Abstract
Strain SN6T is a non-motile and non-spore-forming gram-negative bacterium which was isolated from the stool sample of an Amazonian patient. The optimum growth was observed at 37 °C, pH 7, and 0–5 g/l of NaCl. Based on the 16S rRNA gene sequence similarity, the strain SN6T exhibited 97.5% identity with Vitreoscilla stercoraria strain ATCC_15218 (L06174), the phylogenetically closest species with standing in nomenclature. The predominant fatty acid was hexadecenoic acid (31%). The genomic DNA G + C content of the strain SN6T was 49.4 mol %. After analysis of taxonogenomic data, phenotypic and biochemical characteristics, we concluded that strain SN6T represents a new species of the genus Vitreoscilla for which the name Vitreoscilla massiliensis sp.nov is proposed. The type strain is SN6T (=CSUR P2036 = LN870312 = DSM 100958).
Introduction
This strain was isolated from the stool specimen of an obese Amazonian patient as part of the culturomics study [1] to search for microaerophilic bacteria from human gut. The genus Vitreoscilla was first described by Pringsheim in 1951, after having proposed the family Vitreoscillaceae in 1949. In 1986, Strohl et al. proposed three new species with validated names (Vitreoscilla stercoraria, Vitreoscilla beggiatoides and Vitreoscilla filiformis) of this genus [2]. In 2013, through the use of new-generation sequencing tools, the genus Vitreoscilla was placed in the Neisseriaceae family on the basis of its branching in the 16S rRNA gene tree [3]. Within its clade, members of the genus were the only ones capable of evolving in different habitats.
Since the use of the culturomic concept, the repertoire of bacteria isolated from the human digestive microbiota [4] has expanded considerably. The characterization of these new species is based on a ribosomal RNA sequencing coupled with a taxonogenomics description, a strategy combining a comparison of genomic analysis and phenotypic characteristics, including the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) spectrum. In the present study, we used this approach to facilitate the identification and the description of this novel species named Vitreoscilla massiliensis sp.nov.
Materials and Methods
Vitreoscilla massiliensis SN6T was isolated by cultivation on 5% sheep blood agar under microaerophilic conditions at 37 °C for 48 h and the strain could not be identified by Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
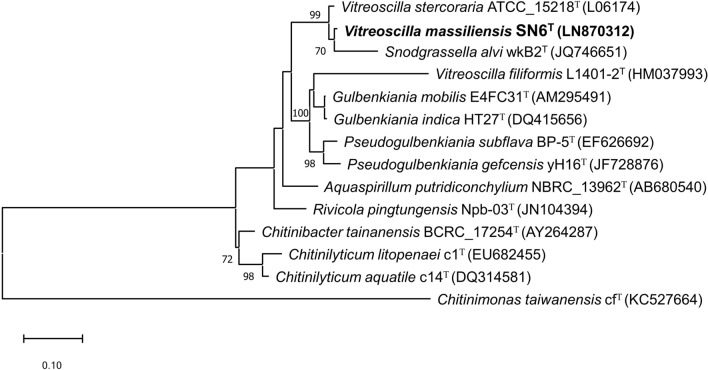
(MALDI-TOF MS). The bacterial spectrum obtained was incremented in our database and its comparison with those of BioTyper database spectra and our own collection did not allow for its identification. Sequencing of 16S rRNA gene of the strain SN6T showed a nucleotide sequence similarity of 97.5% with V. stercoraria strain (ATCC 15218) and V. stercoraria strain Göttingen 1488-6 (NR_025894.1), the phylogenetically closest species with standing in nomenclature (Fig. 1).
Fig. 1.
Phylogenetic tree showing the position of Vitreoscilla massiliensis SN6T relative to other phylogenetically close neighbors. Sequences were aligned using CLUSTALW, and phylogenetic inferences are obtained with kimura two-parameter models using the maximum-likelihood method within the MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1,000 times to generate majority consensus tree. Scale bar indicates 1% nucleotide sequence divergence. The scale bar represents 500 nm
Optimal Growth
Growth at various temperatures (28 °C, 37 °C, 42 °C, 45 °C) in different atmospheres (aerobic, microaerophilic using CampyGen from Thermo Scientific and anaerobic using AnaeroGenTM from bioMérieux) was tested by culture on Columbia agar (bioMérieux) after 48 h of incubation. The salinity acceptance limit of SN6T strain was investigated by culture on a home-made culture medium consisting of a Columbia agar culture medium (Sigma-Aldrich, Saint-Quentin Fallavier, France) modified by adding (per liter) 5 g MgCl2 6H2O, 5 g MgSO4 7H2O, 2 g KCl, 1 g CaCl2 2H2O; 0.5 g NaBr, 0.5 g NaHCO3, and 2 g glucose with various NaCl concentrations 0, 5, 10, 25, 50, and 75 g/L. The pH range (6; 6.5; 7; 8.5) for growth was also determined and pH was adjusted by addition of HCl or NaOH.
Biochemical and Chemotaxonomic Analysis
The abilities of the strain SN6T to use various substrates as sole carbon sources were evaluated using the API 20NE and API 50CH (bioMérieux) and the presence of some enzyme activities using APIZYM following the manufacturer’s instructions. All tests were performed in duplicate. Susceptibility to antimicrobial agents was determined by the disk (i2a, Montpellier, France) diffusion method [5] on Mueller–Hinton agar in a Petri dish (BioMerieux) after 48 h of incubation at 37 °C under aerobic conditions. The interpretation of inhibition diameters to the manual measurement using a ruler was done using a Sirscan system© (i2a, Montpellier, France) according to the criteria proposed by the Comité de l’Antibiogramme of the French Society for Microbiology [6]. Cellular fatty acid methyl ester (FAME) analysis was performed by GC/MS. Two samples were prepared with approximately 65 mg of bacterial biomass per tube harvested from several culture plates. Fatty acid methyl esters were prepared as previously described by Sasser [7]. GC/MS analyses were carried out as previously described by Dione et al. [8]. Briefly, fatty acid methyl esters were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500—SQ 8 S, Perkin Elmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the Standard Reference Database 1A (NIST, Gaithersburg, USA) and the FAMEs mass spectral database (Wiley, Chichester, UK).
Genome Sequencing and Assembly
DNA of strain SN6T was extracted on the EZ1 biorobot (Qiagen) with a EZ1 DNA tissues kit after pretreatment by a lysozyme incubation at 37 °C, as previously described [9]. Genomic DNA (gDNA) was quantified by a Qubit assay with the high-sensitivity kit (Life technologies, Carlsbad, CA, USA) and sequenced on the MiSeq Technology (IlluminaInc, San Diego, CA, USA) with the mate pair strategy, as previously described [9]. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina). The assembly of the genome was carried out with the help of a pipeline that allowed the creation of an assembly with different softwares (Velvet [10], Spades [11] and Soap Denovo [12], on trimmed (MiSeq and Trimmomatic [13] softwares) or untrimmed data (only MiSeq software). For each of the six assemblies performed, GapCloser [12] was used to reduce gaps. Then, contamination with Phage Phix was identified (blastn against Phage Phix174 DNA sequence) and eliminated. Finally, scaffolds whose size was less than 800 bp were removed and scaffolds whose depth value was lower than 25% of the mean depth were removed (identified as possible contaminants). The best assembly was selected using different criteria (number of scaffolds, N50 and number of N). Spades gave the best assembly of this strain, with a depth coverage of 98.
Genome Annotation and Comparison
We used Prodigal as predicting tool of open reading frames (ORFs) [14] with default parameters. The predicted ORFs were excluded if they spanned a sequencing gap region (contained N). Using BLASTP, predicted bacterial protein sequences were blasted against GenBank and clusters of orthologous groups (COG) databases, DNA G + C content was identified by The RAST Server [15], and the tRNAs and rRNAs were predicted using the tRNAScan-SE [16] and RNAmmer tools [17], respectively. SignalP was used for Signal peptides prediction [18], the number of transmembrane helices was predicted using TMHMM [19], ORFans were identified if their BLASTP E-value was lower than 1e03 for alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E-value of 1e-05. Artemis [20] and DNA Plotter [21] were used for data management and visualization of genomic features, respectively. The Average Nucleotide identity at the genome level between V. massiliensis SN6 CZPV00000000.1, V. stercoraria ATCC_15218 (ARNN00000000.1), V. filiformis ATCC_43190 ( CP022423.1), Gulbenkiania mobilis E4FC31 (LIVN00000000.1), Chitinilyticum litopenaei DSM_21440 (ATZJ00000000.1), Chitinilyticum aquatile c14 (AUMS00000000.1), Chitinibacter tainanensis BCRC_17254 (AUCN00000000.1), and Snodgrassella alvi wkB2_wkB2 (CP007446.1) was estimated using Orthologous Average nucleotide identity tool (OAT) [22].
Results and Discussion
Based on the sequence similarity threshold values of the 16S rRNA gene that delineate a new species according to the recommendations of Stackebrandt and Ebers [23], the strain SN6T can, therefore, be classified as a new species of the genus Vitreoscilla and was accordingly named V. massiliensis SN6T [24].
Biochemical and Chemotaxonomic Analyses
API ZYM tests show positive reactions for esterase, esterase lipase, leucine arylamidase, acid phophatase, and naphthol-AS-BI-phosphohydrolase. In API 50CH, no substrate fermentation was observed and in API 20NE assimilation of substrates was observed for L-arginine dihydrolase and potassium gluconate. Some phenotypic characteristics of SN6T with those of closely related species are presented in Table 1. The most abundant fatty acid is hexadecenoic acid (31%). Several hydroxyl fatty acids such as C12:0 3-OH (4.5 ± 1.0) and C14:0 3-OH (2.9 ± 0.1) are described. Other fatty acids such as 9-Hexadecenoic acid (22.0 ± 0.5), Dodecanoic acid (10.2 ± 0.6), 2-hexyl-cyclopropaneoctanoic acid (8.8 ± 0.4), Octadecenoic acid (8.0 ± 0.2), Pentadecanoic acid (5.5 ± 0.2), Tetradecanoic acid (3.3 ± 0.3), and Heptadecanoic acid (1.1 ± 0.1) were detected. The strain SN6T was resistant to Oxacillin and Metronidazole, but susceptible to other antibiotics tested.
Table 1.
Differential characteristics of Vitreoscilla massiliensis SN6T, Vitreoscilla stercoraria ATCC 15218, Vitreoscilla filiformis ATCC 15551, Vitreoscilla beggiatoides B23SS, Gulbenkiania mobilis E4FC31, Chitinibacter tainanensis BCRC 17254, Chitinilyticum litopenaei DSM _21440_c1, and Snodgrassella alvi wkB2 wkB2
| Properties | V.massiliensis | V. stercorariaa | V.filiformisb | V.beggiatoidesc | G. mobilisd | C.litopenaeie | C.tainanensisf | S. alvig |
|---|---|---|---|---|---|---|---|---|
| Cell diameter (µm) | 0.5 | 1.0 | 1.0–1.5 | 2.5–3 | 0.2–0.4 | 0.3–0.5 | 0.5–0.9 | 0.4 |
| Oxygen requirement | Aerobic/Microaerophilic | Aerobic | Aerobic/Microaerophilic | Aerobic/Microaerophilic | Aerobic | Aerobic/Anaerobic | Aerobic | Microarophilic |
| Motility | – | + | + | + | + | + | + | - |
| Endospore formation | – | Na | Na | Na | – | – | Na | Na |
| pH | 7–7.5 | 7.5–7.7 | 7.5 | 7.5 | 5.5–9.0 | 7–11 | 5.5–9 | 6.0–6.5 |
| NaCl % (w/v) | 0–0.5 | Na | 0 | 0.5 | 1.0 | 0–0.75 | Na | Na |
| Indole | – | Na | Na | Na | – | – | – | – |
|
Production of Alkaline phosphatase |
– | Na | Na | Na | Na | + | Na | Na |
| Catalase | – | – | – | – | + | + | + | + |
| Oxidase | – | – | + | + | + | – | + | – |
| Nitrate reductase | – | – | + | + | – | + | – | + |
| Urease | – | Na | Na | Na | – | – | – | + |
| β-galactosidase | – | Na | Na | Na | – | – | – | |
| N-acetyl-glucosamine | – | Na | Na | Na | – | + | + | Na |
|
Acid from L-Arabinose |
+ | – | – | – | – | – | – | – |
| Trehalose | – | – | – | – | – | + | – | Na |
| D-mannose | + | – | – | – | – | + | – | – |
| Mannitol | + | – | – | – | – | – | – | – |
| D-glucose | + | – | + | – | – | + | – | – |
| D-fructose | – | – | – | – | – | + | – | – |
| Maltose | + | – | – | – | – | + | – | – |
| D-lactose | – | – | – | – | – | – | Na | Na |
| D-raffinose | – | Na | – | – | – | – | Na | Na |
| Habitat | Human gut | Dung of Cow | Freshwater sediments | Sandy sediments | Wastewater | Freshwater pond | Soil | Gut of Bees |
Genome Properties
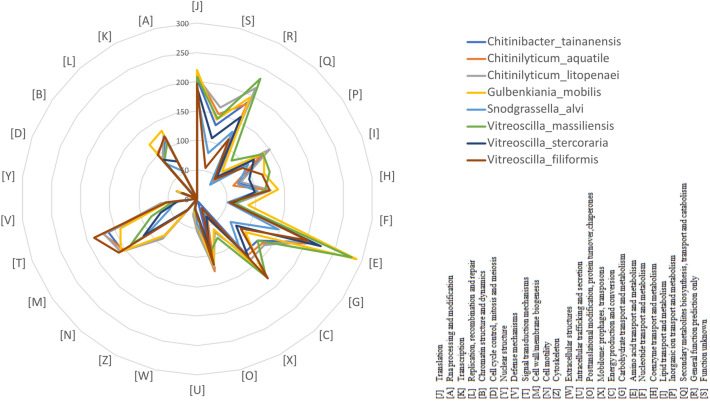
The genome is 3,716,289 bp long with 49.4% GC content (Fig. 2, Table 2). It is composed of 10 scaffolds (composed of 13 contigs). Of the 3 716 predicted genes, 3 627 were protein-coding genes and 89 were RNAs (5 genes are 5S rRNA, 5 genes are 16S rRNA, 5 genes are 23S rRNA and 74 genes are TRNA genes). A total of 2,263 genes (62.3%) were assigned with putative function (by cogs or by NR blast). 475 genes were identified as ORFans (13.1%). The remaining genes were described as hypothetical proteins (744 genes ≥ 20.5%). A summary of the distribution of V. massiliensis genes into the different COGs categories is presented in Table 3.
Fig. 2.
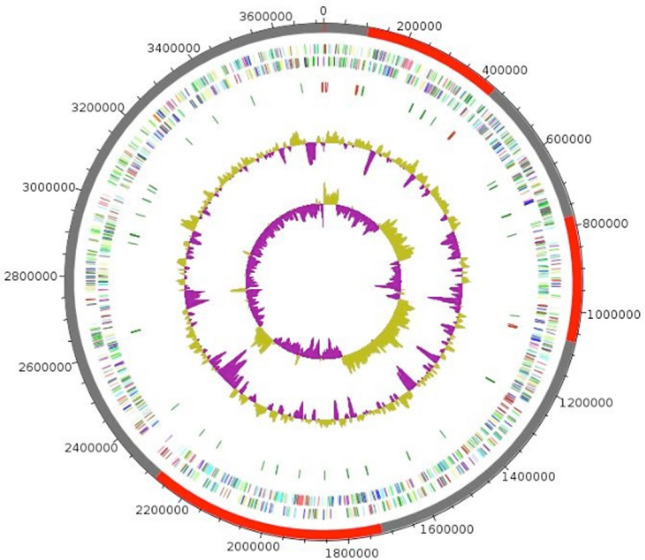
Graphical circular map of the genome of Vitreoscilla massiliensis strain SN6T from outside to the center: genes on the forward strand colored by COG categories (only genes assigned to COG), genes on the reverse strand colored by COG categories (only gene assigned to COG), RNA genes (tRNAs green, rRNAs red), GC content, and GC skew (Color figure online)
Table 2.
Nucleotide content and gene count levels of the genome
| Attribute | Genome (total) | |
|---|---|---|
| Value | % of totala | |
| Size (bp) | 3,716,289 | 100 |
| G+C content (%) | 1,836,063 | 49.42 |
| Coding region (bp) | 3,249,937 | 87.45 |
| Total genes | 3,716 | 100 |
| RNA genes | 89 | 2.39 |
| Protein-coding genes | 3,627 | 100 |
| Genes with function prediction | 2,263 | 62.39 |
| Genes assigned to COGs | 2,184 | 60.21 |
| Genes with peptide signals | 677 | 18.66 |
| Genes with transmembrane helices | 774 | 21.33 |
| Genes associated to virulence | 715 | 19.71 |
| ORFn genes | 475 | 13.09 |
| Genes associated with PKS or NRPS | 20 | 0.55 |
| Genes associated to toxine/antitoxine | 115 | 3.17 |
aThe total is based on either the size of the genome in base pairs or the total number of protein-coding genes in the annotated genome
Table 3.
Number of genes associated with the 25 general COG functional categories
| Code | Value | % of total | Description |
|---|---|---|---|
| [J] | 213 | 5.872622 | Translation |
| [A] | 1 | 0.027570996 | Rna processing and modification |
| [K] | 121 | 3.3360906 | Transcription |
| [L] | 88 | 2.4262476 | Replication, recombination and repair |
| [B] | 4 | 0.110283986 | Chromatin structure and dynamics |
| [D] | 32 | 0.8822719 | Cell cycle control, mitosis and meiosis |
| [Y] | 0 | 0 | Nuclear structure |
| [V] | 44 | 1.2131238 | Defense mechanisms |
| [T] | 85 | 2.3435347 | Signal transduction mechanisms |
| [M] | 140 | 3.8599393 | Cell wall/membrane biogenesis |
| [N] | 29 | 0.7995589 | Cell motility |
| [Z] | 0 | 0 | Cytoskeleton |
| [W] | 19 | 0.52384895 | Extracellular structures |
| [U] | 27 | 0.7444169 | Intracellular trafficking and secretion |
| [O] | 97 | 2.6743865 | Post-translational modification, protein turnover, chaperones |
| [X] | 77 | 2.1229665 | Mobilome, prophages, transposons |
| [C] | 177 | 4.8800664 | Energy production and conversion |
| [G] | 125 | 3.4463744 | Carbohydrate transport and metabolism |
| [E] | 287 | 7.9128757 | Amino acid transport and metabolism |
| [F] | 64 | 1.7645438 | Nucleotide transport and metabolism |
| [H] | 119 | 3.2809484 | Coenzyme transport and metabolism |
| [I] | 133 | 3.6669421 | Lipid transport and metabolism |
| [P] | 136 | 3.7496552 | Inorganic ion transport and metabolism |
| [Q] | 90 | 2.4813895 | Secondary metabolites biosynthesis, transport and catabolism |
| [R] | 234 | 6.4516125 | General function prediction only |
| [S] | 145 | 3.9977942 | Function unknown |
| _ | 1443 | 39.784946 | Not in COGs |
Genome Comparison
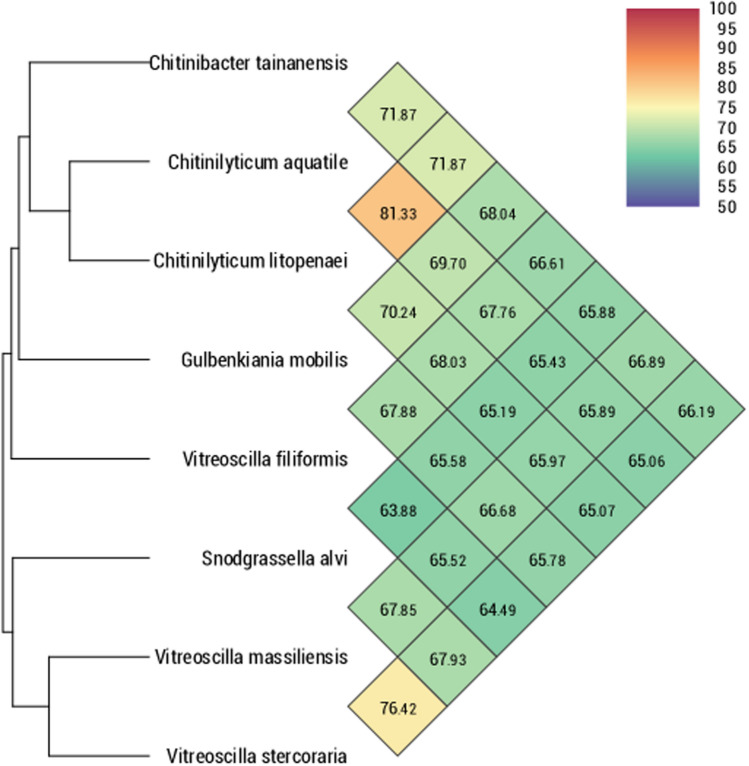
The draft genome sequence and the G + C content of V. massiliensis (3.71 MB and 49.4%) is larger than that of V. stercoraria (2.58 MB and 43.9% respectively). Also, the gene content of V. massiliensis is larger than that of (3,627and 2,440 respectively). The distribution of genes into COG categories was similar in all 7 compared genomes (Fig. 3). All genomes were compared with V. massiliensis using Orthologous average nucleotide identity. The OrthoANI analysis showed that identity nucleotide value is 76.4% with V. stercoraria which is lower than 95% (Fig. 4). Likewise, we obtained similar results for the analysis of the digital DNA-DNA hybridization (dDDH) with 31.60% between V. massiliensis and V. stercoraria (Table 4).
Fig. 3.
Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins
Fig.4.
Heatmap generated with OrthoANI values calculated using the OAT software between Vitreoscilla species and other closely related species with standing in nomenclature
Table 4.
Pairwise comparison of Vitreoscilla massiliensis SN6T with other species using GGDC, formula 2(DNA-DNA hybridization estimates based on identities/HSP length)
| C.tainanensis | C.aquatile | G.mobilis | S.alvi | C.litopenaei | V.stercoraria | V.filiformis | |
|---|---|---|---|---|---|---|---|
| V.massiliensis |
23.20% 2.4± |
30.00% 2.45± |
29.40% 2.45± |
26.80% 2.45± |
30.80% 2.45± |
21.50% 2.35± |
28.30% 2.45± |
| C.tainanensis |
18.80% 2.3± |
19.30% 2.25± |
31.10% 2.45± |
19.20% 2.3± |
19.70% 2.3± |
26.30% 2.4± |
|
| C.aquatile |
18.30% 2.25± |
37.20% 2.5 ± |
25.60% 2.45± |
28.40% 2.45± |
19.50% 2.3± |
||
| G.mobilis |
29.30% 2.45± |
18.7% 2.25± |
31.20% 2.45± |
18.50% 2.3± |
|||
| S.alvi |
33.8% 2.5± |
24.00% 2.35± |
29.60% 2.45± |
||||
| C.litopenaei |
32.9% 2.5± |
18.40% 2.25± |
|||||
| V.stercoraria |
31.60% 2.45± |
V.massiliensis Vitreoscilla massiliensis SN6T, G.mobilis Gulbenkiania mobilis E4FC31, C.litopenaei Chitinilyticum litopenaei DSM _21440_c1, S.alvi Snodgrassella alvi wkB2 wkB2, C.tainanensis Chitinibacter tainanensis BCRC 17254, C.aquatile Chitinilyticum aquatile c14, V.stercoraria Vitreoscilla stercoraria ATCC 15218 and V.filiformis Vitreoscilla filiformis ATCC 15551
Conclusion
Based on the phenotypic characteristics, and phylogenetic and genomic analyses of strain SN6T, we suggest the creation of a new species within the Vitreoscilla genus, for which the name V. massiliensis sp. nov., is proposed.
Description of Vitreoscilla massiliensis sp. nov.
Vitreoscilla massiliensis (mas.si.li.en’sis. L. fem. adj. massiliensis, of Massilia, the Latin name of Marseille where strain SN6T was first isolated).
Cells are Gram-negative (0.5 × 1.5–2 µm), non-motile, non-spore-forming , and often occur in a long chain under electron microscopy. V. massiliensis SN6T grows at 28–37 °C and pH 7–7.5 and does not grow above 0.5% salinity. V. massiliensis SN6T grows under microaerophilic atmosphere and a lower growth was observed under anerobic conditions. On agar plates, colonies were gray, smooth, and hemolytic with 0.5 to 1 mm in diameter after 48 h of incubation under aerobic conditions. They are catalase and oxidase negative. Tests were negative for urease, the reduction of nitrates, indole production, and fermentation of β-galactosidase. API 50CH shows that the carbohydrates provided by this panel were not used. V. massiliensis SN6T is susceptible to Vancomycin (0.5), Cefotaxime (0.94), Tobamycin (0.38 µg), Fosfomycin (16 µg), Teicoplanin (1.5 µg), Rifampicin (0.29 µg), Colistin (0.32 µg), Imipenem (0.23 µg), Erythromycin (0.25 µg), Ceftriaxone (0.32 µg), and resistant to Oxacillin and Metronidazole. Major fatty acids are hexadecanoic acid (C16:00), 9-hexadecenoic acid (C16:1n7), and an unusual cylo fatty acid named 2-hexyl-cyclopropaneoctanoic acid (C16:0 9,10-methylene). The DNA G + C content is about 49.4%.
The type strain is SN6T (=CSUR P2036 = LN870312 = DSM 100958) and was isolated from the stool specimen of an obese Amazonian patient.
Acknowledgements
The authors thank the Xegen Company (http://www.xegen.fr/) for automating the genomic annotation process. They also thank Aurelia Caputo for submitting the genomic sequences to GenBank.
This work has benefited from the French State support, managed by the ‘Agence Nationale pour la Recherche’, including the "Programme d’Investissement d’avenir" under the reference Méditerranée Infection 10-IAHU-03. This work was supported by Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI.
Abbreviations
- CSUR
Collection de Souches de l’Unité des Rickettsies
- DSM
Deutsche Sammlung von Mikroorganismen
- FAME
Fatty acid methyl ester
- GC/SM
Gaz chromatography/mass spectrometry
- OrthoANI
Orthologous average nucleotide identity
- COGs
Clusters of orthologous groups
- NR BLAST
Non-redundant protein sequence basic local alignment search tool
- ORF
Open reading frame
- GGDC
Genome-to-genome distance calculator
Author Contributions
SN, MM, and MR performed the experiments. SN and SK wrote the manuscript. SK, DR, RS, and PEF reviewed the drafts of the manuscript. SK, JCL, and DR are responsible for data interpretation.
Declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lagier J-C, Edouard S, Pagnier I, et al. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strohl WR, Schmidt TM, Lawry NH, et al. Characterization of Vitreoscilla beggiatoides and Vitreoscilla filiformis sp. nov., nom. rev., and comparison with Vitreoscilla stercoraria and Beggiatoa alba. Int J Syst Evol Microbiol. 1986;36:302–313. [Google Scholar]
- 3.Adeolu M, Gupta RS. Phylogenomics and molecular signatures for the order Neisseriales: proposal for division of the order Neisseriales into the emended family Neisseriaceae and Chromobacteriaceae fam. nov. Antonie Van Leeuwenhoek. 2013;104:1–24. doi: 10.1007/s10482-013-9920-6. [DOI] [PubMed] [Google Scholar]
- 4.Lagier J-C, Khelaifia S, Alou MT, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Le Page S, van Belkum A, Fulchiron C, et al. Evaluation of the PREVI® Isola automated seeder system compared to reference manual inoculation for antibiotic susceptibility testing by the disk diffusion method. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2015;34:1859–1869. doi: 10.1007/s10096-015-2424-8. [DOI] [PubMed] [Google Scholar]
- 6.The European Committee on Antimicrobial Susceptibility Testing (2016) Breakpoint tables for interpretation of MICs and zone diameters. http://www.eucast.org/clinical_breakpoints/
- 7.Sasser M. Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME) New York: MIDI, Technical Note; 2006. [Google Scholar]
- 8.Dione N, Sankar SA, Lagier J-C, et al. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier J-C, Ramasamy D, Rivet R, et al. Non-contiguous finished genome sequence and description of Cellulomonas massiliensis sp. nov. Stand Genomic Sci. 2012;7:258–270. doi: 10.4056/sigs.3316719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol J Comput Mol Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyatt D, Chen G-L, Locascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avni D, Biberman Y, Meyuhas O. The 5’ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagesen K, Hallin P, Rødland EA, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: signalp 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 20.Carver T, Thomson N, Bleasby A, et al. DNAPlotter: circular and linear interactive genome visualization. Bioinforma Oxf Engl. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutherford K, Parkhill J, Crook J, et al. Artemis: sequence visualization and annotation. Bioinforma Oxf Engl. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 22.Lee I, Kim YO, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2015;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 23.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 24.Ndongo S, Lagier J-C, Raoult D, Fournier P-E. Gorillibacterium timonense sp. nov., and Vitreoscilla massiliensis sp. nov., two new bacterial species isolated from stool specimens of obese Amazonian patients. New Microbes New Infect. 2018;23:48–51. doi: 10.1016/j.nmni.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayfield DC, Kester AS. Physiological studies on Vitreoscilla stercoraria. J Bacteriol. 1972;112:1052–1056. doi: 10.1128/jb.112.3.1052-1056.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaz-Moreira I, Nobre MF, Nunes OC, Manaia CM. Gulbenkiania mobilis gen. nov., sp. nov., isolated from treated municipal wastewater. Int J Syst Evol Microbiol. 2007;57:1108–1112. doi: 10.1099/ijs.0.64726-0. [DOI] [PubMed] [Google Scholar]
- 27.Chang S-C, Wu M-C, Chen W-M, et al. Chitinilyticum litopenaei sp. nov., isolated from a freshwater shrimp pond, and emended description of the genus Chitinilyticum. Int J Syst Evol Microbiol. 2009;59:2651–2655. doi: 10.1099/ijs.0.005090-0. [DOI] [PubMed] [Google Scholar]
- 28.Chern L-L, Stackebrandt E, Lee S-F, et al. Chitinibacter tainanensis gen. nov., sp. nov., a chitin-degrading aerobe from soil in Taiwan. Int J Syst Evol Microbiol. 2004;54:1387–1391. doi: 10.1099/ijs.0.02834-0. [DOI] [PubMed] [Google Scholar]
- 29.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order “Enterobacteriales” of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]