1 S1A, possibly by affecting protein folding.66
The Middle East Respiratory Syndrome Coronavirus (Betacoronavirus, Lineage C)
MERS-CoV was first found in 2012 as a lethal zoonotic virus in humans in Saudi Arabia and Jordan.67 The clinical characteristics of MERS-CoV infection vary widely, ranging from asymptomatic to influenza-like syndrome, pneumonia, and acute respiratory distress syndrome. The most common symptoms are fever, cough, and difficulty breathing. The RBD of MERS-CoV has a high structural similarity with SARS-CoV in core subunits. Dipeptidyl peptidase 4 (DPP4)/CD26 is the functional receptor of MERS-CoV, which is a multifunctional type II transmembrane glycoprotein that exists on the cell surface as a dimer.68 In addition to DPP4, the S1 subunit of MERS-CoV binds to sialic acid (Sia) to enter the host cell. The MERS-CoV S protein has 25 potential N-linked glycosylation sites (Figure 6). Glycosylation variable regions are mainly located on the S1 subunit.
SARS-CoV-2 (Betacoronavirus, Lineage B)
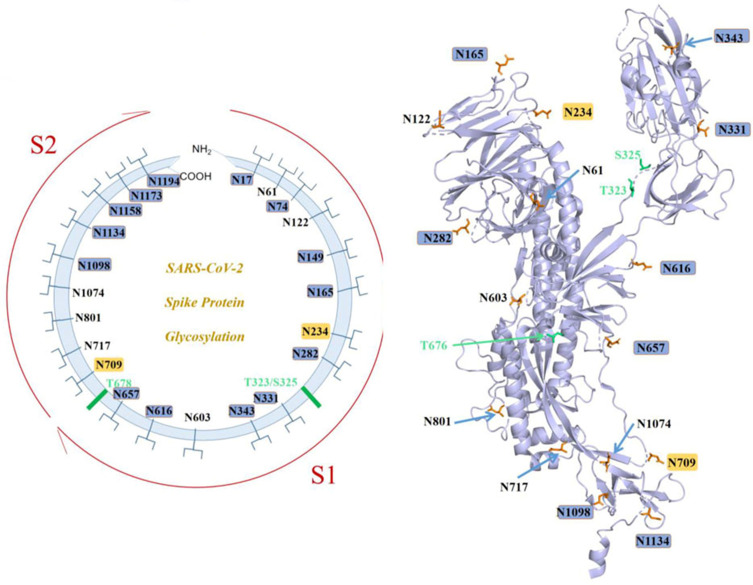
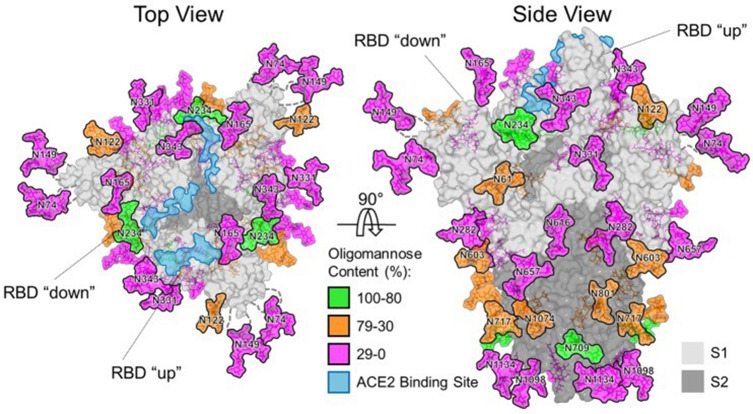
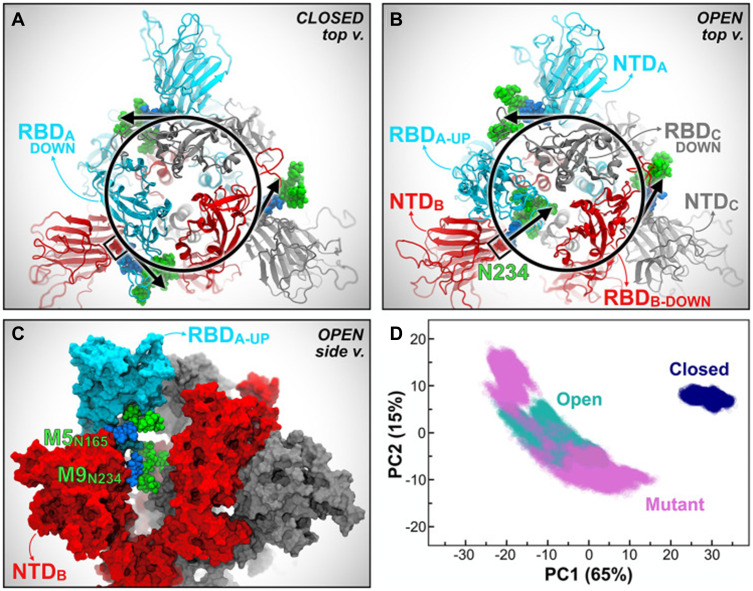
A previously unknown coronavirus, named SARS-CoV-2, was discovered in December 2019 in Wuhan, Hubei province of China. It spreads rapidly from person to person and poses a major threat to global public health. Fever, cough, headache, fatigue, dyspnea, and other symptoms are the main clinical manifestations. SARS-CoV-2 S uses ACE2 to enter cells and might interact with other host targets such as CD26, Ezrin, cyclophilins.69 Unlike other coronaviruses, there is a furin cleavage site at the S1/S2 boundary of SARS-CoV-2, RARR (682–685) site, which can cleave the virus envelope glycoprotein and enhance the fusion of virus and host cell membrane.70 Using liquid chromatography–mass spectrometry technology and cryo-EM, studies have revealed the glycan structure of SARS-CoV-2 S protein. A total of 22 N-glycosylated sites were reported, which are modified by high mannose, hybrid and complex-type glycans (Figure 7). Among them, eight are located in the N-terminal domain, two (N331 and N343) are located in the receptor-binding domain, three are located in the S1 subunit, and nine are located in the S2 subunit.71 Glycan can regulate the up-down transformation of RBD conformation (Figure 8). Lorenzo et al found that the removal of N234 and N165 glycan by mutation would destabilize the “up” conformation of RBD, which is not conducive to the binding of SARS-CoV-2 to the receptor (Figure 9).72 Besides, O-glycosylation has been shown to exist at Thr323 and Ser325 sites in the RBD domain and may play a role in the binding of the virus to ACE2. The S309 MAb targeting the glycopeptide sequence located on the N343 glycosite can not only neutralize SARS-CoV-1 but also SARS-CoV-2.71
Figure 7.
SARS-CoV-2 S N-linked glycans.
Notes: Reprintedfrom Zhao X, Chen H, Wang H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front Mol Biosci. 2021;8:629873 .119 Copyright © 2021 Zhao, Chen and Wang. Creative Commons Attribution License (CC BY; https://creativecommons.org/licenses/by/4.0/legalcode).
Abbreviations: ACE2, angiotensin-converting enzyme 2; RBD, recognition-binding domain.
Figure 8.
Glycan structure of the spike protein of SARS-CoV-2.
Notes: Reprinted with permission from AAAS. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–33.115 Available from: https://science.sciencemag.org/content/369/6501/330.
Abbreviation: N, Sprotein N-glycosylation site.
Figure 9.
Mutations in the S protein N-glycosylation sites of N234A and N165A show increased instability of RBD-A in the “up” state (A–D).
Notes: Reprinted with permission from Casalino LGaieb Z, Goldsmith JA etal Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent Sci. 2020;6(10):1722–1734.72 Copyright © 2020 American Chemical Society. Further permissions related to the material excerpted should be directed to the ACS. Available from: https://pubs.acs.org/doi/10.1021/acscentsci.0c01056.
Glycans in the Diagnosis and Treatment of Viral Infections
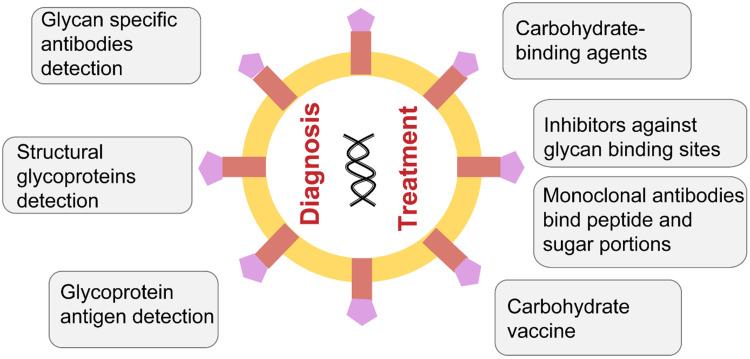
Glycan can be used for the diagnosis and treatment of viruses (Figure 10). Conventional methods for the detection of viruses mainly include serological and molecular biology techniques including immunoassay methods to detect viral antigens or antibodies (eg, ELISA), polymerase chain reaction (PCR)-based techniques, isothermal DNA amplification techniques (eg, loop-mediated isothermal amplification), and next-generation sequencing. These techniques are extremely useful but can be time consuming, expensive, susceptible to false positives and need technical and infrastructure requirements that limit its applications.73 Glycan-based virus diagnostics can represent a potential alternative by detecting structural glycoproteins, early glycoprotein antigen, or the titers of glycan-specific antibodies. In comparison to the existing technologies, this could be cost effective with high specificity and sensitivity. Already, glycoprotein-based IgM and IgG serology have been used to diagnose primary rubella virus infections and SARS.73
Figure 10.
Applications of glycans in the diagnosis and treatment of viral infections.
Viral protein glycosylation is involved in multiple stages of the viral life cycle, and thus targeting virus glycan structures can be a potential therapy option. Carbohydrate-binding agents (CBAs) that interact with the viral-envelope glycans have been reported to be effective antivirals. On the one hand, CBAs can bind to glycans on the surface of the virus and block the virus from entering the host cell. On the other hand, it can make the glycans lose their shielding effect and induce the body’s immune response.74 For example, high doses of mannose-binding lectin are effective against the Ebola virus75 and a lectin isolated from banana fruit called BanLec was identified to interact with HIV-1.76 Other drugs like iminosugars and 2-deoxy sugar derivatives of uridine are used to treat influenza by impairing the production of glycans.77 Tunicamycin can inhibit N-glycans, and it has been reported to affect the synthesis and function of S and M and may be used to treat the SARS-CoV-2. Competitive inhibitors against the glycan-binding sites of glycoproteins may also be an effective option.78
Neutralizing antibodies toward spike proteins play a critical role in protective immunity. Some monoclonal antibodies (mAbs) against protein epitopes have been reported, and more attention should be paid to the glycans attached to them.79,80 mAbs targeting the N-glycan portion on the Env protein surface have been designed, which can resist the glycan shielding effect.81 In addition, antibodies that bind both peptide and sugar portions of spike glycopeptides have also been reported.82
Vaccines are one of the most effective means of controlling the spread of infectious diseases. There are several types of vaccines, including, the whole virus, inactivated vaccines, live attenuated vaccines, subunit vaccines, nucleic acid vaccines, and viral vector vaccines. Considering the immunogenicity, safety, production cost, time, and other factors, each vaccine has its own advantages and disadvantages. For example, subunit vaccines have the problem of low immunogenicity. Glycosylation can affect the immunogenicity of viral proteins.83 Using different glycoprotein expression systems and genetic engineering tools, glycan-based self-adjuvanted vaccines can be designed to enhance immunogenicity and vaccine response. Although no viral carbohydrate-based vaccines have been designed yet, it has been reported that several antibodies directed against viral glycan structures have been isolated.84
Glycans, Nanoparticles, and the Coronavirus Infections
To reduce the impact of coronavirus infections, rapid diagnostics are necessary. Currently, there is a wide variety of tests available for the diagnosis of coronavirus infections based on a limited array of detection methodologies that greatly vary in levels of specificity and sensitivity, detection time, and market distribution. A 2020 systematic review of patents revealed that most of the technologies available to clinicians, and indirectly to the public, for the detection of SARS-CoV, MERS-CoV, and SARS-CoV-2 are primarily based on the use of RT-PCR and/or isothermal amplification principles, which focus on identifying the presence of viral nucleic acids in bodily samples.85 The remaining available technologies are based on the use of ELISA methods that identify either the presence of human serum antibodies against defined or undefined viral proteins or the presence of such viral proteins in bodily samples containing such viral particles.
To further advance current predictive mathematics (eg, on the order of early symptoms onset86 or speed of dissemination87), engineering,88 barrier,89,90 educational,91,92 or pharmacological93 efforts to control the spread of coronavirus, and to advance the effectiveness of treatment initiatives, it is of utmost importance to promote the early diagnostic identification of virus-carriers. Despite the progress94 and the increasing availability of rapid antibody/nucleotide-base tests (eg, novel colloidal gold nanoparticle membranes and monoclonal antibodies),95 it is desirable to further develop highly sensitive technologies and alternative strategies that do not rely on the mounting of immune responses by the host, which require variable time intervals to develop across populations. Delayed effective diagnosis increases the opportunity for (high-risk) asymptomatic viral shedders to disperse viral particles within the community. Lastly, it is important that new technologies improve the level of sensitivity of current methodologies and be more decentralized and readily implementable by the public. Our understanding of glycan biology and how glycosylation defects (such as inflammatory bowel diseases96) or the modulation of glycan metabolism by commensal microorganisms97 or drugs (eg, chloroquine, antimalarial with anti-COVID-19 effect by increasing the endosomal pH required for the virus, cell fusion and interfering with glycosylation of cell receptors98) affect human susceptibility to viral colonization and infections represent a nascent opportunity for diagnostics and therapeutics.
Nanoparticles are widely tested and reported to control viral infections, including coronaviruses.99–104 Metallic nanoparticles (particularly silver and gold nanoparticles), and nanoparticles prepared with antiviral drugs (such as ivermectin) and biomolecules (such as lactoferrin, bilirubin) have shown their ability to inhibit viral entry and replication. Silver and gold nanoparticles interact with the structural proteins or glycan on the surface of viruses, or damage the surface proteins or glycan to affect the structural integrity of virions, thereby inhibiting infection,105,106 while the antiviral activity of other metal nanoparticles, prepared of cerium oxide (ie, nanoceria), is attributed to their enhanced activity in preventing systemic inflammatory complications.107 Recent research studies have proved the efficiency of lactoferrin (LF) to bind to cell surface receptor heparan sulfate proteoglycans (HSPGs), and interfere the entry of coronavirus.108,109 Also, ivermectin-loaded nanoparticles are shown to target glycans to decrease expression of viral spike protein and the angiotensin-converting enzyme 2 (ACE2) receptor.110
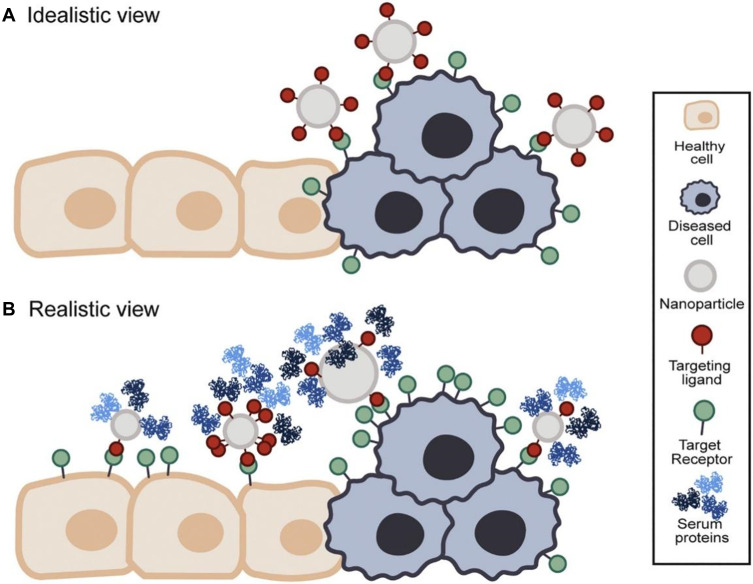
Glycan-based approaches further provide great opportunities for the design and the development of advanced viral therapies. Targeting glycans holds the promise for drug delivery with more specificity and affinity. However, off-target effects remain a challenge for in vivo applications caused by cross-reactivity with other glycan structures that is relatively abundant in most of the biological systems, resulting in possible toxic effects due to the generation of reactive oxygen species and nitric oxide in healthy tissues. Nanoparticles can be used to improve the targeting of glycan structures by increasing the loaded concentration of the targeting moieties. It is worth mentioning that the physicochemical properties of the glycan structures, such as size, shape, and surface charge, can influence their biological activity and functions, and performing more inclusive studies are needed to evaluate the potential use of these properties to enable efficient targeting of glycans and viral drug delivery.111 In addition, multivalent nanomedicines approaches could provide a relatively novel strategy for the development of glycan-based nanoparticles therapeutics for viral infections. Compared with traditional nanomedicine approaches, such approaches have lower cytotoxicity, lower immunogenicity, and higher targeting (Figure 11). Multivalent NPs can be used as an antiviral by blocking the attachment, and entry of the virus particles into host cells, inhibiting viral replication, or selectively triggering the production of reactive oxygen species in the diseased cells.112 Compared with standard monovalent drugs, multivalent NPs can form multivalent ligand receptor pairs and convert inactive NPs into multivalent conjugates. The easy functionalization and bioconjugation of NPs with targeting ligands such as viral receptors, antibodies, peptides, glycan, or proteins can considerably enhance their affinity and tropism for their target, thereby reducing the toxicity and adverse reactions.
Figure 11.
Idealistic versus realistic view of active targeting nanomedicines.
Notes: (A) Schematic presentation of an idealistic view of targeting using nanoparticles. Nanoparticle interacts exclusively with targets on the diseased cells. (B) Schematic presentation of arealistic view of targeting using nanoparticles. The target cell receptor is expressed in the diseased and healthy cells, leading to off-targeting side effects. Reprinted with permission from Woythe L, Tito NB, Albertazzi L.Aquantitative view on multivalent nanomedicine targeting. Adv Drug Deliv Rev. 2021;169:1–21.117 Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode).
Conclusions
Glycans are complex carbohydrate structures that possess several characteristics and advantages over other biomolecules. We have showcased the varied functional and structural features of viral glycans and discussed the particular role of glycans in coronaviruses, including HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS, MERS, and SARS-CoV-2. Glycan-based virus diagnostics and therapeutics have great potential to replace the current methods and provide better options by targeting glycan structures in the viruses. In conclusion, glycans have large and currently underexploited potential applications and studying their nanostructure can lead to novel methods for virus control and eradication.
Disclosure
The authors declare no conflicts of interest for this work.
References
- 1.Reichardt NC, Martin-Lomas M, Penades S. Glyconanotechnology. Chem Soc Rev. 2013;42(10):4358–4376. doi: 10.1039/c2cs35427f [DOI] [PubMed] [Google Scholar]
- 2.Raman R, Tharakaraman K, Sasisekharan V, et al. Glycan–protein interactions in viral pathogenesis. Curr Opin Struct Biol. 2016;40:153–162. doi: 10.1016/j.sbi.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi T, Suzuki T. Role of Glycans in Viral Infection, in Sugar Chains. Springer; 2015:71–93. [Google Scholar]
- 4.Watanabe Y, et al. Exploitation of glycosylation in enveloped virus pathobiology. Biochimica Et Biophysica Acta. 2019;1863(10):1480–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Z, Xu Y, Bao L, et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabasum S, et al. Glycoproteins functionalized natural and synthetic polymers for prospective biomedical applications: a review. Int J Biol Macromol. 2017;98:748–776. [DOI] [PubMed] [Google Scholar]
- 7.Ruhaak LR, Xu G, Li Q, Goonatilleke E, Lebrilla CB. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chemical Reviews. 2018;118(17):7886–7930. doi: 10.1021/acs.chemrev.7b00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levery SB, Steentoft C, Halim A, et al. Advances in mass spectrometry driven O-glycoproteomics. Biochim Biophys Acta. 2015;1850(1):33–42. doi: 10.1016/j.bbagen.2014.09.026 [DOI] [PubMed] [Google Scholar]
- 9.Fulton KM, Li J, Tomas JM, et al. Characterizing bacterial glycoproteins with LC-MS. Expert Rev Proteomics. 2018;15(3):203–216. doi: 10.1080/14789450.2018.1435276 [DOI] [PubMed] [Google Scholar]
- 10.Petrescu AJ, Wormald MR, Dwek RA. Structural aspects of glycomes with a focus on N-glycosylation and glycoprotein folding. Curr Opin Struct Biol. 2006;16(5):600–607. doi: 10.1016/j.sbi.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 11.Roth J, Zuber C. Quality control of glycoprotein folding and ERAD: the role of N-glycan handling, EDEM1 and OS-9. Histochem Cell Biol. 2017;147(2):269–284. doi: 10.1007/s00418-016-1513-9 [DOI] [PubMed] [Google Scholar]
- 12.Reily C, Stewart TJ, Renfrow MB, et al. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15(6):1. doi: 10.1038/s41581-019-0129-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warnecke D, Heinz E. Glycolipid headgroup replacement: a new approach for the analysis of specific functions of glycolipids in vivo. Eur J Cell Biol. 2010;89(1):53–61. doi: 10.1016/j.ejcb.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 14.Faivre V, Rosilio V. Interest of glycolipids in drug delivery: from physicochemical properties to drug targeting. Expert Opin Drug Deliv. 2010;7(9):1031–1048. doi: 10.1517/17425247.2010.511172 [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi Y, Kohyama-Koganeya A, Hirabayashi Y. New insights on glucosylated lipids: metabolism and functions. Biochim Biophys Acta. 2013;1831(9):1475–1485. doi: 10.1016/j.bbalip.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 16.Bertani B, Ruiz N. Function and Biogenesis of Lipopolysaccharides. EcoSal Plus. 2018;8(1). doi: 10.1128/ecosalplus.ESP-0001-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortes-Sanchez Ade J, Hernandez-Sanchez H, Jaramillo-Flores ME. Biological activity of glycolipids produced by microorganisms: new trends and possible therapeutic alternatives. Microbiol Res. 2013;168(1):22–32. doi: 10.1016/j.micres.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 18.Furukawa K, Ohmi Y, Ohkawa Y, et al. New era of research on cancer-associated glycosphingolipids. Cancer Sci. 2019;110(5):1544–1551. doi: 10.1111/cas.14005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida Y, Tanaka K. Cytosolic N-Glycans: triggers for Ubiquitination Directing Proteasomal and Autophagic Degradation: molecular Systems for Monitoring Cytosolic N-Glycans as Signals for Unwanted Proteins and Organelles. Bioessays. 2018;40(3):1700215. doi: 10.1002/bies.201700215 [DOI] [PubMed] [Google Scholar]
- 20.Hebert DN, Lamriben L, Powers ET, et al. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat Chem Biol. 2014;10(11):902–910. doi: 10.1038/nchembio.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe Y, et al. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863(10):1480–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorander S, Mbwana J, Lyamuya E, et al. Mature glycoprotein g presents high performance in diagnosing herpes simplex virus type 2 infection in sera of different Tanzanian cohorts. Clin Vaccine Immunol. 2006;13(6):633–639. doi: 10.1128/CVI.00051-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan GS, Li W, Ye L, et al. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012;8(12):e1003065. doi: 10.1371/journal.ppat.1003065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagae M, Yamaguchi Y, Taniguchi N, et al. 3D Structure and Function of Glycosyltransferases Involved in N-glycan Maturation. Int J Mol Sci. 2020;21(2):437. doi: 10.3390/ijms21020437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73(1):1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752 [DOI] [PubMed] [Google Scholar]
- 26.Bayer K, Banning C, Bruss V, et al. Hepatitis C Virus Is Released via a Noncanonical Secretory Route. J Virol. 2016;90(23):10558–10573. doi: 10.1128/JVI.01615-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiuchi T, Izumi M, Mukogawa Y. Monitoring of Glycoprotein Quality Control System with a Series of Chemically Synthesized Homogeneous Native and Misfolded Glycoproteins. Journal of the American Chemical Society. 2018;140(50):17499–17507. doi: 10.1021/jacs.8b08653 [DOI] [PubMed] [Google Scholar]
- 28.Bagdonaite I, Vakhrushev SY, Joshi HJ, Wandall HH. Viral glycoproteomes: technologies for characterization and outlook for vaccine design. FEBS Letters. 2018;592(23):3898–3920. doi: 10.1002/1873-3468.13177 [DOI] [PubMed] [Google Scholar]
- 29.Planinc A, et al. Glycan characterization of biopharmaceuticals: updates and perspectives. Anal Chim Acta. 2016;921:13–27. doi: 10.1016/j.aca.2016.03.049 [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Nie J, Prochnow C, et al. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology. 2013;10(1):14. doi: 10.1186/1742-4690-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69(1):531–569. doi: 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- 32.Isa P, Arias CF, López S. Role of sialic acids in rotavirus infection. Glycoconj J. 2006;23(1–2):27–37. doi: 10.1007/s10719-006-5435-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gee GV, et al. The role of sialic acid in human polyomavirus infections. Glycoconj J. 2006;23(1–2):19–26. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Hulswit RJG, Widjaja I, et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A. 2017;114(40):E8508–e8517. doi: 10.1073/pnas.1712592114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izquierdo-Useros N, et al. HIV-1 capture and transmission by dendritic cells: the role of viral glycolipids and the cellular receptor Siglec-1. PLoS Pathog. 2014;10(7):e1004146. doi: 10.1371/journal.ppat.1004146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behrens AJ, Vasiljevic S, Pritchard L, et al. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14(11):2695–2706. doi: 10.1016/j.celrep.2016.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quiñones-Kochs MI, Buonocore L, Rose JK. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J Virol. 2002;76(9):4199–4211. doi: 10.1128/JVI.76.9.4199-4211.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu NC, Wilson IA. A Perspective on the Structural and Functional Constraints for Immune Evasion: insights from Influenza Virus. J Mol Biol. 2017;429(17):2694–2709. doi: 10.1016/j.jmb.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walls AC, Tortorici MA, Frenz B, et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23(10):899–905. doi: 10.1038/nsmb.3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su S, Wong G, Shi W, et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pallesen J, Wang N, Corbett KS, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A. 2017;114(35):E7348–e7357. doi: 10.1073/pnas.1707304114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls AC, Park Y-J, Tortorici MA, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henderson R, et al. Glycans on the SARS-CoV-2 Spike Control the Receptor Binding Domain Conformation. bioRxiv. 2020. [Google Scholar]
- 46.Li Z, et al. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirchdoerfer RN, Cottrell CA, Wang N, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tortorici MA, Walls AC, Lang Y, et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26(6):481–489. doi: 10.1038/s41594-019-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe Y, Berndsen ZT, Raghwani J, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):2688. doi: 10.1038/s41467-020-16567-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus Spike Protein and Tropism Changes. Adv Virus Res. 2016;96:29–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonavia A, Zelus BD, Wentworth DE, et al. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J Virol. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reguera J, Santiago C, Mudgal G, et al. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8(8):e1002859. doi: 10.1371/journal.ppat.1002859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, et al. The human coronavirus HCoV-229E S-protein structure and receptor binding. J Virol. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bakkers MJ, Lang Y, Feitsma LJ, et al. Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity. Cell Host Microbe. 2017;21(3):356–366. doi: 10.1016/j.chom.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belouzard S, Millet JK, Licitra BN, et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krokhin O, Li Y, Andonov A, et al. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol Cell Proteomics. 2003;2(5):346–356. doi: 10.1074/mcp.M300048-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie G, Harvey DJ, Feldmann F, et al. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology. 2010;399(2):257–269. doi: 10.1016/j.virol.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyoshi-Akiyama T, Ishida I, Fukushi M, et al. Fully human monoclonal antibody directed to proteolytic cleavage site in severe acute respiratory syndrome (SARS) coronavirus S protein neutralizes the virus in a rhesus macaque SARS model. J Infect Dis. 2011;203(11):1574–1581. doi: 10.1093/infdis/jir084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Hoek L, Pyrc K, Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol Rev. 2006;30(5):760–773. doi: 10.1111/j.1574-6976.2006.00032.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milewska A, Nowak P, Owczarek K, et al. Entry of Human Coronavirus NL63 into the Cell. Journal of virology. 2018;92(3). doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pyrc K, Berkhout B, van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. 2007;81(7):3051–3057. doi: 10.1128/JVI.01466-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woo PC, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X, Dong W, Milewska A, et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J Virol. 2015;89(14):7202–7213. doi: 10.1128/JVI.00854-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan CM, et al. Spike protein, S, of human coronavirus HKU1: role in viral life cycle and application in antibody detection. Exp Biol Med (Maywood). 2008;233(12):1527–1536. [DOI] [PubMed] [Google Scholar]
- 66.Hulswit RJG, Lang Y, Bakkers MJG, et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A. 2019;116(7):2681–2690. doi: 10.1073/pnas.1809667116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 68.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vankadari N, Wilce JA. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerging Microbes & Infections. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walls AC, Park Y-J, Tortorici MA, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183(6):1735. doi: 10.1016/j.cell.2020.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou D, Tian X, Qi R, et al. Identification of 22 N-glycosites on spike glycoprotein of SARS-CoV-2 and accessible surface glycopeptide motifs: implications for vaccination and antibody therapeutics. Glycobiology. 2020. doi: 10.1093/glycob/cwaa052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casalino L, et al. Beyond Shielding: the Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent Sci. 2020;6(10):1722–1734. doi: 10.1021/acscentsci.0c01056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banerjee N, Mukhopadhyay S. Viral glycoproteins: biological role and application in diagnosis. Virusdisease. 2016;27(1):1–11. doi: 10.1007/s13337-015-0293-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balzarini J. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat Rev Microbiol. 2007;5(8):583–597. doi: 10.1038/nrmicro1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Michelow IC, Lear C, Scully C, et al. High-dose mannose-binding lectin therapy for Ebola virus infection. J Infect Dis. 2011;203(2):175–179. doi: 10.1093/infdis/jiq025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hopper JTS, Ambrose S, Grant OC, et al. The Tetrameric Plant Lectin BanLec Neutralizes HIV through Bidentate Binding to Specific Viral Glycans. Structure. 2017;25(5):773–782.e5. doi: 10.1016/j.str.2017.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krol E, Wandzik I, Krejmer-Rabalska M, et al. Biological Evaluation of Uridine Derivatives of 2-Deoxy Sugars as Potential Antiviral Compounds against Influenza A Virus. Int J Mol Sci. 2017;18(8):8. doi: 10.3390/ijms18081700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Desmarets LMB, Theuns S, Roukaerts IDM, et al. Role of sialic acids in feline enteric coronavirus infections. J Gen Virol. 2014;95(Pt 9):1911–1918. doi: 10.1099/vir.0.064717-0 [DOI] [PubMed] [Google Scholar]
- 79.Traggiai E, Becker S, Subbarao K, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10(8):871–875. doi: 10.1038/nm1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berry JD, Hay K, Rini JM, et al. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs. 2010;2(1):53–66. doi: 10.4161/mabs.2.1.10788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong L, Torrents de la Peña A, Deller MC, et al. Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 10):2099–2108. doi: 10.1107/S1399004715013917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinto D, Park Y-J, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 83.Krumm SA, Doores KJ. Targeting Glycans on Human Pathogens for Vaccine Design. Curr Top Microbiol Immunol. 2018. [DOI] [PubMed] [Google Scholar]
- 84.McCoy LE, Burton DR. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol Rev. 2017;275(1):11–20. doi: 10.1111/imr.12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Calvo-Bonacho E, et al. COVID-19 and Sick Leave: an Analysis of the Ibermutua Cohort of Over 1,651,305 Spanish Workers in the First Trimester of 2020. Front Public Health. 2020;8:8. doi: 10.3389/fpubh.2020.580546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larsen JR, Martin MR, Martin JD, et al. Modeling the Onset of Symptoms of COVID-19. Front Public Health. 2020;8:473. doi: 10.3389/fpubh.2020.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin Y-F, Duan Q, Zhou Y, et al. Spread and impact of COVID-19 in China: a systematic review and synthesis of predictions from transmission-dynamic models. Front Med. 2020;7:321. doi: 10.3389/fmed.2020.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Corrêa H, Corrêa DG. Polymer applications for medical care in the COVID-19 pandemic crisis: will we still speak ill of these materials? Front Mater. 2020;7:283. doi: 10.3389/fmats.2020.00283 [DOI] [Google Scholar]
- 89.Rodriguez-Palacios A, Cominelli F, Basson AR, et al. Textile Masks and Surface Covers—A Spray Simulation Method and a “Universal Droplet Reduction Model” Against Respiratory Pandemics. Front Med. 2020;7:260. doi: 10.3389/fmed.2020.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodriguez-Palacios A, Conger M, Cominelli F. Germ-Free Mice Under Two-Layer Textiles Are Fully Protected From Bacteria in Sprayed Microdroplets: a Functional in vivo Test Method of Facemask/Filtration Materials. Front Med. 2020;7:504. doi: 10.3389/fmed.2020.00504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eichler SE, Hopperton AP, Alava JJ, et al. A Citizen science facemask experiment and educational modules to improve coronavirus safety in communities and schools. Front Med. 2020;7:486. doi: 10.3389/fmed.2020.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sassano M, McKee M, Ricciardi W, et al. Transmission of SARS-CoV-2 and Other Infections at Large Sports Gatherings: a Surprising Gap in Our Knowledge. Front Med. 2020;7:7. doi: 10.3389/fmed.2020.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Idda ML, Soru D, Floris M. Overview of the first 6 months of Clinical trials for COVID-19 pharmacotherapy: the most studied drugs. Front Public Health. 2020;8:497. doi: 10.3389/fpubh.2020.00497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu K, et al. Progresses in nucleic acid testing for COVID-19. Am J Transl Res. 2020;12(10):6763. [PMC free article] [PubMed] [Google Scholar]
- 95.Mertens P, De Vos N, Martiny D, et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front Med. 2020;7:225. doi: 10.3389/fmed.2020.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campbell BJ, Yu L-G, Rhodes JM. Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj J. 2001;18:851–858. doi: 10.1023/A:1022240107040 [DOI] [PubMed] [Google Scholar]
- 97.Blandford LE, Johnston EL, Sanderson JD, et al. Promoter orientation of the immunomodulatory Bacteroides fragilis capsular polysaccharide A (PSA) is off in individuals with inflammatory bowel disease (IBD). Gut Microbes. 2019;10(5):569–577. doi: 10.1080/19490976.2018.1560755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heinrich MA, Martina B, Prakash J. Nanomedicine strategies to target coronavirus. Nano Today. 2020;35:100961. doi: 10.1016/j.nantod.2020.100961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Draz MS, Shafiee H. Applications of gold nanoparticles in virus detection. Theranostics. 2018;8(7):1985. doi: 10.7150/thno.23856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Draz MS, Tang Y, Zhang P. Bio-Nanoparticles: Nanoscale Probes for Nanoscale Pathogens, in 21st Century Nanoscience–A Handbook. CRC Press; 2020:20. [Google Scholar]
- 102.Draz MS, Wang Y-J, Chen FF, et al. Electrically oscillating plasmonic nanoparticles for enhanced DNA vaccination against hepatitis C virus. Adv Funct Mater. 2017;27(5):1604139. doi: 10.1002/adfm.201604139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang P, Draz MS. Functional Nanomaterial for Theranostic Medicine. Curr Top Med Chem. 2019;19(27):2447–2448. doi: 10.2174/156802661927191206151729 [DOI] [PubMed] [Google Scholar]
- 104.Draz MS, Fang BA, Zhang P, et al. Nanoparticle-mediated systemic delivery of siRNA for treatment of cancers and viral infections. Theranostics. 2014;4(9):872. doi: 10.7150/thno.9404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jeremiah SS, Miyakawa K, Morita T, et al. Potent antiviral effect of silver nanoparticles on SARS-CoV-2. Biochem Biophys Res Commun. 2020;533(1):195–200. doi: 10.1016/j.bbrc.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mehranfar A, Izadyar M. Theoretical Design of Functionalized Gold Nanoparticles as Antiviral Agents against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J Phys Chem Lett. 2020;11(24):10284–10289. doi: 10.1021/acs.jpclett.0c02677 [DOI] [PubMed] [Google Scholar]
- 107.Allawadhi P, Khurana A, Allwadhi S, et al. Nanoceria as a possible agent for the management of COVID-19. Nano Today. 2020;35:100982. doi: 10.1016/j.nantod.2020.100982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khurana I, Allawadhi P, Khurana A, et al. Can bilirubin nanomedicine become a hope for the management of COVID-19? Med Hypotheses. 2021;149:110534. doi: 10.1016/j.mehy.2021.110534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chang R, Ng TB, Sun WZ. Lactoferrin as potential preventative and adjunct treatment for COVID-19. Int J Antimicrob Agents. 2020;56(3):106118. doi: 10.1016/j.ijantimicag.2020.106118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Surnar B, Kamran MZ, Shah AS, et al. Clinically Approved Antiviral Drug in an Orally Administrable Nanoparticle for COVID-19. ACS Pharmacol Transl Sci. 2020;3(6):1371–1380. doi: 10.1021/acsptsci.0c00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Behzadi S, Serpooshan V, Tao W, et al. Cellular uptake of nanoparticles: journey inside the cell. Chem Soc Rev. 2017;46(14):4218–4244. doi: 10.1039/C6CS00636A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tabish TA, Hamblin MR. Multivalent nanomedicines to treat COVID-19: a slow train coming. Nano Today. 2020;35:100962. doi: 10.1016/j.nantod.2020.100962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen B, Tian E-K, He B, et al. Overview of lethal human coronaviruses. Signal Transduction Targeted Therapy. 2020;5(1):1–16. doi: 10.1038/s41392-020-0190-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Forni D, Cagliani R, Clerici M, et al. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Watanabe Y, Allen JD, Wrapp D, et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shajahan A, Supekar NT, Gleinich AS, et al. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woythe L, Tito NB, Albertazzi L. A quantitative view on multivalent nanomedicine targeting. Adv Drug Deliv Rev. 2021;169:1–21. doi: 10.1016/j.addr.2020.11.010 [DOI] [PubMed] [Google Scholar]
- 118.Artika IM, Dewantari AK, Wiyatno A. Molecular biology of coronaviruses: current knowledge. Heliyon. 2020;6(8):e04743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao X, Chen H, Wang H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front Mol Biosci. 2021;8:629873. [DOI] [PMC free article] [PubMed] [Google Scholar]