Abstract
Accurate and non-invasive monitoring of renal allografts post-transplant is essential for early detection of acute rejection (AR) and to impact the long-term survival of the transplant. We present the development and validation of a noninvasive, spot urine-based diagnostic assay, based on measurements of six urinary DNA, protein, and metabolic biomarkers. The performance of this assay for detecting kidney injury in both native kidneys and renal allografts is presented on a cohort of 601 distinct urine samples. The urinary composite score enables diagnosis of AR, with a receiver-operator characteristic (ROC) curve area-under-the-curve (AUC) of 0.99. In addition, we demonstrate the clinical utility of this assay for predicting AR, prior to a rise in the serum creatinine, enabling earlier detection of rejection than currently possible by standard of care tests. This noninvasive, sensitive, and quantitative approach is a robust and informative method for the rapid and routine monitoring of renal allografts.
One Sentence Summary:
A urinary assay enables the noninvasive diagnosis of kidney transplant rejection in concordance with biopsy histology reports.
Introduction
While kidney transplantation is the treatment of choice for patients with end-stage renal disease, the life of a kidney transplant is limited due to episodes of preventable subclinical and acute rejection. While proper adherence to immunosuppressive medications, as well as proper determination of the therapeutic concentrations on a per-patient basis, can assist in maintaining the stable status of the transplant, ongoing diagnostic monitoring is recognized as essential to monitor the health of the transplanted kidney.
However, current methods for the detection of kidney rejection are suboptimal for several reasons. Regular monitoring of serum creatinine remains the most common method for surveillance after renal transplantation, but it is an insensitive predictor and only increases upon a deficiency in kidney function rather than kidney injury (1). The gold standard of renal biopsy suffers from interobserver variability, complications inherent to the invasive nature of the technique, and relatively high cost (2, 3). Recently, molecular diagnostics based on blood or plasma have been developed based on composite RNA scores (4, 5) or donor-derived cell-free DNA (dd-cfDNA) fractions as determined by massively multiplexed polymerase chain reaction (PCR) (mmPCR) (6) or next-generation sequencing (NGS) (7–9).
While sequencing or single nucleotide polymorphism (SNP)-based methods of quantifying dd-cfDNA from plasma have been used to detect allograft rejection, they remain inconvenient for patients due to the requirement of blood draws, requiring special tubes and kits for collection. As such, urinary biomarkers remain a promising solution to these problems. Because urine is the direct ultrafiltrate of the kidneys, it provides an accurate window into the status of the allograft. Additionally, urine is a readily accessible biofluid, and its collection is truly noninvasive. ddcfDNA has been found in both the urine (10, 11) and serum of renal transplant recipients and has shown initial utility in discriminating acute rejection from stable outcomes. While previous research in this space has mostly relied upon assessment of the urine pellet messenger RNA (mRNA) by PCR-based urine assessment techniques (12, 13), the reproducibility of this assay is limited by failure to obtain an adequate quantity and quality of RNA in 20–30% of cases (12).
We hypothesized that the use of a microwell approach to measure a combination of nucleic acid, metabolite, and protein biomarkers, on raw, untimed urine, with utilization of the urine supernatant alone in a robust, enzyme-linked immunosorbent assay (ELISA)-based approach that avoids mRNA assessments (14–16) would enable a more robust measure for rapid assessment of rejection in a renal allograft. To this end, we first assessed the performance of target markers in a Kidney Injury Test assay for the staged detection of chronic kidney disease (CKD) in the native, non-transplanted kidney (15), and in this study we now demonstrate that the urinary abundance of these same markers can be used to develop a Q-Score for the sensitive and specific detection of acute rejection injury in the renal allograft. We trained and independently validated the Q-Score on urine samples from renal transplant recipients with and without biopsy-confirmed graft rejection. In addition, we also provide a comparison of the kidney injury scores across the same markers in a subset of healthy controls and patients with CKD, to provide an estimate of the baseline renal health in the stable renal transplant samples.
Here we present the results of our custom assay on 6 selected DNA, protein, and metabolite biomarkers consisting of cell-free DNA (cfDNA), methylated cell-free DNA (m-cfDNA), clusterin, total protein, creatinine, and CXCL10 on 601 prospectively collected urine samples from three transplant centers. These samples were collected from both pediatric and adult renal allograft recipients immediately before a renal allograft biopsy. Comparison to the aggregate of paired biopsy results and clinical pathology diagnoses indicated that the developed urine score (the Q-Score) trained and validated in this study can be used to identify grafts undergoing acute rejection, both antibody-mediated rejection (ABMR) and T cell-mediated rejection (TCMR), and can also non-invasively identify allografts that are stable and without any substantive histological injury. These findings suggest that this urine score has the potential to replace the renal biopsy as the gold standard of post-transplant monitoring and that such measurements can be used for the detection of rejection episodes and the proactive management of immunosuppressant titering.
Results
Detailed Patient Sub-Cohorts Assess the Utility of the urine Q-Score for kidney transplant rejection diagnosis
We have designed the study cohorts across 601 urine samples to assess the performance of the urine score, known as Q-score, biomarkers, including CXCL10, clusterin, cfDNA, m-cfDNA, total protein, and creatinine, on the assessment of transplant rejection (data file S1). These biomarkers had previously been identified for use in the Kidney Injury Test, an assay assessing CKD (15). From these biomarkers, we developed and validated the quantitative Q-score on multiple clinical sample groups. This study provided an assessment of the ability to non-invasively diagnose biopsy-confirmed acute renal transplant rejection in 332 distinct adult and pediatric (n=80) kidney transplant recipients with 332 unique biopsy-matched urine samples (where the urine was collected immediately prior to the performance of a biopsy), and an additional 32 biopsy-matched urine samples obtained within 1 week to 8 months before a biopsy confirmed rejection episode in 32 AR patients (Fig. 1). We were able to assess the performance of this urine-based diagnostic assay across a spectrum of recipient age, inclusive of pediatric and adult age groups (age range 10 months to 65 years), recipients of both genders (50% males), various recipient/donor ethnic groups, and with variations in donor gender (49% male) and HLA match grades (Table 1). 8% of recipients had repeat transplants. After transplant, we evaluated the ability of this assay to diagnose rejection in protocol biopsies (55%), when the serum creatinine is stable, or indication biopsies, triggered for graft dysfunction (based on a >20% rise in the serum creatinine from baseline). The timing of the protocol biopsies in this study included biopsies at 1 (5%), 3 (20%), 6 (70%), and 12 (5%) months post-transplant. The mean time for graft dysfunction biopsies was at 22 months post-transplant. Given the size of this dataset, with 103 biopsy-confirmed ARs and the conduct of protocol biopsies at all 3 clinical sites, we were also able to assess the performance of the biopsy to diagnose AR at the time of clinical graft dysfunction (clinical AR or cAR; n=62) as compared to AR diagnosed at the time of protocol biopsies, when there is stable graft function (sub-clinical AR or scAR; n=41).
Fig. 1. Study design.
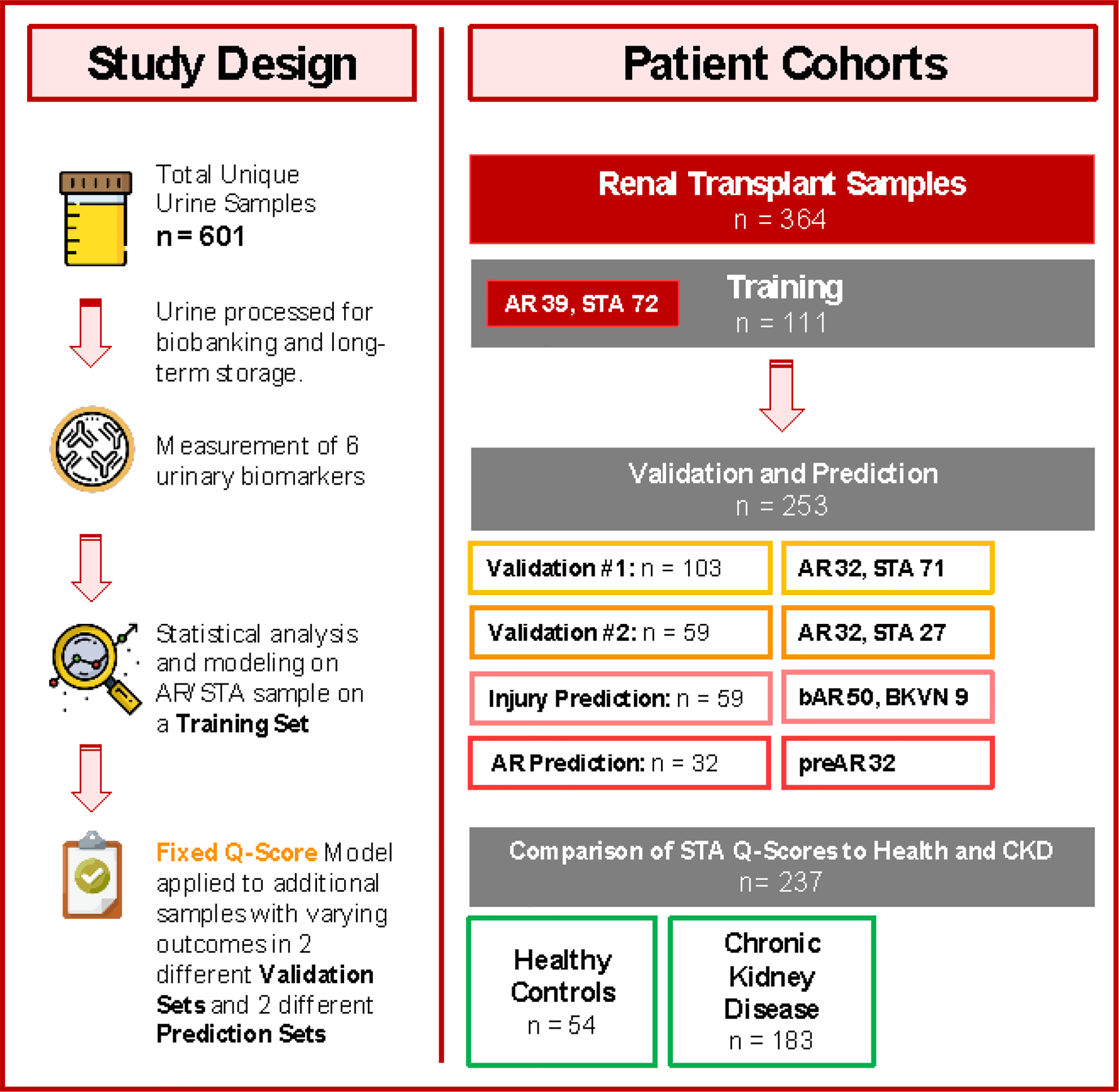
In this multicenter, prospective study, a total of 601 patient samples were assessed. 364 urine samples belonged to renal transplant recipients, where every urine sample was paired with a renal transplant biopsy for phenotype classification into the following diagnoses: stable (STA; n = 170), acute rejection (AR; n = 103), borderline AR (bAR; n = 50), and BK virus nephropathy (BKVN; n = 9). An additional 32 patients with AR had urine samples collected before the rejection episode; these samples were also paired with biopsies. In the context of a stable kidney transplant, we compared patients’ urine scores with a scores run on urine samples from healthy controls without any renal insufficiency (n = 54). In addition, we compared the urine scores from stable kidney transplant patients against the scores from patients with early CKD (stages 1 and 2; n = 183). The selected urinary biomarkers were measured on all urine samples collected, and statistical analysis and modeling were performed on a subset of patients, defined as STA or AR, and split out randomly as a training set (n = 111) for modeling the urine score. The fixed Q-Score model from the training set was then applied to two separate validation sets consisting of STA and AR samples with set sizes of 103 and 59 for validation set 1 and validation set 2, respectively. The fixed urine score was next applied to samples with other transplant injuries (bAR/BKVN; n = 59) and for prediction of rejection in the cohort of patients (preAR samples; n=32) with urine samples collected within 8 months of a biopsy-confirmed AR episode. The preAR samples were collected at a time where the paired biopsy was histologically stable and there was no graft dysfunction.
Table 1. Demographics and characteristics.
Values are reported in the given units with standard deviation in parentheses, and all comparisons between groups were non-significant. Recipients are both adult (>18 years) and pediatric (≤ 18 years) in each cohort. Clinical pathology data are based on the day of biopsy, which is timed with the urine collection, which occurred before performance of the biopsy. ABMR/TCMR split is of AR-confirmed patients, based on the Banff allograft pathology classification (17). All other demographic and clinical information is based on the day of the kidney transplant.
| Phenotype Characteristic | Training (n = 111) | Validation #1 (n = 103) | Validation #2 (n = 59) |
|---|---|---|---|
| Recipient | |||
| • Recipient age, year (SD) | 29 (13.4) | 32 (14.2) | 32 (11.7) |
| • Recipient male/female, % | 48/52 | 49/51 | 42/58 |
| Donor | |||
| • Donor age, year (SD) | 35 (12.4) | 36 (14.4) | 39 (10.0) |
| • Donor male/female, % | 43/57 | 44/56 | 40/69 |
| Ethnicity, % | |||
| • Caucasian | 35 | 39 | 48 |
| • Hispanic | 32 | 25 | 21 |
| • Asian | 26 | 27 | 24 |
| • African American | 3 | 2 | 2 |
| • Other | 4 | 7 | 5 |
| Donor Source, % | |||
| • Deceased | 49.4 | 42.4 | 55% |
| Clinical Pathology | |||
| • AR / STA | 39/72 | 32/71 | 32/27 |
| • ABMR/TCMR (of AR) | 24/15 | 9/23 | 5/27 |
| Cause of ESRD, % | |||
| • Obstructive | 5 | 4 | 5 |
| • Diabetes | 20 | 22 | 18 |
| • Hypertension | 43 | 37 | 42 |
| • Glomerulonephritis | 8 | 7 | 7 |
| • Immune-Mediated | 20 | 23 | 24 |
| • Congenital | 5 | 4 | 2 |
| • Other | 1 | 5 | 2 |
An assessment of all indication biopsies (n=160) showed that only 38.8% of transplants with clinical graft dysfunction had a confirmed histological diagnosis of AR using the established Banff schema (17). For the remainder of the indication biopsies, 20% were histologically read as non-AR injuries [2.3% BK viral nephropathy (BKVN), 17.5% bAR] and 40.6% as stable or no injury (STA). Within the STA group of patients with clinical graft dysfunction, 12 (7.5%) patients went on to develop biopsy confirmed AR at further follow-up (these samples were part of the preAR analysis group in this study). A similar assessment of all protocol biopsies (n=201) showed that 20.4% had stable graft function but still had sub-clinical acute rejection (scAR), histological rejection currently missed by serum creatinine monitoring alone. For the remainder of the protocol biopsies, 19.4% had other injuries (2% BKVN, 17.4% bAR), and 60.2% had no injury (STA); in this latter group, 20 patients went on to develop AR on subsequent follow-up (these samples were also part of the preAR analysis group in this study).
Target Urine Biomarkers can Discriminate Stable Patients from Patients with Acute Rejection
The selected biomarkers of urinary cfDNA, m-cfDNA, creatinine, CXCL10, clusterin, and total protein (15) were assessed on all collected urine samples. To this end, we developed a composite Q-Score ranging from 0 to 100, on all 6 biomarkers, on 111 samples in the training set using a Random Forest boot strap model. The utility of the individual biomarkers in discriminating stable (STA) and acute rejection (AR) outcomes was determined by multivariate nominal logistic regression (Table S1) to find an optimal threshold (≥32) that maximized sensitivity and specificity for detection of rejection in the urine sample paired with an allograft biopsy in the training set of 111 samples. This locked threshold of ≥ 32 was then applied to two independent validation sets to assess the clinical performance of the threshold to discriminate between rejection and no rejection. Model analysis of the urine score demonstrated that each of the biomarkers provided independent value towards the prediction of kidney transplant rejection status with varying significance (Table S2), highlighting that multiple urinary biomarkers had complementary value in discriminating acute renal transplant rejection compared to using a single marker (such as cfDNA) alone. The scores easily distinguished between STA (median 11.2, 95% CI 8.40 – 12.71) and AR patients (median 58.8, 95% CI 50.3 – 69.2) (P < 0.0001) in the training set (Fig. 2A) at a defined threshold of 32. The threshold was chosen because it provided the combination of sensitivity and specificity of 94.9% and 100%, and maximized specificity. A receiver operator characteristic curve of the urine score had an AUC of 0.99 (0.99 – 1.00, P < 0.0001) in the training set (Fig. 2B).
Fig. 2. The selected urinary biomarkers could segregate non-rejection patients from those with acute rejection.
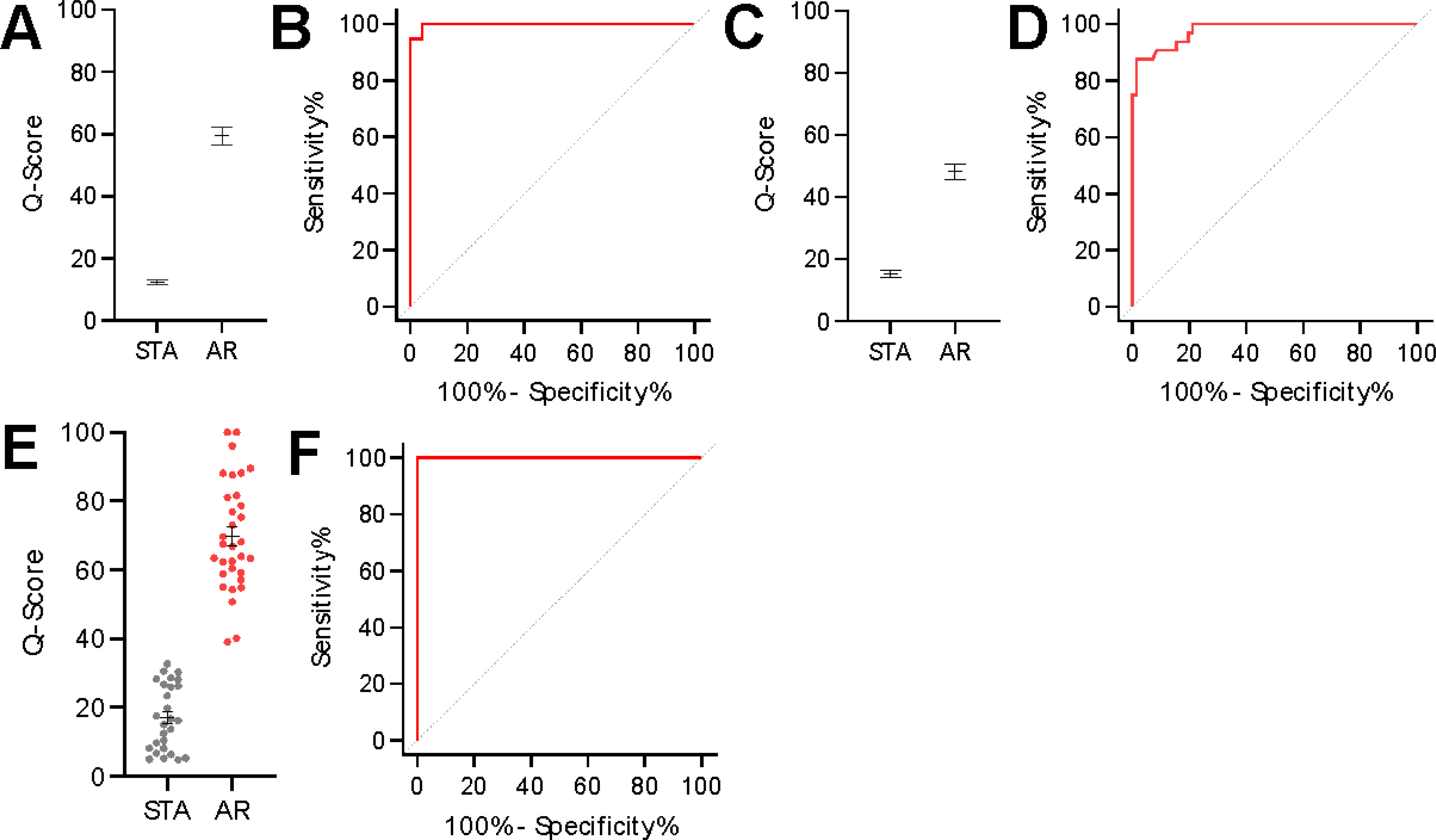
(A) A urine score model was trained on 111 samples consisting of 72 STA and 39 AR samples to generate a scaled Q-score ranging from 0 to 100. The distribution of the STA and AR groups is depicted in the figure. (B) The quiescence threshold was set at 32 with a corresponding sensitivity of 94.9% and specificity of 100%. The AUC of the ROC curve was 0.99 (P < 0.0001). (C) The urine score model was applied to a set of 103 independent samples consisting of 71 STA and 32 AR samples. The median and 95% CI for the STA and AR group were 13.14 (8.75 – 17.94) and 45.16 (40.77 – 57.87), respectively (P < 0.0001). (D) At the pre-determined quiescence threshold, the sensitivity was 90.6% and the specificity was 91.6%. The AUC of the ROC curve was 0.98 (P < 0.0001). (E) The fixed urine score model was applied to a set of 59 independent samples consisting of 27 STA and 32 AR. The median and 95% CI for the STA and AR groups were 16.21 (8.16 – 26.22) and 67.25 (60.46 – 78.73), respectively (P < 0.0001). (F) At the pre-determined quiescence threshold, the sensitivity was 100.0% and the specificity was 96.3%. The AUC of the ROC curve was 1.00 (P < 0.0001).
Applying the same, locked model to independent samples in the first validation set, the urine score distinguished between STA (median 13.1, 95% CI 8.8 – 17.9) and AR patients (median 45.2, 95% CI 40.8 – 57.9) (P < 0.0001) (Fig. 2C). A receiver-operator characteristic curve of the score in the first validation set found that the previously established threshold from the training set had a sensitivity of 90.6%, a specificity of 91.6%, and an AUC of 0.98 (0.96 – 1.00, P < 0.0001) (Fig. 2D). In validation set 2, we could assess the performance of the urine score to again distinguish AR from STA (Fig. 2E). Here, we observed that the scaled score resulted in 100% sensitivity and 96.3% specificity with an AUC of 1.00 (1.00 – 1.00, p<0.0001) (Fig. 2F). Aggregating all AR and STA samples together, the AUC was 0.99 (0.98 – 0.99, P < 0.0001), with a sensitivity and specificity of 95.2% and 95.9% at the established threshold.
We next evaluated the fixed urine score to evaluate other injuries, 50 bAR and 9 BKVN samples, and these samples showed median scores of 38.47 (95% CI 34.71 – 40.80), and 23.70 (95% CI 10.48 – 53.38), respectively (Fig. 3A). A ROC curve of AR vs no-rejection (NR; inclusive of STA, bAR, and BKVN) showed an AUC at 0.96 (95% CI 0.94 to 0.98) (P < 0.0001) (Fig. 3B).
Fig. 3. Application of the urine score to other types of injuries.
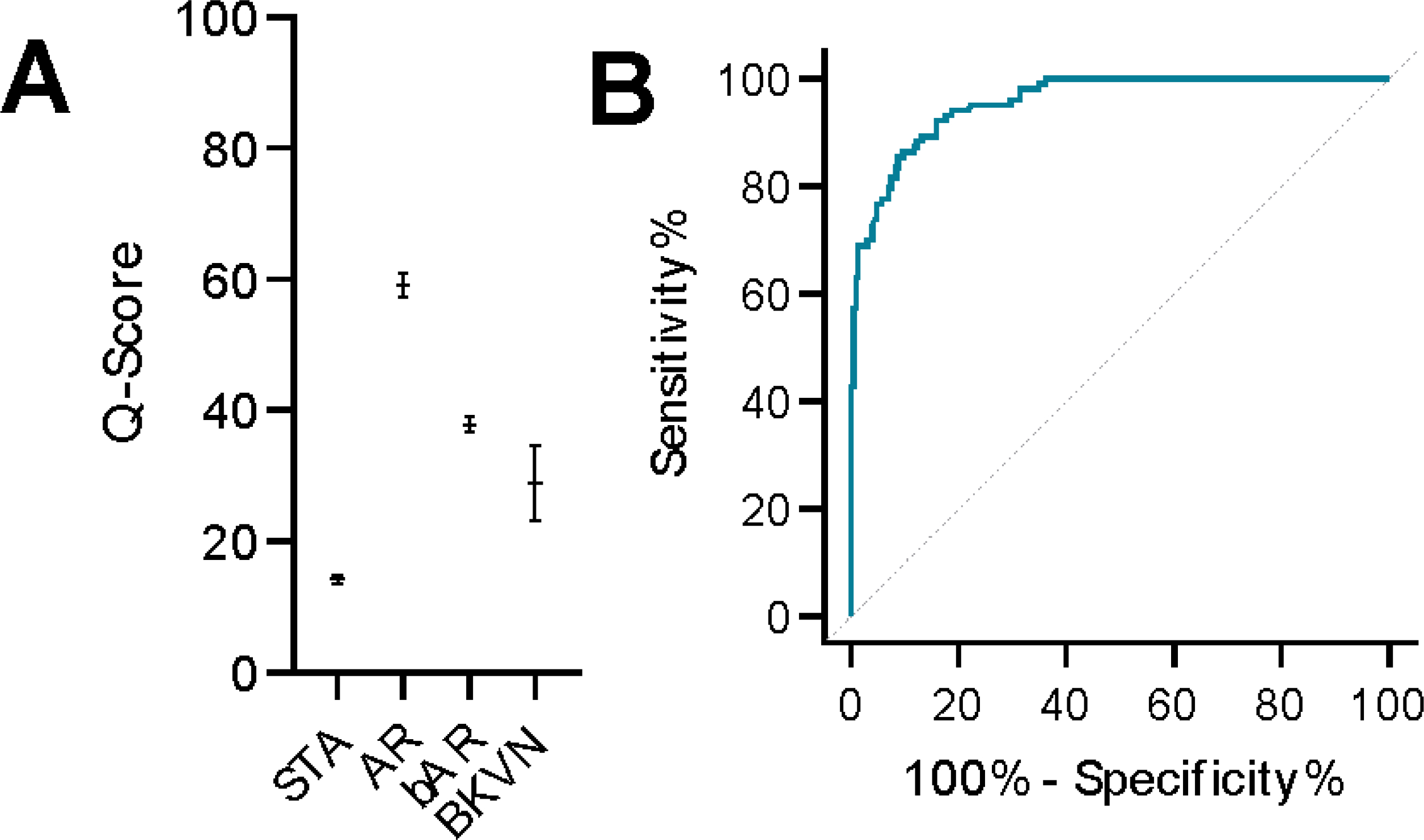
(A) The urine score model was applied to a set of samples consisting of all the previously identified STA and AR along with bAR and BKVN samples. The median and 95% CI for the bAR and BKVN group were 38.57 (34.71 – 40.80) and 23.70 (10.48 – 53.38), respectively (P < 0.0001 for STA vs. AR and STA vs. BKVN, P < 0.01 for AR vs. bAR and AR vs. BKVN). (B) The AUC of the ROC curve for AR vs all other outcomes was 0.96 (P < 0.0001).
We could observe gradations in the urine score across the transplant sample, lowest in STA, moderate in bAR, and highest in AR. The gradation of the scores was also seen when we profiled the same 6 markers in urine samples from 54 healthy individuals (median and 95% CI score 10.80 (7.90 – 12.20)) and in urine samples from 183 patients with stage 1 (median and 95% CI 24.27 (23.10 – 25.12) and stage 2 CKD (median and 95% CI 34.24 (32.17 – 37.12)), using the native kidney injury scores by Watson et al. (15). (Fig. 4).
Fig. 4. Patterns of the urine score in kidney injuries.
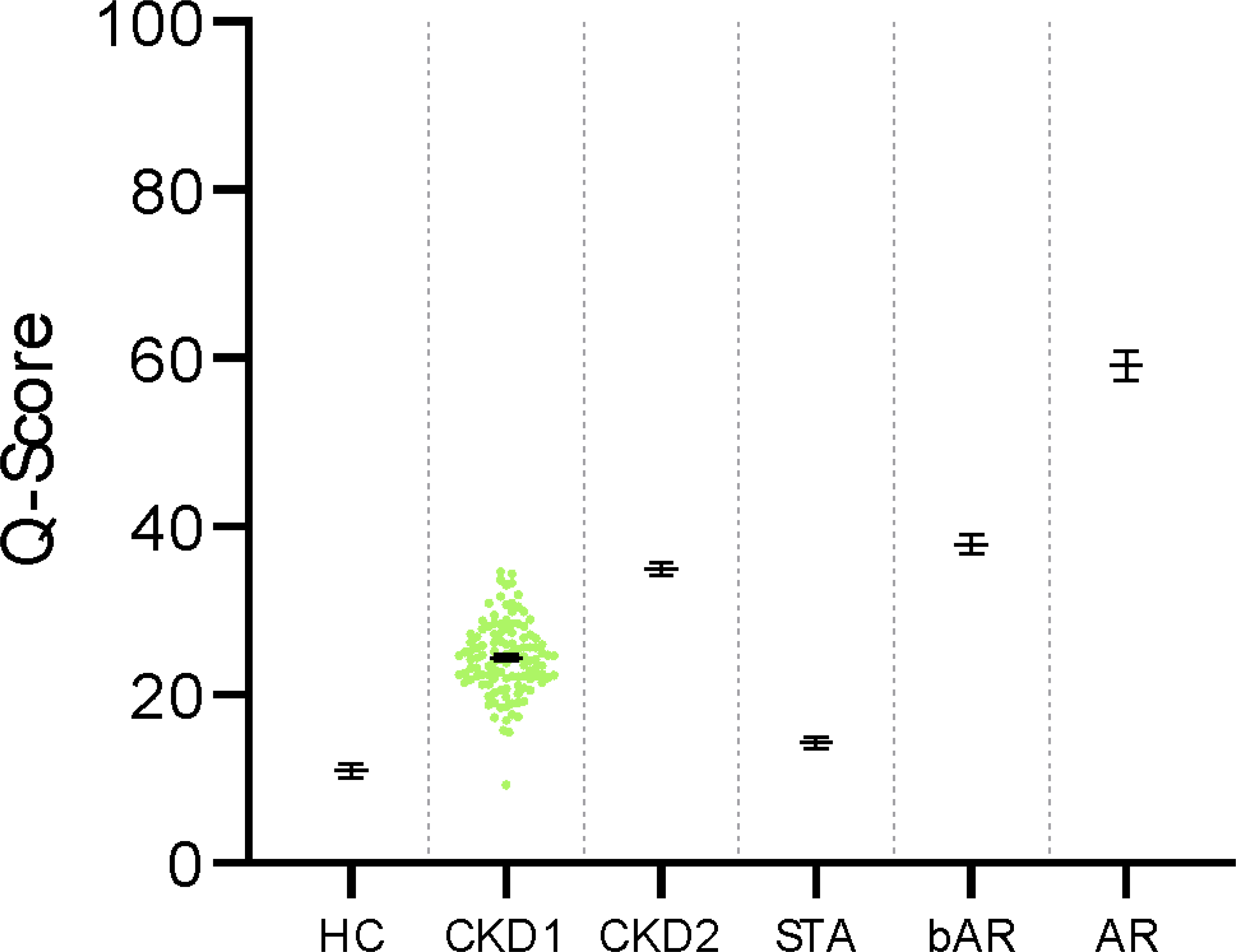
The urine scores were aggregated for different outcomes. HC, healthy control, 10.80 (7.90 – 12.20); CKD1, CKD Stage 1, 24.27 (23.10 – 25.12); CKD2, CKD Stage 2, 34.24 (32.17 – 37.12); STA, Stable, 12.19 (9.75 – 14.39); bAR, borderline AR, 38.47 (34.71 – 40.80); AR, Acute Rejection, 58.76 (54.95 – 63.40). Data are given as median and 95% CI.
Relationship Between the urine score and Clinical Parameters
Aggregating the training and validation sets together to assess the classification performance of AR and no-rejection clinical outcomes, the Q-Score (AUC of 0.99, 95% CI 0.98 – 0.99, P < 0.0001) performed better than proteinuria (Pr/Cr) (AUC of 0.76, 95% CI 0.69 – 0.82, P < 0.0001) and eGFR (AUC of 0.86, 95% CI 0.81 – 0.98, P < 0.0001) (Fig. 5A). Because 60% (62/103) of all AR had clinical graft dysfunction, we would expect the eGFR to perform moderately well, but it failed to pick up all cases of sub-clinical AR, where no decrease in eGFR was expected. At the quiescence threshold of 32, the sensitivity and specificity of the urine score were 95.2% and 95.9%, respectively. Based on a 25% prevalence of rejection in at-risk populations, the positive predictive value (PPV) was projected to be 86.9% and the negative predictive value (NPV) was projected to be 98.4%.
Fig. 5. Comparison of the urine score performance.
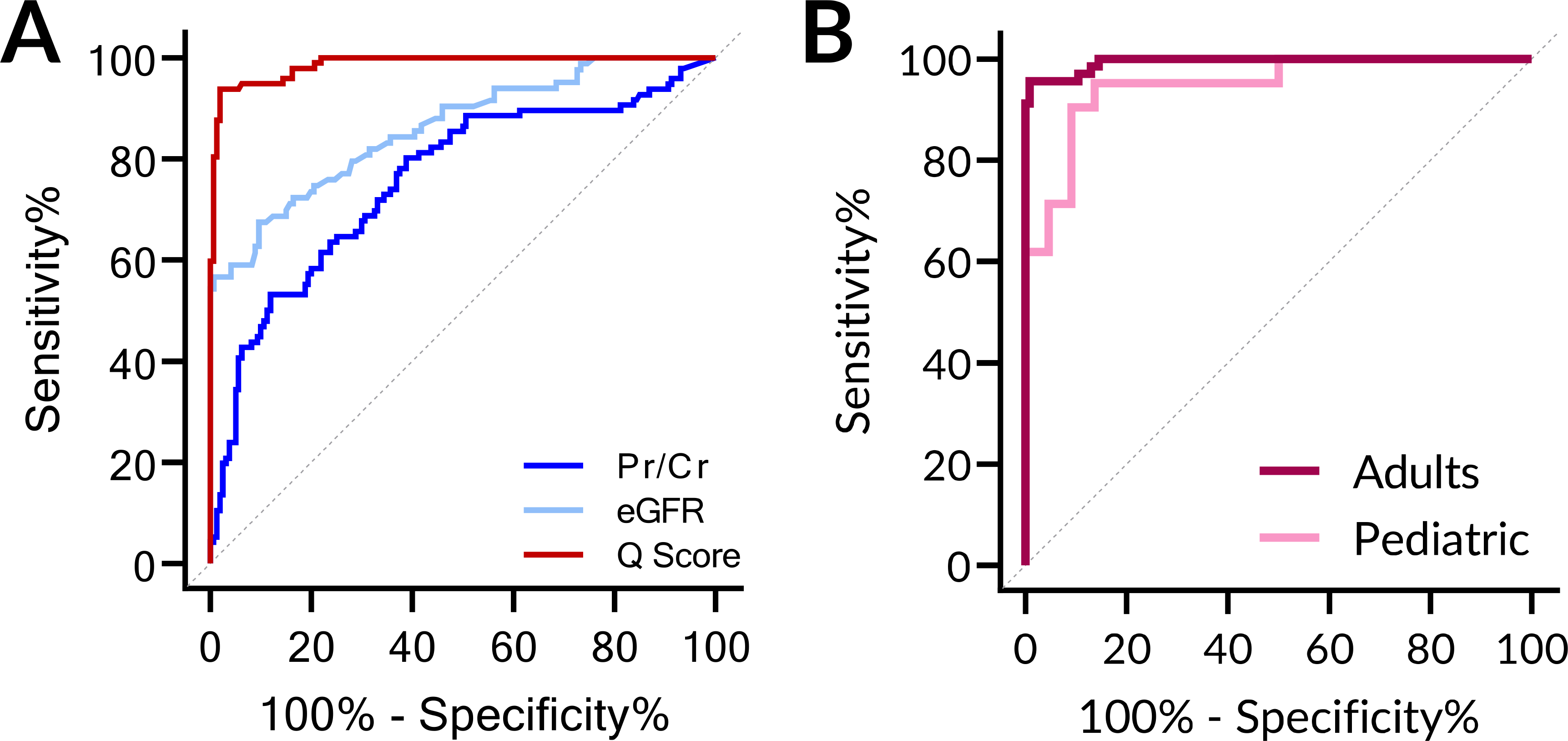
(A) The urine score performed better than eGFR or the urinary protein/creatinine ratio in discriminating AR vs STA. For the entire set of 170 STA and 103 AR, the Q-Score, eGFR, and protein/creatinine (Pr/Cr) ratio were plotted and the AUC calculated. The AUCs were 0.99, 0.86, and 0.76 respectively. (B) Similar performance of the urine score for non-invasive diagnosis of biopsy-confirmed pediatric and adult AR. The urine score performed well for diagnosis of AR in children ≤ 18 yrs. old as well as adult recipients >18 yrs. old: Pediatric AUC = 0.95 (No AR 30, AR 21); Adult AUC = 0.99 (No AR 138, AR 82).
As seen in Fig. 5B, the urine score was able to discern AR, irrespective of recipient age. Notably, the AUCs for the urine score in discriminating STA from AR in both pediatric and adult patients were similar, with AUCs of 0.95 and 0.99, respectively.
Multivariable regression analysis did not show any correlation of the score with donor source (living vs deceased), recipient or donor gender, age, sensitization status, repeat transplantation, or ethnicity (table S3). De novo donor specific antibody (DSA) was detected in 24 of the 38 patients diagnosed with ABMR.
Relationship Between the Q-Score and Acute Rejection Parameters
Based on the biopsy histology grading by Banff (17), we ascertained the correlation of the urine score with various rejection phenotypes in a total of 103 biopsy-confirmed rejections that met histological criteria (17) for diagnosis as either TCMR (n=65) or ABMR (n=38). There was no difference (p=0.776; Fig. 6A) in the scores between TCMR and ABMR. Among the samples with acute rejection, we observed that the scores were generally higher at higher biopsy rejection grades (test for linear trend with one-way ANOVA, p=0.0004, Fig. 6B). A sub-analysis of patients with confirmed AR found that there was no significant difference between the scores for those identified via for-cause biopsies as compared to protocol biopsies (Fig. 6C), highlighting that substantive tissue injury can be masked by normal eGFR.
Fig. 6. The urine score can measure and distinguish between clinically relevant transplant parameters.
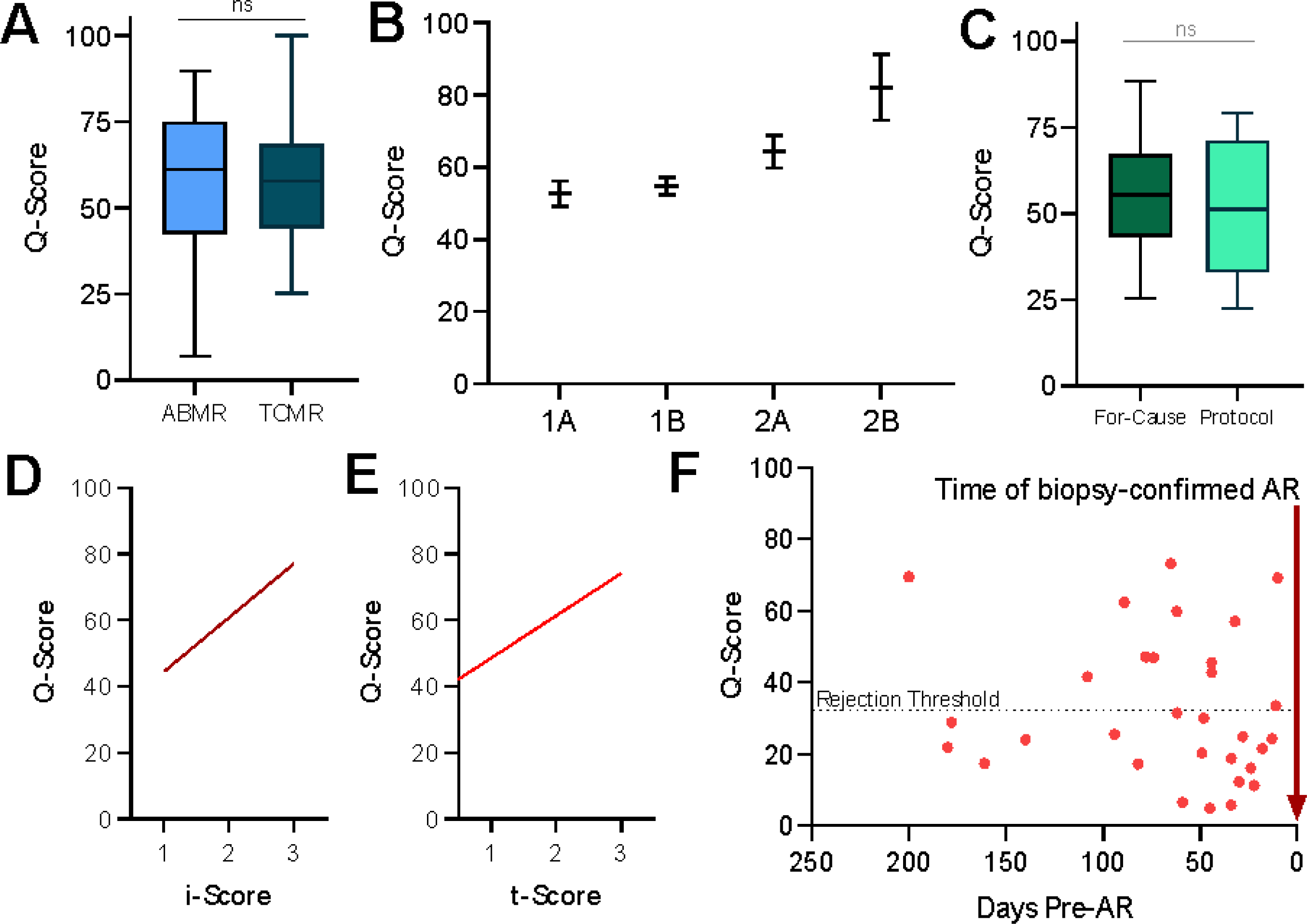
(A) Patients with AR were split into those with ABMR (n = 38) and TCMR (n = 65). There was no significant difference between these two groups (P = 0.776). (B) Patients with AR were split into increasing histology score as per the Banff histology classification. One-way ANOVA with test for linear trend indicated a significant difference (P = 0.0004). (C) The urine scores were no different in patients with biopsy-confirmed AR, whether the AR was diagnosed on a for-cause (n = 47) or protocol biopsy (n = 18) (p = 0.395) (D and E) The urine score correlates with the paired biopsy inflammation (i) score (D, p < 0.0001) and tubulitis (t) score (E, p < 0.0001). (F) When the Q-Score was applied to patients with samples collected before confirmed episodes of AR, 38% had scores above threshold for pre-AR at variable times up to 8 months before the rejection episode.
In addition, there was a correlation of the urine score with graft rejection injury as shown by the inflammation or Banff i-score (Fig. 6D) (R2=0.4271; p<0.0001) and the tubulitis or Banff t-score (Fig. 6E) (R2=0.236; p<0.0001) (17). The variations of the score in urine samples collected at the time of biopsy confirmed BKVN injury in the graft, with the urine score being higher in BKVN patients with higher Banff i-scores (Fig. 6D).
Relationship Between the urine score and its Clinical Utility for Prediction of Rejection Development
The analysis of the urine scores for the 32 urine samples collected before the detection of rejection by biopsy demonstrated that urine scores could be variably high at different times, weeks to months before an AR episode was detected by the current standard of care tests, serum creatinine and biopsy. Because all of these patients went on to develop biopsy-proven acute rejection, we suggest that prediction of rejection with the urine Q-Score may have been possible in 38% of patients in this sub-sampling (Fig. 6F).
Immune Quiescence can be Defined by the Q-Score Threshold
Most (94%) of the samples that had scores above the threshold for acute rejection (≥32; n = 159) had a clinical diagnosis of active acute rejection, early acute rejection as evidenced by borderline changes on the biopsy, or went on to develop biopsy-confirmed acute rejection between weeks to months from the time of sampling (Fig. 7A), and 94% of protocol biopsies with a score (≥32 had histological acute rejection. Conversely, if a patient was scheduled for a protocol biopsy and the urine score was below the threshold, the likelihood of the patient having a Banff-graded acute rejection was only 5%. Based on this observation, a score of less than 32 could provide clinically actionable value by suggesting that the allograft is likely to be in a state of immune quiescence.
Fig. 7. The urine score can measure and distinguish between clinically relevant transplant parameters.
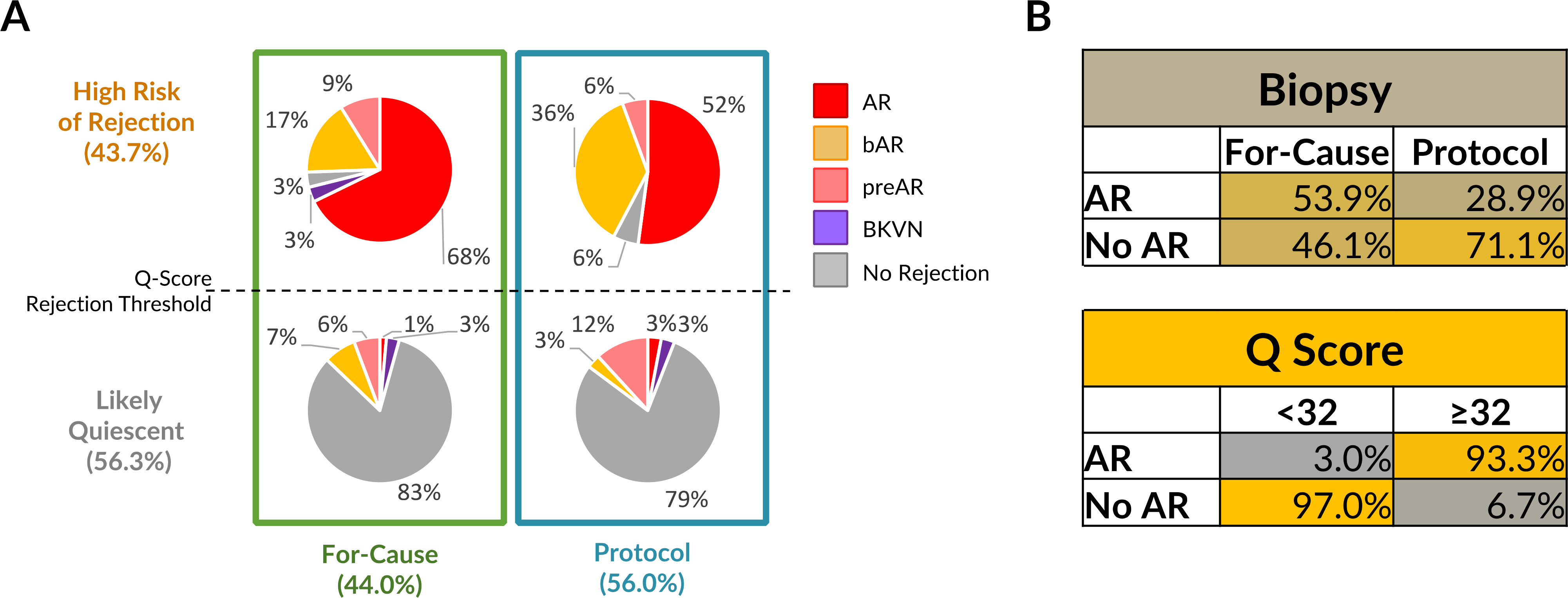
(A) In the total cohort of 364 transplant samples, there were 160 clinically-indicated biopsies, of which 90 had a urine score above the rejection threshold and 70 below the threshold. Out of the 204 protocol biopsies, 71 had a score above the rejection threshold and 133 below. The breakdown of different biopsy diagnoses paired with the sample are shown in the figure as percentages. (B) Accuracy tables for biopsy (for-cause versus protocol) and the Q-Score (score <32 versus ≥32) for AR and STA classification.
In the event of clinical graft dysfunction and a urine score above the rejection threshold, the likelihood that the patient had varying grades of rejection was 93%. In the event of clinical graft dysfunction and a score below the threshold, other causes of graft injury should be clinically explored because the likelihood of Banff-graded acute rejection was less than 1%. Notably, the detection of bAR was greater in the surveillance biopsy group as compared to the for-cause biopsy group. This is not surprising, as there is a lower likelihood of earlier stages of rejection causing clinical graft dysfunction. Further, in 50% (103/204) of the samples, where the graft function was excellent and the patient was clinically stable, a protocol biopsy would not have been necessary if the information from the urine score had been available.
The comparative performance of the two application methods of biopsy (cause versus protocol), and the quiescence versus the rejection threshold of the urine score for AR diagnosis are shown in Fig. 7B, highlighting that the most common indication for performing a renal allograft biopsy at all transplant programs, a drift in the serum creatinine, only results in an accurate diagnosis of rejection ~50% of the time.
Discussion
Renal injury is a spectrum of disease that progresses after the initiating insult, the triggers being varied, including genetic and structural problems (encountered mostly in children), renal inflammation, and more indolent causes of injury due to altered renal hemodynamics. Our earlier study (15) demonstrated that the selected 6 urine markers could detect different stages of renal injury and were more sensitive for the detection of early renal injury, over paired values of eGFR, serum creatinine, or new-onset proteinuria. In this study, we applied the urinary score for analysis of stable, injured, and rejecting renal allografts, and evaluated the performance of the score to detect biopsy-confirmed AR. We also compared these results with non-transplant subjects to compare injury in AR with CKD injury in the native kidney and to compare kidney injury in stable allografts with normal healthy controls. The Q-Score includes six DNA, protein, and metabolite markers, and unlike a simple threshold detection of 1% for AR, as used for dd-cfDNA measurements in blood (6, 8), the Q-Score is quantitative, scaled from 0–100, allowing for sensitive tracking of AR progression and recovery.
AR with graft dysfunction, or clinical rejection (cAR), is the group of rejections that transplant centers report as their annual rejection incidence. In transplant centers where protocol biopsies are not performed, the statistic of sub-clinical AR (scAR) is not available for assessment. Although short-term adverse allograft outcomes including clinical acute rejection episodes have declined over past decades to ~10%, proportionate improvement in long-term allograft survival remains unrealized because much of rejection injury goes unrecognized as it is sub-clinical (18–20) and thus there is lack of effective treatment of rejection and inflammation (21)(22–26)(27). The current standard of care in transplant centers world-wide requires an invasive biopsy for AR diagnosis, a procedure burdened by clinical risks and costs. To obviate these issues, prior studies have tested noninvasive profiling of urinary RNA (12, 13) and blood transcriptomic signatures (28–30), but these assays have not been integrated into clinical care due to issues that relate to sample handling, processing, assay complexity, and/or robustness. We propose that using urine supernatant alone, replacing the need for phlebotomy or extensive urine sample prep and handling (as needed for urine mRNA isolation), and avoiding the need for amplification and sequencing (as used for plasma cfDNA assays), provides the patient and clinician the option of monitoring the allograft with high accuracy and simplicity. Because the assay requires as little as 5 mL of urine, it can also be performed in cases of limited urine volumes, such as graft oliguria or an infant recipient.
Unlike other published studies, where the eGFR has been shown to be a poor measure for detection of rejection (8), in this study we observed somewhat better performance of eGFR as an independent measure to detect AR, as 64/103 rejections in this study had clinical graft dysfunction. Nevertheless, eGFR measurement cannot help in the assessment of sub-clinical rejections, which begs the development of more sensitive AR-specific assays. In this study, we assessed the performance of the urine score for diagnosis of both cAR and scAR and observed similar increases in the urine score during both AR types. An analysis of all the biopsy data highlights that the current use of the serum creatinine drift to guide use of a diagnostic biopsy, or the fixed time point sampling of the allograft by a protocol biopsy, is a weak diagnostic measure for AR. The diagnostic accuracy for the invasive biopsy, combining all clinically indicated and protocol biopsies, for diagnosis of AR in our study is low, at 28.5%. Even for clinically indicated biopsies, the accuracy remains low at 54%.
Interestingly, although the stable samples all clustered below the detection threshold for rejection, we observed that in comparison with the scores in CKD patients, the stable renal transplant may already have some baseline renal injury. Thus it appears that the stable allograft, though without rejection, may carry some baseline burden of renal impairment, possibly due to the impact of early ischemia reperfusion injury, exposure to nephrotoxic drugs such as calcineurin inhibitors (31), or subclinical alloimmune injury (32).
We confirmed that urine samples from biopsy-confirmed AR patients detected both TCMR and ABMR, highlighting the intrinsic overlapping molecular axes in both types of rejection, particularly the presence of monocyte activation and the coexistence of endothelial injury with cellular inflammation in both TCMR and ABMR. The Q-score performed with a high degree of accuracy in the independent validation cohorts with high NPVs and PPVs, considerably better than blood-based assays for rejection diagnosis (6, 8, 18, 29, 33).
The histology-derived Banff i- and t-scores are currently used in the clinical management of patients as a reflection of inflammation and tubular injury in the transplant. The correlations of the scaled urine score with these biopsy histology grades suggest that the quantitative nature of the score could enable noninvasive assessment of rejection severity and immunosuppression therapy choices. Thus, the actual value of the urine score may be able to drive decisions of immunosuppression intensification for rejection, and close monitoring of treatment efficacy and rejection response and resolution. The clinically relevant quantitative nature of the score provides granularity for serial monitoring that may not be possible in blood cfDNA assays that only provide a single threshold for injury (6, 8).
In addition, we observed that if the urine score was below the quiescence threshold (< 32; when a diagnosis of histological STA is highly likely), performance of an invasive biopsy for “rule-out” rejection was not necessary in 55% of patients. Conversely, the high sensitivity of the assay showed that we rarely missed patients who were having active rejection as confirmed by biopsy. In our dataset, 43% or 69/130 clinically indicated biopsies were quiescent, and 65% or 130/201 protocol biopsies were unnecessary invasive procedures. Thus, serial performance of the score in urine samples may be able to reduce patient morbidity from unnecessary transplant biopsies, clinical visits, and related health care costs.
The main focus of this paper was on biopsy-matched cross-sectional samples because the objective was to develop and validate a non-invasive urine score for graft rejection. Besides limitations in overall representation, the clinical utility of graft biopsy is limited by the subjectivity of biopsy reporting, which also emphasizes the potential clinical utility of the urine score even as an adjunct to the biopsy, to avoid pitfalls in the variances in reporting of renal biopsy specimens. This may be especially true in the conduct of multicenter clinical trials, where multiple local pathologists may be involved, and the urine score can provide an unbiased, quantitative readout of graft rejection.
Because the urine sampling of patients in our study was by protocol timed collections, and rejection timing is unexpected, serial urine sampling post-transplant in our center protocols allowed for a review of the trajectory of AR in the patients with paired urine samples before biopsy-confirmed rejection, anywhere from 1 week to 8 months before confirmation of rejection by biopsy. We observed earlier detection of rejection by the urine score on 35% of preAR samples, when biopsy histology was still negative for histological rejection. This may be explained by the fact that renal biopsy specimens represent small 2–3-mm sections of the total core and may not capture a focal phenomenon like allograft rejection. The ability to observe an allograft rejection risk profile over time, such as that made available through urine testing, highlights that natural variances in the tempo and cause of rejection in different individuals may have different causes. As we only have a single urine collection prior to the later biopsy confirmed AR episode in each of the 32 patients, it is possible that repeated urine sampling in preAR patients with initial low urine scores in this study, may have shown an increase in the urine score with increased proximity to the time of biopsy AR diagnosis. The necessity to do serial and repeated monitoring with a non-invasive, rapid turn-around, sensitive and specific assay may be further underscored by obtaining these additional data, and may also provide the background for future studies testing the hypothesis that serial measurements of this urine assay may guide maintenance immunosuppression management.
Other limitations of the current study design include a limited assessment of longitudinal samples, especially those from stable patients, in whom the natural variation of the urine score could be assessed over time and within the same patient. It will be necessary to collect monthly samples from patients from on-going clinical studies to assess this question over time. In addition, this study did not provide an assessment of the changes in the urine score over time after treatment of rejection. Though the assay in this study had been performed at an academic center lab, the assay is being transitioned to run as an analytically validated assay in a College of American Pathologists-accredited, Clinical Laboratories Improvements Act (CLIA)-certified reference laboratory, where urine samples from prospective clinical trials will be able to address the clinical utility of this assay for generalized immune risk serial screening after transplantation.
In conclusion, the Q-Score offers the potential to be used as an immune monitoring tool to guide the use of immunosuppression, with the ultimate goal of controlling subclinical intragraft inflammation and thus prolonging graft survival. Further assessment of the clinical utility of this assay in prospective longitudinal renal transplant studies in both sensitized and unsensitized renal allograft recipients, will help evaluate the potential of the assay to substitute for protocol renal transplant biopsies for the determination of clinical and subclinical rejection. Such studies would allow for early detection and proactive management of graft rejection for the improvement of both patient morbidity and graft survival.
Materials and Methods
Study Design
This was a batched analysis of urine samples prospectively collected between 2010 and 2018 from adult (18–65) and pediatric (1–18 years of age) kidney transplant recipients who had transplant surgeries at Stanford University Medical Center, USA, the University of California at San Francisco (UCSF), USA, or the Instituto Nacional de Ciencias Medicas y Nutricion, Mexico. All urine samples that were biopsy-matched were subsequently selected for this study. Power calculation analyses determined that a sample of 8 from the positive group and 15 from the negative group would have 90% power to detect a difference of 0.28 between the area under the ROC curve (AUC) under the null hypothesis of 0.7 (using eGFR as the comparator in this dataset) and an AUC under the alternative hypothesis of 0.98 using a two-sided z-test at a significance level of 0.05.
The studies were approved by the institutional review boards of all 3 institutions. All patients or their legal guardians provided written informed consent to participate in the research, in full adherence to the Declaration of Helsinki. The clinical and research activities being reported are consistent with the Principles of the Declaration of Istanbul as outline in the Declaration of Istanbul on Organ Trafficking and Transplant Tourism.
Study Populations and Samples
Male and female, adult and pediatric, patients receiving a kidney from related, unrelated living donors, or unrelated deceased donors were included. All patients were on an immunosuppressive regimen consisting of calcineurin inhibitors, mycophenolate mofetil, and steroids. Urine samples were collected before the performance of an indicated or protocol biopsy (at various pre-specified time intervals for each study protocol) and were processed and stored in an existing biorepository, before batch analysis. The selection of study samples was based on adequate urine volume being available and association of the sample with a paired biopsy with sufficient biopsy material for histological information. Healthy control urine samples were collected from volunteers who had good health, normal SCr, no proteinuria, and no identifiable CKD risk factors. For details of patient demographics, see Table 1. The study was divided into different cohorts (Fig. 1). In the training set, the original 6 biomarkers from the study by Watson et al. (15), which developed a Kidney Injury Test score to non-invasively assess patients with chronic kidney disease (CKD), were assessed in biopsy-paired urine samples obtained from kidney transplant recipients, with and without kidney transplant rejection. The score was applied to this dataset to assess renal transplant quiescence vs rejection and called the QiSant score, or Q-Score. The optimal threshold for the Q-Score was determined in the training set to evaluate the score’s performance for transplant rejection diagnosis. The locked Q-Score was next applied to an independent validation set of samples with stable or AR phenotypes, and subsequently to independent samples from a prediction set.
Urine Sample Collection and Processing
Urine samples were collected midstream in sterile containers and centrifuged at 2000×g at 4 °C for 30 minutes within 1 hour of collection. The supernatant was aliquoted and pH adjusted to 7.0 by adding 1 part 1 M Tris-HCl to 10 parts urine supernatant. The urine was stored at −80°C in the Sarwal Lab Biorepository until further use. Samples were processed for the QiSant Assay biomarkers as described below. There were 364 unique urine samples from renal allograft recipients, with 332 cross-sectional samples, each of which was paired with a closed 18G needle allograft biopsy. Thirty-two urine samples collected at protocol time points with paired STA biopsies were from patients who had stable graft function, and were collected proximal to a biopsy-confirmed rejection episode.
Biopsy Samples
All kidney biopsies were analyzed in a blinded manner by the local study pathologist at each site, and all biopsies were also re-evaluated by a central UCSF pathologist (C.C.) and were graded by the Banff classification for acute rejection (AR) (17); intragraft C4d stains were performed to assess for ABMR. AR was defined, at minimum, by the following criteria: (1) TCMR consisting of either a tubulitis (t) score > 2 accompanied by an interstitial inflammation (ii) score > 2 or vascular changes (v) score > 0; (2) C4d positive ABMR consisting of positive donor-specific antibodies (DSA) with a glomerulitis (g) score >0 or peritubular capillaritis score (ptc) > 0 or v > 0 with unexplained acute tubular necrosis / thrombotic micro angiopathy (ATN/TMA) with C4d = 2; or (3) C4d negative ABMR consisting of positive DSA with unexplained ATN/TMA with g + ptc ≥ 2 and C4d = 0 or 1. Normal or stable (STA) allografts were defined by an absence of substantial injury on the matched biopsy pathology and definitions of the inflammation or i-Score and the tubulitis or t-Score. Both chronic allograft injury with borderline acute rejection (bAR) and BK virus nephropathy (BKVN) used standard pathology definitions as described by the Banff Schema (17), on the paired biopsies from each individual urine sample.
Biomarker Selection and Measurement in Urine Samples
Based on legacy urine genomic, metabolomic, and proteomic studies conducted by our group (34–37), we selected biomarkers in urine that reflected kidney injury from different renal sub-compartments as previously described (15). Briefly, cfDNA was selected because it reflects the total apoptotic burden of kidney injury (38, 39), and the methylated fraction was selected to further augment detection of renal parenchymal injury (40). Total protein was selected as a late marker of glomerular injury (41, 42). CXCL10 was selected as an established marker of both renal and transplant inflammation (14, 43, 44). Clusterin was selected as a marker of tubular injury (45, 46). Urine creatinine was used for data normalization and control for hydration status and diurnal variation (47, 48).
For microwell-based measurement of cfDNA, we developed a 5’ biotinylated oligonucleotide complementary chemiluminescent immunoprobe for the measurement of specific target cfDNA fragments, as previously described (15). Streptavidin-HRP (R&D Systems) and SuperSignal ELISA Femto Substrate (Thermo Fisher Scientific) were used for luminescent detection and quantitation. CXCL10, clusterin, and methylated cfDNA (m-cfDNA) were measured using custom-developed assays (NephroSant, Inc.), the reagents for which are available for use by the scientific community for non-commercial purposes, under a material transfer agreement with the Regents, University of California. Total protein was measured using the Pierce Coomassie Plus (Bradford) Assay Kit (Thermo Fisher Scientific). Urinary creatinine was used to normalize the five other biomarkers for urinary concentration and hydration status and was measured using the Creatinine Assay Kit (BioAssay Systems). Microwell plate readings were measured using an iD3 Multi-Mode Microplate Reader (Molecular Devices). All assays were run in duplicates.
Statistical Analysis
Comparisons involving two groups were performed using the nonparametric Mann-Whitney U test. Comparisons involving three groups were performed using the nonparametric Kruskal-Wallis test with Dunn’s post-hoc multiple comparisons correction. Correlations were determined using the Pearson correlation coefficient. To further refine the score for detection of biopsy-confirmed acute rejection, we optimized the assay and the Q-Score for rejection diagnosis. For this purpose, we used 364 biopsy-matched kidney transplant samples from 3 transplant centers, with pediatric and adult allograft recipients, and performed random sampling to split these cohorts with clear outcomes of no rejection or acute rejection into a training set and two independent validation sets (Table 1). The Q-Score was determined using a Bootstrap Random Forest ensemble model. All analyses were performed using either GraphPad Prism 8.3.0 (GraphPad) or JMP 14.3 (SAS Institute).
Supplementary Material
Fig. S1. Correlation Matrix of Biomarkers
Table S1. Multivariate Logistic Regression of Acute Rejection Status as Assessed by Individual Biomarkers
Table S2. Biomarker Log-Likelihood Ratios for the urine score
Table S3. Multivariate Regression of Clinical and Demographic Variables with the urine score
Data File S1. Urine score data
Acknowledgments:
We are grateful for the help from physicians, clinical coordinators, research personnel, patients, and patient families. We thank Evangelos Evangelou, Rekha Mani, Todd Whitson, Lucy Lu, Tyler Schmeckpeper, and Michael Nasr for their contributions to this paper.
Funding:
This work was supported by startup funds for the Sarwal Lab from the Department of Surgery, UCSF to M.M.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests: The IP for this assay has been filed under WO Patent WO 2018/035340 A1 title “A novel immunoprobe-based method to assess organ injury status through a biofluid-based cell-free DNA (cfDNA) assay” and is owned by the Regents, the University of California. M.M.S., J.Y.C.Y., and T.K.S. are founders of NephroSant, Inc. (San Francisco, CA) which has licensed the IP for this assay from the Regents, the University of California. M.M.S. is on the FDA Science Board and consults or has recently consulted or received sponsored research funds from Bristol-Myers Squibb, Natera, Astellas Pharma, Genentech, and Jazz Pharmaceuticals. F.V. consults for eGenesis Bio, Natera, and Sanofi. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Collaborative studies on new patient samples and cohorts with the scientific community for non-commercial purposes are encouraged and facilitated by individual material transfer agreements with KIT Bio, doing business as NephroSant, Inc.
References and Notes:
- 1.Abouchacra S, Chaaban A, Hakim R, Gebran N, El-Jack H, Rashid F, Boobes Y, Muhairi A, Hussain Q, Khan I, Chedid F, Negelkerke N, Renal biomarkers for assessment of kidney function in renal transplant recipients: How do they compare?, Int. Urol. Nephrol. 44, 1871–1876 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Verissimo Veronese F, Cerratti Manfro R, Roman FR, Edelweiss MI, Rush DN, Dancea S, Goldberg J, Goncalves LF, Reproducibility of the Banff classification in subclinical kidney transplant rejection, Clin. Transplant. 19, 518–521 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Furness PN, Taub N, International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP project, Kidney Int. 60, 1998–2012 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Roedder S, Sigdel T, Salomonis N, Hsieh S, Dai H, Bestard O, Metes D, Zeevi A, Gritsch A, Cheeseman J, Macedo C, Peddy R, Medeiros M, Vincenti F, Asher N, Salvatierra O, Shapiro R, Kirk A, Reed E, Sarwal MM, The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study, PLoS Med 11, e1001759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy First M, Pierry D, McNulty M, Kurian SM, Rose S, Whisenant T, Venzon A, Bayat N, Lewis P, Friedewald JJ, Abecassis MM, Lee D, Analytical and Clinical Validation of a Molecular Diagnostic Signature in Kidney Transplant Recipients, J. Transplant. Technol. Res. 07 (2017), doi:10.4172/2161–0991.1000176. [Google Scholar]
- 6.Sigdel T, Archila F, Constantin T, Prins S, Liberto J, Damm I, Towfighi P, Navarro S, Kirkizlar E, Demko Z, Ryan A, Sigurjonsson S, Sarwal R, Hseish S-C, Chan-On C, Zimmermann B, Billings P, Moshkevich S, Sarwal M, Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR, J. Clin. Med. 8, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vlaminck I, a Valantine H, Snyder TM, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Bernstein D, Weisshaar D, Quake SR, Khush KK, Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection., Sci. Transl. Med. 6, 241ra77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, Matas AJ, Mehta S, Mannon RB, Sharfuddin A, Fischbach B, Narayanan M, Jordan SC, Cohen D, Weir MR, Hiller D, Prasad P, Woodward RN, Grskovic M, Sninsky JJ, Yee JP, Brennan DC, Cell-Free DNA and Active Rejection in Kidney Allografts, J. Am. Soc. Nephrol. 28, 2221–2232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Nicolls MR, Cornfield D, Weill D, Valantine H, Khush KK, Quake SR, Noninvasive monitoring of infection and rejection after lung transplantation, Proc. Natl. Acad. Sci. 112, 13336–13341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigdel TK, Vitalone MJ, Tran TQ, Dai H, Hsieh S-C, Salvatierra O, Sarwal MM, A rapid noninvasive assay for the detection of renal transplant injury., Transplantation 96, 97–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, De Vlaminck I, Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma, Sci. Rep. (2016), doi:10.1101/035741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suthanthiran M, Schwartz JE, Ding R, Abecassis M, Dadhania D, Samstein B, Knechtle SJ, Friedewald J, Becker YT, Sharma VK, Williams NM, Chang CS, Hoang C, Muthukumar T, August P, Keslar KS, Fairchild RL, Hricik DE, Heeger PS, Han L, Liu J, Riggs M, Ikle DN, Bridges ND, Shaked A, Clinical Trials in Organ Transplantation 04 (CTOT-04) Study Investigators, Urinary-Cell mRNA Profile and Acute Cellular Rejection in Kidney Allografts, N. Engl. J. Med. 369, 20–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigdel TK, Yang JYC, Bestard O, Schroeder A, Hsieh S-C, Liberto JM, Damm I, Geraedts ACM, Sarwal MM, A urinary Common Rejection Module (uCRM) score for non-invasive kidney transplant monitoring, PLoS One 7, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang JYC, Verleden SE, Zarinsefat A, Vanaudenaerde BM, Vos R, Verleden GM, Sarwal RD, Sigdel TK, Liberto JM, Damm I, Watson D, Sarwal MM, Cell-Free DNA and CXCL10 Derived from Bronchoalveolar Lavage Predict Lung Transplant Survival, J. Clin. Med. 8, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson D, Yang JYC, Sarwal RD, Sigdel TK, Liberto JM, Damm I, Louie V, Sigdel S, Livingstone D, Soh K, Chakraborty A, Liang M, Lin P-C, Sarwal MM, A Novel Multi-Biomarker Assay for Non-Invasive Detection and Quantitative Monitoring of Kidney Injury, J. Clin. Med. 8, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JYC, Sarwal RD, Fervenza FC, Sarwal MM, Lafayette RA, Noninvasive Urinary Monitoring of Progression in IgA Nephropathy, Int. J. Mol 20, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roufosse C, Simmonds N, Clahsen-Van Groningen M, Haas M, Henriksen KJ, Horsfield C, Loupy A, Mengel M, Perkowska-Ptasińska A, Rabant M, Racusen LC, Solez K, Becker JU, A 2018 Reference Guide to the Banff Classification of Renal Allograft Pathology, Transplantation 102, 1795–1814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedewald JJ, Kurian SM, Heilman RL, Whisenant TC, Poggio ED, Marsh C, Baliga P, Odim J, Brown MM, Ikle DN, Armstrong BD, charette jane I., Brietigam SS, Sustento-Reodica N, Zhao L, Kandpal M, Salomon DR, Abecassis MM, Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant, Am. J. Transplant. 19, 98–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parikh C, Ellison DH, Sam R, Leehey DJ, Hariharan S, Stablein D, Improved graft survival after renal transplantation in the United States, 1988 to 1996 [2] (multiple letters) N. Engl. J. Med. 342, 1837–1838 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Hart A, Smith JM, Skeans MA, Gustafson SK, Wilk AR, Robinson A, Wainright JL, Haynes CR, Snyder JJ, Kasiske BL, Israni AK, OPTN/SRTR 2016 Annual Data Report: Kidney HHS Public Access, Am J Transpl. 18, 18–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pirsch JD, Ploeg RJ, Gange S, D’Alessandro AM, Knechtle SJ, Sollinger HW, Kalayoglu M, Belzer FO, Determinants of graft survival after renal transplantation, Transplantation 61, 1581–1586 (1996). [DOI] [PubMed] [Google Scholar]
- 22.El Ters M, Grande JP, Keddis MT, Rodrigo E, Chopra B, Dean PG, Stegall MD, Cosio FG, Kidney allograft survival after acute rejection, the value of follow-up biopsies, Am. J. Transplant. 13, 2334–2341 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Szederkényi E, Iványi B, Morvay Z, Szenohradszki P, Borda B, Marofka F, Kemény É, Lázár G, Treatment of subclinical injuries detected by protocol biopsy improves the long-term kidney allograft function: A single center prospective randomized clinical trial, Transplant. Proc. 43, 1239–1243 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Nankivell BJ, Borrows RJ, Fung CLS, O’Connell PJ, Allen RDM, Chapman JR, Natural history, risk factors, and impact of subclinical rejection in kidney transplantation., Transplantation 78, 242–249 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Shishido S, Asanuma H, Nakai H, Mori Y, Satoh H, Kamimaki I, Hataya H, Ikeda M, Honda M, Hasegawa A, The impact of repeated subclinical acute rejection on the progression of chronic allograft nephropathy, J. Am. Soc. Nephrol. 14, 1046–1052 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Moreso F, Carrera M, Goma M, Hueso M, Sellares J, Martorell J, Grinyó JM, Serón D, Early subclinical rejection as a risk factor for late chronic humoral rejection, Transplantation 93, 41–46 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Mehta R, Bhusal S, Randhawa P, Sood P, Cherukuri A, Wu C, Puttarajappa C, Hoffman W, Shah N, Mangiola M, Zeevi A, Tevar AD, Hariharan S, Short-term adverse effects of early subclinical allograft inflammation in kidney transplant recipients with a rapid steroid withdrawal protocol, Am. J. Transplant. 18, 1710–1717 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Yang JYC, Sarwal MM, Transplant genetics and genomics, Nat. Rev. Genet. 18, 309–326 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Roedder S, Sigdel T, Salomonis N, Hsieh S, Dai H, Bestard O, Metes D, Zeevi A, Gritsch A, Cheeseman J, Macedo C, Peddy R, Medeiros M, Vincenti F, Asher N, Salvatierra O, Shapiro R, Kirk A, Reed E, Sarwal MM, The kSORT Assay to Detect Renal Transplant Patients at High Risk for Acute Rejection: Results of the Multicenter AART Study, PLoS Med. 11 (2014), doi:10.1371/journal.pmed.1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedewald JJ, Kurian SM, Heilman RL, Whisenant TC, Poggio ED, Marsh C, Baliga P, Odim J, Brown MM, Ikle DN, Armstrong BD, charette jane I., Brietigam SS, Sustento-Reodica N, Zhao L, Kandpal M, Salomon DR, Abecassis MM, Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant, Am. J. Transplant. (2018), doi:10.1111/ajt.15011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naesens M, Sarwal MM, Monitoring calcineurin inhibitor therapy: Localizing the moving target Transplantation 89, 1308–1309 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Naesens M, Khatri P, Li L, Sigdel TK, Vitalone MJ, Chen R, Butte AJ, Salvatierra O, Sarwal MM, Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes., Kidney Int. 80, 1364–76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Yi Z, Keung KL, Shang H, Wei C, Cravedi P, Sun Z, Xi C, Woytovich C, Farouk S, Huang W, Banu K, Gallon L, Magee CN, Najafian N, Samaniego M, Djamali A, Alexander SI, Rosales IA, Smith RN, Xiang J, Lerut E, Kuypers D, Naesens M, O’Connell PJ, Colvin R, Menon MC, Murphy B, A Peripheral Blood Gene Expression Signature to Diagnose Subclinical Acute Rejection, J. Am. Soc. Nephrol. 30, 1481–1494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigdel TK, Gao Y, He J, Wang A, Nicora CD, Fillmore TL, Shi T, Webb-Robertson B-J, Smith RD, Qian W-J, Salvatierra O, Camp DG, Sarwal MM, Mining the human urine proteome for monitoring renal transplant injury, Kidney Int. 89, 1244–1252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JYC, Sigdel TK, Sarwal MM, Self-antigens and rejection: a proteomic analysis, Curr. Opin. Organ Transplant. 21, 362–367 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Yang JYC, Sarwal R, Ky K, Dong V, Stoller M, Sarwal M, Chi T, Non-Radiologic Assessment of Kidney Stones by KIT, a Spot Urine Assay, Br. J. Urol. Int. (2019). [DOI] [PubMed] [Google Scholar]
- 37.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O Jr., Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling, N Engl J Med 349, 125–138 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Sigdel TK, Vitalone MJ, Tran TQ, Dai H, Hsieh S-C, Salvatierra O, Sarwal MM, A rapid noninvasive assay for the detection of renal transplant injury., Transplantation 96, 97–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang P, Lo YMD, The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics, Trends Genet. 32, 360–371 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Shiels PG, McGuinness D, Eriksson M, Kooman JP, Stenvinkel P, The role of epigenetics in renal ageing, Nat. Rev. Nephrol. 13, 471–482 (2017). [DOI] [PubMed] [Google Scholar]
- 41.D’Amico G, Bazzi C, Pathophysiology of proteinuria, Kidney Int. 63, 809–825 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Iseki K, Ikemiya Y, Iseki C, Takishita S, Proteinuria and the risk of developing end-stage renal disease, Kidney Int. 63, 1468–1474 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Schaub S, Nickerson P, Rush D, Mayr M, Hess C, Golian M, Stefura W, Hayglass K, Urinary CXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis, Am J Transpl. 9, 1347–1353 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Rabant M, Amrouche L, Lebreton X, Aulagnon F, Benon A, Sauvaget V, Bonifay R, Morin L, Scemla A, Delville M, Martinez F, Timsit MO, Duong Van Huyen J-P, Legendre C, Terzi F, Anglicheau D, Urinary C-X-C Motif Chemokine 10 Independently Improves the Noninvasive Diagnosis of Antibody-Mediated Kidney Allograft Rejection., J. Am. Soc. Nephrol. 26, 2840–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SS, Song SH, Kim JH, Jeon YK, Kim BH, Kang M-C, Chun SW, Hong SH, Chung M, Kim YK, Kim IJ, Kim Y-B, Urine clusterin/apolipoprotein J is linked to tubular damage and renal outcomes in patients with type 2 diabetes mellitus, Clin. Endocrinol. (Oxf). 87, 156–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dieterle F, Perentes E, Cordier A, Roth DR, Verdes P, Grenet O, Pantano S, Moulin P, Wahl D, Mahl A, End P, Staedtler F, Legay F, Carl K, Laurie D, Chibout S-D, Vonderscher J, Maurer G, Urinary clusterin, cystatin C, β2-microglobulin and total protein as markers to detect drug-induced kidney injury, Nat. Biotechnol. 28, 463–469 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Heymsfield SB, Arteaga C, McManus C, Smith J, Moffitt S, Measurement of muscle mass in humans: validity of the 24-hour urinary creatinine method, Am. J. Clin. Nutr. 37, 478–494 (1983). [DOI] [PubMed] [Google Scholar]
- 48.Waikar SS, Sabbisetti VS, Bonventre JV, Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate, Kidney Int. 78, 486–494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Correlation Matrix of Biomarkers
Table S1. Multivariate Logistic Regression of Acute Rejection Status as Assessed by Individual Biomarkers
Table S2. Biomarker Log-Likelihood Ratios for the urine score
Table S3. Multivariate Regression of Clinical and Demographic Variables with the urine score
Data File S1. Urine score data


