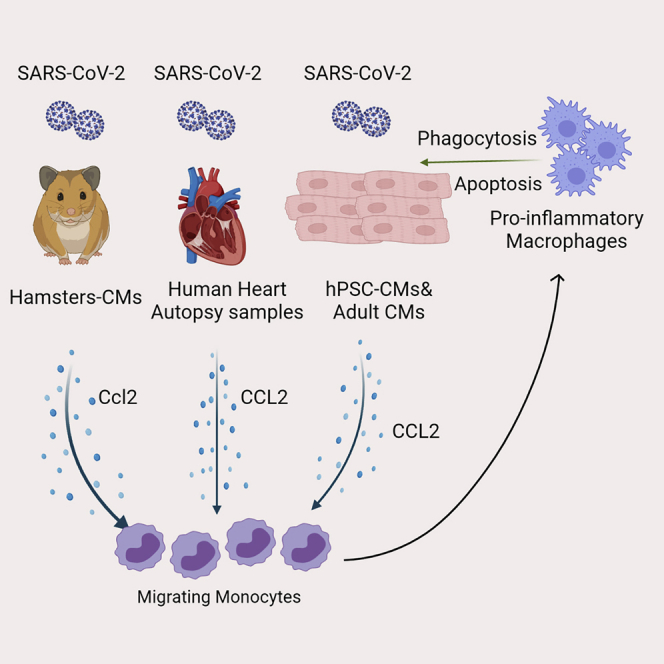
Summary
Heart injury has been reported in up to 20% of COVID-19 patients, yet the cause of myocardial histopathology remains unknown. Here, using an established in vivo hamster model, we demonstrate that SARS-CoV-2 can be detected in cardiomyocytes of infected animals. Furthermore, we found damaged cardiomyocytes in hamsters and COVID-19 autopsy samples. To explore the mechanism, we show that both human pluripotent stem cell-derived cardiomyocytes (hPSC-derived CMs) and adult cardiomyocytes (CMs) can be productively infected by SARS-CoV-2, leading to secretion of the monocyte chemoattractant cytokine CCL2 and subsequent monocyte recruitment. Increased CCL2 expression and monocyte infiltration was also observed in the hearts of infected hamsters. Although infected CMs suffer damage, we find that the presence of macrophages significantly reduces SARS-CoV-2-infected CMs. Overall, our study provides direct evidence that SARS-CoV-2 infects CMs in vivo and suggests a mechanism of immune cell infiltration and histopathology in heart tissues of COVID-19 patients.
Keywords: hamster, COVID-19, immune cell infiltration, cardiomyocyte, hPSC, CCL2
Graphical abstract
Highlights
-
•
SARS-CoV-2 is detected in cardiomyocytes of SARS-CoV-2-infected hamsters
-
•
SARS-CoV-2-infected human cardiomyocytes secrete CCL2
-
•
SARS-CoV-2-infected cardiomyocytes recruit monocytes by secreting CCL2
-
•
Macrophages decrease SARS-CoV-2 infection by engulfing infected CMs
In this article, Dr. Shuibing Chen and colleagues report the direct evidence of SARS-CoV-2 infection of cardiomyocytes in vivo and establish an in vitro model to study immune cell infiltration. They suggest that SARS-CoV-2 infection leads to secretion of CCL2, which recruits monocyte migration.
Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic is caused by the betacoronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). While respiratory failure is a predominant outcome, cardiac involvement is a common feature in hospitalized COVID-19 patients and is associated with poor prognosis. For example, in a Wuhan cohort, 7% of total patients and 22% of critically ill patients suffered myocardial injury, demonstrated by elevated cardiac biomarkers, such as high-sensitivity troponin I (hs-cTnI), or by electrocardiography and echocardiogram abnormalities (Wang et al., 2020). hs-cTnI was reported to be above the 99th percentile upper reference in 46% of non-survivors as opposed to 1% of survivors (Zhou et al., 2020). The mortality risk associated with acute cardiac injury was more significant than age, chronic pulmonary disease, or prior history of cardiovascular disease (Guo et al., 2020; Shi et al., 2020). As emerging cases of COVID-19-related Kawasaki disease-like symptoms are reported, many children with COVID-19 also suffer from cardiac dysfunction (Riphagen et al., 2020). There are also case reports of myocarditis in COVID-19 patients (Gnecchi et al., 2020; Inciardi et al., 2020; Tavazzi et al., 2020). The cause of these cardiac injuries observed in COVID-19 patients is not yet established but could potentially involve increased cardiac stress due to respiratory failure and hypoxemia, direct myocardial infection by SARS-CoV-2, or indirect cardiotoxicity from a systemic inflammatory response.
We and other groups have reported SARS-CoV-2 infection in human pluripotent stem cell-derived cardiomyocytes (hPSC-derived CMs) in vitro (Bojkova et al., 2020; Sharma et al., 2020; Yang et al., 2020). SARS-CoV-2 infection might also cause damage of CMs (Marchiano et al., 2021). Although several studies have detected SARS-CoV-2 RNA in autopsy samples from hearts of COVID-19 patients (Escher et al., 2020; Lindner et al., 2020), the presence of SARS-CoV-2 in CMs remains controversial. Tavazzi et al. (2020) and Lindner et al. (2020) identified SARS-CoV-2 virions in the interstitial cells of the myocardium of COVID-19 patients. Furthermore, Dolhnikoff et al. (2020) observed SARS-CoV-2 virions in cardiac tissues of an 11-year-old child with multisystem inflammatory syndrome in children related to COVID-19 who developed cardiac failure and passed away 1 day after admission to the hospital. Another potential mechanism for cardiac damage could be mediated by immune cells. Although it has been controversial whether CMs are directly infected by SARS-CoV-2 in patients, several studies using COVID-19 postmortem heart samples have consistently identified abnormal inflammatory infiltrates composed of CD11b+ macrophages (Escher et al., 2020), CD68+ macrophages (Lindner et al., 2020; Tavazzi et al., 2020), and, to a lesser extent, T cells (Yao et al., 2020).
Due to inherent challenges to collecting heart biopsies from COVID-19 patients during or after acute infection, we instead systematically examined heart tissues of SARS-CoV-2-infected Syrian hamsters (Mesocricetus auratus), an established and relevant animal model to study COVID-19 in vivo. We show that CMs of intranasally inoculated hamsters are infected leading to altered gene expression profiles associated with impaired CM function and increased reactive oxygen species (ROS), which was further confirmed in heart autopsies of COVID-19 patients. Using an immuno-cardiac co-culture platform with hPSC-derived CMs and monocytes/macrophages, we found that infected CMs recruit monocytes by secretion of CCL2 and that macrophage recruitment limits SARS-CoV-2 infection. This hPSC-based platform thus provides a useful new tool to model CM infection, immune cell infiltration, and cardiac histopathology of COVID-19 patients.
Results
SARS-CoV-2 is detected in the CMs of SARS-CoV-2-infected hamsters
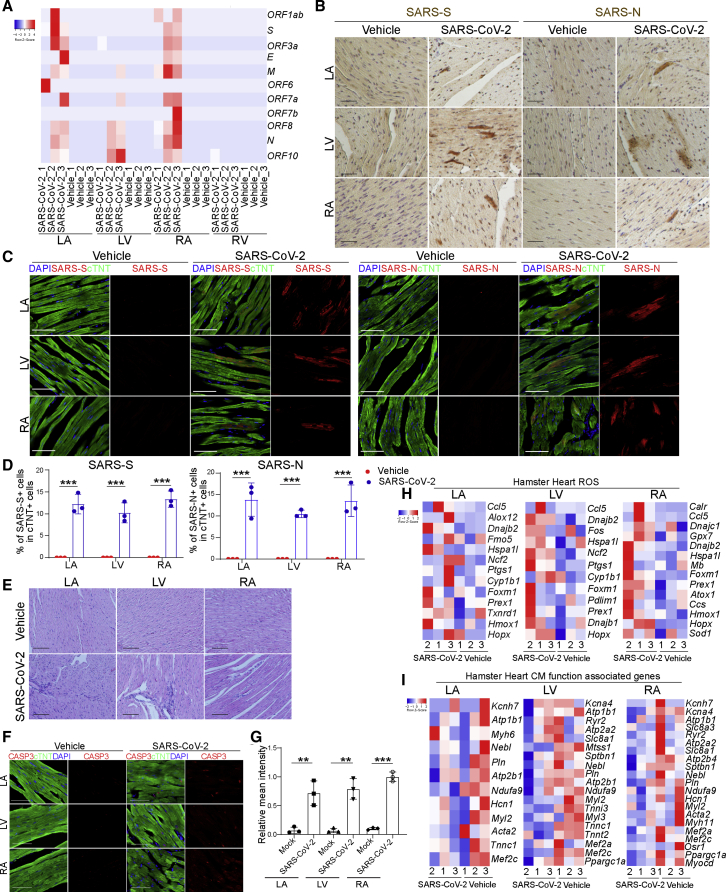
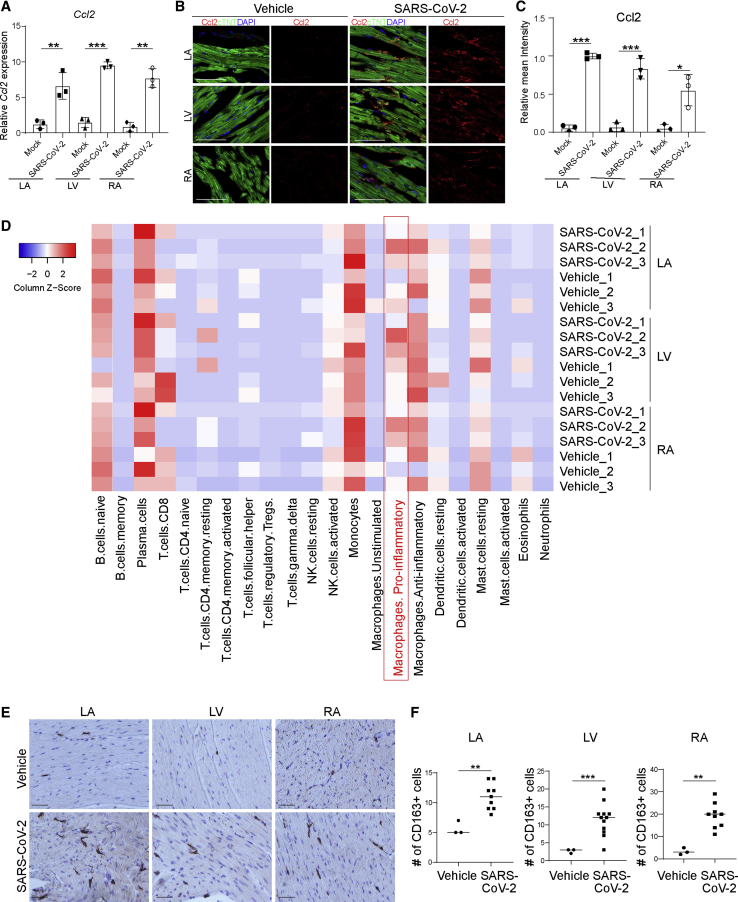
To examine the hearts of SARS-CoV-2-infected hamsters, 3- to 5-week-old male hamsters were intranasally inoculated with SARS-CoV-2. Forty-eight hours post infection (hpi), hamsters were euthanized and hearts were collected and separated into left ventricle (LV), left atrium (LA), right atrium (RA), and right ventricle (RV), followed by RNA sequencing (RNA-seq) analysis. SARS-CoV-2 transcripts were detected in hearts of six out of nine infected hamsters. Interestingly, viral transcripts were detected in LA, LV, and RA, but not RV (Figure 1A). SARS-CoV-2 spike and nucleocapsid protein were detected in CMs by immunohistochemistry (Figure 1B) and immunofluorescence staining (Figures 1C and 1D). Spike protein was also detected in cTNT− non-CMs (Figure S1A). Hematoxylin and eosin staining showed damaged cardiac tissue structure and infiltration of mononuclear cells in heart tissue from infected but not control animals (Figure 1E). Consistent with this tissue damage, we also observed significantly higher relative mean intensities of cleaved caspase-3 (CASP3) in cTNT+ cells of LA, LV, and RA (Figures 1F and 1G), suggesting increased rates of apoptosis in CMs of infected hamsters. RNA-seq analysis revealed transcript profiles consistent with increased expression of ROS-related genes (Figure 1H) and decreased expression levels of CM function-associated genes (Figure 1I) in LA, RA, and LV. The gene expression changes were quite robust, even though a relatively small number of infected CMs and minimal myocardial damage were detected in the immunostaining assays.
Figure 1.
SARS-CoV-2 is detected in CMs of SARS-CoV-2-infected hamsters
(A) Heatmap of SARS-CoV-2 genes in heart tissues obtained from SARS-CoV-2-infected (n = 3) or mock-infected (n = 3) hamsters.
(B) Immunohistochemistry staining of viral spike and nucleocapsid protein in the LA, LV, and RA heart tissues of from uninfected (n = 3) or infected (n = 3) hamsters. Scale bar, 50 μm.
(C and D) Immunofluorescence staining (C) and the percentage (D) of spike+ and nucleocapsid+ cells in the LA, LV, and RA heart tissues from uninfected (n = 3) or infected (n = 3) hamsters. Scale bar, 50 μm.
(E) H&E staining of LA, LV, and RA heart tissues from uninfected (n = 3) or infected (n = 3) hamsters. Scale bar, 50 μm.
(F and G) Immunofluorescence staining (F) and the relative mean intensity of CASP3 (G) in cTNT+ cells in the LA, LV, and RA heart tissues obtained from SARS-CoV-2-infected (n = 3) or mock-infected (n = 3) hamsters. Scale bar, 50 μm.
(H and I) Heatmap of ROS-associated genes (H) and CM function-associated genes (I) in the LA, LV, and RA heart tissues obtained from SARS-CoV-2-infected (n = 3) or mock-infected (n = 3) hamsters.
All heatmap data are presented as the Z score. All graphed data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S1.
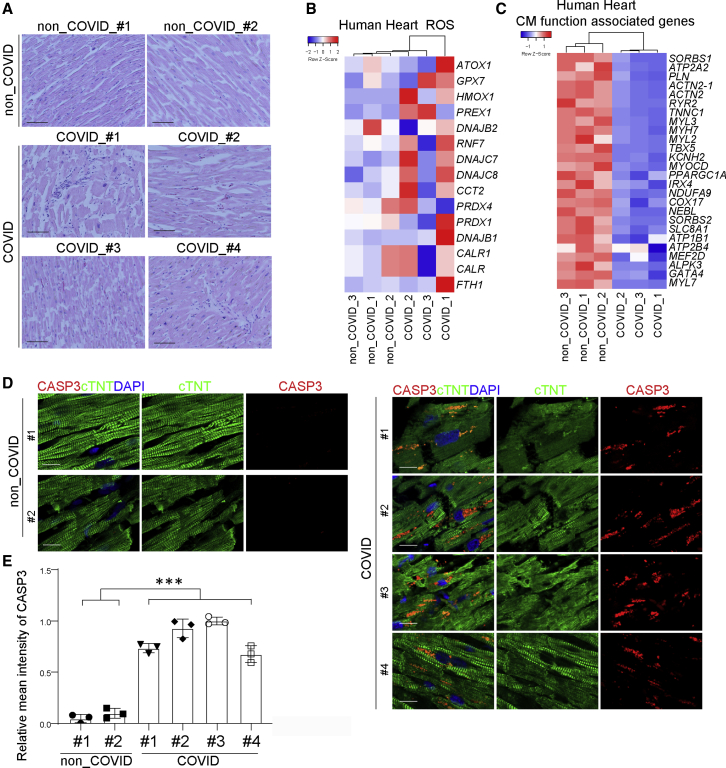
To validate these findings in clinical COVID-19 cases, we compared heart autopsy samples from non-COVID-19 donors and four COVID-19 patients with cardiac symptoms. New EKG findings, including atrial fibrillation/paced rhythm and ST-segment or T-wave changes, were detected in three of the COVID-19 patients. Troponin was detected (0.51–1.83 ng/mL) in all four COVID-19 patients. Consistent with our observations in SARS-CoV-2-infected hamsters, hematoxylin and eosin staining showed damage to cardiac tissues as well as infiltration of mononuclear cells in heart sections of the COVID-19 patients (Figure 2A). Furthermore, increased expression of ROS-associated genes and decreased expression of CM function-associated genes was observed (Figures 2B and 2C). Finally, CASP3 expression in cTNT+ cells of the COVID-19 patient samples was significantly higher than in samples from non-COVID-19 patients (Figures 2D and 2E), suggesting increased apoptosis of CMs in COVID-19 patients. Moreover, we also detected viral transcripts in heart tissue samples of COVID-19 patients (Figure S1B). Together, these data show evidence of SARS-CoV-2 cardiac infection and damage in vivo.
Figure 2.
Analysis of autopsy heart samples of non-COVID-19 and COVID-19 patients
(A) H&E staining of autopsy heart samples of non-COVID-19 and COVID-19 patients (n = 2 non-COVID-19 subjects, n = 4 COVID-19 patients). Scale bar, 50 μm.
(B and C) Heatmap of ROS-associated genes (B) and CM function-associated genes (C) in autopsy heart samples of non-COVID-19 and COVID-19 patients (n = 3 non-COVID-19 subjects, n = 3 COVID-19 patients). Data are presented as the Z score.
(D and E) Immunofluorescence staining (D) and the relative mean intensity of CASP3 (E) in cTNT+ cells in autopsy heart samples of non-COVID-19 and COVID-19 patients (n = 2 non-COVID-19 subjects, n = 4 COVID-19 patients). Scale bar, 50 μm.
Data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student's t test. ∗∗∗p < 0.001.
See also Figure S1.
SARS-CoV-2-infected CMs lose cell identity and secrete CCL2
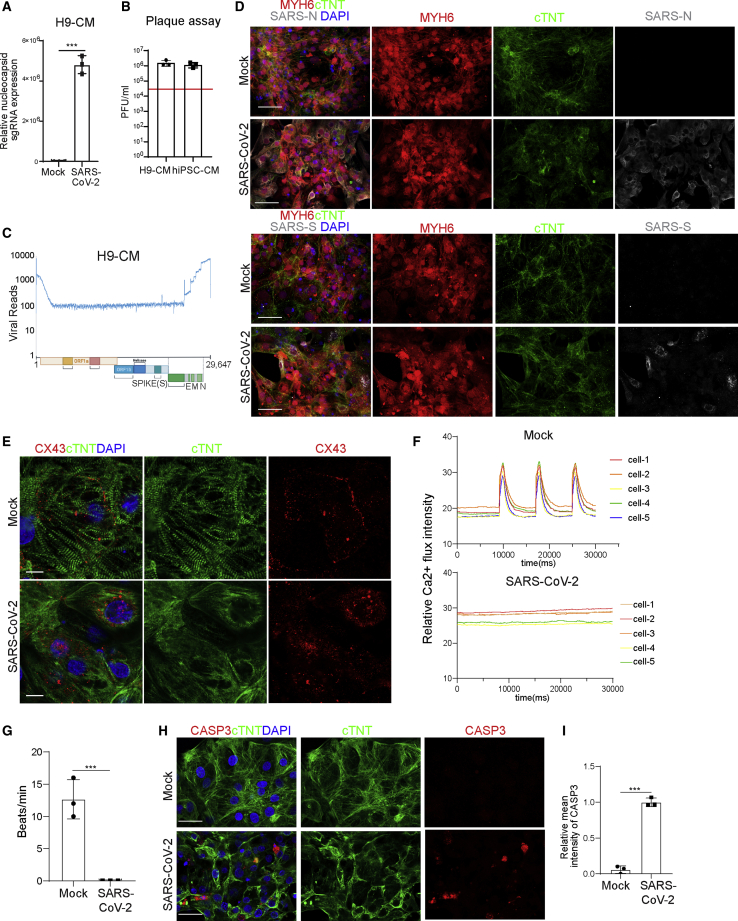
Our previous studies showed that hPSC-derived CMs are permissive to SARS-CoV-2 infection (Yang et al., 2020), which established a platform to model CM cellular response to SARS-CoV-2 infection. CMs were derived from an MYH6:mCherry H9 human embryonic stem cell (hESC) reporter line or an induced PSC (iPSC) line (Tsai et al., 2020) (Figure S2A). Over 90% of these cells expressed mCherry and/or stained positive with antibodies recognizing sarcomeric α-actinin and cTNT (Figure S2B). To better characterize the permissiveness of this hPSC-derived CM platform to SARS-CoV-2, we infected the hESC-derived CMs or iPSC-derived CMs with SARS-CoV-2 (USA-WA1/2020). We observed robust infection of CMs from both sources at 24–48 hpi based on subgenomic transcription (Figures 3A and S3A) and the release of infectious viral particles (Figures 3B and S3B). At 72 hpi, both viral RNA and infectious viral titers decreased, which could be explained by the death of infected cells. Efficient replication in hPSC-derived CMs was demonstrated by full coverage of the viral genome by RNA-seq analysis (Figure 3C) as well as spike and nucleocapsid immunostaining (Figure 3D). In contrast to uninfected CMs, which show organized sarcomeres, SARS-CoV-2-infected CMs lose sarcomere structure (Figure 3E). Connexin 43 (CX43) was detected at the cell membranes of uninfected CMs, while it showed an internalized subcellular localization in SARS-CoV-2-infected CMs, suggesting the loss of gap junctions (Figure 3E). This also corresponded to a loss of Ca2+ influx for infected CMs, leading to the loss of beating (Figures 3F and 3G; Videos S1 and S2). Finally, increased CASP3 staining suggests an increased rate of apoptosis of SARS-CoV-2-infected CMs (Figures 3H and 3I).
Figure 3.
SARS-CoV-2 infection causes CM damage
(A) Relative abundance of viral subgenomic RNA (N) transcription in SARS-CoV-2-infected H9-derived CMs (24 hpi, MOI = 0.1, n = 3 independent experiments).
(B) Plaque assay of SARS-CoV-2-infected H9-derived CMs and iPSC-derived CMs (24 hpi, MOI = 0.1). The red line indicates the input virus: 4 × 104 pfu.
(C) Read alignment to the SARS-CoV-2 genome after RNA-seq analysis of SARS-CoV-2-infected H9-derived CMs (24 hpi, MOI = 0.1, n = 3 independent experiments). Schematic denotes the SARS-CoV-2 genome.
(D) Immunofluorescence staining using viral spike and nucleocapsid antibodies in SARS-CoV-2-infected H9-derived CMs (24 hpi, MOI = 0.1, n = 3 independent experiments). Scale bar, 50 μm.
(E) Immunofluorescence staining using cTNT and CX43 antibodies on mock or SARS-CoV-2-infected H9-derived CMs at 24 hpi (MOI = 0.1, n = 3 independent experiments). Scale bar, 10 μm.
(F) Quantification of Ca2+ flux intensity of mock or SARS-CoV-2-infected H9-derived CMs at 48 hpi (MOI = 0.1, n = 3 independent experiments). Each condition shows five cells.
(G) Beating rate of mock- or SARS-CoV-2-infected H9-derived CMs at 48 hpi (MOI = 0.1, n = 3 independent experiments).
(H and I) Immunofluorescence staining (H) and the relative mean intensity of CASP3 (I) in cTNT+ cells of mock- or SARS-CoV-2-infected H9-derived CMs at 24 hpi (MOI = 0.1, n = 3 independent experiments). Scale bar, 50 μm.
Data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test. ∗∗∗p < 0.001.
See also Figures S2 and S3 and Videos S1 and S2.
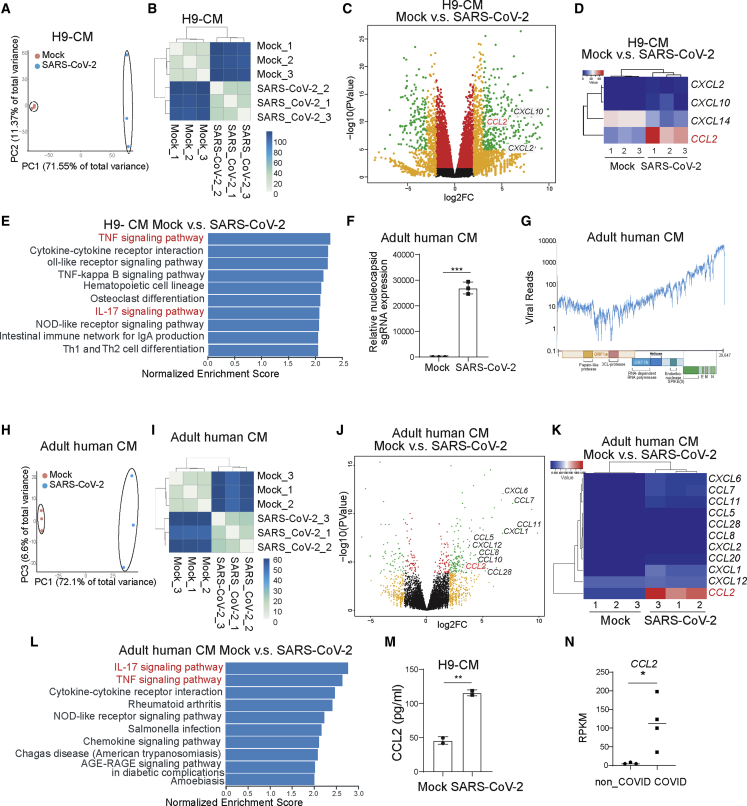
RNA-seq analysis of H9 hESC-derived CMs showed significantly different host transcript profiles of mock and infected CMs (Figures 4A and 4B). Consistent with the host transcriptional response observed in heart autopsies from COVID-19 patients, infected H9 hESC-derived CMs demonstrated a robust induction of chemokines and cytokines, including CCL2 (Figures 4C and 4D). KEGG pathway analysis of differentially expressed genes highlighted pathways involved in inflammatory and immune responses, including the tumor necrosis factor (TNF) signaling pathway, cytokine-cytokine receptor interaction, the nuclear factor κB signaling pathway, and the interleukin-17 (IL-17) signaling pathway (Figure 4E).
Figure 4.
CMs secrete CCL2 upon SARS-CoV-2 infection
(A and B) Principal-component analysis (PCA) plot (A) and heatmap (B) analysis of H9-derived CMs infected with SARS-CoV-2 virus or mock infected at 24 hpi (MOI = 0.1, n = 3 independent experiments).
(C and D) Volcano plot (C) and heatmap (D) analysis of chemokines expressed by H9-derived CMs infected with SARS-CoV-2 virus or mock at 24 hpi (MOI = 0.1, n = 3 independent experiments). Colored dots labeled correspond to chemokines with significant (p < 0.05) and greater than 2-fold expression level changes.
(E) KEGG analysis of H9-derived CMs infected with SARS-CoV-2 virus or mock at 24 hpi (MOI = 0.1, n = 3 independent experiments).
(F) Relative subgenomic RNA (N) transcription in adult human CMs at 24 hpi of SARS-CoV-2 virus at 24 hpi (MOI = 0.1, n = 3 independent experiments).
(G) Alignment of the transcriptome with the viral genome in SARS-CoV-2-infected adult human CMs at 24 hpi (MOI = 0.1, n = 3 independent experiments). Schematic denotes the SARS-CoV-2 genome.
(H and I) PCA plot (H) and heatmap (I) analysis of adult human CMs infected with SARS-CoV-2 virus or mock at 24 hpi (MOI = 0.1, n = 3 independent experiments).
(J and K) Volcano plot (J) and heatmap (K) analysis of chemokines expressed by adult human CMs infected with SARS-CoV-2 virus or mock infected at 24 hpi (MOI = 0.1, n = 3 independent experiments). Colored dots labeled correspond to chemokines with significant (p < 0.05) and greater than 2-fold expression level changes.
(L) KEGG analysis of adult human CMs infected with SARS-CoV-2 virus or mock infected at 24 hpi (MOI = 0.1, n = 3 independent experiments).
(M) ELISA was performed to examine the protein level of CCL2 in H9-derived CMs infected with SARS-CoV-2 virus or mock infected at 24 hpi (MOI = 0.1, n = 3 independent experiments).
(N) CCL2 transcript levels in autopsy heart samples of non-COVID-19 and COVID-19 patients (n = 3 non-COVID-19 subjects, n = 4 COVID-19 patients). RPKM, reads per kilobase of transcript, per million mapped reads.
Data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
We also analyzed the response of adult human CMs to SARS-CoV-2 infection, to confirm responses are not specific to an hPSC-derived platform. Similar to hPSC-derived CMs, robust viral replication was detected in adult CMs (Figures 4F and 4G). In addition, the host transcriptional response to viral infection revealed a similar pattern, with robust induction of chemokines and cytokines, including CCL2 (Figures 4H–4K). KEGG pathway analysis highlighted pathways involved in inflammatory and immune responses, including the IL-17 signaling pathway, the TNF signaling pathway, the cytokine-cytokine receptor interaction, and the chemokine signaling pathway in the infected adult human CMs (Figure 4L). Finally, significantly increased protein levels of CCL2 were confirmed by ELISA in the medium of H9 hESC-derived CMs after SARS-CoV-2 infection, compared with mock-infected cells (Figure 4M), and higher CCL2 expression levels were also detected in heart tissue samples from COVID-19 patients compared with those from non-COVID-19 patients (Figure 4N).
Ccl2 expression and macrophage infiltration in hearts of SARS-CoV-2-infected hamsters
To further investigate the role of Ccl2 in myocardial pathology, we examined Ccl2 expression in the hearts of SARS-CoV-2-infected hamsters. Consistent with our data in hPSC-derived CMs and human samples, the LA, LV, and RA of SARS-CoV-2-infected hamsters showed increased levels of Ccl2 expression (Figure 5A), which was further validated by immunostaining (Figures 5B and 5C). Cell-mixture deconvolution using an LM22 matrix (Vallania et al., 2018) identified the enrichment of pro-inflammatory macrophages in the LA, LV, and RA of SARS-CoV-2-infected hamsters compared with controls (Figure 5D), which is consistent with previous reports of abnormal macrophage infiltration in hearts of COVID-19 patients (Escher et al., 2020; Lindner et al., 2020; Tavazzi et al., 2020; Yao et al., 2020). This was further confirmed by immunohistochemistry analysis showing an increase of CD163+ macrophages in the LA, LV, and RA of SARS-CoV-2-infected hamsters (Figures 5E and 5F).
Figure 5.
Pro-inflammatory macrophages are enriched in hearts of SARS-CoV-2-infected hamsters
(A) Relative expression levels of Ccl2 in LA, LV, and RA heart tissues obtained from SARS-CoV-2-infected hamsters (n = 3) and mock-infected hamsters (n = 3).
(B and C) Immunofluorescence staining (B) and quantification (C) of Ccl2 in LA, LV, and RA heart tissues obtained from SARS-CoV-2-infected hamsters (n = 3) and mock-infected hamsters (n = 3). Scale bar, 50 μm.
(D) Cell-mixture deconvolution identified the enrichment of immune cells in the LA, LV, and RA of SARS-CoV-2-infected hamsters (n = 3) compared with mock-infected hamsters (n = 3).
(E and F) Immunohistochemistry staining (E) and quantification (F) of CD163+ cells in LA, LV, and RA heart tissues obtained from SARS-CoV-2-infected hamsters (n = 3) and mock-infected hamsters (n = 3). Scale bar, 50 μm.
Data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
SARS-CoV-2-infected CMs recruit monocytes by secreting CCL2
Macrophages include tissue-resident macrophages and migrating macrophages (Ginhoux and Jung, 2014). Migrating macrophages are usually derived from monocytes in the blood. During inflammation, circulating monocytes leave the bloodstream and migrate into tissues where, following conditioning by local growth factors, pro-inflammatory cytokines, and microbial products, they differentiate into macrophages (Shi et al., 2020).
We therefore examined the ability of SARS-CoV-2-infected CMs to stimulate migration of monocytes. Monocytes were derived from the same parental H9- or H1-hESC lines following a previously reported protocol (Cao et al., 2019) (Figure S4A) through a stepwise manner, including the generation of mesodermal cells, followed by hematopoietic progenitor cells, monocytes (Figure S4B), and finally CD14+/CD11B+ macrophages (Figure S4C). To determine whether these hPSC-derived macrophages are susceptible to SARS-CoV-2 infection, H9- or H1-hESC-derived macrophages were infected with SARS-CoV-2. However, viral replication was detected neither by qRT-PCR nor plaque assay, suggesting that macrophages are not productively infected by SARS-CoV-2 (Figures S4D and S4E).
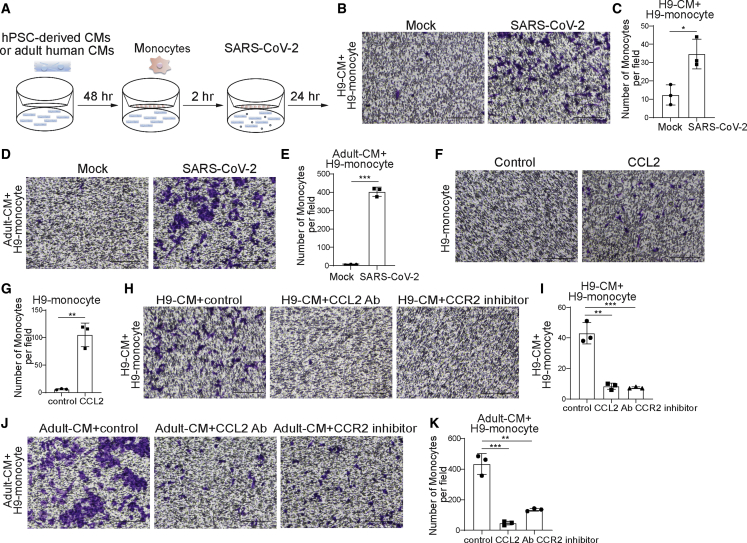
Next, we analyzed the ability of infected CMs to recruit monocytes to the site of infection. To that end, we plated hPSC-derived CMs at the bottom and hPSC-derived monocytes at the top of a transwell plate and infected the CMs at the bottom of the plate (Figure 6A). Monocyte migration was quantified 24 hpi by crystal violet staining. The number of monocytes that migrated was significantly higher when cultured with SARS-CoV-2-infected CMs compared with when cultured with mock-infected hPSC-derived CMs using monocytes derived from either of the two hESC lines, indicating that SARS-CoV-2-infected CMs recruit monocytes (Figures 6B, 6C, S5A, and S5B).
Figure 6.
CMs recruit monocytes following SARS-CoV-2 infection through secreting CCL2
(A) Scheme of the monocyte recruitment assay using hPSC-derived CMs or adult human CMs and hPSC-derived monocytes in the presence of SARS-CoV-2 infection
(B and C) Phase contrast images (B) and quantification (C) of migrated H9-derived monocytes recruited by H9-derived CMs infected with SARS-CoV-2 virus or mock infected in the monocyte migration assay (MOI = 0.1, n = 3 independent experiments). Scale bar, 100 μm.
(D and E) Phase contrast images (D) and quantification (E) of H9-derived monocytes recruited by adult human CMs infected with SARS-CoV-2 virus or mock infected in the monocyte recruitment assay (MOI = 0.1, n = 3 independent experiments). Scale bar, 100 μm.
(F and G) Phase contrast images (F) and quantification (G) of migrated H9-derived monocytes recruited by CCL2 in the monocyte recruitment assay (n = 3 independent experiments). Scale bar, 100 μm.
(H and I) Phase contrast images (H) and quantification (I) of migrated H9-derived monocytes recruited by H9-derived CMs infected with SARS-CoV-2 virus and treated with CCL2-neutralizing antibody or CCR2 inhibitor (RS504393) in the monocyte recruitment assay (MOI = 0.1, n = 3 independent experiments). Scale bar, 100 μm.
(J and K) Phase contrast images (J) and quantification (K) of migrated H9-derived monocytes recruited by adult human CMs infected with SARS-CoV-2 virus and treated with CCL2-neutralizing antibody or CCR2 inhibitor in the monocyte recruitment assay (MOI = 0.1, n = 3 independent experiments). Scale bar, 100 μm.
Data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S4 and S5.
We next tested whether adult human CMs infected with SARS-CoV-2 also recruit monocytes. Consistent with experiments using hPSC-derived CMs, monocytes were recruited at a significantly enhanced rate when cultured with SARS-CoV-2-infected adult human CMs than when cultured with mock-infected adult human CMs (Figures 6D, 6E, S5C, and S5D).
We previously showed that infected CMs secrete significant levels of CCL2. Therefore, to determine whether CCL2 is sufficient to induce monocyte migration, CCL2 was added to the bottom of a transwell plate with monocytes cultured at the top. Indeed, significantly more monocytes were found in CCL2 containing media than in untreated media (Figures 6F, 6G, S5E, and S5F). To determine whether CCL2 is the key driver for monocyte migration, hPSC-derived CMs or adult human CMs were cultured as before with monocytes in transwell plates and SARS-CoV-2 infections were performed in the absence or presence of CCL2-neutralizing antibodies or CCR2 inhibitors (Figures 6H–6K and S5G–S5J). Interestingly, the recruitment of monocytes by infected CMs was significantly inhibited if CCL2 signaling was disrupted, either by neutralizing CCL2 antibodies or by inhibition of the CCL2 receptor CCR2. Together, these data suggest that SARS-CoV-2 infection of CMs induces CCL2 secretion, which recruits monocytes.
Co-culture of hPSC-derived CMs and macrophages reveals that macrophages decrease SARS-CoV-2-infected CMs
We next investigated how recruited macrophages affect viral infection. To model the viral entry process, we created an immunocardiac co-culture platform containing hPSC-derived CMs and hPSC-derived macrophages and infected it with a SARS-CoV-2 spike protein pseudovirus carrying a luciferase (Luc) reporter. Notably, in the presence of macrophages Luc activity was significantly decreased in a dose-dependent manner (Figures S6A and S6B). Immunostaining further confirmed the decrease of Luc+ cells in MYH6:mCherry+ cells (Figures S6C and S6D).
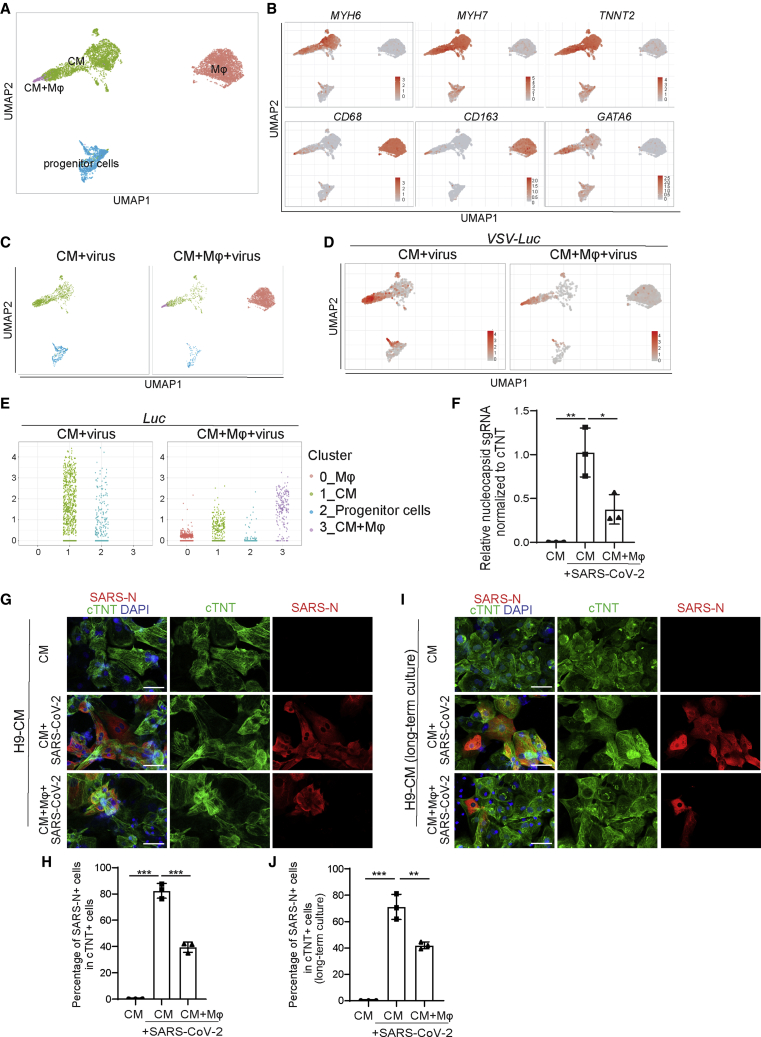
The immunocardiac co-culture was further examined by single-cell RNA-seq (scRNA-seq). The transcript profiling data were projected using Uniform Manifold Approximation and Projection. In the virus-infected immunocardiac co-culture platform, four distinct cell clusters were identified: CMs, macrophages, stem/progenitor cells, and one cluster expressing both CM and macrophage markers (Figure 7A). The expression of marker genes, including MYH6, MYH7, and TNNT2 (CMs), and CD163 and CD68 (macrophages), and GATA6 (progenitor cells) in each cell population confirmed the robustness of the cell-type classification strategy (Figures 7B, S6E, and S6F).
Figure 7.
A virus-immunocardiac co-culture platform reveals that hPSC-derived macrophages limit infection of SARS-CoV-2 CMs
(A) Uniform Manifold Approximation and Projection (UMAP) analysis of the virus-immunocardiac tissue platform containing hPSC-derived CMs and macrophages.
(B) UMAP of hPSC-derived CM and macrophage-related markers differentially expressed in each cluster. Relative expression levels of each marker gene ranged from low (gray) to high (red) as indicated.
(C) UMAP analysis of clusters in hPSC-derived CMs infected with SARS-CoV-2 entry virus (CM + virus) and the virus-immunocardiac tissue platform containing hPSC-derived CMs and macrophages infected with SARS-CoV-2 entry virus (CM + macrophage + virus).
(D) UMAP analysis of Luc expression in hPSC-derived CMs infected with SARS-CoV-2 entry virus (CM + virus) and the virus-immunocardiac tissue platform containing hPSC-derived CMs and macrophages infected with SARS-CoV-2 entry virus (CM + macrophage + virus).
(E) Jitter plot of Luc expression in hPSC-derived CMs infected with SARS-CoV-2 entry virus (CM + virus) and the virus-immunocardiac tissue platform containing hPSC-derived CMs and macrophages and infected with SARS-CoV-2 entry virus (CM + macrophage + virus).
(F) qRT-PCR analysis at 24 hpi of hPSC-derived CMs infected with mock or SARS-CoV-2 in the presence or absence of macrophages (MOI = 0.1, n = 3 independent experiments).
(G and H) Immunostaining (G) and quantification (H) of hPSC-derived CMs at 24 hpi with mock or SARS-CoV-2 in the presence or absence of macrophages for short-time co-culture (24 h, MOI = 0.1, n = 3 independent experiments). Scale bar, 50 μm.
(I and J) Immunostaining (I) and quantification (J) of hPSC-derived CMs at 24 hpi with mock or SARS-CoV-2 in the presence or absence of macrophages for long-time co-culture (7 days, MOI = 0.1, n = 3 independent experiments). Scale bar, 50 μm.
Data are presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S6 and S7.
ACE2, the gene encoding the main cellular receptor for SARS-CoV-2, is mainly expressed in hPSC-derived CMs and cardiac progenitors (Figures S6G and S6H). The effector protease TMPRSS2 (Hoffmann et al., 2020) is not expressed in hPSC-derived CMs and only rarely expressed in hPSC-derived cardiac progenitors (Figures S6G and S6H). However, FURIN, the gene encoding a pro-protein convertase that pre-activates SARS-CoV-2 (Shang et al., 2020), and CTSL, the gene encoding cathepsin L, a proteinase that might be able to substitute for TMPRSS2 (Hoffmann et al., 2020), are highly expressed in both hPSC-derived CMs and cardiac progenitors.
Transcripts deriving from the pseudovirus, including Luc, were detected in infected CMs, but only at very low levels in macrophages (Figure S6I), which is consistent with our previous report (Yang et al., 2020). The cell cluster that expressed both CM and macrophage markers and, in addition, high levels of viral transcripts, likely represents infected CMs engulfed by macrophages (Figure 7C). Luc expression in CMs was significantly lower in the presence of co-cultured macrophages (Figures 7D and 7E). Consistently, infected CMs showed increased expression of CCL2, which was further elevated in the presence of macrophages (Figures S6J and S6K).
To further validate the impact of macrophages on SARS-CoV-2 infection, the immunocardiac co-culture platform containing hPSC-derived CMs and hPSC-derived macrophages was infected with SARS-CoV-2. In agreement with our previous findings, we found that viral sgRNA transcripts were significantly decreased after viral infection in the co-culture relative to the CM marker cTNT (Figure 7F). This was further confirmed by a decrease in viral nucleocapsid expression in cTNT+ cells (Figures 7G, 7H, S6L, and S6M). In hPSC-derived CMs and macrophages that were co-cultured long term for 1 week the same phenomenon was observed (Figures 7I and 7J). Together, these data suggest that macrophages play an active role in limiting the extent of SARS-CoV-2 infection potentially by engulfing infected CMs.
Discussion
Myocardial injury has been reported in COVID-19 patients and is associated with increased mortality (Ruan et al., 2020; Zhou et al., 2020), yet the cause of myocardial injury has not been characterized or elucidated. Recent studies using SARS-CoV-2 hACE2 transgenic mice or hPSC-derived CMs reported the detection of viral RNA in the mouse heart or in infected CMs (Jiang et al., 2020). In addition, viral RNA has been detected in heart autopsies of COVID-19 patients by several groups (Escher et al., 2020; Lindner et al., 2020). However, SARS-CoV-2 virions have largely been detected in interstitial cells of the myocardium in COVID-19 patient samples (Lindner et al., 2020; Tavazzi et al., 2020). A recent study reported the detection of viral antigen and RNA in CMs in hearts of COVID-19 patients (Bulfamante et al., 2020). Here, using intranasally infected hamsters, a relevant animal model for COVID-19, we clearly detected viral protein expression in CMs of infected animals. This provides direct evidence that intranasal exposure of SARS-CoV-2 leads to infection of CMs in vivo. Although CMs and other cardiac cells can be infected by SARS-CoV-2, other mechanisms, such as respiratory failure, hypoxemia, and hyper-inflammation caused by cytokines released by the macrophages recruited to the heart, may also contribute to the heart damage seen in COVID-19 patients.
CCL2 expression levels were significantly upregulated in infected hPSC-derived CMs and adult CMs, and in the hearts of SARS-CoV-2-infected hamsters. We also examined CCL2 expression levels after SARS-CoV-2 infection in hPSC-derived lung organoids (Han et al., 2021) or hPSC-derived pancreatic endocrine cells (Yang et al., 2020), and in lung autopsies of non-COVID-19 versus COVID-19 patients (Han et al., 2021) (Figure S7A). CCL2 levels are upregulated in both SARS-CoV-2-infected cells/organoids and COVID-19 patient lung autopsy samples. However, when we examined CCL2 expression levels in idiopathic dilated cardiomyopathy LV tissue, ischemic LV tissue, or murine encephalomyelitis virus-infected CMs (Figure S7B, GSE57338 and GSE119860), CCL2 expression was not significantly increased compared with non-failing LV tissue or uninfected CMs. This suggests that CCL2 is not upregulated by any cardiac damage but more specifically in the condition of SARS-CoV-2 infection. Moreover, we found that SARS-CoV-2-infected CMs expressed higher levels of CCL2 than non-infected cells in the hamster model according to scRNA-seq data (Figures S7C–S7E).
CCL2, also known as monocyte chemoattractant protein 1, is a chemokine that facilitates the migration and infiltration of monocytes/macrophages to sites of inflammation produced by either tissue injury or infection (Bose and Cho, 2013). Using a transwell platform, we showed that hPSC-derived CMs or adult human CMs infected with SARS-CoV-2 recruit monocytes. This is consistent with previous reports of abnormal macrophage infiltration in hearts of COVID-19 patients (Escher et al., 2020; Lindner et al., 2020; Tavazzi et al., 2020; Yao et al., 2020).
Finally, we created a co-culture platform using hPSC-derived CMs and macrophages to study the impact of macrophages on infected CMs. scRNA-seq data suggested that the presence of macrophages decreases SARS-CoV-2-infected CMs. This might be due to increased apoptosis of SARS-CoV-2-infected CMs or engulfing of virus by macrophages. In the absence of SARS-CoV-2, the presence of macrophages does not affect CM survival (Figures S7F and S7G). However, in the context of COVID-19, macrophages might be considered a “double-edged sword.” When CMs are infected with SARS-CoV-2, recruitment of macrophages can promote secretion of inflammatory cytokines, ROS, and cause CM damage. On the other hand, engulfing of virus by macrophages (and likely engulfment of infected CMs), limits the infection of CMs. Indeed, cell-mixture deconvolution identified the enrichment of pro-inflammatory macrophages in the LA, LV, and RA of SARS-CoV-2-infected hamsters. In addition, ROS was upregulated in the hearts of infected hamsters, suggesting that macrophages recruited by CMs might also contribute to immune-mediated CM inflammatory damage in COVID-19 patients. Together, both direct CM infection and recruitment of immune cells, such as mononuclear cells, might contribute to balancing cardiac pathophysiology in COVID-19 patients. We cannot exclude the possibility that other types of cells in hearts can also be infected by SARS-CoV-2, as discussed in a recent perspective review (Yiangou et al., 2021) and contribute to monocyte recruitment. In addition, systemic conditions (e.g., inflammation, fever, nutrients, dehydration) might also contribute to heart damage. In summary, we report direct evidence of SARS-CoV-2 infection of CMs in vivo and established an in vitro model to study immune cell infiltration and pathophysiology in cardiac tissue of COVID-19 patients.
Experimental procedures
Details are provided in the supplemental experimental procedures.
Propagation and titration of SARS-CoV-2
SARS-CoV-2, isolate USA-WA1/2020 (NR-52281) was deposited by the Center for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH. Full details are available in the supplemental information.
SARS-CoV-2 infections of hamsters
Three- to five-week-old male Golden Syrian hamsters (Mesocricetus auratus) were obtained from Charles River. Hamsters were acclimated to the CDC/USDA-approved BSL-3 facility of the Global Health and Emerging Pathogens Institute at the Icahn School of Medicine at Mount Sinai for 2–4 days. Full details are available in the supplemental information.
Human studies
Tissue samples were provided by the Weill Cornell Medicine Department of Pathology. The Tissue Procurement Facility operates under an institutional review board-approved protocol and follows guidelines set by HIPAA. Full details are available in the supplemental information.
Data and code availability
scRNA-seq and RNA-seq data are available from the GEO repository database under accession number GSE151880.
Author contributions
S.C., T.E., B.R.t., R.E.S., and D.D.H. conceived and designed the experiments. L.Y., Y. Han., F.J., and J.Z. performed CM, macrophage differentiation, co-culture, and immunostaining. M.G. and J.K.L. performed ELISA analysis. P.W. and Y. Huang. provided SARS2-CoV-2 pseudo-entry virus. A.B., Y.B., C.R., and V.C. analyzed human samples. B.E.N.-P., R.M., L.C., S. Horiuchi., and B.R.t. performed SARS-CoV-2-related experiments. J.Z., T.Z., D.R., S. Houghton., and J.X. performed the scRNA-seq and bioinformatics analyses.
Conflicts of interest
S.C. is a member of editorial board of Stem Cell Reports. R.E.S. is on the scientific advisory board of Miromatrix Inc and is a paid consultant and speaker for Alnylam Inc. The other authors have no conflict of interest.
Acknowledgments
This work was supported by the American Heart Association (18CSA34080171 to S.C. and T.E.), NIDDK (R01DK130454, R01DK116075, R01DK119667, R01DK119667-02S1, R01 DK124463, and U01 DK127777, S.C.), American Diabetes Association (7-20-COVID-211 to S.C.), Department of Surgery, Weill Cornell Medicine (to T.E. and S.C.), Bill and Melinda Gates Foundation (S.C., T.E., R.E.S, B.tO,) and (NCI R01CA234614, NIAID 2R01AI107301 and NIDDK R01DK121072 and 1RO3DK117252), Department of Medicine, Weill Cornell Medicine (to R.E.S.), by the Defense Advanced Research Projects Agency (DARPA-16-35-INTERCEPT-FP-006 to B.T.), and by the Jack Ma Foundation (to D.D.H.). S.C. and R.E.S. are supported as Irma Hirschl Trust Research Award Scholars. T.E. is supported by an Outstanding Investigator Award from the NHLBI (R35 HL135778). R.M. is supported by American Heart Association grant #833781. Y.H. is a NYSTEM Stem Cell Biology Scholar.
Published: September 14, 2021; corrected online: September 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.07.012.
Contributor Information
David D. Ho, Email: dh2994@cumc.columbia.edu.
Robert E. Schwartz, Email: res2025@med.cornell.edu.
Benjamin R. tenOever, Email: benjamin.tenoever@mssm.edu.
Todd Evans, Email: tre2003@med.cornell.edu.
Shuibing Chen, Email: shc2034@med.cornell.edu.
Supplemental information
References
- Bojkova D., Wagner J.U.G., Shumliakivska M., Aslan G.S., Saleem U., Hansen A., Luxan G., Gunther S., Pham M.D., Krishnan J., et al. SARS-CoV-2 infects and induces cytotoxic effects in human cardiomyocytes. Cardiovasc. Res. 2020 doi: 10.1093/cvr/cvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S., Cho J. Role of chemokine CCL2 and its receptor CCR2 in neurodegenerative diseases. Arch. Pharm. Res. 2013;36:1039–1050. doi: 10.1007/s12272-013-0161-z. [DOI] [PubMed] [Google Scholar]
- Bulfamante G.P., Perrucci G.L., Falleni M., Sommariva E., Tosi D., Martinelli C., Songia P., Poggio P., Carugo S., Pompilio G. Evidence of SARS-CoV-2 transcriptional activity in cardiomyocytes of COVID-19 patients without clinical signs of cardiac involvement. Biomedicines. 2020;8 doi: 10.3390/biomedicines8120626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Yakala G.K., van den Hil F.E., Cochrane A., Mummery C.L., Orlova V.V. Differentiation and functional comparison of monocytes and macrophages from hiPSCs with peripheral blood derivatives. Stem Cell Reports. 2019;12:1282–1297. doi: 10.1016/j.stemcr.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolhnikoff M., Ferreira Ferranti J., de Almeida Monteiro R.A., Duarte-Neto A.N., Soares Gomes-Gouvea M., Viu Degaspare N., Figueiredo Delgado A., Montanari Fiorita C., Nunes Leal G., Rodrigues R.M., et al. SARS-CoV-2 in cardiac tissue of a child with COVID-19-related multisystem inflammatory syndrome. Lancet Child Adolesc. Health. 2020;4:790–794. doi: 10.1016/S2352-4642(20)30257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher F., Pietsch H., Aleshcheva G., Bock T., Baumeier C., Elsaesser A., Wenzel P., Hamm C., Westenfeld R., Schultheiss M., et al. Detection of viral SARS-CoV-2 genomes and histopathological changes in endomyocardial biopsies. ESC Heart Fail. 2020 doi: 10.1002/ehf2.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- Gnecchi M., Moretti F., Bassi E.M., Leonardi S., Totaro R., Perotti L., Zuccaro V., Perlini S., Preda L., Baldanti F., et al. Myocarditis in a 16-year-old boy positive for SARS-CoV-2. Lancet. 2020;395:e116. doi: 10.1016/S0140-6736(20)31307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inciardi R.M., Lupi L., Zaccone G., Italia L., Raffo M., Tomasoni D., Cani D.S., Cerini M., Farina D., Gavazzi E., et al. Cardiac involvement in a patient with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:819–824. doi: 10.1001/jamacardio.2020.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020 doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner D., Fitzek A., Brauninger H., Aleshcheva G., Edler C., Meissner K., Scherschel K., Kirchhof P., Escher F., Schultheiss H.P., et al. Association of cardiac infection with SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiano S., Hsiang T.-Y., Khanna A., Higashi T., Whitmore L.S., Bargehr J., Davaapil H., Chang J., Smith E., Ong L.P., et al. SARS-CoV-2 infects human pluripotent stem cell-derived cardiomyocytes, impairing electrical and mechanical function. Stem Cell Reports. 2021 doi: 10.1016/j.stemcr.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020 doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Garcia G., Jr., Wang Y., Plummer J.T., Morizono K., Arumugaswami V., Svendsen C.N. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep Med. 2020;1:100052. doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q., et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazzi G., Pellegrini C., Maurelli M., Belliato M., Sciutti F., Bottazzi A., Sepe P.A., Resasco T., Camporotondo R., Bruno R., et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur. J. Heart Fail. 2020;22:911–915. doi: 10.1002/ejhf.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Y., Ghazizadeh Z., Wang H.J., Amin S., Ortega F.A., Badieyan Z.S., Hsu Z.T., Gordillo M., Kumar R., Christini D.J., et al. A human embryonic stem cell reporter line for monitoring chemical-induced cardiotoxicity. Cardiovasc. Res. 2020;116:658–670. doi: 10.1093/cvr/cvz148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallania F., Tam A., Lofgren S., Schaffert S., Azad T.D., Bongen E., Haynes W., Alsup M., Alonso M., Davis M., et al. Leveraging heterogeneity across multiple datasets increases cell-mixture deconvolution accuracy and reduces biological and technical biases. Nat. Commun. 2018;9:4735. doi: 10.1038/s41467-018-07242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., Mou H.M., Wang L.H., Zhang H.R., Fu W.J., et al. A pathological report of three COVID-19 cases by minimally invasive autopsies. Zhonghua Bing Li Xue Za Zhi. 2020;49:E009. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- Yiangou L., Davis R.P., Mummery C.L. Using cardiovascular cells from human pluripotent stem cells for COVID-19 research: why the heart fails. Stem Cell Reports. 2021;16:385–397. doi: 10.1016/j.stemcr.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq and RNA-seq data are available from the GEO repository database under accession number GSE151880.