Abstract
Light is a key environmental cue that fundamentally regulates plant growth and development, which is mediated by the multiple photoreceptors including the blue light (BL) photoreceptor cryptochrome 1 (CRY1). The signaling mechanism of Arabidopsis thaliana CRY1 involves direct interactions with CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)/SUPPRESSOR OF PHYA-105 1 and stabilization of COP1 substrate ELONGATED HYPOCOTYL 5 (HY5). H2A.Z is an evolutionarily conserved histone variant, which plays a critical role in transcriptional regulation through its deposition in chromatin catalyzed by SWR1 complex. Here we show that CRY1 physically interacts with SWC6 and ARP6, the SWR1 complex core subunits that are essential for mediating H2A.Z deposition, in a BL-dependent manner, and that BL-activated CRY1 enhances the interaction of SWC6 with ARP6. Moreover, HY5 physically interacts with SWC6 and ARP6 to direct the recruitment of SWR1 complex to HY5 target loci. Based on previous studies and our findings, we propose that CRY1 promotes H2A.Z deposition to regulate HY5 target gene expression and photomorphogenesis in BL through the enhancement of both SWR1 complex activity and HY5 recruitment of SWR1 complex to HY5 target loci, which is likely mediated by interactions of CRY1 with SWC6 and ARP6, and CRY1 stabilization of HY5, respectively.
Blue light-activated cryptochrome 1 interacts with SWR1 subunits SWC6 and ARP6 to regulate H2A.Z deposition at HY5 target genes promoting cell elongation and photomorphogenesis.
Introduction
Light not only provides an energy source for photosynthesis in plants, but also serves as a pivotal environmental cue that modulates numerous plant developmental processes (Fankhauser and Chory, 1997; Deng and Quail, 1999; Yadav et al., 2020). Multiple photoreceptors have evolved to allow plants to monitor and respond to dynamic changes in wavelengths of light, light direction, and light duration. These include blue/ultraviolet (UV)-A light photoreceptors cryptochromes (CRYs; Cashmore et al., 1999; Wang and Lin, 2020) and phototropins (Briggs and Christie, 2002) and ZTL/FKF1/LKP2 (Ito et al., 2012), red/far-red light (RL/FRL) photoreceptors phytochromes (Quail, 2002), and UV-B light photoreceptor UVB-RESISTANCE 8 (Rizzini et al., 2011). CRYs act as the major blue light (BL) receptors in Arabidopsis to regulate photomorphogenesis, photoperiodic flowering, and stomatal development and closure (Ahmad and Cashmore, 1993; Guo et al., 1998; Mao et al., 2005; Kang et al., 2009). CRYs are present not only in land plants, but also other organisms from algae to humans. In mammals, CRYs act as a critical component of the core oscillator complex of the circadian clock (Kume et al., 1999; van der Horst et al., 1999). In addition, CRYs in the eyes of migratory birds confer their ability to visually detect Earth’s magnetic field and navigate long-distance flying during migration (Gegear et al., 2010).
Arabidopsis has two homologous CRYs, CRY1, and CRY2. CRY1 primarily mediates BL regulation of photomorphogenesis characterized by inhibited hypocotyl elongation and enhanced anthocyanin accumulation (Lin et al., 1998), while CRY2 plays a major role in photoperiodic flowering (Guo et al., 1998). CRY1 and CRY2 each comprise an N-terminal photolyase homologous region domain (CNT1 and CNT2) and a C-terminal extension domain (CCE, or CCT1 and CCT2; Cashmore et al., 1999; Yu et al., 2007). The C-terminal domain of CRYs mediates BL signaling via its direct interaction with CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase that acts as the master negative regulator of photomorphogenesis and flowering time (Deng et al., 1992; Yang et al., 2000; Wang et al., 2001; Yang et al., 2001). This interaction leads to inhibition of COP1 activity and accumulation of transcription factors such as ELONGATED HYPOCOTYL 5 (HY5) and CONSTANS (Osterlund et al., 2000; Jang et al., 2008; Liu et al., 2008b). HY5 is a basic leucine zipper (bZIP) transcription factor that acts as a major positive regulator of photomorphogenesis by directly binding to light-responsive genes to modulate their expression (Oyama et al., 1997; Chattopadhyay et al., 1998). CRY1 also interacts with the COP1 enhancer, SUPPRESSOR OF PHYA-105 1 (SPA1; Seo et al., 2003), to further attenuate COP1 activity (Lian et al., 2011; Liu et al., 2011). It has been demonstrated that CNT1 alone is sufficient to mediate CRY1 signaling independent of CCT1 (He et al., 2015), and several CNT1-interacting proteins have been characterized. These include Aux/IAAs, ARF6/ARF8, BIM1/BES1, HBI1, and TOE1/TOE2, which are transcriptional regulators (Xu et al., 2018; Wang et al., 2018a, 2018b; Du et al., 2020; Mao et al., 2020). The direct interactions of CRYs with transcription regulators such as CIBs, TOE1/TOE2, PIFs, Aux/IAAs, ARF6/ARF8, and BIM1/BES1 can influence their DNA-binding or transcriptional activities to regulate flowering, thermomorphogenesis, shade avoidance, auxin, and brassinosteroids signaling (Liu et al., 2008a; Ma et al., 2016; Pedmale et al., 2016; Xu et al., 2018; Wang et al., 2018a, 2018b; Du et al., 2020; Mao et al., 2020).
H2A.Z is an evolutionarily conserved histone variant identified from yeasts to human species, which plays a critical role in transcriptional regulation (Zlatanova and Thakar, 2008). The exchange of H2A for H2A.Z in the nucleosomes is catalyzed by ATP-dependent SWI2/SNF2-Related 1 chromatin remodeling complex (SWR1-C; Mizuguchi et al., 2004). In yeast, the SWR1 complex contains 14 subunits, in which the Swr1 ATPase is the core catalytic subunit and serves as scaffold protein for the complex assembly (Gerhold and Gasser, 2014). ARP6 and SWC6 subunits interact with Swr1 and act as a bridge between Swr1 and H2A.Z chaperon SWC2 (Wu et al., 2005). Several conserved homologs of SWR1-C subunits have been characterized in Arabidopsis, including ARP6, SWC6/SEF, and PIE1 (homolog of yeast Swr1; Noh and Amasino, 2003; Deal et al., 2005; March-Diaz et al., 2007). Arabidopsis SWC6 and ARP6 interact with each other and are required for H2A.Z deposition (Choi et al., 2007; Deal et al., 2007; March-Diaz et al., 2007). SWR1-C-mediated H2A.Z deposition into chromatin can lead to a repressive chromatin state and repression of gene expression, but also can confer transcriptional competence (Deal et al., 2007; Kumar et al., 2012). In Arabidopsis, H2A.Z deposition at +1 nucleosome and gene body can result in tight wrapping of DNA or promotion of H3K27me3 and prevention of H3K4me3 histone modifications and repression of gene expression (Kumar and Wigge, 2010; Dai et al., 2017). At low temperature, H2A.Z is deposited at a high level which leads to transcription repression at FT and YUC8 loci, and at high-temperature H2A.Z is evicted from the nucleosome at these loci to promote flowering and hypocotyl elongation (Kumar et al., 2012; van der Woude et al., 2019). In mammals, the transcription factors p53 and c-Myc direct the location of H2A.Z occupancy in chromatin (Gevry et al., 2007). In Arabidopsis, the transcriptional factors ELF3 and ESC/SOB3 interact with SWC6 to recruit SWR1-C at PRR7/PRR9 and YUC9 loci to repress their expression to shape oscillatory gene expression and inhibit hypocotyl growth, respectively (Lee and Seo, 2017; Tong et al., 2020).
To date, whether SWR1-C-mediated H2A.Z deposition is involved in light signaling is largely unknown. In this study, we identified SWC6 and ARP6 as CRY1-interacting proteins. We demonstrate through a series of protein–protein interaction assays that Arabidopsis CRY1 interacts with SWC6 and ARP6 in a BL-dependent manner. We further demonstrate through physiological, biochemical, genetic, and molecular studies that SWC6, ARP6, and H2A.Z act to promote photomorphogenesis under BL, RL, and FRL, and that CRYs, SWC6, ARP6, and HY5 play a positive role in regulating H2A.Z deposition at HY5 target genes promoting cell elongation in BL. We also show that both SWC6 and ARP6 physically interact with HY5. Moreover, we demonstrate that BL-activated CRY1 likely enhances the interaction of SWC6 with ARP6. Our findings suggest that SWR1-C is recruited to HY5 target genes, promoting cell elongation and chlorophyll biosynthesis to mediate H2A.Z deposition through direct interactions of SWC6 and ARP6 with HY5, leading to repression of these HY5 target genes expression and promotion of photomorphogenesis under BL. BL-induced interactions of CRY1 with SWC6 and ARP6 and/or CRY1-enchaced SWC6–ARP6 association may contribute to BL promotion of SWR1-C-mediated H2A.Z deposition at HY5 target genes.
Results
CRY1 and CRY2 physically interact with SWC6 in a BL-dependent manner
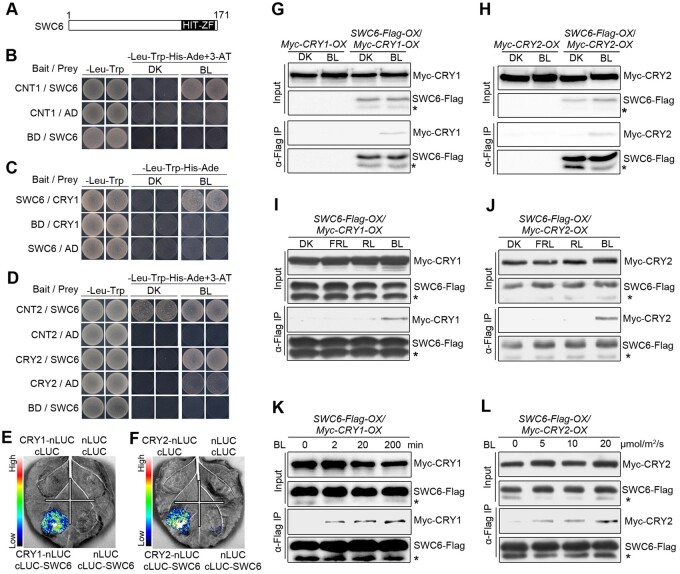
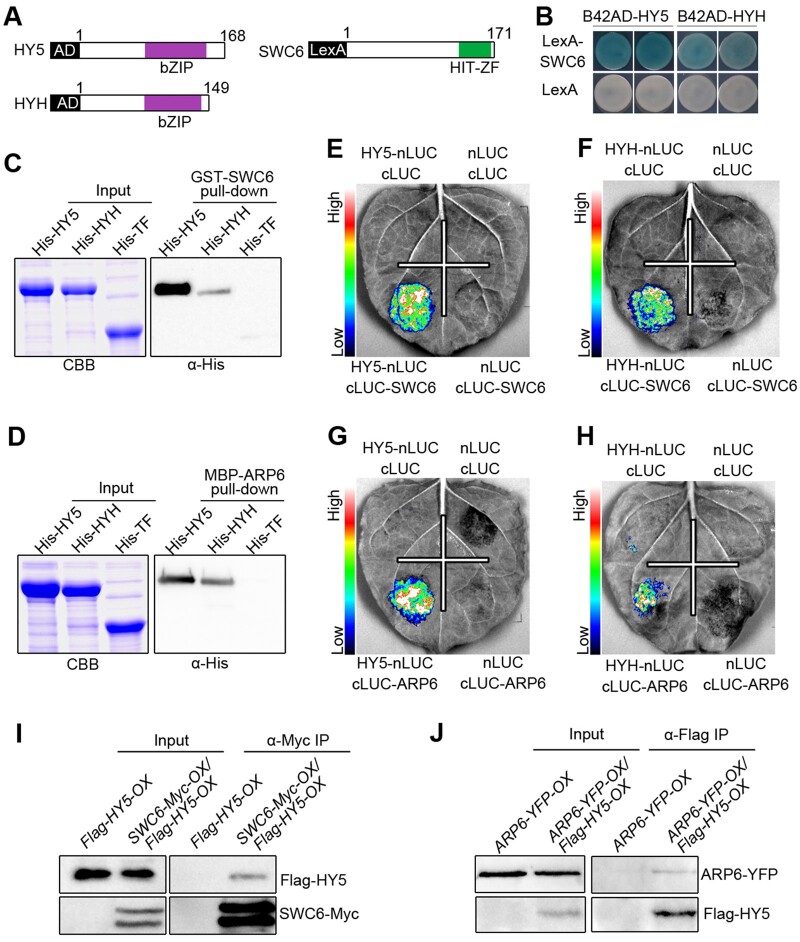
To look for more potential CRY1-interactiong proteins, we performed GAL4 yeast two-hybrid screening using CNT1 as a bait, and identified SWC6, a conserved core subunit of SWR1-C harboring a hit-type zinc finger (HIT-ZF) domain (Figure 1A) responsible for exchange of histone H2A.Z variant in eukaryotic organisms (Mizuguchi et al., 2004; March-Diaz et al., 2007; Cuadrado et al., 2010), which interacts with CNT1 in BL but not in darkness (DK; Figure 1B). We then examined by yeast two-hybrid assay whether the full-length CRY1 would interact with SWC6 and found that CRY1 also interacted with SWC6 in BL but not in the DK (Figure 1C). Furthermore, CNT2 interacted with SWC6 in both BL and DK, while the full-length CRY2 interacted with SWC6 in BL but not in DK (Figure 1D). Moreover, domain mapping assays demonstrated that the C-terminal HIT-ZF-containing domain of SWC6 (amino acids 86–171) mediated the interaction with CRY1 and CRY2 (Supplemental Figure S1). In vitro pull-down assays showed that CNT1, CCT1, CNT2, and CCT2 were pulled down by GST-SWC6 (Supplemental Figure S2, A and B). We then carried out split luciferase (split-LUC) complementation assays in Nicotiana benthamiana leaves and found that the LUC activity was reconstituted when cLUC-SWC6 and CRY1-nLUC or CRY2-nLUC were coexpressed (Figure 1, E and F), indicating interactions of CRY1 and CRY2 with SWC6. Further split-LUC assays confirmed that both N and C termini of CRY1 and CRY2 were able to interact with SWC6 (Supplemental Figure S2, C–F).
Figure 1.
CRY1 and CRY2 interact with SWC6 in a BL-dependent manner. A, Schematic diagram depicting the HIT-ZF domain of SWC6 protein. B, Yeast two-hybrid assays showing the interaction of CNT1 with SWC6. Yeast cells coexpressing the indicated combinations of constructs were grown on SD –Trp–Leu or SD–Trp–Leu–His–Ade medium with 10 mM 3-AT in continuous DK or BL (30 μmol/m2/s). BD, GAL4 DNA-binding domain; AD, GAL4 DNA-activation domain . C and D, Yeast two-hybrid assays showing the interactions of CRY1 with SWC6 (C), and CNT2 and CRY2 with SWC6 (D). E and F, Split-LUC assays indicating the interactions of CRY1 and CRY2 with SWC6 in N. benthamiana cells. G and H, Co-IP assays showing BL-induced interactions of CRY1 and CRY2 with SWC6 in Arabidopsis. Myc-CRY1-OX or Myc-CRY2-OX or SWC6-Flag-OX/Myc-CRY1-OX or SWC6-Flag-OX/Myc-CRY2-OX seedlings were adapted in DK for 2 d and then exposed to BL (30 μmol/m2/s for CRY1 and 20 μmol/m2/s for CRY2) or still adapted in DK for 1 h, followed by IP with an anti-Flag antibody. The IP (SWC6) and co-IP signals (CRY1 and CRY2) were detected by immunoblots probed with anti-Flag and -Myc antibodies. Asterisks denote truncated SWC6-Flag protein. I and J, Co-IP assay showing BL-specific interactions of CRY1 and CRY2 with SWC6 in Arabidopsis. SWC6-Flag-OX/Myc-CRY1-OX or SWC6-Flag-OX/Myc-CRY2-OX seedlings were adapted in DK for 2 d and then exposed to BL (20 μmol/m2/s) or RL (20 μmol/m2/s) or FRL (5 μmol/m2/s) light or still adapted in DK for 1 h, followed by IP with anti-Flag antibody. K, Co-IP assay showing that CRY1–SWC6 interaction is enhanced in response to increasing exposure times of BL. SWC6-Flag-OX/Myc-CRY1-OX seedlings were adapted in DK for 2 d and then exposed to BL (30 μmol/m2/s) for different times. L, Co-IP assay showing that CRY2–SWC6 interaction is enhanced in response to increasing intensity of BL. SWC6-Flag-OX/Myc-CRY2-OX seedlings were adapted in DK for 2 d and then exposed to different fluence rates of BL for 1 h.
To confirm the BL-dependent interactions of CRY1 and CRY2 with SWC6 observed in Saccharomyces cerevisiae AH109 strain cells (Figure 1, C and D), we performed semi-in vivo pull-down assays and found that SWC6 pulled-down Myc-CRY1 and Myc-CRY2 in the extracts prepared from Myc-CRY1-OX and Myc-CRY2-OX seedlings exposed to BL, respectively, but not from those either adapted in DK or exposed to RL or FRL (Supplemental Figure S3, A and C), indicating BL-specific interactions of CRY1 and CRY2 with SWC6. Further semi-in vivo pull-down assays demonstrated that CRY1–SWC6 and CRY2–SWC6 interactions were enhanced in seedlings exposed to either BL for increasing times (Supplemental Figure S3B) or higher fluence rates of BL (Supplemental Figure S3D).
Next, we performed co-immunoprecipitation (co-IP) assays to further confirm whether CRY1 and CRY2 might interact with SWC6 in a BL-dependent manner in Arabidopsis. To do this, we generated double transgenic Arabidopsis plants overexpressing SWC6-Flag and Myc-CRY1 (SWC6-Flag-OX/Myc-CRY1-OX), as well as SWC6-Flag and Myc-CRY2 (SWC6-Flag-OX/Myc-CRY2-OX). Co-IP assays with these transgenic seedlings showed that both Myc-CRY1 and Myc-CRY2 were coimmunoprecipitated with SWC6-Flag in BL, but not in DK (Figure 1, G and H). We then determined the effects of different lights on the interactions of CRY1 and CRY2 with SWC6 by co-IP assays, and found that both CRY1 and CRY2 interacted with SWC6 in BL, but not in DK or RL or FRL (Figure 1, I and J). Further co-IP assay showed that the interactions of CRY1 and CRY2 with SWC6 were enhanced in seedlings exposed to BL for longer times or at higher fluence rates (Figure 1, K and L). Taken together, these results demonstrate that both CRY1 and CRY2 interact with SWC6 in a BL-dependent manner in vivo.
CRY1 and CRY2 physically interact with ARP6 in a BL-dependent manner
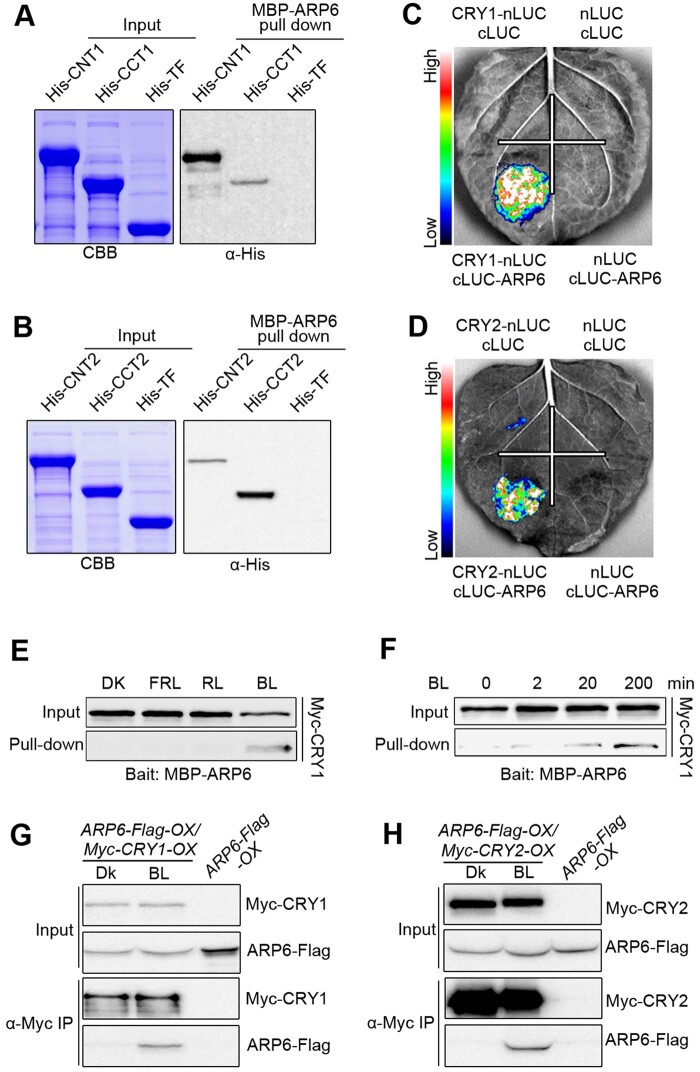
Since SWC6 interacts with ARP6, another core catalytic subunit of SWR1-C, to regulate SWR1-C-mediated H2A.Z deposition (Wu et al., 2005; Choi et al., 2007; March-Diaz et al., 2007), we explored whether CRYs would interact with ARP6. We first performed pull-down assays, and found that CNT1, CCT1, CNT2, and CCT2 interacted with ARP6 (Figure 2, A and B). We then performed split-LUC assays and found that CRY1, CRY2, CNT1, CCT1, CNT2, and CCT2 each interacted with ARP6 in N. benthamiana (Figure 2, C and D; Supplemental Figure S2, G–J). To investigate the effects of BL on the interaction of CRY1 with SARP6, we performed semi-in vivo pull-down assays and found that CRY1 interacted with ARP6 in BL, but not in DK or RL or FRL (Figure 2E). Moreover, the interaction of CRY1 with ARP6 was enhanced in seedlings exposed to BL for increasing times (Figure 2F). We further performed co-IP assays with transgenic Arabidopsis overexpressing ARP6-Flag and Myc-CRY1 (ARP6-Flag-OX/Myc-CRY1-OX) or ARP6-Flag and Myc-CRY2 (ARP6-Flag-OX/Myc-CRY2-OX) and found that both CRY1 and CRY2 interacted with ARP6 in BL, but not in DK (Figure 2, G and H). Taken together, these data demonstrate that both CRY1 and CRY2 also interact with ARP6 in a BL-dependent manner in vivo.
Figure 2.
CRY1 and CRY2 interact with ARP6 in a BL-dependent manner. A and B, In vitro pull-down assays showing the interactions of CNT1 and CCT1 with ARP6 (A), and CNT2 and CCT2 with ARP6 (B). MBP-ARP6 protein served as bait. His-TF, His-TF-CNT1, -CCT1, -CNT2, and -CCT2 served as prey. His-TF was used as negative control. The pulled-down proteins were detected with anti-His antibody. C and D, Split-LUC assays indicating the interactions of CRY1 (C) and CRY2 (D) with ARP6 in N. benthamiana cells. E, Semi-in vivo pull-down assay showing BL-specific interaction of CRY1 with ARP6. Preys were protein extracts from Myc-CRY1-OX seedlings that were DK-adapted for 2 d and then exposed to BL (30 μmol/m2/s) or RL (30 μmol/m2/s) or FRL (5 μmol/m2/s) light or still adapted in DK for 1 h. F, Semi-in vivo pull-down assay showing that CRY1–ARP6 interaction is enhanced in response to increasing exposure times of BL. Preys were protein extracts from Myc-CRY1-OX seedlings that were DK-adapted for 2 d and then exposed to BL (30 μmol/m2/s) for different times. G and H, Co-IP assays showing BL-induced interactions of CRY1 and CRY2 with ARP6 in Arabidopsis. Myc-CRY1-OX Myc-CRY2-OX or ARP6-Flag-OX/Myc-CRY1-OX or ARP6-Flag-OX/Myc-CRY2-OX seedlings were adapted in DK for 2 d and then exposed to BL (30 μmol/m2/s) or still adapted in DK for 1 h, followed by IP with anti-Flag antibody. The IP (ARP6) and co-IP signals (CRY1 and CRY2) were detected by immunoblots probed with anti-Flag and -Myc antibodies.
SWC6, ARP6, and H2A.Z act to inhibit hypocotyl elongation in BL, RL, and FRL, and promote chlorophyll accumulation in BL
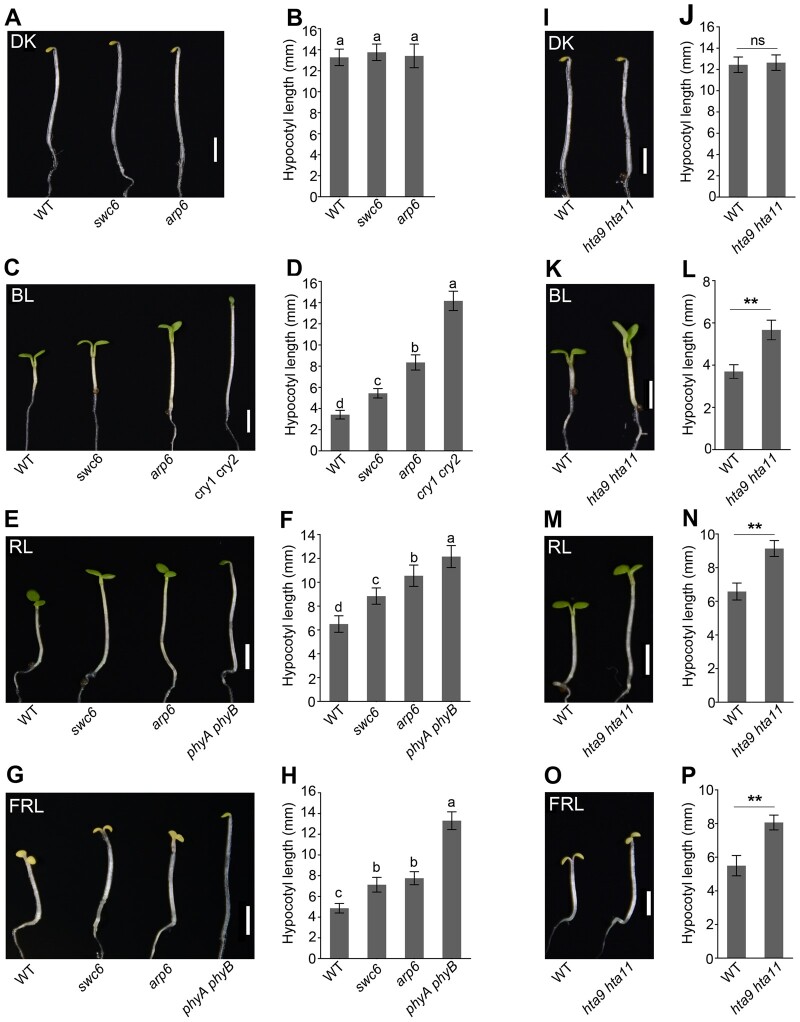
The demonstrations that CRY1 and CRY2 interact directly with SWC6 and ARP6 suggest that SWC6 and ARP6 might be involved in CRY-mediated BL signaling. To explore this possibility, we analyzed the photomorphogenic phenotype shown by hypocotyl lengths of swc6 and arp6 mutants grown in DK and BL, as well as in RL and FRL. Intriguingly, both swc6 and arp6 mutant hypocotyls were as long as those of wild-type (WT) in the DK, but significantly longer than WT not only in BL, but in RL and FRL as well (Figure 3, A–H). Moreover, expression of SWC6-Flag or SWC6-Myc and ARP6-Flag or ARP6-YFP was able to rescue the tall hypocotyl phenotype of swc6 and arp6 mutants in BL, respectively (Supplemental Figure S4). These results demonstrate that both SWC6 and ARP6 may not only be involved in inhibiting hypocotyl elongation in BL, but in RL and FRL signaling as well.
Figure 3.
SWC6, ARP6, and H2A.Z act to inhibit hypocotyl elongation in BL, RL, and FRL. A–H, The swc6, and arp6 mutants show enhanced hypocotyl elongation in BL, RL, and FRL. Seedlings of the indicated genotypes were grown in DK (A), BL (30 μmol/m2/s) (C), RL (50 μmol/m2/s) (E), and FRL (1 μmol/m2/s) (G) for 5 d, and hypocotyl lengths were measured (B, D, F, and H). Bars = 2.5 mm. Letters “a” to “d” indicate statistically significant differences between means for hypocotyl lengths of the indicated genotypes, as determined by ANOVA, followed by LSD test (P < 0.05). I–P, The hta9 hta11 mutant shows enhanced hypocotyl elongation in BL, RL, and FRL. The growth conditions are the same as those in (A–H). Bars = 2.5 mm. Asterisks indicate significant differences between WT and hta9 hta11 seedlings (Student’s t-test: **P < 0.01), and ns denotes no significant differences.
We then examined whether H2A.Z would also be involved in light signaling by using the loss-of-function mutant of Arabidopsis H2A.Z, which comprises three highly homologous genes, HTA8, HTA9, and HTA11 (March-Diaz and Reyes, 2009). We generated the hta9 hta11 double mutant and examined its hypocotyl elongation in different light conditions. The results showed that, like swc6 and arp6 mutants, hta9 hta11 mutant developed significantly taller hypocotyls than WT in BL, RL, and FRL, but not in DK (Figure 3, I–P), indicating that H2A.Z also inhibits hypocotyl elongation in these monochromatic light conditions. We further determined chlorophyll and anthocyanin contents in swc6, arp6, and hta9 hta11 mutants in BL. The results showed that chlorophyll accumulation was clearly reduced to different degrees in these mutants (Supplemental Figure S5A). However, anthocyanin production was dramatically enhanced in swc6 and hta9 hta11mutants, and slightly promoted in arp6 mutant (Supplemental Figure S5B). Taken together, these results suggest that SWC6, ARP6, and H2A.Z act to inhibit hypocotyl elongation and promote chlorophyll accumulation but repress anthocyanin production.
Next, we explored the genetic interaction of CRY1 and CRY2 with ARP6 using the cry1 cry2 arp6 triple mutant created through genetic crossing and comparing the hypocotyl phenotype of this mutant with that of cry1 cry2 and arp6 mutants in BL. The results showed that cry1 cry2 arp6 triple mutant hypocotyls were significantly taller than arp6 single mutant, but similar to cry1 cry2 double mutant (Supplemental Figure S6, A and B), indicating that ARP6 likely acts in the same pathway of CRY1 and CRY2 to regulate hypocotyl elongation in BL.
phyB and phyA may interact with SWC6 and ARP6
Given the demonstrations that CRY1 and CRY2 interact with SWC6 and ARP6 (Figures 1 and 2), and that SWC6, ARP6, and H2A.Z also act to inhibit hypocotyl elongation in RL and FRL (Figure 3, E–H and M–P), we postulated that phytochromes B and A (phyB and phyA) might interact with SWC6 and ARP6. To test this possibility, we performed split-LUC assays in N. benthamiana leaves and found that the LUC activity was detected when cLUC-phyB was coexpressed with SWC6-nLUC and ARP6-nLUC, and phyA-nLUC was coexpressed with cLUC-SWC6 and cLUC-ARP6, respectively (Supplemental Figure S7), indicating possible interactions of phyB and phyA with SWC6 and ARP6.
CRYs and ARP6 coregulate a large number of genes in the same direction in BL
To find the downstream target genes coregulated by CRYs and ARP6, we performed RNA-seq assays using cry1 cry2 and arp6 mutant grown in BL and identified 5,769 CRY-regulated genes (differentially expressed genes [DEGs] between cry1 cry2 vs. WT; fold change (FC) > 1.5, and P-value < 0.05) and 5,017 ARP6-regulated genes (DEGs between arp6 vs. WT; FC > 1.5, and P-value < 0.05; Figure 4A). There were 1,879 genes coregulated by CRYs and ARP6, of which 802 (43%) and 580 (31%) were down-regulated and up-regulated by both CRYs and ARP6, respectively (Figure 4A; Supplemental Data Set S1). We then calculated the Pearson correlation coefficients between CRY- and ARP6-coregulated genes expression and found that there was moderate correlation of expression changes between CRY- and ARP6-coregulated genes (r value = 0.410). A heat map generated by hierarchical clustering analyses using Pearson correlation revealed that 1,382 (74%) of these genes were regulated by CRYs and ARP6 in the same direction (Figure 4B). Next, we performed gene ontology (GO) enrichment analysis and found that the genes coregulated by CRYs and ARP6 were preferentially associated with response to light stimulus and plant hormones, cell wall organization, and chloroplast part (Figure 4C). Further analysis revealed that ARP6-repressed genes were enriched in GO term related to hormone signaling, which are closely related to hypocotyl elongation, while ARP6-activated genes were enriched in GO terms related to photosynthesis and chloroplast part (Figure 4C). The categories of cell wall organization and response to light stimulus were enriched in both ARP6-repressed and ARP6-activated genes (Figure 4C).
Figure 4.
CRYs, ARP6, and HY5/HYH coregulate a large number of genes in the same direction. A, Venn diagrams showing the overlapping genes up- and down-regulated by CRYs and ARP6, which were obtained by analyses of differential expression (FC > 1.5 and P-value < 0.05) from samples of BL-grown WT and cry1 cry2 mutant, and BL-grown WT and arp6 mutant, respectively. B, Hierarchical clustering analyses of the overlapping genes shown in A. Scale bar denotes the log2 value of FC. C, GO analysis showing coordinate regulation of the 1,879 overlapping genes in A by CRYs and ARP6. The numbers on each column denote the percentage of genes in each GO category. Total denotes all Arabidopsis genes. D, Venn diagrams showing the overlapping genes up- and down-regulated by ARP6 and HY5/HYH, which were obtained by analyses of differential expression (FC > 1.5 and P-value < 0.05) from samples of BL-grown WT and arp6 mutant, and BL-grown WT and hy5 hyh mutant, respectively. E, Hierarchical clustering analyses of the overlapping genes shown in D. F, GO analysis showing coordinate regulation of the 2,051 overlapping genes in D by ARP6 and HY5/HYH. The numbers on each column denote the percentage of genes in each GO category.
Given that HY5 is a key transcription factor acting downstream of the multiple photoreceptors to promote photomorphogenesis (Oyama et al., 1997; Chattopadhyay et al., 1998), we asked whether the genes coregulated by CRYs and ARP6 might include the HY5 target genes. To do this, we compared the genes identified in this study with the 5,948 HY5-taget genes characterized previously (Lee et al., 2007; Kurihara et al., 2014). We found that 562 (30%) of the genes coregulated by CRYs and ARP6 were HY5 target genes and included genes promoting cell elongation, such as EXP2, IAA19, XTH32, GH3.5, SAUR41, and BBX31. These results, in conjunction with the pronounced tall-hypocotyl phenotype observed for both cry1 cry2 and arp6 mutants grown in BL (Figure 3, C and D), implying that CRYs and ARP6 may promote photomorphogenesis, at least in part, through repression of the HY5 target genes promoting cell elongation under BL.
ARP6 and HY5 coregulate a large number of genes in the same direction in BL
Given that the genes coregulated by CRYs and ARP6 contain a large number of HY5 target genes, we further performed RNA-seq assays using hy5 hyh mutant seedlings grown in BL, in which HY5 and its close homolog HYH (Holm et al., 2002) were mutated, and analyzed the genes regulated by HY5/HYH in BL. We identified 4,652 HY5/HYH-regulated genes (DEGs between hy5 hyh vs. WT; FC > 1.5, and P-value < 0.05), of which 2,051 genes were also regulated by ARP6 (Figure 4D; Supplemental Data Set S2). Of them, 1,012 (49%) and 799 (39%) were down- and up-regulated by both ARP6 and HY5/HYH, respectively. We then calculated the Pearson correlation coefficients between ARP6- and HY5/HYH-coregulated genes expression and found that there was high correlation of expression changes between ARP6- and HY5/HYH-coregulated genes (r value = 0.749). A heat map generated by hierarchical clustering analyses revealed that 1,811 (88%) of these genes were regulated by ARP6 and HY5/HYH in the same direction (Figure 4E). GO analysis showed that the genes responsive to growth-promoting hormones and regulation of hormone level, as well as those involved in cell wall organization, were highly enriched in the genes that are up-regulated by ARP6 and HY5/HYH (Figure 4F). Importantly, these results confirmed that the genes coregulated by ARP6 and HY5/HYH identified here also included the HY5 target genes promoting hypocotyl elongation such as EXP2, IAAs, XTHs, SAURs, PRE5, and chlorophyll accumulation and photosynthesis such as HEMA1 and RBCS1A. RT-quantitative PCR (RT-qPCR) analyses of the BL-grown cry1 cry2, arp6, and hy5 hyh mutants confirmed that CRYs, ARP6, and HY5/HYH repressed the expression of EXP2, IAAs, XTHs, SAURs, and PRE5 (Supplemental Figure S8), but promoted the expression of HEMA1, RBCS1A, and other genes promoting chlorophyll biosynthesis and photosynthesis (Ang et al., 1998; McCormac and Terry, 2002; Supplemental Figure S9), confirming that CRYs, ARP6, and HY5/HYH regulate hypocotyl elongation and chlorophyll accumulation, at least in part, through regulation of the expression of genes controlling cell elongation and chlorophyll biosynthesis in BL.
CRYs mediate BL-enhanced H2A.Z deposition at the HY5 target genes promoting cell elongation
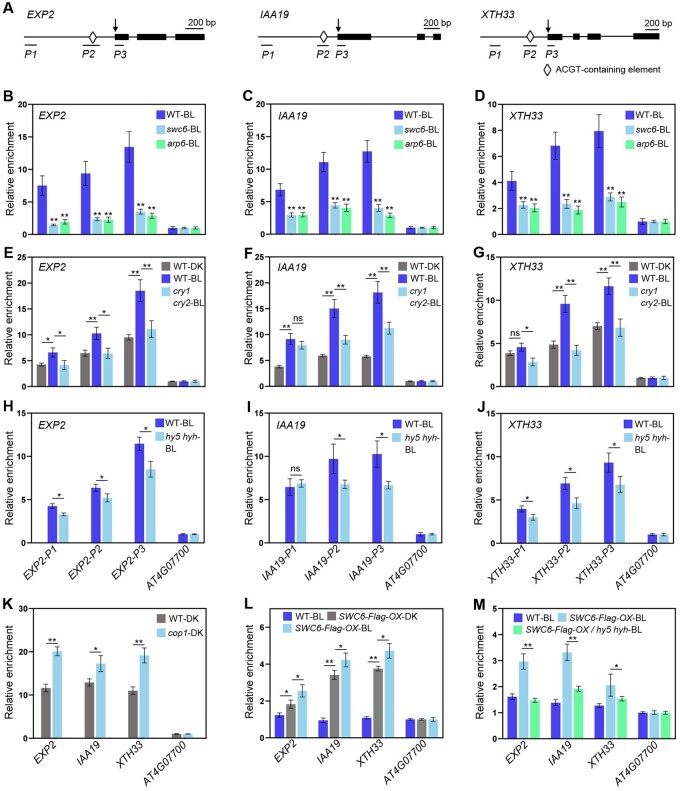
Given that ARP6 and SWC6 are required for SWR1-C-mediated H2A.Z deposition (Deal et al., 2007; March-Diaz et al., 2007) and that they both act to promote photomorphogenesis in BL (Figure 3, C and D), we examined whether SWC6 and ARP6 might promote H2A.Z deposition at the HY5 target genes promoting cell elongation to inhibit their expression under BL. To do this, we performed chromatin IP coupled with qPCR (ChIP-qPCR) assays of three representative HY5 target genes, EXP2, IAA19, and XTH33 (Figure 5A), which are known to promote cell elongation (Jing et al., 2013; Pedmale et al., 2016), using the swc6 and arp6 seedlings to evaluate H2A.Z occupancies at these loci with the anti-HTA9 antibody. The H2A.Z occupancies at EXP2, IAA19, and XTH33 loci were dramatically reduced in either swc6 or arp6 mutant (Figure 5, B–D), indicating that both SWC6 and ARP6 are required for H2A.Z deposition at +1 nucleosomes and HY5-binding sites of these loci containing the ATCG element in BL. Next, we investigated H2A.Z deposition at two more HY5 target genes, HEMA1 and RBCS1A (Ang et al., 1998; McCormac and Terry, 2002; Supplemental Figure S10A), which are involved in chlorophyll biosynthesis and photosynthesis. We found that H2A.Z occupancies at HEMA1 and RBCS1A loci were also significantly reduced in either swc6 or arp6 mutant (Supplemental Figure S10, B and C), suggesting that H2A.Z deposition at these two genes is largely mediated by SWC6 and ARP6. We then evaluated the effects of BL and CRYs on the H2A.Z deposition at these loci using WT and cry1 cry2 seedlings. As shown in Figure 5, E–G, significantly more H2A.Z was enriched at EXP2, IAA19, and XTH33 loci in WT seedlings exposed to BL than in those adapted in DK, while considerably less H2A.Z occupied at these loci in cry1 cry2 seedlings than in WT exposed to BL. Immunoblot analysis demonstrated that H2A.Z protein expressed at a similar level in WT and cry1 cry2 mutant seedlings adapted in the DK or exposed to BL (Supplemental Figure S11, A and B). Taken together, these results demonstrate that BL is able to enhance H2A.Z deposition and that CRYs mediate BL-enhanced H2A.Z deposition at the HY5 target genes promoting cell elongation.
Figure 5.
ChIP-qPCR analyses showing the roles of CRYs, ARP6, SWC6, and HY5/HYH in H2A.Z deposition at HY5 target genes. A, Schematic diagram of EXP2, IAA19, and XTH33 genes with exons indicated as black boxes. Arrowheads denote the transcription start sites. P1–P3 denote the corresponding amplicons for qPCR. B–D, ARP6 and SWC6 are necessary for H2A.Z deposition at +1 nucleosomes of EXP2 (B), IAA19 (C) and XTH33 (D) loci. E–G, CRYs mediate BL-enhanced H2A.Z deposition at the HY5 Target Genes in BL. H–J, HY5 and HYH promote H2A.Z deposition at their target genes in BL. K, H2A.Z deposition at +1 nucleosomes of HY5 target loci is enhanced in cop1-4 mutant. L and M, SWC6 binding to the HY5 target genes promoting cell elongation is dependent on HY5 and HYH in BL. In B–J, L, and M, 5-d-old white light-grown seedlings were adapted in DK for 2 d, and then either exposed to BL (50 μmol/m2/s) or still adapted in DK for 6 h. In K, 5-d-old white light-grown WT and cop1 seedlings were adapted in DK for 2 d, and then harvested and cross-linked. The +1 nucleosome fragments of EXP2, IAA19, XTH33, and AT4G07700 (negative control) were immunoprecipitated by anti-HTA9 or anti-Flag antibody, and then qPCR was performed to quantify the enrichment of the indicated genes, which was normalized to that of AT4G07700. Error bars, SDs of three biological replicates. Asterisks indicate significant differences between different genotypes of plants subjected to different treatments (Student’s t-test: **P < 0.01, *P < 0.05).
HY5 and HYH promote H2A.Z deposition at their target genes
To evaluate whether HY5 and HYH might be required for H2A.Z deposition at their target genes, we performed ChIP-qPCR assays for H2A.Z deposition at EXP2, IAA19, and XTH33 loci using hy5 hyh seedlings adapted in DK and then exposed to BL, respectively. Significantly more H2A.Z was enriched at +1 nucleosomes and HY5-binding sites of these loci in WT than in hy5 hyh mutant seedlings exposed to BL (Figure 5, H–J), immunoblot analysis demonstrated that the H2A.Z protein expressed at a similar level in WT and hy5 hyh mutant seedlings exposed to BL (Supplemental Figure S11C). To further confirm the role for HY5 in promoting H2A.Z deposition, we carried out ChIP-qPCR assays using DK-grown cop1 mutant that is known to accumulate high levels of HY5 (Osterlund et al., 2000). As expected, much more H2A.Z was enriched at EXP2, IAA19, and XTH33 loci in DK-grown cop1 mutant than in DK-grown WT (Figure 5K). Taken together, these results demonstrate that HY5 and HYH act to positively regulate H2A.Z deposition at HY5 target genes promoting cell elongation.
SWC6 association with the HY5 target genes is partially dependent on HY5 in BL
Our demonstrations that HY5 and HYH play a role in promoting H2A.Z deposition and that SWC6 positively regulates H2A.Z deposition led us to explore whether SWC6 might associate with HY5 target genes in BL, and if so, whether HY5 and HYH might be required for its binding. To this end, we generated transgenic lines overexpressing similar levels of SWC6-Flag protein in the WT and hy5 hyh mutant backgrounds, respectively (SWC6-Flag-OX and SWC6-Flag-OX/hy5 hyh; Supplemental Figure S11E). ChIP-qPCR assays using SWC6-Flag-OX seedlings demonstrated that the association of SWC6 to EXP2, IAA19, and XTH33 loci was significantly stronger in SWC6-Flag-OX seedlings exposed to BL than in those adapted in the DK (Figure 5L). Protein blot analysis showed that SWC6-Flag protein was expressed at a similar level in SWC6-Flag-OX seedlings adapted in DK or exposed to BL (Supplemental Figure S11D). Given that HY5 is degraded in the DK but stabilized in the light (Osterlund et al., 2000), these results indicate that BL might promote SWC6 binding to HY5 target genes by stabilizing HY5. To test this possibility, we further performed ChIP-qPCR assays using SWC6-Flag-OX and SWC6-Flag-OX/hy5 hyh seedlings. As anticipated, SWC6 association with EXP2, IAA19, and XTH33 loci was significantly stronger in the WT background than in the hy5 hyh mutant background under BL (Figure 5M). Taken together, these results suggest that SWC6 binding to HY5 target genes requires HY5 and HYH in BL.
HY5 physically interacts with both SWC6 and ARP6
The following demonstrations led us to explore whether HY5 and HYH would interact directly with SWC6 and ARP6 to recruit SWR1-C to promote H2A.Z deposition at the HY5 target genes. (1) ARP6 regulates a large number of HY5 target genes. (2) SWC6 and ARP6 are required for H2A.Z deposition at the HY5 target genes. (3) HY5/HYH act to positively regulate H2A.Z deposition at their target genes. (4) SWC6 association with the H2A.Z deposition at the HY5 target genes requires HY5 and HYH. To test this hypothesis, we first performed LexA yeast two-hybrid assays and found that SWC6 interacted with both HY5 and HYH (Figure 6, A and B). We further demonstrated that the C terminus of SWC6 (amino acids 86–171) mediated the interaction with HY5 (Supplemental Figure S12), and that the C terminus of HY5 (amino acids 78–168) mediated the interaction with SWC6 (Supplemental Figure S13, A and B). We then performed GST pull-down assays and confirmed that SWC6 interacted with HY5 and HYH (Figure 6C), and that the C terminus of HY5 mediated its interaction with SWC6 (Supplemental Figure S13C). Further pull-down assays showed that ARP6 also interacted with HY5 and HYH (Figure 6D). Next, we carried out split-LUC assays, and found that SWC6 and ARP6 interacted with both HY5 and HYH, respectively (Figure 6, E–H). To further confirm the SWC6–HY5 and ARP6–HY5 interactions in vivo, we generated transgenic Arabidopsis overexpressing SWC6-Myc and Flag-HY5 (SWC6-Myc-OX/Flag-HY5-OX) or ARP6-YFP and Flag-HY5 (ARP6-YFP-OX/Flag-HY5-OX) and performed co-IP assays. The results showed that HY5 interacted with both SWC6 and ARP6 in vivo (Figure 6, I and J).
Figure 6.
SWC6 and ARP6 interact with HY5 and HYH. A, Schematic diagram of HY5, HYH, and SWC6 proteins tested in LexA yeast two-hybrid assays. bZIP denotes basic leucine zipper domain. B, LexA yeast two-hybrid assays showing interactions of SWC6 with HY5 and HYH. Yeast cells coexpressing the indicated combinations of constructs were grown on SD –Trp–His–Ura with X-gal. The blue precipitates on the plates represent the GUS activities. C and D, In vitro pull-down assays showing the interactions of HY5 and HYH with SWC6 (C) and ARP6 (D). GST-SWC6 and MBP-ARP6 protein served as baits. His-TF, His-TF-HY5, and -HYH served as preys. Input images show CBB staining. The pulled-down proteins were detected with anti-His antibody. E–H, Split-LUC assays indicating the interactions of SWC6 with HY5 (E) and HYH (F), and the interactions of ARP6 with HY5 (G) and HYH (H). I and J, Co-IP assays showing the interactions of HY5 with SWC6 (I) and ARP6 (J) in Arabidopsis. Flag-HY5-OX and SWC6-Myc-OX/Flag-HY5-OX seedlings were grown in BL (30 μmol/m2/s) for 7 d, followed by IP with anti-Myc antibody. The IP (SWC6) and co-IP signals (HY5) were detected by immunoblots probed with anti-Myc and -Flag antibodies (I). ARP6-YFP-OX and ARP6-YFP-OX/Flag-HY5-OX seedlings were grown in BL (30 μmol/m2/s) for 7 d, followed by IP with anti-Flag antibody. The IP (HY5) and co-IP signals (ARP6) were detected by immunoblots probed with anti-Flag and -YFP antibodies (J).
Given that ARP6 physically interacts with HY5 and HYH (Figure 6), we next examined the genetic interaction between ARP6 and HY5/HYH by constructing the arp6 hy5 hyh triple mutant through genetic crossing. Hypocotyl phenotype analysis showed that the arp6 hy5 hyh triple mutant developed significantly taller hypocotyls than either arp6 or hy5 hyh mutant under BL (Supplemental Figure S6, C and D). These results suggest that ARP6 and HY5 may not only function in the same pathway, but in separate pathways to regulate hypocotyl development as well in BL.
BL-activated CRY1 likely enhances the association of SWC6 with ARP6
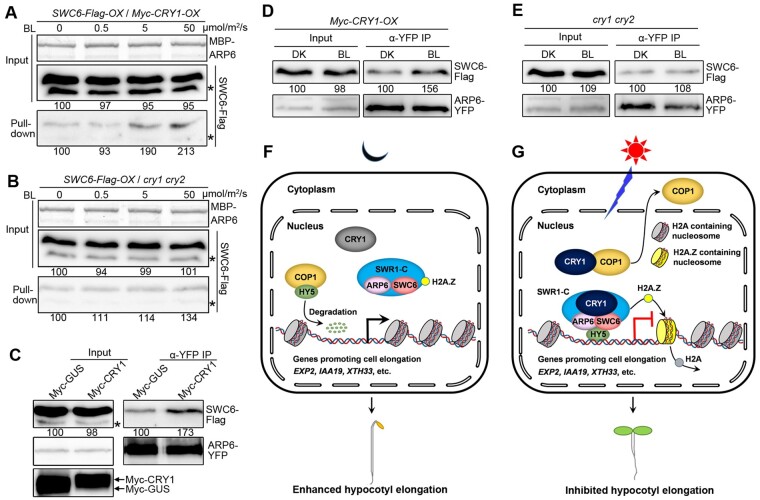
To explore whether SWC–ARP6 interaction would be affected by CRY1, we first performed semi-in vivo pull-down assays using MBP-ARP6 protein as bait, and the Arabidopsis protein extracts containing SWC6-Flag protein plus or minus CRY1 as prey, which were prepared from the DK-adapted seedlings overexpressing SWC6-Flag in Myc-CRY1-overexpressor background (SWC6-Flag-OX/Myc-CRY1-OX) or cry1 cry2 mutant background (SWC6-Flag-OX/cry1 cry2) exposed to different fluence rates of BL. The results showed that the capacity of MBP–ARP6 binding to SWC6-Flag was much more enhanced in SWC6-Flag-OX/Myc-CRY1-OX than in SWC6-Flag-OX/cry1 cry2 seedlings (Figure 7, A and B). We then performed co-IP assays using N. benthamiana leaves expressing SWC6-Flag and ARP6-YFP plus Myc-CRY1, or Myc-β-galactosidase (GUS), and found that SWC6-Flag interacted with ARP6-YFP much more strongly in the presence of Myc-CRY1 than in the presence of Myc-GUS control (Figure 7C). Furthermore, we performed co-IP assays with Myc-CRY1-OX and cry1 cry2 mutant protoplasts coexpressing SWC6-Flag and ARP6-YFP and found that much more SWC6-Flag was coimmunoprecipitated with ARP6-YFP in Myc-CRY1-OX protoplasts irradiated with BL than adapted in DK (Figure 7D), whereas similar amount of SWC6-Flag was coimmunoprecipitated with ARP6-YFP in cry1 cry2 protoplasts either adapted in the DK or exposed to BL (Figure 7E). Taken together, these results demonstrate that BL-activated CRY1 may enhance the association of SWC6 with ARP6 through BL-dependent CRY1–SWC6/ARP6 interactions.
Figure 7.
CRY1 likely enhances the association of SWC6 with ARP6. A and B, Semi-in vivo pull-down assays showing that CRY1 may promote SWC6–ARP6 interaction in a BL fluence rate-dependent manner. MBP-ARP6 protein served as bait. SWC6-Flag protein extracts prepared from SWC6-Flag-OX/Myc-CRY1-OX and SWC6-Flag-OX/cry1 cry2 seedlings adapted in DK for 2 d and then exposed to different fluence rates of BL for 1 h served as preys. C, Co-IP assays showing that CRY1 may promote the interaction of SWC6 with ARP6 in N. benthamiana. SWC6-Flag and ARP6-YFP were coexpressed with Myc-GUS or Myc-CRY1 in N. benthamiana leaves, respectively. ARP6-YFP served as bait, and SWC6-Flag served as prey. Myc-GUS and Myc-CRY1 were detected with anti-Myc antibody. D and E, Co-IP assays showing CRY1 promotion of the association of SWC6 with ARP6 by BL in Arabidopsis protoplasts. SWC6-Flag and ARP6-YFP were coexpressed in Myc-CRY1-OX (D) and cry1 cry2 protoplasts (E). The transformed protoplasts were DK-adapted for 16 h, and then exposed to BL (30 μmol/m2/s) or still adapted to DK for 1 h. SWC6-Flag served as prey and ARP6-YFP served as bait. F and G, A model illustrating how CRY1 mediates regulation of H2A.Z deposition at HY5 target loci. In DK, CRY1 is inactive and unable to interact with COP1 and SWC6/ARP6 to regulate their activities. COP1 is fully functional, while SWR1 complex is not. HY5 undergoes ubiquitination and degradation, and SWR1 complex can hardly be recruited to HY5 target loci to mediate H2A.Z deposition, leading to active expression of HY5 target genes promoting cell elongation and enhanced hypocotyl elongation (F). Upon BL irradiation, CRY1 is activated, and interacts with ARP6 and SWC6 to enhance ARP6–SWC6 interaction, leading to enhanced SWR1 complex activity. At the same time, CRY1 interacts with COP1 to inhibit its activity and promote its translocation from nucleus to cytoplasm, leading to promotion of HY5 accumulation and enhanced recruitment of SWR1 complex to HY5 target loci. These two pathways work together to enhance SWR1 complex-mediated H2A.Z deposition at HY5 target genes to regulate their expression and mediate CRY1 inhibition of hypocotyl elongation (G).
Discussion
Light and temperature are the two key environmental factors that fundamentally influence plant growth and development. Since these factors fluctuate widely, plants have evolved a variety of receptors and regulatory protein complex that enable plants to respond rapidly and accurately to the ambient temperature and light, and fine tune the status of growth and development. They include the multiple photoreceptors that sense light signals, phyB, and ELF3 that sense temperature (Jung et al., 2016; Legris et al., 2016; Jung et al., 2020), and the SWR1 complex that mediates H2A.Z deposition and thermosensory responses (Kumar and Wigge, 2010). Whether SWR1 complex-mediated H2A.Z deposition is involved in light signaling is largely unknown.
In this study, we identified the core subunits of SWR1 complex SWC6 and ARP6 as CRY1-interacting proteins and demonstrate that CRY1 and CRY2 interact with SWC6 and ARP6 in a BL-dependent manner, respectively (Figures 1 and 2; Supplemental Figures S1–S3). The significance of these interactions was tested as follows. (1) SWC6, ARP6, and H2A.Z function to promote photomorphogenesis characterized by inhibited hypocotyl elongation and enhanced chlorophyll accumulation in BL (Figure 3; Supplemental Figure S5A), and ARP6 acts in the same pathway of CRY1/CRY2 to repress hypocotyl elongation under BL (Supplemental Figure S6, A and B). (2) CRYs, ARP6, and HY5/HYH coregulate a large number of genes in the same direction in BL, some of which are associated with cell elongation and chlorophyll biosynthesis (Figure 4; Supplemental Figures S8 and S9). (3) Both ARP6 and SWC6 are essential for H2A.Z deposition at representative HY5-taget genes involved in hypocotyl elongation and chlorophyll biosynthesis under BL (Figure 5, B–D; Supplemental Figure S10), and CRY1/CRY2 are responsible for mediating BL-enhanced H2A.Z deposition at HY5 target genes such as EXP2, IAA19, and XTH33 (Figure 5, E–G). Interestingly, H2A.Z represses the expression of genes promoting cell elongation such as EXP2, IAA19, and XTH33, but promotes the expression of genes involved in chlorophyll biosynthesis and photosynthesis such as HEMA1 and RBCAS1 (Supplemental Figures S8 and S9). Based on previous studies showing that H2A.Z deposition leads to repression of gene expression or constitutive transcriptional competence (Deal et al., 2007; Kumar and Wigge, 2010; Dai et al., 2017; Su et al., 2017), and our results, we postulate that CRY1-mediated BL signaling promotes H2A.Z deposition at HY5 target genes to regulate their expression and promote photomorphogenesis under BL.
How does CRY1 regulate H2A.Z deposition? In mammals, PCI domain-containing protein 2 interacts with ZNHIT1 (homolog of SWC6) to inhibit its interaction with SRCAP (homolog of SWR1), which represses the SRACR activity and H2A.Z deposition (Ye et al., 2017). In yeast, SWC6 and ARP6 subunits interact with each other and are mutually dependent for their associations with the core catalytic subunit of SWR1 complex, Swr1 ATPase (Wu et al., 2005), indicating that the SWC6–ARP6 interaction may affect their interactions with Swr1 to maintain the SWR1 complex activity. The cryo-electron microscopy analysis of the structure of yeast SWR1–nucleosome complex demonstrates that SWC6 and ARP6 form a tight heterodimer for H2A.Z exchange activity (Willhoft et al., 2018). In mammals, the phosphorylation of ZNHIT1 is essential for its interaction with ARP6, and the dephosphorylation of ZNHIT1 abolishes its interaction with ARP6 and H2A.Z deposition (Cuadrado et al., 2010). Based on these reports and the following demonstrations, we speculate that CRY1 promotion of H2A.Z deposition may be mediated through promotion of SWR1 complex activity by BL-dependent interactions of CRY1 with SWC6 and ARP6. (1) Both SWC6 and ARP6 are essential for H2A.Z deposition at the HY5 target genes promoting photomorphogenesis under BL (Figure 5, B–D; Supplemental Figure S10). (2) CRY1 mediates BL-enhanced H2A.Z deposition (Figure 5, E–G). (3) CRY1 interacts with both SWC6 and ARP6 in a BL-dependent manner, which may enhance the association of SWC6 with ARP6 (Figure 7, A–E).
It is interesting to note that SWC6, ARP6, and H2A.Z also act to promote photomorphogenesis in RL and FRL (Figure 3, E–H and M–P), and that phyB and phyA may interact with SWC6 and ARP6, respectively (Supplemental Figure S7). Previous studies have demonstrated that both CRY and phytochrome interact with COP1 and SPAs to regulate photomorphogenesis (Seo et al., 2004; Jang et al., 2010; Lu et al., 2015; Sheerin et al., 2015), and that both CRY and phyB interact directly with AUX/IAA proteins in a BL and RL-dependent manner, respectively, to stabilize these proteins and inhibit auxin signaling (Xu et al., 2018). Furthermore, both CRY and phyB interact with BES1/BZR1 to inhibit brassinosteroid signaling (Wang et al., 2018b; Wu et al., 2019; Dong et al., 2020). Given these reports and the findings obtained in this study, it is possible that phyB and phyA may also interact with SWC6 and ARP6 to regulate H2A.Z deposition at HY5 target genes and photomorphogenesis in RL and FRL.
To mediate H2A.Z deposition at target genes, SWR1 complex has to be recruited to the target genes. It has been demonstrated that transcription factors can interact with SWR1 complex subunits including SWC6 and ARP6 to recruit it to the target gene loci (Lee and Seo, 2017; Su et al., 2017; Tong et al., 2020). Moreover, the chromatin remolding factor PICKEL is recruited by HY5 at its target loci including EXP2 and IAA19 (Jing et al., 2013). In this study, we demonstrate that HY5 is responsible for recruiting SWR1 complex at its target loci under BL to deposit H2A.Z (Figure 5, H–J). A previous study shows that HY5 does not possess the transcription activation or repression domain (Ang et al., 1998), and it has been hypothesized that HY5 may activate and repress target gene expression through binding to its partners, which is regulated by light (Kindgren et al., 2012; Burko et al., 2020). SWC6 and ARP6 may be such partners of HY5, which mediate HY5 repression of its target genes promoting cell elongation by promoting H2A.Z deposition in light.
The loss of function of either SWC6 or ARP6 leads to a severe decrease in H2A.Z deposition at HY5 target loci such as EXP2, IAA19, XTH33, HEMA1, and RBCS1A under BL (Figure 5, B–D; Supplemental Figure S10), indicating that both SWC6 and ARP6 are essential for H2A.Z deposition. By contrast, H2A.Z deposition at HY5 target loci and SWC6 association with HY5 target loci in hy5 hyh mutant background are not severely reduced (Figure 5, H–J and M), implying that HY5 and HYH are not exclusively responsible for H2A.Z deposition at HY5 target loci. Moreover, the genetic interaction analysis showing that arp6 hy5 hyh mutant hypocotyls are significantly longer than those of the hy5 hyh mutant in BL (Supplemental Figure S6, C and D) indicates that other transcription factors may be involved in ARP6-mediated regulation of hypocotyl growth. Indeed, it has been shown that many HY5 target genes including the three representative HY5 target genes used in our ChIP-qPCR assays are also the target genes of other transcriptional factors acting in light and/or phytohormone signaling. For examples, IAA19 is the shared direct target gene of HY5, PIF4, ARF6, and BZR1 which are able to bind T/G-box (CACGTT) and E-box (CAATTG), and its expression is cooperatively regulated by these transcriptional factors (Oh et al., 2012, 2014). EXP2 and XTH33 are also the direct target genes of HY5 and PIF4 (Kurihara et al., 2014; Pedmale et al., 2016). Based on these reports and our findings, we postulate that, besides HY5, other transcriptional factors may participate in the recruitment of SWR1 complex to their shared target loci and mediate H2A.Z deposition. Our results indicate that SWC6, ARP6, and H2A.Z may promote chlorophyll biosynthesis through HY5 (Supplemental Figure S5A). However, consistent with a previous study showing H2A.Z inhibition of anthocyanin accumulation in white light (Cai et al., 2019), they also repress anthocyanin production in BL in this study (Supplemental Figure S5B), indicating that SWC6, ARP6, and H2A.Z repression of anthocyanin biosynthesis may not proceed through HY5. Further studies will be needed to clarify this complication.
In this study, we show that H2A.Z deposition at HY5 target loci is positively correlated with HY5 protein levels (Figure 5, E–G and K). Given that CRY1 can promote HY5 accumulation in response to BL by interacting with COP1 to inhibit its activity on HY5 (; Osterlund et al., 2000; Seo et al., 2003), we propose that CRY1 regulation of H2A.Z deposition at HY5 target genes may be mediated through two simultaneous pathways: CRY1–SWC6/ARP6–HY5 and CRY1–COP1–HY5 (Figure 7, F and G). In DK, CRY1 is inactive and unable to interact with COP1 and SWC6/ARP6 to regulate their activities (Yang et al., 2001; Holtkotte et al., 2017; this report). As a result, COP1 is fully functional, whereas the SWR1 complex is not. HY5 undergoes ubiquitination and degradation, thus barely accumulates. Consequently, the SWR1 complex can hardly be recruited to HY5 target loci to mediate H2A.Z deposition, leading to active expression of HY5 target genes promoting cell elongation and enhanced hypocotyl elongation (Figure 7F). Under BL, CRY1 becomes activated, and it on the one hand may enhance the activity of SWC6 and ARP6 to promote H2A.Z deposition by interacting with SWC6 and ARP6 (this report). On the other hand, it interacts with COP1 to inhibit its activity and promote its translocation from the nucleus to the cytoplasm to promote HY5 accumulation (Osterlund and Deng, 1998; Yang et al., 2001; Holtkotte et al., 2017) and enhance the recruitment of SWR1 complex (this report). These combined effects of the direct interaction of CRY1 with SWR1 complex and increased recruitment of SWR1 complex to HY5 target loci by CRY1 may contribute to the promotion of H2A.Z deposition and subsequent repression of the expression of HY5 target genes and hypocotyl elongation (Figure 7G). How exactly CRY1 regulates SWR1 complex activity is not clear and awaits further investigation in future studies.
Materials and methods
Plant materials and growth conditions
All Arabidopsis thaliana plants used in this study were of the Columbia ecotype. The cry1 cry2 (hy4-104 cry2-1), phyA phyB (phyA-211 phyB-9), hy5 hyh, swc6 (SAIL_536_A05), arp6 (GARLIC_599_G03), hta9 (SALK_054814), hta11 (SALK_031471C), and transgenic lines overexpressing Myc-tagged full-length CRY1 and CRY2 (Myc-CRY1-OX and Myc-CRY2-OX) were described previously (Nagatani et al., 1993; Reed et al., 1993; Bruggemann et al., 1996; Guo et al., 1998; Deal et al., 2005; Mao et al., 2005; Sang et al., 2005; March-Diaz et al., 2007; Lian et al., 2018). The hta9 hta11, cry1 cry2 arp6, and hy5 hyh arp6 mutants were generated by genetic crossing and confirmed by phenotypic analyses and DNA genotyping. All the primers for genotyping are listed in Supplemental Data Set S3.
The expression cassettes 35S::SWC6-Flag and 35S::ARP6-Flag were cloned into pHB (restriction sites used for cloning are described in Supplemental Data Set S3; Mao et al., 2005), which were introduced into WT, swc6, arp6, cry1 cry2, hy5 hyh mutants, Myc-CRY1-OX, and Myc-CRY2-OX backgrounds to generate transgenic lines overexpressing SWC6-Flag and ARP6-Flag (SWC6-Flag-OX, SWC6-Flag-OX/swc6, SWC6-Flag-OX/cry1 cry2, SWC6-Flag-OX/hy5 hyh, SWC6-Flag-OX/Myc-CRY1-OX, and SWC6-Flag-OX/Myc-CRY2-OX, ARP6-Flag-OX, ARP6-Flag-OX/arp6, ARP6-Flag-OX/Myc-CRY1-OX, and ARP6-Flag-OX/Myc-CRY2-OX), respectively. The expression cassette 35S::Flag-HY5 was cloned into pHB, which was introduced into WT and hy5 mutant background to generate Flag-HY5-OX and Flag-HY5-OX/hy5 plants. The expression cassette 35S::SWC6-Myc and 35S::ARP6-YFP were cloned into pKYL71, which were introduced into swc6 and arp6 mutants and Flag-HY5-OX backgrounds to generate SWC6-Myc-OX/swc6, SWC6-Myc-OX/Flag-HY5-OX, ARP6-YFP-OX/arp6, and ARP6-YFP-OX/Flag-HY5-OX plants, respectively. All primers and restriction sites used for vector construction are listed in Supplemental Data Set S3.
The growth condition and light sources were as described previously (Xu et al., 2018). Briefly, sterilizing seeds were kept in 4°C for 2 d and grown on half Murashige and Skoog (MS) medium (Murashige and Skoog, PhytoTech, Overland Park, KS, USA ; M524) plus 1% sucrose with 0.7% agar at 22°C under white light (100 µmol/m2/s). Monochromatic BL, RL, and FRL illumination using in this study were described previously (Wang et al., 2018b). Light intensity was measured with an ILT2400-A quantum photometer (LI-COR Biosciences, Lincoln, NE, USA).
Yeast two-hybrid assays
The Arabidopsis cDNA library cloned into the prey vector pGADT7 (AD; cDNAs were ligated into the SfiI site) was constructed by Shanghai OE BioTech previously. The bait plasmid pGBKT7-CNT1 (BD-CNT1) and the prey library DNA were cotransformed into yeast strain AH109 (S. cerevisiae). The yeast two-hybrid screening and interaction assays were performed as described previously (Wang et al., 2018b). For GAL4 yeast two-hybrid assays, AD-CRY1, BD-CRY2, and BD-CNT1 constructions were described previously (Wang et al., 2018b). The cDNA fragment of SWC6 was cloned into the AD and BD vectors. Transformed yeast cells were spread on Synthetic Drop-out (SD; −Trp−Leu−His−Ade) medium supplemented with 0 or 5 or 10 mM 3-amino-1,2,4-triazole (3-AT), and then one half of these plates were exposed to BL (30 μmol/m2/s) and the other half were kept in the DK for the interaction test. For LexA yeast two-hybrid assays, the cDNA sequences encoding full-length and truncated SWC6 were cloned into pEG202 to generate the corresponding LexA constructs, which were cotransformed with pJG4-5 vectors expressing B42AD-HY5 and B42AD-HYH (Lian et al., 2018) into EGY48 yeast cells. Transformed colonies were selected on SD (−Trp−His−Ura) medium. Six independent clones were selected to grow on SD (−Trp−His−Ura) medium with 80 mg/L X-gal (TaKaRa, Kyoto, Japan) for the interaction test.
Pull-down assays with proteins expressed in Escherichia coli
Pull-down assays were performed as described previously with minor modifications (Mao et al., 2020). The pCold-TF-CNT1, pCold-TF-CCT1, pCold-TF-CNT2, and pCold-TF-CCT2 constructions were described previously (Du et al., 2020). The cDNAs encoding SWC6, ARP6, and HY5/HYH were cloned into pGEX-4T-1 (GE Healthcare Life Sciences, Marlborough, MA, USA) pMAL-c2X (New England Biolabs [NEB], Ipswich, MA, USA), and pCold-TF (TaKaRa), respectively. His-TF-CNT1, His-TF-CCT1, His-TF-CNT2, His-TF-CCT2, His-TF-HY5, His-TF-HYH, His-TF, GST-SWC6, and MBP-ARP6 proteins were expressed in E. coli strain Rosetta. In vitro pull-down assay were as described previously (Xu et al., 2018). For GST pull-down and MBP pull-down assays, MagneGST™ Glutathione Particles (Promega, Madison, WI, USA; V8611) and Amylose Magnetic Beads (NEB; E8035S) were used. Prey proteins were detected with anti-His antibody (GenScript, A00186).
Split-LUC assays
For the split-LUC assays, the constructs expressing the cDNAs encoding HY5/HYH, SWC6/ARP6, phyB, and phyA were cloned into pCambia1300-nLUC and pCambia1300-cLUC (Chen et al., 2008), respectively. These constructs together with those expressing CRY1-nLUC, CRY2-nLUC, CNT1-nLUC, CCT1-nLUC, CNT2-Nluc, and CCT2-nLUC (Du et al., 2020) were cotransformed into Agrobacterium tumefaciens strain GV3101. GV3101 cells transformed with the indicated combination of nLUC- and cLUC-fused proteins were mixed at a ratio of 1:1 and introduced into N. benthamiana leaves. After infiltration for 2–3 d, the N. benthamiana leaves were incubated with 1 mM D-luciferin sodium salt substrate (Yeasen, New York, NY, USA) and treated in DK for 10 min, and then the LUC signal was collected by a luminescence imaging workstation (Tanon 5200).
Semi-in vivo pull-down assays with Arabidopsis protein extracts
For BL specific CRY1/CRY2–SWC6 and CRY1–ARP6 interactions assays, bait GST-SWC6 and MBP-ARP6 proteins were incubated with 15 μL Glutathione Particles (Promega; V8611) and 15 μL Amylose Magnetic Beads (NEB; E8035S) for 2 h and washed 3 times with lysis buffer. The preys were protein extracts from Myc-CRY1-OX and Myc-CRY2-OX seedlings that were DK-adapted for 2 d and then either exposed to 30 μmol/m2/s (Myc-CRY1-OX) or 20 μmol/m2/s BL (MG132-pretreated Myc-CRY2-OX), 30 μmol/m2/s RL, or 10 μmol/m2/s FRL or still adapted in DK for 1 h. For the assays of BL exposure time effects on CRY1–SWC6/ARP6 interactions, the preys were protein extracts were prepared from Myc-CRY1-OX seedlings adapted in the DK for 2 d and then either adapted in DK or exposed to different time of BL (30 μmol/m2/s). For the assays of BL fluence rate effects on CRY2–SWC6 interaction, the preys were protein extracts prepared from Myc-CRY2-OX seedlings adapted in the DK for 2 d and then either exposed to the indicated fluence rates of BL or still adapted in DK for 1 h. The procedures for interaction detection were described previously (Du et al., 2020). Prey proteins were detected using anti-Myc antibody (Millipore, Burlington, MA, USA; 05-724).
Co-IP assays
Co-IP assays using Arabidopsis seedlings were performed as described previously (Mao et al., 2020). For the assays of BL-induced CRY1/CRY2–SWC6/ARP6 interactions, SWC6-Flag-OX/Myc-CRY1-OX, SWC6-Flag-OX/Myc-CRY2-OX, ARP6-Flag-OX/Myc-CRY1-OX, ARP6-Flag-OX/Myc-CRY2-OX, Myc-CRY1-OX, and Myc-CRY2-OX transgenic seedlings were grown in white light (100 μmol/m2/s) for 5 d and then adapted in DK for 2 d. After treated with 100 μM MG132 for 4 h, one half of the seedlings were exposed to 30 μmol/m2/s BL for 1 h, and the other half were adapted in DK for 1 h. Total proteins were extracted with lysis buffer and incubated with 10 μL anti-Flag beads (Sigma, St. Louis, MO, USA) for 1 h at 4°C. The immunoprecipitates were washed 2–3 times with lysis buffer. The precipitates were eluted into 3× Flag peptide (Sigma) and subjected to immunoblot analysis with anti-Flag (Sigma) and anti-Myc (Millipore) antibodies. For the assays of BL-specific CRY1/CRY2–SWC6 interactions, 5-d-old white light-grown SWC6-Flag-OX/Myc-CRY1-OX and SWC6-Flag-OX/Myc-CRY2-OX seedlings were adapted in DK for 2 d and then either exposed to 30 μmol/m2/s (SWC6-Flag-OX/Myc-CRY1-OX) or 20 μmol/m2/s BL (MG132-pretreated SWC6-Flag-OX/Myc-CRY2-OX), 30 μmol/m2/s RL, or 10 μmol/m2/s FRL or still adapted in DK for 1 h. For the assays of the influence of BL exposure time on CRY1–SWC6 interaction, 5-d-old white light-grown SWC6-Flag-OX/Myc-CRY1-OX seedlings were DK-adapted for 2 d and then either adapted in DK or exposed to different time of BL (30 μmol/m2/s). For the assays of the influence of BL intensity on CRY2–SWC6 interaction, 5-d-old white light-grown SWC6-Flag-OX/Myc-CRY2-OX seedlings were DK-adapted for 2 d and then either exposed to the indicated fluence rates of BL or still adapted in DK for 1 h.
For co-IP assay of the interaction of HY5 with SWC6, Flag-HY5-OX, and SWC6-Myc-OX/Flag-HY5-OX seedlings were grown in BL (30 μmol/m2/s) for 7 d, followed by IP with anti-Myc antibody (Millipore). The IP (SWC6) and co-IP signals (HY5) were detected by immunoblots probed with anti-Myc and -Flag antibodies. For co-IP assay of the interaction of HY5 with ARP6, ARP6-YFP-OX, and ARP6-YFP-OX/Flag-HY5-OX seedlings were grown in BL (30 μmol/m2/s) for 7 d, followed by IP with an anti-Flag antibody (Sigma; F3165). The IP (HY5) and co-IP signals (ARP6) were detected by immunoblots probed with anti-Flag and -GFP (Abmart, Shanghai, China; M20009) antibodies.
Hypocotyl length measurements
Sterilized Arabidopsis seeds were grown on half MS plates, kept in 4°C for 2 d, and illuminated for 12 h before transferred to continuous BL condition (30 μmol/m2/s). Five to six days later, the hypocotyl lengths were measured with ImageJ software (http://rsbweb.nih.gov/ij/).
Chlorophyll and anthocyanin measurements
Chlorophyll measurements were performed as described with minor modifications (Burko et al., 2020). About 0.1 g seedlings were extracted in 1.5 mL of ice-cold 80% acetone overnight at 4°C, and then debris was removed by centrifugation. Chlorophyll was measured with a spectrophotometer, and contents were calculated. Anthocyanin measurements were performed as described with minor modifications (Lian et al., 2018). Thirty seedlings were incubated in 600 μL 1% HCl diluted with methanol for 12 h at 4°C. After chloroform extraction, anthocyanin in aqueous phase was used to measure absorbance at 530 and 657 nm and calculated as equation: (A530−A657) g−1 FW. FW denotes the fresh weight of 30 seedlings. Data are represented as the mean values ± SD of triplicates (n = 3).
RNA-Seq data processing
WT, cry1 cry2, hy5 hyh, and arp6 seeds were germinated on half MS plates plus 1% sucrose and placed at 4°C for 3 d and then transferred to white light for 12 h before placed in BL (30 μmol/m2/s) for another 5 d. The RNA-seq was performed and analyzed as described previously (Xu et al., 2018). Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) following the manufacturer’s protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with RNA Integrity Number ≥7 were subjected to the subsequent analysis. The libraries were constructed using TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Then these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten) and 125/150 bp paired-end reads were generated. Raw data (raw reads) were processed using Trimmomatic. The reads containing ploy-N and the low-quality reads were removed to obtain the clean reads. Then the clean reads were mapped to reference genome using hisat. FPKM and read count value of each transcript (protein coding) were calculated using bowtie2 and eXpress. DEGs were identified using the DESeq functions estimateSizeFactors and nbinomTest. Genes with FC > 1.5 or FC < 2/3, and P-value < 0.05 were defined as DEGs. Venn diagram was generated in Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Heatmap was generated with hierarchical clustering analysis by MeV version 4.7 software. GO analysis was performed in agriGO (http://bioinfo.cau.edu.cn/agriGO/).
RT-qPCR
Total RNAs were isolated with RNAprep Plant kit (TIANGEN, DP441) and then reverse-transcribed to cDNA using an iScript cDNA Synthesis kit (BioRad, Hercules, CA, USA; 170-8891). RT-qPCR was performed using TB Green Premix Ex Taq II (TaKaRa; RR820) on a CFX96 connect real-time PCR detection system (Bio-Rad). The reaction program is as follows: one cycle for 30 s at 94°C, 40 cycles for 5 s at 94°C, and then 30 s at 60°C. The relative quantification was calculated with the 2−ΔΔCt method using PP2A as the reference genes. Data are represented as the mean values ± SD of triplicates (n = 3). The primers used are listed in Supplemental Data Set S3.
ChIP-qPCR
ChIP-qPCR assays were performed as described previously with minor modification (Mao et al., 2020). Briefly, 5-d-old white light-grown WT, cry1 cry2, hy5 hyh, arp6, Flag-HY5-OX, SWC6-Flag-OX, and SWC6-Flag-OX/hy5 hyh seedlings were kept in DK for 2 d, and then either exposed to BL (50 μmol/m2/s) or still adapted in DK for 6 h. The seedlings were harvested in dim green safe light and cross-linked with 1% formaldehyde in fixation solution. Chromatin extracts were sheared into 500–1,000 bp fragments with a sonicator (Diagenode, Bioruptor Plus). The sonicated chromatin complex was immunoprecipitated by anti-HTA9 antibody (PhytoAB, PHY0860S, 1:250)-bound Dynabeads Protein G (Life Technologies, Carlsbad, CA, USA; 10004D) or anti-Flag agarose beads (Sigma; M8823). The DNA purification and results analysis were performed as described previously (Zhang et al., 2014). The relative enrichment was normalized to the IP/Input of AT4G07700 (negative control).
Statistical analysis
For multiple comparisons, significant differences between different samples were analyzed by one-way analysis of variance (ANOVA) followed by least significant difference (LSD) test (P < 0.05) using IBM SPSS Statistics software. For Student’s t-test, different lowercase letters above the bars indicate significantly different groups: *P < 0.05 and **P < 0.01. Detailed statistical data with ANOVA data and Student’s t-test for all of the relevant figures are listed in the Supplemental Data Set S4.
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank database or the Arabidopsis Genome Initiative database under the following accession numbers: CRY1 (AT4G08920), CRY2 (AT1G04400), SWC6 (AT5G37055), ARP6 (AT3G33520), HTA9 (AT1G52740), HTA11 (AT3G54560), HY5 (AT5G11260), HYH (AT3G17609), EXP2 (AT5G05290), IAA19 (AT3G15540), XTH33 (AT1G10550), HEMA1 (AT1G58290), RBCS1A (AT1G67090), PP2A (AT1G13320), AT4G07700. RNA-seq data have been deposited into the BioProject with the following accession numbers: BioProject (PRJNA701118), BioSample (SAMN17847966), and SRA (SRR13674696-SRR13674707).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Yeast two-hybrid assays showing the C terminus-mediated interactions of SWC6 with CRY1 and CRY2 (supports Figure 1).
Supplemental Figure S2. Pull-down and split-LUC assays showing that both N and C termini of CRY1 and CRY2 interact with SWC6 and ARP6 (supports Figures 1 and 2).
Supplemental Figure S3. Semi-in vivo pull-down assays showing the BL-specific interactions of CRY1–SWC6 and CRY2–SWC6 (supports Figure 1).
Supplemental Figure S4. SWC6 and ARP6 act to negatively regulate hypocotyl elongation in BL (supports Figure 3).
Supplemental Figure S5. SWC6, ARP6, and H2A.Z act to positively regulate chlorophyll biosynthesis, but negatively regulate anthocyanin accumulation in BL (supports Figure 3).
Supplemental Figure S6. ARP6 may act in the same pathway of CRY1 and CRY2 to regulate hypocotyl elongation in BL via HY5-dependent and -independent pathways (supports Figure 3).
Supplemental Figure S7. phyB and phyA likely interact with SWC6 and ARP6 (supports Figure 3).
Supplemental Figure S8. RT-qPCR analyses showing that expression of genes promoting cell elongation is repressed by CRYs, ARP6, and HY5/HYH (supports Figure 4).
Supplemental Figure S9. RT-qPCR analyses show that expression of genes involved in chlorophyll biosynthesis and photosynthesis is promoted by CRYs, ARP6, and HY5/HYH (supports Figure 4).
Supplemental Figure S10. ChIP-qPCR analyses showing the role of SWC6 and ARP6 in H2A.Z deposition at HEMA1 and RBCS1A loci (supports Figure 5).
Supplemental Figure S11. Immunoblot analyses of nuclear extracts in ChIP-qPCR assays (supports Figure 5).
Supplemental Figure S12. Yeast two-hybrid assays showing the C terminus-mediated interaction of SWC6 with HY5 (supports Figure 6).
Supplemental Figure S13. Yeast two-hybrid and pull-down assays showing the C terminus-mediated interaction of HY5 with SWC6 (supports Figure 6).
Supplemental Data Set S1. List of 1879 genes coregulated by CRYs and ARP6 (supports Figure 4).
Supplemental Data Set S2. List of 2,051 genes coregulated by ARP6 and HY5/HYH (supports Figure 4).
Supplemental Data Set S3. All the primers used in this study.
Supplemental Data Set S4. ANOVA results and Student’s t-test for the data shown in figures.
Supplementary Material
Acknowledgments
We thank Drs Xiaofeng Cao, Aiwu Dong, Sujuan Cui, and Yuda Fang for materials assistance.
Funding
This work was supported by The National Key Research and Development Program of China grant (2017YFA0503802), The National Natural Science Foundation of China grants to H.-Q.Y. (31530085), Z.M. (31900609), W.W. (31900207), and T.G. (32000183), and The Science and Technology Commission of Shanghai Municipality grant (18DZ2260500).
Conflict of interest statement. The authors declare no coflict of interest regarding the publication of this study.
ORCID IDs: 0000-0002-7071-8048 (Z.M.), 0000-0002-4008-2595 (X.W.), 0000-0001-5194-0391 (L.L.), 0000-0002-0477-4201 (P.X.), 0000-0002-5292-8279 (J.Z.), 0000-0001-6490-9553 (W.W.), 0000-0003-0462-3929 (T.G.), 0000-0002-9385-0244 (S.K.), 0000-0002-6163-9751 (W.W.), 0000-0002-2726-7579 (L.M.), 0000-0001-9547-9111 (X.C.), 0000-0002-1859-4842 (J.Z.), 0000-0002-8188-2214 (G.Y.), 0000-0001-6104-6481 (S.Z.), 0000-0002-0213-0242 (H.L.), and 0000-0001-6215-2665 (H.Q.Y.).
H.-Q.Y., Z.M., and L.L. conceived the project; H.-Q.Y. and Z.M. designed the research plan; Z.M., X.W., L.L., P.X., J.Z., W.W., T.G., S.K., W. W., L.M., X.C., J.Z., G.Y., and S.Z. carried out the experiments; Z.M. analyzed the data; Z.M. and H.-Q.Y. wrote the manuscript.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Hong-Quan Yang (hqyang@shnu.edu.cn).
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Bruggemann E, Handwerger K, Essex C, Storz G (1996) Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J 10: 755–760 [DOI] [PubMed] [Google Scholar]
- Burko Y, Seluzicki A, Zander M, Pedmale UV, Ecker JR, Chory J (2020) Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32: 967–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Zhang M, Chai M, He Q, Huang X, Zhao L, Qin Y (2019) Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol 221: 295–308 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Corrado N, Perdiguero E, Lafarga V, Munoz-Canoves P, Nebreda AR (2010) Essential role of p18Hamlet/SRCAP-mediated histone H2A.Z chromatin incorporation in muscle differentiation. EMBO J 29: 2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Bai Y, Zhao L, Dou X, Liu Y, Wang L, Li Y, Li W, Hui Y, Huang X. et al. (2017) H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis. Mol Plant 10: 1274–1292 [DOI] [PubMed] [Google Scholar]
- Deal RB, Kandasamy MK, McKinney EC, Meagher RB (2005) The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH (1999) Signalling in light-controlled development. Semin Cell Dev Biol 10: 121–129 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Dong H, Liu J, He G, Liu P, Sun J (2020) Photoexcited phytochrome B interacts with brassinazole resistant 1 to repress brassinosteroid signaling in Arabidopsis. J Integr Plant Biol 62: 652–667 [DOI] [PubMed] [Google Scholar]
- Du SS, Li L, Li L, Wei X, Xu F, Xu P, Wang W, Xu P, Cao X, Miao L. et al. (2020) Photoexcited cryptochrome2 interacts directly with TOE1 and TOE2 in flowering regulation. Plant Physiol 184: 487–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (1997) Light control of plant development. Annu Rev Cell Dev Biol 13: 203–229 [DOI] [PubMed] [Google Scholar]
- Gegear RJ, Foley LE, Casselman A, Reppert SM (2010) Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463: 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold CB, Gasser SM (2014) INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol 24: 619–631 [DOI] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L (2007) p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev 21: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- He SB, Wang WX, Zhang JY, Xu F, Lian HL, Li L, Yang HQ (2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol Plant 8: 822–825 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkotte X, Ponnu J, Ahmad M, Hoecker U (2017) The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling. PLoS Genet 13: e1007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Imaizumi T (2012) LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F. et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585: 256–260 [DOI] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S. et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Noren L, Lopez Jde D, Shaikhali J, Strand A (2012) Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol Plant 5: 901–913 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Makita Y, Kawashima M, Hamasaki H, Yamamoto YY, Matsui M (2014) Next-generation sequencing of genomic DNA fragments bound to a transcription factor in vitro reveals its regulatory potential. Genes 5: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Seo PJ (2017) Coordination of matrix attachment and ATP-dependent chromatin remodeling regulate auxin biosynthesis and Arabidopsis hypocotyl elongation. PLoS One 12: e0181804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schafer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Lian H, Xu P, He S, Wu J, Pan J, Wang W, Xu F, Wang S, Pan J, Huang J. et al. (2018) Photoexcited CRYPTOCHROME 1 interacts directly with G-protein beta subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol Plant 11: 1248–1263 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008a) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008b) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S. et al. (2020) Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225: 848–865 [DOI] [PubMed] [Google Scholar]
- March-Diaz R, Reyes JC (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2: 565–577 [DOI] [PubMed] [Google Scholar]
- March-Diaz R, Garcia-Dominguez M, Florencio FJ, Reyes JC (2007) SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol 143: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ (2002) Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J 32: 549–559 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Deng XW (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR. et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI. et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Wang S, Zhang F, Zheng H, Liu Y, Huang T, Ding Y (2017) Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A Z deposition. Plant Cell 29: 2197–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong MXZ, Lee K, Ezer D, Cortijo S, Jung J, Charoensawan V, Box MS, Jaeger KE, Takahashi N, Mas P. et al. (2020) The evening complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol 182: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D. et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 627–630 [DOI] [PubMed] [Google Scholar]
- van der Woude LC, Perrella G, Snoek BL, van Hoogdalem M, Novak O, van Verk MC, van Kooten HN, Zorn LE, Tonckens R, Dongus JA. et al. (2019) HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc Natl Acad Sci USA 116: 25343–25354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin C (2020) Mechanisms of cryptochrome-mediated photoresponses in plants. Annu Rev Plant Biol 71: 103–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li L, Xu P, Lian H, Wang W, Xu F, Mao Z, Zhang T, Yang H (2018a) CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J Exp Bot 69: 3867–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu X, Li L, Lian H, Mao Z, Xu P, Guo T, Xu F, Du S, Cao X. et al. (2018b) Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willhoft O, Ghoneim M, Lin CL, Chua EYD, Wilkinson M, Chaban Y, Ayala R, McCormack EA, Ocloo L, Rueda DS. et al. (2018) Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362: eaat7716. [DOI] [PubMed] [Google Scholar]
- Wu J, Wang W, Xu P, Pan J, Zhang T, Li Y, Li G, Yang H, Lian H (2019) phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol 60: 353–366 [DOI] [PubMed] [Google Scholar]
- Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, Wu C (2005) Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol 12: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L. et al. (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit Auxin signaling in Arabidopsis. Mol Plant 11: 523–541 [DOI] [PubMed] [Google Scholar]
- Yadav A, Singh D, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S (2020) Light signaling and UV-B-mediated plant growth regulation. J Integr Plant Biol 62: 1270–1292 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Ye B, Liu B, Yang L, Huang G, Hao L, Xia P, Wang S, Du Y, Qin X, Zhu P. et al. (2017) Suppression of SRCAP chromatin remodelling complex and restriction of lymphoid lineage commitment by Pcid2. Nat Commun 8: 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C (2007) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104: 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, , He SB, , Li L, , Yang HQ ( 2014) Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc Natl Acad Sci USA 111, E3015–3023. 10.1073/pnas.1400542111 25002510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Thakar A (2008) H2A.Z: view from the top. Structure 16: 166–179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.