Abstract
Safe and effective vaccines are urgently needed to stop the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We construct a series of live attenuated vaccine candidates by large-scale recoding of the SARS-CoV-2 genome and assess their safety and efficacy in Syrian hamsters. Animals were vaccinated with a single dose of the respective recoded virus and challenged 21 days later. Two of the tested viruses do not cause clinical symptoms but are highly immunogenic and induce strong protective immunity. Attenuated viruses replicate efficiently in the upper but not in the lower airways, causing only mild pulmonary histopathology. After challenge, hamsters develop no signs of disease and rapidly clear challenge virus: at no time could infectious virus be recovered from the lungs of infected animals. The ease with which attenuated virus candidates can be produced and administered favors their further development as vaccines to combat the ongoing pandemic.
Keywords: coronavirus, SARS-CoV-2, COVID-19, live attenuated vaccine, genome recoding, codon pair deoptimization, synthetic attenuated virus engineering, Syrian hamster, Roborovski dwarf hamster
Graphical abstract
Trimpert et al. construct safe and efficacious live attenuated SARS-CoV-2 vaccine candidates by recoding the SARS-CoV-2 genome. The lead vaccine candidate, sCPD9, is apathogenic in Syrian and Roborovski dwarf hamsters. A single intranasal droplet vaccination with sCPD9 induces strong neutralizing antibody responses and protects hamsters from disease caused by SARS-CoV-2.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 as the causative agent of coronavirus disease 2019 (COVID-19) (Wu et al., 2020; Zhou et al., 2020b). The virus is highly transmissible among humans (Chan et al., 2020). It has spread rapidly around the world within a matter of weeks, and the world is still battling with the ongoing COVID-19 pandemic.
SARS-CoV-2 primarily replicates in the upper respiratory tract (Zou et al., 2020). The infection with SARS-CoV-2 can cause a wide spectrum of clinical manifestations, ranging from asymptomatic to life-threatening disease conditions (Chen et al., 2020; Zhou et al., 2020a). Especially the elderly and patients with pre-existing conditions are at greater risk of developing more severe disease such as pneumonia, acute respiratory distress syndrome, and multiple organ failure (Chen et al., 2020; Garg et al., 2020; Zhou et al., 2020a). The ongoing pandemic imposes an enormous health, psychological, economic, and social burden. To date (June 20, 2021) more than 178 million people have been infected with the virus, of whom more than 3.8 million have died as a result of the infection (https://coronavirus.jhu.edu/map.html) (Dong et al., 2020).
The unprecedented scale and severity of the COVID-19 pandemic prompted the rapid development of novel diagnostic tests, therapeutics, and vaccines that could be used to contain the spread of the virus and limit the pandemic. Globally, 90 vaccines are being tested in clinical trials and 27 of which have reached the final stages of testing (The World Health Organization,)Zimmer et al., 2021). In addition, six vaccines received emergency or limited approval, and eight vaccine received full approval for use in adult humans (The World Health Organization,)(Zimmer et al., 2021). Almost all vaccines that have been or are being evaluated in clinical trials are based either on inactivated or subunit virus preparations (Ella et al., 2021; Gao et al., 2020; Wang et al., 2020; Zhang et al., 2021), replication-defective virus vectors (Emary et al., 2021; Logunov et al., 2021; Solforosi et al., 2021; Voysey et al., 2021b; Zhu et al., 2020), or DNA/RNA molecules (Anderson et al., 2020; Baden et al., 2021; Corbett et al., 2020; Dagan et al., 2021; Jackson et al., 2020; Mulligan et al., 2020; Polack et al., 2020; Sahin et al., 2020; Walsh et al., 2020).
In contrast to most of the vaccines under development, we generated attenuated but replicating SARS-CoV-2 vaccine candidates by genetic modification of the SARS-CoV-2 genome via codon pair deoptimization (CPD) (Coleman et al., 2008). CPD, also known as synthetic attenuation virus engineering, is a virus attenuation strategy that has enabled rapid and highly efficient attenuation of a wide variety of viruses (Broadbent et al., 2016; Coleman et al., 2008; Eschke et al., 2018; Le Nouën et al., 2014; Mueller et al., 2010; Shen et al., 2015; Yang et al., 2013). CPD rearranges the positions of existing synonymous codons in one or more viral genes, without changing the codon bias or amino acid composition of the encoded protein (Coleman et al., 2008; Mueller et al., 2010). Recoded viruses do not produce proteins from the recoded genes as efficiently as their parents and show defects in reproductive fitness, which enables the host to control wild-type virus infection by innate and adaptive immune responses (Mueller et al., 2010; Wimmer et al., 2009). The conserved antigenic identity and replicative potential enable recoded attenuated viruses to fully engage the immune system of the host and provoke strong immune responses.
We generated a series of recoded SARS-CoV-2 mutants, characterized them in cultured cells and also in vivo using Syrian and Roborovski hamster models. We show that a single-dose intranasal immunization with attenuated live viruses can elicit strong immune responses and offer complete protection against SARS-CoV-2 challenge in a robust small animal model of COVID-19.
Results
Vaccine design
Our goal was to generate attenuated SARS-CoV-2 vaccine candidates through large-scale recoding of the SARS-CoV-2 genome by CPD (Coleman et al., 2008; Kunec and Osterrieder, 2016). To achieve virus attenuation in humans, we recoded the genome of SARS-CoV-2 with codon pairs that are the most underrepresented in human genes (Kunec and Osterrieder, 2016). The genetically modified SARS-CoV-2 mutants were produced using a recently established reverse genetics system of SARS-CoV-2 (Thi Nhu Thao et al., 2020) (Figure 1 ). The system relies on 12 subgenomic fragments of the SARS-CoV-2 genome, which are assembled into a single yeast/bacterial artificial chromosome (YAC/BAC) by transformation-associated recombination (TAR) cloning in Saccharomyces cerevisiae (Noskov et al., 2002). The subgenomic fragments are approximately 3,000 bp long, and the neighboring fragments overlap each other by approximately 300 bp to enable the assembly of the SARS-CoV-2 infectious clone by homologous recombination.
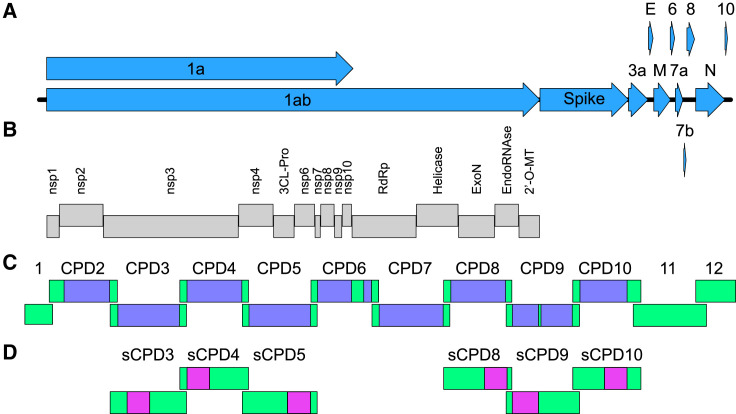
Figure 1.
Structure and recoding of the SARS-CoV-2 genome
(A) The SARS-CoV-2 genome is a single-stranded, positive-sense RNA molecule of about 30,000 nt, which encodes 11 canonical ORFs.
(B) After infection, ORF 1a/1ab is directly translated and cleaved into 15 proteins of the replication-transcription complex.
(C) Recoded SARS-CoV-2 mutants were constructed using a recently established reverse genetics system of SARS-CoV-2, which consists of 12 subgenomic fragments. Fragments 1, 11, and 12 were not recoded. The purple boxes indicate recoded sequences, and the green boxes indicate parental, non-recoded sequences in fragments CPD2-10. The frameshifting element contained in fragment 6 and the transcription regulatory sequence (TRS) of the spike gene in fragment 9 were excluded from the recoding process (green boxes present between two purple boxes in CPD6 and CPD9).
(D) The pink boxes indicate recoded sequences in fragments sCPD3-5 and sCPD8-10. 3CL-Pro, 3C-like proteinase; RdRp, RNA-dependent RNA polymerase; ExoN, 3′-to-5′ exoribonuclease; EndoRNase, endoribonuclease; 2′-O-MT, 2′O-ribose methyltransferase.
To preserve the full compatibility with the available reverse genetics system, we recoded only SARS-CoV-2 sequences that were not present in the overlapping parts of the subgenomic fragments (approximately 2,500 bp in each recoded fragment) (Figure 1; Table S1). This design enabled us to generate a wide variety of SARS-CoV-2 mutants, carrying a single or more recoded fragments.
We recoded nine fragments of the SARS-CoV-2 reverse genetics system (fragments 2–10). Fragments 1 and 12, which are relatively short, 591 and 1,812 bp, respectively, and fragment 11, which contains many short ORFs, were excluded from the recoding. To ensure that mutant viruses are replication-competent, two genomic regions containing essential cis-acting RNA elements were excluded from the recoding: the frameshifting element carried by fragment 6 and the transcription regulatory sequence (TRS) of the spike gene within fragment 9. In addition, the first 500 bp of ORF 1a located in fragment 2 were not recoded either. The characteristics of the parental and recoded sequences are summarized in Table S1.
Recovery of recoded virus mutants
We attempted to rescue infectious progeny from the recoded SARS-CoV-2 constructs by two different methods—directly from DNA in Vero E6 cells and from viral RNA in BHK-21 cells. To enable recovery from DNA, the genome of the SARS-CoV-2 was placed under the control of the immediate-early promoter of the human cytomegalovirus. Despite multiple attempts, we were able to rescue infectious progeny viruses from only two out of eight recoded SARS-CoV-2 constructs, and the rescue occurred only by means of RNA transfection. We rescued infectious viruses that carried recoded fragments 2 and 6 (thereafter labeled as CPD2 and CPD6). It is known that extensive recoding by CPD can lead to a lethal phenotype of recoded mutants (Coleman et al., 2008; Eschke et al., 2018). We suspected that the extent of recoding in constructs that did not produce infectious viruses was too high and, therefore, constructed additional mutants with shorter deoptimized sequences (Figure 1; Table S1). In total, we engineered six SARS-CoV-2 mutant constructs that carried shorter, approximately 1,000-bp-long recoded sequences (sCPD3, sCPD4, sCPD5, sCPD8, sCPD9, and sCPD10). As predicted, reducing the extent of deoptimization led to the rescue of several additional recoded SARS-CoV-2 viruses. Specifically, of the six constructs, we rescued four mutant viruses by means of DNA transfection: sCPD3, sCPD4, sCPD9, and sCPD10.
Characterization of recoded SARS-CoV-2 mutants in cell culture
The morphological changes induced by infection on Vero E6 cells were highly variable among mutant viruses. While CPD6, sCPD3, and sCPD4 viruses produced readily visible plaques on Vero E6 cells within 24 h of infection, sCPD9 and sCPD10 viruses did not produce visible plaques or discernable cytopathic effect (CPE) on Vero E6 cells in the first 72 hpi. Therefore, we determined virus titers and spread of virus mutants in cell-culture monolayers by focus-forming assays. The recoded viruses CPD6, sCPD3, sCPD9 and sCPD10 formed significantly smaller foci of infected cells than the parental virus (Figure 2 A; Figure S1). The foci produced by the sCPD3 and sCPD4 viruses were slightly smaller than those formed by the parental virus. On average, they were about 65% and 80% of the size of those formed by the parental virus. In contrast, foci formed by the CPD6, sCPD9, and sCPD10 viruses were significantly smaller, accounting on average for about 50%, 20%, and 45% of the size of the parental virus, respectively.
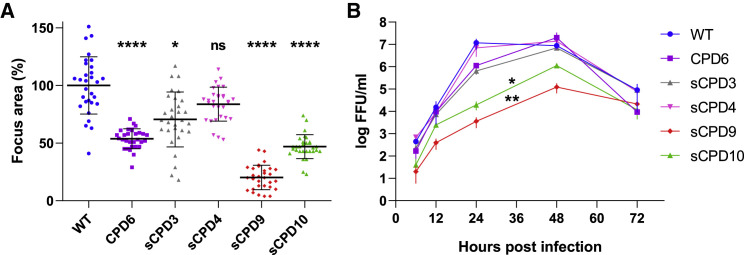
Figure 2.
Areas of infected cell foci and multi-step growth kinetics of the parental and recoded viruses in Vero E6 cells
(A) Relative areas of infected cell foci at 48 hpi. Focus areas were normalized against the average focus size of the parental virus. Bars show geometric mean and SD. p values were calculated using Kruskal-Wallis test with Dunn’s post hoc test, ∗p = 0.0135 and ∗∗∗∗p < 0.0001. See also Figure S1.
(B) Multi-step growth kinetics of the parental and recoded viruses. Vero E6 cells were infected with the parental or recoded viruses at a multiplicity of infection of 0.01. Cell-culture medium was collected 6, 12, 24, 48, and 72 hpi, and virus titers were determined by focus-forming assay. Data are represented as means of three independent biological replicates ± SD. Comparison of growth curves by Friedman test with Dunn’s post hoc test showed that sCPD9 and sCPD10 viruses replicated significantly worse than the parental virus, ∗p = 0.0264 and ∗∗p = 0.0049, respectively.
Multi-step growth kinetics in Vero E6 cells showed that recoded viruses replicated with variable efficiency in Vero E6 cells (Figure 2B). The replication kinetics of the CPD6, sCPD3, and sCPD4 viruses were comparable to those of the parental virus. In contrast, the sCPD9 and sCPD10 viruses replicated at significantly reduced levels at all assessed time points. The mean titers of sCPD9 and sCPD10 viruses were 100–1,000 times lower than those of the parental virus at 24 hpi and 10–100 times lower at 48 hpi.
To assess the genetic stability of the recoded viruses, we passaged the two most attenuated viruses sCPD9 and sCPD10 serially 10 times in Vero E6 cells at a multiplicity of infection (MOI) of 0.1. After 10 passages, the viruses did not appear to have undergone notable phenotypic changes, as they still produced only small foci on Vero E6 cells and did not produce readily observable CPE within 72 hpi. We also analyzed the genomes of the passaged viruses by RT-PCR and Sanger sequencing, and we did not detect any mutations in the recoded regions.
While recoding by codon pair deoptimization does not affect the amino acid sequence of the encoded proteins, it extensively alters the underlying genomic sequence. Suboptimal codon pairs reduce the mRNA stability and translation efficiency of recoded, codon pair deoptimized genes (Groenke et al., 2020; Mueller et al., 2010). This in turn reduces protein production and leads to virus attenuation. To determine whether reduced protein production from recoded genes could be the reason for attenuation of mutant viruses, we examined the production of spike and nucleocapsid proteins in Vero E6 cells infected with wild-type (WT), sCPD9, or sCPD10 viruses. Because the sCPD10 virus carries a codon pair deoptimized spike gene (Figure 1), it is expected to produce comparatively less spike protein than either the WT or sCPD9 viruses. Indeed, spike protein production was significantly reduced in cells infected with sCPD10 virus compared with cells infected with WT or sCPD9 viruses (Figure S2), indicating that CPD reduced protein production of the target gene.
Vaccination of Syrian hamsters and challenge infection setup
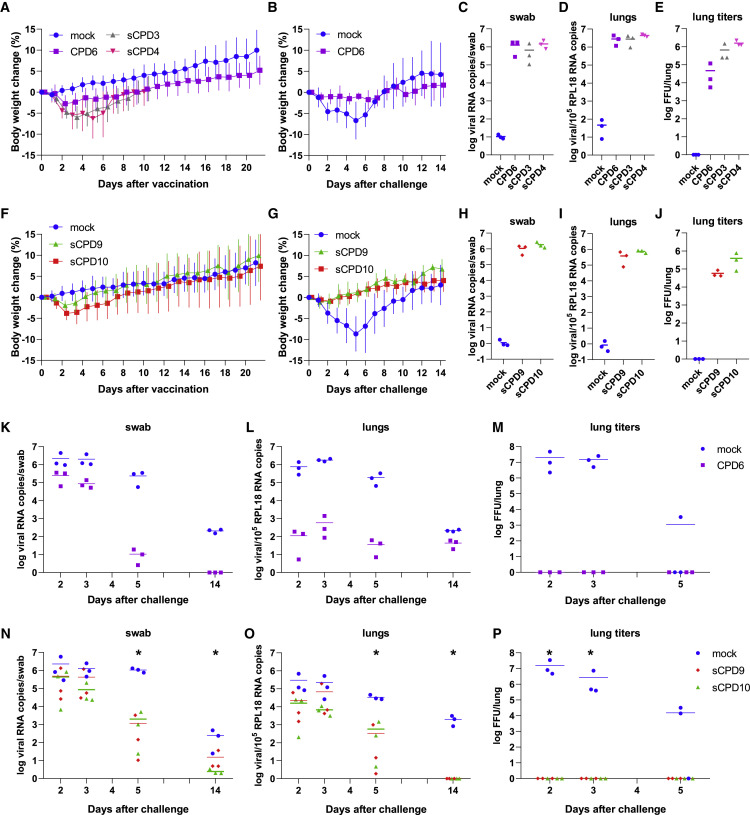
The degree of attenuation of the recoded SARS-CoV-2 mutants was assessed in the Syrian hamster model by a single-dose intranasal infection (Figure 3 ). Hamsters were kept in individually ventilated cages and randomly assigned to four groups of 15 animals. Animals were either mock-vaccinated or vaccinated with 1 × 104 or 1 × 105 focus forming units (FFU) of recoded virus on day 0. Twenty-one days after vaccination, animals were challenged by intranasal infection with 1 × 105 FFU of the WT SARS-CoV-2, variant B.1 (Wölfel et al., 2020). For the duration of the vaccination experiment (35 days), body weights and clinical signs were recorded daily. Three animals of each group were euthanized on day 3 after vaccination, and on days 23, 24, 26, and 35 after vaccination (day 2, 3, 5, and 14 after challenge) to determine the degree of virus replication in different organs and to assess pathological changes in the lungs.
Figure 3.
Attenuation of recoded SARS-CoV-2 mutants in Syrian hamsters
(A–P) The attenuation level of recoded virus vaccine candidates was evaluated in two sequential trials. In the first trial, Syrian hamsters were either mock-vaccinated or vaccinated with CPD6, sCPD3, or sCPD4 viruses (A)–(E) and (K)–(M). In the second trial, Syrian hamsters were either mock-vaccinated, or vaccinated with sCPD9 or sCPD10 viruses (F–J and N–P). Twenty-one days after vaccination, all animals were challenged by infection with WT SARS-CoV-2.
(A, B, F, and G) Change in body weight of Syrian hamsters after vaccination (n = 15); (A) and (F) and challenge (n = 12); (B) and (G). Data are represented as mean ± SD.
(C, D, H, and I) Viral load in the upper (C) and (H) and lower (D) and (H) respiratory tract on day 3 after vaccination.
(E and J) The number of infectious virus particles detected in 50 mg of lung tissue on day 3 after vaccination.
(K, L, N, and O) Viral load in the upper (K) and (N) and lower (L) and (O) respiratory tract of animals on days 2, 3, 5, and 14 after challenge.
(M and P) The number of infectious virus particles detected in 50 mg of lung tissue on days 2, 3, and 5 after challenge. The Kruskal-Wallis test was used to determine whether the differences in viral load among the different groups were significant (N–P), ∗p < 0.05.
Due to space limitations, we examined the attenuation of 5 recoded viruses in two animal trials. In the first trial, hamsters were vaccinated with 1 × 105 FFU of the recoded viruses CPD6, sCPD3, or sCPD4. In the second trial, hamsters were vaccinated with 1 × 104 FFU of the recoded viruses sCPD9, or sCPD10, a 10-fold lower dose than in the first vaccination trial. We used a lower dose because initially we were unable to grow the sCPD9 and sCPD10 mutants to sufficiently high levels, as the peak titers of these mutants in Vero E6 cells were 10–100 times lower than those of the parental virus (Figure 2B).
Recoded viruses CPD6, sCPD9, and sCPD10 are attenuated in Syrian hamsters
On day 3 after vaccination, all tested viruses replicated efficiently in the upper and lower respiratory tract of vaccinated hamsters, as determined by the viral load in oropharyngeal swabs (Figures 3C and 3H) and lung tissues (Figures 3D and 3I). Unexpectedly, despite weaker replication in Vero E6 cells, the recoded viruses sCPD9 and sCPD10 replicated efficiently in the upper respiratory tract on day 3 after vaccination (Figure 3H). In fact, the quantity of detected RNA copies in oropharyngeal swabs (∼106 RNA copies/swab) was comparable among all recoded viruses (Figures 3C and 3H) and is also comparable to the viral loads of the WT virus in mock-infected animals on day 3 after challenge (Figures 3K and 3N). These results indicate that all recoded viruses replicated with comparable efficiency in the upper respiratory tract of infected animals, although the viruses showed markedly different replication capacity in cultured Vero E6 cells and although the animals were vaccinated with dissimilar doses of recoded viruses.
Successful reisolation of replicating virus from the lungs of vaccinated hamsters on day 3 after vaccination confirmed that all recoded viruses also replicated in the lungs of vaccinated animals (Figures 3E and 3J). Virus titers were lower in the lungs of hamsters vaccinated with CPD6 virus than in the lungs of hamsters vaccinated with either sCPD3 or sCPD4 viruses (Figure 3E), indicating that CPD6 was the most attenuated virus of the three viruses examined. We detected less infectious virus in the lungs of hamsters vaccinated with sCPD9 virus than in those vaccinated with sCPD10 virus (Figure 3J), indicating that sCPD9 was more attenuated than sCPD10 virus in vivo. In general, lung virus titers correlated well with virus foci sizes (Figure 2A) and also with lung histopathology on day 3 after vaccination (Figure 4 ).
Figure 4.
Lung histopathology of infected Syrian hamsters 3 days after vaccination
Representative whole cross-sectional scans of left lung lobes (upper row, A–G) and micrographs of bronchial epithelium (bottom row, H–N) of formalin-fixed, paraffin embedded, hematoxylin and eosin-stained tissues. Hamsters were either mock-vaccinated (Mock), infected with SARS-CoV-2 (WT) or vaccinated with viruses CPD6, sCPD3, sCPD4, sCPD9, or sCPD10. Bars: 1 mm (A–G) or 100 μm (H–N).
Hamsters vaccinated with sCPD3 and sCPD4 viruses lost significant body weight from day 2 after inoculation (Figure 3A), and histological examination of the lungs on day 3 revealed a pronounced necrosuppurative bronchointerstitial pneumonia (Figure 4). Four to five days after vaccination, hamsters reached the peak of clinical symptoms and slowly began to recover, resulting in weight gain. As the clinical and histological signs were reminiscent of those caused by infection with pathogenic WT SARS-CoV-2 (Osterrieder et al., 2020), we discontinued the vaccination trial for sCPD3 and sCPD4 viruses on day 10 after vaccination.
The CPD6 virus was moderately attenuated in vivo. Individual hamsters lost up to 6% of their body weight within the first 3 days after vaccination. Vaccinated animals weighed on average 0.4% less on day 1 and 2.8% less on day 2 after vaccination (Figure 3A). From day 3, the animals started to regain the lost weight. In contrast, average daily weight loss was significantly higher in mock-vaccinated hamsters after challenge infection with WT virus in both vaccination trials (Figures 3B and 3G). The average body weight of hamsters decreased by 1.1%, 5.0%, 7.1%, 7.6%, and 9.6% during the first 5 days after challenge infection compared to the day of challenge. In addition, individual hamsters lost up to 14% of their weight during this period (Figures 3B and 3G). In both trials, hamsters began to gain weight from day 5 after challenge.
Because Syrian hamsters were inoculated with a 10-fold lower virus dose in the second animal trial relative to the first trial, the degree of pathogenicity/attenuation of sCPD9 and sCPD10 viruses cannot be directly compared with that of CPD6, sCPD3, or sCPD4 viruses or with that of WT viruses after challenge of mock-vaccinated hamsters in either experiment. Nevertheless, the obtained results show how pathogenic sCPD9 and sCPD10 viruses are to Syrian hamsters when vaccinated with 1 × 104 FFU/animal, and whether this dose can protect vaccinated Syrian hamsters from challenge infection with WT virus.
Syrian hamsters vaccinated with sCPD9 and sCPD10 viruses did not develop any adverse reactions or signs consistent with SARS-CoV-2 infection. Hamsters lost weight during the first 2 days after vaccination, but the average daily weight loss was minor. The weight of animals vaccinated with sCPD9 virus decreased by average of 0.3% on day 1 and 1.9% on day 2 after vaccination (Figure 3F). In hamsters vaccinated with sCPD10 virus, weight loss was more pronounced but still moderate. Animals weighed an average of 1.3% less on day 1 and 3.8% less on day 2 after vaccination compared to baseline weight. We recorded weight gain in both groups on day 3 following vaccination, and when animals had fully recovered, the average daily gains were comparable to those of the mock-vaccinated group.
Vaccination of Syrian hamsters provides complete protection against SARS-CoV-2 challenge
Vaccination with the CPD6 virus induced strong protective immunity against the challenge with the WT SARS-CoV-2. The CPD6-immunized animals did not lose weight (Figure 3B) and showed no signs of disease after the challenge. CPD6-vaccinated animals had lower viral loads in the upper and lower airways on days 2, 3, 5, and 14 after challenge than mock-vaccinated animals (Figures 3K and 3L). To better understand efficacy of vaccination, we also performed virus isolation from lung tissue (Figure 3M). We were able to detect high titers of replicating viruses from lung tissue of mock-vaccinated but not CPD6-vaccinated animals, suggesting that the vaccination with live attenuated CPD6 virus induced an immunity that efficiently prevented replication of the challenge virus in the lungs.
Similarly, vaccination with the sCPD9 and sCPD10 viruses induced strong protective immunity against challenge infection with WT SARS-CoV-2. The vaccinated animals did not lose weight and showed no clinical signs of disease after infection. qRT-PCR analysis indicated active virus replication in the upper airways of infected animals (Figure 3N). On day 5 after challenge, virus loads were approximately 1,000-fold lower when compared to the mock-vaccinated group. Similarly, virus loads in the lower respiratory tract were significantly lower in the vaccinated animals compared to the mock-vaccinated animals (Figure 3O). To evaluate vaccine efficacy, we also tried to isolate the virus from lung tissue of challenged animals. As expected, while we could readily isolate the virus from the tissue samples of the mock-vaccinated animals, no infectious virus was isolated from the lung samples of sCPD9- and sCPD10-vaccinated animals (Figure 3P). However, these animals showed persistent viral loads in the throat, indicating active virus replication (Figure 3N).
Vaccination prevents major tissue damage
Histopathology of lungs 3 days after vaccination identified marked differences in the intensity and distribution of bronchial and lung lesions between animals infected with some of the recoded viruses and the WT virus (Figure 4). However, animals within each group, developed fairly similar lesions, indicating that lungs of animals were effectively infected and that disease outcomes were homogeneous within each group. Whereas sCPD3 and sCPD4 viruses caused degrees of bronchitis and pneumonia that were relatively similar to the WT virus, sCPD10 virus induced less severe pathology, and infection with CPD6 and sCPD9 viruses resulted in weakest pathology and inflammation, both along the bronchi and in the lung tissue (Figure 4).
A more detailed histopathological examination was done to characterize the protective capacity of the attenuated viruses. For this purpose, hamsters vaccinated with CPD6, sCPD9 or sCPD10 viruses and challenged 21 days later with WT SARS-CoV-2 were examined following standard evaluation criteria for the COVID-19 Syrian hamster model (Gruber et al., 2020). The results clearly showed that all three attenuated viruses had overall smaller affected lung areas as well as milder degrees of bronchitis, pneumonia, and other relevant lesions (Figures 5A–5F and 5G–5V). Due to their particular significance in COVID-19 pneumonia, we additionally scored the intensities of endothelialitis as well as perivascular and alveolar edema, which were also markedly reduced in vaccinated hamsters (Figures 5D and 5E). Furthermore, we compared bronchial and alveolar epithelial cell hyperplasia as evidence of tissue regeneration after parenchymal damage. Again, signs indicating the necessity of tissue repair were almost completely missing in hamsters vaccinated with one of the three recoded viruses (Figures 5F and 5O–5R). These observations further supported the notion that all three attenuated viruses had protected hamsters from major tissue damage in a subsequent challenge with the WT virus.
Figure 5.
Lung histopathology of vaccinated Syrian hamsters on days 2–14 after challenge
(A–F) Histopathological evaluation and scoring of lung pathology. Parameters assessed: estimated percentages of affected lung areas (A), degree of bronchitis (B), lung inflammation (C), endothelialitis (D), edema (E), and epithelial hyperplasia (F). Scores and parameters in (B)–(F) were classified as absent (0), minimal (1), mild (2), moderate (3), or severe (4). n = 3 for each treatment at each time point.
(G–V) Representative photomicrographs of formalin-fixed, paraffin-embedded, hematoxylin and eosin-stained lung tissues: bronchial epithelium (G)–(J), air spaces (K)–(V). Insets in (G)–(J) show the distribution of SARS-CoV-2 RNA, visualized by in situ hybridization in lungs of Syrian hamsters on day 2 after challenge. Red signal: viral RNA; blue: hemalaun counterstain. Insets in (O)–(R) show blood vessels affected by endothelialitis on day 5 after challenge. Pathological changes are shown from representative animals that had been either mock-vaccinated (Mock) or vaccinated with CPD6, sCPD9, or sCPD10. Bars: 50 μm (G–V), 1 mm (insets G–J), 100 μm (insets O–R).
In situ hybridization was performed to examine the protective effect of vaccination on the virus load in the lung tissue of hamsters 2 days after challenge, the time when it reaches its peak (Osterrieder et al., 2020). The results clearly demonstrated a complete absence of signals indicative of SARS-CoV-2 RNA throughout the lungs of vaccinated hamsters, whereas a strong positive signal was detected in lungs of mock-vaccinated and challenged hamsters (insets in Figures 5G–5J). It thus seems likely that the drastically reduced lung pathologies in the protected hamsters were due to a markedly diminished initial virus replication in their target cells, which is consistent with the quantity of viral loads in the lungs of vaccinated hamsters as determined by qRT-PCR and virus titration assay (Figures 3K–3P).
Vaccination induces high levels of neutralizing antibodies
SARS-CoV-2 neutralizing antibodies were quantified in sera from vaccinated hamsters by serum virus neutralization assays (Figure 6 ). Analysis showed that mock-vaccinated hamsters had moderate to high titers of neutralizing antibodies as early as day 5 after challenge infection with WT virus (Figure 6). This shows that humoral immunity develops a robust response very quickly after infection with the virus. Moreover, neutralizing titers after challenge infection were very high and remarkably consistent in hamsters that were previously vaccinated with CPD6, sCPD9, or sCP10 viruses (Figure 6). Because the highest dilution we tested was 1:512, we assume individual animals had even higher titers of neutralizing antibodies.
Figure 6.
Titers of SARS-CoV-2 neutralizing antibodies in vaccinated Syrian hamsters
SARS-CoV-2 neutralizing antibodies were quantified in sera of mock-vaccinated hamsters and in hamsters vaccinated with CPD6, sCPD9, and sCPD10 after challenge infection with the WT virus. Sera of naive hamsters were used as a negative control. The dashed lines represents the detection limits of the assay.
Recoded sCPD9 virus is attenuated in Roborovski dwarf hamsters
Because the mutant viruses sCPD9 and sCPD10 replicate with delayed kinetics in Vero E6 cells compared with the parental virus and do not produce visible plaques, we initially were unable to produce virus stocks of the two virus mutants with sufficiently high titers. This drawback prevented us from comparing the pathogenicity of these two viruses with that of parental virus in Syrian hamsters that would be infected with a high virus dose (1 × 105 FFU/animal). To better assess the degree of attenuation of the lead attenuated vaccine candidate sCPD9, we decided to study its pathogenicity in a rodent species that is highly susceptible to severe COVID-19-like disease, the Roborovski dwarf hamster (Phodopus roborovskii) (Trimpert et al., 2020). When infected with WT SARS-CoV-2, Roborovski dwarf hamsters developed fulminant clinical signs 2 to 4 days after infection, with rapid weight loss, pronounced drop of body temperature and signs of respiratory distress (Trimpert et al., 2020).
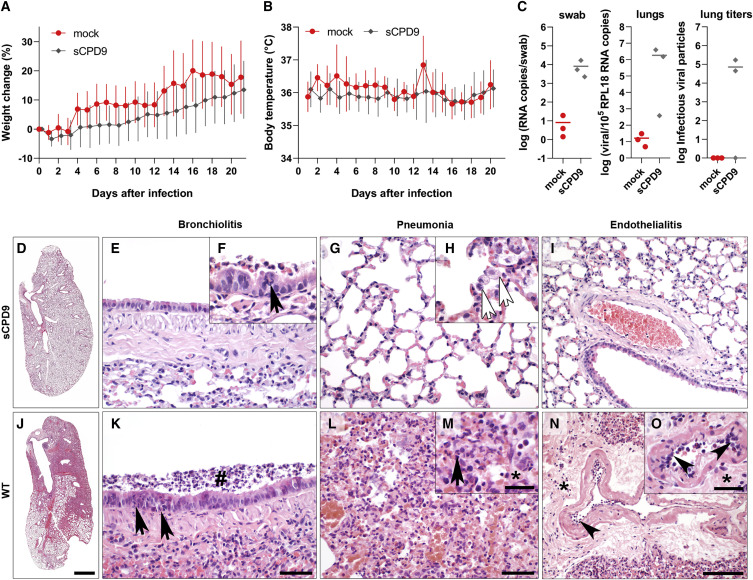
Roborovski dwarf hamsters were randomly assigned to two groups. Twelve hamsters were mock-infected and 30 hamsters were infected with 1 × 105 FFU of the mutant virus sCPD9. Body weight and body temperature were recorded daily for the duration of the experiment (21 days). On average, the sCPD9-infected hamsters lost only little weight on day 1 (3.4%), but began to gain weight already on day 2 and continued to gain weight steadily until the end of the experiment (Figure 7 A). Individual hamsters either lost up to 8.0% weight or gained up to 3.6% weight on day 1 after infection. The mock-infected hamsters did not gain weight during first 3 days after mock infection, but gained weight precipitously on day 4 and steadily thereafter. Daily weight gains were comparable between the two groups, and at the end of this experiment, on day 21, both groups showed comparable weight distribution (Figure 7A). Note that daily weight averages show greater variation in the case of mock-infected hamsters, likely due to the smaller group size. More importantly, no decrease in body temperature (Figure 7B) or other clinical signs, such as forced breathing or apathy, which would indicate severe disease were observed in any of the hamsters vaccinated with the sCPD9 virus (Trimpert et al., 2020). Three animals from each group were euthanized on day 3 after infection, and qRT-PCR analysis confirmed that sCPD9 virus replicated in the upper and lower airways of the infected animals (Figure 7C). Virus reisolation from lung samples was successful in 2/3 of infected sCPD9 animals (Figure 7B).
Figure 7.
Recoded SARS-CoV-2 mutant sCPD9s is strongly attenuated in Roborovski dwarf hamsters
(A) Weights change of mock- (n = 12) and sCPD9-infected hamsters (n = 30). Data show mean ± SD.
(B) Daily body temperature of mock- (n = 12) and sCPD9-infected hamsters (n = 30). Data show mean ± SD.
(C) Viral load in the upper (oropharyngeal swab) and lower (lung) airways and infectious virus particles detected in 50 mg of lung tissue on day 3 after infection (lung titers).
(D–O) Histopathological evaluation of the infection with sCPD9 (D–H) or SARS-CoV-2 variant (WT; I–N) on lung tissues at day 3 after infection. Left lung lobe with mild inflammatory lesions (D). Bronchioli had virtually normal columnar epithelium (E) and only occasional mild necrosuppurative bronchiolitis (F); neutrophil (black arrow). The alveolar septa showed mildly increased numbers of macrophages, and few neutrophils (G) with smaller areas of apparent pneumonia with macrophages (white arrows), neutrophils and necroses of alveolar epithelial cells (H). Normal pulmonary blood vessel (I). Typical lung of hamsters infected with WT virus (J), necrosuppurative bronchiolitis with early hyperplasia of bronchiolar epithelial cells (K), intraluminal cellular debris (hash), infiltrating neutrophils (black arrow). Bronchointerstitial pneumonia (L) with necrosis of alveolar epithelial cells, infiltration by macrophages and neutrophils (M), black arrow) and alveolar edema (M), asterisk). Endothelialitis (arrowhead) with mild to moderate perivascular edema (N, O, asterisk;). Scale bars: 1 mm (D) and (J), 50 μm (E, G, K, and L), 100 μm (I) and (N), 20 μm (insets F, H, M, and O).
Lung histopathology confirms strong attenuation of sCPD9 virus in Roborovski dwarf hamsters
To assess lung pathology induced by sCPD9 infection, three infected Roborovski dwarf hamsters were euthanized on day 3 after infection. Lungs from these animals and Roborovski dwarf hamsters that were euthanized on day 3 after infection with 1 × 105 FFU of the WT SARS-CoV-2 variant B.1 in a previous study (Trimpert et al., 2020) were subjected to a detailed histopathological examination (Figure 7). Although virological parameters confirmed productive viral lower respiratory tract infection in 2/3 of the animals (Figure 7C), the histologic picture was much milder in these animals compared with WT SARS-CoV-2 infection (Figure 7). In Roborovski dwarf hamsters infected with sCPD9 virus, mild inflammatory lesions were visible only in small lung areas (Figure 7D). Of note, pulmonary vessels were devoid of endothelialitis. In sharp contrast, lungs of Roborovski dwarf hamsters infected with the WT SARS-CoV-2 showed extensive inflammation involving up to 70% of the lung tissue (Figure 7J). Detailed analysis revealed that bronchiolitis, pneumonia, and endothelial inflammation were significantly reduced in sCPD9-infected animals (Figures 7E–7I) compared with the WT-infected animals (Figures 7K–7O).
Discussion
Several vaccines have demonstrated efficacy in large-scale phase 3 clinical trials and have been approved for human use, and many others are in the final stages of clinical testing (the most relevant ongoing clinical trials registered with ClinicalTrials.gov are listed in the Table S2) (Baden et al., 2021; Dagan et al., 2021; Ella et al., 2021; Emary et al., 2021; Gao et al., 2020; Logunov et al., 2021; Solforosi et al., 2021; Voysey et al., 2021b; Wang et al., 2020; Zhang et al., 2021; Zhu et al., 2020; (The World Health Organization,)Zimmer et al., 2021). Despite great progress, the majority of the world’s population has not been vaccinated. More than 2.4 billion doses of vaccine have been administered globally as of June 20, 2021, yet there is a wide disparity in vaccination progress between high- and low-income countries. This is because high-income countries, particularly countries in Europe and North America, have secured the majority of the available vaccines. In low- and middle-income countries, only about 1% of the population has been vaccinated, and in some countries not a single dose has been administered (Zimmer et al., 2021). As a result, the COVID-19 pandemic continues to disrupt lives and ravage many different regions of the world. To reduce the social and economic burden of the pandemic, it is necessary to develop, manufacture, and administer billions of safe, effective, and affordable vaccines.
Among the most widely administered vaccines are mRNA vaccines developed by Pfizer–BioNTech (BNT162b2, or Comirnaty) and Moderna (mRNA-1273), non-replicating adenovirus-vectored vaccines developed by the Gamaleya Research Institute (Sputnik V), University of Oxford–AstraZeneca (AZD1222, Vaxzevria or Covishield) and Janssen–Johnson & Johnson (Ad26.COV2.S), inactivated virus vaccines developed by The Beijing Institute of Biological Products–Sinopharm (BBIBP-CorV), Sinovac Biotech (CoronaVac, or PiCoVacc), Wuhan Institute of Biological Products–Sinopharm (unnamed) and the Indian Council of Medical Research–Bharat Biotech (Covaxin) (The World Health Organization,)(Zimmer et al., 2021).
Although no mRNA-based vaccines had been licensed for use in humans previously, they are the most effective and safest of the licensed COVID-19 vaccines. They showed ∼95% efficacy in preventing symptomatic disease, including severe disease, and caused almost no serious side effects in various clinical trials and real-world settings (Baden et al., 2021; Haas et al., 2021; Hall et al., 2021). The disadvantage of mRNA vaccines is their high cost and the need for low storage temperatures (Comirnaty), which makes them unaffordable and logistically impractical for many low-income countries.
Vaxzevria is the most widely administered adenovirus-based vaccine (Zimmer et al., 2021). While the efficacy of the vaccine in preventing symptomatic COVID-19 is significantly lower than the mRNA vaccines at ∼75%, the efficacy against severe disease and hospitalization is 100% (Voysey et al., 2021a). The main advantage of this vaccine is its low, non-profit cost and long stability at standard refrigeration temperatures (2°C –8°C). Very rarely, however, the vaccine has led to the formation of diffuse blood clots with dramatically reduced numbers of platelets (Greinacher et al., 2021). As a result, many countries have paused, restricted, or completely discontinued vaccination with adenovirus-based vaccines, as the adenovirus vector is the suspected trigger of the rare but serious side effect.
Preliminary reports from different clinical trials indicate that inactivated virus vaccines are 50%–80% effective in preventing COVID-19 caused by WT virus (Mallapaty, 2021). The vaccines appear also very safe, as no serious adverse effects have been observed to date. Inactivated virus vaccines are widely administered in China and few other countries. The World Health Organization has approved BBIBP-CorV and CoronaVac for emergency use, which may increase global trust in these vaccines and alleviate the high demand for vaccines, particularly in lower income countries (Mallapaty, 2021).
With the exception of the inactivated virus vaccines, all currently licensed vaccines are based solely on the viral spike glycoprotein. SARS-CoV-2 is evolving rapidly. During the course of the pandemic, a number of viral variants have accumulated the same or similar mutations in the spike protein through convergent evolution. Recent evidence has shown that some of the acquired mutations can compromise the efficacy of licensed vaccines. Viruses from lineages B.1.1.7 (England), B.1.1.28.1 (Brazil), B.1.1.28.3 (Philippines), B.1.525 (West Africa), and B.1.526 (USA) but especially from the lineage B.1.351 (South Africa) have been associated with re-infections and vaccine breakthroughs (Abu-Raddad et al., 2021; Madhi et al., 2021). Widespread vaccination is also expected to increase selective pressure, which could lead to the emergence of SARS-CoV-2 escape variants that cannot efficiently be controlled by vaccination. The emergence of such variants would diminish the efficacy of spike-based vaccines and necessitate vaccine revisions. In fact, Pfizer, AstraZeneca, and Moderna, three of the largest manufacturers of licensed vaccines, are developing updated vaccines that would target the B.1.351 variant (ClinicalTrials.gov Identifier: NCT04785144) (Zimmer et al., 2021).
An important advantage of live attenuated virus vaccines is that they replicate in vaccinated individuals and thus stimulate the immune system to mount an immune response not just against the major surface glycoprotein, but the entire ensemble of virus antigens. Therefore, modified live virus vaccines should theoretically better protect against a wide variety of virus variants, such as the recently emerged B.1.1.7, B.1.351, B.1.1.28.1, and B.1.617.2 variants, because they stimulate a broad antibody and cytotoxic repertoire. Our preliminary data suggest that single-dose vaccination with the sCPD9 mutant indeed induces strong protection. In addition, live attenuated virus vaccines can be administered intranasally and thus induce mucosal immune response directly at the site of virus entry. This may offer better protection of target tissues from infection and could further limit disease severity and virus shedding. For similar reasons, the University of Oxford and CanSino Biologics are launching clinical trials to determine whether nasal administration of the Vaxzevria and Ad5-nCoV vaccines, respectively, would increase protection against infection and limit virus transmission (ClinicalTrials.gov Identifiers: NCT04816019, NCT04840992).
Live attenuated virus vaccines also offer many practical advantages that can play an important role in pandemic response in countries where price, distribution, and administration of the vaccine are important factors. The important advantage of the modified live virus vaccine against SARS-CoV-2 is that the production, storage, distribution, and administration of such a vaccine is relatively simple.
Although the viral stocks of sCPD9 and sCPD10 viruses that we prepared initially had relatively low titers (2 × 105 FFU/mL), we discovered that it was possible to significantly increase the viral yield (1 × 107 FFU/mL) by optimizing the infection of cells and the timing for virus harvest. SARS-CoV-2 replicates rapidly to relatively high titers in Vero E6 cells, allowing large number of vaccine doses to be produced within a short time. In addition, our experiments have shown that vaccination with a relatively low viral dose (2 × 104 FFU/animal) fully protected Syrian hamsters from WT challenge. It is conceivable that an optimal vaccine dose is also relatively low for humans. This would mean that a large number of vaccine doses could be produced very quickly and inexpensively form a small number of cells. Vero cells are easily cultured and have been approved by many regulatory agencies for the production of various human vaccines, including recent inactivated SARS-CoV-2 vaccines (Ella et al., 2021; Zhang et al., 2021). In addition, the biosafety level for highly attenuated viruses could be downgraded from the current level 3 to level 2, which would further facilitate the production and transport of such vaccines. Frozen or freeze-dried virus is stable—the virus titers drop only moderately after thawing or reconstitution—and the vaccination can be easily administered to patients in the form of nasal drops or sprays. A single-dose vaccine against SARS-CoV-2 offers a strong logistical advantage over a two-dose vaccine for mass vaccination campaigns against the COVID-19 pandemic.
The main disadvantage of live attenuated virus vaccines is the risk of reversion to virulence. For example, the oral poliovirus vaccine has 51 mutations, but only 5 of them are attenuating. As a result, this vaccine is prone to reversion, and in rare cases it reverted to virulence and caused paralytic poliomyelitis in vaccinated individuals (Kew et al., 2005). In contrast, recoded viruses contain hundreds of nucleotide mutations, but many of these mutations are attenuating. It is believed that attenuation of CPD viruses occurs through the addition of small defects that each underrepresented “bad” codon pair exerts on gene expression of the recoded gene. Due to the large number of mutations, CPD viruses are considered genetically stable and unlikely to revert to parental virulence. Nevertheless, it is foreseeable that vaccination of large numbers of individuals could increase the chances of reversion. Another theoretical possibility is that recoded mutants could adapt to humans and become eventually endemic in the human population. Further in-depth studies would need to clarify whether recoded SARS-CoV-2 mutants have the propensity to revert to virulence, before live attenuated vaccines can be widely used in humans.
None of the most advanced vaccine candidates is a modified live or attenuated virus vaccine (The World Health Organization,). From the three basic types of viral vaccines, modified live virus vaccines are considered the most efficacious vaccines for healthy individuals and generally outperform inactivated, vectored or subunit vaccines, because they generally provoke broad, strong, and durable immune responses, which are qualitatively identical to those induced by a natural infection with WT strains (Bazin, 2003; Kusters and Almond, 2008; Lauring et al., 2010). Traditionally, modified live virus vaccines have been prepared empirically by the iterative process of serial passage of a virulent virus in cell culture and/or in laboratory animals (Kusters and Almond, 2008; Lauring et al., 2010). The attenuation by CPD represents an alternative to approaches that rely on serendipity as the main attenuating principle. The attenuating mutations are introduced into viral genomes deliberately, based on a rational design (Coleman et al., 2008). In CPD, viral genes are recoded with statistically underrepresented codon pairs, which in turn perturb gene expression and causes attenuation of the recoded virus (Coleman et al., 2008; Groenke et al., 2020). Codon pair bias is a species-specific feature, but phylogenetically closely related species have similar codon pair bias. For example, all mammals have nearly identical codon pair bias (Kunec and Osterrieder, 2016). Thus, mutant viruses carrying genomic sequences that have been codon pair deoptimized on the basis of human codon pair bias should be attenuated not only in human cells but also in all permissive cells derived from different mammalian species (Groenke et al., 2020). Our results are consistent with this assumption, as recoded SARS-CoV-2 mutants were attenuated in cell derived from the African green monkey but also in two different rodent species.
Because CPD substantially alters genomic sequence, additional phenomena may contribute to the attenuation of recoded viruses. For example, recoding can increase the number of CpG and UpA dinucleotides in recoded sequences, which then activate innate host defense mechanisms (Odon et al., 2019). In addition, recoding may destroy unknown cis-regulatory regions, thereby dampening viral replication (Song et al., 2012). Alternative attenuation strategies based on large-scale recoding of viral genomes take advantage of this knowledge and intentionally increase the number of CpG or UpA dinucleotides, rare or “near-stop” codons in viral genomes, as these (interrelated) modifications often lead to the generation of replication-competent but severely attenuated viruses (Lauring et al., 2012; Osterrieder and Kunec, 2018)
Our data demonstrate that a single-dose vaccination with CPD6, sCPD9, or sCPD10 viruses protects vaccinated hamsters from developing disease after challenge infection with WT SARS-CoV-2 (Figures 3, 4, 5, 6, and 7). This was evidenced by histological evaluation of lung pathology, which showed virtually no sign of challenge virus replication after vaccination with CPD6, sCPD9, and sCPD10. Pathological lesions could be detected only in a small part of the lung tissue and generally revealed only mild bronchitis, pneumonia, and edema (Figures 5A–5C). Similarly, epithelial hyperplasia was very limited in vaccinated animals at later times after challenge. This absence of cellular repair and regeneration in the lung strengthens the conclusion that all three vaccine candidates had protected the hamsters from relevant tissue damage caused by challenge virus infection (Figure 5F).
The amount of viral RNA detected in the lungs of challenged animals was very low on days 2, 3, and 5 after the challenge, and, more importantly, we could not isolate a replicating virus from the lungs of the animals after challenge infection (Figures 3K–3P). Our data thus show that, although recoded viruses were unable to protect the upper respiratory tract from reinfection with the virulent WT virus, they did protect the lower respiratory tract very well from infection.
We surmise that a single-dose vaccination with a live modified attenuated virus should provide protective immunity against the infection with pathogenic SARS-CoV-2, as we were unable to detect infectious virus particles in the lungs of any of the vaccinated animals. CPD6-, sCPD9-, and sCPD10-vaccinated animals did not lose any weight after challenge, and histopathological evaluation of the lungs shows clear protective effect of all three vaccine candidates tested. The same data also suggest that a two-dose regimen should also be possible if the development of a more permanent immunity were desirable.
Because sCPD9 was the most attenuated mutant virus examined, but still induced strong protective immunity to disease by the WT virus, we decided to investigate the safety of this virus in a recently identified non-transgenic rodent model of COVID-19, the Roborovski dwarf hamster, which mirrors severe human cases of COVID-19 and thus provides unprecedented sensitivity for preclinical testing of vaccine candidates (Trimpert et al., 2020). This experiment proved that the sCPD9 virus is highly attenuated, as the virus did not induce signs of disease in any of the 30 Roborovski dwarf hamsters and caused only minimal pathological changes in the lungs of infected animals.
Further studies will have to address the durability of protective immunity in vaccinated animals. A known limitation of this study is that we have not evaluated how well the live attenuated viruses can transmit from infected animals to sentinels. While Syrian hamsters readily transmit WT SARS-CoV-2 to co-housed, uninfected contact animals in the first days after infection (Sia et al., 2020), we did not observe any clinical and only limited histological signs of disease in naturally infected contact animals. In our opinion, the Syrian hamster model has limited utility for studying the spread of attenuated viruses and the disease outcomes of infection by natural transmission. Therefore, for the purpose of this study, we focused on characterizing attenuation of the lead vaccine candidate sCPD9 by experimental high-dose infection of a species that is highly susceptible to severe COVID-19-like disease. In addition, we are preparing experiments in ferrets, a well-described model for studying the spread of respiratory viruses (Kutter et al., 2021), to answer questions related to spread of recoded viruses as well as the efficacy against recent SARS-CoV-2 variants of concern in the near future.
In summary, of all the engineered live attenuated viruses tested, sCPD9 showed the best balance between attenuation, safety, immunogenicity, and protective efficacy. We believe that the safety, immunogenicity, and vaccine efficacy of this candidate should be investigated in different animals, non-human primates and possibly also humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monocolonal anti-SARS-CoV-2 nucleocapsid antibody | Sven Reiche, Friedrich Loeffler Institute, Riems, Germany | Cat#4E10A3A |
| Mouse monoclonal anti-SARS-CoV/SARS-CoV-2 (S2) antibody [1A9] | GenTex | Cat#GTX632604; RRID:AB_2864418 |
| Goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#A-11004; RRID:AB_2534072 |
| Goat anti-mouse, horseradish peroxidase conjugated antibody | Merck | Cat#A9917; RRID:AB_258476 |
| Bacterial and virus strains | ||
| Severe acute respiratory syndrome coronavirus 2 | Wölfel et al., 2020 | BavPat1/2020 |
| Escherichia coli strain GS1783 | (Tischer et al., 2010) | N/A |
| Biological samples | ||
| Hamster lungs, swabs, blood, intestines, kidneys | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| high-fidelity polymerase PrimeSTAR GXL | Takara Bio | Cat#R050B |
| Xfect single shots transfection reagent | Takara Bio | Cat#631366 |
| Avicel | FMC BioPolymer | N/A |
| Critical commercial assays | ||
| innuPREP Virus RNA Kit | Analytic Jena | Cat#845-KS-4700250 |
| NEB Luna Universal Probe One-Step RT-qPCR kit | New England Biolabs | Cat#E3006L |
| Monarch DNA Gel Extraction Kit | New England Biolabs | Cat#T1020S |
| Monarch PCR & DNA Cleanup Kit | New England Biolabs | Cat#T1030S |
| mMESSAGE mMACHINE T7 transcription kit | Thermo Fisher Scientific | Cat#AM1344 |
| Amersham ECL prime western blotting detection reagent | Thermo Fisher Scientific | Cat# RPN2236 |
| Experimental models: Cell lines | ||
| African green monkey: Vero E6 cells | ATCC | Cat#CRL-1586 |
| Hamster: BHK-21 [C-13] | ATCC | Cat#CCL-10 |
| Saccharomyces cerevisiae: strain VL6-48N | Noskov et al., 2002 | PMID: 11788734 |
| Experimental models: Organisms/strains | ||
| model organism: Syrian hamster (Mesocricetus auratus) | Janvier Labs | RjHan:Aura |
| model organism: Roborovski dwarf hamster (Phodopus roborovskii) | German pet trade | N/A |
| Oligonucleotides | ||
| RT-qPCR primers and probe | Corman et al., 2020 | N/A |
| Synthetic probes for the in situ-detection of the nucleocapsid RNA of SARS-CoV-2 (NCBI database NC_045512.2, nucleotides 28,274 to 29,533) | Thermo Fisher Scientific | assay ID: VPNKRHM |
| Recombinant DNA | ||
| synthetic reverse genetics platform of SARS-CoV-2 | Thi Nhu Thao et al., 2020 | N/A |
| Plasmid for expression of nucleocapsid of SARS-CoV-2 pVITRO2-EGFP-N | This paper | N/A |
| SARS-CoV-2 mutants containing recoded, codon pair deoptimized sequences | This paper | N/A |
| Deposited data | ||
| SARS-CoV-2 recoded, codon pair deoptimized sequences | GenBank | MZ064531 - MZ064546 |
| Software and algorithms | ||
| GraphPad Prism, Version 8.4 | GraphPad Software | https://www.graphpad.com |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| perl scripts for codon pair deoptimization | Kunec and Osterrieder, 2016 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dusan Kunec (dusan.kunec@fu-berlin.de).
Materials availability
All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cells and viruses
Vero E6 (ATCC CRL-1586) and BHK-21 (ATCC CCL-10) cells were grown in minimal essential medium (MEM) containing 10% fetal bovine serum (PAN Biotech), 100 IU/ml penicillin G and 100 μg/ml streptomycin (Carl Roth) at 37°C and 5% CO2. Parental and mutant SARS-CoV-2 were grown in Vero E6 cells. The ancestral SARS-CoV-2 variant B.1 (SARS-CoV-2/München-1.1/2020/929) (Wölfel et al., 2020) was used as a challenge virus. Virus stocks were stored at −80°C prior to experimental infections.
Ethics statement
In vitro and animal work was done under biosafety conditions in the BSL-3 facility at the Institut für Virologie, Freie Universität Berlin, Germany. All animal experiments were approved by the Landesamt für Gesundheit und Soziales in Berlin, Germany (permit number 0086/20) and done in compliance with relevant national and international guidelines for care and humane use of animals.
Animal husbandry
The animal experiments were done in a certified BSL-3 facility. Six-week-old Syrian hamsters (Mesocricetus auratus; breed RjHan:AURA) were purchased form Janvier Labs, and they were kept in individually ventilated cages (IVCs; Tecniplast), which were equipped with generous enrichment (Carfil). Five- to seven-week-old Roborovski dwarf hamsters (Phodopus roborovskii) were purchased through the German pet trade from a single breeding facility. They were housed in groups of 6 animals in IVCs. The animals had unrestricted access to food and water and were allowed to acclimate to the conditions for seven days prior to infection. During both experiments, cage temperatures ranged from 22 to 24°C and relative humidity ranged from 40 to 55%.
Infection experiments
To study the attenuation and vaccine protection of recoded virus mutants in Syrian hamsters, male and female hamsters were randomly assigned into groups of 15 animals, with each group containing 7 or 8 males and female animals. In the first trial, hamsters were either mock-vaccinated or vaccinated with CPD6, sCPD3 or sCPD4 viruses. Hamsters were mock-vaccinated with 60 μl medium from uninfected Vero E6 cells or vaccinated with 1 × 105 FFU of the mutant virus in 60 μl by intranasal instillation under anesthesia (Osterrieder et al., 2020). In the second trial, hamsters were either mock-vaccinated or vaccinated with sCPD9 or sCPD10 mutant viruses. The vaccination was done as described above, but vaccinated hamsters received only 1 × 104 FFU of the mutant virus. Twenty-one days after vaccination hamsters were challenged by intranasal instillation with 1 × 105 FFU of the challenge virus (variant B.1, strain SARS-CoV-2/München-1.1/2020/929) in 60 μl. During the experiment, all hamsters were monitored twice daily for the clinical signs of disease, and body temperatures and body weight were recorded. Hamsters that had a body weight loss of more than 10% weight over a 72-h period were euthanized in compliance with the animal use protocol. On day 3, and on days 23, 24, 26 and 35 after infection (2, 3, 5 and 14 days after challenge) three hamsters of each group were euthanized (Nakamura et al., 2017). Blood, oropharyngeal swabs and lungs (left and right) were collected for virus titrations, RT-qPCR and/or histopathological examinations. All organs were preserved in 4% formalin for subsequent in-depth histopathological investigations.
To assess the pathogenicity of the recoded mutant virus sCPD9 in the Roborovski dwarf hamster, animals were randomly assigned into two groups, with 60% of the animals in each group being female. Twelve hamsters were mock-infected with cell culture medium and 30 hamsters were infected with 1 × 105 FFU of the mutant virus sCPD9. Anesthetized hamsters were infected as described above, except that the inoculum was 20 μL. Infected hamsters were monitored twice daily for clinical signs of infection. Body weight and temperature was recorded daily. Three hamsters were sacrificed on day 3 after infection to determine virological and histological parameters of infection on day 3 after vaccination. Infected hamsters were monitored twice daily for clinical signs of infection, and body weight and temperature were recorded once daily. Three hamsters from each group were euthanized on the third day after infection to determine virologic and histologic parameters of infection by molecular and virologic assays as described above.
Method details
Recoding of the SARS-CoV-2 genome
Because we planned to make SARS-CoV-2 mutants via TAR cloning, we designed recoded SARS-CoV-2 fragments to be fully compatible with the available reverse genetics system of SARS-CoV-2 (Thi Nhu Thao et al., 2020). All indicated sequence positions in the SARS-CoV-2 genome correspond to the SARS-CoV-2 reference sequence NC_045512.2.
The coding sequences of SARS-CoV-2 were recoded by CPD (Coleman et al., 2008). CPD rearranges the positions of synonymous codons in recoded viral sequences, thereby creating codon-pair combinations that are statistically underrepresented in protein-coding sequences of the virus host. Because our goal was to generate SARS-CoV-2 mutants that should be attenuated for humans, we recoded SARS-CoV-2 sequences using the codon pair score (CPS) calculated for human protein coding genes (Groenke et al., 2020).
The subgenomic fragments were recoded individually, only using the existing codons present in each respective fragment. The creation of novel transcription regulatory sequence (TRS; ACGAAC) and EagI restriction enzyme sites (CGGCCG) in recoded sequences was disallowed. The generation of novel TRS sites was disallowed not to cause aberrant transcription and EagI restriction enzyme sites were reserved for the release of subgenomic fragments from plasmids.
Two genomic sequences containing essential cis-acting RNA elements were excluded from the recoding. The first sequence that was omitted from recoding was the 503 bp sequence (NC_045512.2; nucleotides 13,451-13,953) containing the frameshift stimulation element (FSE). In the SARS-CoV-2 genome the FSE is present between the overlapping genes ORF1a and ORF1b and is essential for −1 programmed ribosomal frameshifting. The 106 bp sequence containing the transcription regulatory sequence (TRS) of the Spike gene (NC_045512.2; nucleotides 21,505-21,610) was also omitted from the recoding. The TRS of the spike gene is located at the end of the ORF1b of the SARS-CoV-2 genome. The non-recoded sequences are carried by fragments 6 and 9 of the yeast-based SARS-CoV-2 reverse genetics system, respectively (Figure 1). The characteristics of the parental and recoded sequences are summarized in Table S1 and sequences of the recoded DNA fragments have been deposited to the NCBI GenBank database (MZ064531-MZ064546).
Recovery of mutant SARS-CoV-2
Infectious viruses were recovered either directly from BAC/YAC SARS-CoV-2 DNA in Vero E6 cells, or from in vitro transcribed viral RNA in BHK-21 cells. To rescue virus progeny from DNA, the immediate-early promoter of cytomegalovirus was inserted upstream of the SARS-CoV-2 genome in BAC/YAC SARS-CoV-2 clones, exactly as described previously (Almazán et al., 2000; Noskov et al., 2002). To promote the recovery of infectious viruses and to monitor transfection efficiency we constructed a dual expression plasmid pVITRO2-EGFP-N which expresses EGFP and SARS-CoV-2 nucleocapsid protein from two different eukaryotic promoters. To recover infectious virus Vero E6 cells were grown to 95% confluence in a T25 cell culture flask and transfected with 4 μg of BAC/YAC DNA and 1 μg pVITRO2-EGFP-N plasmid using the Xfect single shots transfection reagent (Takara Bio Inc.)
To recover infectious viruses in BHK-21 cells, viral RNA was produced by in vitro transcription using the mMESSAGE mMACHINE T7 transcription kit (Thermo Fisher Scientific). Ten μg of in vitro transcribed viral RNA and 1 μg of pVitro2-EGFP-N plasmid were mixed with 1 × 106 BHK-21 cells resuspended in 100 μL OptiPro medium (Thermo Fisher Scientific) and electroporated in a 2 mm gap cuvette by applying one pulse of 140 V for 25 ms in a Gene Pulser Xcell (Bio-Rad Laboratories). Electroporated BHK-21 cells were co-cultured with susceptible Vero E6. Recovered progeny virus was collected from cell culture medium and grown on Vero E6 cells.
Multi-step growth kinetics
To assess the kinetics of virus growth, Vero E6 cells grown in 6-well plates were infected with the parental or recoded viruses at a multiplicity of infection (MOI) of 0.01. After 2 h incubation the viral inoculum was removed and replaced by growth medium. Six, 12, 24, 48 and 72 hours after infection, cell culture medium was collected and virus titers were determined by focus forming assays.
Virus titrations, indirect immunofluorescence and focus-forming assay
To determine virus titers as focus-forming units (FFU) and foci sizes, Vero E6 cells grown in 12-well plates were infected with 100 μl of serial 10-fold dilutions of virus. To determine virus titers in the lung, 50 mg lung tissue was first homogenized with a bead mill (Analytic Jena) and the homogenate was serially diluted. After 2 h of incubation the viral inoculum was removed and cells were overlaid with MEM containing 0.6% microcrystalline cellulose Avicel (FMC BioPolymer). Forty-eight hours after infection cells were fixed with 4% formalin, permeabilized with 0.1% Triton X-100, and blocked with 3% BSA in PBS. Cells were then incubated with a monoclonal mouse anti-SARS-CoV-2 nucleocapsid antibody (provided by Sven Reiche, Friedrich Loeffler Institute, Riems, Germany) for 1 h, and then with goat anti-mouse IgG-Alexa Fluor 568 secondary antibody (Invitrogen) for 45 min. To determine the cell-to-cell spread of mutant viruses in cells, images of 30 randomly selected foci of infected cells were taken at 50-fold magnification using an inverted fluorescence microscope (Axiovert S100, Zeiss). The foci areas were measured using ImageJ software (Schneider et al., 2012), from which the diameters were calculated.
Histopathology and in situ-hybridization
For histopathology and localization of viral RNA by in situ-hybridization (ISH), the left lung lobe was carefully removed and immersion fixed in buffered 4% formalin, pH 7.0, for 48 h. Lungs were embedded in paraffin and cut at 2 μm thickness. Sections were stained with hematoxylin and eosin (HE) and periodic acid-Schiff (PAS) reaction followed by blinded microscopic evaluation by board certified veterinary pathologists (K.D., A.D.G.) (Gruber et al., 2020). For ISH, the ViewRNA ISH Tissue Assay Kit (Thermo Fisher Scientific) was used following the manufacturer’s instructions with minor adjustments. Probe for the detection of the N gene RNA of SARS-CoV-2 (NCBI database NC_045512.2, nucleotides 28,274 to 29,533, assay ID: VPNKRHM) was employed. Lung sections of 2 μm thickness mounted on adhesive glass slides were dewaxed in xylol and dehydrated in graded ethanol. Tissues were incubated at 95°C for 10 min and subsequently protease digested for 20 min. Sections were fixed with 4% paraformaldehyde dissolved in phosphate buffered saline (PBS) and hybridized with the probes. Amplifier and label probe hybridizations were performed according to the manufacturer’s instructions using fast red as chromogen. Sections were counterstained with hematoxylin for 45 s, washed in tap water for 5 min, and mounted with Roti®-Mount Fluor-Care DAPI (4, 6-diaminidino-2-phenylindole; Carl Roth). An irrelevant probe for the detection of streptococcal pneumolysin was used as a control for sequence-specific binding. HE and PAS-stained and ISH slides were analyzed and photographed using an Olympus BX41 microscope with a DP80 Microscope Digital Camera and the cellSens Imaging Software, Version 1.18 (Olympus Corporation-). For overviews with lower magnification, slides were automatically digitized using the Aperio CS2 slide scanner (Leica Biosystems Imaging Inc.) and image files were generated using the Image Scope Software (Leica Biosystems Imaging Inc.).
RNA isolation and RT-qPCR
RNA was extracted from 25 mg lung homogenates and oropharyngeal swabs using the innuPREP Virus RNA kit (Analytik Jena AG). SARS-CoV-2 RNA copies were quantified using the NEB Luna Universal Probe One-Step RT-qPCR kit (New England Biolabs) on a StepOnePlus RealTime PCR System (Thermo Fisher Scientific) as previously described (Corman et al., 2020).
Serum virus neutralization assay
Neutralization assay was performed by two-fold serial dilutions (1:4 to 1:512) of complement inactivated (56°C, 2 h) hamster serum plated on sub-confluent monolayers of Vero E6 cells in 96 well plates. 50 FFU SARS-CoV-2 were added per well and incubated for 72 h at 37°C, fixed with 4% formalin for 24 h and stained with crystal violet (0.75% aqueous solution). Serum neutralization was considered effective in wells that did not show any cytopathic effect, the last effective dilution was counted.
Western blotting
Confluent Vero E6 cells grown in 6-well plates were infected with an MOI of 0.1. After 48 h, infected cells were frozen at −80°C, resuspended in cell culture supernatant and lysed in 2 × Laemmli buffer containing protease inhibitors (Roche). Cell lysates were incubated at 95°C for 30 min, and proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) under reducing conditions. Proteins were transferred to PVDF membranes by semi-dry blotting. Membranes were blocked for 1 h at room temperature in 3% milk in PBS containing 0.05% Tween-20 and then incubated with the primary antibody for 16 h at 4°C. Nucleoprotein was detected with a mouse monoclonal anti-N antibody (see above) and spike protein with a mouse monoclonal anti-S2 antibody (1A9, GeneTex). Horseradish peroxidase-conjugated anti-mouse IgG (1:2000, Merck) was used as a secondary antibody. Antibody binding was visualized using chemiluminescent Amersham ECL prime western blotting detection reagent (Thermo Fisher Scientific).
Quantification and statistical analysis
Statistical tests were performed using GraphPad/Prism. Statistical details of experiments can be found in the figure legends. Exact values of n and statistical tests are indicated in figure legends. P values are indicated in Figures. Data are presented as mean ± SD.
Acknowledgments
We thank Ann Reum, Annett Neubert, Angela Linke, and Michaela Scholz for excellent technical assistance. We thank Sven Reiche and Friedrich-Loeffler-Institut for providing anti-SARS-CoV-2 nucleocapsid antibody. We thank Christa Thöne-Reineke for expert support in animal welfare and husbandry and Carfil Inc. for their support of our animal husbandry. We thank Christian Drosten for bringing together the scientific groups involved in this study. This research was supported by the Deutsche Forschungsgemeinschaft (DFG), grant OS 143/16-1 and COVID-19 grants from Freie Universität Berlin and Berlin University Alliance awarded to N.O., the DFG grant SFB-TR84/Z01b awarded to A.D.G. and J.T., and the Swiss National Science Foundation, grants 31CA30_196644, 4078P0_198473, and 310030_173085 awarded to V.T.
Author contributions
D.K., J.T., J.J., V.T., and N.O. conceived, designed, and supervised the project. D.K., J.T., N.E., T.T.N.T., D.V., S.K., F.L., A.A., A.C., T.H., J.M.A., L.D.B., J.J., and N.O. constructed and characterized viral mutants in cell culture. J.T. and D.V. performed the animal trials. K.D., T.C.F., and A.D.G. performed histopathological analysis. D.K., K.D., A.D.G., and N.O. wrote the manuscript.
Declaration of interests
Freie Universität Berlin and the University of Bern received funding from a commercial partner for research similar to that described in this manuscript.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: July 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109493.
Supplemental information
Data and code availability
Sequences of the recoded SARS-CoV-2 genes have been deposited in the NCBI’s GenBank database (GenBank: MZ064531 - MZ064546). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for COVID-19 Vaccination Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., González J.M., Pénzes Z., Izeta A., Calvo E., Plana-Durán J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., mRNA-1273 Study Group Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin H. A brief history of the prevention of infectious diseases by immunisations. Comp. Immunol. Microbiol. Infect. Dis. 2003;26:293–308. doi: 10.1016/S0147-9571(03)00016-X. [DOI] [PubMed] [Google Scholar]
- Broadbent A.J., Santos C.P., Anafu A., Wimmer E., Mueller S., Subbarao K. Evaluation of the attenuation, immunogenicity, and efficacy of a live virus vaccine generated by codon-pair bias de-optimization of the 2009 pandemic H1N1 influenza virus, in ferrets. Vaccine. 2016;34:563–570. doi: 10.1016/j.vaccine.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernán M.A., Lipsitch M., Reis B., Balicer R.D. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ella R., Reddy S., Jogdand H., Sarangi V., Ganneru B., Prasad S., Das D., Raju D., Praturi U., Sapkal G. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect. Dis. 2021;21:950–961. doi: 10.1016/S1473-3099(21)00070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., Blane B., Bonsall D., Cicconi P., Charlton S., COVID-19 Genomics UK consortium. AMPHEUS Project. Oxford COVID-19 Vaccine Trial Group Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschke K., Trimpert J., Osterrieder N., Kunec D. Attenuation of a very virulent Marek’s disease herpesvirus (MDV) by codon pair bias deoptimization. PLoS Pathog. 2018;14:e1006857. doi: 10.1371/journal.ppat.1006857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S., Kim L., Whitaker M., O’Halloran A., Cummings C., Holstein R., Prill M., Chai S.J., Kirley P.D., Alden N.B. Hospitalization Rates and Characteristics of Patients Hospitalized with Laboratory-Confirmed Coronavirus Disease 2019 - COVID-NET, 14 States, March 1-30, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021;384:2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenke N., Trimpert J., Merz S., Conradie A.M., Wyler E., Zhang H., Hazapis O.G., Rausch S., Landthaler M., Osterrieder N., Kunec D. Mechanism of Virus Attenuation by Codon Pair Deoptimization. Cell Rep. 2020;31:107586. doi: 10.1016/j.celrep.2020.107586. [DOI] [PubMed] [Google Scholar]
- Gruber A.D., Osterrieder N., Bertzbach L.D., Vladimirova D., Greuel S., Ihlow J., Horst D., Trimpert J., Dietert K. Standardization of Reporting Criteria for Lung Pathology in SARS-CoV-2 Infected Hamsters - What Matters? Am. J. Respir. Cell Mol. Biol. 2020;63:856–859. doi: 10.1165/rcmb.2020-0280LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397:1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall V.J., Foulkes S., Saei A., Andrews N., Oguti B., Charlett A., Wellington E., Stowe J., Gillson N., Atti A., SIREN Study Group COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. Lancet. 2021;397:1725–1735. doi: 10.1016/S0140-6736(21)00790-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew O.M., Sutter R.W., de Gourville E.M., Dowdle W.R., Pallansch M.A. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- Kunec D., Osterrieder N. Codon Pair Bias Is a Direct Consequence of Dinucleotide Bias. Cell Rep. 2016;14:55–67. doi: 10.1016/j.celrep.2015.12.011. [DOI] [PubMed] [Google Scholar]
- Kusters I., Almond J.W. In: Vaccine Strategies. In Encyclopedia of Virology. Third Edition. B W J M Editors-in-Chief, M H V v Regenmortel, editors. Academic Press; 2008. pp. 235–243. [Google Scholar]
- Kutter J.S., de Meulder D., Bestebroer T.M., Lexmond P., Mulders A., Richard M., Fouchier R.A.M., Herfst S. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat. Commun. 2021;12:1653. doi: 10.1038/s41467-021-21918-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Jones J.O., Andino R. Rationalizing the development of live attenuated virus vaccines. Nat. Biotechnol. 2010;28:573–579. doi: 10.1038/nbt.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Acevedo A., Cooper S.B., Andino R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe. 2012;12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Nouën C., Brock L.G., Luongo C., McCarty T., Yang L., Mehedi M., Wimmer E., Mueller S., Collins P.L., Buchholz U.J., DiNapoli J.M. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc. Natl. Acad. Sci. USA. 2014;111:13169–13174. doi: 10.1073/pnas.1411290111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., Kovyrshina A.V., Lubenets N.L., Grousova D.M., Erokhova A.S., Gam-COVID-Vac Vaccine Trial Group Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. China’s COVID vaccines are going global - but questions remain. Nature. 2021;593:178–179. doi: 10.1038/d41586-021-01146-0. [DOI] [PubMed] [Google Scholar]
- Mueller S., Coleman J.R., Papamichail D., Ward C.B., Nimnual A., Futcher B., Skiena S., Wimmer E. Live attenuated influenza virus vaccines by computer-aided rational design. Nat. Biotechnol. 2010;28:723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Karakida N., Dantsuka A., Ichii O., Elewa Y.H.A., Kon Y., Nagasaki K.I., Hattori H., Yoshiyasu T. Effects of a mixture of medetomidine, midazolam and butorphanol on anesthesia and blood biochemistry and the antagonizing action of atipamezole in hamsters. J. Vet. Med. Sci. 2017;79:1230–1235. doi: 10.1292/jvms.17-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov V., Kouprina N., Leem S.H., Koriabine M., Barrett J.C., Larionov V. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 2002;30:E8. doi: 10.1093/nar/30.2.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odon V., Fros J.J., Goonawardane N., Dietrich I., Ibrahim A., Alshaikhahmed K., Nguyen D., Simmonds P. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res. 2019;47:8061–8083. doi: 10.1093/nar/gkz581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Kunec D. Attenuation of Viruses by Large-Scale Recoding of their Genomes: the Selection Is Always Biased. Curr. Clin. Microbiol. Rep. 2018;5:66–72. doi: 10.1007/s40588-018-0080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T-cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S.H., Stauft C.B., Gorbatsevych O., Song Y., Ward C.B., Yurovsky A., Mueller S., Futcher B., Wimmer E. Large-scale recoding of an arbovirus genome to rebalance its insect versus mammalian preference. Proc. Natl. Acad. Sci. USA. 2015;112:4749–4754. doi: 10.1073/pnas.1502864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.-L. Pathogenesis and transmission of SARS-CoV-2 virus in golden Syrian hamsters. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solforosi L., Kuipers H., Jongeneelen M., Rosendahl Huber S.K., van der Lubbe J.E.M., Dekking L., Czapska-Casey D.N., Izquierdo Gil A., Baert M.R.M., Drijver J. Immunogenicity and efficacy of one and two doses of Ad26.COV2.S COVID vaccine in adult and aged NHP. J. Exp. Med. 2021;218:e20202756. doi: 10.1084/jem.20202756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Liu Y., Ward C.B., Mueller S., Futcher B., Skiena S., Paul A.V., Wimmer E. Identification of two functionally redundant RNA elements in the coding sequence of poliovirus using computer-generated design. Proc. Natl. Acad. Sci. USA. 2012;109:14301–14307. doi: 10.1073/pnas.1211484109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The World Health Organization Draft landscape of COVID-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- Thi Nhu Thao T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Tischer B.K., Smith G.A., Osterrieder K. In: In Vitro Mutagenesis Protocols. 3rd. Braman Jeff., editor. Humana Press; 2010. En passant mutagenesis: a two step markerless red recombination system; pp. 421–430. [DOI] [PubMed] [Google Scholar]
- Trimpert T., Vladimirova D., Dietert K., Abdelgawad A., Kunec D., Dökel D., Gruber A.D., Bertzbach L., Osterrieder N. The Roborovski dwarf hamster – a highly susceptible model for a rapid and fatal course of SARS-CoV-2 infection. Cell Rep. 2020 doi: 10.1016/j.celrep.2020.108488. Published online December 8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Costa Clemens S.A., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Oxford COVID Vaccine Trial Group Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer E., Mueller S., Tumpey T.M., Taubenberger J.K. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nat. Biotechnol. 2009;27:1163–1172. doi: 10.1038/nbt.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Skiena S., Futcher B., Mueller S., Wimmer E. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc. Natl. Acad. Sci. USA. 2013;110:9481–9486. doi: 10.1073/pnas.1307473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C., Corum J., Wee S.-L. 2021. Coronavirus Vaccine Tracker. The New York Times.https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of the recoded SARS-CoV-2 genes have been deposited in the NCBI’s GenBank database (GenBank: MZ064531 - MZ064546). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.