Abstract
The painted lady butterfly, Vanessa cardui, has the longest migration routes, the widest hostplant diversity, and one of the most complex wing patterns of any insect. Due to minimal culturing requirements, easily characterized wing pattern elements, and technical feasibility of CRISPR/Cas9 genome editing, V. cardui is emerging as a functional genomics model for diverse research programs. Here, we report a high-quality, annotated genome assembly of the V. cardui genome, generated using 84× coverage of PacBio long-read data, which we assembled into 205 contigs with a total length of 425.4 Mb (N50 = 10.3 Mb). The genome was very complete (single-copy complete Benchmarking Universal Single-Copy Orthologs [BUSCO] 97%), with contigs assembled into presumptive chromosomes using synteny analyses. Our annotation used embryonic, larval, and pupal transcriptomes, and 20 transcriptomes across five different wing developmental stages. Gene annotations showed a high level of accuracy and completeness, with 14,437 predicted protein-coding genes. This annotated genome assembly constitutes an important resource for diverse functional genomic studies ranging from the developmental genetic basis of butterfly color pattern, to coevolution with diverse hostplants.
Keywords: PacBio sequencing, de novo genome assembly, RNA-seq, butterfly wing, color patterning
Significance
Vanessa cardui is a widely distributed butterfly species and has emerged as an excellent model for studying color pattern formation, migration, and coevolution. Here, we present a high-quality, annotated reference genome of V. cardui. This new genome assembly will serve as an important tool for genome-scale functional studies in V. cardui and a resource for advancing research in evolution, development, and ecology.
Introduction
The painted lady butterfly, Vanessa cardui (Linnaeus 1758), is one of the most widely distributed butterfly species (Shields 1992). It occurs from sea level to about 5,200 m in elevation on every continent except Antarctica and South America (Shields 1992; Varshney and Smetacek 2015). Vanessacardui is a long-range, seasonal migratory butterfly that undertakes an annual multigenerational migration across most of Europe in spring and summer, and north Africa in autumn and winter (Stefanescu et al. 2007; Stefanescu, et al. 2013; Stefanescu et al. 2016, 2017; Pfeiler and Markow 2017).V. cardui is also actively studied for its hostplant interactions (de la Paz Celorio-Mancera et al. 2016; Gamberale-Stille et al. 2019), visual biology (Briscoe et al. 2003; Briscoe and White 2005; Perry et al. 2016), and thermoregulation (Tsai et al. 2020).
Vanessa cardui has also emerged as an excellent model for studying color pattern formation (Reed and Nagy 2005; Hiyama et al. 2012; Dinwiddie et al. 2014; Connahs et al. 2016). Melanins and ommochromes, the pigment types characteristic of the major butterfly family Nymphalidae, are diverse and abundant in this species, and V. cardui wings display all of the major pattern elements of the Nymphalid Ground Plan (Nijhout 1991). Vanessacardui is also highly accessible for both classroom projects (Martin et al. 2020) and lab studies because it is readily available from commercial vendors and can be reared in large numbers on an artificial diet. Recently, CRISPR/Cas9 genome editing tools have become established in V. cardui, which allows for straightforward experimental validation of gene function. CRISPR/Cas9 knockout studies carried out in V. cardui have identified color patterning (optix, WntA, distal-less, spalt) (Zhang and Reed 2016; Mazo-Vargas et al. 2017; Zhang et al. 2017b) and pigmentation genes (pale, Ddc, yellow, yellow-d, yellow, ebony, black) (Perry et al. 2016; Zhang et al. 2017a). In sum, V. cardui is attracting increasing attention in the field of developmental genetics, ecology, and evolutionary biology as a model for connecting genotypes to diverse phenotypes and is thus a powerful addition to comparative studies.
Lepidoptera are a diverse order of insects with complex morphological and behavioral traits, and work on this group will benefit from more and better genomic resources. Vanessacardui belongs to the Nymphalidae, which is the largest family of butterflies. There are currently seven annotated nymphalid genomes accessible on the public genome browser Lepbase (Challi et al. 2016) ( http://lepbase.org/, May 18, 2021): Heliconius erato (Lewis et al. 2016; Van Belleghem et al. 2017), Heliconius melpomene (Dasmahapatra et al. 2012), Bicyclus anynana (Nowell et al. 2017), Melitaea cinxia (Blande et al. 2020), Calycopis cecrops (Cong, Shen, Borek, et al. 2016), Junonia coenia (van der Burg et al. 2019), and Danaus plexippus (Zhan et al. 2011). This paper adds to this list by reporting a high-quality V. cardui genome assembly, generated using PacBio long-read sequencing technology. The final assembly was 425.4 Mb in length, with a contig N50 of 10.3 Mb. We further performed deep transcriptomic sequencing and analyzed 29 RNA-seq data sets across multiple tissues and developmental stages. Using the genome assembly and transcriptomic resources, we annotated protein-coding genes and repeat sequences. The resulting genome assembly, annotation, and wing development expression profiles will provide a valuable resource for future studies of the painted lady butterfly and for butterfly and insect biology in general.
Results and Discussion
High-Quality Genome Assembly
A total of 36.53 Gb of PacBio long reads (coverage of 84×) were generated from 55 SMART cells. The total length of the genome assembly of V. cardui was 425.41 Mb with a contig N50 of 10.30 Mb (table 1). We further generated V. cardui pseudochromosomes using a high-quality chromosomal assembly from M. cinxia (v2) (Blande et al. 2020), which is the closest related nymphalid with a high-quality assembly. The final pseudochromosome assembly contained 143 contigs with the N50 of 15.35 Mb ( fig. 1a). The completeness of our assembly was assessed by Benchmarking Universal Single-Copy Orthologs (BUSCO). Using Lepidoptera-specific single-copy orthologs (lepidoptera_odb10), 96.9% and 0.7% of 5,286 BUSCOs were complete and partially assembled, respectively, with only 0.3% duplicated. Overall, all evidence suggests that the V. cardui assembly is a high-quality genome assembly that can be used for further downstream analyses.
Table 1.
Vanessa cardui Genome Assembly and Annotation Summary
| Genome assembly statistics | |
| Total length (bp) | 425,413,715 |
| Contig N50 length (bp) | 10,297,021 |
| Contig N90 length (bp) | 1,988,721 |
| Longest contig length (bp) | 15,944,461 |
| Number of contigs | 205 |
| Number of contigs larger than N50 | 16 |
| Number of contigs larger than N90 | 54 |
| Genome characteristics | |
| GC content | 33.37% |
| Number of protein-coding genes | 14,437 |
| Average transcript length (bp) | 7,947.27 |
| Average CDS length (bp) | 1,285.78 |
| Average exon length | 208.90 |
| Average exons per gene | 6.26 |
| Repetitive sequences (% of genome) | |
| DNA (bp) | 26,747,187 (6.29%) |
| LINE (bp) | 44,319,571 (10.42%) |
| SINE (bp) | 36,688,707 (8.62%) |
| LTR (bp) | 7,782,116 (1.83%) |
| Simple repeat (bp) | 7,080,895 (1.66%) |
| Unknown (bp) | 23,180,775 (5.45%) |
| Total (bp) | 142,884,949 (33.59%) |
| Gene annotations (% of all genes) | |
| SwissProt | 13,751 (95.25%) |
| KEGG | 8,153 (56.47%) |
| GO | 9,563 (66.24%) |
| PFAM | 12,000 (83.12%) |
| InterProScan | 10,533 (72.96%) |
| Total | 14,097 (97.64%) |
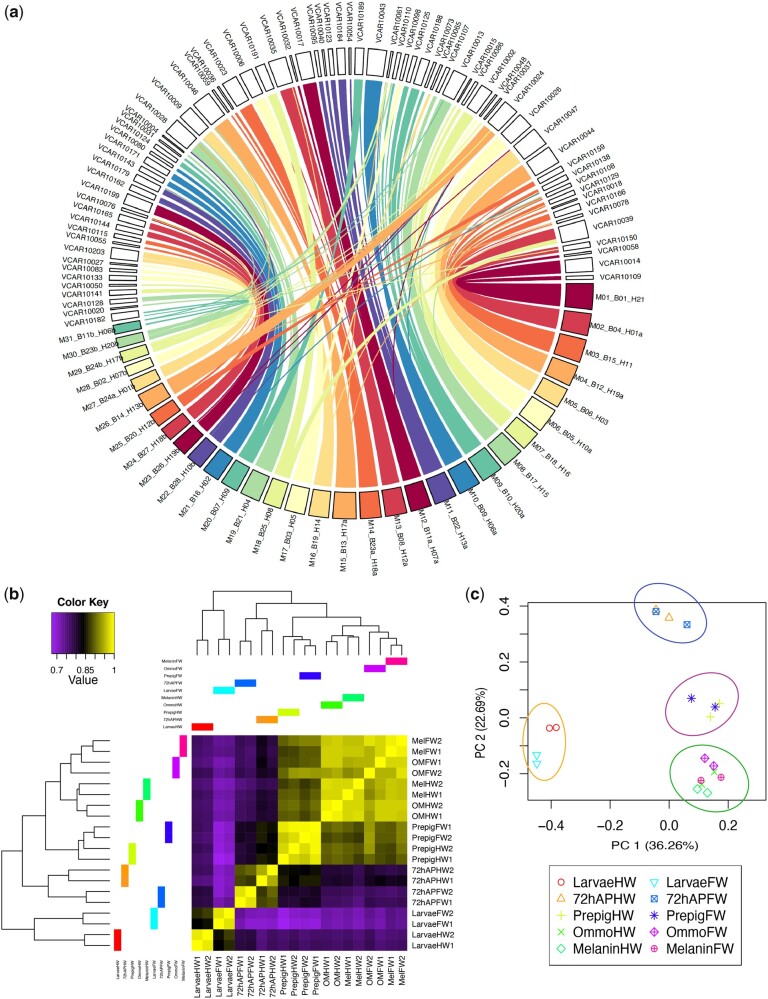
Fig. 1.
Vanessa cardui genome synteny and transcript clustering. (a) Synteny of corresponding chromosomes between V. cardui and Melitaea cinxia. Homologous regions of the genome assemblies are connected by colored lines that represent syntenic regions identified by MUMmer. (b) Heatmap of gene expression clustering by replicate (1, 2), tissue type (FW: forewing, HW: hindwing), and developmental stage (last instar larvae, 72 h after pupation, prepigmentation, ommochrome stage, melanin stage). (c) Principal component analysis of gene expression.
Repeat and Gene Annotation
We identified a total length of 144,928,423 bp repeat sequences, accounting for 34.07% of V. cardui genome (table 1). The most abundant of the transposable and repetitive element type was long interspersed nuclear elements (LINE), representing 44.32 M (10.42%) of the genome. A gene set of 14,437 protein-coding genes was generated with a mean of 6.16 exons per gene (table 1). A total of 14,097 protein-coding genes (97.64%) were successfully annotated for at least one function term by searching against functional databases (SwissProt, gene ontology [GO], Kyoto Encyclopedia of Genes and Genomes [KEGG], PFAM, and InterProScan) (table 1). In order to test the quality of gene annotation, we compared ortholog hit ratios between our final V. cardui annotation with that from Bombyx mori and D. plexippus. More than 90% of the 14,439 B. mori query proteins had orthologous alignments against annotations from both V. cardui and D. plexippus, suggesting both annotations are very complete (supplementary fig. S1 and S2, Supplementary Material online).
Phylogenetic Analysis
To confirm the phylogenetic position of V. cardui and estimate divergence times using whole-genome data, we analyzed the orthologous gene relationships between V. cardui and 12 other lepidopterans. The phylogenetic analysis suggests that butterflies originated from moths around 85–177 Myr and Nymphalidae started diversifying around 85–131 Myr. These results broadly agree with a previous study’s confidence intervals (Espeland et al. 2018). Of the species examined, V. cardui is most closely related to M. cinxia, and the two species diverged from the H. melpomene lineage ∼73–84 Myr (supplementary fig. S3, Supplementary Material online).
Gene Expression Analysis
To explore the molecular basis of the butterfly wing developmental process, we generated a comprehensive profile of gene expression across wing developmental stages from both forewings and hindwings (supplementary table S1, Supplementary Material online, and fig. 1b). The first principal component explained 36.36% of the variance in gene expression and showed strong separation at larval and pupal stages, highlighting the different development processes occurring at these wing developmental stages (fig. 1c). We further performed differential gene expression analysis by comparing consecutive developmental stages. Overall, we identified 2,305 genes significantly differentially expressed (false discovery rate [FDR] < 0.001) (supplementary fig. S4, Supplementary Material online) including 1,692 genes identified from forewing and 1,806 from hindwing transcriptomes (supplementary table S3, Supplementary Material online). The gene set provides a useful resource to further explore the molecular genetic underpinnings of butterfly wing pattern evolution.
Materials and Methods
Sample Collection and Sequencing
Vanessa cardui butterflies were purchased from Carolina Biological Supply. They were fed on a multispecies artificial diet (Southland) and maintained in a 16:8 h light/dark cycle at 28 °C. Total genomic DNA of a single female V. cardui was extracted from a prepigmentation stage pupa using a QIAGEN Genomic-tip kit. We applied PacBio single-molecule, real-time (SMRT) sequencing system for DNA library construction and sequencing.
Vanessa cardui whole-body and wing tissue samples were collected for RNA library construction and sequencing. Vanessacardui were first sampled at multiple developmental stages, including early embryonic development (<12 h postoviposit), late embryonic to early larval development (12–52 h postoviposit), and hatched larva (mixture with early, middle-, and late-stage larvae). Vanessacardui pupal tissues were also collected along the anterior–posterior body axis (head, thorax, and abdomen, respectively) from both early stage (i.e., 3 days after pupation) and late melanin-stage pupae (i.e., ∼6 days after pupation when black melanin pigments began to show up). Second, forewings from five different wing developmental stages of V. cardui were sampled (supplementary table S1, Supplementary Material online), including last instar larvae, 3 days after pupation, prepigmentation stage (∼5 days after pupation), ommochrome development (∼5.5 days after pupation when red–orange ommochrome pigments started to show up), and melanin development pupae. Hindwings across multiple wing developmental stages were previously sampled (Zhang et al. 2017a). Two biological replicates of each wing developmental stage were prepared. Total RNA was extracted from each sample with an Ambion Purelink RNA Mini Kit (Life Technologies). RNA libraries were constructed using the NEBNext Ultra RNA Library Prep kit for Illumina (New England Biolabs).
Genome Assembly and Assessment
Whole-genome SMRT data of V. cardui was first passed through TANmask and REPmask modules from the Damasker suite. The initial error-corrected reads were then processed by the overlap portion of the FALCON pipeline (Chin et al. 2016) using a length cutoff of 5,000bp. After assembly, the genome was polished by Quiver using the original raw reads. HaploMerger2 (Huang et al. 2017) was run to produce an improved, deduplicated assembly. In addition, we aligned the V. cardui genome against M. cinxia genome reference for chromosome assembly. Using MUMmer alignment package (Marçais et al. 2018), we generated one-to-one alignments of best hits between these two genomes with an alignment identity of between 80% and 90%, for regions of at least 200 bp in length, for scaffolds of ≥1 Mb in length. A circle plot of the alignment was made using custom R scripts, with packages tidyverse v1.3.0 (Wickham et al. 2019), circlize v0.4.10 (Gu, et al. 2014) and RColorBrewer v1.1-2. We used BUSCO (Simão et al. 2015) to evaluate the genome completeness. We compared the assembled and structural annotation metrics of V. cardui with those of other butterfly species for further evaluation (supplementary table S2, Supplementary Material online).
Annotation of Repetitive Elements
Genome sequences were analyzed with RepBase (v20181026) (Bao et al. 2015) to identify repeats using RepeatMasker (v4.0.6) (Bergman and Quesneville 2007) and RepeatProteinMask (-noLowSimple P value 0.0001). Tandem repeat finder (v4.09) (Benson 1999) was used to identify tandem repeats. In addition, RepeatModeler (v1.0.9) (Flynn et al. 2020) was employed to construct a de novo repeat library. This species-specific library was subsequently utilized to detect repeat sequences with RepeatMasker in the V. cardui genome.
Gene Prediction, Functional Annotation, and Assessment
We employed three different approaches to predict protein-coding genes. First, homology-based annotation was performed by TBLASTN (Camacho et al. 2009) using protein sequences from six related species including Heliconius erato (Lewis et al. 2016), H. melpomene (Davey et al. 2016), B. anynana (Nowell et al. 2017), D. plexippus (Zhan et al. 2011), Phoebis sennae (Cong, Shen, Warren, et al. 2016), and Papilio xuthus (Li et al. 2015). GeneWise v2.4 (Birney et al. 2004) was then employed to align against the matching protein for the accurate spliced alignment and gene structure prediction. Second, transcriptome-based annotation was applied by both de novo and reference-guided approaches. With the 34.24 Gb of RNA sequence data generated from the 29 samples described above (supplementary table S1, Supplementary Material online), de novo transcript assembly was performed by Trinity pipeline v2.4.0 (Grabherr et al. 2011). For the reference-guided approach, RNA reads were mapped onto the V. cardui genome assembly using Tophat v2.1.1 (Trapnell et al. 2009). Subsequently, Cufflinks v2.2.1 (Trapnell et al. 2010) and cuffmerge were employed to assemble the mapped reads and predict the structure of all transcribed reads with the default parameters. The predicted gene sets generated from de novo and reference-guided approaches were then integrated to produce nonredundant empirical transcript evidence by Program to Assemble Spliced Alignment v2.0.2 (Haas et al. 2003). Third, ab intio gene prediction were carried out on the repeat-masked V. cardui genome assembly using Scalable Nucleotide Alignment Program v 2006-07-28 (Korf 2004) and Augustus v3.2.3 (Stanke and Waack 2003). Gene models from homology-based and transcriptome-based annotation were trained for gene prediction. Finally, MAKER v 2.31.8 (Campbell, et al. 2014) was used to combine homology, transcriptome, and ab intio gene models to form a comprehensive and non-redundant reference gene set.
Gene function annotation of protein-coding genes was performed by BLASTP (with an e-value threshold of 1e−5 against SwissProt, Apweiler et al. 2004), GO (Gene Ontology Consortium 2017), KEGG (Kanehisa et al. 2014), PFAM (Finn et al. 2016), and InterProScan (Jones et al. 2014) databases, respectively.
We tested the quality of the final V. cardui annotation using an ortholog hit ratio analysis (OHR) modified from O’Neil, et al. (2010), which quantified the number and similarity of homologous proteins between our V. cardui annotation and a high-quality B. mori annotation (NCBI B.mori annotation release 102). We identified complete transcripts in the V. cardui annotation with gffread of the Cufflinks (Trapnell et al. 2010), collapsed both the B. mori and V. cardui proteins to nonredundant representative sequences with CD-HIT (Fu et al. 2012), and searched the collapsed B. mori proteins against a BLASTP (Camacho et al. 2009) database of the V. cardui annotation. For each B. mori protein, the OHR was calculated as the proportion of the B. mori protein covered by the longest orthologous hit. For each of these hits, we also analyzed the amino acid similarity (% identity) reported in the BLASTP output. We further compared the V. cardui OHR analysis results with that from another published butterfly D. plexippus (Danaus_plexippus.Dpv3.48.gff3.gz, updated July 11, 2020).
Phylogenetic and Molecular Clock Analysis
To confirm the evolutionary position of V. cardui, OrthoFinder v1.0.6 (Li et al. 2003) was used to cluster gene families. Protein data sets from V. cardui and 12 related species were used for phylogenetic tree construction, including M. cinxia, H. melpomene, B. anynana, D. plexippus, C. cecrops, P. sennae, Lerema accius, P. xuthus, B. mori, Plutella xylostella, D. melanogaster, and Anopheles gambiae. All butterfly data were downloaded from LepBase (updated January 1, 2019). All-to-all BLASTP was carried out with an e-value threshold of 1e−5. Single-copy orthologs were subsequently aligned by MUSCLE v3.8.31 (Edgar 2004a, b). Guided by the protein multisequence alignment, the alignment of coding sequences (CDSs) for these single-copy genes were concatenated for the final data set. jModelTest v2.1.7 (Posada 2008) was used to select the best-fit model for this data set. The clade with D. melanogaster and A.gambiae was set as outgroup. RAxML v8.2.12 (Stamatakis 2015) was used to construct the phylogenetic relationships with the GTR + G + I model. MCMCtree program in PAML v4.7a (Yang 2007) was used to estimate the divergence time with the options “correlated molecular clock” and “JC69” model. Divergence time was calculated according to the fossil records, one for the split of Diptera and Lepidoptera with 290–417 Myr (Douzery, et al. 2004) and the other for the common ancestor of D. melanogaster and A. gambiae (238.5–295.4 Myr) (Benton and Donoghue 2007).
Transcriptome Analyses
The cleaned paired-end reads were aligned to the reference genome using Tophat (Trapnell et al. 2009), and reads uniquely matched to the genome were counted by htseq-count v0.13.5 (Anders et al. 2015). Global gene expression for transcripts was quantified by fragments per kilobase of transcript per million mapped reads (FPKM) using cuffquant v2.2.1 and subsequently normalized by cuffnorm v2.2.1. The principal component analysis and heatmap was performed using the PtR package of the Trinity pipeline. The average normalized FPKM value represented the corresponding quantitative gene expression level at each sample. Differential gene expression between developmental stages was measured using edgeR (Robinson et al. 2010) with biological replicates and a cutoff FDR of 0.001.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by United States National Science Foundation (Grant Nos. IOS-1656514 and IOS-1753559 to R.D.R.), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB42000000 to L.Z.), National Natural Science Foundation of China (Grant No. 41976088 to L.Z.), Pilot National Laboratory for Marine Science and Technology (Grant No. YJ2019NO01 to L.Z.), Key Development Project of Centre for Ocean Mega-Research of Science, Chinese Academy of Science (Grant No. COMS2019R01 to L.Z.), Carl Tryggers Stiftelse anslag (CTS 18:415 to C.W.W. and R.A.S.), and Swedish Research Council (2017-04386 to C.W.W).
Author Contributions
L.Z. and R.D.R. conceived the study. L.Z. performed bench work and data analysis. R.S. and C.W.W. performed synteny and gene annotation assessment analyses. L.Z. and R.D.R. wrote the manuscript.
Data Availability
The raw PacBio sequence data (SRA, SRR12619592-SRR12619646) and final genome assembly have been deposited in NCBI Sequence Read Archive under BioProject accession PRJNA661999. The Illumina RNA-sequencing data generated in this study were deposited under SRA accession SRR12619933–SRR12619941 and SRR12620007–SRR12620015. The assembly and gene predictions are also available on LepBase (http://lepbase.org/) and the Reed Lab genome server (http://butterflygenome.org).
Literature Cited
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, et al. 2004. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32(Database Issue):D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC.. 2007. Paleontological evidence to date the tree of life. Mol Biol Evol. 24(1):26–53. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H.. 2007. Discovering and detecting transposable elements in genome sequences. Brief Bioinform. 8(6):382–392. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blande D, et al. 2020. Improved chromosome level genome assembly of the Glanville fritillary butterfly (Melitaea cinxia) based on SMRT Sequencing and linkage map. bioRxiv: 2020.2011.2003.364950. doi: 10.1101/2020.11.03.364950. [DOI] [PMC free article] [PubMed]
- Briscoe AD, Bernard GD, Szeto AS, Nagy LM, White RH.. 2003. Not all butterfly eyes are created equal: rhodopsin absorption spectra, molecular identification, and localization of ultraviolet‐, blue‐, and green‐sensitive rhodopsin‐encoding mRNAs in the retina of Vanessa cardui. J Comp Neurol. 458(4):334–349. [DOI] [PubMed] [Google Scholar]
- Briscoe AD, White RH.. 2005. Adult stemmata of the butterfly Vanessa cardui express UV and green opsin mRNAs. Cell Tissue Res. 319(1):175–179. [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M.. 2014. Genome annotation and curation using MAKER and MAKER‐P. Curr Protoc Bioinformatics. 48(1):4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challi RJ, Kumar S, Dasmahapatra KK, Jiggins CD, Blaxter M.. 2016. Lepbase: the Lepidopteran genome database. bioRxiv 056994. doi: 10.1101/056994. [DOI]
- Chin C-S, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13(12):1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Borek D, et al. 2016. Complete genomes of Hairstreak butterflies, their speciation and nucleo-mitochondrial incongruence. Sci Rep. 6(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Warren AD, et al. 2016. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol Evol. 8(3):915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connahs H, Rhen T, Simmons RB.. 2016. Transcriptome analysis of the painted lady butterfly, Vanessa cardui during wing color pattern development. BMC Genomics 17(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra KK, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, et al. 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3: Genes, Genomes, Genetics 6: 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Paz Celorio-Mancera M, et al. 2016. Evolutionary history of host use, rather than plant phylogeny, determines gene expression in a generalist butterfly. BMC Evol Biol. 16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinwiddie A, et al. 2014. Dynamics of F-actin prefigure the structure of butterfly wing scales. Dev Biol. 392(2):404–418. [DOI] [PubMed] [Google Scholar]
- Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H.. 2004. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci U S A. 101(43):15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004a. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004b. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeland M, et al. 2018. A comprehensive and dated phylogenomic analysis of butterflies. Curr Biol. 28(5):770–778.e775. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, et al. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberale-Stille G, Schäpers A, Janz N, Nylin S.. 2019. Selective attention by priming in host search behavior of 2 generalist butterflies. Behav Ecol. 30(1):142–149. [Google Scholar]
- Gene Ontology Consortium. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B.. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30(19):2811–2812. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31(19):5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama A, Taira W, Otaki JM.. 2012. Color-pattern evolution in response to environmental stress in butterflies. Front Genet. 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Kang M, Xu A.. 2017. HaploMerger2: rebuilding both haploid sub-assemblies from high-heterozygosity diploid genome assembly. Bioinformatics 33(16):2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42(Database Issue):D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JJ, van der Burg KR, Mazo-Vargas A, Reed RD.. 2016. ChIP-Seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Rep. 16(11):2855–2863. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. 2015. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat Commun. 6(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, et al. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 14(1):e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Wolcott NS, O'Connell LA.. 2020. Bringing immersive science to undergraduate laboratory courses using CRISPR gene knockouts in frogs and butterflies. Journal of Experimental Biology. 223(Suppl_1): jeb208793. [DOI] [PubMed] [Google Scholar]
- Mazo-Vargas A, et al. 2017. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci U S A. 114(40):10701–10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout HF. 1991. The development and evolution of butterfly wing patterns. Smithson. Inst. 293: [Google Scholar]
- Nowell RW, et al. 2017. A high-coverage draft genome of the mycalesine butterfly Bicyclus anynana. Gigascience. 6(7):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M, et al. 2016. Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature. 535(7611):280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiler E, Markow TA.. 2017. Population connectivity and genetic diversity in long-distance migrating insects: divergent patterns in representative butterflies and dragonflies. Biological Journal of the Linnean Society. 122(2):479–486. [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256. [DOI] [PubMed] [Google Scholar]
- Reed RD, Nagy LM.. 2005. Evolutionary redeployment of a biosynthetic module: expression of eye pigment genes vermilion, cinnabar, and white in butterfly wing development. Evol Dev. 7(4):301–311. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields O. 1992. World distribution of the Vanessa cardui group (Nymphalidae). J Lepid Soc. 46:235–238. [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2015. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics. 51(1):6–14. [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S.. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Stefanescu C, Alarcón M, Àvila A.. 2007. Migration of the painted lady butterfly, Vanessa cardui, to north-eastern Spain is aided by African wind currents. J Anim Ecol. 76(5):888–898. [DOI] [PubMed] [Google Scholar]
- Stefanescu C, et al. 2013. Multi‐generational long‐distance migration of insects: studying the painted lady butterfly in the Western Palaearctic. Ecography 36(4):474–486. [Google Scholar]
- Stefanescu C, et al. 2017. Back to Africa: autumn migration of the painted lady butterfly Vanessa cardui is timed to coincide with an increase in resource availability. Ecol Entomol. 42(6):737–747. [Google Scholar]
- Stefanescu C, Soto DX, Talavera G, Vila R, Hobson KA.. 2016. Long-distance autumn migration across the Sahara by painted lady butterflies: exploiting resource pulses in the tropical savannah. Biol Lett. 12(10):20160561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- T O'Neil S, et al. 2010. Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC Genomics. 11:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-C, et al. 2020. Physical and behavioral adaptations to prevent overheating of the living wings of butterflies. Nat Commun. 11(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem SM, et al. 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 1(3):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg KR, et al. 2019. Contrasting roles of transcription factors Spineless and EcR in the highly dynamic chromatin landscape of butterfly wing metamorphosis. Cell Rep. 27(4):1027–1038. e1023. [DOI] [PubMed] [Google Scholar]
- Varshney R, Smetacek P.. 2015. A synoptic catalogue of the Butterflies of India: Butterfly Research Centre, Bhimtal & Indinov Publishing.
- Wickham H, et al. 2019. Welcome to the Tidyverse. JOSS. 4(43):1686. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM.. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell. 147(5):1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. 2017a. Genetic basis of melanin pigmentation in butterfly wings. Genetics. 205(4):1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Mazo-Vargas A, Reed RD.. 2017b. Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proc Natl Acad Sci U S A. 114(40):10707–10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Reed RD.. 2016. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun. 7:11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw PacBio sequence data (SRA, SRR12619592-SRR12619646) and final genome assembly have been deposited in NCBI Sequence Read Archive under BioProject accession PRJNA661999. The Illumina RNA-sequencing data generated in this study were deposited under SRA accession SRR12619933–SRR12619941 and SRR12620007–SRR12620015. The assembly and gene predictions are also available on LepBase (http://lepbase.org/) and the Reed Lab genome server (http://butterflygenome.org).