Abstract
The architecture of brain networks has been extensively studied in multiple species. However, exactly how the brain network reconfigures when a local region, particularly a hub region, stops functioning remains elusive. By combining chemogenetics and resting-state functional magnetic resonance imaging (rsfMRI) in an awake rodent model, we investigated the causal impact of acutely inactivating a hub region (i.e. the dorsal anterior cingulate cortex) on brain network properties. We found that suppressing neural activity in a hub could have a ripple effect that went beyond the hub-related connections and propagated to other neural connections across multiple brain systems. In addition, hub dysfunction affected the topological architecture of the whole-brain network in terms of the network resilience and segregation. Selectively inhibiting excitatory neurons in the hub further changed network integration. None of these changes were observed in sham rats or when a non-hub region (i.e. the primary visual cortex) was perturbed. This study has established a system that allows for mechanistically dissecting the relationship between local regions and brain network properties. Our data provide direct evidence supporting the hypothesis that acute dysfunction of a brain hub can cause large-scale network changes. These results also provide a comprehensive framework documenting the differential impact of hub versus non-hub nodes on network dynamics.
Keywords: DREADD, Resting-state fMRI, Graph theory, Awake, Rat
1. Introduction
The mammalian brain is a highly inter-connected network system with distributed brain regions orchestrating to support normal function and mediate complex behavior (Bullmore and Bassett, 2011). The brain network is organized to maintain an optimal balance of information segregation and integration, which simultaneously maximizes the efficiency of distal communication via long-range connections and minimizes the wiring cost through clustered local processing ((Bullmore and Sporns, 2009, 2012), but also see (Betzel and Bassett, 2018)). Such organization is well conserved in multiple species including rodents, primates and humans (Liang et al., 2011; Vincent et al., 2007; Wang et al., 2010).
It is becoming increasingly clear that altered brain network organization is tightly linked to brain disorders (Bassett and Bullmore, 2009; Fornito et al., 2015; Liu et al., 2008). However, the neural substrates causing these large-scale network changes remain unknown. A key hypothesis is the dysfunction of brain hub regions. Hubs are brain regions that have high degrees of connections with the rest of the brain (Rubinov and Sporns, 2010; van den Heuvel and Sporns, 2013). Because of their central roles, dysfunction of hub nodes can change global integrative process, and has been hypothesized to be a direct cause of altered brain network properties and pathophysiology of brain disorders (Alstott et al., 2009; Crossley et al., 2014; Takahashi et al., 2009). For instance, brain network analysis in patients with Alzheimer’s Disease showed that amyloid-beta deposition mainly accumulated in functional hubs (Buckner et al., 2009). Therefore, comprehensively understanding the causal relationship between hub activity and brain network organization is critical. However, directly testing this hypothesis in humans is challenging, as selectively altering activity in a hub and dissecting its causal impact on brain networks are difficult.
This obstacle can be overcome in experimental animals using neuroscience tools like Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) and resting state functional magnetic resonance imaging (rsfMRI). DREADDs introduce genetically encoded modified muscarinic G-protein-coupled receptors into living neurons, which allow reversible in vivo manipulation of neuronal activity of a defined brain region for hours (Armbruster et al., 2007). rsfMRI measures brain-wide resting-state functional connectivity (RSFC), and provides comprehensive assessment of brain network properties (Biswal et al., 1995; Liang et al., 2011). Thus, combining these two techniques offers a system that allows us to characterize the reconfiguration of the brain network as the neural activity of a network node is selectively perturbed. Given the highly conserved brain network architecture in the mammalian brain (Liang et al., 2011; Vincent et al., 2007; Wang et al., 2010), the information obtained might be translatable to humans.
Here we investigate the causal impact of manipulating a hub region — the dorsal anterior cingulate cortex (dACC) — on brain network organization using an awake rodent model established in our lab (Dopfel et al., 2019; Liang et al., 2014; Liang et al., 2013; Ma et al., 2018; Ma and Zhang, 2018; Zhang et al., 2010). Imaging awake rodents avoids the confounding effects of anesthesia (Gao et al., 2016; Hamilton et al., 2017; Liang et al., 2012b; Liang et al., 2015a; Ma et al., 2017). The dACC is selected as it was identified as a functional hub in the awake rat brain in our previous study (Ma et al., 2018), based on a well-established method that defines the hubness of brain nodes (van den Heuvel et al., 2010). In this method, hubs are brain regions that meet at least three out of four criteria in a network: i) 20% highest degree; ii) 20% highest betweenness centrality; iii) lowest 20% characteristic path length; and iv) lowest 20% local clustering coefficient. Consistent with our finding, the dACC was reported as a functional and anatomical hub in both the human and rat brain (Bush et al., 2000; Ma et al., 2018; van den Heuvel et al., 2016; van den Heuvel and Sporns, 2013).
Our data show that suppressing all neurons in the brain hub impacts the network topology including network resilience and segregation, but not network integration. Interestingly, selectively inhibiting excitatory neurons in the dACC not only reduces the network resilience and segregation, but also lowers the network integration, suggesting that excitatory neurons in a brain hub might play a key role in global information integration. All these changes are absent in sham rats or when a non-hub region is suppressed. Taken together, this study provides direct measurement of the brain’s response to targeted attack at a hub region, and presents a comprehensive framework demonstrating the pivotal role of hubs in the brain network.
2. Methods and materials
2.1. Animals
44 adult male Long-Evans rats (300-500g) were used in the present study. Data from the all-neuron DREADD group, sham group and non-hub group (35 rats) were also used in a previous publication (Tu et al., 2021), but were reanalyzed for the purpose of the present study. Animals were housed in Plexiglas cages. Food and water were provided ad libitum. The housing room was maintained at an ambient temperature (22-24 °C) with a 12h light:12h dark cycle. All experiments were approved by the Pennsylvania State University Institutional Animal Care and Use Committee.
2.2. Surgery
Aseptic stereotaxic surgery was conducted to infuse the virus into the brain. Rats were anesthetized by intramuscular (IM) injections of ketamine (40 mg/kg) and xylazine (12 mg/kg). Dexamethasone (0.5 mg/kg) and Baytril (2.5 mg/kg) were injected subcutaneously to prevent tissue inflammation and bacterial infections. During the surgery, respiration was controlled by a ventilator with natural oxygen gas. The heart rate was monitored by a pulse oximetry (MouseSTAT® Jr., Kent Scientific Corporation), and the body temperature was maintained at 37 °C with a warming pad (PhysioSuite, Kent Scientific Corporation). DREADD viral vectors with the pan-neuron promotor (AAV8-hSyn-hM4Di-mCherry, 1 μL at titer ≥ 3×1012 vg/mL, Addgene, Watertown, MA) were bilaterally injected into the dACC (coordinates: AP +2, ML +/−0.5, DV −1) and the primary visual cortex (coordinates: AP −6.5, ML +/−3.5, DV −1) for the hub (n = 19) and non-hub (n = 8) groups, respectively. DREADD viral vectors with the CaMKII promoter (AAV8-CaMKII-hM4Di-mCherry, 1 μL at titer ≥ 3×1012 vg/mL, Addgene, Watertown, MA, n=9) were bilaterally injected into the dACC (coordinates: AP +2, ML +/−0.5, DV −1) to selectively inhibit excitatory neurons. Sham rats were injected with control null viral vectors (AAV8-hSyn-GFP, 1 μL at titer ≥ 3×1012 vg/mL, Addgene, Watertown, MA, n = 8) into the dACC. Rats were allowed to recover for at least four weeks.
2.3. fMRI experiments
Before imaging, animals were acclimated to the MRI environment for 7 days to minimize stress and motion during imaging. To better adapt the animal to the restrainer and scanning environment, the acclimation period was gradually increased from 15 min on Day 1 to 30 min on Day 2, 45 min on Day 3, and maintained at 60 min/day for Days 4-7. Detailed procedures for acclimation can be found in our previous publications (Dopfel and Zhang, 2018; Liang et al., 2012a). A similar approach to imaging awake animals has also been adopted by other research groups in multiple species (Bergmann et al., 2016; Chang et al., 2016; Yoshida et al., 2016). Clozapine-N-Oxide (CNO, 1mg/kg in saline, dissolved in DMSO, Sigma-Aldrich, St. Louis, MO) or saline (with DMSO) was IP injected 30 min before rsfMRI data acquisition. Imaging sessions with CNO and saline injections were randomized with at least three days apart.
All imaging sessions were conducted at the High Field MRI Facility at the Pennsylvania State University on a 7T Bruker 70/30 BioSpec running ParaVision 6.0.1 (Bruker, Billerica, MA). rsfMRI data were acquired at the awake state using a T2*-weighted echo-planar-imaging (EPI) sequence with the following parameters: repetition time = 1000ms; echo time = 15ms; matrix size = 64×64; field of view = 3.2 × 3.2 cm2 ; slice number = 20; slice thickness = 1mm; flip angle = 60; 600 volumes each run. Three runs were acquired each rsfMRI session. Raw EPI images were displayed in Fig. S1.
In a subgroup of animals (n = 9), we simultaneously measured the respiratory signal with rsfMRI data in both the CNO and saline conditions (Fig. S2). The mean respiratory rates were similar to previous studies in awake rats at the resting state (Kabir et al., 2010; Stenroos et al., 2018). In addition, there was no significant difference in the respiration rate between the CNO and saline conditions (Fig. S2).
2.4. Data analysis
Before image preprocessing, rsfMRI volumes with relative framewise displacement (FD) > 0.25 mm and their preceding and following volumes were removed (Power et al., 2012). The motion censoring threshold chosen was half of the voxel size, which was routinely used in previous rodent fMRI studies (Bajic et al., 2017; Liang et al., 2012b). In addition, the first 10 volumes of each rsfMRI scan were discarded to ensure steady state of magnetization. rsfMRI scans with > 15% volumes discarded were removed from further analysis.
All acquired imaging data were aligned to a defined atlas using Medical Image Visualization and Analysis software (MIVA), and then subjected to motion correction (SPM12), spatial smoothing (Gaussian kernel, full-width at half-maximum = 1 mm), voxel-wise nuisance regression of motion parameters, as well as the signals from the white matter and ventricles, and bandpass filtering (0.01-0.1Hz). The band-pass filtering was applied as it is known that the coherence between regional rsfMRI signals was dominant in the low-frequency range (Cordes et al., 2001; Fox and Raichle, 2007). Fig. S1 shows the power spectra of rsfMRI time courses before bandpass filtering in several representative brain regions, confirming that the dominant rsfMRI power was below 0.1Hz.
The whole-brain RSFC network was constructed by parcellating the rat brain into 68 anatomical regions of interest (ROIs) based on the Swanson Atlas (Swanson, 2004), which were grouped into 10 functional systems including amygdala complex, striatum, hypothalamus, hippocampus, thalamus, sensory-motor cortex, polymodal association cortex, brainstem, pallidum, and retro-hippocampal regions. RSFC between each pair of ROIs was calculated by Pearson correlations of regionally averaged time series of the ROIs. Fisher transformation was applied to convert r values to z values, so that correlation coefficients follow normal distributions required for statistical comparisons.
To assess brain network properties at each condition, we calculated the hub scores of individual nodes as well as topological parameters of the whole-brain network. The hub score of each node ranging from 0-4 was determined by the number of the following four criteria the node met: (1) 20% highest degree; (2) 20% highest betweenness centrality; (3) 20% lowest characteristic path length; (4) 20% lowest local clustering coefficient (van den Heuvel et al., 2010).
Local topological parameters including local efficiency, characteristic path length and eigenvector centrality for the dACC were calculated using the brain connectivity toolbox (BCT) (Rubinov and Sporns, 2010) based on weighted whole-brain graphs with inter-ROI RSFC as the weight, defined as follows:
Local efficiency (Latora and Marchiori, 2001)
where N is the set of all nodes; wxy is the connectivity weight (i.e. RSFC) between nodes x and y; represents the weighted shortest path length between y and z based on the Dijkstra’s algorithm. The distance between two ROIs is quantified by the inverse of their corresponding RSFC (1/RSFC). kx is the node degree of x.
Characteristic path length (Watts and Strogatz, 1998)
where N is the set of all nodes; n is the total number of nodes; is the weighted shortest path length between x and y. The characteristic path length for weighted graphs is an estimate of proximity.
Eigenvector centrality (Newman, 2008)
where n is the total node number; λ1 is the largest eigenvalue of the connectivity matrix A; μ1 (y) is the y-th component of the first eigenvector of A; axy is the connectivity weight between nodes x and y.
Global topological parameters of the whole-brain network including the assortativity, modularity and global efficiency were calculated based on weighted whole-brain network graphs using BCT (Rubinov and Sporns, 2010), defined below:
Assortativity (Newman, 2002)
where are the weighted degrees of nodes x and y, respectively; wxy is the connectivity weight between x and y; L is the set of all edges within the network, and l is the total number of edges.
Modularity (Newman, 2004)
where lw is the sum of weights in the network; wxy is the connectivity weight between nodes x and y; is the weighted degrees of nodes x and y, respectively.
Global efficiency (Latora and Marchiori, 2001)
where N is the set of all nodes; n is the total number of nodes; is the weighted shortest path length between nodes x and y.
All topological parameters were determined for each scan. Changes in each parameter were determined by two-sample t-tests with a linear mixed model between the CNO injection and saline injection conditions. The effect size determined by Cohen’s d was reported in Table S2.
To control for any potential non-specific effects of CNO, between-group comparisons for individual topological parameters were performed using two-way ANOVA with the groups of DREADD and sham groups and the conditions of CNO and saline injection.
2.5. Histology and immunofluorescence imaging
After fMRI imaging, the animals were perfused with saline followed by 4% PFA solution. The brain was taken out carefully and stored in solution with 4% paraformaldehyde and 20% sucrose. After fixation, the brain was sliced into 60 μm slices. Fluorescent expression in the injection site was evaluated under microscopy. Immunofluorescent staining was performed to further confirm the AAV expression. After being washed with phosphate buffered saline (PBS), sections were treated with 1% Triton X-100 for 15 minutes followed by 1-hour blocking with 10% goat serum (ThermoFisher Scientific) at room temperature, then incubated with primary antibodies (1:50, anti-NeuN, Abcam; 1:20, anti-GLS, Proteintech) at 4 degree overnight, and followed by secondary antibody (1:200, goat-anti-rabbit-Cy5 and goat-anti-mouse-488, ThermoFisher Scientific) incubation for 1 hour at room temperature. Lastly, immunostained sections were stained with DAPI and mounted for fluorescent confocal microscopy.
3. Results
Experimental animals were stereotactically injected with adeno-associated viruses (AAVs) expressing inhibitory G-protein coupled hM4Di receptor with a pan-neuronal synapsin promoter (AAV8-hSyn-hM4Di-mCherry) or a CaMKII promoter (AAV8-CaMKII-hM4Di-mCherry). The selective expression of AAV8-CaMKII-hM4Di in excitatory neurons in the dACC was confirmed as shown in Fig. S3 as well as in a previous study (Koike et al., 2016). Sham animals were injected with control null viral vectors (AAV8-hSyn-GFP). After 4-6 weeks of recovery and protein expression (Fig. S3), animals received either CNO or saline 30 min before rsfMRI scanning. Saline and CNO sessions were separated by at least 3 days in a random order. There was no difference in motion level during rsfMRI scanning between CNO and saline injection conditions in both the DREADD and sham groups (Fig. S4). The experimental procedure is summarized in Fig. 1. The inhibitory effect of DREADDs was confirmed by electrophysiology recordings in our previous publication (Tu et al., 2021), and also evidenced by lower fMRI activity in the dACC (Fig. 3 in (Tu et al., 2021)) and V1(Fig. S5). Consistent with our published data (Tu et al., 2021), previous awake behavioral studies using the same DREADDs showed a comparable regional inhibition level between the V1 and ACC (Gao et al., 2020; Wang et al., 2021).
Fig. 1.

Schematic diagram of the experimental paradigm.
Fig. 3. Suppressing the dACC changes RSFC in connections widespread the whole brain.
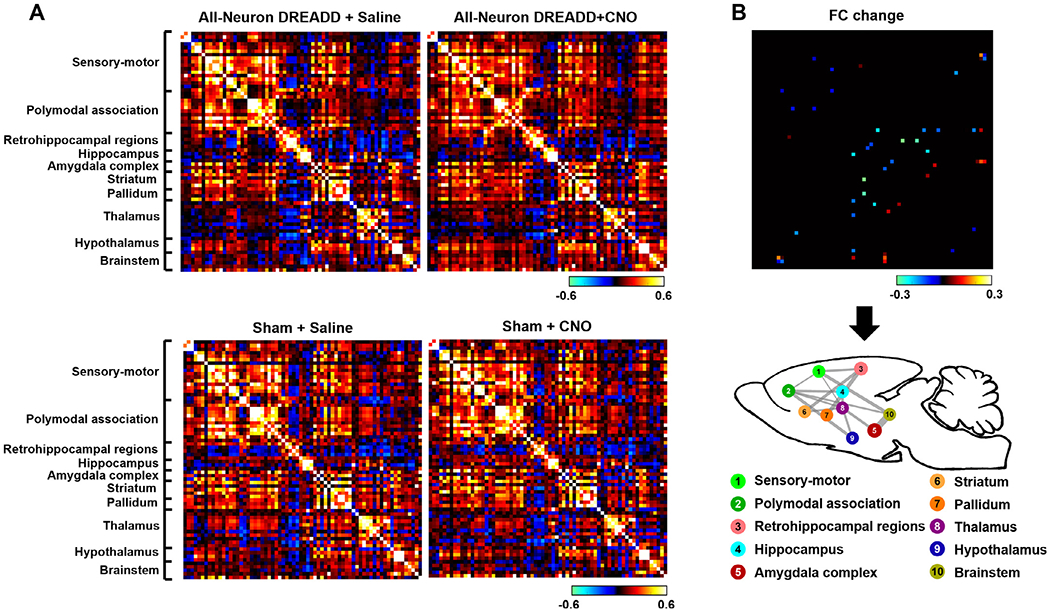
A) Top: averaged ROI-wise RSFC matrices after saline and CNO injections in the all-neuron DREADD group, respectively. Bottom: averaged ROI-wise RSFC matrices after saline and CNO injections in the sham group, respectively. B) Top: connections exhibiting significant RSFC changes after saline and CNO injections (two-way ANOVA, linear mixed model, pinteraction < 0.05, FDR corrected). RSFC changes between the CNO and saline conditions in the DREADD group were color coded with cold colors representing reduced RSFC and hot colors representing increased RSFC. Bottom: Connections with significant functional connectivity changes were grouped into multiple brain systems. Line thickness represents the total number of connections with significant RSFC changes between the two systems.
3.1. Suppressing the dACC reduced its FC strength, communication efficiency and local information processing
We first examined the local impact of suppressing the activities of all neurons in the dACC. Fig. 2A shows that DREADD suppression of the dACC reduced its FC strength, quantified by the sum of the absolute FC amplitude between the dACC and all other ROIs relative to saline injection. Fig. S6A shows dACC seedmaps in the all-neuron DREADD group for both the saline and CNO conditions, also indicating a generally reduced RSFC with dACC after CNO injection. In addition, suppressing the dACC led to impaired communication efficiency, reflected by longer characteristic path length between the dACC and other ROIs (Fig. 2B). Besides communication directly related to the dACC, we found that suppressing the dACC affected information processing in its neighbors, evidenced by reduced local efficiency, which measures communication efficiency among the neighbors of dACC (Fig. 2C). Taken together, these data suggest that suppressing activities of all neurons in the dACC not only changes its own FC strength and communication efficiency, but also impacts local information processing in its neighboring regions.
Fig. 2. Suppressing the dACC affects dACC FC strength, characteristic path length, and local efficiency.

A) dACC FC strength, B) dACC characteristic path length, and C) dACC local efficiency, with CNO or saline injection in the all-neuron DREADD group (top) and sham group (bottom). DREADD: n = 19, 78 scans; sham: n = 8, 41 scans. Linear mixed model; ***:p<0.05; NS: not significant. Error bar: standard error of the mean.
None of these changes were observed in sham animals as shown in Fig. 2 (bottom row) and Fig. S6B, suggesting unappreciable non-specific effects of CNO in the system. This conclusion is corroborated by two-way ANOVA tests (linear mixed model) in topological parameters between all-neuron DREADD and sham groups in both the CNO and saline conditions (FC strength: pinteraction = 0.017; Characteristic path length: pinteraction = 0.023; Local efficiency: pinteraction = 0.016; Table S1; effect size; FC strength: cohen’s d (all-neuron DREADD) = 0.85, cohen’s d (sham) = 0.063; Characteristic path length: cohen’s d (all-neuron DREADD) = 0.998, cohen’s d (sham) = 0.01; Local efficiency: cohen’s d (all-neuron DREADD) = 0.804, cohen’s d (sham) =0.098; Table S2).
Notably, DREADD suppression of the dACC did not change the overall connectivity of the brain, assessed by the averaged FC strength from all ROIs (Fig. S7).
3.2. Suppressing dACC affected connections widespread the whole brain
We then assessed the influences of suppressing activities of all neurons in the dACC on RSFC in brain-wide connections. Averaged RSFC matrices obtained after saline and CNO injections in both the DREADD and sham groups were shown in Fig. 3A. Multiple distributed connections exhibited altered RSFC (Fig. 3B, two-way ANOVA, pinteraction < 0.05, linear mixed model, FDR corrected) with lower RSFC in the majority of these connections (i.e. blue elements in the RSFC difference matrix, Fig. 3B), indicating that the impact of suppressing neural activity in a hub can go beyond the hub itself and propagate to neural circuits across the whole brain. To better characterize the brain connections affected, all ROIs were grouped into 10 anatomical-based brain systems including sensory-motor cortical regions, polymodal association cortical regions, retrohippocampal regions, hippocampus, amygdala complex, striatum, pallidum, thalamus, hypothalamus and brainstem. Connections with significant RSFC changes were overlaid on a glass rat brain, based on their brain systems (Fig. 3B). Prominent changes in polymodal association cortical regions were observed including reduced cortical-cortical connectivity between polymodal association cortical regions and sensory-motor cortical regions, along with reduced cortical-subcortical connectivity between polymodal association coritical regions and multiple subcortical systems such as the thalamus, hypothalamus, and brainstem (Fig. 3B). These changes indicate that disrupting a hub can lead to a reorganization of the polymodal association cortical regions, consistent with the view that polymodal association cortical regions are information integration centers in the brain.
Suppressing a hub region not only affected the polymodal association system to which the dACC belonged, it also disrupted communication among other brain systems, manifested by reduced cortical-subcortical RSFC between sensory-motor regions and thalamus and brain stem, as well as reduced RSFC between separate subcortical systems. A similar ripple effect was observed in human patients with focal brain lesions to critical locations (Gratton et al., 2012).
3.3. Suppressing a hub node changed whole-brain network organization
We investigated the effect of suppressing activities of all neurons in the dACC on the organization of the whole-brain network architecture in three aspects: network resilience, functional segregation, and functional integration.
Assortativity measures network resilience. Brain networks with high assortativity are more resilient to local attack such as lesion and neurological degeneration (Newman, 2002; Rubinov and Sporns, 2010). Fig. 4A shows that suppressing activities of all neurons in the dACC considerably reduced the network assortativity, and as a consequence, the whole-brain network became more vulnerable. Functional segregation, quantified by modularity, measures the network’s ability of specialized processing. We found that dACC suppression decreased the modularity of the global brain network, suggesting reduced functional segregation (Fig. 4B). Global efficiency measures functional integration, which assesses the ability to integrate specialized information from distributed brain regions. Surprisingly, suppressing activities of all neurons in the dACC did not seem to affect the global efficiency (Fig. 4C). No topological changes were observed in sham rats (Fig. 4, bottom row). In summary, we observed that suppressing activities of all neurons in the dACC affected global brain topology including network resilience and network segregation, but not network integration.
Fig. 4. Suppressing a hub node changed whole-brain network topology.
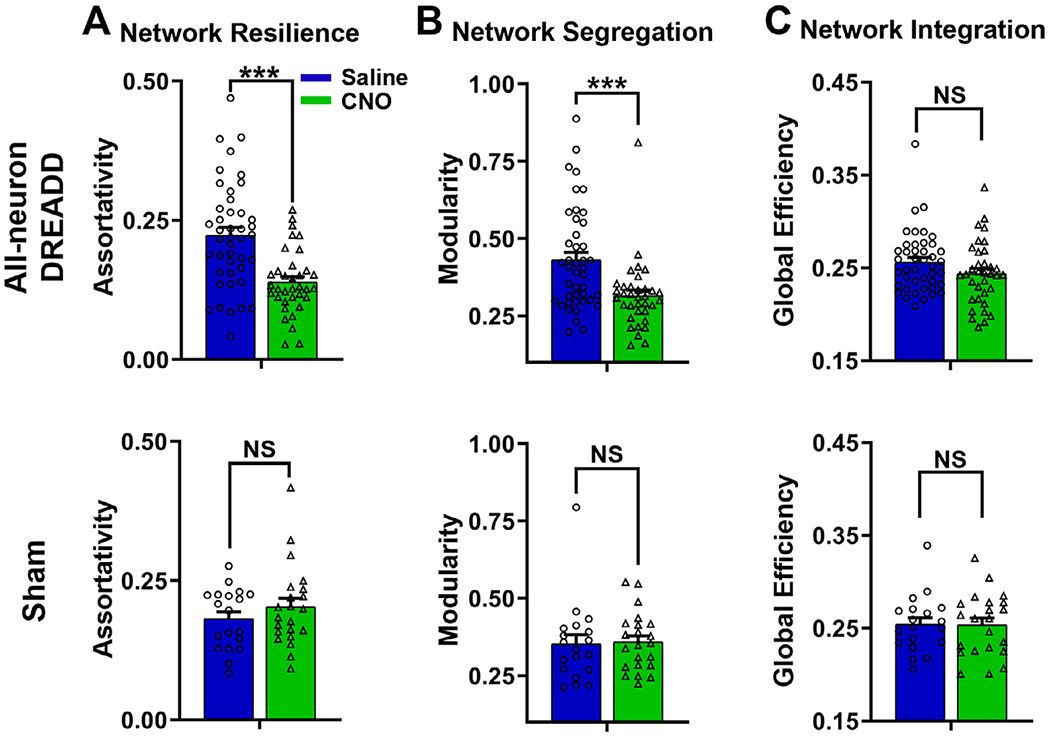
A) Network resilience with CNO or saline injection in the all-neuron DREADD group and sham group; B) Network segregation with CNO or saline injection in the all-neuron DREADD group and sham group; C) Network integration with CNO or saline injection in the all-neuron DREADD group and sham group. Linear mixed model; all-neuron DREADD: n = 19, 78 scans; sham: n = 8, 41 scans. ***:p<0.005; NS: not significant. Error bar: standard error of the mean.
3.4. New hub emerged after an existing hub was inactivated
Given the central role of hub nodes, we ask whether a new hub can emerge after an existing hub is inactivated. The hubness of a brain region was quantified by its hub score described in Methods, and was also assessed using node eigenvector centrality, which is often used to characterize the pivotal role of a node in the network. A node with a large eigenvector centrality value tends to strongly connect with other nodes that also have high centrality in the network (Lohmann et al., 2010).
As expected, DREADD suppression of the dACC significantly lowered its hub score and centrality (Fig. 5A & B), suggesting that dACC lost its hubness in the network. Interestingly, the centrality of the ventral retro-splenial cortex (vRSC) was significantly increased after dACC suppression (Fig. 5A & B). Again, no change in the centrality of the dACC or vRSC was observed in sham rats (Fig. S8). These results indicate that a new hub can emerge when an existing hub stops functioning, likely due to the compensatory mechanism.
Fig. 5. New hub emerged after the inactivation of an existing hub.

A) Hub scores without and with dACC inactivation, respectively; B) Centrality of the dACC and vRSC without and with dACC inactivation (linear mixed model; n = 19, 78 scans). **: p < 0.01; ***: p < 0.005. Error bar: standard error of the mean.
3.5. Test of reproducibility
To test the reproducibility of our results, and also to control for the potential impact of different sample size in the all-neuron DREADD (n = 19) and sham (n = 8) groups, we separated the all-neuron DREADD group into two subgroups (n = 9 for subgroup 1, and n = 10 for subgroup 2). All analyses were repeated in both subgroups. Fig. S9 shows that all results obtained were highly reproducible between the two subgroups and consistent with our main findings, supporting the rigor of our findings.
3.6. Selectively suppressing excitatory neurons in dACC lowered network integration
Considering that pan-neuronal inhibition could suppress inhibitory interneurons (i.e. disinhibition), which might have excitatory effects, we selectively inhibited excitatory neurons in the dACC using inhibitory DREADDs with a CaMKII promoter (AAV8.CaMKII.hM4Di.mCherry). Consistent with data obtained under pan-neuronal inhibition, we observed reduced dACC FC strength and local efficiency, as well as increased dACC characteristic path length (Fig. 6), suggesting the dominant role of excitatory neurons in dACC-related local information processing.
Fig. 6. Suppressing dACC excitatory neurons affects dACC FC strength, characteristic path length, and local efficiency.

A) dACC FC strength with CNO or saline injection in the excitatory-neuron DREADD group. B) Characteristic path length of dACC with CNO or saline injection in the excitatory-neuron DREADD group. C) Local efficiency of dACC with CNO or saline injection in the excitatory-neuron DREADD group. Linear mixed model; excitatory-neuron DREADD group: n = 9, 56 scans. ***:p<0.005. Error bar: standard error of the mean.
Fig 7A displays averaged RSFC matrices obtained after saline and CNO injections, respectively, in the excitatory-neuron DREADD group, as well as connections exhibiting significant RSFC changes between the two conditions (FC change matrix, two-way ANOVA, pinteraction < 0.05, linear mixed model, FDR corrected). Connections showing significantly altered RSFC were organized into brain systems and overlaid on a glass rat brain (Fig. 7B). Prominent changes were again observed in polymodal association cortical regions, including reduced cortical-cortical RSFC with the sensory-motor regions, as well as reduced cortical-subcortical RSFC with the hippocampus and hypothalamus but increased RSFC with the amygdala. Interestingly, increased polymodal association-amygdala RSFC was also observed in patients with major depressive disorder (Zhang et al., 2020) that was related to reduced excitation/inhibition ratio (Page and Coutellier, 2019), which was not found when all dACC neurons were suppressed (Fig. 3). Furthermore, inhibiting excitatory neurons in the hub had a similar ripple effect, which reduced cortical-subcortical RSFC between sensory-motor cortical regions and the thalamus and retrohippocampal regions, as well as altered subcortical-subcortical RSFC between retrohippocampal and hippocampal regions, although specific connections affected were different from the case when all neurons in the dACC were suppressed.
Fig. 7. Suppressing dACC excitatory neurons changes RSFC in widespread connections and whole-brain network topology.

A) Left: averaged ROI-wise RSFC matrices after saline and CNO injections in the excitatory-neuron DREADD group, respectively. Right: connections exhibiting significant RSFC changes after saline and CNO injections (two-way ANOVA, linear mixed model, pinteraction < 0.05, FDR corrected). RSFC changes between the CNO and saline conditions in the DREADD group were color coded with cold colors representing reduced RSFC and hot colors representing increased RSFC. These connections are grouped into B) multiple brain systems and overlaid on a glass rat brain. Line thickness represents the total number of connections with significant RSFC changes between the two systems. C) Suppressing dACC excitatory neurons changes whole-brain network topology including network resilience (left), network segregation (middle), and network integration (right). Linear mixed model; DREADD-CaMKII: n = 9, 56 scans. *: p<0.05; ***:p<0.005. Error bar: standard error of the mean.
Consistent with the results of pan-neuronal inhibition, whole-brain network topological properties including the network resilience and network segregation were reduced after the suppression of excitatory neurons in the dACC (Fig. 7C). Intriguingly, unlike the situation of pan-neuronal inhibition, the network integration was significantly reduced with selective inhibition of dACC excitatory neurons (Fig. 7C). This result is consistent with previous findings that long-distance pathways are dominant by excitatory projections, while inhibitory neurons are mainly involved in local circuits (Dehaene et al., 1998; Somogyi et al., 1998). Collectively, our data suggest that selective suppression of excitatory neurons might have more appreciable impact on long-distance projections, and thus significantly affect the information integration.
3.7. Suppression of a non-hub region did not change whole-brain network properties
To determine whether aforementioned network changes depend on the specific role of the node (i.e. hub versus non-hub), we also suppressed the activity in a non-hub region—primary visual cortex (V1, bilateral) (Ma et al., 2018) using the same pan-neuron inhibitory DREADDs. We found that suppression of V1 did not cause any whole-brain topological changes including network resilience, functional segregation, or functional integration (Figs. 8A-8C). These results were confirmed using two-way ANOVA tests (linear mixed model) on topological parameters between V1 DREADD and sham groups in the CNO and saline conditions (Assortativity: pinteraction = 0.16, F(1,103) = 2.03; Modularity: pinteraction = 0.4, F(1,103) = 0.72; Global efficiency: pinteraction = 0.86, F(1,103) = 0.032). The effect size was calculated based on Cohen’s d (Assortativity: cohen’s d = 0.239; Modularity: cohen’s d = 0.283; Global efficiency: cohen’s d = 0.1226). Taken together, these data suggest that attack on separate brain regions can have differential impact on brain network properties, and brain networks are particularly vulnerable to targeted attack at hub nodes.
4. Discussion
By combining DREADDs and rsfMRI in an awake rodent model, we have established a system that enables us to manipulate the activity in any brain region, and measure the corresponding changes in brain-wide networks. This system allows for mechanistically dissecting the causal relationship between a (or multiple) local brain region(s) and brain network properties. In this study we specifically investigated the role of a hub region (i.e. dACC) in brain networks. We found that suppressing neural activity in the hub could have a ripple effect that went beyond the hub-related connections and propagated to other neural circuits across the whole brain (Figs. 3 & 7). In addition, hub dysfunction affected the topological architecture of the whole-brain network in terms of the network resilience and segregation (Fig. 4). Selectively suppression of excitatory neurons in the hub region further impaired the network integration (Fig. 7), suggesting a key role of excitatory neurons in long-distance communication. None of these changes were observed in sham rats (Figs. 3 & 4) or when a non-hub region (i.e. V1) was suppressed (Fig. 8). These data offer direct evidence supporting the hypothesis that acute dysfunction of a brain hub can cause large-scale network changes. They also document the differential impact of inactivating a hub versus a non-hub node on brain network dynamics.
Fig. 8. Suppressing a non-hub node did not change whole-brain network topology.

A) Network resilience with CNO or saline injection; B) Network segregation with CNO or saline injection; C) Network integration with CNO or saline injection (linear mixed model; n = 8, 66 scans). NS: not significant. Error bar: standard error of the mean.
4.1. Tight relationship between local regions and brain networks
The activity in local brain regions can be manipulated using tools like chemogenetic, optogenetic, pharmacological and lesion methods. Such perturbations are often used to study the role of a specific brain region in behavior (Whissell et al., 2016). Meanwhile, the capacity of local-region manipulations can be tremendously expanded when combining with neuroimaging approaches with a global field of view (e.g. fMRI), as the causal impact of local region dysregulation on the rest of the interconnected network can be monitored across the whole brain (Liang et al., 2017; Liang et al., 2015b). For instance, using DREADDs and rsfMRI, Grayson and colleagues showed that inactivating the amygdala disrupted communication throughout the cortex in anesthetized monkeys (Grayson et al., 2016). This observation echoes human patient studies in which lesions of critical focal brain locations caused widespread disruption of the brain modular structure (Gratton et al., 2012; Warren et al., 2014). These results collectively suggest tight linkage between regional activity and large-scale network properties, and highlight the importance of combining local perturbations and global network measurement. This notion has indeed been applied in studies examining the role of local regions in special functional networks (Folloni et al., 2019). For example, in the study by Del Ferraro et al., long-term potentiation in the hippocampus was used to identify the memory-related functional network and essential integrators in the network (Del Ferraro et al., 2018). It showed that suppression of the essential integrator disrupted the memory-related functional network. In the present study we went beyond specific neural circuits and functional networks (Tu et al., 2021), and showed that dysfunction of hub nodes can be a direct cause of altered whole-brain network properties (Alstott et al., 2009; Crossley et al., 2014; Takahashi et al., 2009). Interestingly, in a recent study Peeters et. al. unilaterally inhibited the ACC and examined the corresponding changes in rsfMRI signal amplitude and functional connectivity in regions across the brain (Peeters et al., 2020). Consistent with our results, disrupted functional connectivity across multiple brain regions was observed, indicating the hub role of the ACC in both rats and mice. However, in contrast to increased centrality of the RSC observed in our study, reduced functional connectivity in the RSC was reported. This discrepancy could be attributed to the difference in species (rats vs mice) and animal preparation (awake vs anesthetized conditions), as well as the fact that different subregions of ACC were suppressed (unilateral ACC vs bilateral dACC), which are known to have distinct connectivity profiles and function (van Heukelum et al., 2020).
Notably, all data were collected in awake animals in our study, which avoids the confounding effects of anesthesia on brain network function, and permits direct association of network features and possible behavioral changes. Awake animal imaging also makes the findings translatable to humans, considering numerous reports of large-scale brain network changes in human brain disorders.
4.2. Hubs and brain network topology
The topological changes resulting from hub inactivation, including reduced network resilience and segregation, are important for our understanding of the role of individual regions in the global brain network organization. A brain network can be modeled as a graph composed of spatially distributed neuronal components as nodes and anatomical or functional associations between components as edges (Bullmore and Sporns, 2009, 2012). Topological features of the brain graph are critical in understanding inter-areal information exchange in health and disease. Although the architecture of brain networks has been extensively studied in multiple species (Bassett and Bullmore, 2006; Belcher et al., 2013; Liang et al., 2011; Liska et al., 2015), it remains elusive whether different types of nodes, such as hubs versus non-hubs, play distinct or similar roles in the topology of the global brain network. Our data revealed that changes in a hub, but not a non-hub, can significantly alter brain network topology. These results provide critical insight into understanding the pathogenesis of neurological and psychiatric disorders, which indicate that altered brain topological properties reported in these brain disorders might start from dysfunction of certain hub nodes (Seeley et al., 2009).
Interestingly, our data showed that network integration was intact after inactivating all dACC neurons, likely due to compensatory effects of the so-called ‘rich club organization’ in the rat brain (Ma et al., 2018). Indeed, we observed that a new hub (vRSC) emerged after the inactivation of dACC (Fig. 5). Additionally, inhibition of inhibitory neurons (i.e. disinhibition) has excitatory effect, which could potentially counteract some inhibitory effect on excitatory neurons. To further examine this issue, we selectivity inactivated excitatory neurons, and observed reduced network integration. Excitatory pyramidal neurons, especially those in layer 2-3, mainly integrate information for long-range communication (Dehaene et al., 1998), and therefore, selective inactivation of excitatory neurons can specifically impact long-distance projections and affect information integration directly.
4.3. Potential pitfalls
It is known that physiologic fluctuations, such as respiration, can affect rsfMRI signal (Dehaene et al., 1998). In the present study, we cannot completely rule out the potential impact of the physiologic noise on our results. However, we believe this impact was very minor, if any. First, relative to human data, the influence of physiologic noise on rodent rsfMRI data is considerably lower. Kalthoff and colleagues showed that in rats the relative contributions from cardiac and respiratory noise to the rsfMRI signal were 1% and 5%, respectively (Kalthoff et al., 2011). Second, in our image preprocessing we regressed out the signals from the white matter and ventricles, which are known to be dominated by physiologic noise (Chang and Glover, 2009). Third, to further elucidate the possible effect of respiration on voxel-wise rsfMRI signal, we calculated the voxel-wise correlations between respiratory variation and rsfMRI signals, a method used in a recent human study (Chen et al., 2020). Specifically, we calculated the standard deviation of the respiratory waveform in 7-sec sliding windows (step size = 1 sec), with each window center corresponding to a TR. The correlations between the respiratory variation and voxel-wise rsfMRI signals were computed to generate a respiration-related correlation map. We did not find any region exhibiting significant correlations (one-sample t test, Fig. S2) in the map, again confirming that the impact of physiology fluctuations on rsfMRI signal is minimal in our study.
The findings of our study resulted from acute, reversible suppression of a hub brain region, which might closely relate to immediate functional network changes in response to acute neural modulations by TMS or tDCs in humans (Guo et al., 2017). Given the clinical achievement with these neural modulation tools (Loo and Mitchell, 2005), acute modulations of neural activity using techniques such as DREADDs and optogenetics could potentially lead to treatment of neuropsychiatric and neurological disorders in the future. However, our study may not correspond to functional network changes due to long-term suppression or permanent loss of function of a local region, such as chronic lesion, which can cause neural compensation and/or network reorganization (Adam et al., 2020; Grefkes and Fink, 2011). Interestingly, a longitudinal study for one year showed that post-stroke patients exhibit a gradually less segregated network during the recovery (Siegel et al., 2018). It will be of our great interest to examine how chronical hub suppression affects whole-brain networks in a longitudinal manner in the future.
Supplementary Material
Acknowledgments
We would like to thank Yucong Ma, Yikang Liu and David Dopfel for their technical support. The present study was partially supported by National Institute of Neurological Disorders and Stroke Grant R01NS085200 (PI: Nanyin Zhang, PhD) and National Institute of Mental Health Grant RF1MH114224 (PI: Nanyin Zhang, PhD).
Footnotes
Declaration of Competing Interest
The Authors declare no competing interests.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2021.118219.
Data availability
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request.
References
- Adam R, Johnston K, Menon RS, Everling S, 2020. Functional reorganization during the recovery of contralesional target selection deficits after prefrontal cortex lesions in macaque monkeys. Neuroimage 207, 116339. [DOI] [PubMed] [Google Scholar]
- Alstott J, Breakspear M, Hagmann P, Cammoun L, Sporns O, 2009. Modeling the impact of lesions in the human brain. PLoS Comput. Biol 5, e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL, 2007. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. U. S. A 104, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic D, Craig MM, Mongerson CRL, Borsook D, Becerra L, 2017. Identifying rodent resting-state brain networks with independent component analysis. Front. Neurosci 11, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E, 2006. Small-world brain networks. Neuroscientist 12, 512–523. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET, 2009. Human brain networks in health and disease. Curr. Opin. Neurol 22, 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher AM, Yen CC, Stepp H, Gu H, Lu H, Yang Y, Silva AC, Stein EA, 2013. Large-scale brain networks in the awake, truly resting marmoset monkey. J. Neurosci 33, 16796–16804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann E, Zur G, Bershadsky G, Kahn I, 2016. The Organization of Mouse and Human Cortico-Hippocampal Networks Estimated by Intrinsic Functional Connectivity. Cereb. Cortex 26, 4497–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Bassett DS, 2018. Specificity and robustness of long-distance connections in weighted, interareal connectomes. Proc. Natl. Acad. Sci. U. S. A 115, E4880–E4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS, 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med 34, 537–541. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA, 2009. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J. Neurosci 29, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci 10, 186–198. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2012. The economy of brain network organization. Nat. Rev. Neurosci 13, 336–349. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Bassett DS, 2011. Brain graphs: graphical models of the human brain connectome. Annu. Rev. Clin. Psychol 7, 113–140. [DOI] [PubMed] [Google Scholar]
- Bush G, Luu P, Posner MI, 2000. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn. Sci 4, 215–222. [DOI] [PubMed] [Google Scholar]
- Chang C, Glover GH, 2009. Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. Neuroimage 47, 1448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PC, Procissi D, Bao Q, Centeno MV, Baria A, Apkarian AV, 2016. Novel method for functional brain imaging in awake minimally restrained rats. J. Neurophysiol 116, 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JE, Lewis LD, Chang C, Tian Q, Fultz NE, Ohringer NA, Rosen BR, Polimeni JR, 2020. Resting-state “physiological networks”. Neuroimage 213, 116707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Carew JD, Turski PA, Moritz CH, Quigley MA, Meyerand ME, 2001. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. AJNR Am. J. Neuroradiol. 22, 1326–1333. [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, Bullmore ET, 2014. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137, 2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaene S, Kerszberg M, Changeux JP, 1998. A neuronal model of a global workspace in effortful cognitive tasks. Proc. Natl. Acad. Sci. U. S. A 95, 14529–14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Ferraro G, Moreno A, Min B, Morone F, Perez-Ramirez U, Perez-Cervera L, Parra LC, Holodny A, Canals S, Makse HA, 2018. Finding influential nodes for integration in brain networks using optimal percolation theory. Nat. Commun 9, 2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopfel D, Perez PD, Verbitsky A, Bravo-Rivera H, Ma Y, Quirk GJ, Zhang N, 2019. Individual variability in behavior and functional networks predicts vulnerability using an animal model of PTSD. Nat. Commun 10, 2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopfel D, Zhang N, 2018. Mapping stress networks using functional magnetic resonance imaging in awake animals. Neurobiol. Stress 9, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folloni D, Verhagen L, Mars RB, Fouragnan E, Constans C, Aubry JF, Rushworth MFS, Sallet J, 2019. Manipulation of Subcortical and Deep Cortical Activity in the Primate Brain Using Transcranial Focused Ultrasound Stimulation. Neuron 101, 1109–1116 e1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M, 2015. The connectomics of brain disorders. Nat. Rev. Neurosci 16, 159–172. [DOI] [PubMed] [Google Scholar]
- Fox MD, Raichle ME, 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci 8, 700–711. [DOI] [PubMed] [Google Scholar]
- Gao SH, Shen LL, Wen HZ, Zhao YD, Chen PH, Ruan HZ, 2020. The projections from the anterior cingulate cortex to the nucleus accumbens and ventral tegmental area contribute to neuropathic pain-evoked aversion in rats. Neurobiol. Dis 140, 104862. [DOI] [PubMed] [Google Scholar]
- Gao YR, Ma Y, Zhang Q, Winder AT, Liang Z, Antinori L, Drew PJ, Zhang N, 2016. Time to wake up: Studying neurovascular coupling and brain-wide circuit function in the un-anesthetized animal. Neuroimage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Nomura EM, Perez F, D’Esposito M, 2012. Focal brain lesions to critical locations cause widespread disruption of the modular organization of the brain. J. Cogn. Neurosci 24, 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DS, Bliss-Moreau E, Machado CJ, Bennett J, Shen K, Grant KA, Fair DA, Amaral DG, 2016. The rhesus monkey connectome predicts disrupted functional networks resulting from pharmacogenetic inactivation of the Amygdala. Neuron 91, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefkes C, Fink GR, 2011. Reorganization of cerebral networks after stroke: new insights from neuroimaging with connectivity approaches. Brain 134, 1264–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Li C, Wang J, 2017. Updated review on the clinical use of repetitive transcranial magnetic stimulation in psychiatric disorders. Neurosci. Bull 33, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C, Ma Y, Zhang N, 2017. Global reduction of information exchange during anesthetic-induced unconsciousness. Brain Struct. Funct 222, 3205–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir MM, Beig MI, Baumert M, Trombini M, Mastorci F, Sgoifo A, Walker FR, Day TA, Nalivaiko E, 2010. Respiratory pattern in awake rats: effects of motor activity and of alerting stimuli. Physiol. Behav 101, 22–31. [DOI] [PubMed] [Google Scholar]
- Kalthoff D, Seehafer JU, Po C, Wiedermann D, Hoehn M, 2011. Functional connectivity in the rat at 11.7T: Impact of physiological noise in resting state fMRI. Neuroimage 54, 2828–2839. [DOI] [PubMed] [Google Scholar]
- Koike H, Demars MP, Short JA, Nabel EM, Akbarian S, Baxter MG, Morishita H, 2016. Chemogenetic inactivation of dorsal anterior cingulate cortex neurons disrupts attentional behavior in mouse. Neuropsychopharmacology 41, 1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V, Marchiori M, 2001. Efficient behavior of small-world networks. Phys. Rev. Lett 87, 198701. [DOI] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2011. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J. Neurosci 31, 3776–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2012a. Anticorrelated resting-state functional connectivity in awake rat brain. Neuroimage 59, 1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2012b. Intrinsic organization of the anesthetized brain. J. Neurosci 32, 10183–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2014. Neuroplasticity to a single-episode traumatic stress revealed by resting-state fMRI in awake rats. Neuroimage 103, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Li T, King J, Zhang N, 2013. Mapping thalamocortical networks in rat brain using resting-state functional connectivity. Neuroimage 83, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Liu X, Zhang N, 2015a. Dynamic resting state functional connectivity in awake and anesthetized rodents. Neuroimage 104, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Ma Y, Watson GDR, Zhang N, 2017. Simultaneous GCaMP6-based fiber photometry and fMRI in rats. J. Neurosci. Methods 289, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Watson GD, Alloway KD, Lee G, Neuberger T, Zhang N, 2015b. Mapping the functional network of medial prefrontal cortex by combining optogenetics and fMRI in awake rats. Neuroimage 117, 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liska A, Galbusera A, Schwarz AJ, Gozzi A, 2015. Functional connectivity hubs of the mouse brain. Neuroimage 115, 281–291. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liang M, Zhou Y, He Y, Hao Y, Song M, Yu C, Liu H, Liu Z, Jiang T, 2008. Disrupted small-world networks in schizophrenia. Brain 131, 945–961. [DOI] [PubMed] [Google Scholar]
- Lohmann G, Margulies DS, Horstmann A, Pleger B, Lepsien J, Goldhahn D, Schloegl H, Stumvoll M, Villringer A, Turner R, 2010. Eigenvector centrality mapping for analyzing connectivity patterns in fMRI data of the human brain. PLoS One 5, e10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo CK, Mitchell PB, 2005. A review of the efficacy of transcranial magnetic stimulation (TMS) treatment for depression, and current and future strategies to optimize efficacy. J. Affect. Disord 88, 255–267. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hamilton C, Zhang N, 2017. Dynamic connectivity patterns in conscious and unconscious brain. Brain Connect. 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Perez P, Ma Z, Liu Y, Hamilton C, Liang Z, Zhang N, 2018. Functional atlas of the awake rat brain: a neuroimaging study of rat brain specialization and integration. Neuroimage 170, 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Zhang N, 2018. Temporal transitions of spontaneous brain activity. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ME, 2002. Assortative mixing in networks. Phys. Rev. Lett 89, 208701. [DOI] [PubMed] [Google Scholar]
- Newman ME, 2004. Fast algorithm for detecting community structure in networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys 69, 066133. [DOI] [PubMed] [Google Scholar]
- Newman MEJ, 2008. The mathematics of networks. In: Durlauf SN, Blume LE (Eds.), The New Palgrave Encyclopedia of Economics, 2nd ed. Palgrave Macmillan, Bas-ingstoke. [Google Scholar]
- Page CE, Coutellier L, 2019. Prefrontal excitatory/inhibitory balance in stress and emotional disorders: Evidence for over-inhibition. Neurosci. Biobehav. Rev 105, 39–51. [DOI] [PubMed] [Google Scholar]
- Peeters LM, Hinz R, Detrez JR, Missault S, De Vos WH, Verhoye M, Van der Linden A, Keliris GA, 2020. Chemogenetic silencing of neurons in the mouse anterior cingulate area modulates neuronal activity and functional connectivity. Neuroimage 220, 117088. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE, 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD, 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron 62, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Seitzman BA, Ramsey LE, Ortega M, Gordon EM, Dosenbach NUF, Petersen SE, Shulman GL, Corbetta M, 2018. Re-emergence of modular brain networks in stroke recovery. Cortex 101, 44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Tamas G, Lujan R, Buhl EH, 1998. Salient features of synaptic organisation in the cerebral cortex. Brain Res. Brain Res. Rev 26, 113–135. [DOI] [PubMed] [Google Scholar]
- Stenroos P, Paasonen J, Salo RA, Jokivarsi K, Shatillo A, Tanila H, Grohn O, 2018. Awake rat brain functional magnetic resonance imaging using standard radio frequency coils and a 3D printed restraint kit. Front. Neurosci 12, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW, 2004. Brain Maps: Structure of the Rat Brain. Elsevier. [Google Scholar]
- Takahashi T, Wood SJ, Yung AR, Soulsby B, McGorry PD, Suzuki M, Kawasaki Y, Phillips LJ, Velakoulis D, Pantelis C, 2009. Progressive gray matter reduction of the superior temporal gyrus during transition to psychosis. Arch. Gen. Psychiatry 66, 366–376. [DOI] [PubMed] [Google Scholar]
- Tu W, Ma Z, Ma Y, Dopfel D, Zhang N, 2021. Suppressing anterior cingulate cortex modulates default mode network and behavior in awake rats. Cereb. Cortex 31, 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Stam CJ, Kahn RS, Hulshoff Pol HE, 2010. Aberrant frontal and temporal complex network structure in schizophrenia: a graph theoretical analysis. J. Neurosci 30, 15915–15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, de Reus MA, 2016. Topological organization of connectivity strength in the rat connectome. Brain Struct. Funct 221, 1719–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O, 2013. Network hubs in the human brain. Trends Cogn. Sci 17, 683–696. [DOI] [PubMed] [Google Scholar]
- van Heukelum S, Mars RB, Guthrie M, Buitelaar JK, Beckmann CF, Tiesinga PHE, Vogt BA, Glennon JC, Havenith MN, 2020. Where is cingulate cortex? A cross-species view. Trends Neurosci. 43, 285–299. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME, 2007. Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83–86. [DOI] [PubMed] [Google Scholar]
- Wang BS, Bernardez Sarria MS, An X, He M, Alam NM, Prusky GT, Crair MC, Huang ZJ, 2021. Retinal and callosal activity-dependent chandelier cell elimination shapes binocularity in primary visual cortex. Neuron 109, 502–515 e507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zuo X, He Y, 2010. Graph-based network analysis of resting-state functional MRI. Front. Syst. Neurosci 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DE, Power JD, Bruss J, Denburg NL, Waldron EJ, Sun H, Petersen SE, Tranel D, 2014. Network measures predict neuropsychological outcome after brain injury. Proc. Natl. Acad. Sci. U. S. A 111, 14247–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH, 1998. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442. [DOI] [PubMed] [Google Scholar]
- Whissell PD, Tohyama S, Martin LJ, 2016. The use of DREADDs to deconstruct behavior. Front. Genet 7, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Mimura Y, Ishihara R, Nishida H, Komaki Y, Minakuchi T, Tsurugizawa T, Mimura M, Okano H, Tanaka KF, Takata N, 2016. Physiological effects of a habituation procedure for functional MRI in awake mice using a cryogenic radiofrequency probe. J. Neurosci. Methods 274, 38–48. [DOI] [PubMed] [Google Scholar]
- Zhang A, Yang C, Li G, Wang Y, Liu P, Liu Z, Sun N, Zhang K, 2020. Functional connectivity of the prefrontal cortex and amygdala is related to depression status in major depressive disorder. J. Affect. Disord 274, 897–902. [DOI] [PubMed] [Google Scholar]
- Zhang N, Rane P, Huang W, Liang Z, Kennedy D, Frazier JA, King J, 2010. Mapping resting-state brain networks in conscious animals. J. Neurosci. Methods 189, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request.


