Abstract
Heliconius butterflies (Lepidoptera: Nymphalidae) are a group of 48 neotropical species widely studied in evolutionary research. Despite the wealth of genomic data generated in past years, chromosomal level genome assemblies currently exist for only two species, Heliconius melpomene and Heliconius erato, each a representative of one of the two major clades of the genus. Here, we use these reference genomes to improve the contiguity of previously published draft genome assemblies of 16 Heliconius species. Using a reference-assisted scaffolding approach, we place and order the scaffolds of these genomes onto chromosomes, resulting in 95.7–99.9% of their genomes anchored to chromosomes. Genome sizes are somewhat variable among species (270–422 Mb) and in one small group of species (Heliconius hecale, Heliconius elevatus, and Heliconius pardalinus) expansions in genome size are driven mainly by repetitive sequences that map to four small regions in the H. melpomene reference genome. Genes from these repeat regions show an increase in exon copy number, an absence of internal stop codons, evidence of constraint on nonsynonymous changes, and increased expression, all of which suggest that at least some of the extra copies are functional. Finally, we conducted a systematic search for inversions and identified five moderately large inversions fixed between the two major Heliconius clades. We infer that one of these inversions was transferred by introgression between the lineages leading to the erato/sara and burneyi/doris clades. These reference-guided assemblies represent a major improvement in Heliconius genomic resources that enable further genetic and evolutionary discoveries in this genus.
Keywords: Heliconius, genome assembly, structural variation, inversions, introgression
Introduction
Advances in sequencing technology have revolutionized the field of evolutionary biology. Generating short-read genomic data sets is now common practice, enabling investigation of fundamental evolutionary processes including the genetic basis of adaptive traits, dynamics of selection on particular alleles, and demographic histories of populations. In order to exploit the power of low-cost short-read data, a common strategy is to align reads to a reference genome.
The availability of high-quality reference genomes can determine the breadth and power of comparative and population genomic analyses in evolutionary studies. For instance, placing genome scaffolds on chromosomes allows one to contrast patterns between autosomes and sex chromosomes, and this has been important for understanding speciation (Coyne and Orr 1989; Prowell 1998; Masly and Presgraves 2007; Ellegren et al. 2012; Fontaine et al. 2015; Coyne 2018; Seixas et al. 2018; Martin et al. 2019). Anchoring scaffolds to chromosomes can also enable discovery of divergence and gene flow along chromosomes and how it is modified by recombination rate variation (Schumer et al. 2018; Martin et al. 2019). Furthermore, chromosome-level assemblies greatly improve the power and resolution of genome-wide association and QTL studies (Markelz et al. 2017; Benevenuto et al. 2019). However, high-quality, chromosome-level, contiguous reference genome assemblies are often limited to one or a few species in many groups of taxa, especially in nonmodel organisms. Generating chromosome-level assemblies often uses a mixture of lower fidelity long-read sequencing data (such as PacBio or Nanopore) complemented with high fidelity short-read sequencing data, typically Illumina, genetic linkage mapping, optical (restriction site) mapping, and/or chromatin interaction frequency data (Hi-C) (Deschamps et al. 2018; Ghurye and Pop 2019; Rice and Green 2019; Yu et al. 2019; Wei et al. 2020; Yang et al. 2020). These methods can be expensive and time consuming, especially for multiple species in entire clades.
With 48 described species, Heliconius butterflies are a prime example of an adaptive radiation where multiple chromosome-level reference assemblies could improve evolutionary analyses. Currently, published high-contiguity genome assemblies (hereafter, reference genomes) exist for only two species—Heliconius melpomene melpomene (Davey et al. 2017) and Heliconius erato (H. erato lativitta [Lewis et al. 2016] and H. erato demophoon [Van Belleghem et al. 2017]). Although these chromosome-level reference assemblies are essential tools for genomic studies in Heliconius, each has limitations. At 275 Mb, H. melpomene has the most compact Heliconius genome assembled to date (Edelman et al. 2019). Mapping short-read sequencing data from other species with larger genomes to this reference genome likely results in the loss of information, due to loss of ancestral orthologous sequence in the H. melpomene genome, and spurious read mapping to similar but nonorthologous regions. In contrast, the two H. erato reference genomes (383 and 418 Mb) are among the largest Heliconius genomes assembled to date. However, although these might be appropriate for studies focusing on closely related species (e.g., species within the erato clade), mapping accuracy decreases in more divergent species (Prüfer et al. 2010) and better results are obtained when mapping to closer reference genomes (Gopalakrishnan et al. 2017). Also, as we move from comparative (e.g., phylogenomic) toward more functional genetics studies (Lewis et al. 2016; Lewis and Reed 2019; Pinharanda et al. 2019), this genus could benefit greatly from higher-quality species-specific genomic resources.
Recently, de novo draft genomes of 16 Heliconius species (supplementary table 1, Supplementary Material online) have been assembled (Edelman et al. 2019). The experimental protocol included two species, H. melpomene melpomene and H. erato demophoon, for which good reference genomes already existed, for comparison. The draft genomes were generated from Illumina PCR-free libraries sequenced at deep coverage (at least 60× coverage) using paired-end 250-bp reads on the Illumina Hi-Seq 2500 and assembled using w2rap (Clavijo et al. 2017), an extension of the DISCOVAR de novo genome assembly method (Weisenfeld et al. 2014; Love et al. 2016). This strategy results in high-quality genomes in terms of read accuracy, contiguity within scaffolds, and genome completeness (87.5–97.3% complete single copy core BUSCO genes present; Edelman et al. 2019). Nonetheless, because these assemblies (hereafter, w2rap assemblies) employ only short-read data, they were considerably more fragmented (contig N50 = 11–49 kb; scaffold N50 = 23–106 kb) than the Heliconius reference genomes. Furthermore, scaffolds were not assigned to chromosomes.
A cost-effective approach for improving the contiguity of existing draft genomes is to use synteny-based methods that identify potentially adjacent scaffolds from multispecies alignments. Such methods are particularly efficient if high-quality reference genome assemblies of closely related species are available, and especially if there is high synteny between the genomes of the draft and reference assemblies (Alonge et al. 2019), as in Heliconius. Although a limited number of genomic rearrangements have been identified in Heliconius (Davey et al. 2017; Jay et al. 2018; Edelman et al. 2019; Meier et al. 2020), even species as divergent as H. melpomene and H. erato, which last shared a common ancestor over 10 Ma, remain highly collinear (Davey et al. 2017). Synteny-based assembly should thus be especially effective within this genus.
Here, we exploit the chromosome-mapped assemblies of the H. melpomene melpomene and H. erato demophoon reference genomes to guide improvement of contiguity of the w2rap draft genome assemblies of 16 Heliconius species. The w2rap scaffolds were ordered, oriented and anchored onto chromosomes, resulting in a level of completeness of the scaffolded w2rap assemblies similar to that of reference genomes. A potential weakness of our synteny-based assembly method is that it can miss structural variation among species where it occurs. However, we use these scaffolded w2rap assemblies (hereafter, reference-guided assemblies) to identify clade-specific local genomic expansions due to local duplications with potentially functional consequences. To estimate how much structural variation we might be missing, we also carry out a systematic search for candidate inversions in the genus using the original w2rap scaffolds to detect break points, and demonstrate that the results can be used to investigate phylogenetic uncertainty and gene flow deep in the tree of Heliconius species.
Materials and Methods
Genome Merging and Scaffolding
We used the draft genome scaffolder MEDUSA (Bosi et al. 2015) for reference-aided assembly of the existing DISCOVAR de novo/w2rap genomes (Edelman et al. 2019). MEDUSA relies on reference genomes from closely related species to determine the correct order and orientation of the draft genome scaffolds, assuming collinearity between reference and the lower contiguity genome. The w2rap genome assemblies of 16 Heliconius species produced by Edelman et al. (2019)—supplementary table 1, Supplementary Material online—and high-quality reference genome assemblies of two Heliconius species—H. melpomene (Hmel2.5) and H. erato demophoon (Heliconius_erato_demophoon_v1) - were downloaded from Lepbase (http://lepbase.org/, last accessed April 21, 2021). Before the reference-scaffolding step, alternative haplotypes present in the w2rap assemblies were collapsed using the HaploMerger2 pipeline (version 20180603) (Huang et al. 2017). Repetitive elements and low complexity regions in the w2rap assemblies were first soft-masked using WindowMasker (Morgulis et al. 2006) with default settings. A score matrix for LASTZ (used within HaploMerger) was generated for each w2rap assembly. This was done using the lastz_D_Wrapper.pl script with identity = 90 and splitting the w2rap assemblies into two sets of scaffolds (scaffolds greater or smaller than 150 kb). HaploMerger2 batch scripts A and B were then run using default settings. Finally, MEDUSA was used with default parameters to place and orient the w2rap assembly scaffolds based on either of the two reference genomes, placing 100 Ns between adjacent pairs of scaffolds mapping to the same reference chromosome/scaffold. This resulted in two scaffolded assemblies per species (one based on mapping to H. melpomene and another based on mapping to H. erato demophoon reference genomes).
Reference-guided assemblies were then re-aligned to the H. melpomene and H. erato reference genomes using the Mashmap aligner as implemented in D-GENIES v1.2.0 online tool (Cabanettes and Klopp 2018) to assess collinearity. Scaffolds in the reference-guided assemblies aligning to reference assembly scaffolds anchored to chromosomes were renamed to reflect their association to chromosomes and order within chromosomes (as in the reference genomes). Also, when necessary, scaffold sequences were reverse complemented to maintain the same orientation as in the reference.
Mitochondrial Genome Assembly
To assemble the mitochondrial genomes of the 16 Heliconius species analyzed here, we first subsampled 1 million read pairs from the original reads used to produce the w2rap assemblies. We then used ABySS 2.0 (Jackman et al. 2017) to assemble the reads, using 5 different k-mer sizes (64, 80, 96, 112 and 128 bp) and requiring a minimum mean unitig k-mer coverage of 10. All other parameters were left as default. Because of the higher number of mtDNA copies relative to nuclear DNA, resulting in higher mtDNA coverage, we were able to recover the mitochondrial genome as a single large contig (about the size of the complete mitogenome) whereas any nuclear contigs should be small. In Heliconius, the sizes of the mitogenomes sequenced so far are approximately 15,300 bp, thus only contigs larger than 15 kb were retained. These where then blasted to the NCBI Nucleotide collection (nr/nt) to confirm that they corresponded to the mitochondrial genome. Finally, for each species, only the largest contig (after removing Ns) was retained. The mitochondrial sequences were aligned using MAFFT v7.407 (Katoh and Standley 2013), with default parameters and a maximum-likelihood (ML) tree was estimated using IQ-TREE v1.6.10 (Nguyen et al. 2015) (fig. 1a). Model selection was performed using ModelFinder (Kalyaanamoorthy et al. 2017) and branch support was assessed with 1,000 ultrafast bootstraps (Hoang et al. 2018), as implemented in IQ-TREE. We also used this approach to recover the mitogenome of Eueides tales (accession number: SRS4612550) to use as an outgroup.
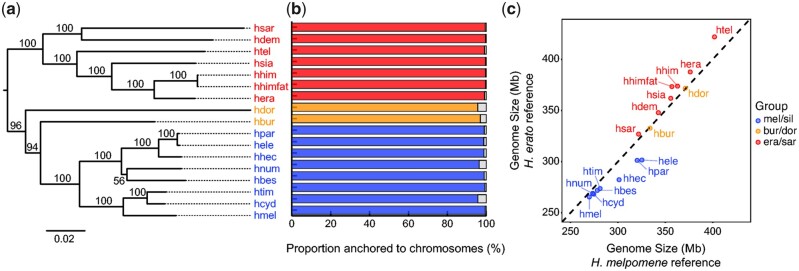
Fig. 1.
Reference-guided assemblies. (a) ML tree from whole mitochondrial genomes assembled here. Bootstrap values are shown next to the branches. The tree was rooted using the E. tales mitochondrial genome. (b) Proportion of the reference-scaffolded assemblies length anchored to chromosomes. The results are shown for the reference-guided assemblies mapped to the closest reference (either H. melpomene or H. erato demophoon; see supplementary table 2, Supplementary Material online). For a complete report of the results, see supplementary figure 37, Supplementary Material online. (c) Reference-scaffolded genome sizes using either H. melpomene (x axis) or H. erato demophoon (y axis) as the reference. The dashed line represents the expectation if there was a 1:1 correspondence. In all panels, subclade memberships are represented by different colors—melpomene/silvaniform (blue), burneyi + doris (yellow), erato/sara (red). Species codes for all the new reference-guided assemblies are as follows: hmel—H. melpomene; hcyd—H. cydno; htim—H. timareta; hbes—H. besckei; hnum—H. numata; hhec—H. hecale; hele—H. elevatus; hpar—H. pardalinus; hbur—H. burneyi; hdor—H. doris; hera—H. erato; hhimfat—H. himera; hhim—H. himera; hsia—H. hecalesia; htel—H. telesiphe; hdem—H. demeter; hsar—H. sara.
Scaffolded Assemblies Quality Assessment
Basic statistics (e.g., scaffold N50, cumulative length, proportion of missing sequence) of the reference-guided scaffolded genome assemblies were calculated using QUAST v5.0.2 (Gurevich et al. 2013). Assembly completeness was assessed using BUSCO_V3 (Simão et al. 2015), which looks for the presence (complete, partial, or duplicated) or absence (missing) of core arthropod genes (arthropoda-odb9 data set; available from https://busco-archive.ezlab.org/v3/datasets/eukaryota_odb9.tar.gz, last accessed April 21, 2021).
Gene Annotation
We used the Liftoff tool (Shumate and Salzberg 2020) to map gene annotations from the reference genomes to the new reference-guided assemblies. This software aligns gene sequences, as annotated in a reference genome, to a target genome and finds the alignments of the exons that maximize sequence identity while preserving the transcript and gene structure. We thus used either the H. melpomene (Hmel2.5.gff3) or H. erato demophoon (Heliconius_erato_v1_-_genes.gff.gz) gene annotations (downloaded from www.butterflygenome.org and http://lepbase.org/, respectively; last accessed April 21, 2021), depending on the reference genome used for the scaffolding of the reference-guided assemblies. We ran Liftoff setting the maximum distance between two nodes to be either 1) twice the distance between two nodes in the reference genome (i.e., distance scaling factor of 2) or 2) 20 kb distance between in the target, depending on which of these distances is greater. In order to improve mapping of exons at the ends of genes we extended gene sequences by 20% of the gene length, to include flanking sequences on each side (-flank 0.2). Given the w2rap scaffolds were ordered, oriented, and anchored to chromosomes using the reference genomes as the backbone, and thus we know the association between scaffolds in the reference genomes and in the reference-guided assemblies, we have also enabled the option to first align genes chromosome by chromosome. All other parameters were set as default.
Mapping and Genotype Calling of Resequencing Data
Mapping efficiency of the original w2rap reads to the reference-guided assemblies was compared with mapping efficiency of the same reads to the reference genomes. Reads were first filtered for Illumina adapters using cutadapt v1.8.1 (Martin 2011) and then mapped to their respective reference-guided genome assemblies, the H. melpomene and H. erato demophoon reference genomes using BWA mem v0.7.15 (Li 2013), with default parameters and marking short split hits as secondary. Mapped reads were sorted and duplicate reads removed using sambamba v0.6.8 (Tarasov et al. 2015). Realignment around indels was performed with the Genome Analysis Toolkit (GATK) v3.8 RealignerTargetCreator and IndelRealigner modules (McKenna et al. 2010; DePristo et al. 2011), in order to reduce the number of indel miscalls. Mapping statistics and mean read depth were calculated in nonoverlapping sliding windows of 25 kb using the flagstat and depth modules implemented in sambamba v0.6.8, respectively.
Genotype calling was also performed for reads mapped to either of the two reference genomes and for each individual separately with bcftools v1.5 (Li et al. 2009) mpileup and call modules (Li 2011), using the multiallelic-caller model (call -m) and requiring a minimum base and mapping qualities of 20. Genotypes were filtered using the bcftools filter module. Both invariant and variant sites were required to have a minimum quality score (QUAL) of 20. Furthermore, individual genotypes were filtered to have a depth of coverage (DP) ≥ 8 (except for the Z-chromosome of females for which the minimum required depth was 4) and genotype quality ≥ 20. All genotypes not fulfilling these requirements or within 5 bp of an indel (–SnpGap) were recoded as missing data.
Copy Number Variation and Selection Tests
Copy number variation (CNV) of genes within repeat regions of interest was estimated using two different approaches. The first relies on mapping exonic sequences of genes annotated in the H. melpomene reference within regions of interest onto the reference-guided assemblies. The reference-guided assemblies were split back into the original haplotype merged scaffolds by breaking apart regions separated by 100 consecutive Ns, in order to avoid potential mismappings over scaffold breakpoints while retaining information regarding chromosome assignment. Exon sequences were mapped to these scaffolds using minimap2 v2.9 (Li 2018), with default settings (except that, as we were interested in repeats, we allowed a much larger threshold of up to 1,000 different alignments). Only alignments for which ≥50% of the length of the exon was mapped were considered. Copy number of each exon was then estimated based on the number of alignments to these genomes. The second approach is based on read coverage of the original w2rap read data, mapped to the H. melpomene reference genome using BWA as described above. For each species, the mean read coverage within an exon (based on the coordinates of exons as annotated in H. melpomene) was calculated using the sambamba v0.6.8 depth module (Tarasov et al. 2015). Exon coverage was then normalized dividing by the median genomic coverage (calculated in nonoverlapping windows of 25 kb along the genome as described above) to estimate copy number. This second approach was also used to estimate CNV in Amazon and extra-Amazonian populations of H. hecale, H. elevatus, and H. pardalinus (supplementary table 12, Supplementary Material online).
We further investigated whether CNV in specific genes resulted in potentially functional copies or pseudogenization by analyzing signals of codon-based selection and looking for the presence or absence of stop codons. For each gene, we examine each exon independently since different exons can show different copy number. Sequences of the different putative copies were extracted from the reference-guided assemblies, based on the coordinates obtained by aligning the reference H. melpomene exon sequences to the reference-guided assemblies (as described above in this section). When shorter than the exon length, coordinates were extended to match the total exon length. Exon sequences including 10 consecutive Ns (introduced during the w2rap assembly process) were excluded from this analysis to avoid artificial sequence frameshifts. The remaining exonic sequences of all species were then aligned to the H. melpomene reference genome using MAFFT v7.407 (Katoh and Standley 2013), with default parameters and allowing reverse complementing of sequences when necessary. Bases before the start and after the end of the H. melpomene reference sequence were removed from the alignment since these could have been erroneously included when extending sequences to match the total exon length (see above). Also, alignments including frameshift mutations (determined based on the H. melpomene sequence) were excluded. We then calculated the ratio of nonsynonymous versus synonymous changes (dN/dS) for each pairwise comparison between exon copies detected in the reference-guided assemblies and the reference H. melpomene sequence, using Li’s method (Li 1993) implemented in the “seqinr” package in R. Finally, we checked for the presence of stop codons using a custom script.
Repeat Annotation
Repeat content within and outside the repeat regions of interest was characterized using RepeatMasker v4.0.9 (Smit et al. 2013). We specified the Heliconius repeat library, which includes de novo TE annotations produced using the original w2rap assemblies (Ray et al. 2019), and applied the most sensitive search setting (-s). Only scaffolds greater than 1 Mb were considered. Enrichment of overall repeat content within the repeat regions for each species in the trio hecale, elevatus, and pardalinus was evaluated by proportion of repeat annotations inside and outside the repeat regions and compared with the same proportion in H. melpomene. We further tested for the enrichment of particular repeat families within the repeat regions of species in the trio in two ways: 1) as compared with the homologous repeat regions in the H. melpomene reference; 2) as compared with the genomic background of the same species. In both cases, we counted the number of repeats of a given repeat family (Ri) and total number of repeats (R) within the repeat regions and compared with that in H. melpomene (Mi and M) or in the genomic background (Bi and B), where i is a specific repeat family. We then calculated the fold change in repeat content of a given repeat family as (Ri / R)/(Mi / M) or (Ri / R)/(Bi / B). In all comparisons, we performed a Fisher’s exact test to determine the significance.
Detection of Inversions in the w2rap Assemblies
In order to detect potential inversions in relation to the reference genomes, we mapped the w2rap scaffolds (after filtering with HaploMerger2; see above) onto the reference genomes. We also included the haplotype merged w2rap assembly of an outgroup species, Eueides tales, to determine the ancestral and derived orientations of the inversions. Scaffolds of at least 5 kb were mapped to the H. m. melpomene and the H. erato demophoon reference genomes using minimap2 (Li 2018) with default settings. Only primary alignments (tp:A:P), at least 1 kb long, with mapping quality ≥60 and with less than 25% approximate per-base sequence divergence (dv) to the reference were kept. Mappings of scaffolds spanning inversion breakpoints in the reference genome should result in split alignments to different strands. We thus considered scaffolds as potentially informative for inversions if they had at least two alignments to the same chromosome (split alignments) and at least one alignment to each strand as potentially informative for inversions. Same-scaffold alignments mapping to the same strand, partially overlapping or not more than 50 kb apart were concatenated. If less than 20% of the length of the scaffold aligned to the reference, the scaffold was excluded. Furthermore, any scaffolds for which both forward and reverse alignments to the reference 1) come from overlapping scaffold regions (overlap greater than 5 kb), 2) overlap in the reference by more than 5 kb, or 3) in which the alignment in one strand is completely within the alignment to the other strand, were removed as these likely represent spurious alignments, perhaps due to repeats. Candidate inversions less than 50 kb from scaffold boundaries within chromosomes of the reference genome were also excluded. Finally, we considered any two informative scaffolds to support the same candidate inversion if they overlapped by at least 75% of the maximum length of the two. We also mapped the two reference genomes against each other (and also the H. erato lativitta onto both) using minimap2 and inferred candidate inversions by looking for alignments, within a scaffold, to the reverse strand. Only alignments with a MQ ≥ 10 and to the same chromosome in the reference were considered. Entire scaffolds aligning to the reverse strand are possibly misoriented and were not considered to be inversions.
For each candidate inversion we made sequence alignments for a subset of species (H. melpomene, H. numata, H. doris, H. burneyi, H. erato, and H. hecalesia, using Eueides tales as an outgroup) based on the original w2rap sequencing data mapped to both H. melpomene and H. erato reference genomes. We then estimated ML trees for these candidate regions using IQ-TREE v1.6.10 (Nguyen et al. 2015). Model selection was performed using ModelFinder (Kalyaanamoorthy et al. 2017) and branch support was assessed with 1000 ultrafast bootstraps (Hoang et al. 2018), as implemented in IQ-TREE.
We used Patterson’s D statistic (Green et al. 2010; Durand et al. 2011) to test 1) which branching pattern best describes the relationships between H. doris, H. burneyi, the erato/sara, and the melpomene/silvaniform groups and 2) whether the alternative clustering of H. doris and H. burneyi with either of the two groups (both patterns were observed in the inversions) could be explained by introgression. We used the ABBABABAwindows.py script (available from github.com/simonhmartin/genomics_general, last accessed April 21, 2021) to estimate the D statistic in non-overlapping windows of 1 Mb, discarding all windows with fewer than 100 informative sites. The mean and variance of the D statistic were calculated using a 1-Mb block jackknifing approach, allowing a test of whether D differed significantly from zero. We have also used the internal branch length based approach QuIBL (Edelman et al. 2019), which uses the distribution of internal branch length and calculates the likelihood that the triplet topologies discordant from the species tree are due to introgression rather than ILS alone. For this analysis, we sampled 10 kb windows along the genome (50 kb apart) and for each we estimated ML trees using the phyml_sliding_windows.py (available from github.com/simonhmartin/genomics_general, last accessed April 21, 2021). Only alignments with less than 5% of the sites genotyped were discarded. We then ran QuIBL on the filtered data set with default parameters and adjusting the number of steps to 50. In both Patterson’s D and QuIBL analyses, Eueides tales was used as outgroup.
In order to detect local signals of introgression, we also calculated the fdM statistic (Malinsky et al. 2015), which, like the fd statistic (Martin et al. 2015), checks for imbalance in the number of shared variants between the inner outgroup population and one of two ingroup populations, and was developed specifically to investigate introgression of small genomic regions. Unlike the fd statistic, it simultaneously tests for an excess of shared variation between the inner outgroup population and either ingroup population, at each genomic window. Again, we used the ABBABABAwindows.py script (available from github.com/simonhmartin/genomics_general, last accessed April 21, 2021) to estimate the fdM in nonoverlapping windows of 100 kb, discarding all windows with fewer than 100 informative sites. Because a local excess of derived alleles could also be explained by retention of ancestral polymorphism (incomplete lineage sorting—ILS), we calculated the divergence (DXY) between both H. doris and H. burneyi to H. erato, normalized by divergence to H. melpomene (i.e., relative node depth, RND), to control for variation in substitution rate across the genome. DXY was calculated in 100 kb nonoverlapping windows using the popgenWindows.py script (available from github.com/simonhmartin/genomics_general, last accessed April 21, 2021). Finally, we also used QuIBL to estimate the probability that gene trees within the chromosome 13 inversion were generated by introgression.
Gene Expression Analyses
Ovaries were dissected from adult females of H. melpomene rosina and H. pardalinus butleri at 2 weeks post-eclosion, divided into developmental stages, and stored in RNALater. Ovaries were blotted dry with kimWipes to remove excess RNALater solution. Tissue was then transferred to TRIZOL and homogenized with the PRO200 tissue homogenizer (PRO Scientific). RNA was extracted with the Direct-zol RNA miniprep kit (Zymo R2051). mRNA libraries were prepared by the Harvard University Bauer Core with the KAPA mRNA HyperPrep kit, with mean fragment insert sizes of 200–300 bp. mRNA was sequenced with the NovaSeq S2, producing an average of 49 million paired-end, 50 bp reads.
RNASeq reads were mapped to the H. melpomene v2.5 transcriptome (Pinharanda et al. 2019) using kallisto (Bray et al. 2016). Analysis was carried out in R using the Sleuth package (Pimentel et al. 2017). Significant differences in expression levels between H. melpomene and H. pardalinus were assessed with a likelihood ratio test, comparing expression as a function of developmental stage to expression as a function of developmental stage + species identity.
Results
Reference-Guided Genome Assemblies and Annotation
Alternative haplotype scaffolds in the w2rap assemblies were first merged using HaploMerger2 (Huang et al. 2017), reducing the numbers of scaffolds by 31.3–64.6% and total assembly length by 3.4–25.9% (supplementary table 2, Supplementary Material online). These haplotype-merged scaffolds were then assembled using a reference guided approach (Bosi et al. 2015). Standard metrics for the resulting assemblies can be found in supplementary table 2, Supplementary Material online. Contiguity of all assemblies was considerably improved, with a reduction in the numbers of scaffolds to 0.9–16.8% of the original w2rap assemblies (supplementary table 2; supplementary figs. 1 and 2, Supplementary Material online). N50 length values were 14.2–20.0 Mb when using the H. melpomene genome as reference (the N50 of the H. melpomene reference genome is ∼14.3 Mb) and 7.1–11.5 Mb when using the H. erato demophoon genome (H. erato demophoon reference genome N50 is ∼10.7 Mb). In general, scaffolds in the reference anchored to chromosomes have a single corresponding scaffold in each of our reference-guided assemblies (supplementary figs. 3–36, Supplementary Material online). Overall, 94.5–99.5% and 91.6–99.7% of nucleotide positions in each reference-guided assembly were anchored to chromosomes using the H. melpomene and H. erato references, respectively (supplementary table 2, Supplementary Material online; fig. 1b; supplementary fig. 37, Supplementary Material online). For each species, the proportion of reference-guided assembly length anchored to chromosomes was higher in assemblies guided by the genome of the phylogenetically closest species, H. doris being the only exception. This species is distant from both reference genomes, but has been inferred to be phylogenetically closer to H. melpomene (Kozak et al. 2015, 2018; Edelman et al. 2019). However, it shows a 0.2% higher proportion of the assembly length included in scaffolds anchored to chromosomes using H. erato demophoon as the reference, likely because the larger genome of H. erato contains ancestral sequence that was lost by the smaller H. melpomene genome but retained in the early branching H. doris.
Genome sizes, considering only scaffolds anchored to chromosomes, varied between approximately 270–422 Mb (fig. 1c; supplementary table 2, Supplementary Material online). Phylogeny is a predictor of genome size: species within the erato/sara clade have larger genomes (327–422 Mb) than species in the melpomene/silvaniform group (270–325 Mb; fig. 1c), whereas genome sizes of H. burneyi and H. doris (334 and 371 Mb, respectively), which branched early in the melpomene/silvaniform clade, are more typical of those of the erato/sara group. The genome size of the H. melpomene reference-guided assembly (270; 271 Mb including all scaffolds) is similar to that of the reference assembly (273; 275 Mb total; Davey et al. 2017), but both are smaller than estimates based on flow cytometry (292 ± 2.4 Mb; Jiggins et al. 2005). The genome size of the H. erato demophoon reference-guided assembly (388; 391 Mb total) is a little larger than that of the reference assembly (383 Mb; Van Belleghem et al. 2017) but both are smaller than flow cytometry estimates (396–397 Mb, misnamed as H. e. petiverana; Tobler et al. 2005). Despite the difference in genome sizes of the reference genome used to guide scaffolding, genome sizes of our assemblies (considering only scaffolds anchored to chromosomes) did not depend strongly on which reference genome was used (Spearman’s rank correlation test ρ = 0.99; P ≪ 0.01; linear regression slope = 0.81; fig. 1c). Likewise, individual chromosome lengths of the species assemblies scaffolded using the two different references differed little and were highly correlated (Spearman’s rank correlation test, ρ = 0.94–0.99; P ≪ 0.01; linear regression slope = 0.80–1.05; supplementary fig. 38; supplementary table 3, Supplementary Material online).
Assembly completeness was evaluated by the presence of core arthropod genes in BUSCO. The proportion of detected orthologs varied between 98.6% and 99.6%, values similar to those reported by Edelman et al. (2019) for the original w2rap genomes (supplementary fig. 39; supplementary table 4, Supplementary Material online). There are however improvements (1–10% increase) in terms of the percentage of complete single copy BUSCOs which were previously recovered as complete duplicated, fragmented and missing BUSCOS. These improvements are a consequence of 1) decreased scaffold redundancy (due to collapsing of alternative haplotype scaffolds during the haplotype merging step) which helps reducing the number of complete duplicated BUSCOs whereas 2) the increased contiguity resulting from the reference guided scaffolding step reduces the number of fragmented BUSCOs (supplementary table 4, Supplementary Material online). The latter improvement provides strong evidence that the reference aided assembly was largely correct.
There was no significant correlation between the quality of the original w2rap assemblies (e.g., number of scaffolds and scaffold N50) and final metrics for the reference-guided assemblies (e.g., number of scaffolds, genome size estimates and proportion of the assembly anchored to chromosomes) (supplementary fig. 40 and supplementary table 2, Supplementary Material online). For example, we included two assemblies (hhimfat and hhim) of the same species, H. himera. These two individuals should have similar genomes and so we expect their final metrics to be similar. Although the original w2rap assemblies of these two individuals differed in quality (e.g., number of scaffolds, scaffold N50 and assembly length), the final assemblies were very similar in terms of total assembly length, chromosome sizes, scaffold N50, assembly completeness and number of annotated genes. Overall, the initial quality of the assemblies does not seem to greatly affect the quality of the final assemblies, at least in our data set.
Gene annotation of H. melpomene and H. erato demophoon reference genomes was mapped onto the reference-guided assemblies using the annotation lift-over tool Liftoff (Shumate and Salzberg 2020). We considered only transcripts with ORFs (i.e., start and stop codon, no frame-shift mutation and no internal stop codons) as successful mappings. Out of the 21,656 transcripts from 20,096 H. melpomene annotated genes and 20,118 transcripts from 13,676 H. erato demophoon annotated genes, we were able to successfully map 5,817–14,838 H. melpomene genes (6,217–16,007 transcripts) and 4,530–9,780 H. erato demophoon genes (6,139–14,472 transcripts)—supplementary table 5, Supplementary Material online. The success of the gene annotation lift-over approach decreased with phylogenetic distance to the reference. Although some of the genes that were not successfully lifted-over could potentially represent misannotations in the reference, this could also reflect differences in the structure of these genes or differences in gene composition between species. In fact, Liftoff is designed to map annotations between assemblies of the same or closely related species and assumes gene structure is conserved between target and reference assemblies. Species-specific de novo gene annotation using transcriptome data would be needed to obtain a more comprehensive annotation for all species.
Whole Mitochondrial Genome Assemblies
The de novo assembly of Heliconius mitochondrial genomes enables the recovery of near-complete mitochondrial sequences (∼15 kb, typical of Heliconius—see, e.g., Massardo et al. 2020) for all 16 species, including part of the mitochondrial DNA control region. A genealogy based on these mitochondrial genomes (fig. 1a) did not differ from that for the mitochondrial genomes assembled using reference-aided approaches (Kozak et al. 2015; Massardo et al. 2020), partially validating our de novo approach.
Improved Mapping Efficiency Using the Reference-Guided Assemblies
Mapping the original w2rap Illumina short read sequence data to reference-guided genome assemblies of their own species resulted in 0.45–12.77% more mapped reads and 0.60–40.77% more properly paired reads than when mapping to the closest reference genome (supplementary table 6, Supplementary Material online). These mappings also show an increase in depth of coverage (1.02–2.29 times the coverage obtained when mapped to the closest reference; supplementary table 6, Supplementary Material online), and uniformity of coverage along chromosomes (supplementary figs. 41 and 42, Supplementary Material online). The largest increases in depth of coverage were observed for H. burneyi and H. doris, which are the Heliconius species sequenced here that are phylogenetically most distant to both reference genomes. Increases in depth of coverage tend to be larger in species in the erato/sara clade (1.06–1.97 times more coverage) than in species in the melpomene/silvaniform clade (1.02–1.35 times more coverage). This is expected since species sampled within the erato/sara clade were typically more divergent from H. erato than species in the melpomene/silvaniform group are from H. melpomene. These results show how studies focusing on Heliconius species with deeper divergence to both H. melpomene and H. erato will benefit from mapping resequence data to the reference-guided assemblies generated here. Also, the greater uniformity of coverage along chromosomes when mapping reads to the reference-guided assemblies suggests that they should better capture fine-scale structural variation. This likely reflects the ability of the high sequencing fidelity of the original w2rap assemblies to resolve short imperfect repeats (<500 bp long) (Love et al. 2016; Edelman et al. 2019) that differ between species.
Genome Expansions and Gene Duplications
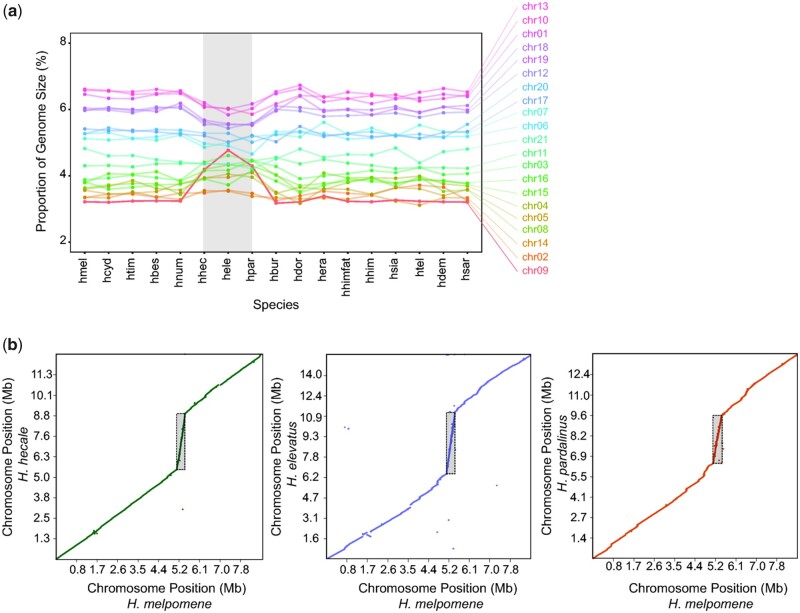
Although genome sizes vary among Heliconius species, relative but not absolute sizes of chromosomes were generally conserved (fig. 2a and supplementary fig. 43, Supplementary Material online). The three closely related species with the largest genomes in the melpomene/silvaniform group (H. hecale, H. elevatus, and H. pardalinus, all from the Amazon basin) are exceptions. Upon closer inspection, variation in chromosome size in these three species is particularly accentuated on chromosome 9 (fig. 2a and supplementary fig. 43, Supplementary Material online). Alignment of reference-guided assemblies of these three species to the H. melpomene reference genome suggests that the increase in size of chromosome 9 mainly corresponds to a single genomic region in H. melpomene (Hmel209001o:5125000–5450000, fig. 2b). This region is approximately 325 kb long in H. melpomene but the scaffolds that map to it total over 10× as long (3.350–4.125 Mb) in the hecale/elevatus/pardalinus trio.
Fig. 2.
Chromosome size variation and local genomic expansions. The results are shown for the reference-guided assemblies mapped to the closest reference (either H. melpomene or H. erato demophoon; see supplementary table 2, Supplementary Material online). (a) Chromosome sizes in proportion to the genome size across the different species for the reference-guided assemblies mapped to the H. melpomene reference genome. Chromosome relative sizes are generally similar across species, with the exception of H. hecale, H. elevatus, and H. pardalinus, particularly chromosome 9. (b) Genome to genome alignment showing the repeat region on chromosome 9 (highlighted by the gray rectangles) in the species trio: H. hecale, H. elevatus and H. pardalinus.
We investigated whether other genomic regions also underwent an increase in size in these three species. When mapping reads to the H. melpomene reference genome, four regions show exceptionally high coverage in these three species (at least 5-fold local increase in the hecale/elevatus/pardalinus trio and less than 2-fold local increase in every other species, in at least two consecutive 25 kb windows). These included the region on chromosome 9 discussed above and three other regions on chromosome 2 (Hmel202001o:4075000–4125000), chromosome 4 (Hmel204001o:5650000–5875000), and chromosome 8 (Hmel208001o:3300000–3475000) (supplementary fig. 44 and supplementary table 7, Supplementary Material online). In contrast, mapping reads onto the reference-guided assemblies of the same species resulted in more uniform coverage in these regions (supplementary figs. 45–48, Supplementary Material online). This suggests the repeats are divergent enough so that they could be largely resolved in the w2rap assemblies.
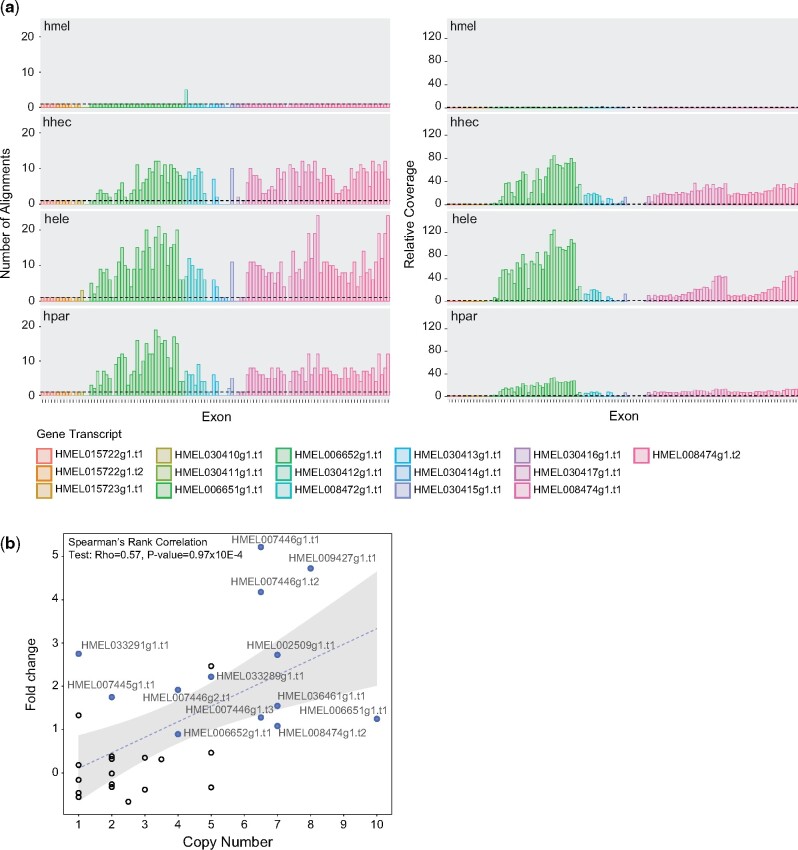
All four repeat regions harbor protein coding genes (supplementary table 7, Supplementary Material online), as annotated in the H. melpomene reference genome, and thus structural variation in these regions could have resulted in gene CNV with potential functional consequences. To address this, we first estimated exon copy numbers based on the number of valid alignments of H. melpomene exon sequences onto the reference-guided assemblies. We opted to estimate copy numbers using exon alignments instead of gene alignments to account for the possibility that entire gene copies are not present in the reference-guided assemblies within these regions. This could be due either to misassembly or to partial gene duplications. Although this could be problematic when aligning entire gene sequences, it should not affect or affect to a lesser degree the alignment of smaller exonic sequences. We have also estimated exon copy numbers based on the normalized mean per base coverage for each exon, mapping H. hecale, H. elevatus, and H. pardalinus resequencing data to the H. melpomene reference. Copy number estimates based on number of exon sequence alignments are generally lower than estimates based on read coverage (fig. 3a and supplementary figs. 49–51, Supplementary Material online), possibly due to over-merging of scaffolds that represent true duplicate haplotypes during the haplotype merging step. Nevertheless, for both measures, exonic copy number is much larger in hecale/elevatus/pardalinus trio than in H. melpomene, suggesting duplications of the corresponding genes. It should also be noted that copy number is variable between exons of the same gene and, and whereas it can probably be attributed to different alignment efficiency due to variation in exon sequence length (Li 2018), it might also be due to partial duplications of some of these genes. However, given the fragmented nature of the w2rap genomes, we could not assess whether genes were wholly or partially duplicated, nor whether the duplications were translocated elsewhere in the genome or are located in the same region as in H. melpomene. Long-read sequencing data would be required to resolve this.
Fig. 3.
CNV and increased expression levels of genes in the repeat region on chromosome 9. (a) CNV for each exon of genes in the chromosome 9 repeat region. The number of alignments (left panel) and relative coverage (right panel) were used as proxies of copy number. The number of alignments was obtained by aligning exon sequences, as annotated in the H. melpomene reference genome, to the reference guided assemblies. Relative coverage was calculated by dividing exon coverage by the median genomic coverage, based on mappings to the H. melpomene reference. Dashed horizontal lines on both plots represent a copy number of one. Our new H. melpomene assembly was also included as a control. (b) Change in expression level in H. pardalinus compared with H. melpomene (y axis) as a function of H. pardalinus transcript copy number (x axis). For each transcript, copy number was calculated as the median number of alignments across exons for the H. pardalinus sample. Full blue circles represent transcripts for which the levels of expression in H. pardalinus were significantly higher than in H. melpomene. The best fit linear model regression line and confidence intervals are depicted by the dashed line and gray band, respectively. Species codes are as in figure 1.
These gene duplications could result in pseudogenes, in which case we might expect to find stop codons within exons and a relaxation of selection. In general, we find high exon copy numbers even after excluding exon copies with stop codons (10–14% exon copies have a stop codon; supplementary figs. 52–55, Supplementary Material online). Also, dN/dS estimates are overall close to zero, suggestive of purifying selection (supplementary figs. 56–59, Supplementary Material online). RNA-Seq shows a significant correlation between gene copy number and expression levels and that many of these genes have significantly higher expression in H. pardalinus than in H. melpomene (fig. 3b). Together, these results suggest that many of the gene copies are functional and that CNV at these genes resulted in altered gene dosage.
Inversions Fixed between the Two Heliconius Major Clades
Reference-guided assemblies will inevitably be ineffective at detecting inversions or translocated regions, so it seems important to quantify potential drawbacks of our approach. Here, we make a systematic search for small to medium sized inversion differences among Heliconius species, focusing on those 50 kb–2 Mb long. At the broad scale, the genome structure of the reference-guided assemblies is constrained by the reference genome, so we returned to the w2rap scaffolds (after collapsing alternative haplotypes with HaploMerger2), mapping these to the H. melpomene and the H. erato reference genomes to infer inversion breakpoints. In total, and after filtering, we found 2,560 and 3,829 scaffolds for which one end aligns to the positive strand of the reference genome and the other end maps to the negative strand, using the H. melpomene and H. erato, respectively. Of these, 900 and 1,786 support inversions 50 kb–2 Mb long, yielding 345 and 741 unique candidate inversions across all species (mapping to H. melpomene and H. erato demophoon, respectively), supported by at least one scaffold per species, some of which were shared by multiple species (supplementary table 8, Supplementary Material online).
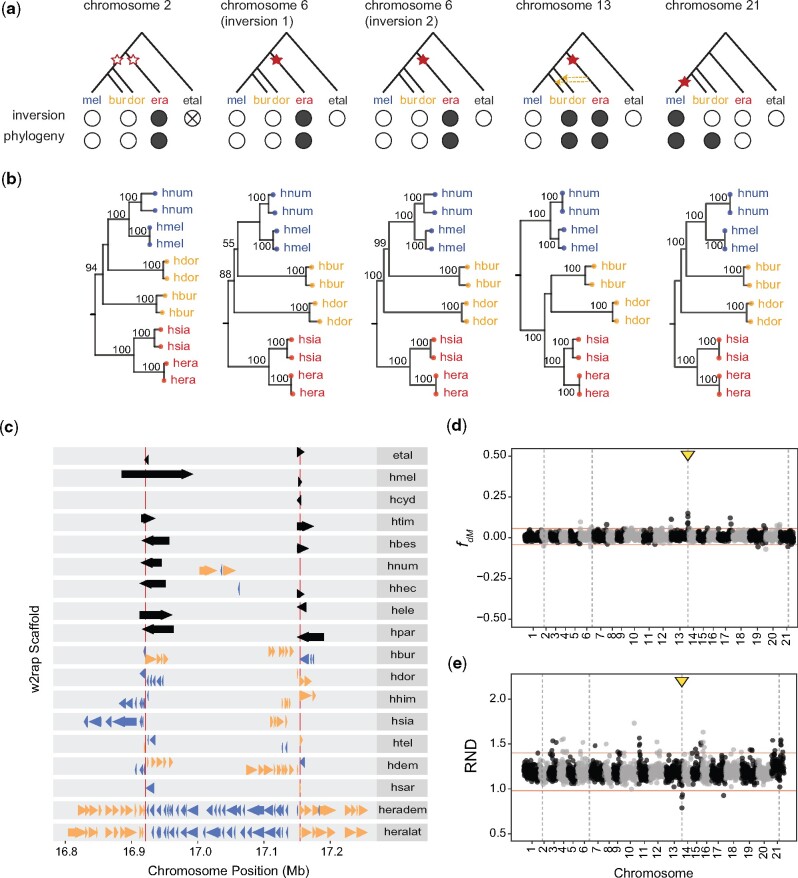
Our systematic search confirmed previous findings of two independent but overlapping introgressed inversions around a color patterning locus on chromosome 15 (one shared by H. sara, H. demeter, H. telesiphe, and H. hecalesia and the other shared by H. pardalinus and H. numata) and another inversion on chromosome 2 (shared by H. erato and H. hecalesia) (Jay et al. 2018; Edelman et al. 2019). In addition, we found five moderately large inversions, previously identified as inversion candidates based on alignments between H. melpomene and H. erato reference genomes (Davey et al. 2017), to be fixed between the two major clades of the Heliconius phylogeny (fig. 4a). Such shared inversions occur on chromosome 2 (supplementary fig. 60, Supplementary Material online), chromosome 6 (supplementary fig. 61, Supplementary Material online), chromosome 13 (fig. 3a and supplementary fig. 62, Supplementary Material online) and the Z chromosome, chromosome 21 (supplementary fig. 63 and supplementary table 9, Supplementary Material online). The two inversions on chromosome 6 occur in tandem and are further supported by linkage maps in H. melpomene and H. erato (Davey et al. 2017).
Fig. 4.
Large inversions fixed between the melpomene/silvaniform and erato/sara clades. (a) Possible scenario for the origin and sharing of the inversion. Stars represent inversions and the branch in which these likely took place. Empty stars are used when the inversion could have occurred in either of two branches. Introgression between branches is represented by arrows, the direction of the arrow indicating directionality. Full and empty circles represent the orientation of the inversion (empty circle with a cross indicates that the orientation could not be inferred) and the groupings based on the phylogeny. (b) ML phylogenies of each major inversion estimated using IQ-TREE, based on mapping of resequence data to the H. melpomene reference. (c) Alignments of w2rap haplotype merged scaffolds to the H. melpomene reference genome supporting the inversion on chromosome 13. Inversion breakpoints are depicted by the vertical red lines. Scaffold alignments are represented by the arrows, the direction and color of the arrows representing whether the alignments are to the forward strand (yellow rightwards arrows) or the reverse strand (blue leftward arrows). Black arrows represent alignments spanning the inversion breakpoints. (d, e) fdM and relative node depth (RND) statistics along the genome. Both statistics were calculated in 25 kb nonoverlapping windows across the genome, based on mapping of resequencing data to the H. melpomene reference. Chromosomes are shown with alternating gray and black colors. The location of inversions is given by the dashed vertical lines whereas horizontal red lines represent ±3 SD from the mean fdM and RND values. Outlier windows overlapping the chromosome 13 inversion are indicated by the yellow arrows. Positive fdM values (and lowered RND) indicate an excess of shared variation between H. burneyi with H. erato and negative values of fdM represent an excess of shared with H. melpomene. In this test, H. melpomene and H. burneyi were considered to be the ingroup species and H. erato the inner outgroup. Derived alleles were determined using E. tales (et al.). Species codes are as in figure 1.
The placement of H. doris and H. burneyi in the Heliconius phylogeny remains contentious. These two species have been inferred to be more closely related to the melpomene/silvaniform clade than to the erato/sara clade (see also the mitochondrial tree of fig. 1a), but node supports are relatively weak and the internal branches leading to H. doris and H. burneyi are short (Kozak et al. 2015). We here test whether homologous inversions can be used as a phylogenetic character to resolve their placement. Both H. burneyi and H. doris group with the melpomene/silvaniform clade based on the orientation of the three inversions on chromosomes 2 and 6, but with the erato/sara clade based on the inversions on chromosomes 13 and Z (fig. 1a). These groupings are further confirmed based on ML phylogenetic analysis of the inversion regions using a subset of species (fig. 4b). The only exception is that H. doris and H. burneyi both group with melpomene/silvaniform species for the inversion on the Z chromosome in the ML phylogeny, rather than with erato/sara (fig. 4b), as might be expected solely based on presence/absence of the inversion (supplementary fig. 63, Supplementary Material online). This apparent contradiction can be reconciled if the melpomene/silvaniform clade is sister both to H. doris and to H. burneyi, but if the Z chromosome inversion occurred in the melpomene/silvaniform ancestor after it split from the burneyi and doris lineages. In this scenario, the sharing of the inversion between the burneyi/doris and erato/sara clades on chromosome 13 must be explained by secondary transfer via introgression, perhaps soon after the initial separation of the two major clades, or through incomplete lineage sorting of an inversion polymorphism at the base of Heliconius. Previously, reticulation involving H. burneyi and H. doris and the erato/sara group had been hypothesized, although different phylogenomic methods gave different results (Kozak et al. 2018). The phylogenetic discordance in precise inversion breakpoints we observe here adds weight to the argument for gene flow at the base of Heliconius.
To test for introgression genome-wide, we used Patterson’s D statistic (Green et al. 2010; Durand et al. 2011). Specifically, we calculated D for all possible topologies of the triplets (H. erato—H. melpomene—H. doris) and (H. erato—H. melpomene—H. burneyi), in each case using Eueides tales as an outgroup. For a given triplet of species, the minimum absolute whole genome Patterson’s D statistic should result for the topology that best describes the relationships between species. We found that this is the case when H. erato is the inner outgroup in both triplets, implying that H. burneyi and H. doris are more closely related to H. melpomene. Yet, Patterson’s D statistics are still significantly different from zero (Patterson’s D = 0.037 and 0.060 for H. doris and H. burneyi, respectively) based on block jackknifing, providing evidence of introgression among lineages leading to H. burneyi, H. doris, and H. erato. We also used an alternative branch length-based approach, QuIBL (quantifying introgression via branch lengths; Edelman et al. 2019), which further corroborated these results (supplementary table 10, Supplementary Material online). To understand which specific genomic regions were shared by introgression between these species, we estimated the excess of shared derived mutations between H. doris and H. burneyi with either H. melpomene or H. erato, using the fdM statistic (Malinsky et al. 2015). The fdM estimates in windows overlapping the chromosome 13 inversion show a significant deviation from the genomic average, with an excess of shared variation between H. erato and both H. doris and H. burneyi (fig. 4d and supplementary fig. 64, Supplementary Material online). Likewise, relative divergence between H. erato to both H. burneyi and H. doris is significantly reduced in this inversion (fig. 4d and supplementary fig. 65, Supplementary Material online). We also used QuIBL with the triplets (H. erato—H. melpomene—H. doris) and (H. erato—H. melpomene—H. burneyi) to calculate the likelihood that the discordant phylogenies at the chromosome 13 inversion were due to introgression. For both triplets, the average internal branch of gene trees within the chromosome 13 inversion is larger than the genome-wide average, corresponding to a 90.1% and 86.7% probability of introgression, respectively (supplementary fig. 66, Supplementary Material online). We found no significant fdM or relative divergence estimates for any of the other four inversions, including the Z chromosome inversion. These results strongly support the argument that the chromosome 13 inversion of H. doris and H. burneyi results from introgression from the common ancestor of the erato/sara clade.
Discussion
Genome Assembly Improvements and Limitations
Here, we implement a purely in silico reference-guided scaffolding approach to improve draft genome assemblies of 16 species from across the genus Heliconius. The contiguity of our new assemblies is similar to that of the reference genomes. For instance, the H. melpomene reference genome assembly has 38 scaffolds anchored to chromosomes (99.1% of the assembly length), and the reference-guided assemblies scaffolded based on this reference have 31–36 scaffolds anchored to chromosomes representing 83.8–99.1% of the total assembly. Similarly, the H. erato reference has 195 scaffolds anchored to chromosomes (100% of the assembly length), and the reference-guided assemblies scaffolded based on this reference have 94–168 scaffolds anchored to chromosomes representing 83.2–99.9% of the total assembly.
Our reference-guided assembly strategy assumes that the orientation and order of the new scaffolds in our genomes is the same as the reference. Clearly, it may not fully represent the structure of these genomes. Although small genomic rearrangements spanned by the original scaffolds (rearrangements in relation to the reference present within w2rap scaffolds) are recovered in our reference-guided assemblies, larger genomic rearrangements relative to the reference not spanned by a single w2rap scaffold can be missed. One such example is the case of the approximately 400 kb inversion around a color pattern locus known from H. numata and H. pardalinus on chromosome 15 (Jay et al. 2018) which is not recovered in our reference-guided assemblies, in either species. This is also the case for the five large inversions we discovered that are fixed between the two Heliconius major clades, depending on the reference genome used to guide scaffolding. For instance, for species in the melpomene/silvaniform group, all reference-guided assemblies mapped to the H. melpomene reference have the correct orientation for all five inversions, but not when mapped to the H. erato reference. The same logic applies for species in the erato/sara group, when mapped to different references. For H. burneyi and H. doris however, neither of the two alternative reference-guided assemblies recovers the correct orientation of all five inversions, since these two species share the same orientation as H. melpomene for the inversions on chromosome 2 and 6, but not for chromosomes 13 and 21 (for which they have the same orientation as H. erato). Long-read sequence data and/or linkage mapping could better resolve the genome structure of species-specific assemblies.
Nevertheless, our reference-guided assemblies represent a major improvement over mapping short-read data directly to existing reference genomes, and researchers that use these and other reference-guided assemblies for this purpose will see marked improvement in their data quality. Mapping the original w2rap Illumina reads back to the reference-guided assembly of their own species resulted in more than doubling of the median genomic coverage in some species and in a more uniform depth of coverage along the genome than when mapping to the closest reference genome. Mapping efficiency improves in all species studied here (supplementary table 6, Supplementary Material online), but we see the greatest benefits in H. burneyi and H. doris, the two Heliconius species studied here that are most divergent from either reference genome assembly. In these two species, the proportion of properly mapped reads increases from 53.6% and 49.9% (for H. burneyi and H. doris, respectively) when mapped to the H. melpomene reference genome, to 90.7% and 90.6% when mapped to their own reference-guided assembly. In another study (Rosser et al., in preparation), a linkage map produced from backcrosses of F1 male hybrids, between H. pardalinus butleri and H. p. sergestus, to the parental H. p. butleri population contained approximately 29% more markers when RADseq data were mapped to the new H. pardalinus reference-guided assembly than to the H. melpomene reference. The use of reference-guided assemblies of the closest species thus greatly improves the efficiency of mapping resequencing data over mapping to the currently available reference genomes.
The more uniform depth of coverage when mapping to reference-guided assemblies also leads to improvements in discovery of species-specific genomic variation and in resolving imperfect repeat regions. Indeed, given variation in genome sizes among Heliconius species (275–418 Mb), the new genomes are helpful in mapping variation that is otherwise lost or mapped to similar but nonorthologous regions of more divergent reference genomes. Variations in depth of coverage along the genome, if not properly filtered, could lead to biased estimates of diversity and divergence. For example, partially divergent repeats mapping to the same region in the reference genome (resulting in unusually high coverage) could inflate local estimates of diversity. This is especially likely in studies focusing on Heliconius species with larger genomes when mapping reads to the H. melpomene reference, the smallest genome assembled here. On the other hand, if regions with abnormal coverage are filtered out, information could be lost by discarding genomic regions with potentially relevant biological signals. For example, highly divergent regions may result in abnormally low coverage, even though such regions could be important for diversification of the group.
Overall, our reference-guided assemblies extend the number of applications for which these genomes can be used. By ordering, orienting, and anchoring scaffolds onto chromosomes, the new reference-guided assemblies enable improved chromosome-scale analyses and genome scans.
Prevalence of Structural Variants in Heliconius Butterflies
Chromosomal rearrangements can play a major role in adaptation and speciation (Feulner and De-Kayne 2017; Wellenreuther and Bernatchez 2018). By reducing recombination, inversions can facilitate the build-up of associations between loci involved in traits responsible for reproductive isolation, and thus could play a role in establishing or reinforcing species barriers (Noor et al. 2001). Inversions can also be favored by selection by maintaining adaptive combinations of locally adapted alleles (Christmas et al. 2019; Faria et al. 2019; Todesco et al. 2020).
In Heliconius, a previous study focusing on two closely related species (H. melpomene and H. cydno) found no evidence for major inversions that might have aided speciation (Davey et al. 2017). Thus, Heliconius appeared to have low rates of chromosomal rearrangement, and selection without the help of chromosomal rearrangements was believed to maintain the differences between these two species. In another species, H. numata, the tandem inversion complex that forms the supergene locus P allows the maintenance of a multiallele color pattern polymorphism of mimicry morphs (Joron et al. 2011). The first inversion in the tandem supergene was most likely transferred to H. numata via introgression from H. pardalinus (Jay et al. 2018). An independently derived inversion has since been found for the same color pattern determination region in four species in the erato/sara clade (H. telesiphe, H. hecalesia, H. demeter, and H. sara). This inversion was also inferred to have been shared via introgression, this time between H. telesiphe and H. sara subclades (Edelman et al. 2019). In parallel hybrid zones of H. erato and H. melpomene, 14 and 19 polymorphic inversions were detected within each species, respectively (Meier et al. 2020). Most of these inversion polymorphisms did not differ across the hybrid zones of either species. The frequency of only one inversion on chromosome 2 (different to the inversion on chromosome 2 reported here) differed strongly across the hybrid zone between highland H. e. notabilis and lowland H. e. lativitta races, and may be associated with ecological adaptation to altitude (Meier et al. 2020).
In the 16 species studied here, we systematically searched for inversions. We found several candidates in all 16 species (17–61 and 40–126 inversions per species, compared with H. melpomene and H. erato, respectively), including some described previously (Davey et al. 2017; Jay et al. 2018; Edelman et al. 2019). However, the strategy we implemented to search for inversions, that is, split alignment of w2rap scaffolds to forward and reverse strands of the reference genomes, is liable to false positives because small interspersed duplications and translocations (e.g., due to transposable element activity) might generate a similar signal. This is particularly likely in highly repetitive regions where we find many different, partially overlapping candidate inversions in many or all species (supplementary fig. 67, Supplementary Material online). It is thus difficult to assess, solely based on these results, how pervasive inversions are among Heliconius species. Although it is possible that inversions in this group occur more frequently than earlier studies indicated (The Heliconius Genome Consortium 2012; Davey et al. 2017), long-read or linked-read sequencing, preferably with a larger set of individuals per species, will ultimately be needed to answer this question.
However, by focusing on phylogenetically informative inversions, we were able to verify five candidate inversions that occurred deep in the Heliconius phylogeny. We searched for inversions fixed between the melpomene/silvaniform and erato/sara clades. We are confident that these were correctly identified for two reasons. First, the inversions are supported in multiple species, with breakpoint coordinates consistent among species. Second, although a misassembly in the reference genome could generate a misleading signal of inversion, this is unlikely to happen for the same candidate inversion when mapping to two or more different genomes. All five of these inversions were supported in multiple species when mapping scaffolds to either reference genome, the orientation of the inversion being mirrored depending on the reference used. Furthermore, the inversion orientation shows a phylogenetic signal (fixed between clades) that is unexpected if due to misassembly in one of the reference genomes.
The most parsimonious scenario that explains both the orientation and the phylogenetic pattern, taking all five inversions into account, supports the hypothesis that H. burneyi and H. doris are more closely related to the melpomene/silvaniform group than to the erato/sara group (fig. 4), in line with previous studies (Kozak et al. 2018; Edelman et al. 2019). The relationships of the inversion on chromosome 13, which groups H. burneyi, H. doris, and the erato/sara clade is then explained by introgression between the ancestor of the latter group and both H. burneyi and H. doris (supplementary figs. 64–66, Supplementary Material online). Introgression almost certainly occurred from the erato/sara clade into H. burneyi and H. doris, since the relative divergence between H. erato and both H. burneyi and H. doris is reduced at the chromosome 13 inversion when compared with the rest of the genome (fig. 4e), but not between H. erato and H. melpomene as expected if introgression took place in the other direction (supplementary fig. 68, Supplementary Material online). Interestingly, H. burneyi has been inferred to be on a separate branch from H. doris, although the two branches were connected by introgression (Kozak et al. 2018, 2015). This suggests that introgression of the chromosome 13 inversion occurred twice. Either there were two separate introgression events from the erato/sara ancestor to H. burneyi and to H. doris, or the inversion first passed from the erato/sara ancestor to one of these two species which then passed it to the other. Altogether, and in line with previous studies (Kozak et al. 2018; Edelman et al. 2019), this inversion supports a hypothesis that hybridization and introgression among species occurred early in the radiation of Heliconius, as well as later, between more closely related species extant within each major subgroup. Alignment issues have previously made it hard to interpret evidence for introgression so deep in the phylogeny. Although we still do not know whether it has functional implications, our finding of transfer of this chromosome 13 inversion provides stronger support for introgression deeper in the Heliconius phylogeny than was available earlier.
Species may also differ in gene copy number. Copy number can affect the phenotype by altering gene dosage, altering protein sequence, or by creating paralogs that can diverge and gain new functions (Iskow et al. 2012). CNV has been implicated in ecological adaptation—for example, insecticide resistance in Anopheles mosquitoes (Lucas et al. 2019), climate adaptation in white spruce (Prunier et al. 2017) and polar bears (Rinker et al. 2019), and resistance to malaria in humans (Leffler et al. 2017). Gene copy number may also be involved in reproductive barriers among species—for example, hybrid lethality in Mimulus sympatric species (Zuellig and Sweigart 2018). Gene duplications within specific gene families in the branch leading to Heliconius have been linked to evolution of visual complexity, development, immunity (The Heliconius Genome Consortium 2012), and female oviposition behavior (Briscoe et al. 2013). Within the genus, gene CNV is plausibly associated with species divergence between H. melpomene and H. cydno (Pinharanda et al. 2017).
Here we show that the genomes of different Heliconius species vary in size, with each chromosome typically showing similar directional changes in size between species. Thus, genome expansions and reductions in size seem typically to involve all chromosomes, so that the relative sizes of chromosomes are conserved. Our study of the Heliconius butterfly radiation conforms, on a much more restricted phylogenetic scale, to the pattern of relative chromosome size across eukaryotes: across many orders of magnitude of genome size, relative chromosome sizes can be predicted based on chromosome number and are almost always between approximately 0.4× and approximately 1.9× the mean (Li et al. 2011).
We find that, in Heliconius, genomic expansion is at least partially driven by small genomic regions that became hotspots of repeat accumulation. Amplified regions tend to be conserved among closely related species and are more frequent toward chromosome ends (supplementary fig. 69, Supplementary Material online). However, in a subclade of three closely related species (H. hecale, H. elevatus, and H. pardalinus), we found four small genomic regions with highly aberrant increases in size and exon copy number compared with related species. These three species therefore provide an exception to the more or less orderly pattern across chromosomes in the rest of the genus. Our approach for detecting exceptional repeat regions relies on the H. melpomene genomic arrangement as a backbone. Hence, we do not know whether the additional copies we found were translocated to other regions of the genomes of these three species, or whether they remained clustered as tandem copies at a single genomic location. Transposable element activity is one possible mechanism responsible for these repeats (Bourque et al. 2018), and rapid divergent transposable element evolution has already been found among Heliconius species (Ray et al. 2019). In fact, we found a significant increase in transposable elements content in the repeat regions on chromosomes 8 and 9 in all three species of the trio (H. hecale, H. elevatus, and H. pardalinus) when compared with the H. melpomene reference genome (supplementary table 11, Supplementary Material online). We also found an increase (≥5%) of particular transposable element families in these regions, although none was consistently significant in all three species when compared with the homologous regions in H. melpomene and to the genomic background (supplementary table 11, Supplementary Material online). Hybridization could also spread variation in copy number among the species. Heliconius hecale, H. elevatus, and H. pardalinus are sympatric in the Amazon where they are known to hybridize occasionally (Mallet et al. 2007; Rosser et al. 2019). We found significantly higher copy numbers in the Amazon than in extra-Amazonian populations of these species (supplementary fig. 70, Supplementary Material online). The correlations of copy number among species in an area suggests that hybridization and introgression among these closely related species might indeed have been involved.
Genes within highly amplified regions had significantly higher expression levels in H. pardalinus than in H. melpomene (fig. 3b), which suggests that this gene copy variation could have functional significance. The orthologs of genes within these regions in Drosophila are involved in important functions such as cytoskeletal processes and oogenesis (i.e., Dhc64C, sima, shotgun, and capicua; supplementary table 7, Supplementary Material online). Evaluating how variation in these critical genes impacts phenotypes in H. pardalinus, H. elevatus, and H. hecale will advance our understanding of the role of CNV in evolution.
The full extent to which inversions and CNV play a role in the evolution of Heliconius butterflies remains to be examined. However, the current work suggests that the types of structural variation examined here could be relevant to diversification. The characterization of intra- and interspecific structural variation in this group could thus be an especially promising avenue for future studies particularly now that improvements in sequencing technology allow for more detailed, rigorous and cost-effective detection of structural variants (Wellenreuther et al. 2019; Logsdon et al. 2020).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Harvard FAS Research Computing team for their support, A. Shumate for her guidance using the Liftoff software, J. Davey and D. Ray for their valuable inputs to our thinking around genome structural variation, and N. Rosser for the helpful discussions on Heliconius. This project was funded by a SPARC Grant from the Broad Institute of Harvard and MIT and funds from Harvard University.
Data Availability
The reference-guided genome assemblies, gene annotations and mitochondrial genomes assemblies generated in this study are available in Zenodo (https://doi.org/10.5281/zenodo.4708442). All custom scripts used in this study are available on the GitHub repository: https://github.com/FernandoSeixas/HeliconiusReferenceGuidedAssemblies.
Literature Cited
- Alonge M, et al. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20(1):224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevenuto J, Ferrão LV, Amadeu RR, Munoz P.. 2019. How can a high-quality genome assembly help plant breeders? Gigascience 8(6):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi E, et al. 2015. MeDuSa: a multi-draft based scaffolder. Bioinformatics 31(15):2443–2451. [DOI] [PubMed] [Google Scholar]
- Bourque G, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34(5):525–527. [DOI] [PubMed] [Google Scholar]
- Briscoe AD, et al. 2013. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9(7):e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanettes F, Klopp C.. 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6:e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas MJ, et al. 2019. Chromosomal inversions associated with environmental adaptation in honeybees. Mol Ecol. 28(6):1358–1374. [DOI] [PubMed] [Google Scholar]
- Clavijo B, et al. 2017. W2RAP: a pipeline for high quality, robust assemblies of large complex genomes from short read data. bioRxiv. doi: 10.1101/110999. [DOI]
- Coyne JA. 2018. “Two rules of speciation” revisited. Mol Ecol. 27(19):3749–3752. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr AH.. 1989. Two rules of speciation. In: Otte D, Endler JA, editors. Speciation and its consequences. Sunderland (MA): Sinauer Associates. p. 180–207. [Google Scholar]
- Davey JW, et al. 2017. No evidence for maintenance of a sympatric Heliconius species barrier by chromosomal inversions. Evol Lett. 1(3):138–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S, et al. 2018. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat Commun. 9(1):4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28(8):2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman NB, et al. 2019. Genomic architecture and introgression shape a butterfly radiation. Science 366(6465):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491(7426):756–760. [DOI] [PubMed] [Google Scholar]
- Faria R, et al. 2019. Multiple chromosomal rearrangements in a hybrid zone between Littorina saxatilis ecotypes. Mol Ecol. 28(6):1375–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feulner PGD, De-Kayne R.. 2017. Genome evolution, structural rearrangements and speciation. J Evol Biol. 30(8):1488–1490. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217):1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Pop M.. 2019. Modern technologies and algorithms for scaffolding assembled genomes. PLoS Comput Biol. 15(6):e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, et al. 2017. The wolf reference genome sequence (Canis lupus lupus) and its implications for Canis spp. population genomics. BMC Genomics. 18(1):495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Kang M, Xu A.. 2017. HaploMerger2: rebuilding both haploid sub-assemblies from high-heterozygosity diploid genome assembly. Bioinformatics 33(16):2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, Gokcumen O, Lee C.. 2012. Exploring the role of copy number variants in human adaptation. Trends Genet. 28(6):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman SD, et al. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27(5):768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P, et al. 2018. Supergene evolution triggered by the introgression of a chromosomal inversion. Curr Biol. 28(11):1839–1845. [DOI] [PubMed] [Google Scholar]
- Jiggins CD, et al. 2005. A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics. 171(2):557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477(7363):203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak KM, et al. 2015. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst Biol. 64(3):505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak KM, McMillan O, Joron M, Jiggins CD.. 2018. Genome-wide admixture is common across the Heliconius radiation. bioRxiv. 414201. doi: 10.1101/414201. [DOI] [PMC free article] [PubMed]
- Leffler EM, et al. 2017. Resistance to malaria through structural variation of red blood cell invasion receptors. Science 356(6343):eaam6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JJ, van der Burg KRL, Mazo-Vargas A, Reed RD.. 2016. ChIP-Seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Rep. 16(11):2855–2863. [DOI] [PubMed] [Google Scholar]
- Lewis JJ, Reed RD.. 2019. Genome-wide regulatory adaptation shapes population-level genomic landscapes in Heliconius. Mol Biol Evol. 36(1):159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 00:3. Available from: http://arxiv.org/abs/1303.3997 (accessed July 10, 2014).
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 36(1):96–99. [DOI] [PubMed] [Google Scholar]
- Li X, et al. 2011. Chromosome size in diploid eukaryotic species centers on the average length with a conserved boundary. Mol Biol Evol. 28(6):1901–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Eichler EE.. 2020. Long-read human genome sequencing and its applications. Nat Rev Genet. 21(10):597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love RR, Weisenfeld NI, Jaffe DB, Besansky NJ, Neafsey DE.. 2016. Evaluation of DISCOVAR de novo using a mosquito sample for cost-effective short-read genome assembly. BMC Genomics. 17:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas ER, et al. 2019. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 29(8):1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, et al. 2015. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science 350(6267):1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Beltrán M, Neukirchen W, Linares M.. 2007. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol Biol. 7(1):28.7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markelz RJC, et al. 2017. Using RNA-Seq for genomic scaffold placement, correcting assemblies, and genetic map creation in a common Brassica rapa mapping population. G3 Genes Genomes Genet. 7:2259–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17(1):10. [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the Use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Salazar C, Jiggins CD.. 2019. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17(2):e2006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC.. 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5(9):e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massardo D, et al. 2020. The roles of hybridization and habitat fragmentation in the evolution of Brazil’s enigmatic longwing butterflies, Heliconius nattereri and H. hermathena. BMC Biol. 18(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JI, et al. 2020. Haplotype tagging reveals parallel formation of hybrid races in two butterfly species. bioRxiv. doi: 10.1101/2020.05.25.113688. [DOI] [PMC free article] [PubMed]
- Morgulis A, Michael Gertz E, Schä AA, Agarwala R.. 2006. WindowMasker: window-based masker for sequenced genomes. Bioinformatics 22(2):134–141. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J.. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 98(21):12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L.. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 14(7):687–690. [DOI] [PubMed] [Google Scholar]
- Pinharanda A, et al. 2019. Sexually dimorphic gene expression and transcriptome evolution provide mixed evidence for a fast-Z effect in Heliconius. J Evol Biol. 32(3):194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinharanda A, Martin SH, Barker SL, Davey JW, Jiggins CD.. 2017. The comparative landscape of duplications in Heliconius melpomene and Heliconius cydno. Heredity 118(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowell PD. 1998. Sex linkage and speciation in Lepidoptera. In: Howard DJ, Berlocher SH, editors. Endless forms. Species and speciation. New York: Oxford University Press. p. 309–319. [Google Scholar]
- Prüfer K, et al. 2010. Computational challenges in the analysis of ancient DNA. Genome Biol. 11(5):R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier J, et al. 2017. Gene copy number variations in adaptive evolution: the genomic distribution of gene copy number variations revealed by genetic mapping and their adaptive role in an undomesticated species, white spruce (Picea glauca). Mol Ecol. 26(21):5989–6001. [DOI] [PubMed] [Google Scholar]
- Ray DA, et al. 2019. Simultaneous TE Analysis of 19 heliconiine butterflies yields novel insights into rapid TE-based genome diversification and multiple SINE births and deaths. Genome Biol Evol. 11(8):2162–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice ES, Green RE.. 2019. New approaches for genome assembly and scaffolding. Annu Rev Anim Biosci. 7:17–40. [DOI] [PubMed] [Google Scholar]
- Rinker DC, Specian NK, Zhao S, Gibbons JG.. 2019. Polar bear evolution is marked by rapid changes in gene copy number in response to dietary shift. Proc Natl Acad Sci USA. 116(27):13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser N, et al. 2019. Geographic contrasts between pre‐ and postzygotic barriers are consistent with reinforcement in Heliconius butterflies. Evolution 73(9):1821–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 3684:eaar3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas FA, Boursot P, Melo-Ferreira J.. 2018. The genomic impact of historical hybridization with massive mitochondrial DNA introgression. Genome Biol. 19(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumate A, Salzberg SL.. 2020. Liftoff: accurate mapping of gene annotations. Bioinformatics. doi: 10.1093/bioinformatics/btaa1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P.. 2013. RepeatMasker Open-4.0. Available from: www.repeatmasker.org.
- Simão FA, Waterhouse R, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P.. 2015. Genome analysis Sambamba: fast processing of NGS alignment formats. Bioinformatics 31(12):2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler A, et al. 2005. First-generation linkage map of the warningly colored butterfly Heliconius erato. Heredity 94(4):408–417. [DOI] [PubMed] [Google Scholar]
- Todesco M, et al. 2020. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584(7822):602–607. [DOI] [PubMed] [Google Scholar]
- Van Belleghem SM, et al. 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 1:0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Yang Y, Yin T.. 2020. The chromosome-scale assembly of the willow genome provides insight into Salicaceae genome evolution. Hortic Res. 7(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, et al. 2014. Comprehensive variation discovery in single human genomes. Nat Genet. 46(12):1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L.. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 33(6):427–440. [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Mérot C, Berdan E, Bernatchez L.. 2019. Going beyond SNPs: the role of structural genomic variants in adaptive evolution and species diversification. Mol Ecol. 28(6):1203–1209. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2020. Chromosome-level reference genome assembly and gene editing of the dead-leaf butterfly Kallima inachus. Mol Ecol Resour. 20(4):1080–1092. [DOI] [PubMed] [Google Scholar]
- Yu A, et al. 2019. Application of a high-resolution genetic map for chromosome-scale genome assembly and fine QTLs mapping of seed size and weight traits in castor bean. Sci Rep. 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuellig MP, Sweigart AL.. 2018. Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS Genet. 14(4):e1007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The reference-guided genome assemblies, gene annotations and mitochondrial genomes assemblies generated in this study are available in Zenodo (https://doi.org/10.5281/zenodo.4708442). All custom scripts used in this study are available on the GitHub repository: https://github.com/FernandoSeixas/HeliconiusReferenceGuidedAssemblies.