Abstract
Signaling events triggered by hydrogen peroxide (H2O2) regulate plant growth and defense by orchestrating a genome-wide transcriptional reprogramming. However, the specific mechanisms that govern H2O2-dependent gene expression are still poorly understood. Here, we identify the Arabidopsis Mediator complex subunit MED8 as a regulator of H2O2 responses. The introduction of the med8 mutation in a constitutive oxidative stress genetic background (catalase-deficient, cat2) was associated with enhanced activation of the salicylic acid pathway and accelerated cell death. Interestingly, med8 seedlings were more tolerant to oxidative stress generated by the herbicide methyl viologen (MV) and exhibited transcriptional hyperactivation of defense signaling, in particular salicylic acid- and jasmonic acid-related pathways. The med8-triggered tolerance to MV was manipulated by the introduction of secondary mutations in salicylic acid and jasmonic acid pathways. In addition, analysis of the Mediator interactome revealed interactions with components involved in mRNA processing and microRNA biogenesis, hence expanding the role of Mediator beyond transcription. Notably, MED8 interacted with the transcriptional regulator NEGATIVE ON TATA-LESS, NOT2, to control the expression of H2O2-inducible genes and stress responses. Our work establishes MED8 as a component regulating oxidative stress responses and demonstrates that it acts as a negative regulator of H2O2-driven activation of defense gene expression.
Arabidopsis MED8 suppresses oxidative stress-induced gene expression and phytohormone signaling pathways.
Introduction
Reactive oxygen species (ROS) are key regulators of plant growth, development, stress responses, and cell death, and their role in modulating gene expression is well documented (Mhamdi and Van Breusegem, 2018; Waszczak et al., 2018). Different types of ROS, such as hydrogen peroxide (H2O2), superoxide, and singlet oxygen, can trigger specific changes in the transcriptome (Gadjev et al., 2006; Willems et al., 2016). Genome-wide transcriptome analyses revealed that H2O2 metabolism and photorespiration have a prominent influence on gene expression (Vandenabeele et al., 2003, 2004; Vanderauwera et al., 2005, 2011; Mhamdi et al., 2010a; Queval et al., 2012; Kerchev et al., 2016). H2O2 and H2O2-triggered signaling events interact with various other signaling pathways, such as phytohormone signaling. In particular, perturbations in H2O2 homeostasis affect pathways involving hormones, such as salicylic acid (SA), jasmonic acid (JA), auxin, ethylene (ET), and abscisic acid (ABA; Tognetti et al., 2010; Chaouch et al., 2010; Noctor et al., 2015, 2018; Kerchev et al., 2015).
Changes in gene expression triggered by ROS are mediated by stress-responsive cis-regulatory promoter elements, redox regulation of transcription regulators, and upstream signaling cascades, such as mitogen-activated kinase modules (Dietz, 2014; He et al., 2018). Posttranslational oxidative modifications of transcription factors (TFs) and regulators either govern conformational switches, nuclear-cytosolic partitioning, and assembly with coregulators or affect DNA-binding capacities (He et al., 2018). In contrast, ROS- and redox-dependent regulation of the core transcriptional machinery (such as the RNA polymerase II [RNAP II]) is less studied and data from plants are scarce (Shaikhali et al., 2015).
Interactions between RNAP II, general and specific TFs, coregulators, and chromatin modifiers determine gene expression activities in a given developmental and environmental context. The multisubunit Mediator complex is an evolutionarily conserved transcriptional coregulator in eukaryotes (Bourbon, 2008) and is involved in various transcription steps, including initiation, pausing, elongation, and reinitiation (reviewed in Allen and Taatjes, 2015). The best-characterized mediator function is to facilitate the preinitiation complex assembly. It bridges specific TFs with the RNAP II machinery and, thus, converges different signaling pathways before channeling the transcriptional instructions to the RNAP II machinery. In this manner, the Mediator complex enables TF-dependent regulation of transcription and helps to decode biological cues (e.g. external and internal stimuli) into physiological responses (Fondell et al., 1996; Holstege et al., 1998). The structure of the Mediator complex was first determined by electron microscopy in yeast, revealing that the complex consists of three distinct domains, annotated as Head, Middle, and Tail (Asturias et al., 1999; Dotson et al., 2000). This “modular” organization was further confirmed by a biochemical approach, assigning specific functions to each module (Kang et al., 2001). The Head and Middle modules interact with RNAP II and general TFs and the Tail module with sequence-specific TFs (Myers et al., 1999; Kang et al., 2001). In addition to the three core modules, a separate regulatory module, designated the kinase module, has been isolated (Borggrefe et al., 2002) and was shown to act mainly as a negative transcriptional regulator by associating with the core Mediator, thereby preventing the interaction with RNAP II and transcription initiation (Elmlund et al., 2006; Tsai et al., 2013). Biochemical purification, tandem mass spectrometry, and subsequent bioinformatics analyses revealed that the Arabidopsis (Arabidopsis thaliana) Mediator complex comprises at least 34 subunits of which six are specific to plant lineage (Bäckström et al., 2007; Mathur et al., 2011). To date, homologs of the Arabidopsis Mediator complex in rice (Oryza sativa), tobacco (Nicotiana sp.), wheat (Triticum aestivum), and tomato (Solanum lycopersicum) have been identified and conserved functions for some subunits have been discovered (Mathur et al., 2011; Wang et al., 2011; Li et al., 2015; Samanta and Thakur, 2015, 2017; Dolan and Chapple, 2017; Pérez-Martin et al., 2018; Dwivedi et al., 2019; Liu et al., 2019; Hiebert et al., 2020).
Analyses of loss-of-function mutants in Mediator subunits revealed that the plant Mediator complex regulates transcription in a specific manner. Whereas the Mediator subunits might not be all required for general gene transcription, they are implicated in the control of various pathways (Kidd et al., 2009; Elfving et al., 2011; Hemsley et al., 2014; Dolan et al., 2017; Ruiz-Aguilar et al., 2020; Liu et al., 2020). So far, Mediator subunits have been reported to play roles in organ and tissue development, hormone signaling, flowering transition, immune response, and abiotic stresses (Kidd et al., 2009; Wang et al., 2011; Buendía-Monreal and Gillmor, 2016; Crawford et al., 2020; Zhu et al., 2020). Several mutants of the Mediator subunits, including med8, med12, med13, med14, med15, med17, med18, and med20a have developmental and growth defects; yet, the underlying mechanisms are still only partly established (reviewed in Samanta and Thakur, 2015; Buendía-Monreal and Gillmor, 2016). Development phenotypes reported for med8 such as reduced growth (Kidd et al., 2009; Xu and Li, 2012) are probably driven by differential expression of genes involved in the regulation of organ size and cell division and/or expansion rates (Li et al., 2008). MED16 and MED25 have been reported to act as negative regulators of organ growth (Xu and Li, 2011; Liu et al., 2019) and both med16 and med25 mutants exhibited enlarged organs, which is the opposite of med8 mutants. One of the best-characterized subunits is MED25 and, in particular, its role in plant immunity through interaction with JA signaling regulators and receptors, such as the basic helix–loop–helix (bHLH) MYC2 TF, jasmonate–ZIM (JAZ) domain protein transcriptional repressors, and the F-box protein CORONATINE INSENSITIVE 1 (COI1) receptor (reviewed in Zhai and Li, 2019). Like MED25, MED8 is also involved in plant immunity and is required for the expression of the JA-inducible PLANT DEFENSIN (PDF) genes (Kidd et al., 2009; Zhang et al., 2012; Wang et al., 2015, 2016; Li et al., 2018); however, insights into the mechanisms that could be in play are lacking.
To identify negative regulators of H2O2-dependent gene expression, we performed a forward genetic screen for mutations that induce a luciferase (LUC) reporter gene under the control of an H2O2-inducible promoter. We identified MED8 as a suppressor of H2O2-triggered gene expression and characterized its functions in stress responses and signaling. We show that MED8 modulates oxidative stress responses by negatively regulating the expression of genes associated with ROS, phytohormones, and defense. Analysis of the Mediator interactome revealed that the MED8 function might be achieved by more than one mechanism involving interactions with other Mediator subunits, transcription repressors, and components implicated in miRNA biogenesis.
Results
A forward genetic screen identifies MED8 as a negative regulator of an early H2O2-responsive gene
To identify negative regulators of H2O2-responsive genes, we mutagenized transgenic reporter line expressing the firefly LUC reporter gene under the control of the promoter of photorespiratory H2O2-responsive transcripts. Transgenic Arabidopsis plants containing the promoter of the EMBRYONIC ABUNDANT PROTEIN-LIKE-RELATED 4 (EAL4; AT4G22530) gene were used, hereafter referred to as ProEAL4:LUC plants (Supplemental Figure 1A), because EAL4 transcripts accumulate rapidly and highly after the onset of a photorespiratory H2O2 stress triggered by catalase deficiency (Inzé et al., 2012; Supplemental Figure 1B). To assess the reporter specificity, we monitored the LUC activity under different conditions known to increase H2O2 levels. The LUC activity in ProEAL4:LUC seedlings was strongly induced by methyl viologen (MV) and H2O2 treatments, but to a minor extent by ABA, salt, and mannitol treatments, indicating that the EAL4 promoter is indeed a sensitive marker of increased levels of ROS and, in particular, of H2O2 (Supplemental Figure 1C). Seeds of the homozygous ProEAL4:LUC line were EMS mutagenized and M2 seedlings were screened for mutants with a constitutive enhanced LUC activity under control conditions, without compromised photosystem II (PSII) efficiency (Fv′/Fm′). This workflow aimed to avoid the isolation of mutants with induced LUC activity due to mutations causing oxidative stress and subsequently an impaired photosynthetic efficiency (Supplemental Figure 1D). The retained mutants were designated continuously induced ProEAL4:LUC (ceal).
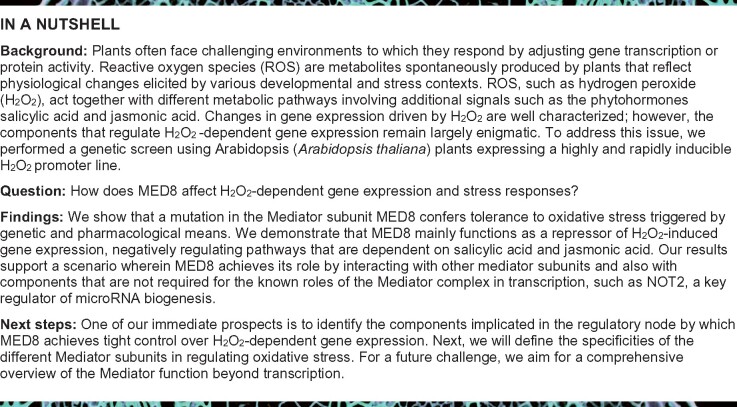
One of the validated ceal mutants, ceal5, was subjected to a detailed analysis. Quantitative analysis of luminescence intensities demonstrated that the LUC activity in ceal5 seedlings was significantly higher than that of ProEAL4:LUC under both control and H2O2 treatment (Figure 1, A and B). In addition, the LUC transcript levels in the ceal5 mutant were approximately four-fold higher than those in ProEAL4:LUC, pointing to enhanced EAL4 promoter activity (Figure 1C). In the absence of stress, the ceal5 mutant exhibited slightly lower Fv‘/Fm’ values compared to ProEAL4:LUC, yet retained significantly higher Fv‘/Fm’ values following treatment with 10 mM H2O2 for 6 h (Figure 1D).
Figure 1.
Identification of MED8 as a negative regulator of EAL4 expression. A, Photographs of 10-day-old seedlings grown in multiwell plates (left), PSII efficiency Fv′/Fm′ (middle), and luminescence images (right) of ProEAL4:LUC and ceal5 seedlings before and after treatment with 10 mM H2O2 for 6 h. The Fv′/Fm′ and luminescence were visualized with color scales from low to high. B, Relative LUC activity of 10-day-old ProEAL4:LUC and ceal5 seedlings with and without H2O2 treatment for 6 h. C, RT-qPCR analysis of the relative expression of LUC gene in ProEAL4:LUC and ceal5 seedlings under control conditions (relative to ARP7 and UBIQUITIN). D, Quantified Fv‘/Fm’ of 10-day-old ProEAL4:LUC and ceal5 seedlings with and without H2O2 treatment for 6 h. Data are means ± SE of four biological replicates. E, Schematic gene and protein model of MED8. The ceal5 mutation and two T-DNA insertion sites are shown. Numbers indicate the nucleotide/amino acid (aa) position of the mutation and/or the insertion. Gray and black blocks denote untranslated region and exons, respectively, with the introns indicated by lines. MED8 protein contains a glutamine (Q)-rich domain at the C-terminus. F, Phenotypes of 4-week-old plants grown under long day and moderate light. G, Genetic complementation tests between the ceal5 and med8 mutations. The LUC activation of F1 plants from crosses between Col-0×ceal5 or med8×ceal5 was measured. Data are means ± SE of eight biological replicates. H, Relative expression of EAL4 in Col-0 and med8 plants. I, Seeds development in immature siliques (top) and mature siliques (bottom) of wild-type and heterozygous med8-2 (med8-2+/−) mutant. Arrows indicate aborted embryos. Bar = 0.5 mm. Unless stated otherwise, data are means ± se of three biological replicates. FC, fold change. Significant differences **P≤ 0.01, *P ≤ 0.05 (Student’s t test). Mutants or transgenic lines were compared to wild-type in the same conditions.
In the progeny of backcrossed plants from the M3 generation (with the nonmutagenized reporter line), constitutively induced LUC activity segregated in a 1:3 ratio, indicating a single and recessive causative mutation. Whole-genome sequencing of F2-segregating population and subsequent SHOREmap analysis revealed that a C-to-T transition at nucleotide 2102 of MEDIATOR SUBUNIT 8 (MED8; AT2G03070) was the causative mutation, resulting in a nonsense mutation (premature stop codon at glutamine [Q] position 377) in the C-terminal Q-rich domain (76 Q residues between amino acids 291 and 524). Hence, ceal5 plants most probably express a truncated (377 amino acid long) MED8 protein lacking most of its Q-rich domain (Figure 1E).
To validate the outcome of the SHOREmap analysis, we characterized two med8 T-DNA insertion lines, SALK_092406 (med8; Kidd et al., 2009) and GABI_270C11 (med8-2). The med8 T-DNA insertion resides in the ninth exon of MED8, in close proximity to the EMS-induced mutation in ceal5. Ceal5 plants exhibited a growth phenotype similar to that of med8, with a more compact and flat rosette, an increased number of leaves, and shorter petioles than Col-0 (Figure 1F). F1 seedlings from a med8×ceal5 cross failed to rescue the ceal5 molecular phenotype, with seedlings still showing a high LUC activity, unlike F1 seedlings of a Col-0×ceal5 cross (Figure 1G). Moreover, EAL4 transcript levels in med8 were approximately three-fold higher than those of Col-0 (Figure 1H), demonstrating that ceal5 is a mutant allele of med8. Genotyping the progeny of a heterozygous med8-2 mutant, with a T-DNA insertion in the first exon of MED8, revealed a 1:2 (WT:heterozygotes) segregation ratio, indicating that med8-2 homozygous mutants are embryo lethal. Consistent with that conclusion, approximately 25% of the seeds were aborted in the siliques of heterozygous med8-2 plants (Figure 1I). Hence, these data demonstrate that med8-2 contains a recessive and embryo-lethal mutation in MED8, unlike med8 and ceal5 that represent viable mutant alleles.
Truncation of MED8 activates SA biosynthesis and modulates photorespiratory H2O2-triggered phenotypes
To explore the role of MED8 in H2O2 signaling, we used a genetic approach in which oxidative stress intensity is easily manipulated in a noninvasive manner. The catalase-deficient mutant (cat2) is an experimental model where intracellular H2O2 is produced in a physiologically relevant manner, making it a very useful genetic background to mimic stress responses (Vanderauwera et al., 2005; Queval et al., 2007; Mhamdi et al., 2010a; Chaouch et al., 2012; Noctor et al., 2015). When grown under ambient air and moderate light intensities, cat2 mutants display a conditional stress phenotype that depends on the day length. Increased H2O2 availability triggers upregulation of defense pathways and activates pathogenesis responses, and development of hypersensitive response-like lesions in long days (cell death-permissive conditions), but not in short days (nonpermissive conditions; Chaouch et al., 2010; Mhamdi et al, 2010a). We crossed the med8 mutation into the cat2 background and analyzed the morphological, molecular, and biochemical phenotypes in the double cat2 med8 mutants.
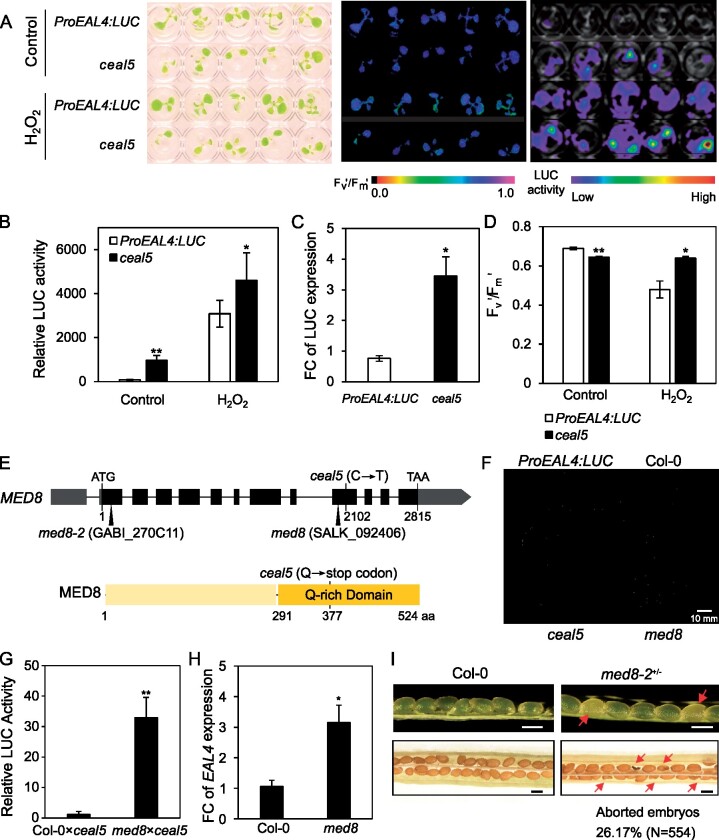
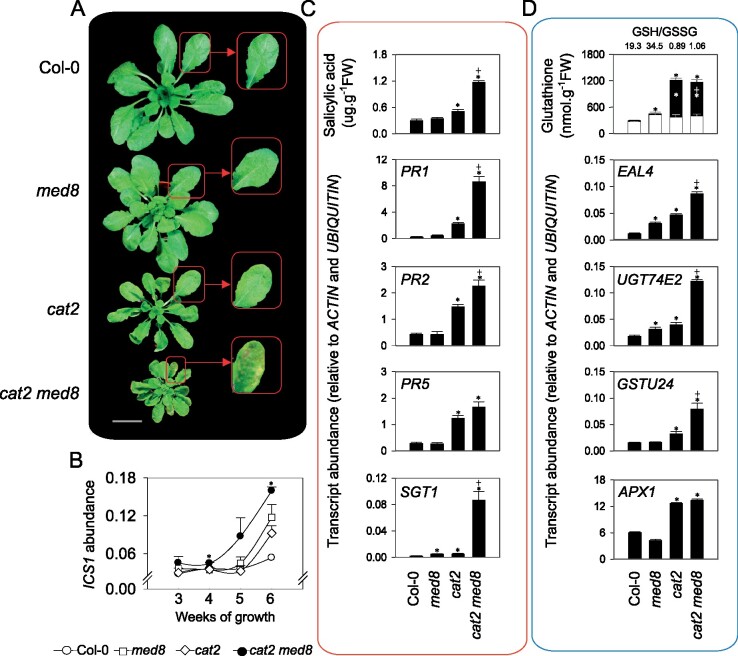
Plant phenotypes in long days were determined at different light intensities allowing photorespiratory H2O2 fluxes to be manipulated. Under moderate light (200 µmol·m−2·s−1 irradiance at leaf level), cat2 med8 plants exhibited a very severe developmental phenotype consisting of growth arrest a few days after emergence of the first four leaves and exacerbated lesion formation when compared to the single cat2 mutants (Figure 2A). However, when grown under low light (50 µmol·m−2·s−1), cat2 and med8 single and double mutants were smaller than Col-0, and the cat2 med8 mutants were able to survive and had rosette phenotypes similar to those of cat2 (Figure 2B). To circumvent interference from the severe growth inhibition in the double mutants, we designed a transfer experiment in which plants were first grown for 18 days in low light before transfer to moderate light (Figure 2C). Four days after the transfer, no lesions developed on cat2 leaves (Figure 2, C and D); however, cat2 med8 developed lesions and exhibited a more pronounced decrease in Fv′/Fm′ particularly in older leaves (Figure 2D). Because the SA pathway is key to the cat2-dependent lesion development and long-day induction of pathogenesis responses (Chaouch et al., 2010; Noctor et al., 2015), we quantified transcripts of the SA biosynthesis gene ISOCHORISMATE SYNTHASE1 (ICS1), and two SA-dependent transcripts, PATHOGENESIS-RELATED 1 (PR1) and PR2. PR1, PR2, and ICS1 transcripts were markedly induced in cat2 med8, above the cat2 levels, following transfer from low light to moderate light (Figure 2E, see fold-change induction displayed above the bars). Seven days after transfer to photorespiratory promoting conditions, SA levels were similar in Col-0 and med8 but accumulated approximately to eight-fold higher in cat2. SA accumulation was strongly enhanced in cat2 med8 (about 16-fold higher than Col-0; Figure 2F).
Figure 2.
The med8 mutation promotes H2O2-dependent cell death and associated response in long days. A and B, Representative photographs of 3-week-old plants grown under moderate (200 µmol· m−2·s−1) and low light (50 µmol·m−2·s−1) in a long-day photoperiod (16-h light/8-h dark), respectively. C, Phenotypes induced by the onset of oxidative stress. Plants were grown under low light (50 µmol·m−2·s−1) for 18 days and then transferred to moderate light irradiance (200 µmol·m−2·s−1) to induce oxidative stress in the cat2 backgrounds. Photographs were taken 4 days after transfer. Arrows indicate foliar lesion formation. D, Leaf series (top) and corresponding Fv′/Fm′ (bottom) from a representative rosette taken 4 days after transfer. Whole-plant leaf series from representative rosettes are displayed to highlight the occurrence of cell death on older leaves. Leaf sets are preceded by two cotyledons, with leaf 1 at the third position from the left. E, SA-related gene expression in Col-0, med8, cat2, and cat2 med8 plants under the conditions described in (C). ICS1, PR1, and PR2 transcript levels were quantified by RT-qPCR, relative to ACTIN and UBIQUITIN, in plants transferred from low-to-moderate light at the indicated days. Values are means ± SE of biological triplicates. Numbers above the bars indicate fold-change relative to control conditions (Day 0) in the respective genotypes. Only fold-change values higher than 2 are displayed. Significant differences are *P ≤0.05 (comparison between mutants and Col-0) and +P ≤0.05 (comparison between double mutants and cat2; Student’s t test). F, Total SA levels were determined 7 days after transfer from low-to-moderate light. Values are means ± SE of three biological repeats. Significant differences are *P ≤0.05 (comparison between mutants and Col-0) and +P ≤0.05 (comparison between double mutants and cat2; Student’s t test).
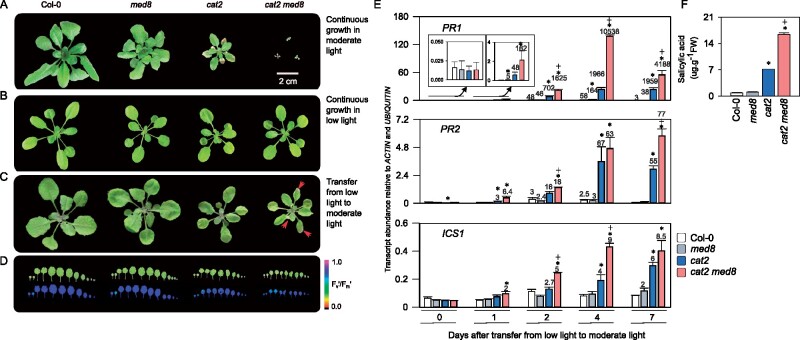
When plants were grown in short days, under cell death nonpermissive conditions (at an irradiance of 200 µmol·m−1·s−2), cat2 and med8 displayed decreased rosette growth, but no lesions were observed (Figure 3A). Interestingly, the cat2 med8 double mutants showed reduced growth and lesion development within 5 weeks, a phenotype that was accompanied by marked induction of ICS1 transcripts (Figure 3B). Moreover, analysis of cat2 med8 phenotypes in short days indicated, consistent with ICS1 induction, accumulation of SA and significant induction of PR as well as the SA-inducible SA GLUCOSYLTRANSFERASE SGT1 transcripts (Figure 3C). In plant tissues, H2O2 levels, like those of other ROS, are difficult to quantify in a specific and direct manner; therefore, we assessed redox homeostasis by alternative, more reliable proxies (Noctor et al., 2015, 2016). First, we quantified reduced (GSH) and oxidized (GSSG) glutathione as biochemical markers of intracellular thiol redox status. Increased accumulation of glutathione is a well-described response in cat2 plants and is key to activate SA biosynthesis and signaling (Queval et al., 2007; Mhamdi et al., 2010a, 2010b; Han et al., 2013a). This analysis revealed that the med8 mutation did not further promote perturbation of the redox state of glutathione, as both oxidized and reduced glutathione were similar in cat2 and cat2 med8 (Figure 3D). Secondly, we quantified four oxidative signaling marker transcripts known to be induced by H2O2 (Noctor et al., 2016; Mhamdi et al., 2017). Three of these transcripts were highly induced in cat2 med8 plants, hinting at a stronger activation of oxidative signaling in these conditions, above the cat2 level (Figure 3D). Taken together, our data define MED8 as a negative regulator of H2O2-induced gene expression and signaling in both short-day and long-day photoperiods.
Figure 3.
The med8 mutation activates the H2O2-dependent cell death response in nonpermissive short-day conditions. A, Representative pictures of 5-week-old plants grown under moderate light (200 µmol·m−2·s−1) in a short-day photoperiod (8-h light/16-h dark). Red arrows indicate lesion formation on the double mutant leaves. Bar = 2 cm. B, Time course analysis of ICS1 expression. ICS1 transcripts were quantified in short-day grown plants at the indicated weeks. C, SA levels and related gene expression in Col-0, cat2, med8, and cat2 med8 plants grown in short days for 6 weeks. The PR1, PR2, PR5, and SGT1 transcripts were quantified by RT-qPCR, relative to ACTIN and UBIQUITIN. D, Glutathione and redox marker transcript levels in Col-0, cat2, med8, and cat2 med8 plants grown in short days for 6 weeks. In the top graph (glutathione), white and black bars correspond to reduced glutathione (GSH) and oxidized glutathione (GSSG), respectively. Numbers above the bars indicate GSH/GSSG ratios. In (B), (C), and (D), values are means ± SE of three biological repeats. Significant differences are *P ≤0.05 (comparison between mutants and Col-0) and +P ≤0.05 (comparison between double mutants and cat2; Student’s t test).
MED8 regulates responses to oxidative stress triggered pharmacologically
To further investigate the physiological consequences of MED8 mutation on oxidative stress responses, we phenotyped 2-week-old seedlings exposed to oxidative stress-inducing agents. First, we tested the effects of the herbicide 3-aminotriazole (3-AT), a catalase inhibitor that triggers a cell death phenotype and accumulation of glutathione as GSSG (May and Leaver, 1993; Gadjev et al., 2006). Growth in liquid medium supplemented with 1 mM 3-AT caused a decrease in Fv′/Fm′ in both Col-0 and med8 seedlings, although the effect was less marked in med8 (Supplemental Figure 2A). Spraying soil-grown plants with 2 mM 3-AT provoked severe bleaching and a drastic decrease of Fv′/Fm′ within 2 days of treatment; however, the effects were significantly reduced in med8 (Supplemental Figure 2B). The differential response of med8 to 3-AT was not correlated with an altered redox homeostasis as glutathione levels and oxidation state was similar in Col-0 and med8 plants (Supplemental Figure 2C).
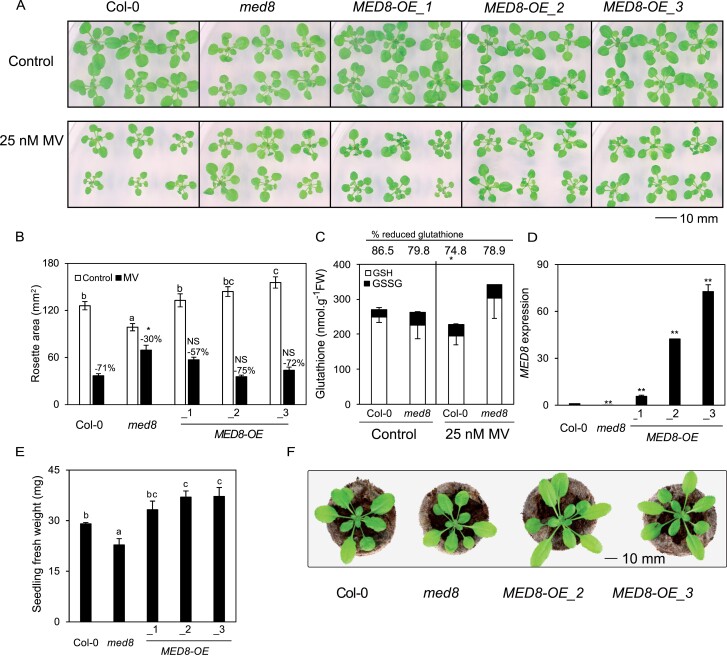
Second, we tested the effects of the herbicide MV, which initially leads to superoxide production in the chloroplasts and mitochondria and, subsequently, to the formation of other ROS molecules. Remarkably, when grown on medium containing 25-nM MV, med8 seedlings displayed milder stress symptoms relative to Col-0, with a less pronounced reduction of rosette area in med8 (30%) compared to wild-type seedlings (71%; Figure 4, A and B). Total glutathione levels did not differ between Col-0 and med8 grown in control conditions, although, interestingly, GSSG was higher in med8 than in Col-0. In the presence of MV, glutathione levels decreased in Col-0 plants, whereas med8 plants maintained higher glutathione levels similar to control conditions (Figure 4C). We complemented the med8 mutant by overexpressing the MED8 gene and included the complemented lines into our experiments. The med8-driven tolerance to MV was abolished in three independent overexpressor lines MED8-OE1/2/3 (Figure 4, A and B). The MED8-OE lines had larger rosettes and an increased biomass under control conditions, even when compared to Col-0 plants, in particular in the MED8-OE2 and MED8-OE3 lines (with high MED8 expression levels; Figure 4, D–F). Together, our results demonstrate that MED8 truncation confers resistance to oxidative stress while ectopic overexpression promotes rosette growth.
Figure 4.
Truncation of MED8 Q-rich domain confers tolerance to MV. A, Phenotypes of 3-week-old Col-0, med8, and MED8 overexpression lines (MED8OE-1/2/3). Plants were grown on ½ MS medium with or without 25 nM, in a long-day photoperiod (16-h light/8-h dark) and under a light intensity of 80 μmol·m−2·s−1. B, Rosette area quantification of med8 and MED8OE lines grown in the same condition as in (A). The data are means ± SE of 16 biological replicates. The different letters indicate statistically significant difference between Col-0, med8, and MED8OE lines under control conditions analyzed with one-way ANOVA followed by Duncan’s test (P ≤0.05). Numbers above the bars indicate the reduction percentage of the rosette areas in the respective genotypes. Asterisks indicate a significant difference in MV resistance between mutant and Col-0 analyzed with two-way ANOVA (P ≤0.05). NS, no significant differences. C, Glutathione content in med8 mutants in response to MV-induced oxidative stress. Glutathione content in 3-week-old Col-0 and med8 plants grown on ½ MS medium with or without 25-nM MV. White and black bars indicate GSH and glutathione disulfide (GSSG) content, respectively. Numbers above the bars indicate glutathione reduction states in Col-0 and med8 plants under control conditions and MV stress. The data are means ± se of biological triplicates. Asterisk indicates a significant difference at P ≤ 0.05 (Student’s t test) between Col-0 and the med8 mutant. D, MED8 transcript levels, relative to ARP7 and UBIQUITIN, in the respective overexpression lines. The data are means ± SE of three biological replicates. E, Fresh weight of 3-week-old Col-0, med8, and MED8OE plants grown on ½ MS medium. The data are means ± SE of eight biological replicates. The different letters indicate statistically significant difference analyzed with one-way ANOVA followed by Duncan’s test (P ≤ 0.05). F, Phenotypes of 3-week-old plants grown in soil at an irradiance of 100 µmol·m−2·s−1 in long days (16-h light/8-h dark).
In addition to oxidative stress, we also studied the phenotypes med8 in response to several abiotic stresses. The salt stress sensitivity (50-mM NaCl) was lower in med8 seedlings than in Col-0, but no differences were scored when plants were grown on 25-mM mannitol (Supplemental Figure 3, A and B). When exposed to 1.3 µmol·m−2·s−1 of UV-B, the med8 mutant retained slightly higher Fv′/Fm′ values than the Col-0 plants (Supplemental Figure 3C).
Tolerance to MV in med8 is associated with upregulation of defense-related genes
To assess changes in gene expression associated with the med8-driven tolerance to MV, we performed a transcriptome analysis through RNA-sequencing (RNA-seq) using 2-week-old Col-0 and med8 seedlings grown on control or MV-supplemented medium.
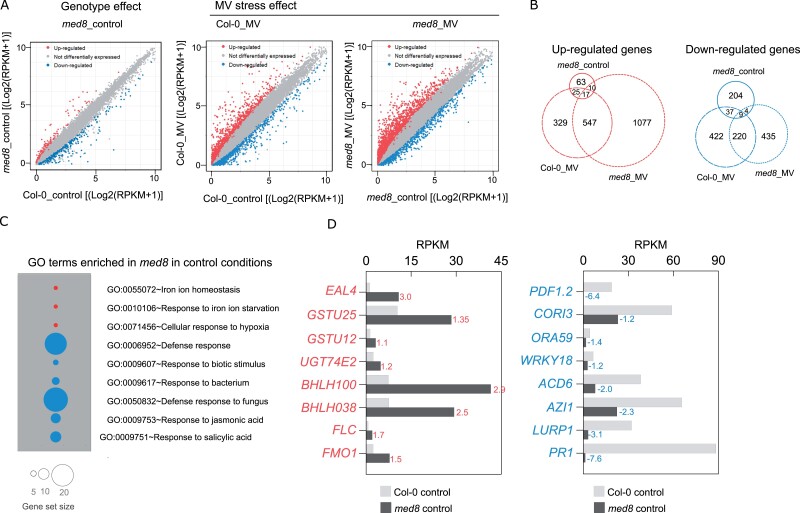
In the absence of MV, 115 upregulated and 254 downregulated transcripts were identified in the med8 mutant relative to Col-0, indicating that truncation of MED8 does not have an extensive genome-wide effect on gene expression (Figure 5A;Supplemental Data Set 1). Gene ontology (GO) enrichment analysis of upregulated genes revealed enrichment of genes involved in stress responses (Supplemental Data Set 1; Figure 5, C and D). For instance, four bHLH TFs that function in iron homeostasis were among the genes that were strongly induced (Supplemental Data Set 1) as well as genes involved in glutathione metabolism or known to be induced by H2O2, including, besides EAL4, UDP-GLUCOSYLTRANSFERASE 74E2 (UGT74E2), GLUTATHIONE S-TRANSFERASE (GSTU25), and GSTU12. Stress-responsive genes were also significantly enriched among the downregulated genes (Figure 5C). In particular, response to biotic stimulus and defense responses were the most significantly over-represented, including genes encoding the PLANT DEFENSIN genes PDF1.2, PDF1.3, and PDF1.4, PATHOGENESIS-RELATED proteins PR1 and PR4, VEGETATIVE STORAGE PROTEIN genes VSP1 and VSP2, and several TFs, such as WRKY18, WRKY60, and ORA59. Most of the defense genes downregulated in med8 under control conditions are involved in the JA, SA, or immune responses (Figure 5D, left). The expression levels of a few selected genes were validated by quantitative reverse transcription PCR RT-qPCR analysis (Supplemental Figure 4).
Figure 5.
Overview of changes in the med8 transcriptome under control and MV-induced oxidative stress. A, Scatter plot visualization of expression levels of all transcripts analyzed in RNA-seq showing med8_control versus Col-0_control (left), Col-0_MV versus Col-0_control (middle), and med8_MV versus med8_control (right). Differentially expressed genes (absolute value of log2FC>1, FDR<0.01) are indicated by red (upregulated) or blue (downregulated). ctrl, control. B, Venn diagrams for comparison of differentially expressed genes shown in (A). Red and blue circles refer to upregulated and downregulated genes, respectively. C, Gene ontology enrichment analysis in med8 control conditions. Red and blue circles indicate gene sets that are upregulated or downregulated, respectively. D, Examples of genes induced or repressed in med8 under control conditions. Numbers next to each bar indicate log2FC (med8_control versus Col-0_control).
Next, we mined the dataset to identify genes differentially expressed in a stress context. Overall transcriptional changes in the presence of MV were higher in med8 (1651 up- and 668 downregulated) than those in Col-0 (918 up- and 688 downregulated; Figure 5A;Supplemental Data Sets 2 and 3). We compared differentially expressed transcripts in med8 and Col-0 in control and stress conditions and found an overlap of 42 up- and 46 downregulated genes, respectively (Figure 5B). Analysis of MV-induced gene expression in med8 revealed that MED8 truncation suppresses the induction of half of the MV-responsive genes in Col-0. Only 33 genes showed opposite expression patterns in MV-grown Col-0 and med8 (Table 1). Notably, most of them are involved in SA signaling pathways and dependent responses (e.g. ACCELERATED CELL DEATH 6, FLAVIN-DEPENDENT MONOOXYGENASE 1, GLUTAREDOXIN 480, PR1, WRKY54, and WRKY70).
Table 1.
List of genes showing opposite regulation in med8 and Col-0 in response to oxidative stress induced by MV.
| Gene ID | Description | Expression |
Category | |
|---|---|---|---|---|
| Col-0 | med8 | |||
| AT2G14610 | Pathogenesis-related gene 1 (PR1) | −5.02 | 4.21 | a |
| AT4G14400 | Accelerated cell death 6 (ACD6) | −1.34 | 1.98 | a |
| AT2G14560 | Late Upregulated in Response to Hyaloperonospora parasitica 1 (LURP1) | −2.71 | 3.41 | a |
| AT5G54610 | Ankyrin/Bian Da 2 (BDA1) | −1.72 | 2.04 | a |
| AT3G22235 | Cysteine-rich transmembrane module 8 (HCYSTM8) | −3.09 | 1.95 | b |
| AT3G22231 | Pathogen and Circadian Controlled (PCC1) | −3.56 | 2.05 | a |
| AT5G45080 | Phloem Protein 2-A6 (PP2-A6) | −1.45 | 1.15 | b |
| AT1G49860 | Glutathione S-transferase (class phi) 14 (GSTF14) | −1.56 | 1.38 | b |
| AT5G03350 | SA-INDUCED LEGUME LECTIN-LIKE PROTEIN 1 (SAI-LLP1) | −1.82 | 1.86 | a |
| AT3G56400 | WRKY DNA-binding protein 70 (WRKY70) | −2.24 | 2.04 | a |
| AT2G40750 | WRKY DNA-binding protein 54 (WRKY54) | −1.91 | 2.25 | a |
| AT1G28480 | Glutaredoxin (GRX480) | −1.55 | 2.49 | a |
| AT2G45760 | BON association protein 2 (BAP2) | −3.87 | 3.58 | b |
| AT2G13810 | AGD2-like defense response protein 1 (ALD1) | −2.91 | 1.51 | a |
| AT1G19250 | Flavin-dependent monooxygenase 1 (FMO1) | −2.87 | 1.40 | a |
| AT2G41090 | Calmodulin like 10 | −2.07 | 1.07 | b |
| AT5G01600 | Ferretin 1 (FER1) | −2.28 | 1.02 | b |
| AT4G25490 | C-repeat/DRE binding factor 1 (CBF1)/DREB1B | −2.50 | 2.92 | b |
| AT4G25480 | C-repeat/DRE binding factor 3 (CBF3)/DREB1A | −1.31 | 1.10 | b |
| AT3G23170 | PROLINE/SERINE-RICH PROTEIN (PRP) | −1.64 | 1.37 | b |
| AT5G39670 | CALMODULIN-LIKE 46 (CML46) | −1.45 | 1.15 | a |
| AT1G05880 | Ariadne 12 (ARI12) | −1.16 | 2.54 | b |
| AT1G08090 | Nitrate transporter 2:1 (NRT2:1) | −1.11 | 4.20 | b |
| AT3G21500 | 1-deoxy-d-xylulose 5-phosphate synthase 1 (DXPS1) | −3.01 | 2.84 | |
| AT5G66620 | DA1-related Protein 6 (DAR6) | −2.32 | 1.03 | |
| AT1G21140 | Vacuolar Iron Transporter (VIT) family protein | −1.85 | 1.35 | |
| AT3G60420 | Phosphoglycerate mutase family protein | −1.77 | 1.86 | |
| AT3G49130 | SWAP (Suppressor-of-White-APricot) | −2.13 | 1.63 | |
| AT2G26400 | Acireductone Dioxygenase 3 (ARD3) | −1.75 | 3.23 | |
| AT5G41730 | Protein kinase family protein | −2.09 | 2.13 | |
| AT1G13310 | Endosomal targeting BRO1-like domain-containing protein | −3.79 | 2.73 | |
| AT5G66640 | DA1-related protein 3 (DAR3) | −1.67 | 3.43 | |
| AT1G80470 | F-box/RNI-like/FBD-like domains-containing protein | −5.34 | 3.48 | |
Salicylic acid signaling pathways and dependent responses.
Defense- and stress-related genes.
Additionally, among the 859 genes previously defined as ROS-induced genes (Willems et al., 2016; ROS wheel Cluster V) and detectable in our dataset, 252 genes were upregulated in med8 under MV stress versus only 62 genes in Col-0 (Supplemental Figure 5 and Supplemental Data Set 4). Moreover, the ROS transcripts were markedly more abundant in the stress-exposed med8 mutant than in Col-0 (Supplemental Figure 5 and Supplemental Data Set 4). However, among the 309 ROS-repressed genes (Willems et al., 2016; ROS wheel Cluster V), only 17 and 34 transcripts were downregulated by MV stress in Col-0 and med8, respectively. Thus, med8 was overall more responsive than Col-0.
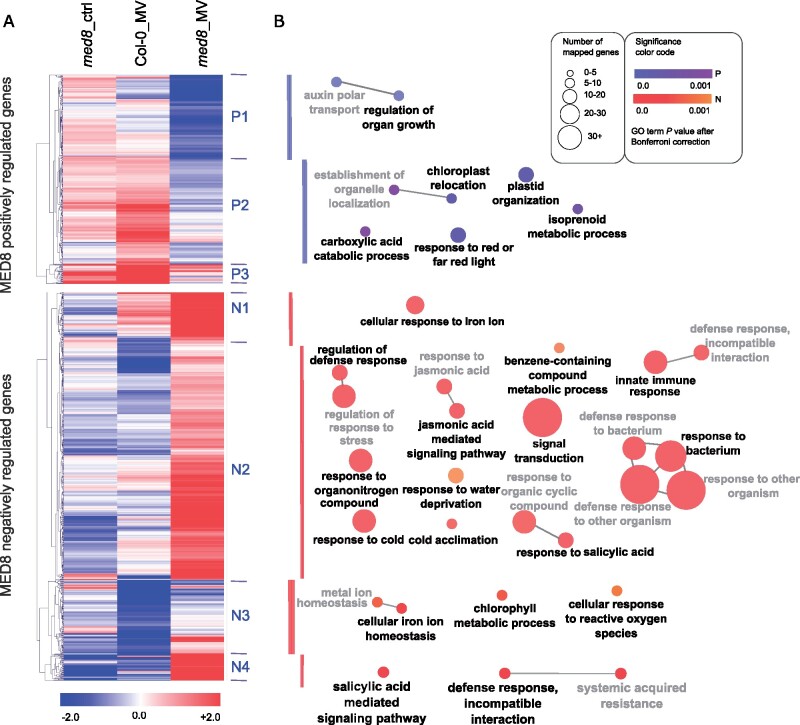
To gain further insights into the MED8-dependent gene expression under MV stress, a two-way ANOVA analysis (treatment×genotype) was performed. Whereas the MV induction decreased in 415 genes in med8 (referred to as MED8 positively regulated genes), it increased in 769 genes (referred to as MED8 negatively regulated genes; Figure 6;Supplemental Data Set 5). Hierarchical clustering of these two groups identified seven major clusters that organized the positively (P) and negatively (N) regulated MED8 genes into three and four clusters, respectively (Figure 6A). Genes in each cluster were functionally classified by GO term enrichment analysis as indicated with a color code (Figure 6B). Cluster P1 (163 genes) contained genes associated with regulation of organ growth and auxin transport and were strongly repressed in response to MV in med8, but not in Col-0. Cluster P2 (213 genes) had similar expression characteristics and were enriched for genes associated with plastid organization and response to red or far-red light. In the smaller Cluster P3 (39 genes) without a significant GO enrichment, transcripts were highly induced in Col-0 in response to MV, but largely repressed or not altered in the med8 mutants. A much larger set of genes was more strongly induced in the med8 mutant in response to MV than in Col-0 (Figure 6, MED8 negatively regulated genes). Whereas Clusters N1 and N3 (89 and 145 genes, respectively) were enriched in genes associated with response to iron, chlorophyll metabolic process, and response to ROS, the majority of the MED8 negatively regulated genes (Cluster N2 and N4; 480 and 55 genes, respectively) were associated with GO terms related to defense, response to bacterium and innate immune responses, dependent on phytohormone pathways (Supplemental Data Set 5). In particular, JA- and SA-mediated signaling pathways were enriched together with TFs and their upstream potential regulators, including MYB51, WRKY18, WRKY70, SARD1, ERF1, MYC2, and its interacting JAZ1 and JAZ5 were induced in a stress specific manner when MED8 was truncated. It is worth emphasizing that Clusters N2 and N4 accounted for almost half of the ANOVA significant genes and were clearly the most evidently enriched compared to other clusters (see the size of the nodes reflecting the number of genes in Figure 6; Supplemental Data Set 5). Therefore, we conclude that the med8 ‘signature’ in oxidative stress-induced gene expression is mainly to repress defense and hormone pathways.
Figure 6.
Transcriptomic signature of med8 in response to MV-induced oxidative stress. A, Heat map visualizing the expression of 1,184 genes that are either positively (P) or negatively (N) regulated by MED8 under MV stress based on two-way ANOVA analysis (Supplemental Data Set 5). Hierarchical clustering with tMEV identified seven main clusters. Expression is given as Log2FC. From left to right: med8_control versus Col-0_control, Col-0_MV versus Col-0_control, and med8_MV versus med8_control. B, GO term enrichment analysis for each cluster displayed in (A) by ClueGo 2.5. The size of the nodes reflects the number of genes in each GO term, and the fill color intensity reflects the corresponding P value for enrichment after Bonferroni correction. GO terms were grouped based on their similarity (edge thickness represents the kappa score of similarity between two GO terms), and the most significant terms in each group are shown in bold.
Tolerance to oxidative stress in med8 requires SA biosynthesis
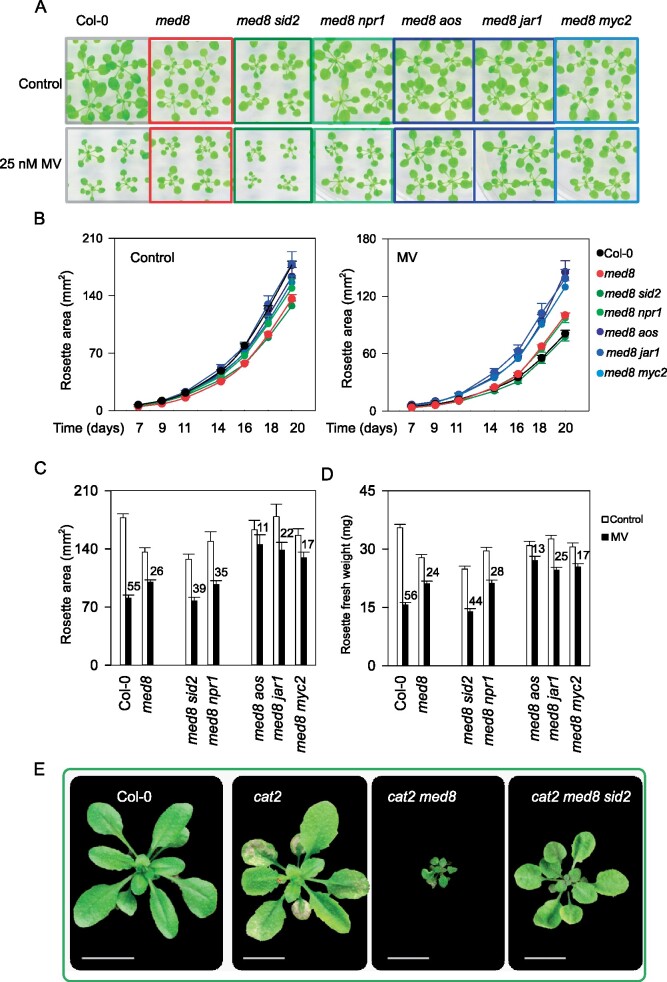
Due to the substantial effect of med8 mutation on SA- and JA-related gene expression, we introduced several mutations (SA induction-deficient2 (sid2), nonexpressor of pathogenesis-related1 (npr1), allene oxide synthase (aos), jasmonate response1 (jar1), and myc2) into the med8 background to genetically address the involvement of SA and JA biosynthesis and signaling in the oxidative stress-related med8 phenotypes. First, we analyzed the effect on med8 tolerance to MV-induced oxidative stress. Analysis of the single mutant phenotypes showed no compromised responses, with the exception of aos mutants that were more tolerant to MV relative to Col-0 (Supplemental Figure 6). Also, our data show that med8 sid2 and med8 npr1 rosette growth was more severely reduced by MV than the med8 single mutants (Figure 7, A–D). To some extent, both mutants exhibited a reduction in rosette areas and fresh weight resembling more that of Col-0 than of med8 (Figure 7, A–D). In contrast, the double mutants med8 aos, and med8 myc2 plants were even less affected than med8 plants, while med8 jar1 tend to have a response similar to med8. These mutants were hence less sensitive to MV stress and exhibited a reduction in rosette areas and fresh weight improved relative to med8 (Figure 7, A–D). Taken together, these observations indicate that SA biosynthesis and signaling are, at least in part, required for the med8 tolerance to MV, whereas JA synthesis and signaling genes AOS, JAR1, and MYC2 are seemingly imposing growth restrictions during long term oxidative stress exposure.
Figure 7.
Genetic manipulation of the med8 oxidative stress tolerance phenotypes by introduction of secondary mutations in SA or JA biosynthesis and signaling. A, Phenotypes of 3-week-old Col-0, med8, and med8 double mutant plants grown on ½ MS medium with or without 25 nM MV. Mutations of SA or JA biosynthesis and signaling were achieved by introduction of the sid2 and npr1 or aos, jar1, and myc2 mutations in the med8 background. B, Time-course quantification of rosette areas of Col-0, med8, and SA- or JA-related mutants. All lines were grown in vitro for 20 days under control or stress conditions as described in (A). The data are means ± SE. C, Quantification of rosette areas of all genotypes grown under the conditions as in (A) at 20 days. Number above the bars indicates the rosette growth reduction in respective genotypes. The data are means ± SE. D, Quantification of rosette fresh weight of all genotypes grown in conditions as in (A) at 20 days. Number above the bars indicates the reduction in fresh weight in the respective genotypes. The data are means ± SE. In (A), (C), and (D) 30 rosettes were used for quantification. E, Phenotypes of 3-week-old soil-grown Col-0, cat2, cat2 med8, and cat2 med8 sid2 plants at an irradiance of 200 μmol·m−2·s−1 in long days (16-h light/8-h dark).
Next, we assessed the influence of SA on med8-dependent phenotypes caused by photorespiratory H2O2 in the catalase-deficient background. To establish whether the phenotypes of cat2 med8 depended on SA, triple mutants were produced that additionally carried the sid2 mutation. As described above, under long-day conditions, lesion formation and severe growth restriction occur in cat2 med8. However, in the triple cat2 med8 sid2 both the growth restriction and the cell death phenotypes were largely reverted (Figure 7E). These results demonstrate that the phenotypes of cat2 med8 depend on SA biosynthesis via ICS1.
The MED8 glutamate-rich C-terminal domain is not required for Mediator complex assembly and does not affect the interaction repertoire of the Mediator complex
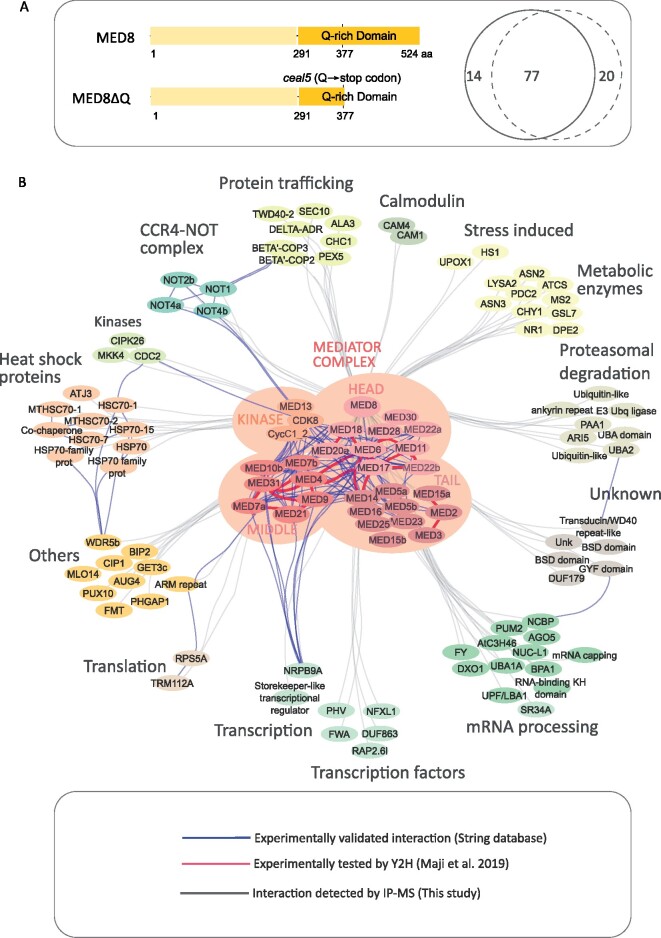
In ceal5 and med8 mutants, MED8 is truncated at the C-terminus, leading to a shortened Q-rich domain. Q-rich domains have features of intrinsically disordered regions (IDRs), lacking a rigid or folded stable structure and might contribute to the formation of an interface, interacting with multiple partners and, hence, acting as hubs in protein interaction networks (Ding et al., 2006; Wright and Dyson, 2015; Nagulapalli et al., 2016). Consequently, the MED8 IDR truncation might result in an altered interactome. We hypothesized that in the full-length MED8 the Q-rich domain, through its interaction partners, facilitates the suppression of stress and defense genes and that hence in a truncated MED8 that capacity is abolished.
To test this hypothesis, we performed an immunoprecipitation and mass spectrometry analysis (IP-MS) on Arabidopsis cells expressing a GFP-tagged full-length MED8 or the truncated version, MED8ΔQ, in which a large part of the Q-rich domain was deleted, to mimic the ceal5 mutation (Figure 8A). We identified 92 and 98 co-purified proteins with MED8 and MED8ΔQ respectively, which were enriched in the GFP samples relative to the wild-type (Supplemental Data Set 6). In total, 77 proteins were co-purified with both baits (Figure 8A), suggesting only a minor influence of the Q-rich domain on the protein–protein interaction capacity and repertoire. Both baits pulled down 28 Mediator subunits and analysis of the protein abundance revealed only minor differences. These results demonstrate that the truncated MED8 can integrate the Mediator complex similar to the full-length protein.
Figure 8.
Mediator complex-interacting proteins identified in IP-MS. A, Venn diagram showing the overlap between Mediator complex interactors identified by IP-MS, in MED8 (solid line) and MED8ΔQ (dashed line), respectively (see Supplemental Data Set 6 for full lists of interactors). Schematic protein models for MED8 and MED8ΔQ expressed in Arabidopsis cells are presented. The MED8 protein contains a glutamine (Q)-rich domain at the C-terminus with features of IDRs. The Q-rich domain truncation in MED8ΔQ mimics the ceal5 mutation. Numbers indicate the amino acid (aa) position. B, Interactome of the Mediator complex in Arabidopsis. The Mediator complex network was constructed with the Cytoscape software. The gray edges represent interactions detected by our IP-MS experiments, the blue dashed lines indicate interactions reported in plants (String database) and the red edges indicate the one-to-one interaction detected by Maji et al. (2019) within the mediator complex.
We used the ensemble of 112 proteins identified in the MED8 and MED8ΔQ pull-downs to generate a protein interactome network, entailing Arabidopsis and all orthologous, experimentally validated, protein–protein interactions reported in the String database (Figure 8B, blue edges; Supplemental Data Set 7). In addition, we plotted the yeast two-hybrid (Y2H)-based Mediator protein interaction map (Maji et al., 2019; Figure 8B, red edges; Supplemental Data Set 7). Mining of the MED8 and MED8ΔQ interactomes revealed potential previously unrecognized roles for the Mediator complex. Several transcriptional regulators and TFs were identified, including NRPB9A, which belongs to the RNAP II core complex. Interestingly, one of the TFs identified here was the NF-X1-TYPE ZINC FINGER1 (NFXL1) protein that negatively regulates defense-related genes (such as WRKYs and ICS1) via a SA-dependent signaling pathway (Asano et al., 2008). One prominent functional category of interactors was RNA-binding proteins. Proteins involved in mRNA processing, such as SR34A and UBA1A immunoprecipitated with the Mediator complex, supporting its role in various transcription steps. Interestingly, we identified NEGATIVE ON TATA-LESS 1 (NOT1), NOT2b, and two RNA-binding proteins, recently referred to as NOT4a (AT2G28540) and NOT4b (AT3G45630; Zhou et al., 2020), all components of the CARBON CATABOLITE REPRESSION4 (CCR4)-NOT complex, a key regulator of nuclear gene expression (reviewed in Collart, 2016). Important to note is that the interaction with NOT2b and NOT4b was weaker when MED8ΔQ was used as a bait, indicating that the C-terminal IDR of MED8 is required for the interaction of the Mediator complex with the CCR4-NOT complex.
In addition, proteins involved in posttranslational modification, including protein ubiquitination and phosphorylation, were identified (Supplemental Data Set 6; Figure 8B). For instance, an interaction between the CYCLIN-DEPENDENT KINASE8 (CDK8), component of the kinase module, and the CELL DIVISION CONTROL 2 (CDC2) had been tested previously by Bimolecular Fluorescence Complementation, revealing its nuclear localization (Van Leene et al., 2010). As such, the interactome analysis points to the interconnection of MED8 and the Mediator complex as a whole with other pathways and might help understanding the Mediator function beyond transcription.
MED8 interacts with the transcriptional regulator NOT2 to control EAL4 expression
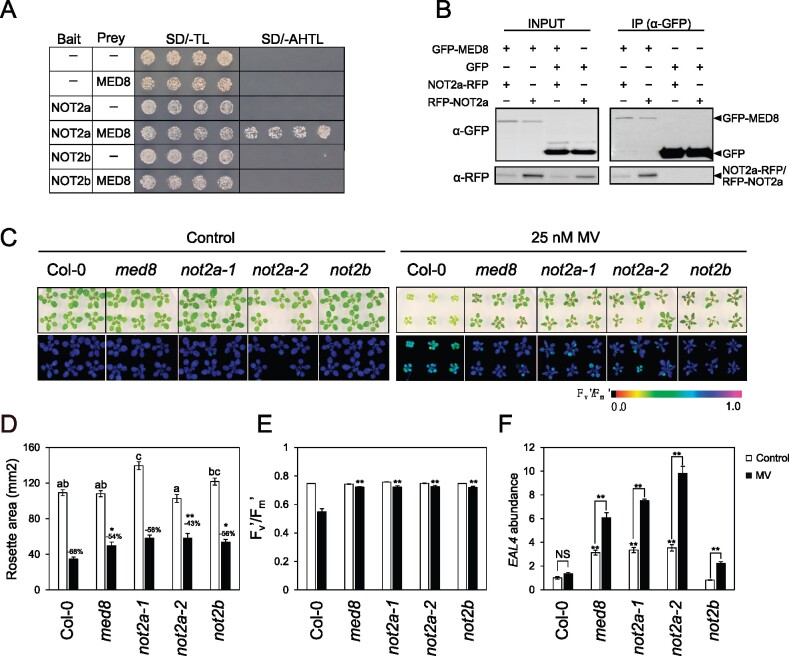
As a complementary approach to the IP-MS experiments, a Y2H screen was performed to identify direct interactors using the full-length MED8 as a bait and a cDNA library enriched for stress-responsive genes (Jaspers et al., 2009). Interestingly, six independent positive clones were identified, encoding the transcription regulator NOT2a (AT1G07705). NOT2 that belongs to the CCR4-NOT complex had also been detected as Mediator interactor by the pull-down approach (Figure 8). The Arabidopsis genome has two NOT2 homologs, NOT2a and NOT2b, which promote miRNA biogenesis by interacting with other microRNA biogenesis factors. Inactivation of both NOT2 genes is lethal due to severe defects in male gametophyte development (Wang et al., 2013).
To substantiate the interaction between MED8 and NOT2a, we introduced the full-length coding sequence of NOT2a into the prey vector (AD-NOT2a) and coexpressed it with DBD-MED8 in yeast. Direct interaction between MED8 and NOT2a was confirmed in yeast (Figure 9A). We further examined whether MED8 also interacted with NOT2b but did not detect direct interaction in yeast. In planta, the interaction was examined by co-immunoprecipitation (co-IP) assays. GFP-fused MED8 (GFP-MED8) and RFP-fused NOT2a (RFP-NOT2a and NOT2a-RFP) were transiently coexpressed in Nicotiana benthamiana leaves through agro-infiltration. The RFP-fused NOT2a proteins co-immunoprecipitated with MED8-GFP, but not with the GFP control (Figure 9B). These results indicate that MED8 associates with NOT2a in planta.
Figure 9.
NOT2 proteins associate with MED8 and regulate oxidative stress phenotypes and EAL4 expression. A, GAL4-based Y2H assays. Bait and prey represent the plasmids encoding the fusions to the GAL4 activation domain (AD) and the DNA-binding domain (DBD), respectively. Cotransformed yeast colonies were spotted on the selective SD medium minus Trp and Leu (-TL), then grown on SD medium minus Adenine, His, Trp, and Leu (-AHTL). Growth on SD-AHTL medium indicates a positive interaction. An empty vector was used as a negative control. B, Co-IP assays. Pro35S:GFP-MED8 was transiently coexpressed with Pro35S:NOT2a-RFP or Pro35S:RFP-NOT2a in N. benthamiana plants. Total protein extracts were immunoprecipitated with α-GFP beads. Input and immunoprecipitation (IP) samples were resolved by SDS–PAGE and the immunoblots were probed with α-GFP and α-RFP antibodies separately. C, Representative bright-field (top) and color-coded Fv′/Fm′ (bottom) images of 3-week-old plants germinated and grown under control (½ MS medium) and MV stress (25 nM MV) conditions. D, Quantification of rosette areas of plants grown in with and without MV stress. The data are means ± se (n =16). Asterisks indicate a significant difference between Col-0 and the mutants (**P ≤0.01; Student’s t test) and the different letters indicate statistically significant differences under control conditions analyzed by one-way ANOVA followed by Duncan’s test (P ≤0.05). The reduction (%) in rosette areas is indicated. E, Quantification of Fv′/Fm′. The data are means ± se (n = 16). Asterisks indicate a significant difference in MV resistance between Col-0 and mutants analyzed by two-way ANOVA (**P ≤ 0.01). D and E, Plants were grown in conditions described in (C) and 16 rosettes were quantified. F, Relative EAL4 expression. mRNA was extracted in triplicate samples from plants grown under control conditions and MV stress. EAL4 transcripts were quantified relative to ARP7 and UBIQUITIN. The data are means ± se of biological triplicates. Asterisks indicate a significant difference (**P ≤ 0.01).
As the interaction between MED8 and NOT2 might be relevant to define the oxidative stress responses, we investigated whether NOT2 was similarly involved in the oxidative stress response and associated gene expression by characterizing and phenotyping the T-DNA insertion mutants, not2a-1 (SALK_062057), not2a-2 (GABI_104B08), and not2b (SALK_058645). NOT2 expression was abolished in the respective mutants (Wang et al., 2013; Supplemental Figure 7A). The phenotype of not2 mutants grown in control or under MV stress was evaluated (Figure 9C). Under control conditions, rosettes of not2a-2 plants were significantly smaller relative to Col-0, whereas the not2a-1 and not2b mutants displayed slightly larger rosettes (Figure 9D). When grown in the presence of MV, the rosettes of both not2a and not2b mutants were visibly larger and the growth reduction was less evident, reminiscent of the med8 phenotype. The Fv′/Fm′ values were similar to Col-0 in the not2 mutants in the absence of stress, whereas under MV stress, Fv′/Fm′ values of all not2 mutants were significantly higher than those of Col-0, suggesting a decreased sensitivity to MV. Collectively, these data show that similar to MED8, NOT2 negatively affects responses to oxidative stress.
Interestingly, EAL4 expression was significantly enhanced by not2a mutations relative to Col-0 in both control and stress conditions, an effect that mimics the effect of med8 mutation (Figure 9E). However, not2b exhibited induction of EAL4 only under stress conditions, suggesting a divergent function for the two homologs. Our data demonstrate that, similar to MED8, NOT2 negatively affects responses to oxidative stress and that a MED8-NOT2A interaction is in play to regulate EAL4 expression.
Given the known role of NOT2 proteins in miRNA biogenesis, we hypothesized that MED8 may also function in the transcription of miRNA genes. Therefore, we assessed the levels of eight primary miRNAs (pri-miRNAs) by RT-qPCR. Consistent with previous observations (Wang et al., 2013), the levels of pri-miRNAs were reduced in not2a mutants (Supplemental Figure 7B). Similarly, med8 showed reduced pri-miRNA levels compared with the Col-0 levels, which was even more pronounced than in not2a mutants. Thus, the MED8–NOT2a interaction may play a role in the regulation of miRNA biogenesis in Arabidopsis.
Discussion
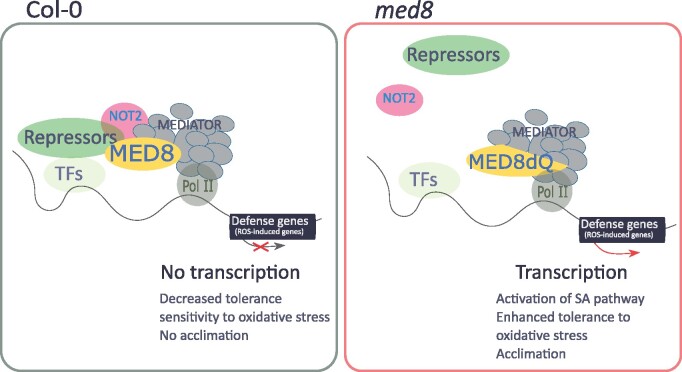
Oxidative stress activates several defense pathways, including SA- and JA-associated signaling. However, our understanding of the mechanisms underlying the transcriptional activation of defense pathways by H2O2 is limited. Here, we show that the MED8 subunit of the Mediator complex is a component of H2O2 signaling. The polyQ tail of MED8 negatively controls H2O2-dependent activation of defense pathways and represses SA-dependent signaling (Figure 10).
Figure 10.
Proposed mechanism by which MED8 functions in oxidative stress signaling. In the wild-type background, the full-length MED8 associates with NOT2 as well as with other unknown “repressors” to suppress defense genes expression and decrease tolerance to oxidative stress as a consequence. In the med8 background, the lack of the MED8 C-terminal region potentially drives the dissociation from the repressor module, the expression of defense genes, and the activation of phytohormone-related pathways (for instance, the salicylic acid pathway), thus enhancing the tolerance to oxidative stress.
Insights into MED8 functions in response to oxidative stress
The plant Mediator complex is predominantly known for its role in immunity and until now, only a few Mediator subunits have been linked to abiotic stress responses (Dhawan et al., 2009; Kidd et al., 2009; Çevik et al., 2012; Canet et al., 2012; Lai et al., 2014; An et al., 2017; Li et al., 2018; Crawford et al., 2020; Zhu et al., 2020). In this context, MED25 positively regulates germination under salt stress by interacting with DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A (DREB2A), ZINC FINGER HOMEODOMAIN1, and MYB-like TFs, while it negatively regulates drought tolerance by interacting with the repressor domain of DREB2A (Elfving et al., 2011). MED16, also known as SENSITIVE TO FREEZING 6, as well as MED2 and MED14, positively regulate cold acclimation by modulating the recruitment of the whole Mediator complex to C-repeat/DRE-binding factor (CBF)-responsive cold-regulated genes (Knight et al., 2009; Hemsley et al., 2014). Here, we demonstrate that MED8 regulates the transcriptional response to oxidative stress. In the med8 mutants, responses to oxidative stress, triggered both by chemical and genetic means, (Figures 2–4) are induced. At first sight, the cat2 med8 photorespiration-sensitive phenotype does not seem in line with the decreased sensitivity to MV observed in med8 seedlings. However, biochemical and molecular analyses revealed that a similar mechanism is involved (Figures 2–4). In both cases, a hyper-activation of SA biosynthesis and SA-dependent transcripts was in place. Recently, a positive role for SA in MV tolerance was supported by the fact that pretreatment with SA alleviates MV-induced oxidative stress (Cui et al., 2019). Therefore, the decreased sensitivity of med8 to MV might be due to activated SA synthesis and signaling, a hypothesis supported by the decreased MV tolerance in the med8 sid2 mutant (Figure 7). On the other hand, it is known that SA biosynthesis is required for cell death induced by photorespiratory H2O2 (Chaouch et al., 2010). Hence, the combination of activation of the SA pathway in both cat2 and med8 mutant backgrounds leads to a hyper accumulation in cat2 med8. This, in combination with increased H2O2 signaling in cat2 leads to accelerated cell death, a phenotype abolished in cat2 med8 sid2 triple mutants (Figure 7). Consequently, within the Mediator complex, the MED8 subunit acts as a negative regulator of SA biosynthesis or stability, at least when plants are exposed to conditions inducing oxidative stress.
In plants, the oxidative stress outcomes are conditional and dependent on the photoperiod (Queval et al., 2007, 2012; Vollsnes et al., 2009; Dghim et al., 2013). The cat2 med8 mutant phenotypes include a severe growth arrest and extensive cell death associated with hyper-accumulation of SA (Figures 2, 3, and 7), a response that is known to occur in long days but not in short days in cat2 plants (Chaouch et al., 2010; Queval et al., 2012, Han et al., 2013a, 2013b). Remarkably, the med8-triggered phenotypes in the cat2 background were dependent on the light intensity but not on the photoperiod, suggesting that MED8 has a higher hierarchical position in repressing the H2O2 signaling pathway and acts upstream of potential factors linking photoperiod to oxidative stress responses. Hence, MED8 might cooperate with components involved in shutting down SA pathways in short days, a role that was not previously recognized. Genetic factors that might determine outcomes of oxidative stress in short days are largely unknown. Indeed, 2A-type protein phosphatase subunit PP2A-B'γ is the only regulator currently established to be involved in suppressing the daylength-dependent pathogenesis responses (Trotta et al., 2011; Li et al., 2014). Like med8, the pp2a-b'γ mutation promoted lesion development, PR1 induction, and SA accumulation in response to photorespiratory H2O2. Roles of PP2A-B'γ are possibly linked to phytochrome pathways as PHYTOCHROME A expression levels were decreased (Li et al., 2014). Our data suggest that MED8 also influences the regulatory nexus that links oxidative stress, day length, and defense responses.
MED8 opposes H2O2-driven activation of the SA pathway
Emerging evidence indicates that the Mediator complex is a key component of SA-dependent pathways. Several subunits function as positive regulators, including the Tail module subunits MED14, MED15, MED16, and MED5b as well as the Kinase module subunits CDK8, MED12, and MED13. Although different subunits employ distinct mechanisms, overall, these appear to diverge from MED8 function. Plants defective in MED14, MED15, and MED16 exhibited an attenuated response to SA and pathogens and failed to induce PR genes (Canet et al., 2012; Wathugala et al., 2012; Zhang et al., 2012, 2013; Wang et al., 2016). Similarly, cdk8 as well as med12 show decreased basal SA levels and compromised systemic acquired resistance (Huang et al., 2019). CDK8 directly regulates the expression of ICS1 and ENHANCED DISEASE SUSCEPTIBILITY5 and counteracts the hyper-accumulation of SA in a semi-dominant mutation of MED5b (Huang et al., 2019; Mao et al., 2019). Furthermore, CDK8 interacts with NPR1, the key regulator of SA signaling, to control the expression of NPR1 and its targets (Chen et al., 2019). Hence, this raises the question whether the med8-driven hyper-activation of SA pathways (in response to oxidative stress) is also dependent on CDK8 protein or activity levels. Such a mechanism would involve a physical interaction between MED8 and the Kinase module, the latter occurring in MED8, but also in MED8ΔQ (Supplemental Data Set 6). Our preliminary data do not support such a scenario, because the cdk8 mutation reverts the cell death phenotype in the cat2 background, which is opposite to med8 (Supplemental Figure 8). The two Head subunits MED18 and MED20 play a negative role in SA responses. The med18 and med20 mutants displayed increased induction of PR genes in response to Fusarium oxysporum. Both subunits have been proposed to form a subdomain within Mediator to control the balance between SA and JA (Fallath et al., 2017). In med18 plants, growth phenotypes and gene expression were associated with the accumulation of defense metabolites, such as SA and the phytoalexin camalexin (Davoine et al., 2017), to levels that are expected only in pathogen-infected tissues. Although the action mechanisms of MED18 and MED20 are not fully defined, MED8 might be part of this subdomain. The exact position of each of the Mediator modules in the SA signaling pathway and their specific interactors or targets remain largely unknown, but MED8 seems to mainly negatively regulate the stress-induced expression of SA-related genes.
The C-terminal IDR feature of MED8 is required for interaction with components involved in transcription and miRNA biogenesis
Both ceal5 and med8 mutants contain a mutation in the Q-rich domain, most likely resulting in a truncated MED8 protein. One question is, therefore, whether deletion of this IDR affects MED8 function and affects the topology of the Mediator complex. The IDR feature occurs in many subunits of the Mediator complex and can accommodate dynamic interactions with proteins and nucleic acids (Ding et al., 2006; Nagulapalli et al., 2016; Wright and Dyson, 2015). This may be one mechanism by which Mediator is made sufficiently plastic to create “conformational ensembles” that are required for transient association with a diverse array of TFs or other transcriptional regulators (Cooper and Fassler, 2019). In agreement, the Q-rich domain of MED25 is necessary for its interaction with COI1 and JAZ1 (An et al., 2017; Zhai et al., 2018). Our in planta interactome data revealed that both MED8 and its truncated version interact with 26 known Mediator subunits, including the scaffolds MED14 and MED17 (Maji et al., 2019), and hence indicating no major topological changes in the truncated mutant version (Figure 8). This in contrast with a, most likely, drastic conformational change or even dissolving complex in the nonviable full knockout alleles. Nevertheless, given the increased response to oxidative stress in med8, a truncated Q-rich domain might affect Mediator complex plasticity and affect the responsiveness to transcriptional activator or repressor proteins. Our interactome study expands our knowledge on potential roles of the Mediator complex beyond transcription. We identified proteins involved in RNA binding, processing, and posttranslational modification processes. Interaction with RNA-binding proteins was recently reported in neural cells (Quevedo et al., 2019). Worth noting is that several of the RNA-processing enzymes identified here are involved in the plant immune responses. One of these is DECAPPING AND EXORIBONUCLEASE1 (DXO1), which enhances posttranscriptional gene silencing by increasing siRNA levels. The dxo1 mutation results in upregulation of defense-related genes. Interestingly, the dxo1-dependent autoimmunity phenotype is also suppressed by the manipulation of SA signaling through npr1 or eds1 mutation (Pan et al., 2020). The interaction of MED8 and the Mediator complex with the CCR4-NOT complex as well as its interactor PUMILIO HOMOLOG2 (Arae et al., 2019) now also links MED8 to miRNA biogenesis. NOT2 proteins regulate protein-coding and noncoding gene expression, and, similar to MED8, they negatively regulate EAL4 expression (Figure 9). Low accumulation of miRNAs in med17, med18, and med20a was linked to reduced RNAP II occupancy at several miRNA transcription start sites in med20a (Kim et al., 2011). The Arabidopsis microRNA miR164 and NAC4 have been shown to regulate cell death by targeting LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA1, WRKY40, and WRKY54, which act as negative regulators of transcription and cell death processes (Lee et al., 2017). Besides its function in miRNA biogenesis, NOT2b modulates the expression of genes involved in Agrobacterium-mediated plant transformation as well as abiotic stresses (Raman et al., 2019).
MED8 functions involve genetic and physical interaction with other repressors
How does MED8 affect oxidative stress responses? As stated above, a more in-depth mechanistic understanding of the genetic or physical interactions between MED8 and specific transcriptional regulators will be needed. Our data point to a genetic interaction between MED8 and the JA pathway, analysis of med8 aos1 and med8 myc2 revealed that JA exerts growth restriction when plants are subjected to a long-term oxidative stress (Figure 7). Inhibition of vegetative growth by JA is part of the wound response and was shown to depend on JA synthesis genes, upstream of AOS1, such LIPOXYGENASE LOX3 and LOX4 (Yang et al., 2020). Both LOX3 and LOX4 are negatively regulated by med8, pointing to an interaction between MED8 and JA synthesis at multiple levels. In JA signaling, the well-studied MED25 acts through interactions JA receptor COI1, transcriptional activator MYC2, and its homolog JA-ASSOCIATED MYC2-LIKE (JAM) protein, and the HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY1, thereby targeting different routes in the JA pathways (Çevik et al., 2012; Chen et al., 2012; An et al., 2017; Zhai et al., 2018; Zhai and Li, 2019; Liu et al., 2019). Similarly, MED8 action could involve several repressors, besides NOT2 (Figure 10). Analysis of double med8 med25 mutants under biotic stress revealed additive effects of both mutations, pointing to parallel mechanisms for MED8 and MED25 in the regulation of JA-dependent defenses (Kidd et al., 2009; Li et al., 2018). Although we demonstrated here that MED8 negatively affects the transcription of MYC2 and JAZs, and that med8 growth phenotypes depend on activation of JA signaling (Figure 7), this is unlikely to be mediated by a physical interaction with MYC2, because a direct interaction between MED8 and MYC2 has not been detected by Y2H (Li et al., 2018). However, the MED8 function has been shown to depend on another bHLH TF, FAMA, that directly binds to promoters of genes that act in JA signaling (for instance, ORA59) hinting at a potential mechanism by which MED8 might regulate downstream JA genes (Li et al., 2018). Repressors or repressor complexes might conditionally interact with the MED8 Q-rich domain (Figure 10). These could be repressors such as JAMs upstream of MYC2 in JA signaling and NPR3 and NPR4 upstream of WRKY70 and SARD1 in SA signaling (Van der Does et al., 2013; Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Ding et al., 2018; Liu et al., 2019). Our data suggest a scenario in which MED8 conditionally evolved to control the activation or repression of transcriptional regulators and their targets (for instance control condition versus oxidative stress). In such a scenario, the MED8 Q-rich domain would be required to interact with (yet unknown) repressors, hence fine-tuning the activation of SA and JA pathways.
Materials and methods
Plant materials
Arabidopsis (Arabidopsis thaliana L. Heynh.) Columbia-0 (Col-0) accession was used in this study. T-DNA lines med8 (SALK_092406), med8-2 (GABI_270C11), not2a-1 (SALK_062057), not2a-2 (GABI_104B08), and not2b-1 (SALK_058645) were obtained from the Nottingham Arabidopsis Stock Centre. Homozygotes were identified by genomic DNA-PCR with primers flanking the T-DNA insertions (Supplemental Table 1) and the left border primers. The cat2 med8 double mutants were generated by crossing med8 into the cat2 background (Queval et al., 2007) and double homozygotes identified by PCR genotyping of the segregating F2 populations. Other mutant lines were produced by crossing sid2, npr1, aos, jar1, and myc2 into the med8 background and the homozygous double mutants were identified by PCR using gene-specific primers or by genomic DNA sequencing (Supplemental Table 1).
Growth conditions and stress treatments
For in vitro experiments, sterilized seeds were first incubated at 4°C for 2 days and then sown on half-strength Murashige and Skoog (½ MS) agar (pH 5.7). Plants were germinated and grown at 21°C in long day photoperiod (16-h light/8-h dark) and a light intensity of ∼80 µmol·m−2·s−1. White light was supplemented by Fluorescent lamp Spectralux®Plus (Radium, NL-T8 36W/840/G13). For stress treatment, plants were germinated and grown in ½ MS supplemented with 25-mM mannitol, 50-mM NaCl, or 25nM MV. To investigate the response to 3-aminotriazole (3-AT), seedlings were grown in 96-well plates in liquid ½ MS. After 10 days, 3-AT was added to each well to a final concentration of 1 mM. For soil-based growth and stress phenotyping, plants were grown for 2 weeks at 21°C with a 16-h/8-h light/dark photoperiod and then sprayed with 2-mM 3-AT for 2 days or were transferred to a growth chamber with white light (40 W m−2) supplemented with UV-B (0.4 W m−2). The cat2 and cat2 med8 phenotypes were analyzed either in long (16-h light/8-h dark) or short (8-h light/16-h dark) days in a controlled-environment growth chamber at the specified irradiance. Each of the phenotyping experiments (in vitro and in soil grown plants) was repeated at least twice. Only one representative experiment was shown, with number of biological repeats indicated where relevant. Unless stated otherwise, for each of the phenotyping experiment a minimum of 16 plants was used.
For rosette area quantification, plants were photographed and images were subsequently analyzed with Image J v1.45. The Fv′/Fm′ images were generated by an Imaging PAM series (MAXI version) chlorophyll fluorometer and ImagingWin software application (Walz; Effeltrich, Germany) was used for signal quantification.
Mutant screens and identification of ceal5
Generation of pEAL4:LUC transgenic plants and mutant screening
The 839-bp region upstream of the EAL4 start codon was amplified from Col-0 genomic DNA by PCR with Platinum Taq High Fidelity DNA polymerase (Invitrogen). The PCR product was cloned into pDONR221 to generate the entry vector and was subsequently transferred into the destination vector pKGWL7, generating a promoter-LUC reporter construct. The construct was then transformed into Col-0 plants by means of the Agrobacterium floral dip. Homozygous events with a single insertion locus were selected on ½MS medium supplemented with 35-mg L−1 kanamycin.
Seeds of the homozygous pEAL4:LUC transgenic plants were mutagenized with a 0.3% (v/v) EMS solution for 8 h and subsequently washed with water. Approximately 94,000 M2 seedlings were grown on liquid ½ MS medium in 96-well plates. Eight wells in the first column containing nonmutagenized pEAL4:LUC seeds were used as a negative control. After 10 days of growth, the PSII efficiency (Fv′/Fm′) was determined. Subsequently, 100 µL of luciferin solution (Promega ONE-Glo™ luciferase assay system) was added to individual wells. After an 8-min dark incubation, a bioluminescence signal was acquired with a LUMI star Galaxy luminometer (BMG Labtech, Ortenberg, Germany). The isolated mutants were transferred to soil to set seeds and their progeny seeds were rescreened to eliminate false positives.
Mapping and identification of causative mutations
The M3 ceal5 mutant was backcrossed to the pEAL4:LUC progenitor line and F2 mutants with constitutive LUC activity were selected, pooled, and used for extraction of nuclear DNA based on the method of Schneeberger et al. (2009). The samples were sequenced on Illumina HiSeq 2000 in a 100-bp paired-end run at ∼36-fold genome coverage. Reads were aligned to the Arabidopsis reference genome (The Arabidopsis Information Resource [TAIR] version 10) with the Burrows-Wheeler Alignment v.0.6.1 (Li and Durbin, 2009). A SHOREmap backcross strategy was used for consensus and variant calling (Schneeberger et al., 2009). With a mutation frequency cut-off of 90%, a nonsense mutation in AT2G03070 (MED8) was identified as the sole mutation located in a genic region.
LUC activity measurement and imaging
Plants were grown for 10 days in 96-well plate containing 150-µL ½ MS medium with 0.5% (w/v) sucrose and treated with H2O2, NaCl, mannitol, ABA, or MV. After 6, 12, or 24 h of treatment, 90-µL luciferin was added to each well, followed by a 6-min dark incubation. Luminescence was measured with a LUMI star Galaxy luminometer (BMG Labtech). Photos showing LUC signals were taken using IndiGO software that controls the charge-coupled device (CCD) camera in a NightShade plant imager (Berthold Technologies).
Generation of MED8 and NOT2A full-length and truncated variants
The coding sequences of MED8 and MED8ΔQ were amplified by PCR from Col-0 Arabidopsis cDNA and cloned by means of the Gateway-based cloning system. PCR products were first cloned into the pDONR221 vector and then transferred to the destination vectors pB7FWG2 (35S:MED8-GFP and 35S:MED8ΔQ-GFP), pB7WGF2 (35S:GFP-MED8), and pB7WG2 (35S:MED8) for plants or pK7FWG2 (35S:MED8-GFP and 35S:MED8ΔQ-GFP) and pK7WGF2 (35S:GFP-MED8 and 35S:GFP-MED8ΔQ) for cell suspensions. All constructs were verified by SANGER sequencing and transformed into Agrobacterium tumefaciens strain C58C1.
The constructs with the expression cassettes 35:MED8, 35:MED8-GFP, 35:GFP-MED8, 35:MED8ΔQ-GFP were transformed into med8 plants and in soil-grown bialaphos-resistant T1 transformants were selected. For cell culture transformation, cell suspension cultures (PSB-L) were cocultivated with Agrobacterium containing the N- and C-terminal GFP fusions (Van Leene et al., 2007). Cultures expressing the bait proteins were cultivated in fresh MSMO medium with gentle agitation (130 rpm).
Gene expression analysis
RNA isolation, reverse transcription, and quantitative PCR
Total RNA was isolated in triplicates from Arabidopsis seedlings or leaf tissues with the TRIzol reagent (Thermo Fisher Scientific), purified with RNeasy mini spin column (Qiagen, Hilden, Germany), and treated with DNase. RNA concentration was measured and 1 µg of RNA was used for reverse transcription (RT), carried out with the cDNA Synthesis Kit (Bio-Rad), according to the manufacturer’s instructions. Five µL of the 1:8 diluted cDNA was used as a template for qPCR with gene-specific primers (Supplemental Table 1). The data were normalized to two reference genes (as indicated in the corresponding Figure legends). Unless stated otherwise, the qPCR experiments were repeated at least twice with a minimum of three biological repeats. For each of the experiments at least 12 plants per genotype and per condition were used.
RNA-seq and data analysis
For global transcript analysis, we harvested 14-day-old Col-0 and med8 seedlings grown on ½MS (1% [w/v] sucrose) with or without 25-nM MV. In this experiment, 60 seedlings in each condition were used per genotype. Three samples were collected for each genotype and treatment and total RNA was extracted as described above. Libraries were prepared and sequenced at the Nucleomics Core (VIB, Leuven, Belgium) with Illumina Nextseq 500, resulting in ∼30 million 75-bp single-end reads per sample. Adapter sequences and low-quality base pairs (less than 20 bp) were trimmed with Trim Galore v0.3.3 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), retaining high-quality reads of at least 50 bp in length. Quality-filtered reads were aligned to the TAIR10 Arabidopsis reference genome with the spliced aligner TopHat2 v2.1.0 (Kim et al., 2013). The number of reads per gene was quantified with the featureCounts function as implemented in the Subread package v1.4.6 (Liao et al., 2014). Read mapping to genes annotated as rRNA, tRNA, and other RNA forms (TAIR10 annotation) were not considered for further analysis. As expected, in the med8 mutants, no reads were mapped to the region following the T-DNA insertion site in the MED8 locus (Supplemental Figure 9A), confirming the truncation of MED8. Differentially expressed genes were identified with the R v3.1.2 software package edgeR (Robinson et al., 2010). Genes with expression values greater than 0.12 counts per million (corresponding to five read counts) in at least three samples were retained and trim mean of M-values normalization was applied by means of the calcNormFactors function. Variability in the data set was assessed with a MDSplot employing the 3,000 top genes to calculate pairwise distances (Supplemental Figure 9B). To test user-defined hypotheses, a no-intercept single-factor model was defined combining genotype and treatment factors, such as med8_MV. Dispersions were estimated with the estimateGLMRobustDisp function. A negative binomial regression model was used to model the overdispersed counts for each gene separately with fixed values for the dispersion parameter as outlined (McCarthy et al., 2012) and as implemented in the glmFit function using the above-described model. Hypothesis testing was based on likelihood ratio tests. Contrasts of interest were the response between med8 and Col-0 under control conditions, the effect of MV stress in each genotype, and the interaction effect of MV stress and genotype. False discovery rates of the P-values were adjusted as described by Benjamini and Hochberg (1995).
Venn diagrams were made with the online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) or BioVenn (http://www.biovenn.nl/; Hulsen et al., 2008). Hierarchical clustering was generated with the MultiExperimentViewer v4.9. Gene ontology (GO) enrichment was analyzed by means of ClueGO in Cytoscape (Bindea et al., 2009) and agriGO (Tian et al., 2017).
Metabolite measurements
Oxidized and reduced glutathione were determined by spectrophotometric assays as described by Noctor et al. (2016). Briefly, 100 mg rosette tissue was ground in liquid nitrogen and then extracted with 0.2 M HCl. After centrifugation at 16,000g for 10 min at 4°C, the supernatant was neutralized to pH 5.0 and reduced and oxidized forms of glutathione were quantified by a plate-reader method (Queval and Noctor, 2007). SA was quantified by HPLC-fluorescence as in Chaouch et al. (2010). Glutathione and SA samples were harvested in triplicates, and six plants were used for each genotype per replicate.
Protein extraction and immunoblot analysis
Total proteins were extracted to homogenized material (seedlings or cells) with radioimmunoprecipitation assay (RIPA) buffer containing 1% (v/v) NP40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) sodium dodecyl sulfate, 10% (v/v) glycerol supplemented with 1-mM phenylmethane sulfonyl fluoride. After two rounds of centrifugations at 16,000g and 4°C for 20 min, the supernatant was transferred to a fresh tube and its concentration determined by the Bradford assay (Bio-Rad). For immunoblot analysis, 20-µg soluble protein was separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE; 12% Mini-PROTEAN® TGXTM precast protein gels, Bio-Rad) and blotted on a polyvinylidene fluoride (PVDF) membrane (Trans-Blot® TurboTM Mini PVDF Transfer; Bio-Rad) according to the manufacturer’s instructions. Afterward, blotted PVDF membranes were incubated with the primary antibody, 1:5,000 rabbit polyclonal anti-GFP antibody (AB290, Abcam) and the secondary antibody, 1:10,000 anti-rabbit IgG linked with horseradish peroxidase (NA934, GE Healthcare) at room temperature on an orbital shaker. The signal was captured on the chemiluminescent substrates from the Western lightning plus enhanced chemiluminescence kit (PerkinElmer).
Analysis of protein–protein interactions
Protein extraction and immunoprecipitation
The coding sequence (CDS) of NOT2a was cloned into the pK7WGR2 and pK7RWG2 vectors to generate the 35S:RFP-NOT2a and 35S:NOT2a-RFP constructs, respectively. Agrobacterium (GV3101) carrying the different constructs were used together with the p19 strain for coinfiltration into Nicotiana benthamiana leaves, while the Agrobacterium strains carrying the 35S:GFP constructs were used as a negative control. Samples were harvested 3 days after agroinfiltration, ground in liquid nitrogen, and extracted with extraction buffer containing 50-mM Tris–HCl (pH 7.5), 150-mM NaCl, 1% (v/v) NP40, 10% (v/v) glycerol supplemented with protease inhibitor cocktail (Roche). A 100 µL aliquot of total protein extract was reserved for immunoblot analysis as input. The remaining samples were incubated for 2 h at 4°C with 20-μL GFP-Trap coupled to agarose beads (Chromotek). After incubation, the beads were collected and washed three times with the wash buffer (20-mM Tris–HCl, pH 7.5, 150-mM NaCl, 0.5% (v/v) NP40). Finally, beads were resuspended in loading buffer and heated at 95°C for 10 min to dissociate immune complexes from the beads. Total (input) and immunoprecipitated proteins were analyzed by immunoblotting with anti-GFP (Roche, 1:1,000) and anti-RFP (Chromotek, 1:1,000) antibodies.
To identify potential interactors, a GFP pull-down strategy was adapted to cells expressing full-length and truncated variants of MED8 fused to GFP (Wendrich et al., 2017). Approximately 2 g of cells were harvested and ground to powder in liquid nitrogen and then solubilized in extraction buffer (50-mM Tris–HCl, pH 7.5, 0.15-M NaCl, 1 Protease inhibitor tablet/50 mL (Roche), and 1% (v/v) NP40). Protein extracts were sonicated, incubated on ice for 30 min, and NP40 diluted to 0.2%. Samples were cleared twice by 15-min centrifugation at 41,657g and 4°C and by filtering through a 40-µm cell strainer. After incubation with 100-µL anti-GFP-coated microbeads (μMACS, Miltenyi) for 2 h at 4°C, protein extracts were applied on the μColumns placed in the magnetic field of the μMACS Separator (μMACS, Miltenyi). When all the samples had been run through the columns, the columns were washed twice and after removal from the separator, the samples eluted with 50 µL of preheated ammonium bicarbonate buffer.
Sample preparation and mass spectrometry analysis
The 50-µL eluate was reduced with 500-mM dithiotreitol, alkylated by incubation with 550-mM iodocetamide. The alkylation was stopped by adding 1 µL of 200-mM cysteine. Samples were digested overnight at 20°C with 0.5-µg μL−1 trypsin sequencing grade (MS Gold, Promega) and 10% (v/v) trifluoroacetic acid was added to the samples to adjust the pH to 3.0. Subsequently, the samples were loaded onto a homemade microcolumn (C18 Empore) to remove uncut proteins and salts. Peptides were eluted with 50 µL of 50% (v/v) acetonitrile and 50% (v/v) formic acid and concentrated in a vacuum concentrator (SpeedVack). The sample concentration was determined, and equal amounts of peptides (600 ng) were injected into the column and analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) with a tandem UltiMate 3000 RSLCnano system in-line connected to an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific).
MS/MS data processing and analysis
Raw files from the LC–MS/MS runs were processed with the MaxQuant software version 1.6.9.0. Spectra were searched against the TAIR10 database containing the Arabidopsis forward/reverse sequences and frequently observed contaminants with the build-in Andromeda search engine in the label-free quantification mode. The resulting “peptides.txt” and “proteingroups.txt” output tables were used for differential protein testing by means of the MSqRob package (Goeminne et al., 2016). Peptides matching reverse/contaminant proteins or with solely a single peptide-to-spectrum match were filtered. Peptide intensities were preprocessed by quantile normalization and log2 transformation. Preprocessed peptide intensities were used to fit a robust ridge model for each protein with genotype (wild-type, MED8, or MED8ΔQ) as fixed effect, and MS/MS run, replicate, and peptide sequence as random effects with empirical Bayes variance estimator and M-estimation with Huber weights as described (Goeminne et al., 2016). Hereafter, each condition was pairwise tested, and differential proteins were defined as having a |log2 Fold Change | ≥ 1 and Q value ≤ 0.05. Besides significant quantitative differences, “singleton proteins” were selected in a qualitative manner as proteins identified either by matching or MS/MS in at least three out of six replicates in either MED8 or MED8ΔQ (MaxQuant “proteinGroups.txt”), but not identified in the wild-type.
Interactome data analysis
All 112 protein interactors (Supplemental Data Set 6) were used to create a protein-interaction network in the String database (version 11; https://string-db.org/;Szklarczyk et al., 2019). Only “Experiments” were selected as an active interaction resource by means of the default medium confidence of 0.400 as interaction score threshold. All filtered experimental interactions were exported and appended manually with pairwise interactions between the Mediator subunits (Maji et al., 2019). This interaction network was loaded in Cytoscape (version 3.8.0; Shannon et al., 2003), which was used to export graphics.
Y2H assays
The Y2H library was screened according to the HybridHunter instructions (Invitrogen). The CDS of MED8 was cloned into the pHybLex/Zeo vector (bait) and then transformed into the L40 yeast strain. The Arabidopsis cDNA library in the pYESTrp2 vector (prey) used had been cloned (Jaspers et al., 2009). The cDNA library was transformed into the bait vector-containing L40 strain and selected on SD/–Trp–His–Ura supplemented with 5-mM 3-AT to suppress autoactivation. The CDSs of positive clones were isolated by PCR and subjected to Sanger sequencing.
To verify the interaction between MED8 and NOT2a, the CDSs of MED8 and NOT2a were cloned into the pDEST32 (bait) and pDEST22 (prey) vectors, respectively. Primers used for generating these constructs are listed in Supplemental Table 1. The prey and bait constructs were cotransformed into the yeast strain pJ69-4a and selected on SD/–Leu/–Trp. To assess the protein interactions, the transformed yeasts were suspended in liquid SD/–Leu/–Trp/–His and placed on SD/–Leu/–Trp/–His/–Ade plates. The interactions were observed after 3 days of incubation at 30°C.
Statistical analyses
Samples were statistically analyzed with data analysis tools (Student’s t test: two sample assuming equal variances). Two-way ANOVA was analyzed with SPSS Statistics v25. Homogeneity of variance was verified with Levene’s test and significant differences were separated with the Tukey’s honestly significant difference test.
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: EAL4 (AT4G22530), MED8 (AT2G03070), CAT2 (AT4G35090), NOT2A, (AT1G07705), and NOT2B (AT5G59710). RNA sequencing data have been submitted to the Gene Expression Omnibus repository and have been assigned GEO accession number GSE141547. The interactome data have been submitted to the ProteomeXchange consortium via PRIDE and have been assigned the identifier number PXD016615.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure 1. Selection of H2O2-responsive luciferase reporter lines and EMS screen workflow.
Supplemental Figure 2. Response of med8 to 3-AT.
Supplemental Figure 3. Phenotype of med8 mutant in response to salt, osmotic, and UV-B stress.
Supplemental Figure 4. Validation of the RNA-sequencing results by RT-qPCR.
Supplemental Figure 5. ROS-related gene expression in the med8 RNA-seq dataset.
Supplemental Figure 6. Oxidative stress tolerance in SA- and JA-related mutants.
Supplemental Figure 7. Expression of NOT2 genes and abundance of Pri-miRNAs in med8.
Supplemental Figure 8. The cdk8 mutation prevents cell death in cat2.
Supplemental Figure 9. Overview of the med8 mutant RNA-sequencing data.
Supplemental Table 1. List of primer sequences used in this study.
Supplemental Data Set 1. List of genes differentially regulated by med8 relative to Col-0.
Supplemental Data Set 2. List of genes differentially regulated in Col-0 under MV stress relative to control conditions.
Supplemental Data Set 3. List of genes differentially regulated by med8 under MV stress relative to control conditions.
Supplemental Data Set 4. Expression analysis of ROS-responsive genes (Cluster V-ROS wheel; Willems et al., 2016) in the different genotypes and conditions.
Supplemental Data Set 5. List of MED8 positively and negatively regulated genes identified by two-way ANOVA.
Supplemental Data Set 6. List of putative Mediator complex interactors identified using MED8 and MED8ΔQ as baits by GFP pull-down experiments in Arabidopsis cell suspension.
Supplemental Data Set 7. List of putative interactors of the Mediator complex, and experimentally validated interactions from Maji et al. (2019) or the STRING database.
Supplementary Material
Acknowledgments
We are grateful to Joris Messens (VUB, Belgium) for access to the HPLC facility, Khadija Wahni for technical assistance, Inge de Clercq (Ghent University-VIB, Belgium), Pinja Jaspers, Tuomas Puukko, and Jaakko Kangasjärvi (University of Helsinki, Finland) for the kind gift of the Y2H cDNA library, and Martine De Cock for help in preparing the manuscript.
Funding
This work was supported by the Research Foundation-Flanders (The Excellence of Science [EOS] Research project 30829584), the Agency for Innovation by Science and Technology (Industrial R&D project no. 100555 and Phoenix project 070347), Ghent University (‘Bijzonder Onderzoeks Fonds’ grant no. 01J11311). H.H. is indebted to the China Scholarship Council for a PhD fellowship (CSC no. 201406350070).
Conflict of interest statement. The authors declare no conflict of interest.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Amna Mhamdi (amna.mhamdi@psb.vib-ugent.be) and Frank Van Breusegem (frank.vanbreusegem@psb.vib-ugent.be).
H.H., M.A.H., F.V.B., and A.M. designed the research. H.H. and A.M. developed the experimental strategy, performed most of the experiments, analyzed the data and prepared the figures. R.P. helped with plant growth, genotyping and stress phenotyping. J.D. and K.V.D.K. performed the mutant screen and the SHOREmap analysis. P.W. processed RNA-seq data and helped with interaction network analysis. S.Y.P. performed the Y2H screen. D.V. injected IP-MS samples and collected raw data. A.M. wrote the manuscript, with input from H.H. and F.V.B., and produced the final version. K.V.D.K., F.V.B., and A.M. supervised the research. All authors reviewed and approved the submitted manuscript.
References
- Allen BL, Taatjes DJ (2015) The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Li L, Zhai Q, You Y, Deng L, Wu F, Chen R, Jiang H, Wang H, Chen Q, et al. (2017) Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc Natl Acad Sci USA 114: E8930–E8939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arae T, Morita K, Imahori R, Suzuki Y, Yasuda S, Sato T, Yamaguchi J, Chiba Y (2019) Identification of Arabidopsis CCR4-NOT complexes with pumilio RNA-binding proteins, APUM5 and APUM2. Plant Cell Physiol 60: 2015–2025 [DOI] [PubMed] [Google Scholar]
- Asano T, Masuda D, Yasuda M, Nakashita H, Kudo T, Kimura M, Yamaguchi K, Nishiuchi T (2008) AtNFXL1, an Arabidopsis homologue of the human transcription factor NF-X1, functions as a negative regulator of the trichothecene phytotoxin-induced defense response. Plant J 53: 450–464 [DOI] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD (1999) Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283: 985–987 [DOI] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S (2007) Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Stat Methodol 57: 289–300 [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD (2002) A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J Biol Chem 277: 44202–44207 [DOI] [PubMed] [Google Scholar]
- Bourbon H-M (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendía-Monreal M, Gillmor CS (2016) Mediator: a key regulator of plant development. Dev Biol 419: 7–18 [DOI] [PubMed] [Google Scholar]
- Canet JV, Dobón A, Tornero P (2012) Non-recognition-of-BTH4, an Arabidopsis Mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24: 4220–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, Holub EB, Cahill DM, Manners JM, Schenk PM, et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol 160: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G (2012) AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, Mhamdi A, Vandorpe M, Langlois-Meurinne M, Van Breusegem F, Saindrenan P, Noctor G (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Mohan R, Zhang Y, Li M, Chen H, Palmer IA, Chang M, Qi G, Spoel SH, Mengiste T, et al. (2019). NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol 181: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors .Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA (2016) The Ccr4-Not complex is a key regulator of eukaryotic gene expression. WIREs RNA 7: 438–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DG, Fassler JS (2019) Med15: glutamine-rich Mediator subunit with potential for plasticity. Trends Biochem. Sci 44: 737–751 [DOI] [PubMed] [Google Scholar]
- Crawford T, , Karamat F, , Lehotai N, , Rentoft M, , Blomberg J,, Strand Å,, Björklund S (2020) Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Scientific Reports 10: 5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Brosché M, Shapiguzov A, He X-Q, Vainonen JP, Leppälä J, Trotta A, Kangasjärvi S, Salojärvi J, Kangasjärvi J, et al. (2019) Interaction of methyl viologen-induced chloroplast and mitochondrial signalling in Arabidopsis. Free Radic Biol Med 134: 555–566 [DOI] [PubMed] [Google Scholar]
- Davoine C, Abreu IN, Khajeh K, Blomberg J, Kidd BN, Kazan K, Schenk PM, Gerber L, Nilsson O, Moritz T, et al. (2017) Functional metabolomics as a tool to analyze Mediator function and structure in plants. PLoS ONE 12: e179640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dghim AA, Mhamdi A, Vaultier M-N, Hasenfratz-Sauder M-P, Le Thiec D, Dizengremel P, Noctor G, Jolivet Y (2013) Analysis of cytosolic isocitrate dehydrogenase and glutathione reductase 1 in photoperiod-influenced responses to ozone using Arabidopsis knockout mutants. Plant Cell Environ 36: 1981–1991 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, AbuQamar S, Du H-N, Briggs SD, Mittelsten Scheid O, Mengiste T (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the Mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K-J (2014). Redox regulation of transcription factors in plant stress acclimation and development. Antioxid Redox Signal 21: 1356–1372 [DOI] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467 [DOI] [PubMed] [Google Scholar]
- Ding Y-H, Liu N-Y, Tang Z-S, Liu J, Yang W-C (2006) Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan WL, Chapple C (2017) Conservation and divergence of Mediator structure and function: insights from plants. Plant Cell Physiol 58: 4–21 [DOI] [PubMed] [Google Scholar]
- Dolan WL, Dilkes BP, Stout JM, Bonawitz ND, Chapple C (2017) Mediator complex subunits MED2, MED5, MED16, and MED23 genetically interact in the regulation of phenylpropanoid biosynthesis. Plant Cell 29: 3269–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ (2000) Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci USA 97: 14307–14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi N, Maji S, Waseem M, Thakur P, Kumar V, Parida SK, Thakur JK (2019) The Mediator subunit OsMED15a is a transcriptional co-regulator of seed size/weight-modulating genes in rice. Biochim Biophys Acta 1862: 194432. [DOI] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle G, et al. (2011) The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc Natl Acad Sci USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJB, Holmberg S, Hebert H, Gustafsson CM (2006) The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA 103: 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallath T, Kidd BN, Stiller J, Davoine C, Björklund S, Manners JM, Kazan K, Schenk PM (2017) MEDIATOR18 and MEDIATOR20 confer susceptibility to Fusarium oxysporum in Arabidopsis thaliana. PLoS ONE 12: e0176022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA 93: 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeminne LJE, Gevaert K, Clement L (2016) Peptide-level robust ridge regression improves estimation, sensitivity, and specificity in data-dependent quantitative label-free shotgun proteomics. Mol Cell Proteomics 15: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G (2013a) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Mhamdi A, Chaouch S, Noctor G (2013b) Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ 36: 1135–1146 [DOI] [PubMed] [Google Scholar]
- He H, Van Breusegem F, Mhamdi A (2018) Redox-dependent control of nuclear transcription in plants. J Exp Bot 69: 3359–3372 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis Mediator complex subunits MED16, MED14, and MED2 regulate Mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert CW, Moscou MJ, Hewitt T, Steuernagel B, Hernández-Pinzón I, Green P, Pujol V, Zhang P, Rouse MN, Jin Y, et al. (2020) Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nat Commun 11: 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Huang J, Sun Y, Orduna AR, Jetter R, Li X (2019) The Mediator kinase module acts as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J 98: 842–852 [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W (2008) BioVenn- a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé A, Vanderauwera S, Hoeberichts FA, Vandorpe M, Van Gaever T, Van Breusegem F (2012) A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ 35: 308–320 [DOI] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, et al. (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim Y-J (2001) The structural and functional organization of the yeast Mediator complex. J Biol Chem 276: 42003–42010 [DOI] [PubMed] [Google Scholar]
- Kerchev P, Mühlenbock P, Denecker J, Morreel K, Hoeberichts FA, Van Der Kelen K, Vandorpe M, Nguyen L, Audenaert D, Van Breusegem F (2015) Activation of auxin signalling counteracts photorespiratory H2O2-dependent cell death. Plant Cell Environ 38: 253–265 [DOI] [PubMed] [Google Scholar]
- Kerchev P, Waszczak C, Lewandowska A, Willems P, Shapiguzov A, Li Z, Alseekh S, Mühlenbock P, Hoeberichts FA, Huang J, et al. (2016) Lack of GLYCOLATE OXIDASE1, but not GLYCOLATE OXIDASE2, attenuates the photorespiratory phenotype of CATALASE2-deficient Arabidopsis. Plant Physiol 171: 1704–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The Mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Mugford SG, Ülker B, Gao D, Thorlby G, Knight MR (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun D-J, Mengiste T (2014) MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat Commun 5: 3064. [DOI] [PubMed] [Google Scholar]
- Lee M-H, Jeon HS, Kim HG, Park OK (2017) An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol 214: 343–360 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mhamdi A, Trotta A, Kangasjärvi S, Noctor G (2014) The protein phosphatase subunit PP2A-B'γ is required to suppress day length-dependent pathogenesis responses triggered by intracellular oxidative stress. New Phytol 202: 145–160 [DOI] [PubMed] [Google Scholar]
- Li W, Yoshida A, Takahashi M, Maekawa M, Kojima M, Sakakibara H, Kyozuka J (2015) SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J 81: 282–291 [DOI] [PubMed] [Google Scholar]
- Li X, Yang R, Chen H (2018) The Arabidopsis thaliana Mediator subunit MED8 regulates plant immunity to Botrytis cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS ONE 13: e0193458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu Q, Bischof S, Harris CJ, Zhong Z, Zhan L, Nguyen C, Rashoff A, Barshop WD, Sun F, Feng S, et al. (2020) The characterization of Mediator 12 and 13 as conditional positive gene regulators in Arabidopsis. Nat Commun 11: 2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, Lu Y, Wang Q, Li C, Zhai Q (2019) MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 31: 106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji S, Dahiya P, Waseem M, Dwivedi N, Bhat DS, Dar TH, Thakur JK (2019) Interaction map of Arabidopsis Mediator complex expounding its topology. Nucleic Acids Res 47: 3904–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Kim JI, Wheeler MT, Heintzelman AK, Weake VM, Chapple C (2019) Mutation of Mediator subunit CDK8 counteracts the stunted growth and salicylic acid hyperaccumulation phenotypes of an Arabidopsis MED5 mutant. New Phytol 223: 233–245 [DOI] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK (2011) The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Van Breusegem F (2018) Reactive oxygen species in plant development. Development 145: dev164376. [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou J-P, et al. (2010b) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Kerchev PI, Willems P, Noctor G, Van Breusegem F (2017) Measurement of transcripts associated with photorespiration and related redox signaling. Methods Mol Biol 1653: 17–29 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G (2010a) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61: 4197–4220 [DOI] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD (1999) Mediator protein mutations that selectively abolish activated transcription. Proc Natl Acad Sci USA 96: 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagulapalli M, Maji S, Dwivedi N, Dahiya P, Thakur JK (2016) Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res 44: 1591–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJK, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M (2013) A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Lelarge-Trouverie C, Mhamdi A (2015) The metabolomics of oxidative stress. Phytochemistry 112: 33–53 [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Foyer CH (2016) Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant Cell Environ 39: 1140–1160 [DOI] [PubMed] [Google Scholar]
- Noctor G, Reichheld J-P, Foyer CH (2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80: 3–12 [DOI] [PubMed] [Google Scholar]
- Pan S, Li K-e, Huang W, Zhong H, Wu H, Wang Y, Zhang H, Cai Z, Guo H, Chen X, Xia Y (2020) Arabidopsis DXO1 possesses deNADding and exonuclease activities and its mutation affects defense-related and photosynthetic gene expression. J Integr Plant Biol 62: 967–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín F, Yuste-Lisbona FJ, Pineda B, García-Sogo B, del Olmo I, de Dios Alche J, Egea I, Flores FB, Pineiro M, Jarillo JA, et al. (2018) Developmental role of the tomato Mediator complex subunit MED18 in pollen ontogeny. Plant J 96: 300–315 [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Queval G, Neukermans J, Vanderauwera S, Van Breusegem F, Noctor G (2012) Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ 35: 374–387 [DOI] [PubMed] [Google Scholar]
- Quevedo M, Meert L, Dekker MR, Dekkers DHW, Brandsma JH, van den Berg DLC, Ozgür Z, van IJcken WFJ, Demmers J, Fornerod M, et al. (2019) Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat Commun 10: 2669 [Erratum Nat. Commun. 10: 3318] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman V, Anand A, Vasudevan B, Morsy MR, Pant BD, Lee H-K, Tang Y, Mysore KS (2019) Overexpression of VIRE2-INTERACTING PROTEIN2 in Arabidopsis regulates genes involved in Agrobacterium-mediated plant transformation and abiotic stresses. Sci Rep 9: 13503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Aguilar B, Raya-González J, López-Bucio JS, Reyes de la Cruz H, Herrera-Estrella L, Ruiz-Herrera LF, Martínez-Trujillo M, López-Bucio J (2020) Mutation of MEDIATOR 18 and chromate trigger twinning of the primary root meristem in Arabidopsis. Plant Cell Environ 43: 1989–1999 [DOI] [PubMed] [Google Scholar]
- Samanta S, Thakur JK (2015) Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front Plant Sci 6: 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S, Thakur JK (2017) Characterization of Mediator complex and its associated proteins from rice. Methods Mol Biol 1629: 123–140 [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K (2013) Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol 163: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Høgh Petersen A, Lehmann Nielsen K, Jørgensen J-E, Weigel D, Uggerjøh Andersen S (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Shaikhali J, Davoine C, Brännström K, Rouhier N, Bygdell J, Björklund S, Wingsle G (2015) Biochemical and redox characterization of the mediator complex and its associated transcription factor GeBPL, a GLABROUS1 enhancer binding protein. Biochem J 468: 385–400 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45: W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, et al. (2010) Perturbation in indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UTG74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A, Wrzaczek M, Scharte J, Tikkanen M, Konert G, Rahikainen M, Holmström M, Hiltunen H-M, Rips S, Sipari N, et al. (2011) Regulatory subunit B'γ of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol 156: 1464–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K-L, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ (2013) A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction. Nat Struct Mol Biol 20: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, et al. (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al. (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inzé D, Van Breusegem F (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Suzuki N, Miller G, van de Cotte B, Morsa S, Ravanat J-L, Hegie A, Triantaphylidès C, Shulaev V, Van Montagu MCE, et al. (2011) Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci USA 108: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollsnes AV, Erikson AB, Otterholt E, Kvaal K, Oxaal U, Futsaether C (2009) Visible foliar injury and infrared imaging show that daylength affects short‐term recovery after ozone stress in Trifolium subterraneum. J Exp Bot 60: 3677–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Du X, Mou Z (2016) The Mediator complex subunits MED14, MED15, and MED16 are involved in defense signaling crosstalk in Arabidopsis. Front Plant Sci 7: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins JA, Mou Z (2015) The Arabidopsis Mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol 169: 856–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wei H, Tong Z, Zhang X, Yang Z, Lan T, Duan Y, Wu W (2011) Knockdown of NtMed8, a Med8-like gene, causes abnormal development of vegetative and floral organs in tobacco (Nicotiana tabacum L.). Plant Cell Rep 30: 2117–2129 [DOI] [PubMed] [Google Scholar]
- Wang L, Song X, Gu L, Li X, Cao S, Chu C, Cui X, Chen X, Cao X (2013) NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25: 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J (2018) Reactive oxygen species in plant signaling. Annu Rev Plant Biol 69: 209–236 [DOI] [PubMed] [Google Scholar]
- Wathugala DL, Hemsley PA, Moffat CS, Cremelie P, Knight MR, Knight H (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol 195: 217–230 [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Boeren S, Möller BK, Weijers D, De Rybel B (2017) In vivo identification of plant protein complexes using IP-MS/MS. Methods Mol Biol 1497: 147–158 [DOI] [PubMed] [Google Scholar]
- Willems P, Mhamdi A, Stael S, Storme V, Kerchev P, Noctor G, Gevaert K, Van Breusegem F (2016) The ROS wheel: refining ROS transcriptional footprints. Plant Physiol 171: 1720–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Li Y (2011) Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 454–4554 [DOI] [PubMed] [Google Scholar]
- Xu R, Li Y (2012) The Mediator complex subunit 8 regulates organ size in Arabidopsis thaliana. Plant Signal Behav 7: 182–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TH, Lenglet-Hilfiker A, Stolz S, Glauser G, Farmer EE (2020) Jasmonate precursor biosynthetic enzymes LOX3 and LOX4 control wound-response growth restriction. Plant Physiol 184: 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li C (2019) The plant Mediator complex and its role in jasmonate signaling. J Exp Bot 70: 3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li L, An C, Li C (2018) Conserved function of mediator in regulating nuclear hormone receptor activation between plants and animals. Plant Signal Behav 13: e1403709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z (2012) The Arabidopsis Mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z (2013) The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J 75: 484–497 [DOI] [PubMed] [Google Scholar]
- Zhou H-R, Lin R-N, Huang H-W, Li L, Cai T, Zhu J-K, Chen S, He X-J (2020) The CCR4-NOT complex component NOT1 regulates RNA-directed DNA methylation and transcriptional silencing by facilitating Pol IV-dependent siRNA production. Plant J 103: 1503–1515 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Huang P, Guo P, Chong L, Yu G, Sun X, Hu T, Li Y, Hsu CC, Tang K, et al. (2020) CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. DOI: 10.1111/nph.16787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.