Abstract
Enhancers located in introns are abundant and play a major role in the regulation of gene expression in mammalian species. By contrast, the functions of intronic enhancers in plants have largely been unexplored and only a handful of plant intronic enhancers have been reported. We performed a genome-wide prediction of intronic enhancers in Arabidopsis thaliana using open chromatin signatures based on DNase I sequencing. We identified 941 candidate intronic enhancers associated with 806 genes in seedling tissue and 1,271 intronic enhancers associated with 1,069 genes in floral tissue. We validated the function of 15 of 21 (71%) of the predicted intronic enhancers in transgenic assays using a reporter gene. We also created deletion lines of three intronic enhancers associated with two different genes using CRISPR/Cas. Deletion of these enhancers, which span key transcription factor binding sites, did not abolish gene expression but caused varying levels of transcriptional repression of their cognate genes. Remarkably, the transcriptional repression of the deletion lines occurred at specific developmental stages and resulted in distinct phenotypic effects on plant morphology and development. Clearly, these three intronic enhancers are important in fine-tuning tissue- and development-specific expression of their cognate genes.
Genome-wide analysis and CRISPR/cas editing of intronic enhancers in Arabidopsis thaliana provide insights into tissue-specific gene expression.
Introduction
Plant development is governed by precise patterns of gene expression that are achieved by coordinated binding of regulatory proteins to promoters, enhancers, and other cis-regulatory elements (CREs). Enhancers contain short DNA sequences that serve as binding sites for transcription factors (TFs). In turn, TFs recruit transcriptional cofactors that modify nearby chromatin and lead to transcriptional activation (Shlyueva et al., 2014). Enhancers drive transcription independent of their distance and location relative to their cognate promoter, and this allows a gene to be regulated by multiple distal enhancers with different spatiotemporal activities (Long et al., 2016). Divergence of enhancer sequence and activities is thought to play an important role in inter- and intra-species phenotypic variation (Long et al., 2016). Enhancers had long been difficult to identify due to their unpredictable genomic positions. However, enhancers can be predicted using different types of genomic data sets, such as open chromatin (or chromatin accessibility) data sets, histone modifications, such as H3K27ac, and transcriptional co-activators, such as P300/CBP (Jiang, 2015; Weber et al., 2016; Marand et al., 2017; Catarino and Stark, 2018; Li et al., 2019a). Such data have been used for enhancer prediction and functional validation in a number of animal and plant species (Heintzman et al., 2009; Creyghton et al., 2010; Hnisz et al., 2013; Smemo et al., 2014; Zhu et al., 2015; Zhao et al., 2018; Lu et al., 2019; Ricci et al., 2019; Yan et al., 2019; Zicola et al., 2019).
Enhancers located in introns were first discovered in the mouse immunoglobulin heavy chain gene (Banerji et al., 1983; Gillies et al., 1983; Mercola et al., 1983). Early studies based on this intronic enhancer showed that its activity is cell-type specific and is independent of its position and orientation relative to the promoter (Banerji et al., 1983; Gillies et al., 1983). These interesting characteristics were later found to be commonly associated with most developmental enhancers. The abundance of intronic enhancers in mammalian species was not realized until the genomics era. Human genes contain an average of 7.8 introns that have an average size of 5.4 kb (Sakharkar et al., 2004). Strikingly, intronic sequences account for ∼25% of the human genome and are 4 − 5 times the size of exons (Sakharkar et al., 2004). The massive amount of intronic sequences provides a hidden bed of various CREs, including enhancers (Jo and Choi, 2015). For example, a total of 36,589 enhancers were predicted in HeLa cells based on chromatin signatures, of which, 37.9% were located in introns (Heintzman et al., 2009). In Drosophila melanogaster, enhancers associated with developmental core promoters are predominantly localized in introns (Zabidi et al., 2015). Intronic enhancers are generally presumed to regulate their cognate genes. However, an enhancer located in one gene can sometimes regulate the expression of another gene(s) megabases away (Smemo et al., 2014).
Regulation of plant gene expression by intronic sequences was first reported 30 years ago (Callis et al., 1987; Vasil et al., 1989). Since then, intronic enhancers have been reported in a number of plant species (Fu et al., 1995; Rose, 2002; Jeong et al., 2007; Sekhon et al., 2007; Weise et al., 2008; Giani et al., 2009; Broeckling et al., 2016; Sicard et al., 2016; Friede et al., 2017; Gallegos and Rose, 2017; Xie et al., 2018). However, these reports have been sporadic and it is unknown whether intron-mediated regulation of gene expression is common in plants. Systematic analysis of intronic sequences on plant gene expression has been performed solely based on bioinformatics (Rose et al., 2008; Parra et al., 2011; Rose, 2019). Intronic enhancers associated with genes related to flower development in Arabidopsis thaliana have recently been predicted based on open chromatin signatures revealed using DNase-seq analyses (Yan et al., 2019). It is interesting to note that the structures of plant genes are substantially different from those of mammalian species. Genes in two model plant species, A. thaliana and Oryza sativa (rice), contain an average of 6 and 5 introns, respectively. The average sizes of the introns are merely 170 bp in A. thaliana (Kaul et al., 2000, TAIR 10) and 447 bp in rice (Kawahara et al., 2013, IRGSP-1.0); significantly smaller than those in humans. In addition, the lack of mutations and deletions of known intronic enhancers have prohibited in-depth studies of the functional roles of intronic CREs in plants.
We performed a genome-wide search of candidate intronic enhancers in A. thaliana using open chromatin signatures associated with hypersensitivity to DNase I digestion (Zhang et al., 2012). We identified 941 and 1,271 candidate intronic enhancers that are potentially functional in seedling and floral tissues, respectively. We validated the function of 15 out of 21 (71%) candidate enhancers using a transgenic assay with the β-glucuronidase (GUS) reporter gene. In addition, we created transgenic lines containing homozygous deletions of predicted intronic enhancers associated with two different genes using CRISPR/Cas-based genome editing. Using these lines, we demonstrate that deletions of the candidate enhancers result in varying levels of transcriptional repression of the cognate genes at specific developmental stages.
Results
Prediction of intronic enhancers based on chromatin accessibility
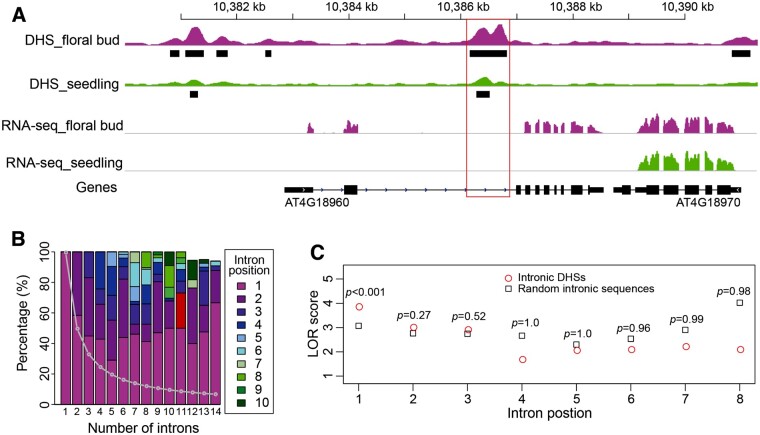
DNase I hypersensitive sites (DHSs) were previously used to successfully predict enhancers located in intergenic regions of A. thaliana (Zhu et al., 2015). We used a similar strategy to assess the frequency and function of enhancers located in the introns of A. thaliana genes. We first conducted a genome-wide search of DHSs that overlap with introns. DHS peaks were identified using F-seq (Boyle et al., 2008; Zhang et al., 2012), and we found 6,053 and 9,343 intron-overlapping DHSs in seedling and floral bud DHS data sets, respectively. We defined these as an “intronic DHS” if a minimum of 80% of the DHS sequence overlapped with an intron. Using this criterion, we identified 941 intronic DHSs associated with 806 genes in seedling tissue, and 1,275 intronic DHSs associated with 1,069 genes in floral bud tissue (Supplemental Figure S1A). For example, a DHS was found in the second intron of the AGAMOUS (AG; AT4G18960) gene (Figure 1A). The function of this intron in regulating AG expression was previously demonstrated by reporter gene-based transgenic assays (Sieburth and Meyerowitz, 1997). A construct containing only the promoter, but without this intron, conferred a GUS staining pattern that deviates from normal AG expression (Sieburth and Meyerowitz, 1997). Similarly, a DHS was identified in the first intron of AGL11 (At4g09960), also known as STK (Pinyopich et al. 2003; Supplemental Figure S1B). The regulatory role of this intron was previously validated by transgenic assay and tethered particle motion analysis (Kooiker et al., 2005). Interestingly, we identified an additional DHS in the second intron (1,576 bp) of AGL11 (Supplemental Figure S1B). We also analyzed Assay for Transposase-Accessible Chromatin (ATAC-seq) data generated from seedling tissue (Lu et al., 2017) and identified 689 intronic peaks. We found that 373 (54.5%) of these ATAC-seq peaks overlap with intronic DHSs. The remaining ATAC-seq peaks were generally in low scores (Supplemental Figure S1C).
Figure 1.
Characteristics of intronic DHSs in A. thaliana. A, A DHS associated with the second intron of AGAMOUS (AT4G18960). Black bars represent DHS sites. The peak height indicates the DNase-seq read enrichment. The red box includes the intronic region containing the DHSs. B, Positional preference of introns containing DHSs. The Arabidopsis genes were grouped based on the number of introns. The positions of introns in each gene were assigned as follows: 1 represents the first intron, 2 represents the second intron, and so on. The percentages of introns contain DHSs at each position were calculated and plotted. The gray line represents the expected percentage of introns at a particular position. The x-axis is the number of introns in genes. The y-axis is the percentage of introns at each position. C, DHS enrichment in different introns. The LOR scores of intronic DHSs (circles) and random intronic sequences (squares) at different intron positions were calculated and plotted. Random intronic sequences have the same numbers and lengths as intronic DHSs except that the intron positions of random intronic sequences are random. The x-axis represents intron positions 1 − 8. The y-axis represents LOR scores. The P-values of enrichment are shown at each intron position.
We found that 221 (22.8%) seedlings and 289 (22.1%) floral bud intronic DHSs overlapped with conserved noncoding sequences in Arabidopsis (Van de Velde et al., 2016). Genes containing intronic DHSs tended to have more introns compared to genes without intronic DHSs (Supplemental Figure S1D). Introns with DHSs tended to be closer to the promoter, especially for genes containing more introns (Figure 1B). For example, ∼50% of intronic DHSs were located in the first intron for genes containing 10 introns. Furthermore, 73% of the first introns containing DHSs were also the largest or second largest intron of that gene. These results agree with previous reports that introns close to promoters are more likely to have enhancer function (Rose et al., 2008; Parra et al., 2011).
It is known that the first intron of Arabidopsis genes tends to be larger than downstream introns (Bradnam and Korf, 2008). To examine whether it is the size or position of an intron that is the main factor for DHS enrichment in the first intron, we calculated the log odds ratio (LOR) to test DHS enrichment at different intron positions (see Methods). Compared to random intronic sequences, the first intron was significantly enriched for DHSs independent of intron size (Figure 1C). Intronic DHSs are infrequent for downstream introns, especially after the third intron. These results suggest that the physical position of the first intron is a favorable location for intronic enhancers.
Association of intronic DHSs with the expression of cognate genes
We investigated the potential impact of intronic DHSs on the expression of their cognate genes. We found that 78.7% (634 of 806) leaf and 80.6% (862 of 1,069) floral genes containing intronic DHSs also contain DHSs at their promoter regions (1 kb upstream of the transcription start site [TSS]) in the same tissue. Genes containing both intronic and promoter DHSs showed higher expression levels than genes with only intronic DHSs or only promoter DHSs (Supplemental Figure S2A). We found that 17.5% (165 of 941) of the intronic DHSs in seedlings were absent in floral buds, and this was a significantly higher levels than the 8.3% (2,685 of 32,456) of all seedling DHSs that were absent in floral buds (P-value <2.2 × 10−16, Fisher’s exact test; Supplemental Figure S2B). Similarly, 39.1% (499 of 1,275) intronic DHSs in floral buds were absent in seedlings, again higher than the 27.7% (11,615 of 41,878) of all DHSs that were absent in seedlings (P-value <2.2 × 10−16, Fisher’s exact test). These results indicate that intronic DHSs are generally more tissue-specific compared to all DHSs. Consistent with this, the expression levels of genes containing intronic DHSs showed more dynamic changes between seedling and floral tissues than genes without intronic DHSs (Supplemental Figure S2C).
Validation of the predicted intronic enhancers using transgenic assays
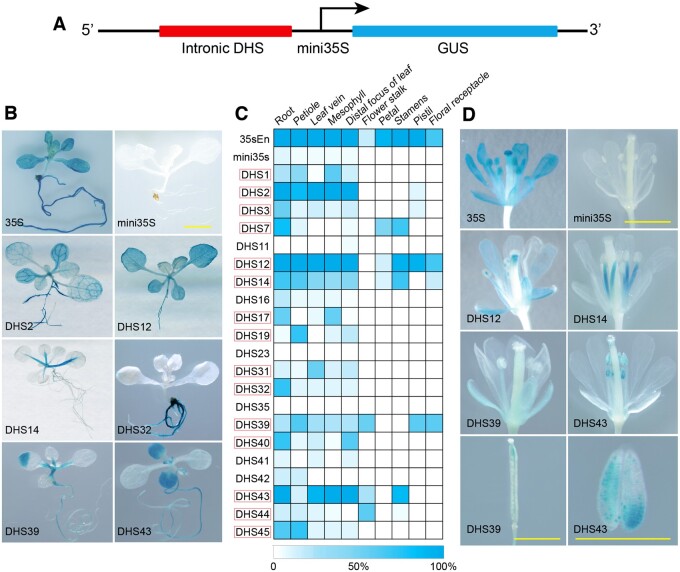
We selected 21 intronic DHSs for functional validation (see Methods) and then used a GUS-based reporter system to confirm the enhancer activity of each candidate. Briefly, the target DHSs, ranging in size from 173 to 778 bp (Supplemental Table S1), were either amplified from genomic DNA or synthesized, and were subsequently inserted into the pKGWFS7.0 vector containing a minimal cauliflower mosaic virus (CaMV) 35S promoter (mini35S, −50 to −2 bp) and a GUS reporter gene (Zhu et al., 2015; Figure 2A). Due to an inability of the minimal CaMV35S to drive expression alone, GUS expression can be detected in transgenic plants only if the inserted DHS has enhancer function. Constructs with only the mini35S promoter or the full length 35S promoter were used as negative and positive controls, respectively. We selected eight additional intronic DNA fragments as negative controls. These intronic sequences, ranging in size from 219 bp to 627 bp (Supplemental Table S1), were not associated with DHSs and also cloned into the pKGWFS7.0 vector. All constructs were used to transform A. thaliana (Col-0) plants. We obtained 14–31 independent transgenic lines from each construct. GUS assays were performed at various developmental stages for each transgenic plant. A DHS was considered to be validated as a functional enhancer only if more than 50% of the transgenic plants showed consistent GUS signals in the same tissue(s). Through this approach, 15 (71%) of the 21 intronic DHSs were validated as functional enhancers (Figure 2C, Supplemental Table S2). Interestingly, this proportion was similar to our previous experiments of intergenic candidate enhancer assays, which validated 71% (10 out of 14) of the DHSs located in intergenic regions (Zhu et al., 2015). GUS signals were detected at various tissues/organs; including leaves, roots, and different floral organs from the transgenic plants (Figure 2, B−D). We did not detect any unambiguous signals from the transgenic plants created from the eight controls (non-DHS) intronic DNA fragments as only inconsistent and faint GUS signals were observed from a few transgenic lines from these constructs (Supplemental Table S2).
Figure 2.
Validation of predicted intronic enhancers and representative GUS expression patterns from transgenic plants transformed with different enhancer constructs. A, A diagram illustrating the structure of the GUS fusion construct used for enhancer validation. B, Representative transgenic seedlings showing GUS signals in leaves and roots. The enhancer construct used in transformation is marked on each seedling image. 35S represents the full 35S promoter. The bar indicates 2 mm for all images. C, A heat map of GUS expression in different tissues of transgenic plants transformed with all enhancer constructs. White indicates no GUS expression was detected in any plants. Dark blue indicates that GUS expression was detected in 100% of the transgenic plants. Red boxes mark the 15 DHSs that were validated as functional enhancers. D, Representative transgenic plants at flowering stage showing GUS signals in different parts of flowers. The enhancer construct used in transformation is marked on each image. All bars indicate 2 mm.
We further made constructs where the sequence was inserted in the reverse orientation, for seven of the 15 validated intronic DHSs (Supplemental Table S2). We obtained 12–24 independent transgenic lines from each of these reversed constructs. Transgenic plants from all seven constructs showed similar GUS signal patterns, including both signal strength and tissue specificity, as did the transgenic plants derived from the respective forward constructs (Supplemental Figure S3; Supplemental Table S2).
Compound enhancer activity of DHS7 is associated with gene TRY
DHS7 (323 bp in length) is located in the second intron (426 bp in length) of the gene TRIPTYCHON (TRY; AT5G53200; Figure 3A), an MYB-related TF in A. thaliana. This DHS was observed in the floral bud data set, but not in the seedling data set (Figure 3A). TRY functions as a negative regulator of trichome formation and a positive regulator of root hair formation (Schellmann et al., 2002; Yu et al., 2010). Interestingly, TRY was also implicated to play a role in flowering (Tominaga-Wada et al., 2013; Wada and Tominaga-Wada, 2015). Transgenic assays using a GUS fusion construct with the 1.4 kb upstream region of TRY (TRYpro:GUS) revealed ubiquitous expression in young leaves, but was exclusively expressed in the trichome initiation zone and trichome cells of old leaves (Schellmann et al., 2002). GUS activities of TRYpro:: GUS were also detected in the inflorescence stem and floral organs from transgenic lines (Yu et al., 2010). The TRY promoter is active in the root epidermis (Tominaga et al., 2008). However, very weak GFP signals were observed in the root epidermis of TRY-GFP (TRYpro:TRY-GFP) transgenic plants (Tominaga-Wada and Wada, 2017). These results highlight a complex regulatory architecture underlying TRY expression.
Figure 3.
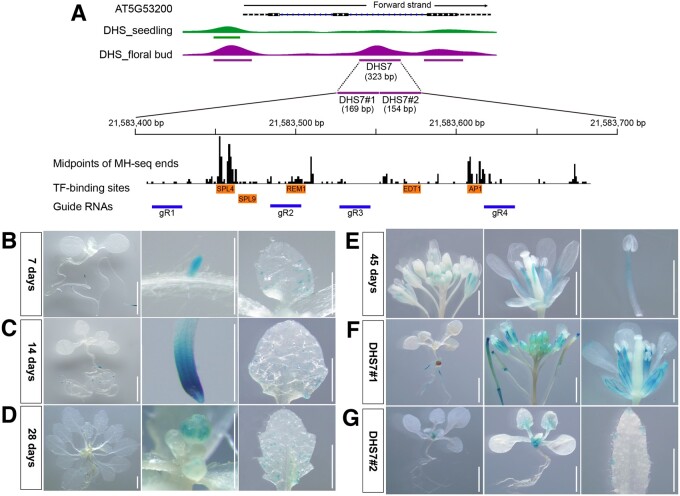
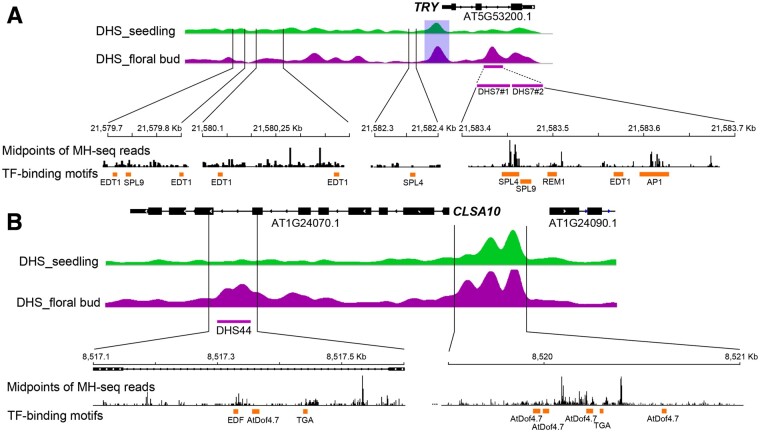
Validation of the DHS7 enhancer using transgenic assays. A, Integrative Genomics Viewer (IGV) tracks of TRY (AT5G53200) and its associated DHSs. DHS7 (323 bp) was identified in the flower data set and is located in the second intron. The DHS7 region is expanded to show the sequence positions of DHS7#1 (169 bp) and DHS7#2 (154 bp), and the locations of midpoints of all sequence reads derived from MNase hypersensitivity sequencing (MH-seq). The positions of binding motifs of five TFs and the four guide RNAs used for CRISPR/Cas editing are also marked. B, GUS expression of 7-d-old transgenic plants derived from DHS7. GUS expression was observed in the main root tips, emerging lateral root tips, and in the incipient trichome at the base of first rosette leaf. C, GUS expression of 14-d-old transgenic plants derived from DHS7. GUS signals were detected in root tips and the trichome-forming zone of young leaves. D, GUS expression of 28-d-old transgenic plants derived from DHS7. GUS signals were detectable in the flower buds as well as in the root tip and the trichome-forming zone of young leaves. E, GUS expression of 45-d-old transgenic plants derived from DHS7. Robust GUS signals were detected in the stamen filaments and faintly in the petals. F, Representative GUS expression patterns in transgenic plants derived from DHS7#1. GUS signals were faintly expressed in roots and robustly expressed in stamen filaments and sepals. G, Representative GUS expression pattern of transgenic plants derived from DHS7#2. GUS activity was detected in roots, rosette, and cauline leaves. All white bars represent 2 mm.
To better understand the regulatory mechanisms that govern TRY expression, we created transgenic lines with a construct containing DHS7 and the mini35S promoter and examined GUS signals throughout development. GUS signals were mainly detected in primary root tips and emerging lateral root tips of transgenic plants 7 d after germination (Figure 3B). In addition, weak signals were observed in incipient trichomes at the base of first rosette leaf (Figure 3B). At 14 d, GUS signals were detected in trichome initiation zones in parallel to enhanced GUS expression in root tips (Figure 3C). At 28 d, GUS signals were detected in floral buds and the trichome forming zone on young leaves (Figure 3D). At 45 d, unambiguous signals were observed in the stamen filaments and petals (Figure 3E). Thus, the CREs contained within DHS7 can drive reporter gene expression in multiple tissues and developmental stages in a manner similar to the promoter of TRY.
GUS signals detected in roots, trichomes, and floral organs from transgenic plants suggest that DHS7 is likely a compound enhancer containing multiple CREs. We recently generated a high-resolution open chromatin data set from Arabidopsis seedling tissue using micrococcal nuclease (MNase) hypersensitivity sequencing (MH-seq; Zhao et al., 2020). MH-seq is based on digestion of chromatin with MNase and can be used to identify open chromatin regions that are not accessible to DNase I (Zhao et al., 2020). We used this MH-seq data for fine mapping of the open chromatin region associated with DHS7 by mapping the midpoints of all MH-seq reads within this region. The midpoints of these reads represent the genomic regions that are protected by the regulatory proteins from MNase digestion. Interestingly, analysis of midpoints revealed multiple peaks within DHS7 (Figure 3A). Sequence analysis uncovered several associated TF-binding motifs, including SPL9, SPL4, REM1, EDT1, and AP1, within DHS7 (Figure 3A). SPL9 and SPL4 are master regulators of phase transition and promote flowering (Wang et al., 2009; Yu et al., 2010). EDT1 (HDG11) belongs to the HD-ZIP IV family and is involved in trichome branching (Nakamura et al., 2006; Khosla et al., 2014).
To establish whether the CREs within DHS7 are modular, we divided the DHS7 fragment (323 bp) into two smaller fragments. The DHS7#1 (169 bp) fragment spanned the putative SPL4- and SPL9-binding motifs, whereas the DHS7#2 fragment (154 bp) spanned the EDT1-binding motif (Figure 3A). These two fragments were separately cloned into the pKGWSF7.0 vector. Transgenic lines using DHS7#1 showed robust GUS activity in the stamen filaments and sepal, and sporadically in a few sections of the root (Figure 3F). In contrast, transgenic lines using DHS7#2 showed GUS signals in roots, rosette ,and cauline leaves, but not in flowers (Figure 3G). These results confirmed that DHS7 contains at least two independent enhancers.
Deletions of DHS7 alter TRY transcription
To further validate the functions of DHS7#1 and DHS7#2 enhancers in vivo, we used CRISPR/Cas9-mediated genome editing to create DHS7#1 and DHS7#2 single deletion lines and DHS7#1#2 double deletion lines (Supplemental Figure S4A). We also made TRY CRISPR/Cas9 knockout mutants using a single short guide RNA (sgRNA) targeting the first exon. We created homozygous lines for three DHS7#1 deletions, five DHS7#2 deletions, two DHS7#1#2 deletions, and two TRY knockout mutants (Supplemental Figure S4, B and C). We chose two homozygous deletion lines, labeled “a” and “b,” respectively (Figure 4), from each group for further analysis. Both knockout mutants, TRYa and TRYb, were single nucleotide insertions, which resulted in frame shift mutations (Supplemental Figure S4B).
Figure 4.
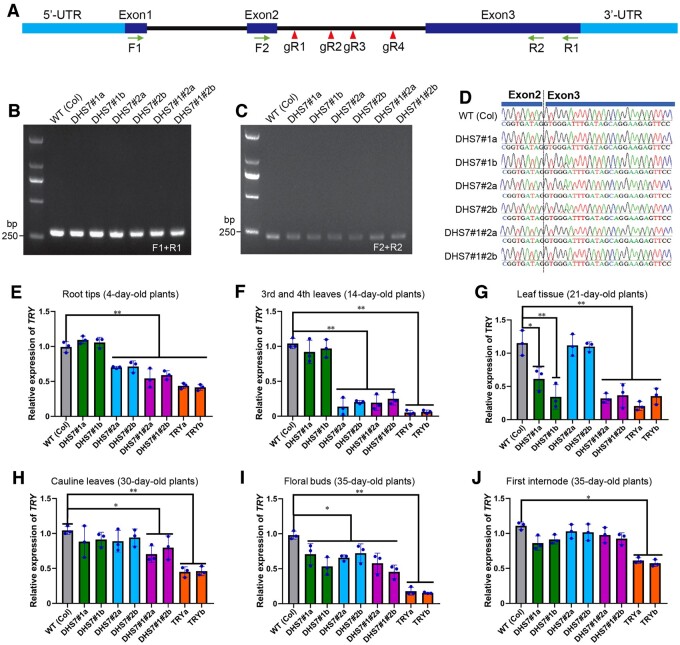
Transcription of TRY in CRISPR/Cas lines. A, A diagram illustrating the structure of TRY. The positions of four PCR primers (arrows) and four sgRNAs (arrowheads) are shown. B, RT-PCR analysis of six CRISPR/Cas lines using primers F1 and R1, which span all three exons. All deletion lines produced a PCR product with the expected size (280 bp). C, RT-PCR analysis of six CRISPR/Cas lines using primers F2 and R2, which span the junction of exon 2 and exon 3. All deletion lines produced a PCR product with the expected size (234 bp). D, Sequencing of RT-PCR products of (C) confirmed the presence of a normal junction between exon 2 and exon 3 in all deletion lines. E–J, Real-time RT-PCR analysis of TRY expression in: (E) root tips of 4-d-old deletion lines; (F) third and fourth leaves of 14-d-old deletion lines; (G) leaf tissues of 21-d-old deletion lines; (H) cauline leaves of 30-d-old deletion lines; (I) flower buds of 35-d-old deletion lines; and (J) the first internode of 35-d-old deletion lines. Values are the average from three biological replicates. Ten plants per line were used in all independent experiments, with the exception of “4-day-old root tips” in which 100 plants were used from each per line. Error bars show the standard deviation (sd) from the three biological replicates. Statistically significant differences are indicated (*P < 0.05, **P < 0.01, Supplemental File S1).
We first conducted PCR-based analyses to examine whether the homozygous deletion lines maintained normal splicing after the loss of part of the second intron. We prepared cDNAs from all deletion lines and found that RT-PCR using primers (F1 and R1, Figure 4A) spanning exon 1 to 3 produced a DNA fragment identical in length across all lines (Figure 4B). These RT-PCR products were cloned into the pGEM-T vector, and 20 clones derived each amplicon were sequenced. The sequencing results showed that transcripts from all deletion lines were normally spliced products containing all three exons. We further analyzed transcripts spanning exons 2–3 using primers F2 and R2 (Figure 4A). Similarly, intron 2 was precisely spliced out in all deletion lines (Figure 4, C and D). These results confirmed that partial deletions of the intron did not alter the splicing of the TRY gene.
We then analyzed the expression levels of TRY in the deletion lines using real-time RT-PCR. In roots from 4-d-old plants, the expression of TRY was reduced by ∼30% in DHS7#2 single deletion lines and DHS7#1#2 double deletion lines compared to wild type (Figure 4E). However, TRY expression in the DHS7#1 single deletion lines was unchanged (Figure 4E). In rosette leaves from 14-d-old plants, expression of TRY was further reduced by ∼70% in DHS7#2 and DHS7#1#2 deletion lines (Figure 4F). In rosette leaves from 21-d-old plants, the expression of TRY in DHS7#1 and DHS7#1#2 deletion lines was significantly reduced (Figure 4G). Remarkably, the expression of TRY returned to wild-type levels in the DHS7#2 deletion lines at Day 21 (Figure 4G). In cauline leaves from 30-d-old plants, significant repression was observed only in DHS7#1#2 deletion lines (Figure 4H). In flowers from 35-d-old plants, expression of TRY in all deletion lines was reduced by ∼25% (Figure 4I). In the first internode of the main stems from 35-d-old plants, expression of TRY of DHS7#1 and DHS7#1#2 deletion lines was slightly decreased compared to wild type (Figure 4J). These results confirm that the DHS7#1 and DHS7#2 enhancers regulate the expression of TRY in different tissues and different developmental stages.
The DHS7#1 enhancer regulates flowering
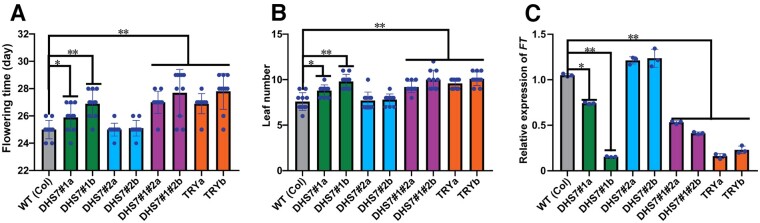
We observed several distinct phenotypes associated with deletion lines. Both DHS7#1a (69 bp deletion) and DHS7#1b (81 bp deletion) lost the putative SPL4- and SPL9-binding motifs (Figure 3A). Under long-day condition (16 h light), both DHS7#1a and DHS7#1b flowered later than wild-type plants. DHS7#1b plants flowered slightly later than DHS7#1a plants (Figure 5A). The TRY knockout mutants also showed a late-flowering phenotype (Figures 5A and 6A). The rosette leaf numbers of DHS7#1 deletion lines and TRY mutants increased under long-day conditions (Figure 6B). The DHS7#1#2 double deletion lines (187 bp and 166 bp deletions, respectively, Supplemental Figure S4) showed a similar late-flowering phenotype as the DHS7#1 deletion lines (Figure 5A). However, the two DHS7#2 deletion lines showed normal flowering time (Figure 5A).
Figure 5.
Phenotypes of DHS7 CRISPR/Cas9 lines and TRY mutants. A, Images of seedlings revealing their different flowering times. DHS7#2a and DHS7#2b deletion lines showed normal flowering time compared to the wild-type plants. DHS7#1 deletions, DHS7#1#2 double deletions, and TRY knockout mutants showed delayed flowering. All plants were of the same age and grown in parallel under long-day conditions (16 h light 8 h dark). Images were taken from 30-d-old plants. Bar = 2 cm. B, C, Trichome phenotypes. Increased percentage of trichome with altered numbers of branches or trichome clusters were associated with DHS7#2 deletions, DHS7#1#2 double deletions, and TRY mutants. Images were taken from the third leaf of 2-week-old seedlings. Bar = 1 mm in (B), 0.5 mm in (C).
Figure 6.
Transcription of FT and its related phenotypic changes of the DHS7 CRISPR/Cas9 lines and TRY mutants. A, Flowering times (days) of the DHS7 CRISPR/Cas9 lines and TRY mutants. Plants were the same age and grown in parallel under long-day conditions (16 h light 8 h dark). B, Leaf numbers of the DHS7 CRISPR/Cas lines and TRY mutant lines. C, Expression of FT in the leaf tissues of 21-d-old CRISPR/Cas lines. Data for flowering time and leaf number were collected from a minimum of 10 plants per line. Values are average from three biological replicates. Ten plants of each line were used in every independent experiment. Error bars in (A)–(C) show the sd from three biological replicates. Statistically significant differences are indicated (*P < 0.05, **P < 0.01, Supplemental File S1).
It was previously reported that TRY mutation can delay the flowering of the cpl3 mutant, which has an early flowering phenotype. It was proposed that TRY may compete with CPL3 in the activation of CONSTANS (CO) and increase FLOWERING LOCUS T (FT) expression, which alters flowering time (Tominaga-Wada et al., 2013; Wada and Tominaga-Wada, 2015). FT is produced in the leaves and moves to the shoot apical meristem, where it evokes the transition from vegetative to reproductive meristem identity (Corbesier et al., 2007). Strikingly, the transcription of FT (AT1G65480; Figure 6C) and TRY (Figure 4G) were similarly repressed in the leaves of 21-d-old DHS7#1a and DHS7#1b plants compared to wild type plants. In addition, both genes showed a higher level of repression in DHS7#1b than DHS7#1a, which correlated with the larger deletion in DHS7#1b (81 bp) than in DHS7#1a (69 bp) as well as their flowering time (Figure 6A). Collectively, these results showed that the DHS7#1 enhancer regulates TRY transcription in late development stages and impacts the expression level of FT, which regulates flowering.
The DHS7#2 enhancer regulates trichome development
The DHS7#2a (56 bp) and DHS7#2b (68 bp) deletions lost the putative EDT1-binding motif (Figure 3A). Homozygous DHS7#2 deletion lines and the TRY mutants showed slight reductions in the length of main roots, although the reductions were not statistically significant compared to wild-type plants (Supplemental Figure S5). In leaf epidermis, the density and spacing of the trichomes were very uniform in wild-type plants, but were often irregular in DHS7#2a and DHS7#2b deletion lines (Figure 5B). The trichomes of wild-type plants have two or three branches (Figure 5, B and C), while four or more branches were rare (∼1.7%; Supplemental Table S3; Khosla et al., 2014). Interestingly, ∼10% of the trichomes from the DHS7#2 deletion lines showed four or more branches, and ∼1% trichomes formed as part of a cluster (Figure 5, B and C; Supplemental Table S3), and this phenotype was not observed in wild type plants. Thus, the DHS7#2 enhancer appears to play a minor role in the regulation of development and distribution of trichomes in the leaf epidermis.
The DHS7#1#2 double deletion lines largely mirrored the branched and clustered trichome characteristics of the DHS#2 deletion lines (Figure 5, B and C). However, a unique aspect of the DHS7#1/#2 deletion lines was an increased frequency of trichome clusters and four-branched trichomes on the margin of cauline leaves (Supplemental Figure S6). This is in contrast to the DHS#2 deletion lines, in which we did not detect any four-branched trichomes or trichome clusters on the margin of cauline leaves (Supplemental Table S4).
The DHS7#2a and DHS7#2b deletion lines also lost the binding motif of AP1 (Figure 3A), a TF associated with floral development (Irish and Sussex, 1990). However, DHS7#2a and DHS7#2b plants did not show altered flowering time (Figure 5, B and C) or abnormal floral development. Thus, this AP1-binding motif within DHS7 does not appear to be functional under long-day conditions. We examined the expression of two genes flanking TRY (AT5G53190 and AT5G53210) and found that the transcription levels of these two flanking genes were not altered in the deletion lines compared to wild type plants (Supplemental Figure S7, A−D). Thus, the intronic enhancers appeared to only regulate the expression of TRY but no other nearby genes.
Enhancer activity of DHS44 is associated with gene CSLA10
DHS44 (423 bp) is located in the seventh intron (426 bp) of the gene CELLULOSE SYNTHASE LIKE A10 (CSLA10; AT1G24070; Figure 7A), one of the nine members of the CSLA gene family (Goubet et al., 2009), which is responsible for synthesis of β-linked mannan and glucomannan polysaccharides. Mannan is a type of hemicellulosic polysaccharide found in plant cell walls (Liepman et al., 2005, 2007). Previous analysis of single mutants for all nine genes showed no obvious phenotypic abnormalities, with the exception of the CSLA7 mutant, which was embryo lethal (Goubet et al., 2003, 2009). Transgenic assays using a GUS fusion construct with the 1.1 kb CSLA10 upstream region (CSLA10pro:GUS) revealed GUS signals in cotyledons, leaves, petals, stamens, style, and gynophore (floral organ abscission zone) of juvenile siliques (Skinner, 2012).
Figure 7.
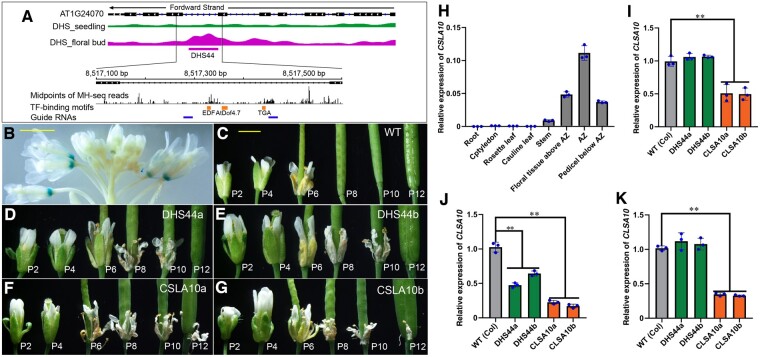
Functional characterization of the DHS44 enhancer. A, IGV tracks of CSLA10 (AT1G24070) and the DHS data sets. DHS44 is 423 bp long and is located in intron 7. DHS44 is expanded to show positions in the genomic sequence and the locations of midpoints of all MH-seq reads. The positions of binding motifs for three TFs and for the two guide RNAs used for CRISPR/Cas editing are also shown. B, Flower tissues from a 5-week-old transgenic plant-derived from construct DHS44. GUS signals were detected exclusively in the floral organ abscission zone of old flowers. Bar = 2 mm. C–G, Floral organ abscission phenotypes of wild type plants (C), deletion line DHS44a (D), deletion line DHS44b (E), mutant CSLA10a (F), mutant CSLA10b (G). Organ abscission occurs between positions 6 and 8 (P6 and P8) of flowers in wild-type plants, between P10 and P12 of flowers in DHS44a and DHS44b lines, and between P12 and P14 of flowers in CSLA10a and CSLA10b mutants. Bar for (C)–(G) = 2 mm. H, The relative expression levels of CSLA10 in different tissues. I, Expression of CSLA10 in floral organs above the abscission zone. J, Expression of CSLA10 in floral organ abscission zone. K, Expression of CSLA10 in pedicels below the abscission zone. Values are the average of three biological replicates. Statistically significant differences are indicated (**P < 0.01, Supplemental File S1). Error bars in (H)–(K) show the sd from three biological replicates.
We created 15 transgenic lines using a forward DHS44 enhancer construct, and 20 transgenic lines using a reverse DHS44 construct. Strikingly, GUS signals were observed exclusively in the floral organ abscission zone from Position 6 and older flowers (the first flower showing visible white petals was referred as Position 1 flower and then counted down along the inflorescence sequence) in all transgenic lines from both constructs (Figure 7B, Supplemental Figure S3). No signals were observed in any other tissues. This result illustrated a distinct function of DHS44 compared to the promoter of CSLA1 and suggested a potential role of DHS44 in floral organ abscission.
DHS44 deletions resulted in delayed floral organ abscission
We analyzed the distribution of midpoints of MH-seq reads associated with DHS44 and identified four peaks in this region (Figure 7A). Interestingly, several sequence motifs located close to the two middle peaks were identified to be associated with TFs related to senescence and abscission, including EDF, AtDOF4.7, and TGA (Wei et al., 2010; Wu et al., 2012; Chen et al., 2015). Therefore, we attempted to create CRISPR/Cas9 deletion lines targeting the region spanning these motifs (Figure 7A). Two independent homozygous deletion lines, named DHS44a (159 bp deletion, bases 130–288 within the 426-bp intron) and DHS44b (160 bp deletion, bases 130–289 in the intron; Supplemental Figure S8B). We also created two CSLA10 knockout mutants using CRISPR/Cas9, named CSLA10a and CSLA10b, both carrying a single bp insertion in exon 2, resulting in a frame shift mutation (Supplemental Figure S8C). Both DHS44a and DHS44b showed normal splicing of intron 7 based on RT-PCR and sequencing analysis (Supplemental Figure S8, E−G).
We observed a delay in floral organ abscission phenotype for both DHS44a and DHS44b, similar to the phenotypes of the knockout mutants (Figure 7, D−E). Organ abscission of petals, sepals, and stamens in the DHS44 deletion lines occurred from the siliques at position 10 − 13 flowers in DHS44a and DHS44b, but at position 5 − 8 flowers for the control plants (Supplemental Figure S8H). Senescence and abscission of floral organs usually occur after pollination and are well coordinated in wild-type Arabidopsis plants (Chen et al., 2015). The floral organs of DHS44a and DHS44b showed a normal progression of senescence, with sepals beginning to yellow at positions 3–4 flowers, and sepals, petals, and stamens withering at position 5–6 flowers, similar to that of the wild type (Figure 7C). These results suggest that the deletion of DHS44 has a significant effect on floral organ abscission but not on senescence.
We analyzed the expression of CSLA10 in various tissues. CSLA10 transcripts were only detected in stems and floral tissues but not in other tissues of wild-type plants and were most abundant in the floral abscission zone (Figure 7H). We then analyzed the expression of CSLA10 in different parts of young siliques from position 5–8 flowers among the deletion lines. In the floral abscission zone, the expression of CSLA10 was reduced to 55% in DHS44 deletion lines and 22% in CSLA10 mutants (Figure 7J). In the floral organs above the abscission zone and pedicel below the abscission zone, reduction of CSLA10 transcripts was observed only in CSLA10 mutants (Figure 7, I−K). We also examined the expression of the two genes flanking CSLA10 (AT1G24062 and AT5G24090). The transcription levels of these two genes were not altered in the deletion lines compared to wild-type plants (Supplemental Figure S7, E−G). Collectively, these results showed the DHS44 enhancer specifically regulates the expression of CSLA10 in the floral abscission zone and shows no impact on nearby genes.
Distinct features of TF-binding motifs in the intronic enhancers vs. the promoters of TRY and CSLA10
One intriguing question was—why did deletion of the intronic enhancers not completely abolish the tissue-specific expression of TRY and CSLA10? We wondered whether the promoters of these two genes had the capacity to attract similar TFs as those targeting the intronic enhancers. Interestingly, the TRY promoter is marked by a DHS detectable in both seedling and floral tissues. This DHS does not contain any of the TF motifs (SPL4, SPL9, and EDT1) that were detected in DHS7 (Figure 8A). Instead, an SPL4-binding motif was found upstream of the promoter DHS, 409 bp away from the TSS of TRY. In addition, an SPL9-binding motif and four copies of the EDT1-binding motif were found further upstream, 2,394–2,056 bp away from the TSS. These motifs were not associated with any DHSs in seedling and floral tissues. However, we cannot exclude the possibility that these motifs are associated with DHSs in other tissues, such as roots and stems, not yet analyzed by MH-seq/DNase-seq.
Figure 8.
Distribution of TF-binding motifs in the promoter regions of genes TRY and CSLA10. A, TF-binding motifs identified in the 5′ region of TRY. A DHS located in the promoter is shaded by a light blue box. The SPL4-binding motif upstream of the promoter is 409 bp away from the TSS of TRY. The SPL9-binding motif and four copies of the EDT1-binding motif are 2394–2056 bp from the TSS. Similar TF-binding motifs were not found in the TRY promoter. B, TF-binding motifs in the promoter of CSLA10. A single TGF- and four copies of AtDof4.7-binding motifs were identified in the promoter. An EDF-binding motif is identified in DHS44, but not in the promoter.
The promoter of CSLA10 is also marked by a DHS detectable in both seedling and floral tissues. This promoter DHS contains a TGA-binding motif and four copies of the AtDof4.7-binding motif, both TFs being related to senescence and abscission (Figure 8B). However, a binding motif for EDF, a repressor of senescence and abscission (Chen et al., 2015), was found in DHS44, but not in the CSLA10 promoter (Figure 8B). These analyses revealed that the TF-binding motifs identified within the DHS7 and DHS44 enhancers are different from those located in the promoter regions. These differences are manifested by presence/absence of a specific motif, the copy number of a motif, and the distance from the motif to the promoter (Figure 8).
Functional assays of the SPL9- and EDT1-binding motifs that were identified in the DHS7 enhancer
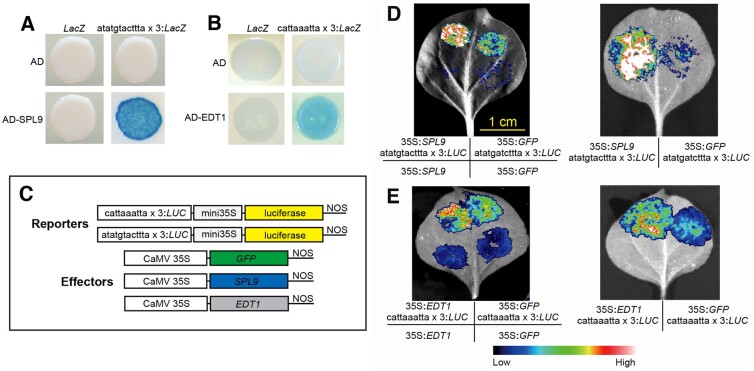
SPL9 is known to play a role in an endogenous flowering pathway and to activate TRY expression by binding to its promoter (Wang et al., 2009; Yu et al., 2010). We identified an ATATGTACTTTA sequence within the DHS7#1 enhancer, a sequence similar to the consensus SPL9-binding motif NNYGTACKKHH (O'Malley et al., 2016). We performed yeast one-hybrid (Y1H) experiments to determine whether SPL9 can bind to the ATATGTACTTTA sequence (see Methods). The Y1H assay confirmed that SPL9 bound to the ATATGTACTTTA sequence in yeast cells and strongly activated the transcription of the downstream LacZ reporter gene (Figure 9A).
Figure 9.
Functional assays of the SPL- and EDT1-binding motifs identified in the DHS7#1 and DHS7#2 enhancers. A, Y1H assay of the SPL9-binding motif (atatgtacttta) identified in the DHS7#1 enhancer. SPL9 can bind to the “atatgtactttta × 3” motif in yeast and strongly activates LacZ transcription. B, Y1H assay of the EDT1-binding motif (cattaaatta) identified in the DHS7#2 enhancer. EDT1 can bind to the “cattaaatta × 3” motif in yeast, and moderately activated LacZ transcription. AD: empty pB42AD; LacZ: empty vector pLacZi. C, Schematic diagrams of the effector and reporter constructs used in luciferase bioluminescence assay. D, Luciferase bioluminescence assay showing the activation of the atatgtactttta-driven luciferase reporter gene by SPL9 in N. benthamiana leaves. Bar for (D) and (E) = 1 cm. E, Luciferase bioluminescence assay showing the activation of cattaaatta-driven luciferase reporter gene by EDT1 in N. benthamiana leaves. A. tumefaciens strain GV3101 carrying different combination of constructs was infiltrated into different regions of a N. benthamiana leaf. Images of luciferase activities were taken 2 d after the infiltration.
EDT1 (AT1G73360) is known to negatively regulate trichrome branching (Nakamura et al., 2006; Khosla et al., 2014). EDT1 mutants (Khosla et al., 2014) show a phenotype similar to the DHS7#2 deletion lines. We identified a CATTAAATTA sequence within the DHS7#2 enhancer, a sequence similar to the consensus EDT1-binding motif HMWTWAATGH (Weirauch et al., 2014). Our Y1H assays revealed that EDT1 can bind to the CATTAAATTA sequence and moderately activate the transcription of the LacZ reporter gene (Figure 9B).
We then made luciferase reporter vectors harboring three copies of the predicted SPL9-binding motif ATATGTACTTTA or the EDT1-binding motif CATTAAATTA (Figure 9C). We examined the ability of SPL9 or EDT1 to activate reporter gene expression in Nicotiana benthamiana leaves. Transient expression assays showed that the ATTTGTACTTTA sequence was strongly activated by SPL9 (Figure 9D), and the CATTAAATTA sequence was activated by EDT1 at a modest, but significant level (Figure 9E). Collectively, these results showed that the predicted SPL9- and EDT1-binding motifs identified in the DHS7#1 and DHS7#2 enhancers are functional. Thus, these motifs represent the functional units of CREs underlying the regulation of TRY gene expression.
Discussion
Transgenic assays using reporter genes have been among the most commonly used techniques to validate the function of predicted enhancers in both animals (Visel et al., 2008; Kvon et al., 2014; Osterwalder et al., 2018) and plants (Zhu et al., 2015; Yan et al., 2019). However, transgenic assays do not reveal the endogenous activity of the candidate enhancers. The random integration of reporter constructs into the host genome can strongly influence reporter gene transcription (Akhtar et al., 2013). Thus, careful phenotypic evaluation of multiple transgenic lines from the same reporter construct is required to obtain reliable information regarding the function of a candidate enhancer. A key question not addressed by transgenic assays is whether and how the candidate enhancer affects the expression of its cognate gene in vivo (Yao et al., 2015). The advent of CRISPR/Cas-based genome editing techniques has made it possible to mutate or delete specific enhancers, enabling evaluation and quantification of endogenous enhancer activities. Several cases of enhancer editing using CRISPR/Cas have been reported in animals (Hnisz et al., 2015; Shin et al., 2016; Tak et al., 2016; Moorthy et al., 2017; Rogers et al., 2017; Sancisi et al., 2017; Will et al., 2017; Barakat et al., 2018; Miao et al., 2018). Generally, deletions of targeted enhancers result in reduced expression of associated genes in mammalian species (Hnisz et al., 2015; Moorthy et al., 2017). In the current study, we demonstrate that deletions of three intronic enhancers did not abolish gene expression but resulted in aberrant levels of transcriptional repression of their cognate genes, a pattern similar to that reported in mammalian species.
One of the most intriguing questions of enhancer function is whether and how intronic enhancers affect gene expression differently from intergenic enhancers. Genome-wide analysis showed that the first introns are favorable locations of intronic enhancers in Arabidopsis (Figure 1C). The close distance to the TSSs may be favorable for such intronic enhancers to cooperate with the promoters in regulating gene expression. For instance, activation of Fgf5 expression in mice upon exit from naive murine pluripotency is regulated by one enhancer located in the first intron and four intergenic enhancers located at the downstream of the gene (Thomas et al., 2021). Interestingly, deletion of the intronic enhancer caused a significantly reduced Fgf5 expression compared to the deletions of the four intergenic enhancers (Thomas et al., 2021). All five enhancers were transcribed, but the levels of RNA Pol II at the intronic enhancer were 5- to 10-fold higher compared to the four intergenic enhancers. In addition, the levels of RNA Pol II at the enhancers correlate with the distance to the closest promoter rather than intrinsic enhancer strength (Thomas et al., 2021). A recent study in Arabidopsis using several known enhancers revealed that enhancers are most active immediately upstream of the promoter, and their activity levels are reduced when they are placed away from the promoter (Jores et al., 2020). These results showed that the distance of an enhancer to the promoter is a key factor for its function in Arabidopsis. Thus, introns, especially the first introns, are ideal locations for enhancers.
Intronic enhancers presumably regulate their cognate genes, although there are rare examples in which an intronic enhancer can regulate the expression of a different gene through long-range interactions (Smemo et al., 2014). Deletions of the DHS7 and DHS44 enhancers caused exclusive repression of their cognate genes, TRY and CSLA10, but had no impact on the transcription of genes flanking TRY and CSLA10. In contrast, we previously demonstrated that insertion of a 32-kb T-DNA near an intergenic enhancer can cause alteration of transcription of multiple flanking genes, suggesting that intergenic enhancers may regulate multiple flanking genes (Zhu et al., 2015). Data from more intronic and intergenic enhancers are required to tell whether the results from currently available enhancer studies represent a general trend.
Introns can elevate gene expression via other mechanisms than serving as a dock for TF binding, often referred to as intron-mediated enhancement (IME). Specifically, the presence of introns can increase the steady-state levels of mature mRNAs in the cytosol or the efficiency of mRNA translation (Shaul, 2017; Rose, 2019). Nevertheless, it is unclear if the functions of traditionally identified IME introns are in fact associated with TFs or other regulatory proteins. This question can now be addressed by analyses of genomic and epigenomic data sets derived from chromatin.
We predicted 941 intronic enhancers associated with 806 genes in seedling tissue and 1,271 associated with 1,069 genes in floral tissue. These numbers are likely an underestimate. First, we used a threshold of at least 80% of the DHS located in an intron to avoid calling DHSs associated with exons or UTR regions as intronic enhancers. Thus, some intronic DHSs may be neglected by our computational pipeline. Second, we recently showed that some open chromatin regions are not sterically accessible to DNase I or Tn5 and will not be recovered by DNase-seq or ATAC-seq (Zhao et al., 2020). Last and most importantly, our DHS data sets were generated from seedlings and floral buds, both containing many cell types as well as cells at different developmental stages. Signals of open chromatin specifically associated with a minority cell type are likely masked by bulk profiling. For example, GUS signals are detected in trichome cells in transgenic plants made using both DHS7 and DHS#2 enhancer constructs (Figure 3). However, open chromatin signals associated with DHS7 were not detected in the seedling tissue, indicating that the signals associated with trichomes were not detectable. Therefore, production of a complete enhancer map will rely on open chromatin data sets associated with more specific cell or tissue types as well as tissues at different developmental stages.
Methods
Enhancer validation using transgenic assays
Raw DHSs were identified using F-seq (Boyle et al., 2008) with parameters “-l 300 –t 4.” DHS data sets associate with 2-week seedling and flower bud tissues were generated and analyzed previously (Zhang et al., 2012). A total of 21 intronic DHSs (Supplemental Table S1) were selected for validation using a β-GUS reporter-based system (Zhu et al., 2015). Each of the selected DHSs was associated with an expressed cognate gene. In addition, the function of the cognate gene for selected DHSs has been previously studied. The DHS sequences were either amplified from A. thaliana genomic DNA or synthesized, and subsequently cloned into the pKGWFS7.0 vector containing a minimal CaMV35S promoter (mini35S, −50 to −2 bp) and the GUS reporter (Zhu et al., 2015). Constructs were delivered into Agrobacterium tumefaciens strain GV3101, and were used to transform A. thaliana ecotype Col-0. Seeds were harvested and selected on solid MS plates containing 50 mg mL−1 kanamycin. Positive transformants were transferred to soil and grown under normal greenhouse conditions (18°C–22°C, 16/8 h light/dark, light intensity of 70 µmol m−2 s−1).
GUS activity of transgenic lines was examined according to the published protocol (Zhu et al., 2015). Seedlings and inflorescences of independent transgenic lines were mixed in a single tube and soaked in GUS-staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% [v/v] Triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.05% [w/v] X-Gluc), vacuum infiltrated for 30 min, and then incubated in dark at 37°C overnight. The samples were then washed in 80% ethanol several times until the green color was eliminated. The cleared samples were observed directly using a microscope. The images were collected using an EPSON Perfection 4180 scanner and a LEICA MZ16F microscope. GUS signals were recorded for each transgenic plant. A DHS is considered to be validated as a functional enhancer only if more than 50% positive transgenic plants derived from the DHS construct showed consistent signals in the same tissue(s).
Creation of deletion lines using CRISPR/Cas-based genome editing
We used the Yao promoter-based CRISPR/Cas9 system (Yan et al., 2015) to delete the DHS7#1, DHS7#2, and DHS7#1#2 enhancers in AT5g53200 or the DHS44 enhancer in AT1G24070. In brief, two short guide RNAs flanking DHS7#1, DHS7#2, DHS7#1#2, or the DHS44 enhancer were designed (http://skl.scau.edu.cn/;Xie et al., 2017; Supplemental Figures S4 and S8). The first sgRNA was inserted into the BsaI site between the AtU6-26 promoter and the sgRNA of SK vector. The resulting vector was then double-digested with SpeI and NheI, and the fragment containing the first sgRNA expression cassettes was purified. The second sgRNA was also inserted into the AtU6-26-sgRNA-SK vector and then linearized by SpeI. The purified first sgRNA expression cassette was inserted into the linearized vector so that the two sgRNAs were cloned into AtU6-26-sgRNA-SK vector (Yan et al., 2015). The two sgRNA expression cassettes were then transferred into pCAMBIA1300-pYAO:Cas9 vector (a gift from Dr. Qi Xi, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China) using SpeI and NheI (Yan et al., 2015). The sgRNAs of TRY and CLSA10 were assembled into the pCAMBIA1300-pYAO:Cas9 vector to produce the TRY and CLSA10 knockout lines. All oligomers for sgRNAs used in the CRISPR/Cas9 vectors are listed in Supplemental Table S5. All plasmid vectors were introduced into A. tumefaciens GV3101 (Gold Biotechnology, Missouri, USA), and were used to transform wild type Arabidopsis (Col-0) plants using the floral dip method (Clough and Bent, 1998). Seeds from the T0 generation were screened on MS plates containing 20 µg L−1 hygromycin and 150 µg L−1 carbenicillin.
Genomic DNA was extracted from ∼4-week-old T1 transgenic plants via the cetyltrimethylammonium bromide method (Allen et al., 2006). PCR was conducted with Primer STAR GXL DNA polymerase (Takara, Cat. # R051S) to amplify the genomic regions surrounding the target sites. Primer sequences are listed in Supplemental Table S6. T1 lines showing the expected smaller PCR products were further confirmed by the Sanger sequencing. To identify Cas9-negative T2 plants, PCR was performed to amplify the Cas9 gene (Q99ZW2.1) from Streptococcus pyogenes. Approximately 100 independent T2 plants of each transgenic line were screened by PCR for the absence of SpCas9. Genomic DNA from the Cas9-negative T2 plants was then used to identify homozygous deletions.
Phenotyping of CRISPR/Cas deletion lines
Trichomes and root hairs associated with the CRISPR/Cas9 lines were analyzed and photographed using a LEICA MZ16F microscope as described previously (Wang et al., 2007). Seeds of all CRISPR/Cas9 lines were treated with 15% (w/v) NaClO for 5 min and washed 5 times with sterile double-distilled water. The surface-sterilized seeds were grown on solid MS medium in vertically oriented Petri plates. For each CRISPR/Cas9 line, at least ten individual 4-d-old seedlings were assayed for root hair and root length. For leaf trichome analysis, at least ten plants were examined for each line, counting the trichomes on the third and fourth leaves of 2-week-old plants. For flowering time analysis, 7-d-old seedlings were transplanted to soil and kept under long-day conditions (16 h day/8 h night) at 22°C. Developmental uniformity was controlled by selecting the 10 most uniform plants at ∼7 d after transplanting and rotating the trays 3 times every week (Wada and Tominaga-Wada, 2015).
Seeds of DHS44 deletion lines and CSLA10 knockout mutants were sown on soil, stratified for 2 d at 4°C, and transferred to growth chambers (16-h day/8-h night at 22°C). Floral organ abscission was assessed on inflorescences containing 13 or more open flowers before floral meristem arrest as described previously (Bleecker and Patterson, 1997). The flower position was counted from the first flower with visible white petals (P1) at the top of the inflorescence (Butenko et al., 2003). The inflorescences of all of the plants were brushed gently to remove unattached floral organs. Individual plants in which petals abscised after position 10 were selected as floral delayed abscission mutants and further characterized (Bleecker and Patterson, 1997).
Gene transcription and splicing assays
RNAs were extracted from 4-d-old root tips, 14- and 21-d-old rosette leaves, 30-d-old cauline leaves, 35-d-old flowers, and the first internode of main stems using the RNeasy Plant Mini Kit (Qiagen, Cat. # 74904). In addition, young siliques from position 5–8 flowers in the DHS44 deletion lines and CSLA10 knockout mutants were collected. Floral organ abscission zones were dissected by taking a 2-mm section of floral tissue comprised of 2/3 mm pedicel and 4/3 mm receptacle (Taylor and Walker, 2018). The floral organs above the abscission zone and pedicel below the abscission zone were also collected for RNA extraction. Complementary DNAs were synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Cat. # RR047A) according to the manufacturer’s instructions. qPCR was performed with TB Green® Fast qPCR Mix (Takara, Cat. # RR430A) on Roche LightCycler® 480 real-time PCR system (Roche, Switzerland) using the following program: 95°C for 1 min, followed by 5 s at 95°C and 1 min at 60°C for 40 cycles for TRY, FT, CSLA10, and ACT2 (AT3G18780; Czechowski et al., 2005). Relative expression levels were calculated by a formula using LightCycler 480 Software v1.5.0 (Roche, Switzerland) and normalized to the concentration of ACT2 mRNA. Experiments were performed in triplicates in three independent experiments and the results are presented as standard error of means of the biological replicates. We used 10 plants per line for all independent experiments, with the exception of “4-day-old root tips” in which we used 100 plants per line.
For splicing assays, we prepared cDNAs from leaf tissues of all DHS7 deletion lines or flower tissues of all DHS44 deletion lines. We use the TRY primers (F1: 5′-CGTCGTCGCCGTCGTAAGCA-3′ R1: 5′-GGTGAGGCTTGGTATGTTTG-3′, 280 bp) and CSLA10 primers (F3: 5′-GACAAAGCCAACTCCAGAT-3′ R3: 5′-CTCTAATGTTCCATCGAGG-3′, 1,590 bp) to amplify the full-length transcript of TRY and CSLA10. The exon 2 to exon 3 of TRY was amplified using primers (F2: 5′-CTCCATGACTCTGAAGAAGTG-3′ R2: 5′-CAGCAAAGCCTTCACTGTTTC-3′, 234 bp). Similarly, exon 7 to exon 8 of CSLA10 was amplified using primers (F4: 5′-CGTTCGTGCTGGTCTTCTC-3′ R4: 5′-CCCATTCGTTCACTCTTTG-3′, 436 bp). The RT-PCR products were cloned into pGEM-T vector (Promega, Wisconsin, USA) and 20 clones were randomly selected for Sanger sequencing (Li et al., 2019b).
Y1H assay and transient expression analyses
The coding sequences of SPL9 and EDT1 were cloned in the pB42AD vector (Clontech, California, USA; pB42AD-SPL9, pB42AD-EDT1). Three copies of the predicted binding motif for SPL9 (ATATGTACTTTA) and EDT1 (CATTAAATTA) were synthesized and fused with the Placzi reporter vector (Clontech; pLacZi-SPL9, pLacZi-EDT1). The pB42AD gene construct and the Placzi vector containing three copies of the binding motif were co-transformed into the EGY48 yeast strain (Clontech) according to the Yeast Protocols Handbook. Yeast strains containing the empty pB42AD in combination with Placzi vectors or pB42AD gene vector combined with an empty Placzi vector were used as negative controls. The transformants were grown on SD/-Trp/-Ura for 48 h, and then spotted on the plate containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) for blue color development (Lin et al., 2007).
Transient expression assay was performed as described previously with only minor modifications (Hellens et al., 2005). The reporter construct pGreenII 0800-LUC (Creative Biogene, New York, USA) containing the luciferase reporter gene driven by three copies of TF binding motif sequences linked to the CaMV 35S minimal promoter and the effector construct pGreenII 62-SK containing SPL9 or EDT1 driven by the CaMV 35S promoter. The effector construct pGreenII 62-SK was also used to express GFP as a negative control. The reporter construct, transformed into Agrobacterium strain GV3101, was mixed with a Agrobacterium strain GV3101 carrying an effector construct and co-infiltrated into Nicotiana benthamiana leaves. As a negative control, the effector construct 35Spro:GFP was mixed with the respective reporter and infiltrated into the same leaf. Luciferase activities were recorded in these regions 2 d after infiltration.
Analyses of intronic sequences
To examine whether the size (length) or position of the introns is the main factor for DHS enrichment in the first introns, we calculated the LOR to test the DHS enrichment at different intron positions using:
The LOR score accounts for the sizes of the introns at different positions. To generate a background distribution of LOR, we also created a set of random intronic sequences by randomly permuting the intronic DHSs among all introns. The LOR scores of these random intronic sequences were calculated by the same formula. We used 1,000 times bootstrap to determine the p-values.
DHSs and ATAC-seq peaks were obtained from GSE34318 and GSE85203, respectively. A. thaliana gene annotation of Araport11 (Cheng et al., 2017) was used in overlaping analysis. Overlaps between DHS/ATAC-seq peaks and introns were analyzed using BEDTools (Quinlan and Hall, 2010). Entropy was calculated using published methods (Schug et al., 2005). Fisher’s exact test and correlation analysis were conducted using R. Motif database were obtained from MEME website (http://meme-suite.org/). Motifs in enhancers and promoters were identified using FIMO (Grant et al., 2011) against motif database “CIS-BP,” “ArabidopsisDAPv1,” and “ArabidopsisPBM_20140210” using default parameters.
Accession numbers
The accession numbers of genes inlcuded in this research: AT4G18960 (AG), AT4G09960 (AGL11/STK), AT1G24070 (CSLA10), AT1G73360 (EDT1), AT1G65480 (FT), AT2G42200 (SPL9) and AT5G53200 (TRY). The accession number of SpCas9 is Q99ZW2.1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analyses of intronic DHSs identified in A. thaliana.
Supplemental Figure S2. Expression of genes containing intronic DHSs.
Supplemental Figure S3. Representative images of GUS staining patterns from transgenic plants derived from seven reverse constructs.
Supplemental Figure S4. Characterization of CRISPR/Cas lines associated with DHS7.
Supplemental Figure S5. Root phenotypes of DHS7 CRISPR/Cas lines.
Supplemental Figure S6. Trichome phenotypes associated with mature cauline leaves from DHS7 CRISPR/Cas lines.
Supplemental Figure S7. Expression of genes flanking TRY or CSLA10 in CRISPR/Cas lines.
Supplemental Figure S8. Characterization of CRISPR/Cas lines associated with DHS44.
Supplemental Table S1. Genomic locations and primers used for the selected intronic DHSs and intronic non-DHS controls for functional validation.
Supplemental Table S2. GUS expression patterns of transgenic lines derived from intronic DHSs and intronic non-DHS controls.
Supplemental Table S3. Trichome number and cluster formation on leaves.
Supplemental Table S4. Trichome number and cluster formation on the margin of cauline leaves.
Supplemental Table S5. List of sgRNAs used in CRISPR/Cas editing.
Supplemental Table S6. List of primers for detection of CRISPR/Cas9 deletions.
Supplemental File S1. Statistical analysis tables.
Supplementary Material
Acknowledgments
We are grateful to Dr. Qi Xie for providing the Yao promoter-based CRISPR/Cas vectors.
Funding
F.M. was supported by National Key R & D Project for 14 Crop Breeding (2016YFD0100304) and Natural Science Foundation of Heilongjiang Province (C2018012). B.Z. was supported by Sichuan Science and Technology Program (2020YFH0147). This research is supported by NSF grant MCB-1412948 and MSU startup funds to J.J.
Conflict of interest statement. None declared.
F.M. and J.J. conceived the experiments. F.M., B.Z., M.Y., Y.L., and Y.H. conducted the experiments. F.M., H.Z., T.Z., and J.J. analyzed the data. F.M., H.Z., and J.J. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plcell) is: Fanli Meng (mengfanli@neau.edu.cn).
References
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B (2013) Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927 [DOI] [PubMed] [Google Scholar]
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W (1983) A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33: 729–740 [DOI] [PubMed] [Google Scholar]
- Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, Chambers I (2018) Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell 23: 276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE (1997) Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS (2008) F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Korf I (2008) Longer first introns are a general property of eukaryotic gene structure. Plos One 3: e3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling BE, Watson RA, Steinwand B, Bush DR (2016) Intronic sequence regulates sugar-dependent expression of Arabidopsis thaliana production of anthocyanin pigment-1/MYB75. PLoS One 11: e0156673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Gene Dev 1: 1183–1200 [DOI] [PubMed] [Google Scholar]
- Catarino RR, Stark A (2018) Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Gene Dev 32: 202–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Li PF, Chen MK, Lee YI, Yang CH (2015) FOREVER YOUNG FLOWER negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol 168: 1666–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C. et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al . (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friede A, Zhang BP, Herberth S, Pesch M, Schrader A, Hulskamp M (2017) The second intron is essential for the transcriptional control of the Arabidopsis thaliana GLABRA3 gene in leaves. Front Plant Sci 8: 1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HY, Kim SY, Park WD (1995) A potato Sus3 sucrose synthase gene contains a context-dependent 3' element and a leader intron with both positive and negative tissue-specific effects. Plant Cell 7: 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos JE, Rose AB (2017) Intron DNA sequences can be more important than the proximal promoter in determining the site of transcript initiation. Plant Cell 29: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani S, Altana A, Campanoni P, Morello L, Breviario D (2009) In trangenic rice, alpha- and beta-tubulin regulatory sequences control GUS amount and distribution through intron mediated enhancement and intron dependent spatial expression. Transgenic Res 18: 151–162 [DOI] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S (1983) A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33: 717–728 [DOI] [PubMed] [Google Scholar]
- Goubet F, Barton CJ, Mortimer JC, Yu XL, Zhang ZN, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P (2009) Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J 60: 527–538 [DOI] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang ZN, Twell D, Dupree P (2003) AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al . (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA (2013) Super-enhancers in the control of cell identity and disease. Cell 155: 934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA (2015) Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 58: 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YM, Mun JH, Kim H, Lee SY, Kim SG (2007) An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tissue-specific expression in transgenic Arabidopsis (Arabidopsis thaliana). Plant J 50: 230–239 [DOI] [PubMed] [Google Scholar]
- Jiang JM (2015) The ‘dark matter’ in the plant genomes: non-coding and unannotated DNA sequences associated with open chromatin. Curr Opin Plant Biol 24: 17–23 [DOI] [PubMed] [Google Scholar]
- Jo BS, Choi SS (2015) Introns: the functional benefits of introns in genomes. Genomics Inf 13: 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores T, Tonnies J, Dorrity MW, Cuperus JT, Fields S, Queitsch C (2020) Identification of plant enhancers and their constituent elements by STARR-seq in tobacco leaves. Plant Cell 32: 2120–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito MI, Lin XY, et al . (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG, et al . (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Paper JM, Boehler AP, Bradley AM, Neumann TR, Schrick K (2014) HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26: 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiker M, Airoldi CA, Losa A, Manzotti PS, Finzi L, Kater MM, Colombo L (2005) BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yanez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A (2014) Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95 [DOI] [PubMed] [Google Scholar]
- Li ZJ, Wang MY, Lin KD, Xie YL, Guo JY, Ye LH, Zhuang YL, Teng W, Ran XJ, Tong YP, et al . (2019a) The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol 20: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZX, Xiong XY, Wang FZ, Liang JP, Li JF (2019b) Gene disruption through base editing-induced messenger RNA missplicing in plants. New Phytol 222: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Nairn CJ, Willats WGT, Sorensen I, Roberts AW, Keegstra K (2007) Functional genomic analysis supports conservation of function among cellulose synthase-like a gene family members and suggests diverse roles of mannans in plants. Plant Physiol 143: 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci U S A 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J (2016) Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167: 1170–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZF, Hofmeister BT, Vollmers C, DuBois RM, Schmitz RJ (2017) Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZF, Marand AP, Ricci WA, Ethridge CL, Zhang XY, Schmitz RJ (2019) The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat Plants 5: 1250–1259 [DOI] [PubMed] [Google Scholar]
- Marand AP, Zhang T, Zhu B, Jiang JM (2017) Towards genome-wide prediction and characterization of enhancers in plants. BBA-Gene Regul Mech 1860: 131–139 [DOI] [PubMed] [Google Scholar]
- Mercola M, Wang XF, Olsen J, Calame K (1983) Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science 221: 663–665 [DOI] [PubMed] [Google Scholar]
- Miao YF, Ajami NE, Huang TS, Lin FM, Lou CH, Wang YT, Li S, Kang J, Munkacsi H, Maurya MR, et al . (2018) Enhancer-associated long non-coding RNA LEEN Eregulates endothelial nitric oxide synthase and endothelial function. Nat Commun 9: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy SD, Davidson S, Shchuka VM, Singh G, Malek-Gilani N, Langroudi L, Martchenko A, So V, Macpherson NN, Mitchell JA (2017) Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res 27: 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T (2006) Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol 141: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Huang SSC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissieres V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu YW, Plajzer-Frick I, Pickle CS, et al . (2018) Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554: 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39: 5328–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci WA, Lu ZF, Ji LX, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejia-Guerra MK, Colome-Tatche M, et al . (2019) Widespread long-range cis-regulatory elements in the maize genome. Nat Plants 5: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers WA, Goyal Y, Yamaya K, Shvartsman SY, Levine MS (2017) Uncoupling neurogenic gene networks in the Drosophila embryo. Gene Dev 31: 634–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8: 1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB (2019) Introns as gene regulators: A brick on the accelerator. Front Genet 9: 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar MK, Chow VT, Kangueane P (2004) Distributions of exons and introns in the human genome. In Silico Biol 4: 387–393 [PubMed] [Google Scholar]
- Sancisi V, Manzotti G, Gugnoni M, Rossi T, Gandolfi G, Gobbi G, Torricelli F, Catellani F, do Valle TF, Remondini D, et al . (2017) RUNX2 expression in thyroid and breast cancer requires the cooperation of three non-redundant enhancers under the control of BRD4 and c-JUN. Nucleic Acids Res 45: 11249–11267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. Embo J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ (2005) Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6: R33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Peterson T, Chopra S (2007) Epigenetic modifications of distinct sequences of the p1 regulatory gene specify tissue-specific expression patterns in maize. Genetics 175: 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O (2017) How introns enhance gene expression. Int J Biochem Cell Biol 91: 145–155 [DOI] [PubMed] [Google Scholar]
- Shin HY, Willi M, Yoo KH, Zeng XK, Wang CC, Metser G, Hennighausen L (2016) Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet 48: 904–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15: 272–286 [DOI] [PubMed] [Google Scholar]
- Sicard A, Kappel C, Lee YW, Wozniak NJ, Marona C, Stinchcombe JR, Wright SI, Lenhard M (2016) Standing genetic variation in a tissue-specific enhancer underlies selfing-syndrome evolution in Capsella. Proc Natl Acad Sci U S A 113: 13911–13916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M (2012) Characterization of the expression patterns of CELLULOSE SYNTHASE-LIKE A (CSLA) genes in Arabidopsis thaliana using reporter promoter-fusions and immunolocalization. Master's Theses and Doctoral Dissertations. University of Michigan.
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al . (2014) Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YG, Hung YL, Yao LJ, Grimmer MR, Do A, Bhakta MS, O'Geen H, Segal DJ, Farnham PJ (2016) Effects on the transcriptome upon deletion of a distal element cannot be predicted by the size of the H3K27Ac peak in human cells. Nucleic Acids Res 44: 4123–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, Walker JC (2018) Transcriptomic evidence for distinct mechanisms underlying abscission deficiency in the Arabidopsis mutants haesa/haesa-like 2 and nevershed. BMC Res Notes 11: 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas HF, Kotova E, Jayaram S, Pilz A, Romeike M, Lackner A, Penz T, Bock C, Leeb M, Halbritter F, et al . (2021) Temporal dissection of an enhancer cluster reveals distinct temporal and functional contributions of individual elements. Mol Cell 81. 10.1016/j.molcel.2020.1012.1047 [DOI] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Wada T (2017) Extended C termini of CPC-LIKE MYB proteins confer functional diversity in Arabidopsis epidermal cell differentiation. Development 144: 2375–2380 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Nukumizu Y, Wada T (2013) Flowering is delayed by mutations in homologous genes CAPRICE and TRYPTICHON in the early flowering Arabidopsis cpl3 mutant. J Plant Physiol 170: 1466–1468 [DOI] [PubMed] [Google Scholar]
- Van de Velde J, Van Bel M, Vaneechoutte D, Vandepoele K (2016) A collection of conserved noncoding sequences to study gene regulation in flowering plants. Plant Physiol 171: 2586–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Clancy M, Ferl RJ, Vasil IK, Hannah LC (1989) Increased gene expression by the first Intron of maize Shrunken-1 locus in grass species. Plant Physiol 91: 1575–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA (2008) Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tominaga-Wada R (2015) CAPRICE family genes control flowering time through both promoting and repressing CONSTANS and FLOWERING LOCUS T expression. Plant Sci 241: 260–265 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang SC, Kwak SH, Zeng QN, Ellis BE, Chen XY, Schiefelbein J, Chen JG (2007) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Weber B, Zicola J, Oka R, Stam M (2016) Plant enhancers: a call for discovery. Trends Plant Sci 21: 974–987 [DOI] [PubMed] [Google Scholar]
- Wei PC, Tan F, Gao XQ, Zhang XQ, Wang GQ, Xu H, Li LJ, Chen J, Wang XC (2010) Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol 153: 1031–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al . (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Lalonde S, Kuhn C, Frommer WB, Ward JM (2008) Introns control expression of sucrose transporter LeSUT1 in trichomes, companion cells and in guard cells. Plant Mol Biol 68: 251–262 [DOI] [PubMed] [Google Scholar]
- Will AJ, Cova G, Osterwalder M, Chan WL, Wittler L, Brieske N, Heinrich V, de Villartay JP, Vingron M, Klopocki E, et al . (2017) Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat Genet 49: 1539–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XM, Yu Y, Han LB, Li CL, Wang HY, Zhong NQ, Yao Y, Xia GX (2012) The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol 159: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XR, Ma XL, Zhu QL, Zeng DC, Li GS, Liu YG (2017) CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol Plant 10: 1246–1249 [DOI] [PubMed] [Google Scholar]
- Xie YY, Zhang YL, Han JL, Luo JK, Li GS, Huang JL, Wu HB, Tian QW, Zhu QL, Chen YL, et al . (2018) The intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of ELONGATED UPPERMOST INTERNODE1 in rice. Mol Plant 11: 720–735 [DOI] [PubMed] [Google Scholar]
- Yan LH, Wei SW, Wu YR, Hu RL, Li HJ, Yang WC, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yan WH, Chen DJ, Schumacher J, Durantini D, Engelhorn J, Chen M, Carles CC, Kaufmann K (2019) Dynamic control of enhancer activity drives stage-specific gene expression during flower morphogenesis. Nat Commun 10: 1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao LJ, Berman BP, Farnham PJ (2015) Demystifying the secret mission of enhancers: linking distal regulatory elements to target genes. Crit Rev Biochem Mol 50: 550–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Cai WJ, Wang SC, Shan CM, Wang LJ, Chen XY (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A (2015) Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WL, Zhang T, Wu YF, Jiang JM (2012) Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang W, Chen L, Wang L, Marand AP, Wu Y, Jiang JM (2018) Proliferation of regulatory DNA elements derived from transposable elements in the maize genome. Plant Physiol 176: 2789–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HN, Zhang WL, Zhang T, Lin Y, Hu YD, Fang C, Jiang JM (2020) Genome-wide MNase hypersensitivity assay unveils distinct classes of open chromatin associated with H3K27me3 and DNA methylation in Arabidopsis thaliana. Genome Biol 21: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zhang WL, Zhang T, Liu B, Jiang JM (2015) Genome-wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 27: 2415–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicola J, Liu LY, Tanzler P, Turck F (2019) Targeted DNA methylation represses two enhancers of FLOWERING LOCUS T in Arabidopsis thaliana. Nat Plants 5: 300–307 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.