Abstract
Coronaviruses rely on host membranes for entry, establishment of replication centers, and egress. Compounds targeting cellular membrane biology and lipid biosynthetic pathways have previously shown promise as antivirals and are actively being pursued as treatments for other conditions. Here, we test small molecule inhibitors that target the PI3 kinase VPS34 or fatty acid metabolism for anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) activity. Our studies determine that compounds targeting VPS34 are potent SARS-CoV-2 inhibitors. Mechanistic studies with compounds targeting multiple steps up- and downstream of fatty acid synthase (FASN) identify the importance of triacylglycerol production and protein palmitoylation as requirements for efficient viral RNA synthesis and infectious virus production. Further, FASN knockout results in significantly impaired SARS-CoV-2 replication that can be rescued with fatty acid supplementation. Together, these studies clarify roles for VPS34 and fatty acid metabolism in SARS-CoV-2 replication and identify promising avenues for the development of countermeasures against SARS-CoV-2.
Keywords: antiviral, autophagy, coronavirus, COVID-19, SARS coronavirus 2, fatty acid, membranes, phosphoinositol 3 kinase
Graphical abstract
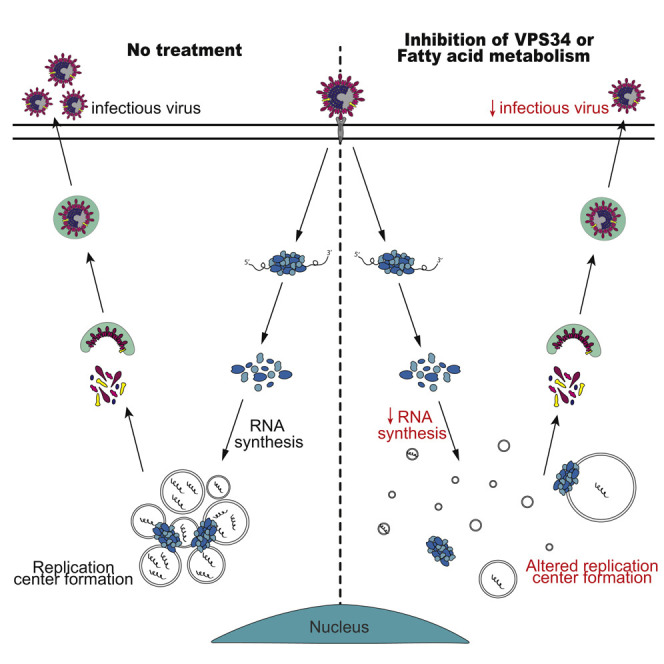
Williams et al. employ pharmacological inhibitor and genetic knockout approaches to demonstrate critical roles for the class III phosphatidylinositol 3-kinase VPS34 and fatty acid metabolism in SARS-CoV-2 growth. The data further implicate triacylglycerol production and protein palmitoylation as critical functions related to fatty acid metabolism that are needed by SARS-CoV-2.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), is an enveloped positive-sense RNA virus of the Betacoronavirus genus (Holshue et al., 2020; Lundstrom, 2020; Zhu et al., 2020). Since its emergence, SARS-CoV-2 is responsible for the most significant pandemic in the last century and has resulted in worldwide social and economic disruption. The severity of the pandemic has prompted urgent efforts to understand the requirements of the viral life cycle and identify potential therapeutic strategies (Bouhaddou et al., 2020; Gordon et al., 2020c; Hoffmann et al., 2021; Wang et al., 2020). Repurposing drugs developed for other diseases and conditions may provide a shortcut to antiviral development (García-Serradilla et al., 2019; Li and De Clercq, 2020; Pizzorno et al., 2019; Saini et al., 2020). The use of compounds known to target specific host factors may also elucidate key pathways and processes utilized in SARS-CoV-2 replication.
Coronaviruses (CoVs) interact with host cell membranes and membrane machinery at many different stages of their life cycle including entry, genome replication, and virion maturation and release (Hagemeijer et al., 2014; Prentice et al., 2004; Reggiori et al., 2010, 2011; Snijder et al., 2020; V’kovski et al., 2021). One of the most striking features of CoV infection is the reorganization and recruitment of host cell membranes to form replication organelles. These rearrangements lead to formation of double-membrane vesicles (DMVs), double-membrane spherules (DMSs), convoluted membranes (CMs), and vesicle packets (VPs) (Knoops et al., 2008; Snijder et al., 2020). Although there are several existing models that suggest these various membranous structures originate from modified endoplasmic reticulum (ER) membranes and that DMVs are the primary sites of replication, the mechanistic basis of their formation and CoV utilization remains incompletely understood (Angelini et al., 2013; Knoops et al., 2008; Oudshoorn et al., 2017; Reggiori et al., 2010; Snijder et al., 2020). Previous studies have observed DMVs colocalizing with LC3, a protein with well-known functions in autophagy (Reggiori et al., 2010, 2011). However, studies on mouse hepatitis virus (MHV), a model betacoronavirus, have found conflicting data on whether or not autophagy is necessary for replication (Prentice et al., 2004; Zhao et al., 2007). Alternate models for membrane remodeling implicate EDEMosomes, vesicles originating from the ER that associate with non-lipidated LC3 (Reggiori et al., 2010, 2011). Additionally, other positive-sense RNA viruses that induce comparable membrane rearrangements and DMV formation utilize host cell lipid droplets, cellular organelles that store neutral lipids, and lipid metabolism as an underlying platform and energy source for replication (Cloherty et al., 2020; Heaton and Randall, 2011).
Inhibitors of metabolic and biosynthesis pathways related to membranes and their precursors inhibit the replication of a number of viruses. One example of this is the inhibition of VPS34, a class III phosphoinositol-3 kinase (PI3K) that plays roles in autophagy, endosomal trafficking, and other aspects of membrane biology, which has been shown to impair hepatitis C virus (HCV), tombusvirus (TBSV), and coronavirus infection (Feng et al., 2019; Su et al., 2011; Wang et al., 2021).
Targeting enzymes involved in late steps of the neutral lipid synthesis pathway, such as long chain acyl-CoA synthetase (ACS) or diglyceride acyltransferases (DGATs) has been shown to inhibit rotavirus (RV) and HCV, possibly reflecting a dependence of these viruses on lipid droplets (Cheung et al., 2010; Kim et al., 2012; Liefhebber et al., 2014). Targeting upstream enzymes in fatty acid metabolism such as acetyl-CoA carboxylase (ACC) or fatty acid synthase (FASN) also decreases replication of several different viruses including Coxsackievirus B, Chickungunya virus (CHIKV), and several flaviviruses (Ammer et al., 2015; Gaunt et al., 2013; Hitakarun et al., 2020; Merino-Ramos et al., 2015; Tongluan et al., 2017).
In this study, we sought out to determine the susceptibility of SARS-CoV-2 to modulators of VPS34 and fatty acid metabolism. Compounds targeting VPS34 strongly impaired SARS-CoV-2 replication in vitro. Additionally, orlistat and triacsin C, inhibitors of FASN and ACS, respectively (Carvalho et al., 2008; Igal et al., 1997; Kridel et al., 2004), effectively inhibited SARS-CoV-2. Time-of-addition studies revealed that each of these inhibitors exert antiviral effects post-entry, with the VPS34 inhibitors also inhibiting an early step in the replication cycle. Experiments using additional inhibitors of enzymes upstream and downstream of FASN and ACS point to palmitoylation and neutral lipid production as necessary for virus replication. Immunofluorescence studies of compound-treated cells demonstrate disruption of double-stranded RNA (dsRNA)-positive viral replication centers, and the same inhibitors also impact viral RNA synthesis. Studies in FASN knockout Caco2 cells confirmed the critical role of fatty acid biosynthesis in SARS-CoV-2 replication. Taken together, the data presented here demonstrate that specific fatty acid and lipid metabolism pathways are critical for SARS-CoV-2 replication and provide novel mechanistic insights that may serve as a basis for the potential repurposing and development of therapeutics targeting these pathways for the treatment of COVID-19.
Results
Development of 96-well format impedance-based assay to measure SARS-CoV-2 cytopathic effects
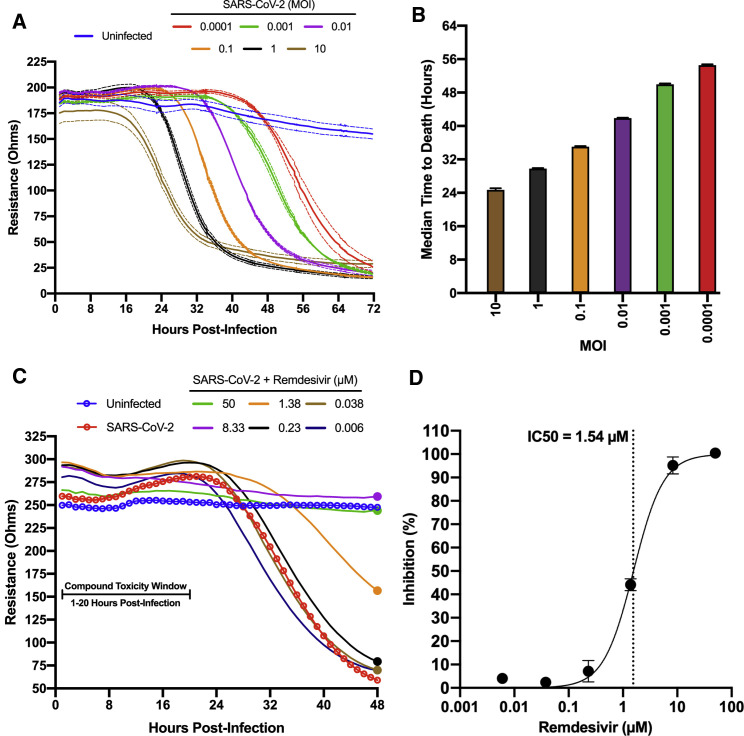
SARS-CoV-2 induces significant cytopathic effects in infected Vero E6 cells. Based on this property, we standardized a 96-well format assay that provides continuous real-time, label free monitoring of the integrity of cell monolayers as a direct correlation of virus growth and infection. This assay was standardized using the Maestro Z platform (Axion BioSystems, Atlanta, GA), an instrument that uses plates containing electrodes in each well (CytoView-Z plates) to measure electrical impedance across the cell monolayer every minute throughout the course of the experiment. As SARS-CoV-2-induced cytopathic effects damage the cell monolayer, impedance measurements decrease over time providing a detailed assessment of infection kinetics. To determine the capacity of the system to differentiate levels of virus replication, confluent Vero E6 monolayers in CytoView-Z plates were infected with SARS-CoV-2 at multiple MOIs (0.0001 to 10), and resistance measurements were acquired for 72 h post-infection (hpi) (Figure 1 A). The progression of infection at each MOI was clearly distinct. A decrease in resistance could be observed as early as 18–20 hpi at an MOI of 1 and 10, and as late as 56 hpi at an MOI of 0.0001. All sample signals reached their lowest point between 32 to 72 hpi in an MOI-dependent manner. The raw kinetic data was used to determine the median time to cell death for each MOI that shows a direct correlation with a decrease in resistance (Figure 1B). MOI of 0.01 was chosen for antiviral assays based on its desirable infection kinetics.
Figure 1.
Standardization of an electrical resistance-based assay as a measure of SARS CoV-2-induced CPE and anti-SARS-CoV-2 activity
Vero E6 cells were seeded into a CytoView-Z 96-well plate and allowed to stabilize overnight, as measured by electrical resistance.
(A) Resistance was measured every minute over the course of 72 hours in wells that were mock infected or infected with SARS-CoV-2 in 10-fold dilutions ranging from an MOI of 10-0.0001. Solid lines indicate the mean, dotted lines indicate the standard error of three replicates.
(B) Median time-to-death calculations based on raw resistance data for each MOI.
(C and D) Remdesivir was titrated in 6-fold dilutions ranging from 50–0.006 μM. (C) After infection at an MOI of 0.01, resistance was monitored for 48 hpi and (D) percent inhibition for remdesivir based on the data from the 48-h time point is presented (solid circles). Resistance measurements were monitored between 1–20 hpi for potential compound-induced cytopathic effects prior to SARS-CoV-2 induced cytopathic effects, as indicated in (C). The data represent the mean and SE of three replicates.
To establish the Maestro Z as a potential instrument for screening of anti-SARS-CoV-2 therapeutics, we first tested remdesivir, a well-described inhibitor of SARS-CoV-2 that has been granted emergency use authorization (EUA) for the treatment of COVID-19 (Gordon et al., 2020a; Wu et al., 2020). Vero E6 cells were seeded on a CytoView-Z plate, incubated overnight to allow cells to stabilize, pretreated with 6-fold dilutions of remdesivir for 1 h and infected with SARS-CoV-2. Resistance measurements were recorded for 48 hpi with potential compound induced cytopathic effect being assessed within the first 20 h, prior to SARS-CoV-2 induced cytopathic effect (Figure 1C). In agreement with previous studies, we determined a 50% inhibitory concentration (IC50) for remdesivir of 1.54 μM (Figure 1D; Gordon et al., 2020a). Taken together, these data validate the impedance-based assay described as a tool for screening of potential SARS-CoV-2 therapeutics.
Inhibitors of VPS34 activity impair SARS-CoV-2 growth
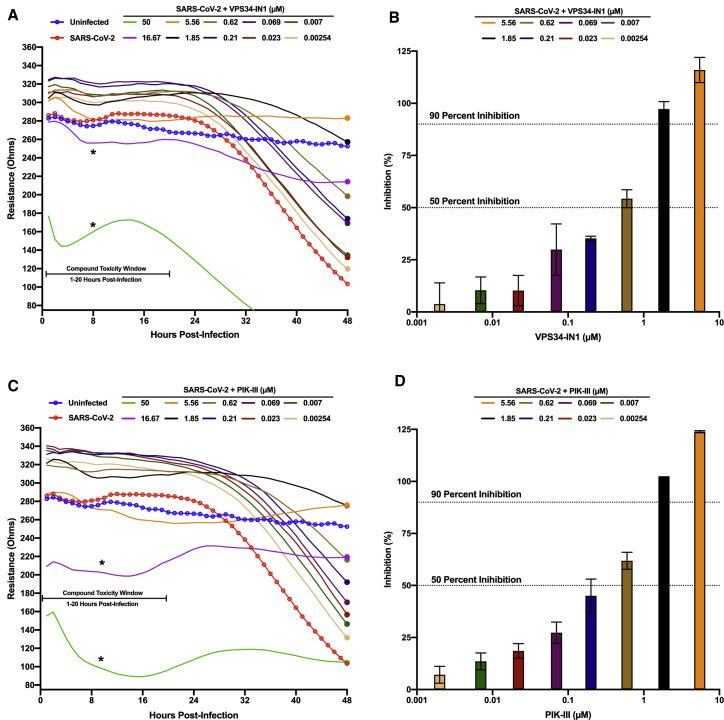
VPS34 is a multifunctional protein involved in autophagy and membrane trafficking (Jaber and Zong, 2013; Ohashi et al., 2020). Because of the significant role of membrane rearrangements in coronavirus replication, we wanted to determine if VPS34 activity was essential for SARS-CoV-2 growth. We tested two well characterized VPS34 inhibitors, VPS34-IN1 and PIK-III (Bago et al., 2014; Dowdle et al., 2014; Figure 2 ). This was done over a 10-point dose response using the resistance assay described above. Briefly, pre-plated Vero E6 cells were treated with compound 1 h prior to infection with SARS-CoV-2. Both VPS34-IN1 and PIK-III induced rapid cytotoxicity at 50 μM and 16.67 μM as indicated by a rapid decrease in resistance measurements between 1 and 20 hpi (Figures 2A and 2C). At the remaining concentrations, no toxicity was observed. For several concentrations, the integrity of the monolayer was preserved relative to the mock-treated infected control indicating an antiviral effect of both VPS34-IN1 and PIK-III. Based on normalized resistance measurements at 48 hpi for non-toxic doses, we estimated IC50 values of <600 nM for VPS34-IN1 and PIK-III (Figures 2B and 2D, respectively). These data suggest that the VPS34 kinase plays a significant role in SARS-CoV-2 replication.
Figure 2.
VPS34 inhibitors exhibit anti-SARS-CoV-2 activity
Vero E6 cells were seeded into a CytoView-Z 96-well plate and allowed to stabilize overnight. Cells were pre-treated with serial half-log10 dilutions of VPS34-IN1 (A and B) or PIK-III (C and D) and infected with SARS-CoV-2 at an MOI of 0.01. Resistance (A and C) was measured every minute over the course of 48 h and percent inhibition (B and D) was determined at the 48-h time point (solid circles) as compared to the infected DMSO-treated control (red). Uninfected cells are indicated in blue. Resistance measurements were monitored between 1–20 hpi for potential compound-induced cytopathic effects prior to SARS-CoV-2 induced cytopathic effects, as indicated in (A) and (C). The data represent the mean and SE of three replicates.
Inhibition of fatty acid metabolism impairs SARS-CoV-2 growth
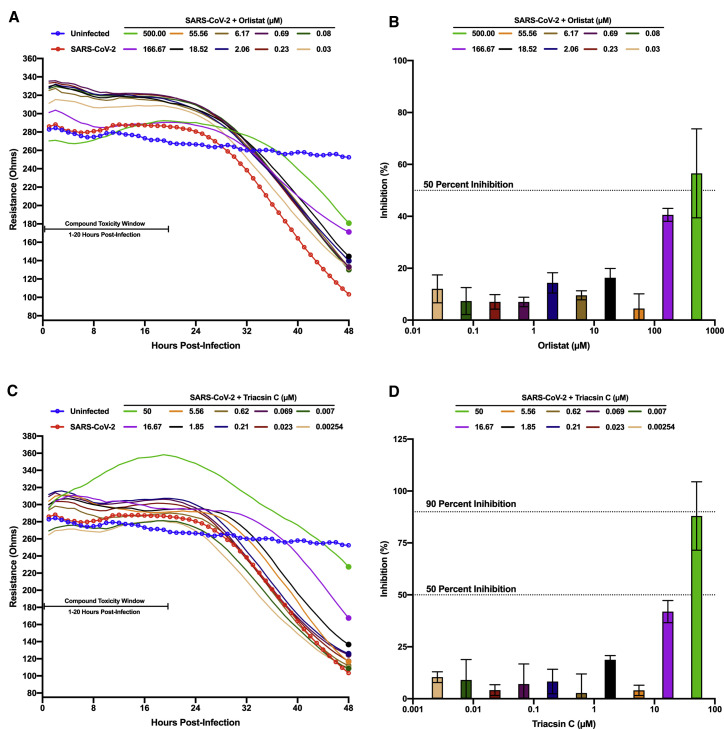
Fatty acid metabolism contributes to various host processes including production of lipid-based molecules such as triglycerides, phospholipids, and cholesterol, as well as protein modifications, such as palmitoylation and myristoylation (Baenke et al., 2013; Chen et al., 2018; Wakil and Abu-Elheiga, 2009). Modulation of fatty acid metabolism has been shown to impact replication and virion maturation for numerous flaviviruses, enteroviruses, and alphaviruses (Ammer et al., 2015; Bakhache et al., 2019; Cheung et al., 2010; Gaunt et al., 2013; Hitakarun et al., 2020; Liefhebber et al., 2014; Merino-Ramos et al., 2015; Nasheri et al., 2013; Tongluan et al., 2017). Two well-described compounds that inhibit fatty acid metabolism are orlistat, a Food and Drug Administration (FDA)-approved drug that inhibits gastric lipases and FASN, and triacsin C, an inhibitor of long chain ACS, both of which have been shown to have antiviral activity (Ammer et al., 2015; Hitakarun et al., 2020; Kim et al., 2012; Nasheri et al., 2013). To test the efficacy of these compounds against SARS-CoV-2 infection, Vero E6 cells were pre-seeded onto a CytoView-Z plate, allowed to stabilize, and then pre-treated with orlistat or triacsin C for 1 h prior to infection (Figure 3 ). Based on the toxicity window of 1–20 h post-treatment (hpt) determined with the VPS34 inhibitors, neither orlistat nor triacsin C induced early cytotoxic effects, even at the highest concentrations of 500 μM and 50 μM, respectively (Figures 3A and 3C). Both compounds exhibited inhibition of viral cytopathic effects at higher concentrations, although complete inhibition was not achieved even with 500 μM of orlistat. Based on normalized resistance measurements at 48 hpi, we estimated IC50s of ∼500 μM for orlistat and ∼20 μM for triacsin C (Figures 3B and 3D). These data suggest that fatty acid metabolism and neutral lipid synthesis likely play an important role in SARS-CoV-2 infection.
Figure 3.
Screening of fatty acid inhibitors for anti-SARS-CoV-2 activity
Vero E6 cells were seeded into a CytoView-Z 96-well plate and allowed to stabilize overnight. Cells were pre-treated with serial half-log10 dilutions of orlistat (A and B) or triacsin C (C and D) and infected with SARS CoV-2 at an MOI of 0.01. Resistance (A and C) was measured every minute over the course of 48 h and percent inhibition (B and D) was determined at the 48-h time point (solid circles) as compared to the infected DMSO-treated control (red). Uninfected cells are indicated in blue. Resistance measurements were monitored between 1–20 hpi for potential compound-induced cytopathic effects prior to SARS-CoV-2-induced cytopathic effects, as indicated in (A) and (C). The data represent the mean and SE of three replicates.
VPS34 and fatty acid metabolism inhibitors exhibit activity on post-entry steps of the viral life cycle
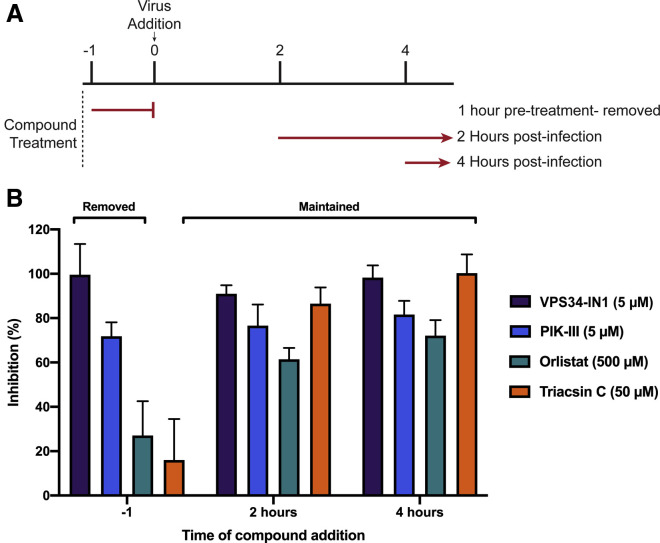
Next, we performed time-of-addition studies with VPS34-IN1, PIK-III, orlistat, or triacsin C. This allowed us to determine if the anti-viral activity of each compound impacted the viral life cycle at steps pre- or post-entry. As indicated in Figure 4 A, three conditions were tested: (1) single treatment 1 h prior to viral infection, with compound removed prior to infection; (2) dosing at 2 hpi; and (3) dosing at 4 hpi. We observed that a single 5 μM treatment of VPS34-IN1 or PIK-III prior to infection inhibited SARS-CoV-2 replication, and inhibition could be observed even when added up to 4 hpi (Figure 4B). In contrast, orlistat or triacsin C showed minimal efficacy when removed prior to infection but remained inhibitory when added up to 4 hpi. Altogether, these data demonstrate that the VPS34 inhibitors likely act on viral entry and at later steps in the replication cycle, and inhibition by orlistat and triacsin C occurs post-entry.
Figure 4.
Time-of-addition studies
VeroE6 cells were seeded into a CytoView-Z 96-well plate and allowed to stabilize overnight.
(A) Timeline for the time-of-addition experiment.
(B) VeroE6 cells were either pre-treated for one hour with compound removed prior to infection (−1) or treated at 2 (+2) or 4 (+4) hours post-infection. All conditions were infected with an MOI of 0.01. Resistance was measured every minute over the course of 48 h and percent inhibition was determined at the 48-h time point. The data represent the mean and SE of three replicates.
Attenuation of VPS34 kinase activity and fatty acid metabolism inhibit SARS-CoV-2 in a human airway epithelial cell line
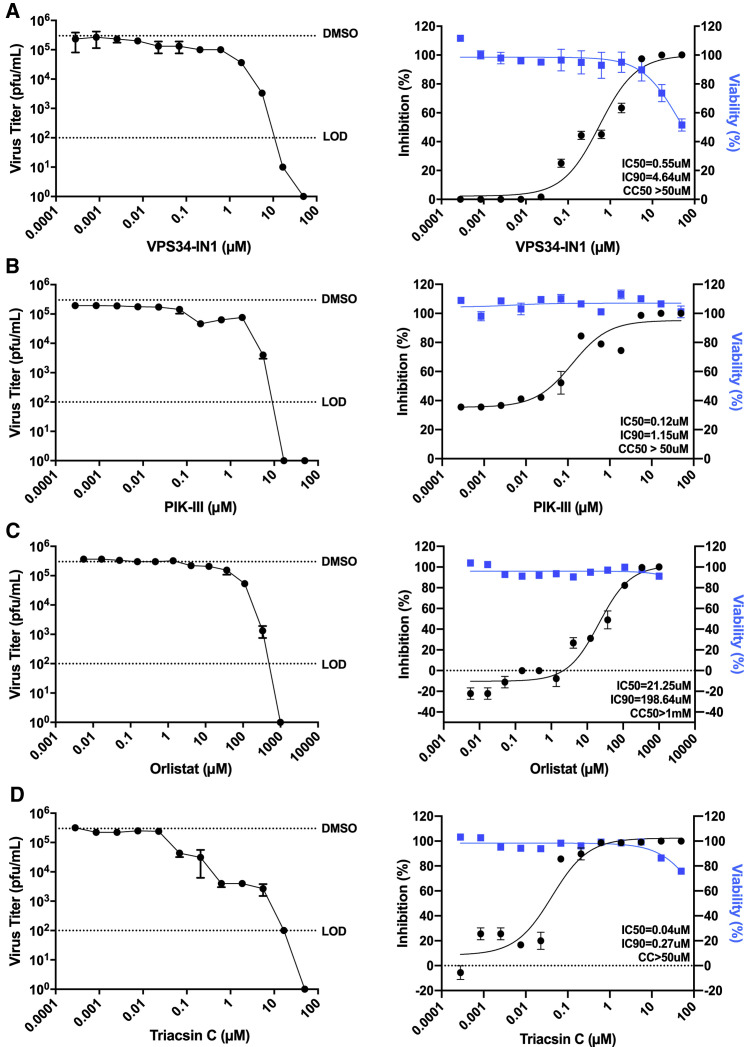
To determine whether or not the previously observed inhibition could be maintained in a cell line more functionally relevant to human infection by SARS-CoV-2, we directly measured the impact of compounds on production of infectious virus and cell viability in Calu-3 cells. These cells are derived from human airway epithelium and are highly susceptible to infection, establishing them as a standard for infection studies with SARS-CoV-1, Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 (Chu et al., 2020; Sheahan et al., 2020; Tseng et al., 2005). For these studies, Calu-3 cells were plated onto 96-well plates and allowed to reach confluency. Cells were either pre-treated with a range of concentrations of VPS34-IN1, PIK-III, triacsin C and orlistat, DMSO, or mock treated with media alone for 1 h, then infected with SARS-CoV-2 at an MOI of 0.01. Supernatants were collected at 48 hpi and viral titer determined by plaque assay. Cytotoxicity of the compounds was determined in parallel. Consistent with results from infected Vero E6 cells, each of the compounds tested above inhibited production of infectious virus, and cytotoxicity was only observed in Calu-3 cells at the highest concentrations of some compounds (Figure 5 ). We observed IC50s of 0.55 μM (VPS34-IN1; Figure 5A), 0.12 μM (PIK-III; Figure 5B), 21.25 μM (orlistat; Figure 5C), and 0.04 μM (triacsin C; Figure 5D). Notably, the IC50s measured for VPS34-IN1 and PIK-III by plaque assay are in close agreement with estimated IC50s determined in Vero E6 cells using the resistance-based assay. The IC50s for orlistat and triacsin C were substantially lower than those estimated in VeroE6 cells.
Figure 5.
Attenuation of VPS34 kinase activity and fatty acid metabolism inhibit SARS-CoV-2 replication in human airway epithelial cell line
Calu-3 cells were plated onto a 96-well plate and allowed to reach confluency. Cells were then pre-treated with a series of 3-fold dilutions of VPS34-IN1 (A), PIK-III (B), orlistat (C), triacsin C (D), DMSO, or mock-treated with media alone for 1 h, then infected with SARS-CoV-2 at an MOI of 0.01. Supernatants were collected at 48 hpi, and virus was quantified by plaque assay on VeroE6 cells. The data are reported as plaque forming units per milliliter (pfu/mL) (left panels) and percent inhibition (right panels). Cell viability over 48 h was determined in parallel and plotted with the percent inhibition data. IC50 and IC90 were calculated from the plaque assay data and are indicated on the curves. For the plaque assay data, the dotted lines labeled DMSO and LOD indicate the level of virus growth in the DMSO control and the limit of detection, respectively. The data represent the mean and SE of three replicates. See also Figure S1.
To determine the specificity of inhibition to the class III PI3 kinase VPS34, we extended our Calu-3 compound study to include two additional VPS34 inhibitors: SAR405 and compound 19 (Honda et al., 2015; Pasquier, 2015; Ronan et al., 2014). SAR405 and compound 19 each inhibited with IC50s of 0.376 μM and 5.843 μM, respectively (Figures S1A and S1B). These data further support the importance of the class III PI3 kinase during SARS-CoV-2 replication. To confirm that the compounds were able to inhibit VPS34 activity, we assessed in Huh7 cells the effect of the inhibitors on GFP-2xFYVE localization to endosomes, indicating a loss of phosphoinositol-3 phosphorylation by VPS34, (Gillooly et al., 2000, 2001; Kutateladze, 2007; Ronan et al., 2014). Huh7 cells were chosen for their favorable characteristics, such as transfectability and large cytoplasm that facilitates imaging. Although DMSO or orlistat did not affect GFP-2xFYVE localization, the four VPS34 inhibitors disrupted GFP-2xFYVE puncta, consistent with loss of PI-3 phosphorylation (Figure S1C).
Protein palmitoylation and triacylglycerol production are implicated in SARS-CoV-2 infection
Several recent reports have implicated pathways related to lipid metabolism and cholesterol homeostasis as being required for SARS-CoV-2 replication; however, the precise mechanism by which these pathways contribute to the SARS-CoV-2 replication cycle remains unclear (Abu-Farha et al., 2020; Daniloski et al., 2021; Schneider et al., 2021; Zang et al., 2020). Given that our data suggest a role for fatty acid metabolism, we set out to further clarify the enzymatic steps required for SARS-CoV-2 replication.
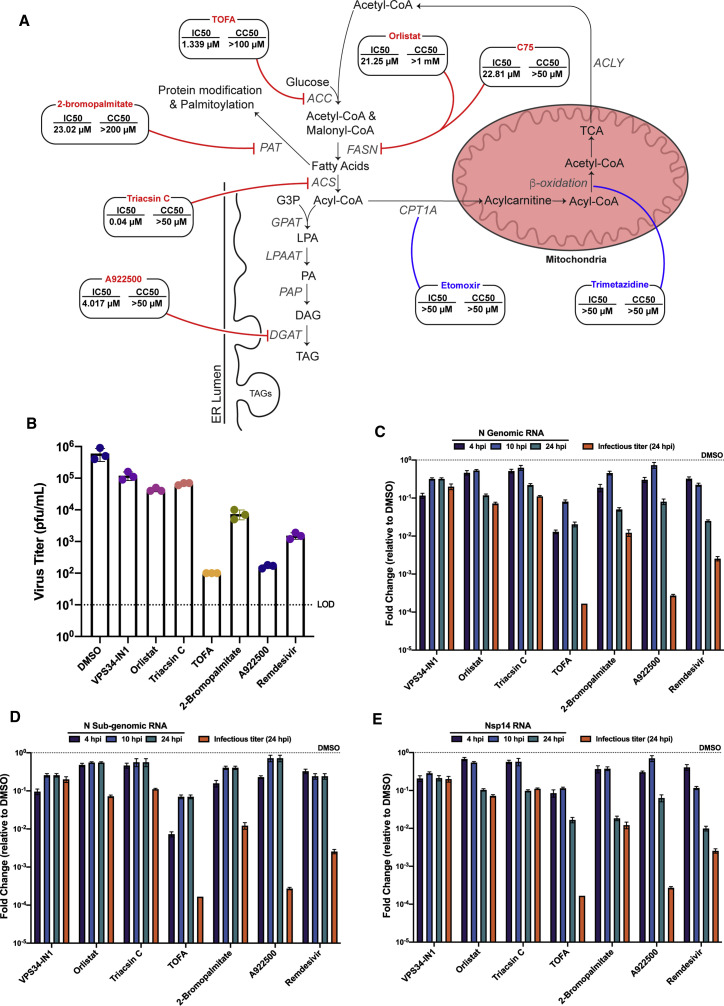
To further evaluate the importance of de novo fatty acid synthesis, TOFA, a competitive inhibitor of acetyl-CoA carboxylase (ACC), the enzyme directly upstream of FASN, and C75, an additional inhibitor of FASN, were used (Halvorson and McCune, 1984; Kuhajda et al., 2000). Both compounds exhibited activity against SARS-CoV-2 infection with IC50s of 1.339 μM and 22.81 μM, respectively (Figures 6 , S2A, and S2B).
Figure 6.
Mechanistic characterization of anti-SARS-CoV-2 activity
To determine which steps involved in fatty acid metabolism contribute to the observed anti-SARS-CoV-2 activity, Calu-3 cells were pre-treated with DMSO or a series of 3-fold dilutions of the indicated compounds, or mock-treated with media alone for 1 h, then infected with SARS-CoV-2 at an MOI of 0.01.
(A–D) Supernatants were collected at 48 hpi and virus was quantified by focus forming assay on VeroE6 cells. Cytotoxicity assays were performed in parallel. IC50 and CC50 values were calculated for each compound (A). To discern whether or not genomic and subgenomic RNA levels are affected by compound treatment, Calu-3 cells were pre-seeded in 24-well format, allowed to grow to confluency, and infected at an MOI of 1. Two hours post-infection, cells were treated with VPS34-IN1 (5 μM), orlistat (500 μM), triacsin C (5 μM), TOFA (50 μM), 2-bromopalmitate (50 μM), A922500 (30 μM), remdesivir (1 μM), or DMSO. At 4, 10, and 24 hpi, total RNA was extracted from the cell monolayers, and 24 hpi supernatants were harvested for viral titers. Virus titers were determined by plaque assay (B).
(C–E) Levels of genomic RNA, subgenomic N RNA, and NSP14 RNA were quantified via qPCR. Data are represented as fold change of RNA levels in infected compound treated samples versus infected DMSO-treated samples. The virus titer at 24 hpi for compound treated cells (orange bars) are plotted alongside the qPCR data and represented as fold-change compared to titers from DMSO-treated cells. The data represent the mean and SE of three replicates.
See also Figures S2, S3, and S4.
Inhibition of fatty acid synthesis at early enzymatic steps like FASN can lead to dysregulation of three main downstream pathways: fatty acid-based protein modification, fatty acid β-oxidation in the mitochondria, and neutral lipid synthesis (Baenke et al., 2013). To further determine which branches and enzymatic steps of fatty acid metabolism contribute to anti-SARS-CoV-2 activity, a series of additional compounds were tested for inhibitory activity and cytotoxicity (Figure 6).
To assess the contribution of decreased protein palmitoylation, 2-bromoplamitate, an inhibitor of palmitoyl acyltransferases (PAT), was used (Davda et al., 2013). 2-Bromopalmitate inhibited infectious virus production at concentrations greater than 10 μM with an IC50 of 23.02 μM (Figures 6A and S2C). To determine if fatty acid β-oxidation in the mitochondria contributes to inhibition of SARS-CoV-2, two compounds were tested: etomoxir, a compound that targets carnitine palmitoyltransferase 1A (CPT1A), blocking translocation of fatty acids into the mitochondria, and trimetazidine, an inhibitor of long-chain 3-ketoacyl-CoA thiolase (Ma et al., 2020). Neither compound showed any inhibition, indicating that fatty acid β-oxidation is not required for SARS-CoV-2 replication (Figures 6A, S2D, and S2E). Last, the importance of the terminal steps of the fatty acid metabolism pathways was assessed by inhibiting neutral lipid production and lipid droplet formation using A922500, a potent inhibitor of diacylglycerol acyltransferase 1 (DGAT1) (Zhao et al., 2008). Treatment with A922500 inhibited SARS-CoV-2 with an IC50 value of 4.017 μM (Figures 6A and S2F). All together, these data suggest that protein palmitoylation and neutral lipid synthesis are needed for efficient SARS-CoV-2 replication.
Inhibition of VPS34 kinase activity and fatty acid metabolism alters SARS-CoV-2 replication centers
Because time-of-addition studies implicated post-entry steps as targets of the compounds, we sought to assess the status of sites of viral RNA synthesis, which can be detected with anti-dsRNA antibodies (Knoops et al., 2008; Reggiori et al., 2010; Weber et al., 2006). Foci of dsRNA staining in coronavirus infected cells reflect the formation of membrane-associated viral replication compartments (Knoops et al., 2008; Oudshoorn et al., 2017; Prentice et al., 2004; Reggiori et al., 2010). Calu-3 cells were seeded onto fibronectin-coated glass coverslips and allowed to reach partial confluency. Cells were infected with SARS-CoV-2 at a MOI of 1, treated at 2 hpi with concentrations of VPS34-IN1, PIK-III, orlistat, triacsin C, TOFA, 2-bromopalmitate, A922500, or remdesivir determined to result in significant inhibition without cytotoxicity, or with DMSO. At 24 hpi, cells were fixed, permeabilized, and stained for indirect immunofluorescence using primary antibodies against SARS-CoV-2 N and dsRNA (Figure S3).
DMSO-treated control cells displayed distinct puncta positive for dsRNA consistent with CoV replication centers (Knoops et al., 2008; Reggiori et al., 2010). These structures were also positive for the SARS-CoV-2 nucleocapsid protein (N). Treatments with VPS34-IN1, orlistat, TOFA, or A922500 resulted in a loss of these distinct puncta and dispersion of dsRNA and SARS-CoV-2 N. When treated with triacsin C and 2-bromopalmitate, cells exhibited fewer but larger foci positive for both markers. Treatment with a non-sterilizing concentration of remdesivir (1μM) served as a control and yielded a distribution of N and dsRNA similar to the DMSO control. Taken together, these data suggest inhibition VPS34 activity and fatty acid metabolism may affect SARS-CoV-2 replication by affecting viral replication centers (Figures S3A and S3B).
Inhibition of VPS34 and fatty acid metabolism impacts genomic and subgenomic RNA levels
The effects on dsRNA and N distribution suggested possible effects on viral RNA synthesis. To determine if the production of viral RNA was affected, Calu-3 cells were infected at an MOI of either 0.01 or 1. Two hours post-infection, cells were treated with DMSO or concentrations of each inhibitor determined to result in significant inhibition at an MOI of 0.01 without toxicity. At 4, 10, and 24 hpi, levels of viral RNA were quantified by RT-PCR using three distinct primer pairs. Supernatants corresponding to 24 hpi were used to quantify viral titers to ensure inhibition was achieved. Remdesivir was used as a positive control compound that inhibits SARS-CoV-2 RNA synthesis.
Compound treatments resulted in a 1- to 4-log10 reduction of viral titers at both MOIs at 24 hpi (Figures 6B and S4A). Consistent with the decrease in viral titers, all compounds reduced the amounts of genomic RNA at the 24 h time point. Remdesivir inhibited RNA synthesis at earlier time points as well. At 4 and 10 hpi at MOI of 1, the inhibition was more apparent with the N genomic RNA and NSP14 primers as compared with primers designed to amplify N sub-genomic RNA. The degree of inhibition was magnified at an MOI of 0.01 (Figures 6C–6E and S4B–S4D). Similarly, treatment with VPS34-IN1, orlistat, triacsin C, and 2-bromopalmitate reduced RNA levels at both MOI 0.01 and 1. In general, there were minimal differences in fold-inhibition at 4 versus 10 hpi (Figures 6C–6E and S4B–S4D). The inhibition of RNA synthesis resulting from treatment with TOFA and A922500 followed a similar pattern to the other compounds at each MOI and time point. However, the reduction in RNA synthesis at 24 hpi was significantly less substantial than the reduction in viral titers (Figures 6C–6E and S4B–S4D). This discordance suggests that these compounds have additional effects beyond viral RNA synthesis that explain their inhibition of virus growth.
Genetic knockout confirms a critical role of de novo fatty acid synthesis for SARS-CoV-2 replication
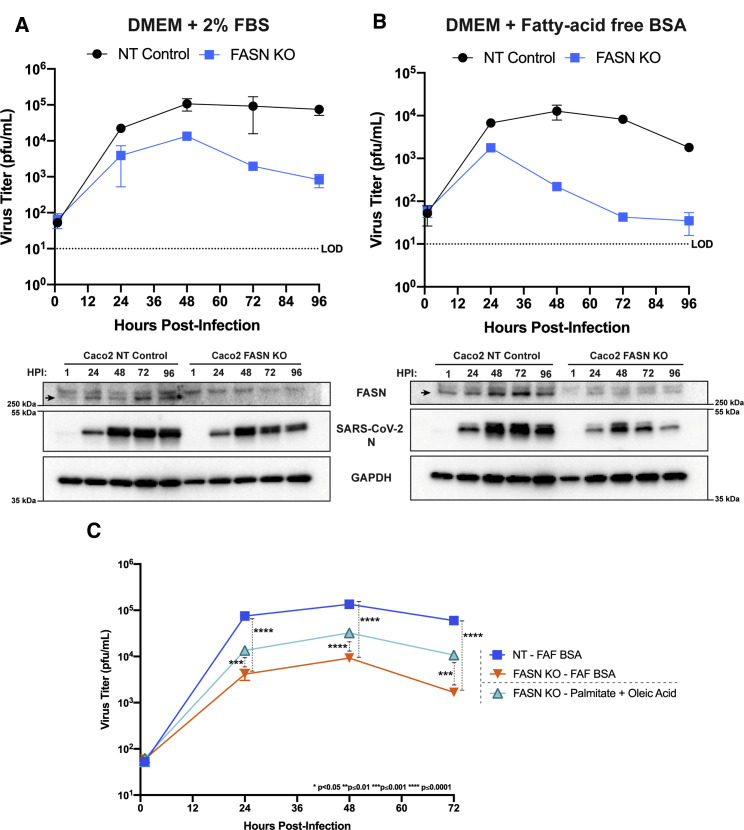
As a genetic correlate to the studies described above, we compared the growth of SARS-CoV-2 in non-targeting control (NT) and FASN knockout Caco2 (FASN KO) cells in the presence of 2% fetal bovine serum (FBS) or 1% fatty acid free bovine serum albumin (FAF-BSA), with the latter condition intended to address the possible contribution of exogenous fatty acids supplied by the FBS. FASN KO Calu3 and Caco2 cells were generated. The FASN KO Calu3 cells exhibited a slow and unpredictable growth rate, whereas the FASN KO Caco2 cells exhibited more consistent and predictable growth characteristics (data now shown). Therefore, Caco2 FASN KO cells were used for infection studies. Consistent with the previous compound data, SARS-CoV-2 replicated to substantially lower titers in the FASN KO cells as compared to non-targeting control (NT) cells when the cells were maintained in medium with 2% FBS (Figure 7 A). The inhibition was enhanced when cells were maintained in 1% FAF-BSA media, reaching an almost 2-log10 reduction in titer as early as 48 hpi (Figure 7B). For each experiment, FASN knockout was confirmed by western blot. In addition, SARS-CoV-2 N protein levels were decreased in FASN KO cells as compared to NT cells. At later time points, N protein levels in FASN KO cells declined consistent with the changes in virus titer. The diminished replication and protein production in cells lacking FASN confirms our earlier findings and suggests that fatty acid metabolism is necessary for efficient, sustained replication during SARS-CoV-2 infection.
Figure 7.
Fatty acid metabolism is essential for efficient SARS-CoV-2 replication
FASN CRISPR KO Caco2 cells and corresponding NT Caco2 cells were pre-seeded in 96-well plates, grown to confluency, and then infected at an MOI 0.01 in minimal media. Post-adsorption, cells were maintained in either 2% FBS DMEM (A) or 1% fatty-acid free (FAF)-BSA DMEM (B). Supernatants were collected at 1, 24, 48, 72, and 96 hpi and viral titers were determined by plaque assay. Protein samples were obtained from cell monolayers and analyzed by western blot to confirm FASN knockout and look for changes in SARS-CoV-2 N levels. FASN KO and WT cells were pre-seeded and infected as previously described. Post adsorption, inoculum was removed and replaced with 2% FAF-BSA DMEM or 2% FAF-BSA DMEM containing 250 μM palmitic acid + 250 μM oleic acid (C). Supernatants were collected at 1, 24, 48, and 72 hpi and viral titers were determined via plaque assay. The data represent the mean and SE of three replicates.
Based on our observations that the reduction in protein palmitoylation and neutral lipid synthesis are likely responsible for the inhibition seen when blocking fatty acid metabolism, we asked whether viral replication in FASN knockout cells would be restored by supplementing with substrates that feed into these two sub-pathways. Supplementation was achieved by infecting FASN KO Caco2 cells in minimal media and adding back media containing 2% FAF-BSA pre-complexed to a combination of 250 μM palmitic acid and 250 μM oleic acid. Cell supernatants were collected at 1, 24, 48, and 72 hpi, and viral titers were determined by plaque assay. Consistent with our previous data, growth in FASN KO cells was significantly reduced as compared to NT control cells. Supplementation with palmitate and oleic acid partially rescued virus growth further confirming the importance of fatty acid metabolism in SARS-CoV-2 replication (Figure 7C).
Discussion
Engagement with, and usurpation of, host membranes is central to coronavirus biology. We therefore sought to identify potential host factors and pathways relevant to lipid metabolism that are critical for SARS-CoV-2 replication. In addition to providing insights into virus-host interactions, such approaches might also suggest new therapeutic strategies. Our results demonstrate the importance of both the class III PI3 kinase VPS34 and enzymes involved in fatty acid metabolism for SARS-CoV-2 replication. Treatment of infected cells with inhibitors against VPS34 and various steps of fatty acid metabolism impairs virus growth, reduces viral RNA synthesis, and perturbs the formation of dsRNA-positive replication centers. Compounds targeting ACC and DGAT1 likely also inhibit additional steps in the virus replication cycle.
VPS34 is a class III phosphoinositide 3-kinase that plays important roles in autophagy and endosomal trafficking, as well as other cellular functions (Backer, 2016; Ohashi et al., 2020). In our inquiry into host factors relevant for SARS-CoV-2 replication, we tested two well characterized specific and structurally similar inhibitors of VPS34: VPS34-IN1 and PIK-III (Bago et al., 2014; Dowdle et al., 2014). Both inhibitors showed potent inhibition of viral titers at sub-micromolar concentrations in both Vero E6 and Calu-3 cells as measured by resistance assay and plaque assay, respectively. To further confirm the specificity to VPS34 activity, we tested the anti-SARS-CoV-2 activity of two additional inhibitors, compound 19 and SAR405, the latter of which is structurally distinct from the others (Honda et al., 2015; Pasquier, 2015; Ronan et al., 2014). Each compound inhibited virus growth, further validating the importance of VPS34 for virus replication, consistent with recent reports (Wang et al., 2021; Yuen et al., 2021). Mechanistically, our data point to early steps, potentially viral entry, and later steps, including viral RNA synthesis, as sensitive to VPS34 inhibition.
Although our inhibitor studies do not differentiate between the various functions of VPS34 that might be involved in SARS-CoV-2 replication, studies in different cell types suggest autophagy is not essential for MHV growth, and more recent studies suggest that coronaviruses interfere with autophagy, and activation of autophagy can inhibit replication of SARS-CoV, MERS CoV, and SARS-CoV-2 (Gassen et al., 2019; Guo et al., 2016; Prentice et al., 2004; Zhao et al., 2007). Given that inhibition of VPS34 results in the inhibition of autophagy, it would be expected that inhibition of VPS34 would eliminate these anti-CoV effects of autophagy and promote SARS-CoV-2 replication. At the same time, host protein TMEM41B that is also implicated in autophagy has been demonstrated to facilitate SARS-CoV-2 growth, suggesting that the virus may usurp some autophagy functions (Schneider et al., 2021). It will therefore be of interest to determine whether the disruptions in SARS-CoV-2 replication due to VPS34 targeting reflect inhibition of autophagy or non-autophagy related functions.
Separate from autophagy, VPS34 has several other roles, most notably in vesicular trafficking and sorting. VPS34 mediated formation of phosphatidylinositol 3-phosphate allows for recruitment of various proteins for endosomal fusion and tethering, recruitment of the endosomal sorting complex required for transport (ESCRT) proteins for the formation of multivesicular bodies, and proper functioning of the Retromer complex for endosome-to-Golgi transport (Backer, 2016; Ohashi et al., 2020). For members of the positive-sense RNA tombusviruses, VPS34 has been shown to be recruited to replication compartments providing increased levels of phosphoinositol-3-phosphate, which allows for recruitment of Rab5-positive early endosomes, providing phosphatidylethanolamine-enriched membranes for replication center formation (Feng et al., 2019). Based on our observation that the VPS34 inhibitor VPS34-IN1 interferes with the formation of dsRNA-positive SARS-CoV-2 replication centers, it is possible that VPS34 functions to facilitate membrane availability for SARS-CoV-2 replication organelle formation. Disruption of endocytic trafficking might also explain our observation that pre-treatment followed by removal of VPS34 inhibitors alone had significant effects on early phases of SARS-CoV-2 replication. Alternatively, the significant effects on SARS-CoV-2 replication observed by pre-treatment alone could also be due to an extended in vitro half-life of these compounds or irreversible binding to VPS34 leading to inhibition of later steps in the virus life cycle without continuous treatment. Further studies will be required to fully deconvolute these possibilities.
Fatty acid metabolism is co-opted by viruses for a variety of purposes including as a means to provide energy for replication, the establishment of membrane-associated replication centers, and the proper assembly and budding of newly synthesized viral particles (Heaton and Randall, 2011). Orlistat, an FDA-approved inhibitor of FASN, and triacsin C, an inhibitor of long chain ACS have previously been examined for their antiviral activities against viruses the utilize fatty acids and neutral lipids for replication (Ammer et al., 2015; Cheung et al., 2010; Gaunt et al., 2013; Hitakarun et al., 2020; Kim et al., 2012; Liefhebber et al., 2014; Tongluan et al., 2017). In our study on SARS-CoV-2, we found that orlistat and triacsin C both exhibited antiviral activity but only when maintained throughout infection, consistent with the post-entry inhibition previously described for other viruses.
Inhibition of upstream steps in the metabolic pathway, like FASN and ACS, can lead to dysregulation of multiple downstream pathways. In this case, the viral inhibition we observe from orlistat and triacsin C could be the result of downregulating fatty acid protein modification, β-oxidation of fatty acids in the mitochondria, or neutral lipid synthesis and lipid droplet formation, (Baenke et al., 2013). In HCV infection, NS4B and the core protein both require protein palmitoylation for their function in replication and virus particle formation, respectively (Majeau et al., 2009; Yu et al., 2006). Protein palmitoylation of the spike proteins of MHV and SARS-CoV have also been shown to be critical to virion assembly, cell-cell fusion, and infectivity (McBride and Machamer, 2010; Petit et al., 2007; Thorp et al., 2006). Palmitoylation of coronavirus envelope proteins has also been described (Boscarino et al., 2008; Tseng et al., 2014). DENV activates pathways that trigger breakdown of lipid droplets and neutral lipids to redirect fatty acids to the mitochondria for β-oxidation and energy production (Heaton and Randall, 2011). Conversely to this, rotavirus directly utilizes lipid droplets as a platform for replication center formation and a protected environment for virion maturation (Cheung et al., 2010; Lever and Desselberger, 2016). To gain further mechanistic insight into the role of fatty acid metabolism in the SARS-CoV-2 life cycle, we tested a series of compounds that targeted enzymatic steps involved in each one of the aforementioned pathways downstream of FASN. Our results showed that the PAT inhibitor 2-bromopalmitate, which should block protein palmitoylation, led to a substantial reduction in SARS-CoV-2 replication. This may reflect a requirement for spike modification and/or palmitoylation of envelope and host proteins. Additionally, A922500, a specific inhibitor of DGAT1, resulted in significant reduction of SARS-CoV-2. This inhibition is indicative of a need for neutral lipid synthesis and suggests a role for lipid droplets, which have previously been implicated in SARS-CoV-2 growth and suggested to provide a platform for viral assembly (Dias et al., 2020). Our data with A922500 are consistent with a role for lipid droplets in assembly, given that its effects on production of infectious virus were substantially greater than on viral RNA synthesis.
Previous studies tracking newly synthesized RNA during CoV infection show that RNA synthesis occurs at foci referred to as replication centers that correspond to double membrane vesicles produced during coronavirus infection (Snijder et al., 2020). We therefore assessed the impacts of our inhibitors on replication center formation. Under the conditions tested, viral RNA synthesis and viral replication were reduced but not abrogated by the compounds. It was striking that treatment of infected cells with VPS34-IN1 and inhibitors of different aspects of fatty acid metabolism all resulted in aberrant dsRNA and N localization whereas treatment with remdesivir did not. The distribution of dsRNA and N varied, depending on the compound used. These data suggest VPS34 and various aspects of fatty acid metabolism play a role in proper formation SARS-CoV-2 replication centers and possibly in virally induced membrane rearrangements.
VPS34-IN1, triacsin C, orlistat, and 2-bromopalmitate all reduced RNA detected by primers designed to amplify genomic N and NSP14 RNA. The magnitude of the effect roughly correlated with the reduction in viral titer, while signal from primers designed to detect subgenomic N was only moderately affected in comparison. Determining whether these compounds selectively impact viral genome replication versus subgenomic mRNA production will be of interest. Treatment with TOFA and A922500 reduced genomic N and NSP14 RNA to a substantially lesser degree than the reductions in viral titers. The discrepancy between the reduction in RNA synthesis and infectious virus is consistent with what has been proposed for rotavirus, where RNA synthesis and virion formation utilize specialized replication organelles. Disruption of these structures moderately reduces RNA synthesis but has a much larger impact on virus assembly and release (Cheung et al., 2010; Gaunt et al., 2013; Kim et al., 2012; Lever and Desselberger, 2016).
The demonstration that SARS-CoV-2 replication is impaired in FASN KO cells as compared to cells that received a non-targeting guide RNA further confirms the requirement for fatty acids. In cells lacking FASN, there was a significant reduction in viral titers that was sustained throughout infection. That this attenuation is due to loss of fatty acid production is supported by the increased attenuation when the cells were maintained in the presence of fatty acid free medium as compared to medium containing FBS that might provide exogenous fatty acids. The consumption of free fatty acid pools unable to be replaced due to loss of FASN likely explains the rapid decline in N protein production observed during infection. To further address the role of fatty acids in inhibition, supplementation was performed with two primary substrates downstream of FASN, palmitic acid and oleic acid (Ackerman et al., 2018; Alsabeeh et al., 2018; Baenke et al., 2013). This resulted in a significant rescue of SARS-CoV-2 replication in FASN KO cells when compared to cells without supplementation. The ability to rescue infection using fatty acid metabolism products provides a potentially useful approach that could be exploited to determine the mechanisms by which fatty acid metabolism contributes to the SARS-CoV-2 life cycle.
Cumulatively, these data support lipid metabolism as a potential therapeutic target for SARS-CoV-2 infection. Additionally, the identification of fatty acid metabolism, and more specifically protein palmitoylation and lipid droplet production, as key components for replication provides important insights into SARS-CoV-2-host interaction. Limitations of the study include the facts that the precise mechanisms by which VPS34 promotes SARS-CoV-2 replication as well as the mechanistic basis by which inhibitors of VSP34 and fatty acid metabolism impair replication warrant further investigation. In addition, although the inhibitors of VPS34 and fatty acid metabolism alter the morphology and distribution of viral replication centers, to what extent these changes are related to inhibition of virus growth remains to be determined.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SARS-CoV Nucleoprotein | Novus Biological | Cat# NB100-56576; RRID:AB_838838 |
| Anti-dsRNA, clone RJ2 | EMD Millipore | Cat# MABE1134; RRID:AB_2819101 |
| Anti-FASN | Invitrogen | Cat #MA5-31491; RRID:AB_2787122 |
| HRP anti-rabbit secondary | Cell Signaling | Cat# 7074; RRID:AB_2099233 |
| Anti-Rabbit AlexaFluor 488 | ThermoFisher | Cat# A32731; RRID:AB_2633280 |
| Anti-mouse AlexaFluor 647 | ThermoFisher | Cat# A32728; RRID:AB_2633277 |
| Bacterial and virus strains | ||
| SARS-CoV-2 USA/WA/1/2020 | World Reference Collection for Emerging Viruses and Arboviruses – UTMB - Galveston | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| VPS34 IN-1 | Cayman Chemical | Cat# 17392 |
| PIK-III | Cayman Chemical | Cat# 17002 |
| Triacsin C | Cayman Chemical | Cat# 10007448 |
| Orlistat | Cayman Chemical | Cat# 10005426 |
| TOFA | Cayman Chemical | Cat# 10005263 |
| C75 | Cayman Chemical | Cat# 10005270 |
| Etomoxir | Cayman Chemical | Cat# 11969 |
| Trimetazidine | Cayman Chemical | Cat# 18165 |
| A922500 | Cayman Chemical | Cat# 10012708 |
| Remdesivir | TargetMol | Cat# T7766 |
| 2-Bromopalmitate | Sigma-Aldrich | Cat# 21604 |
| Microcrystalline colloidal cellulose | Sigma-Aldrich | Cat# 435244 |
| TrueBlue HRP substrate | SeraCare | Cat# 5510-0030 |
| Trizol | Thermo Fisher | Cat# 15596026 |
| ezDNase | Thermo Fisher | Cat# 11766051 |
| Superscript IV | Thermo Fisher | Cat# 18090010 |
| AmpliTaq Gold 360 | Thermo Fisher | Cat# 439881 |
| PerFeCTa SYBR Green FastMix | VWR | Cat# 101414-272 |
| Blocker FL Fluroescent Blocker | Thermo Fisher | Cat# 37565 |
| Fatty Acid Free BSA | Sigma-Aldrich | Cat# A8806-5G |
| Palmitic Acid | Sigma-Aldrich | Cat# P0500 |
| Oleic Acid | Sigma-Aldrich | Cat# O1008 |
| Lipofectamine 2000 | Invitrogen | Cat# 11668500 |
| Streptococcus pyogenes NLS-Sp.Cas9-NLS (SpCas9) nuclease | Aldevron | Cat# 9212 |
| SF Buffer | Lonza | Cat# V5SC-2002 |
| Critical commercial assays | ||
| CellTox Green Cytotoxicity Assay | Promega | Cat# G8741 |
| CytoView-Z 96-well electrode plates | Axion BioSystems | Cat# Z96-IMP-96B |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | Cat# CRL-1586 |
| Calu-3 | ATCC | Cat# HTB-55 |
| Caco-2 | ATCC | Cat# HTB-37 |
| Caco-2 Non-Targeting sgRNA Control cells | This paper | N/A |
| Caco-2 FASN KO cells | This paper | N/A |
| Oligonucleotides | ||
| FASN sgRNA 1: 5′ - ACAUACCCGCCUUCCAGCGA | This paper | N/A |
| FASN sgRNA 2: 5′ - UUCUGGGACAACCUCAUCGG | This paper | N/A |
| FASN sgRNA 3: 5′ - UGGUGAUUGCCGGCAUGUCC | This paper | N/A |
| FASN Forward sequencing primer: 5′ - TAGGCCACTCTGGTGCAATG | This paper | N/A |
| FASN Reverse sequencing primer: 5′ - GTGTCCACCTGTTCTGGGT | This paper | N/A |
| SARS-CoV-2 N Genomic Forward: 5′ - TAATC AGACAAGGAACTGATTA | Schneider et al., 2021 | N/A |
| SARS-CoV-2 N Genomic Reverse: 5′ - CGAAG GTGTGACTTCCATG | Schneider et al., 2021 | N/A |
| SARS-CoV-2 N Subgenomic Forward: 5′ - GTTTATACCTTCCCAGGTAACAAACC | Schneider et al., 2021 | N/A |
| SARS-CoV-2 N Subgenomic Reverse: 5′ - GTAGAAATACCATCTTGGACTGAGATC | Schneider et al., 2021 | N/A |
| SARS-CoV-2 NSP14 Forward: 5′ - TGG GGYTTTACGGGTAACCT | Schneider et al., 2021 | N/A |
| SARS-CoV-2 NSP14 Reverse: 5′ - AACGCGCTTAACAAAGCACTC | Schneider et al., 2021 | N/A |
| RPS11 Forward: 5′ - GCCGAGACTATCT GCACTAC | Schneider et al., 2021 | N/A |
| RPS11 Reverse: 5′ - ATGTCCAGCCTC AGAACTTC | Schneider et al., 2021 | N/A |
| Recombinant DNA | ||
| pEGFP-2xFYVE | Addgene | Cat# 140047 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Christopher F. Basler (cbasler@gsu.edu).
Materials availability
The Caco-2 NTC and Caco-2 FASN KO cell lines may be requested from the lead contact upon request.
Experimental model and subject details
Vero E6 (ATCC# CRL-1586), Calu-3 (ATCC# HTB-55), and Caco-2 (ATCC# HTB-37) were maintained in DMEM (Corning) supplemented with 10% heat inactivated fetal bovine serum (FBS; GIBCO). Cells were kept in a 37°C, 5% CO2 incubator without antibiotics or antimycotics. Non-targeting wild-type (NT-WT) Caco-2 and FASN KO Caco-2 cells were produced by CRISPR editing (Synthego, Redwood City, CA) and maintained the same as their Caco2 parental cell line. SARS-CoV-2, strain USA_WA1/2020, was obtained from the World Reference Collection for Emerging Viruses and Arboviruses at the University of Texas Medical Branch-Galveston.
Method details
sgRNA Selection
sgRNAs were designed according to Synthego’s multi-guide gene knockout. Briefly, two or three sgRNAs are bioinformatically designed to work in a cooperative manner to generate small, knockout-causing, fragment deletions in early exons. These fragment deletions are larger than standard indels generated from single guides. The genomic repair patterns from a multi-guide approach are highly predictable based on the guide-spacing and design constraints to limit off-targets, resulting in a higher probability protein knockout phenotype.
sgRNA Synthesis
RNA oligonucleotides were chemically synthesized on the Synthego solid-phase synthesis platform, using CPG solid support containing a universal linker. 5-Benzylthio-1H-tetrazole (BTT, 0.25 M solution in acetonitrile) was used for coupling, (3-((Dimethylamino-methylidene)amino)-3H-1,2,4-dithiazole-3-thione (DDTT, 0.1 M solution in pyridine) was used for thiolation, dichloroacetic acid (DCA, 3% solution in toluene) for used for detritylation. Modified sgRNA were chemically synthesized to contain 2′-O-methyl analogs and 3′ phosphorothioate nucleotide interlinkages in the terminal three nucleotides at both 5′ and 3′ ends of the RNA molecule. After synthesis, oligonucleotides were subject to series of deprotection steps, followed by purification by solid phase extraction (SPE). Purified oligonucleotides were analyzed by ESI-MS.
RNP Formation and Transfection
To deliver CRISPR-Cas9 ribonucleoprotein (RNP) complexes, 10 pmol Streptococcus pyogenes NLS-Sp.Cas9-NLS (SpCas9) nuclease (Aldevron Cat. #9212) was combined with 30 pmol total synthetic sgRNA (Synthego,10 pmol each sgRNA) to form RNPs in 20uL total volume with SF Buffer (Lonza Cat #V5SC-2002) and allowed to complex at room temperature for 10 minutes. Cells were dissociated into single cells using TrypLE Express (GIBCO), as described above, resuspended in culture media and then counted. 100,000 cells per nucleofection reaction were pelleted by centrifugation at 100 xg for 3 minutes. Following centrifugation, cells were resuspended in transfection buffer according and diluted to 2∗104 cells/μL. 5 μL of cell solution was added to preformed RNP solution and gently mixed. Nucleofections were performed on a Lonza 96-well nucleofector shuttle system using program CM-150. Immediately following nucleofection each reaction was transferred to a tissue-culture treated 96-well plate containing 100μL normal culture media and seeded at a density of 50,000 cells per well. Transfected cells were incubated following standard protocols.
Genomic Analysis
Two days post-nucleofection, DNA was extracted from using DNA QuickExtract (Lucigen Cat. #QE09050). Briefly, cells were lysed by removal of the spent media followed by addition of 50 μL of QuickExtract solution to each well. Once the QuickExtract DNA Extraction Solution was added, the cells were scraped off the plate into the buffer. Following transfer to compatible plates, DNA extract was then incubated at 68°C for 15 minutes followed by 95°C for 10 minutes in a thermocycler before being stored for downstream analysis.
Amplicons for indel analysis were generated by PCR amplification AmpliTaq Gold 360 polymerase (Thermo Fisher Scientific Cat. #4398881) according to the manufacturer’s protocol. Primers were designed to create amplicons between 400 - 800bp, with both primers at least 100bp distance from any of the sgRNA target sites. PCR products were cleaned-up and analyzed by Sanger sequencing (Genewiz). Sanger data files and sgRNA target sequences were input into Inference of CRISPR Edits (ICE) analysis (https://ice.synthego.com:443/) to determine editing efficiency and to quantify generated indels (Hsiau et al., 2019). Percentage of alleles edited is expressed an ice-d score. This score is a measure of how discordant the Sanger trace is before versus after the edit. It is a simple and robust estimate of editing efficiency in a pool, especially suited to highly disruptive editing techniques like multi-guide.
Compounds
VPS34 IN-1 (#17392), PIK-III (#17002), Triacsin C (#10007448), Orlistat (#10005426), TOFA (#10005263), C75 (#10005270), Etomoxir (#11969), Trimetazidine (#18165), and A-922500 (#10012708) were purchased from Cayman Chemical (Ann Arbor, Michigan, USA). Remdesivir was purchased from Target Molecule Corp. (T7766, Boston, Massachusetts, USA). 2-bromopalmitate was purchased from Sigma-Aldrich (#21604, St. Louis, MO, USA). All compounds were resuspended in dimethylsulfoxide (DMSO).
Virus Propagation and Viral Titering
A lyophilized ampule of SARS-CoV-2 was initially resuspended in DMEM supplemented with 2% FBS. Vero E6 cells were inoculated in duplicate with a dilution of 1:100 with an adsorption period of 1 hour at 37°C and shaking every 15 minutes. Cells were observed for cytopathic effect (CPE) every 24 hours. Stock SARS-CoV-2 virus was harvested at 72 hours post infection (hpi) and supernatants were collected, clarified, aliquoted, and stored at −80°C.
Plaque assays were performed as previously described (Jureka et al., 2020). Briefly, Vero E6 cells were seeded onto 24-well plates 24 hours before infection. 100ul of SARS-CoV-2 serial dilutions were added and adsorbed for 1 hour at 37°C with shaking at 15-minute intervals. After the absorption period, 1 mL of 0.6% microcrystalline cellulose overlay (MCC; Sigma-Aldrich 435244, St. Louis, MO, USA) was added and plates were returned to the incubator for the indicated amount of time. To stain plaque assays, MCC was removed by aspiration and 10% neutral buffered formalin (NBF) added for one hour at room temperature. Formalin was removed and monolayers were washed with water and stained with 0.4% crystal violet. Plaques were quantified and recorded as plaque forming units (PFU)/mL.
Focus forming assays were performed as previously described (Gordon et al., 2020b; Jureka et al., 2020). Briefly, Vero E6 cells were pre-seeded in 96-well plates and grown to confluency. 50 μL of SARS-CoV-2 serial dilutions were added and adsorbed for 1 hour at 37°C. Post-adsorption, 50 μL of 2.4% microcrystalline cellulose overlay (MCC; Sigma-Aldrich 435244, St. Louis, MO, USA) was added and plates were returned to the incubator for 24 hours. Cell monolayers were inactivated with 10% NBF for one hour at room temperature, washed with deionized water and fixed/permeabilized with ice cold methanol containing 0.3% hydrogen peroxide for 10 minutes at −20°C followed by 20 minutes at room temperature. Prior to primary antibody addition, plates were washed with PBS and blocked for one hour at room temperature with 5% milk. Primary antibody detection with SARS N diluted in milk occurred overnight at 4°C. The next day, plates were washed with PBS and anti-rabbit-HRP diluted in milk was added for 1 hour at room temperature. After final washing, plates were developed using TrueBlue HRP substrate (SeraCare, Milford, MA, USA). Foci were quantified and graphed as focus forming units (FFU)/mL.
Maestro Z Impedance Experiments
Prior to cell plating, CytoView-Z 96-well electrode plates (Axion BioSystems, Atlanta, GA, USA) were coated with 5 μg/mL human fibronectin (Corning, Tewksbury, MA, USA) for 1 hr at 37°C. After coating, fibronectin was removed and 100 μL of DMEM containing 10% FBS was added to each well. The plate was then docked into the Maestro Z instrument to measure impedance electrode baseline. Vero E6 cells were then plated to confluency (∼75,000 cells/well) in the coated CytoView-Z plates and left at room temperature for 1 hour to ensure even coverage of the well. Plates containing Vero E6 cells were docked into the Maestro Z for 24 hours at 37°C with 5% CO2 to allow the cells to attach and the monolayer to stabilize, as measured by resistance, a component of impedance. The Maestro Z was used to monitor the resistance of the monolayer as it formed, very similar to transepithelial electrical resistance (TEER) (Benson et al., 2013). In this study, resistance was measured at 10 kHz, which reflects both cell coverage over the electrode and strength of the barrier formed by the cell monolayer. For compound treatments, media was removed from wells of the CytoView-Z plates and 195 μL of pre-warmed DMEM containing 2% FBS was added with the indicated concentration of compound. Infections with SARS-CoV-2 at an MOI of 0.01 were carried out by directly adding 5 μL of virus to each well. Plates were then docked within the Maestro Z and resistance measurements were continuously recorded for 48-72 hours post infection. All plates contained media only, full lysis, uninfected, and SARS-CoV-2 infected controls. For calculation of percent inhibition, raw resistance values for each well were normalized to the value at 1 hour post infection within the Axis Z software, and percent inhibition was calculated with the following formula: Percent Inhibition = 100∗(1-(1- average of treated cells)/(1-average of infected control)). Median time to death calculations were performed by fitting the Boltzmann sigmoid equation to raw kinetic resistance data in GraphPad Prism. Fifty percent maximum velocity (V50) values obtained from the Boltzmann sigmoid fits were used to determine median time to death for each MOI.
Cell viability assay
Vero E6 or Calu-3 cells were seeded in 96-well black walled microplates and incubated overnight. Media was replaced with fresh DMEM containing 2% FBS and CellTox Green Dye (Promega, Madison, WI, USA) as described in the manufacturer’s protocol. Cells were then treated with compound in parallel with the corresponding experiment and put back in the incubator for the indicated duration. Fluorescence (Excitation: 485nm, Emission: 520nm) was measured at the designated end point. Percent viability was determined using the minimum fluorescence obtained from media only cells and the maximum value obtained by wells lysed with the included lysis solution.
Quantification of vRNA and mRNA
Calu-3 cells were seeded in 24-well plates and allowed to grow to confluency. Infection with SARS-CoV-2 was carried out at an MOI of 0.01 and 1.0. Media containing the indicated compound was added 2 hours post infection. Supernatants were collected at 24 hours post infection for titering, and RNA was extracted from cell monolayers using TRIzol reagent (ThermoFisher). Post extraction, RNA was DNase treated using ezDNase (ThermoFisher, Waltham, MA, USA) and subjected to first strand synthesis using SuperScript IV (ThermoFisher, Waltham, MA, USA) using the included random hexamer primers. qPCR was performed using PerfeCTa SYBR Green FastMix (VWR, Radnor, PA, USA) and primers for SARS-CoV-2 N (genomic and subgenomic), SARS-CoV-2 NSP14, and RPS11 as an internal control (Schneider et al., 2021). Each assay was performed in triplicate with three technical replicates, and each assay contained no-template controls. Data were analyzed by the ΔΔCt method with RPS11 serving as the housekeeping gene and uninfected DMSO treated Calu3 cells as a mock control.
Confocal microscopy
For confocal microscopy analysis, cells were pre-seeded in 24-well plates on fibronectin coated glass coverslips and allowed to grow to confluency. Cells were infected with SARS-CoV-2 at an MOI of 1 and media containing compound treatments was added 2 hours post infection. At 24 hours post infection, the supernatants were removed, and samples were fixed with 10% NBF for 1 hour at room temperature. Post-fixation, the cells were washed with PBS and permeabilized with sterile filtered 0.1% Saponin and 0.15% glycine in PBS. Cells were blocked with 0.1% Saponin in 1x Fluorescent Blocker (ThermoFisher, Waltham, MA, USA) for 1 hour at room temperature. Primary antibodies were diluted in blocking buffer, added to the wells and incubated overnight at 4°C. The following day, coverslips were washed with PBS and incubated with secondary antibodies diluted in 0.1% saponin in PBS for 1 hour. AlexaFluor 488 and 647 conjugated secondary antibodies (ThermoFisher, Waltham, MA, USA) were used. Coverslips were mounted using ProLong Glass Antifade mountant with NucBlue stain (ThermoFisher, Waltham, MA, USA). Samples were imaged on a Zeiss LSM800 Confocal and images were rendered in ZenBlue (Zeiss, White Plains, NY, USA).
Supplementation
Supplementation with fatty acids and lipids was achieved by pre-complexing the supplements to fatty acid free bovine serum albumin (FAF-BSA). Briefly, FAF-BSA was dissolved in tissue culture grade water to achieve a final concentration of 10%. Stock solutions of palmitic acid (150 mM in ethanol, Sigma-Aldrich St. Louis, MO, USA) and oleic acid (3 M, Sigma-Aldrich St. Louis, MO, USA) were diluted to 2.5 mM in 10% FAF-BSA and incubated at 37°C with frequent mixing for 30 minutes. The pre-complexed solutions were then added to DMEM at a final concentration of 2% FAF-BSA and 0.25 mM supplement. For the infection, NT-WT and FASN KO Caco2 cells were pre-seeded in 96 well plates and allowed to grow to confluency. Infection with SARS-CoV-2 was performed at an MOI of 0.01 in the serum-free media. One hour post adsorption, virus inoculum was removed and medium containing the designated serum/lipid treatment was added. Cells were incubated at 37C with 5% CO2 until the designated end point. Supernatants were collected and quantified by plaque assay.
GFP-2xFYVE assay
Huh7 cells were seeded in 96-well black walled plates and transfected with a pEGFP-2xFYVE plasmid obtained from Addgene (#140047) using Lipofectamine 2000 (Invitrogen St. Louis, MO, USA). 24 hours post transfection (hpt) media was removed and replaced with media containing either DMSO, VPS34-IN1 (2 μM), PIK-III (5 μM), SAR405 (5 μM), Compound 19 (5 μM), or Orlistat (200 μM). 24 hours post treatment cells were imaged using a BioTek Cytation 5 to determine the localization pattern of eGFP-2xFYVE.
Quantification and statistical analysis
All data are representative of the mean and standard of error of three replicates. IC50s were determined by fitting percent inhibition data with a four-parameter non-linear regression. IC90s were interpolated from the fit data generated from IC50 calculations using the online ECanything calculator (GraphPad). Statistical significance was determined using one-way ANOVA with p < 0.05 being considered as significant. Levels of significance are indicated within the figures. Generation of plots and statistical analysis was all performed within Prism 9 unless otherwise indicated (GraphPad).
Acknowledgments
We would like to thank the Georgia State University High Containment team Natasha Griffith, Martin Wildes, and Robert “Mike” Walsh for their continuous support. This work was supported by NIH grants R01AI125453 and P01AI120943 (Amarasinghe), by a Fast Grant for COVID-19 from the Emergent Ventures program at the Mercatus Center of George Mason University, and by the Augusta University-Georgia State University Seed Grant program (to C.F.B.).
Author Contributions
C.G.W., A.S.J., J.A.S., and C.F.B. designed the experiments. C.G.W., A.S.J., and J.A.S. performed the experiments. C.G.W., A.S.J., J.A.S., A.M.N., S.A.C., and C.F.B. analyzed the data. A.M.N. and S.A.C. provided key equipment and supplies. J.C.-S., J.O., and K.H. generated and provided the FASN knockout cells. C.W.G., A.S.J., J.A.S., and C.F.B. wrote the paper with input from the other authors.
Declaration of Interests
A.M.N. and S.A.C. are employees of Axion BioSystems who provided the Axion Maestro Z instrument used in these studies. C.F.B. serves on the scientific advisory board of Axion BioSystems. J.C.S., J.O., and K.H. are employees of Synthego Corporation who generated and provided the FASN knockout cells.
Published: July 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109479.
Supplemental information
Data and code availability
The published articled includes all datasets generated during this study. Raw data are available from the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Farha M., Thanaraj T.A., Qaddoumi M.G., Hashem A., Abubaker J., Al-Mulla F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020;21:3544. doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman D., Tumanov S., Qiu B., Michalopoulou E., Spata M., Azzam A., Xie H., Simon M.C., Kamphorst J.J. Triglycerides Promote Lipid Homeostasis during Hypoxic Stress by Balancing Fatty Acid Saturation. Cell Rep. 2018;24:2596–2605.e5. doi: 10.1016/j.celrep.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsabeeh N., Chausse B., Kakimoto P.A., Kowaltowski A.J., Shirihai O. Cell culture models of fatty acid overload: Problems and solutions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:143–151. doi: 10.1016/j.bbalip.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammer E., Nietzsche S., Rien C., Kühnl A., Mader T., Heller R., Sauerbrei A., Henke A. The anti-obesity drug orlistat reveals anti-viral activity. Med. Microbiol. Immunol. (Berl.) 2015;204:635–645. doi: 10.1007/s00430-015-0391-4. [DOI] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.M. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 2016;473:2251–2271. doi: 10.1042/BCJ20160170. [DOI] [PubMed] [Google Scholar]
- Baenke F., Peck B., Miess H., Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013;6:1353–1363. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago R., Malik N., Munson M.J., Prescott A.R., Davies P., Sommer E., Shpiro N., Ward R., Cross D., Ganley I.G., Alessi D.R. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhache W., Neyret A., McKellar J., Clop C., Bernard E., Weger-Lucarelli J., Briant L. Fatty acid synthase and stearoyl-CoA desaturase-1 are conserved druggable cofactors of Old World Alphavirus genome replication. Antiviral Res. 2019;172:104642. doi: 10.1016/j.antiviral.2019.104642. [DOI] [PubMed] [Google Scholar]
- Benson K., Cramer S., Galla H.J. Impedance-based cell monitoring: barrier properties and beyond. Fluids Barriers CNS. 2013;10:5. doi: 10.1186/2045-8118-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82:2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M.A., Zecchin K.G., Seguin F., Bastos D.C., Agostini M., Rangel A.L., Veiga S.S., Raposo H.F., Oliveira H.C., Loda M. Fatty acid synthase inhibition with Orlistat promotes apoptosis and reduces cell growth and lymph node metastasis in a mouse melanoma model. Int. J. Cancer. 2008;123:2557–2565. doi: 10.1002/ijc.23835. [DOI] [PubMed] [Google Scholar]
- Chen B., Sun Y., Niu J., Jarugumilli G.K., Wu X. Protein Lipidation in Cell Signaling and Diseases: Function, Regulation, and Therapeutic Opportunities. Cell Chem. Biol. 2018;25:817–831. doi: 10.1016/j.chembiol.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W., Gill M., Esposito A., Kaminski C.F., Courousse N., Chwetzoff S., Trugnan G., Keshavan N., Lever A., Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloherty A.P.M., Olmstead A.D., Ribeiro C.M.S., Jean F. Hijacking of Lipid Droplets by Hepatitis C, Dengue and Zika Viruses-From Viral Protein Moonlighting to Extracellular Release. Int. J. Mol. Sci. 2020;21:7901. doi: 10.3390/ijms21217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davda D., El Azzouny M.A., Tom C.T., Hernandez J.L., Majmudar J.D., Kennedy R.T., Martin B.R. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 2013;8:1912–1917. doi: 10.1021/cb400380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S.S.G., Soares V.C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., Teixeira L., Nunes da Silva M.A., Barreto E., Mattos M. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16:e1009127. doi: 10.1371/journal.ppat.1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle W.E., Nyfeler B., Nagel J., Elling R.A., Liu S., Triantafellow E., Menon S., Wang Z., Honda A., Pardee G. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- Feng Z., Xu K., Kovalev N., Nagy P.D. Recruitment of Vps34 PI3K and enrichment of PI3P phosphoinositide in the viral replication compartment is crucial for replication of a positive-strand RNA virus. PLoS Pathog. 2019;15:e1007530. doi: 10.1371/journal.ppat.1007530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Serradilla M., Risco C., Pacheco B. Drug repurposing for new, efficient, broad spectrum antivirals. Virus Res. 2019;264:22–31. doi: 10.1016/j.virusres.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Cheung W., Richards J.E., Lever A., Desselberger U. Inhibition of rotavirus replication by downregulation of fatty acid synthesis. J. Gen. Virol. 2013;94:1310–1317. doi: 10.1099/vir.0.050146-0. [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Simonsen A., Stenmark H. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 2001;355:249–258. doi: 10.1042/0264-6021:3550249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., QCRG Structural Biology Consortium. Zoonomia Consortium Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yu H., Gu W., Luo X., Li R., Zhang J., Xu Y., Yang L., Shen N., Feng L., Wang Y. Autophagy Negatively Regulates Transmissible Gastroenteritis Virus Replication. Sci. Rep. 2016;6:23864. doi: 10.1038/srep23864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J., Reggiori F., de Haan C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458-459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorson D.L., McCune S.A. Inhibition of fatty acid synthesis in isolated adipocytes by 5-(tetradecyloxy)-2-furoic acid. Lipids. 1984;19:851–856. doi: 10.1007/BF02534514. [DOI] [PubMed] [Google Scholar]
- Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitakarun A., Khongwichit S., Wikan N., Roytrakul S., Yoksan S., Rajakam S., Davidson A.D., Smith D.R. Evaluation of the antiviral activity of orlistat (tetrahydrolipstatin) against dengue virus, Japanese encephalitis virus, Zika virus and chikungunya virus. Sci. Rep. 2020;10:1499. doi: 10.1038/s41598-020-58468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Schneider W.M., Sanchez-Rivera F.J., Luna J.M., Ashbrook A.W., Soto-Feliciano Y.M., Leal A.A., Le Pen J., Ricardo-Lax I., Michailidis E. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Washington State 2019-nCoV Case Investigation Team First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A., Harrington E., Cornella-Taracido I., Furet P., Knapp M.S., Glick M., Triantafellow E., Dowdle W.E., Wiedershain D., Maniara W. Potent, Selective, and Orally Bioavailable Inhibitors of VPS34 Provide Chemical Tools to Modulate Autophagy in Vivo. ACS Med. Chem. Lett. 2015;7:72–76. doi: 10.1021/acsmedchemlett.5b00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T., Conant D., Rossi N., Maures T., Waite K., Yang J., Joshi S., Kelso R., Holden K., Enzmann B.L. Inference of CRISPR Edits from Sanger Trace Data. bioRxiv. 2019 doi: 10.1101/251082. [DOI] [PubMed] [Google Scholar]
- Igal R.A., Wang P., Coleman R.A. Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. Biochem. J. 1997;324:529–534. doi: 10.1042/bj3240529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N., Zong W.X. Class III PI3K Vps34: essential roles in autophagy, endocytosis, and heart and liver function. Ann. N Y Acad. Sci. 2013;1280:48–51. doi: 10.1111/nyas.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jureka A.S., Silvas J.A., Basler C.F. Propagation, Inactivation, and Safety Testing of SARS-CoV-2. Viruses. 2020;12:622. doi: 10.3390/v12060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., George D., Prior A.M., Prasain K., Hao S., Le D.D., Hua D.H., Chang K.O. Novel triacsin C analogs as potential antivirals against rotavirus infections. Eur. J. Med. Chem. 2012;50:311–318. doi: 10.1016/j.ejmech.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel S.J., Axelrod F., Rozenkrantz N., Smith J.W. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64:2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- Kuhajda F.P., Pizer E.S., Li J.N., Mani N.S., Frehywot G.L., Townsend C.A. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc. Natl. Acad. Sci. USA. 2000;97:3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T.G. Mechanistic similarities in docking of the FYVE and PX domains to phosphatidylinositol 3-phosphate containing membranes. Prog. Lipid Res. 2007;46:315–327. doi: 10.1016/j.plipres.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A., Desselberger U. Rotavirus replication and the role of cellular lipid droplets: New therapeutic targets? J. Formos. Med. Assoc. 2016;115:389–394. doi: 10.1016/j.jfma.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Liefhebber J.M., Hague C.V., Zhang Q., Wakelam M.J., McLauchlan J. Modulation of triglyceride and cholesterol ester synthesis impairs assembly of infectious hepatitis C virus. J. Biol. Chem. 2014;289:21276–21288. doi: 10.1074/jbc.M114.582999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. Coronavirus Pandemic-Therapy and Vaccines. Biomedicines. 2020;8:109. doi: 10.3390/biomedicines8050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wang W., Devarakonda T., Zhou H., Wang X.Y., Salloum F.N., Spiegel S., Fang X. Functional analysis of molecular and pharmacological modulators of mitochondrial fatty acid oxidation. Sci. Rep. 2020;10:1450. doi: 10.1038/s41598-020-58334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau N., Fromentin R., Savard C., Duval M., Tremblay M.J., Leclerc D. Palmitoylation of hepatitis C virus core protein is important for virion production. J. Biol. Chem. 2009;284:33915–33925. doi: 10.1074/jbc.M109.018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Machamer C.E. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology. 2010;405:139–148. doi: 10.1016/j.virol.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino-Ramos T., Vázquez-Calvo Á., Casas J., Sobrino F., Saiz J.C., Martín-Acebes M.A. Modification of the Host Cell Lipid Metabolism Induced by Hypolipidemic Drugs Targeting the Acetyl Coenzyme A Carboxylase Impairs West Nile Virus Replication. Antimicrob. Agents Chemother. 2015;60:307–315. doi: 10.1128/AAC.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasheri N., Joyce M., Rouleau Y., Yang P., Yao S., Tyrrell D.L., Pezacki J.P. Modulation of fatty acid synthase enzyme activity and expression during hepatitis C virus replication. Chem. Biol. 2013;20:570–582. doi: 10.1016/j.chembiol.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Tremel S., Masson G.R., McGinney L., Boulanger J., Rostislavleva K., Johnson C.M., Niewczas I., Clark J., Williams R.L. Membrane characteristics tune activities of endosomal and autophagic human VPS34 complexes. eLife. 2020;9:e58281. doi: 10.7554/eLife.58281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio. 2017;8 doi: 10.1128/mBio.01658-17. e01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy. 2015;11:725–726. doi: 10.1080/15548627.2015.1033601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Chouljenko V.N., Iyer A., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Terrier O., Rosa-Calatrava M. Drug Repurposing Approaches for the Treatment of Influenza Viral Infection: Reviving Old Drugs to Fight Against a Long-Lived Enemy. Front. Immunol. 2019;10:531. doi: 10.3389/fimmu.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., de Haan C.A., Molinari M. Unconventional use of LC3 by coronaviruses through the alleged subversion of the ERAD tuning pathway. Viruses. 2011;3:1610–1623. doi: 10.3390/v3091610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan B., Flamand O., Vescovi L., Dureuil C., Durand L., Fassy F., Bachelot M.F., Lamberton A., Mathieu M., Bertrand T. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- Saini K.S., Lanza C., Romano M., de Azambuja E., Cortes J., de Las Heras B., de Castro J., Lamba Saini M., Loibl S., Curigliano G. Repurposing anticancer drugs for COVID-19-induced inflammation, immune dysfunction, and coagulopathy. Br. J. Cancer. 2020;123:694–697. doi: 10.1038/s41416-020-0948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W.C., Chao T.C., Huang Y.L., Weng S.C., Jeng K.S., Lai M.M. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J. Virol. 2011;85:10561–10571. doi: 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp E.B., Boscarino J.A., Logan H.L., Goletz J.T., Gallagher T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006;80:1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongluan N., Ramphan S., Wintachai P., Jaresitthikunchai J., Khongwichit S., Wikan N., Rajakam S., Yoksan S., Wongsiriroj N., Roytrakul S., Smith D.R. Involvement of fatty acid synthase in dengue virus infection. Virol. J. 2017;14:28. doi: 10.1186/s12985-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Tseng J., Perrone L., Worthy M., Popov V., Peters C.J. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J. Virol. 2005;79:9470–9479. doi: 10.1128/JVI.79.15.9470-9479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.T., Wang S.M., Huang K.J., Wang C.T. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. J. Biomed. Sci. 2014;21:34. doi: 10.1186/1423-0127-21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil S.J., Abu-Elheiga L.A. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 2009;50(Suppl):S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. Agents. 2020;55:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wu B., Lai T. Compassionate Use of Remdesivir in Covid-19. N. Engl. J. Med. 2020;382:e101. doi: 10.1056/NEJMc2015312. [DOI] [PubMed] [Google Scholar]
- Yu G.Y., Lee K.J., Gao L., Lai M.M. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 2006;80:6013–6023. doi: 10.1128/JVI.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Wong W.M., Mak L.F., Wang X., Chu H., Yuen K.Y., Kok K.H. Suppression of SARS-CoV-2 infection in ex-vivo human lung tissues by targeting class III phosphoinositide 3-kinase. J. Med. Virol. 2021;93:2076–2083. doi: 10.1002/jmv.26583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Case J.B., Yutuc E., Ma X., Shen S., Gomez Castro M.F., Liu Z., Zeng Q., Zhao H., Son J. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. USA. 2020;117:32105–32113. doi: 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W., 4th Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Zhao G., Souers A.J., Voorbach M., Falls H.D., Droz B., Brodjian S., Lau Y.Y., Iyengar R.R., Gao J., Judd A.S. Validation of diacyl glycerolacyltransferase I as a novel target for the treatment of obesity and dyslipidemia using a potent and selective small molecule inhibitor. J. Med. Chem. 2008;51:380–383. doi: 10.1021/jm7013887. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published articled includes all datasets generated during this study. Raw data are available from the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.