Abstract
MicroRNAs (miRNAs) control various biological processes by repressing target mRNAs. In plants, miRNAs mediate target gene repression via both mRNA cleavage and translational repression. However, the mechanism underlying this translational repression is poorly understood. Here, we found that Arabidopsis thaliana HYPONASTIC LEAVES1 (HYL1), a core component of the miRNA processing machinery, regulates miRNA-mediated mRNA translation but not miRNA biogenesis when it localized in the cytoplasm. Cytoplasmic HYL1 localizes to the endoplasmic reticulum and associates with ARGONAUTE1 (AGO1) and ALTERED MERISTEM PROGRAM1. In the cytoplasm, HYL1 monitors the distribution of AGO1 onto polysomes, binds to the mRNAs of target genes, represses their translation, and partially rescues the phenotype of the hyl1 null mutant. This study uncovered another function of HYL1 and provides insight into the mechanism of plant gene regulation.
The nuclear miRNA biogenesis factor HYL1 also localizes to the cytoplasm to modulate miRNA-mediated translational repression.
Introduction
MicroRNAs (miRNAs) are a class of 21–24 nucleotide small RNAs that regulate target gene expression through complementary base pairing at the post-transcriptional level (Bartel, 2004). They are generated from hairpin-containing primary miRNAs (pri-miRNAs) transcribed by RNA polymerase II (Cai et al., 2004; Lee et al., 2004; Xie et al., 2005; Kim et al., 2011). In animals, pri-miRNAs are first cleaved into miRNA precursors (pre-miRNAs) in the nucleus by Drosha and DiGeorge Critical Region 8 (Lee et al., 2003; Gregory et al., 2004; Han et al., 2004a). The pre-miRNAs are then exported to the cytoplasm and cleaved by Dicer and Transactivation Response Element RNA-binding Protein to release miRNA: miRNA* (asterisk indicates the passenger strand) duplexes (Bernstein et al., 2001; Lee et al., 2002; Zhang et al., 2004; Park et al., 2011; Lee and Doudna, 2012). In plants, the Dicer-like enzyme, DCL1, performs both steps of cutting on pri-miRNAs to generate mature miRNAs in the nucleus (Park et al., 2002; Reinhart et al., 2002; Kurihara and Watanabe, 2004). HYPONASTIC LEAVES1 (HYL1) and SERRATE (SE) influence DCL1 by increasing the efficiency and accuracy of miRNA processing (Vazquez et al., 2004; Han et al., 2004b; Grigg et al., 2005; Hiraguri et al., 2005; Kurihara et al., 2006; Lobbes et al., 2006; Yang et al., 2006a; Song et al., 2007; Dong et al., 2008; Yang et al., 2010; Manavella et al., 2012; Yang et al., 2014). As great efforts have been made to decipher the mechanism underlying this canonical miRNA biogenesis, several new proteins, such as DAWDLE (Yu et al., 2008; Zhang et al., 2018) and TOUGH (Ren et al., 2012), have been identified to interact with the DCL1 complex and enhance the processing of pri-miRNAs. Furthermore, additional factors, including cap-binding protein (CBP) complex (Gregory et al., 2008; Kim et al., 2008; Laubinger et al., 2008), Modifier Of Snc1, 2 (Wu et al., 2013), and RACK1 (Speth et al., 2013), also regulate miRNA biogenesis to produce miRNAs more efficiently. After cleavage, miRNA/miRNA* duplexes are methylated by HUA ENHANCER 1, which protects them from degradation (Yu et al., 2005; Yang et al., 2006b).
After being generated, miRNAs are then loaded into an ARGONAUTE1 (AGO1)-dominant RNA-induced silencing complex (RISC) and repress their target mRNAs mainly in two ways: cleavage and translational repression. In plants, RISC-catalyzed mRNA cleavage was described before mRNA-mediated translational repression (Llave et al., 2002; Tang et al., 2003; Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005). After cleavage, the 5′ fragments produced could be uridylated by HESO1 (Ren et al., 2014), and degraded by the SKI2 complex, nonstop decay factors Pelota/Hbs1, or RISC-interacting clearing 3′–5′ exoribonucleases (Branscheid et al., 2015; Zhang et al., 2017b; Szadeczky-Kardoss et al., 2018). For a long time, mRNA cleavage was thought to be the major function of plant miRNAs (Jones-Rhoades et al., 2006) since they always share a high complementation to their target mRNAs.
However, in recent years, emerging evidence suggests that translational repression also plays a vital role in miRNA-mediated post-transcriptional regulation. Mainly recognized as the disproportionate effects of miRNA on its target mRNA versus protein level, translational repression activity has been confirmed for several plant miRNAs (Aukerman and Sakai, 2003; Chen, 2004; Arteaga-Vazquez et al., 2006; Gandikota et al., 2007; Brodersen et al., 2008; Dugas and Bartel, 2008; Lanet et al., 2009; Zhu et al., 2009; Beauclair et al., 2010; Li et al., 2013a, 2013b). Using pulse labeling experiments, Li et al. provide direct evidence that two plant miRNAs, miR165/166 and miR398, inhibit the protein synthesis of their target genes (Li et al., 2013b). All these studies have revealed a widespread translational repression by plant miRNAs and the common coexistence of their two regulation modes (cleavage and translational repression) on the same target gene. However, how each mode is decided remains unclear. Specifically, an in vitro study using a catalytic AGO1 mutant has demonstrated that plant miRNAs require near-perfect base pairing for their translational repression activity (Iwakawa and Tomari, 2013), but with this almost fully complementary binding sequence, how these translationally repressed target mRNAs avoid being cleaved in vivo is largely unknown.
Furthermore, multiple factors involved in miRNA-mediated translational repression have been identified (Brodersen et al., 2008; Yang et al., 2012; Li et al., 2013b; Reis et al., 2015). In particular, ALTERED MERISTEM PROGRAM1 (AMP1) is an integral membrane protein associated with AGO1 on the endoplasmic reticulum (ER). In the amp1 lamp1 double mutant, target mRNA accumulation was enhanced on the membrane-bound polysomes (MBPs) but not on the total polysomes, suggesting that the ER is the site of miRNA-mediated translation repression (Li et al., 2013b). Moreover, the identification of VARICOSE (VCS; homolog of the mammalian decapping activator Ge-1, Brodersen et al., 2008), SUO (a GW-repeat protein that colocalizes with the decapping activator DCP1, Yang et al., 2012), KATANIN 1 (KTN1; a subunit of the microtubule-severing enzyme, Brodersen et al., 2008), and DRB2 (a double-stranded RNA-binding protein, Reis et al., 2015) as required translational repression factors suggests that multiple cellular processes, such as microtubule dynamics and mRNA decapping, contribute to the repression of target protein synthesis. However, the molecular roles of these factors and their crosstalk during translational repression merit closer investigation.
As a double-stranded RNA-binding protein, Arabidopsis thaliana HYL1 is involved in many aspects of plant development (Lu and Fedoroff, 2000; Vazquez et al., 2004; Wu et al., 2007; Liu et al., 2011; Li et al., 2012; Lian et al., 2013; Sacnun et al., 2020), and its close homolog BcpLH determines the leaf curvature and properties of the leafy head in Chinese cabbage (Ren et al., 2020). HYL1 achieves these developmental regulations mainly through controlling miRNA biogenesis in the nucleus, and its protein modification (e.g. phosphorylation/dephosphorylation and ubiquitination) is critical for this function (Manavella et al., 2012; Cho et al., 2014; Karlsson et al., 2015; Raghuram et al., 2015; Su et al., 2017; Yan et al., 2017). In addition, several studies have shown that HYL1 can also localize to the cytoplasm and its translocation by Ketch1 is important for nuclear pri-miRNA processing (Han et al., 2004b; Cho et al., 2014; Yang et al., 2014; Zhang et al., 2017a). However, whether HYL1 functions in the cytoplasm and, if so, how it does so remain poorly understood.
Here, we revealed another function of HYL1 in the cytoplasm. HYL1 localized on the ER, and associated with AGO1, the core factor of RISC, and AMP1. The cytoplasmic HYL1 facilitated the loading of AGO1 onto polysomes and miRNA-mediated translational repression. Our findings demonstrated another function of HYL1 in the miRNA pathway and provided insight into the mechanisms underlying miRNA-mediated translational repression. HYL1 can be defined as a factor with dual contributions to miRNA biogenesis and function.
Results
HYL1 localizes in both the nucleus and cytoplasm
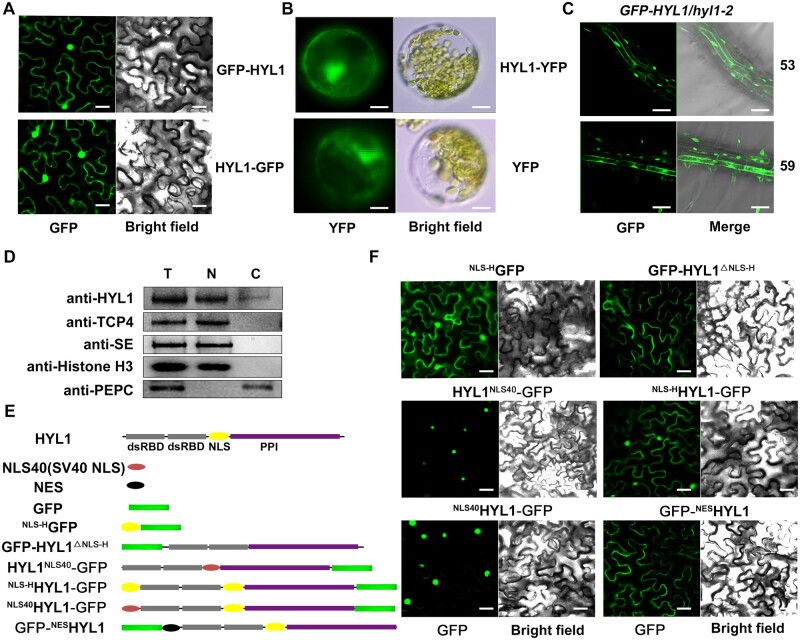
In a previous study, in which protein interactions were examined using bimolecular fluorescence complementation, we found that the fluorescence signals of HYL1-nYFP/DCL1-cYFP and HYL1-nYFP/SE-cYFP appeared only in the nucleus, while the signal of HYL1-nYFP/HYL1-cYFP was in both the nucleus and cytoplasm (Yang et al., 2014). Since HYL1 acts as an important partner of DCL1 to accurately cleave the pri-miRNAs in the nucleus (Han et al., 2004b; Hiraguri et al., 2005; Kurihara et al., 2006; Song et al., 2007; Dong et al., 2008), this finding raised the question of whether (and if so, then how) cytoplasmic HYL1 functions in the miRNA pathway. To verify its cytoplasmic localization, HYL1 was fused with a GFP tag at either the N- (GFP-HYL1) or C-terminus (HYL1-GFP) and transiently expressed in Nicotiana benthamiana. Both fusion proteins were present in the nucleus and cytoplasm (Figure 1A). When the HYL1-YFP fusion protein was transiently expressed in Arabidopsis protoplasts, the fluorescence signals were also found in both the nucleus and cytoplasm (Figure 1B). In addition, the introduction of Pro35S:GFP-HYL1 (hereafter referred to as GFP-HYL1) into the hyl1-2 null mutant generated GFP-HYL1 fluorescence signals in both the nucleus and cytoplasm (Figure 1C), and the mutant phenotypes were fully rescued (Supplemental Figure S1A), suggesting that the fusion protein was functional. To further investigate the subcellular accumulation of HYL1 directly in vivo, we performed a cell-fractionation assay. HYL1 was detected in both cytoplasmic and nuclear fractions extracted from wild-type (WT) Arabidopsis (Columbia [Col] ecotype; Figure 1D). As controls, histone H3 and phosphoenolpyruvate carboxylase (PEPC) were only detected in the nuclear or cytoplasmic aliquots, respectively. Importantly, another two nuclear proteins, SE and TEOSINTE BRANCHED1/CYCLOIDEA/PCF family transcription factor 4 (TCP4; approximately two-fold and 5.3-fold at the mRNA level compared to HYL1 in the seedling, respectively, Ran et al., 2020), were both restricted to the nuclear fraction (Figure 1D), suggesting that the cytoplasmic HYL1 detected was not due to the nuclear leakage during fraction preparation. Together, these results revealed that HYL1 localizes to the cytoplasm as well as the nucleus.
Figure 1.
Subcellular localization of HYL1 and its various chimeric proteins. A, Subcellular localization of GFP-HYL1 and HYL1-GFP in N. benthamiana leaves. B, Subcellular localization of HYL1-YFP in Arabidopsis protoplasts. C, Subcellular localization of HYL1-GFP in the root of two independent Pro35S:GFP-HYL1/hyl1-2 transgenic lines. D, Immunoblot analysis to detect HYL1 protein in the nuclear and cytoplasmic fractions of WT plant. T, N, and C represent total, nuclear, and cytoplasmic aliquots, respectively. E, Schematic diagrams of various HYL1 chimeric proteins. Grey block, HYL1 double-stranded RNA-binding domain; Yellow oval, NLS of HYL1 (NLS-H); Purple block, PPI domain of HYL1; Red oval, NLS of SV40 (NLS40); Black oval, NES; Green block, GFP protein. F, Localization of various HYL1 fusion constructs from (E) in N. benthamiana leaves. Scale bar, 30 μm in (A) and (F); 5 μm in (B); and 100 μm in (C).
Nuclear HYL1 is not fully functional in plant development
HYL1 contains a putative nuclear localization signal (NLS) in the middle of the protein. Because of the dual localizations of this protein, we next determined whether this NLS (hereafter referred to as NLS-H) is responsible for the localization of HYL1 (Figure 1E). Firstly, GFP was fused with NLS-H at its C-terminus and transiently expressed in N. benthamiana. The fluorescence signal was observed in both the nucleus and cytoplasm (Figure 1F), which indicated that NLS-H might not be a nuclear-specific localization signal. Then, the NLS-H domain was deleted, and the fluorescence signal of HYL1△NLS-H-GFP was seen only in the cytoplasm, suggesting that NLS-H is required for the nuclear localization of HYL1 but is too weak to restrict all HYL1 proteins to the nuclei. To evaluate the function of nuclear HYL1, we excluded the cytoplasmic HYL1 by replacing NLS-H with a strong NLS signal, SV40 NLS (PKKKRKV, hereafter referred to as NLS40; Kalderon et al., 1984a, 1984b; Adam and Gerace, 1991; Jouannet et al., 2012), and the fluorescence signal of HYL1NLS40-GFP chimera was seen only in the nucleus (Figure 1F). To eliminate any positional effects of the NLS, we independently fused HYL1-GFP with NLS-H (NLS-HHYL1–GFP) or NLS40 (NLS40HYL1–GFP) at the N-terminus (Figure 1E). The presence of NLS40 resulted in the nuclear-specific localization of HYL1-GFP, while NLS-H did not (Figure 1F). Thus, compared with NLS-H, the function of NLS40 appeared stronger and was sufficient to confine the nuclear-specific localization of HYL1.
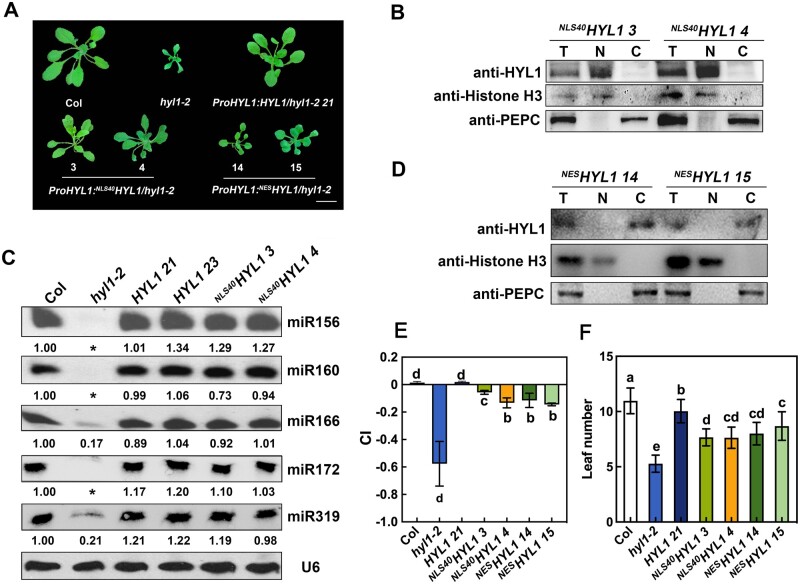
To investigate the roles of the nuclear-localized HYL1, HYL1 and NLS40HYL1 were introduced into the hyl1-2 mutant under the control of its native promoter, respectively. Transgenic plants with similar HYL1 expression levels to that of WT Arabidopsis were chosen for further analysis (Figure 2, A–B; Supplemental Figure S1, B–C). Compared to hyl1-2, both HYL1 and NLS40HYL1 restored the accumulation of miRNAs almost back to the WT levels (Figure 2C), suggesting that nucleus-only NLS40HYL1 is sufficient for miRNA biogenesis. However, the NLS40HYL1 plants only showed a partial rescue of the hyl1-2 phenotype, including leaf number, leaf incurvature (curvature index [CI]; Liu et al., 2010), and plant stature, while the HYL1 plants showed almost complete rescue (Figure 2, A, E, and F). This result suggested that, compared to the native HYL1 with dual localizations, NLS40HYL1 is not fully functional in plant development. Thus, we hypothesized that cytoplasmic HYL1 may also play roles in miRNA-mediated gene regulation and plant development.
Figure 2.
Both nuclear and cytoplasmic HYL1 partially rescue the hyl1-2 phenotype. A, Phenotypes of transgenic plants expressing different HYL1 constructs at the rosette stage. Arabic numbers represent different transgenic lines. Scale bar, 1 cm. B, Immunoblot analysis to detect HYL1 protein in the nuclear and cytoplasmic fractions from the seedling of NLS40HYL1 transgenic plants. T, N, and C represent total, nuclear, and cytoplasmic aliquots, respectively. C, Northern blotting showing the accumulation of miRNAs in WT, hyl1-2, HYL1, and NLS40HYL1 transgenic plants. Asterisks repredcsents no band was observed. D, Immunoblotting to detect HYL1 protein in the nuclear and cytoplasmic fractions from the seedling of NESHYL1 transgenic plants. E, Quantification (Mean ± SD) showing the leaf CI of transgenic plants expressing different HYL1 constructs. The sixth rosette leaves of 4-week-old WT and transgenic plants were selected for CI measurement. n = 20. F, Quantification (mean ± SD) showing the leaf number of WT and different transgenic plants. n > 15. Statistically significant differences between groups were indicated by different letters. ANOVA, P < 0.05.
Cytoplasmic HYL1 functions in plant development but not in miRNA biogenesis
To determine how cytoplasmic HYL1 functions, we fused HYL1 with a strong nuclear export signal (NES; ALPPLERLTL, Fischer et al., 1995; Wen et al., 1995; Jouannet et al., 2012) at its N-terminus (Figure 1E). When transiently expressed in N. benthamiana leaves, GFP-NESHYL1 was localized only in the cytoplasm (Figure 1F), indicating that NES caused the specific cytoplasmic localization. Then GFP-NESHYL1 was introduced into the hyl1-2 mutant under the control of its native promoter. Cell-fractionation of ProHYL1:NESHYL1/hyl1-2 (hereafter referred to as NESHYL1) showed that the HYL1 fusion protein was detected only in the cytoplasm (Figure 2D), and transgenic plants with similar expression levels to WT were chosen for further analysis (Supplemental Figure S1, D and E). Compared to hyl1-2, NESHYL1 showed partial rescue in many aspects, including plant stature, leaf incurvature, and leaf numbers, to different degrees (Figure 2, A, E, and F). Together, these results suggested that NESHYL1 participates in plant development, presumably through miRNA-mediated gene regulation.
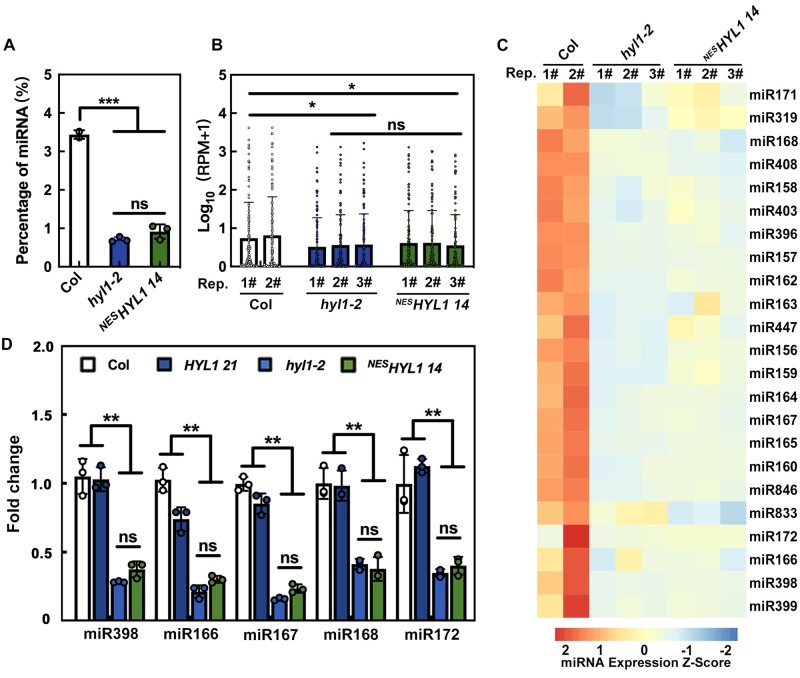
To investigate the function of cytoplasmic HYL1, we first explored whether it is also involved in miRNA biogenesis. To this end, small RNA sequencing was employed to detect the miRNA abundance in WT (n = 2), hyl1-2 (n = 3), and NESHYL1 14 (n = 3) plants. In hyl1-2 and NESHYL1 14, the proportion of miRNA among the total sRNA population (Figure 3A; Supplemental Figure S2A) and the global miRNA expression levels (i.e. [Log10 (reads per million [RPM] + 1)]; Figure 3B) were almost equal, and were much lower than in Col, suggesting that cytoplasmic HYL1 does not rescue the defect of hyl1-2 in miRNA biogenesis. We then analyzed the abundance of individual miRNAs. As shown in Figure 3C and Supplemental Figure S2B, most of the miRNAs accumulated similarly between hyl1-2 and NESHYL1 14, supporting the notion that cytoplasmic HYL1 is not sufficient for miRNA biogenesis. Lastly, to confirm the results obtained from high-throughput sequencing, we checked the accumulation of several miRNAs by reverse transcription quantitative PCR (RT-qPCR) . As shown in Figure 3D, in HYL1 plants, the accumulation of miRNAs was almost restored to WT levels. By contrast, the miRNA levels in NESHYL1 were almost the same as those in the hyl1-2 mutant (Figure 3D). These results, together with the fact that other miRNA processers (such as DCL1 and SE) only localized in the nucleus, suggested that cytoplasmic HYL1 is not sufficient for miRNA biogenesis. Instead, cytoplasmic HYL1 may affect plant development through post-miRNA-biogenesis steps.
Figure 3.
Cytoplasmic HYL1 is not sufficient for miRNA biogenesis. A, The percentage of miRNA reads in total clean reads of sRNA sequencing. B, The global version of miRNA abundance in WT, hyl1-2, and NESHYL1 14 plants. C, The heat map showing the z-score (accumulation level, see “Materials and methods” section for details) of some general miRNAs in WT, hyl1-2, and NESHYL1 14 plants. D, RT-qPCR showing the relative expression levels of miRNA in WT and different transgenic plants. P-values were calculated with a two-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. ns, no significance. Rep, replicates. Quantification is presented as mean ± SD.
Cytoplasmic HYL1 is involved in miRNA-mediated translational repression
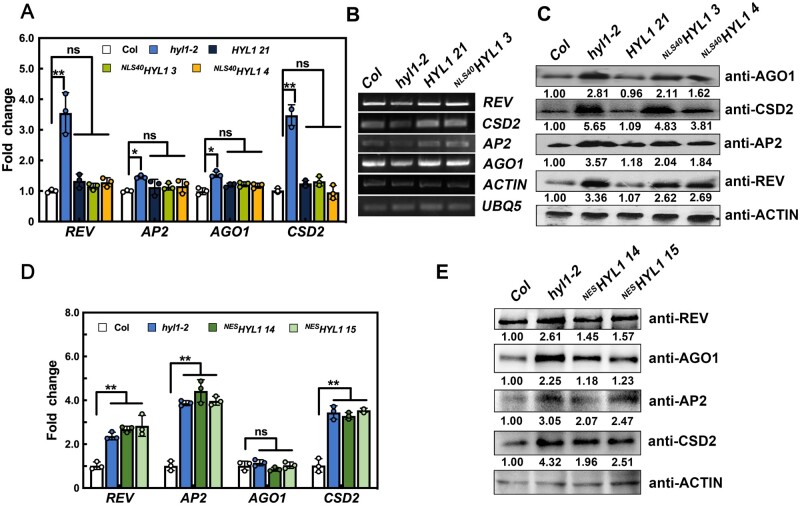
What could be the function of cytoplasmic HYL1? Since NLS40HYL1 only partially rescued the phenotype of hyl1-2 with the almost complete recovery of miRNA accumulation, we hypothesized that gene silencing may be disrupted when cytoplasmic HYL1 is absent. Therefore, we first examined the expression levels of several miRNA target genes in WT, hyl1-2, and transgenic plants. As shown in Figure 4A, the expression levels of REV (target of miR165/166), CSD2 (target of miR398), AP2 (target of miR172), and AGO1 (target of miR168) were higher in the hyl1-2 mutant, but were similar among WT,NLS40HYL1, and HYL1 plants. Furthermore, when detected by 5′ RACE RT-PCR (Li et al., 2013b), the abundance of their 3′ cleavage products was reduced in the hyl1-2 mutant, but showed no obvious difference among WT,NLS40HYL1, and HYL1 plants (Figure 4B). Together, these data suggested that miRNA-mediated target mRNA cleavage is not impaired without cytoplasmic HYL1.
Figure 4.
Translational inhibition of miRNA-targeted genes by cytoplasmic HYL1. A, The relative expression levels of miRNA-targeted mRNAs in WT, hyl1-2, HYL1, and NLS40HYL1 transgenic plants. B, 5′ RACE RT-PCR showing the accumulation of the 3′ fragments generated by miRNA-guided cleavage of target mRNAs. UBQ5 and ACTIN were used as loading controls. C, Immunoblotting showing the levels of target proteins in WT, hyl1-2, HYL1, and NLS40HYL1 transgenic plants. D, The relative expression levels of miRNA-targeted mRNAs in WT, hyl1-2, and NESHYL1 transgenic plants. E, Immunoblotting showing the levels of target proteins in WT, hyl1-2, and NESHYL1 transgenic plants. P-values were calculated with a two-tailed Student’s t test, *P < 0.05, **P < 0.01. ns, no significance. Quantification of the RT-qPCR is presented as Mean ± SD.
Subsequently, the protein levels of different target genes, including REV, CSD2, AP2, and AGO1, were examined using immunoblotting. These target proteins accumulated to higher levels in the hyl1-2 mutant than in the WT, and to almost the same levels in HYL1 and WT plants (Figure 4C). In NLS40HYL1 plants, however, the levels of these proteins showed only partial reduction and were still higher than in the WT (Figure 4C). This result, in conjunction with the largely unaltered target mRNA levels, indicated that miRNA-guided target mRNA translational repression was compromised in NLS40HYL1 plants. To further confirm that cytoplasmic HYL1 is indeed involved in translational repression, we then examined the mRNA and protein levels of target genes in NESHYL1. Compared to hyl1-2, the protein levels in NESHYL1 were reduced to different levels, while the mRNA levels were similar (Figure 4, D and E). Together, these results indicated that cytoplasmic HYL1 is responsible for translational repression of these target proteins.
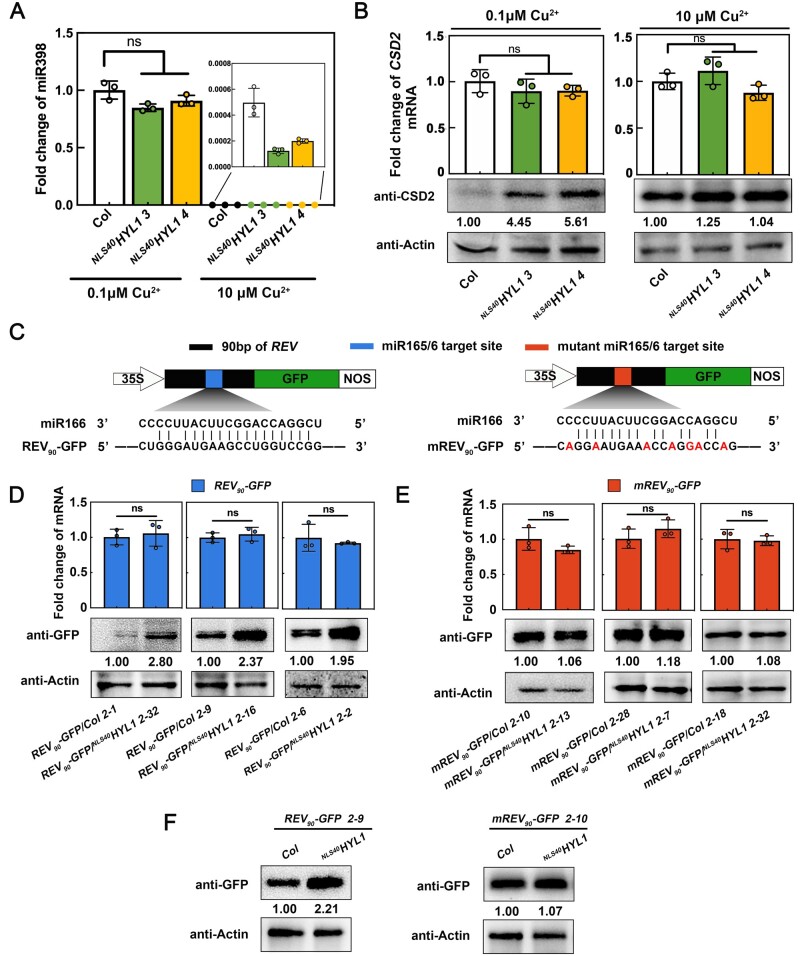
To rule out the possibility that cytoplasmic HYL1 is generally involved in protein translation, we tested whether its function required the activity of miRNA. To this end, we took advantage of the fact that miR398 is strictly regulated by Cu2+ levels (Yamasaki et al., 2007; Li et al., 2013b), and examined CSD2 protein levels under different Cu2+ conditions. As shown in Figure 5, A–B, CSD2 protein levels were higher in NLS40HYL1 than in the WT when grown in normal MS0 media containing 0.1 μM Cu2+, but showed no difference when miR398 was depleted by growing seedlings in Cu2+-replete conditions (10 μM). Similar results have been observed between hyl1-2 and NESHYL1 plants (Supplemental Figure S3, A and B). These results suggested that the translational repression on CSD2 by cytoplasmic HYL1 requires the activity of miR398.
Figure 5.
Cytoplasmic HYL1 influences translational repression in a miRNA-dependent manner. A, RT-qPCR showing the relative expression levels of miR398 in WT and NLS40HYL1 plants under different Cu2+ conditions. B, The CSD2 mRNA and protein levels in WT and NLS40HYL1 plants under different Cu2+ conditions. C, The schematic diagram of reporter gene construction. Black block, 90 bp of REV; Blue block, miR165/166 target site; Red block, the mutant miR165/166 target site; White arrow, 35S promoter; Green block, GFP; White block, Terminator. D, The mRNA and protein levels of REV90-GFP in different background transgenic plants. E, The mRNA and protein levels of mREV90-GFP in different background transgenic plants. F, The protein levels of REV90-GFP or mREV90-GFP in WT or NLS40HYL1 plants carrying a transgene at the same genomic location. Quantification of the RT-qPCR is presented as Mean ± SD. P-values were calculated with two-tailed Student’s t test, *P < 0.05, **P < 0.01. ns, no significance.
To further demonstrate that cytoplasmic HYL1 regulates translational repression in a miRNA-dependent manner, we constructed reporter genes to directly compare the target protein expression levels under different backgrounds. In this experiment, a 90-base pair (bp) REV gene sequence that contained the miR165/6 target site was fused with GFP (REV90-GFP; Figure 5C), and then transformed into WT and NLS40HYL1 plants, respectively. Two transgenic plants with almost identical mRNA levels between the WT and NLS40HYL1 background were picked as one group, and three groups with independent transgenic events were analyzed. As shown in Figure 5D, in all three groups, the REV90-GFP protein levels were higher in NLS40HYL1 plants than in the WT. By contrast, when the miRNA binding site was mutated (mREV90-GFP; Figure 5C), miR165/6-resistant mREV90-GFP protein always accumulated to similar levels in the WT and NLS40HYL1 background plants (Figure 5, C and E). The same results were obtained when another reporter gene, SPL9 (SPL975-GFP versus mSPL975-GFP), was evaluated (Supplemental Figure S3, C–E).
Lastly, to ensure that these results were not due to the variation caused by random transgene integration in different transgenic plants, we picked two independent lines (REV90-GFP/NLS40HYL1 2-9 and mREV90-GFP/NLS40HYL1 2-10) and crossed them with a WT plant. By doing this, we were able to obtain homozygous lines that contain the same genomic locus transgene under WT or NLS40HYL1 backgrounds. As shown in Figure 5F, the REV90-GFP protein level was higher in NLS40HYL1 plants than in the WT, while mREV90-GFP levels were similar between these two genotypes. Together, all these results suggested that cytoplasmic HYL1 indeed regulates translational repression in a miRNA-dependent manner.
HYL1 co-localized with AGO1 and AMP1 on the ER
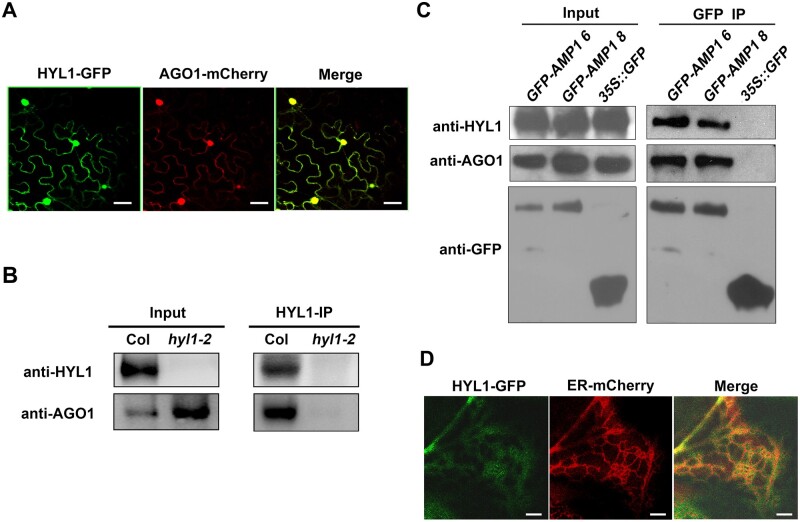
How is cytoplasmic HYL1 involved in this translational repression? Since AGO1 is the key protein of the gene silencing complex in Arabidopsis, we speculated that HYL1 may function together with AGO1. To investigate this hypothesis, HYL1-GFP and AGO1-mCherry were transiently co-expressed in N. benthamiana. AGO1-mCherry co-localized with HYL1-GFP both in the nucleus and cytoplasm (Figure 6A). This association was then analyzed by co-immunoprecipitation (Co-IP). After pulling down using HYL1 antibody, AGO1 was detected by immunoblotting in the Co-IP products of the WT (Figure 6B) but not in those of hyl1-2. This suggested that HYL1 associates with AGO1 in vivo.
Figure 6.
Co-localization of HYL1 with AGO1 and AMP1 on the ER. A, The co-localization of HYL1-GFP and AGO1-mCherry in N. benthamiana leaves. B, Immunoblotting showing the interaction between HYL1 and AGO1 using Co-IP products from WT and hyl1-2. C, Immunoblotting showing the interactions among HYL1, AGO1, and GFP-AMP1 using Co-IP products from the transgenic lines 6 and 8 of Pro35S:GFP-AMP1/Col transgenic plants. D, The co-localization of HYL1-GFP and ER-mCherry in N. benthamiana leaves. Scale bar, 30 μm in (A) and 5 μm in (D);
Because AMP1 is required for miRNA-mediated translational repression and associates with AGO1 (Li et al., 2013b), we next determined whether HYL1 and AMP1 are associated in vivo. To this purpose, Pro35S:GFP-AMP1 was transformed into Arabidopsis plants, and a Co-IP of GFP-AMP1 was performed using GFP antibodies. Immunoblotting showed that either HYL1 or AGO1 was enriched in the IP products of Pro35S:GFP-AMP1 plants, but not in that of Pro35S:GFP plants (Figure 6C). This result suggested that HYL1 is also associated with AMP1. miRNAs inhibit the translation of target mRNAs on the ER where AMP1 and AGO1 are localized (Li et al., 2013b). To examine whether HYL1 is associated with the ER, HYL1-GFP, and ER-mCherry (an ER marker; Nelson et al., 2007; Jouannet et al., 2012) were transiently co-expressed in N. benthamiana. The proteins were found to co-localize in a mesh-like pattern, which is typical for the ER (Figure 6D). This result suggested that HYL1 is associated with the ER.
Cytoplasmic HYL1 modulates the distribution of AGO1 on polysomes
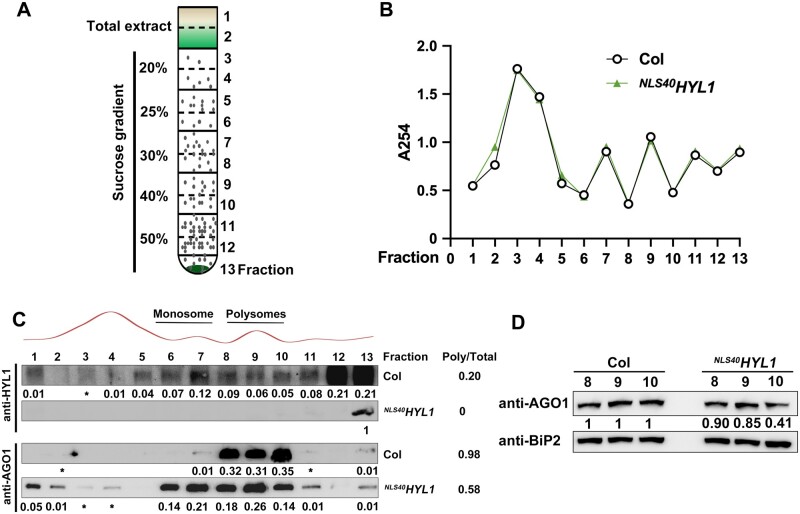
The translational repression of target mRNA is associated with the presence of miRNAs and AGO1 in polysomes (Lanet et al., 2009; Li et al., 2013b). Since HYL1 is associated with AGO1, we next determined whether cytoplasmic HYL1 regulates the distribution of AGO1 on polysomes. To examine this, the cell lysate was extracted from WT andNLS40HYL1 plants and resolved on sucrose density gradients (Figure 7A). After centrifugation, 13 fractions were collected along the gradients. The A254 absorption profiles from these two plants were largely similar (Figure 7B), further supporting the notion that NLS40HYL1 does not affect translation in general.
Figure 7.
Effect of cytoplasmic HYL1 on the enrichment of AGO1 in polysomes. A, A diagram showing the sucrose gradients used for the isolation of polysomes from wild-type and NLS40HYL1 plants. Sucrose gradients from 50% to 20% were injected into the tubes, and 13 fractions were collected for further analysis. B, The A254 absorption profiles of the 13 sucrose gradient fractions. C, Immunoblotting showing the accumulation of HYL1 and AGO1 proteins in the 13 sucrose gradient fractions. * indicates the intensity of the band is ˂0.01 compared to total abundance. D, Immunoblotting showing the accumulation of AGO1 proteins in fractions 8, 9, and 10 of sucrose gradients.
We then determined the amount of proteins in each fraction by immunoblotting (Figure 7C). In WT plants, HYL1 protein was detected in both monosome and polysome fractions, while AGO1 was mainly detected in the polysome fractions (Fractions 8–10). The distribution of HYL1 and AGO1 in the polysomes overlapped, consistent with the fact that these proteins are associated in vivo. It is also of note that HYL1 has a broader distribution than AGO1, suggesting that it may have other functions in the cytoplasm. In NLS40HYL1 samples, HYL1 proteins were detected only in the pellet fraction (Fraction 13). AGO1 was enriched in the other fractions in addition to those containing polysomes, indicating that the strict localization of HYL1 in the nucleus leads to the redistribution of AGO1 in the other fractions beyond polysomes. Also, the ratio of AGO1 protein in polysome fractions (0.58) was decreased in NLS40HYL1 compared with that of the WT (0.98). To rule out the possibility that the shift of AGO1 distribution in NLS40HYL1 was due to its higher AGO1 protein level (Figure 4C), immunoblotting was performed to examine the accumulation of AGO1 in polysome fractions directly (Figure 7D). An abundant ER-targeted protein-immunoglobulin binding protein 2 (BiP2) was used as a loading control, and the protein levels of AGO1 decreased in the polysome fractions of NLS40HYL1, compared to that of the WT. Together, these results indicated that cytoplasmic HYL1 facilitates the loading of AGO1 onto polysomes.
HYL1 binds to mRNAs of miRNA-targeted genes in the cytoplasm
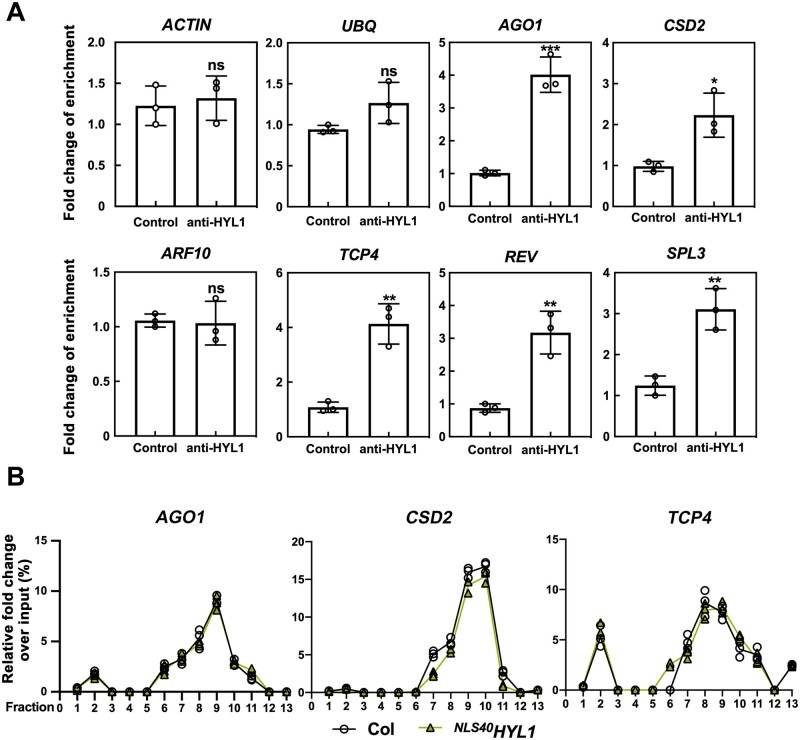
Since cytoplasmic HYL1 promotes the miRNA-directed repression of mRNA translation, HYL1 should form a complex together with target mRNAs in the cytoplasm. To test this hypothesis, we performed RNA immunoprecipitations (RIPs) in WT plants. After cell-fractionation, the nuclear and cytoplasmic aliquots of WT plants were incubated with HYL1 antibody, respectively, and the mRNA levels in the RIP products were further examined by RT-qPCR. Compared to the negative control without adding antibody, most target mRNAs (except ARF10) examined were significantly enriched in the HYL1-IP products from cytoplasmic aliquots (Figure 8A), while other abundant nontarget mRNAs, such as ACTIN or UBQ5, showed no difference. In the nuclear aliquots, however, no enrichments of these target mRNAs were observed (Supplemental Figure S4). These results suggested that cytoplasmic HYL1 specifically binds to miRNA-targeted mRNAs.
Figure 8.
The binding of cytoplasmic HYL1 to miRNA-targeted mRNAs. A, RT-qPCR showing the enrichment of the mRNAs in cytoplasmic HYL1-IP product. Samples lacking antibodies were used as the controls. Quantifications are presented as Mean ± SD. P-values were calculated with two-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. B, RT-qPCR showing the distribution of mRNAs along the sucrose gradients. Quantifications are presented as Mean ± SD.
Next, we determined whether cytoplasmic HYL1 plays a role in the accumulation of target mRNAs on polysomes. The polysomes were extracted from NLS40HYL1 and WT plants by the sucrose density gradient fractionation. Thirteen fractions were also collected after centrifugation, and the accumulations of mRNAs in each fraction were quantified by RT-qPCR. The three target mRNAs examined, AGO1, CSD2, and TCP4, were similarly distributed along with fractions in WT and NLS40HYL1 samples (Figure 8B), indicating that cytoplasmic HYL1 has no effect on the accumulation of target mRNAs on polysomes.
Discussion
HYL1 plays dual roles in mRNA biogenesis and translational repression
HYL1 is an important partner of DCL1 in miRNA processing and plays a key role in miRNA biogenesis in the nucleus. However, there are some clues indicating that HYL1 proteins are not only restricted to the nucleus (Han et al., 2004b; Cho et al., 2014; Yang et al., 2014; Zhang et al., 2017b), while its cytoplasmic function is largely unknown. Combining genetics with other approaches, such as biochemistry and high-throughput sequencing, we determined the role of cytoplasmic HYL1 in miRNA-mediated translational repression in this research.
In a recent study, Reis et al. also investigated the potential function of DRB1 (HYL1) in translational repression (Reis et al., 2015). They found that, compared to the WT, the mRNA and protein levels of six target genes they checked in the drb1 (hyl1) mutant were either both up-regulated or both unchanged. Thus, they concluded that DRB1 (HYL1) is not responsible for miRNA-guided translational repression. Out of these six target genes, three (AGO1, CSD2, and AP2) were also tested in our study, and the changes of target mRNA and protein levels in hyl1-2 were largely similar (Figure 4, D–E). However, although the target protein levels are up-regulated in both studies, it is unclear whether this is due to the impairment of translational repression. The defective miRNA biogenesis in the hyl1 null mutant result in the reduction of mRNA cleavage (Figure 4B) and the accumulation of target mRNAs in both studies, making it hard to distinguish the contribution of translational repression from mRNA cleavage.
To separate its cytoplasmic function from pri-miRNA processing, we generated NES/NLS40HYL1 transgenic plants. With similar miRNA and target mRNA levels (NLS40HYL1 versus WT, Figures 2C and 4A; NESHYL1 versus hyl1-2, Figures 3 and 4D), the target protein levels were clearly up-regulated in the absence of cytoplasmic HYL1 (NLS40HYL1 versus WT, Figure 4C) or partially rescued in its presence (NESHYL1 versus hyl1-2, Figure 4E). We further confirmed that this translational repression requires miRNA activities (Figure 5; Supplemental Figure S3). These results strongly suggested that cytoplasmic HYL1 is involved in miRNA-guided translational repression. HYL1 is a factor with dual roles in both miRNA biogenesis and function. Furthermore, since only translational repression but not mRNA cleavage was affected, NLS40HYL1 may serve as a good genetic material to investigate why plant miRNAs can use two different modes (cleavage and translational repression) to regulate the same target mRNA, and how each mode is decided.
Cytoplasmic HYL1 facilitates the enrichment of AGO1 in the polysome
Despite its important role in gene regulation, the mechanism underlying miRNA-guided translational repression is poorly understood. Particularly, although AGO1, as the core of miRNA-mediated translational repression, is known to localize to the polysomes (Lanet et al., 2009), how this localization is regulated remains largely unclear. In this study, we found that HYL1 associates with AGO1 in vivo and participates in the translational repression pathway, presumably by facilitating the loading of AGO1 on polysomes (Figure 9). Without cytoplasmic HYL1, AGO1 is redistributed to other fractions, thus leading to impaired translational repression. Interaction between AGO1 and HYL1 has also been found in foxtail millet (Setaria italica (L.) P. Beauv) (Liu et al., 2016), suggesting that this might be a conserved mechanism across the plant kingdom. Interestingly, recent studies have shown that AGO1 can shuttle between the cytoplasm and nucleus, and CARP9 facilitates the nuclear miRNA loading into AGO1, likely through stabilizing the HYL1–AGO1 interaction (Bologna et al., 2018; Tomassi et al., 2020). Whether this nuclear interaction is important for their cytoplasmic functions requires further investigation.
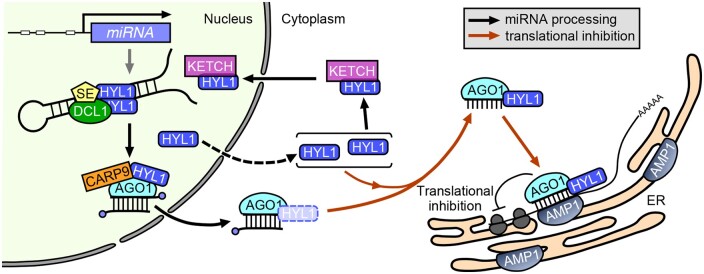
Figure 9.
An updated model for the functions of HYL1 in miRNA-mediated gene silencing. In plant, HYL1 functions in two distinct pathways: miRNA processing and translational repression. In the nucleus, HYL1 participates in miRNA processing along with DCL1 and SE. HYL1 protein is localized to polysomes on the ER where it associates with AGO1 and AMP1, and binds to the miRNAs and mRNAs of miRNA-targeted genes to form a miRNA-mediated effector complex.
The result that cytoplasmic HYL1 specifically binds to miRNA-targeted mRNAs (Figure 8A) suggests that it may have other functions in addition to facilitating the localization of AGO1, although we cannot rule out the possibility that this protein–RNA interaction is mediated by other components from the co-immunoprecipitated complex. The biological function of this interaction is unclear: it does not seem to be the determinant of which mode (cleavage or translational repression) miRNA is going to act, because the 3′ cleavage products of target mRNAs are not changing (Figure 4B) in NLS40HYL1. Moreover, it is also not critical for the accumulation of target mRNAs on the polysomes (Figure 8B). Since cytoplasmic HYL1 can localize to ER and associates with AMP1, it may work together with AMP1 to sequester miRNA target transcripts from MBPs, which still needs close investigation. Further study on the interrelation between cytoplasmic HYL1 and other factors, such as AMP1 or AGO1, may provide more mechanistic insights about what molecular role it is playing in the miRNA-guided translational repression pathway.
The crucial role of cytoplasmic HYL1 in plant development and its potential crosstalk with other components
Mutations in most of the known factors required for miRNA-guided translational repression, such as KTN1(Brodersen et al., 2008), VCS (Brodersen et al., 2008), SUO (Yang et al., 2012), and AMP1 (Li et al., 2013b), all result in a pleiotropic phenotype, indicating that translational repression is an essential activity for plant miRNAs. Consistently, in this study we showed that NLS40HYL1 does not fully rescue the developmental defect of hyl1-2 despite its intact target mRNA cleavage activity, suggesting that cytoplasmic HYL1 is also crucial for plant development. Further investigation showed that cytoplasmic HYL1 is involved in miRNA-guided translational repression. NESHYL1 can partially rescue target protein levels and the hyl1-2 phenotype without affecting mRNA levels, suggesting its important role in plant development through miRNA-guided translational repression.
It is also of note that although cytoplasmic HYL1 and AMP1 are associated with each other, compared to NLS40HYL1, amp1 has a stronger developmental defect. Moreover, among the mutants in the miRNA-guided translational repression pathway, amp1, amp1 lamp1, and vcs give the most severe developmental phenotype, while other mutants, including drb2, ktn1, suo, and NLS40HYL1, seem to be only mildly to moderately defective (Brodersen et al., 2008; Yang et al., 2012; Li et al., 2013b; Reis et al., 2015; this study). Why are these genes in the same pathway so different in plant development? Three possibilities may explain this difference: (1) some factors may have miRNA-independent functions that can also contribute to the pleiotropic developmental defects; (2) some factors, such as AMP1 or VCS, may play more dominant functions in this pathway; (3) redundancy may exist in the translational repression pathway, making the mutation of certain components less severe. For example, in NLS40HYL1, the loading of AGO1 onto polysomes is not completely abolished, suggesting that besides cytoplasmic HYL1, some other factors may also be involved in this step. Interestingly, as double-stranded RNA-binding proteins, both HYL1(DRB1) and DRB2 are found to participate in the miRNA biogenesis and miRNA-guided translational repression pathways (Han et al., 2004b; Vazquez et al., 2004; Eamens et al., 2012; Reis et al., 2015; this study), and they can regulate the translation of the same targets (e.g. AGO1, AP2, and CSD2; Reis et al., 2015; and this study). Furthermore, HYL1 protein and mRNA levels were elevated in drb2 plants, and the drb1 drb2 double mutant exhibited a much stronger developmental defect than both drb1 and drb2 single mutants (Reis et al., 2015), suggesting the potential redundancy of DRB1 and DRB2 in translational repression, although the effects from enhanced miRNA biogenesis deficiency also need to be considered. Their divisions as well as cooperation in the miRNA pathway are worth investigating.
Materials and methods
Plant materials and growth conditions
All A. thaliana plants in this study were in the Col background, and hyl1-2 (SALK 064863; Col ecotype) mutants were used. Seeds were surface-sterilized in 70% ethanol for 30 s and then in 0.1% HgCl2 for 10 min. This was followed by four washes with sterile distilled water. For CSD2 immunoblots, Murashige and Skoog medium (MS0) was used to allow the addition or omission of 10 μM CuSO4. For the in vitro tissue culture, the seeds were sown on solid MS0 containing 1% sugar, incubated at 4°C in darkness for 2 d, and then moved to a growth chamber at 22.8°C for 16 h in the light (150 umol/m2/s). For phenotypic observations, seeds were sown in pots with peat soil and grown in 22.5°C growth chambers.
DNA constructs for transgenes
An HYL1 promoter region (1,240 bp upstream of the translational start site) and a full-length coding sequence (1,257 bp) were amplified from Arabidopsis Col seedlings. Then, both sequences were cloned into pCAMBIA1301 binary vectors to obtain the ProHYL1:HYL1 constructs. HYL1 with NLS40 and NES at the N terminal under the HYL1 promoter was cloned into pCAMBIA1301, producing ProHYL1:NLS40HYL1 and ProHYL1:NESHYL1. N or C terminal GFP-tagged HYL1 was cloned independently into pCAMBIA1301 under the CaMV 35S promoter, producing Pro35S: GFP-HYL1 or Pro35S: HYL1-GFP. N terminal GFP-tagged AMP1 was cloned into pCAMBIA1301 under the 35S promoter, to obtain Pro35S: GFP-AMP1. For reporter gene construction, the 90 bps of the REV gene sequence and 75 bp of the SPL9 gene sequence that contained the miRNA target site were fused with GFP and then cloned into pCAMBIA3301, producing Pro35S:REV90-GFP and Pro35S:SPL75-GFP. Meanwhile, Pro35S:mREV90-GFP and Pro35S:mSPL75-GFP, the miRNA binding sites of which were synonymously mutated, were constructed. The Arabidopsis plants were transformed using the flower dip method (Clough and Bent, 1998). Transgenic seeds were sterilized and germinated on agar medium containing 50 mg/mL hygromycin or 20 mg/mL phosphinothricin. The primers used for construction are listed in Supplemental Data Set S1.
RNA analysis
Total RNA samples were extracted from whole 20-d-old seedlings using TRIzol (Invitrogen, Carlsbad, CA, USA ), extracted with phenol:chloroform:isoamylalcohol(25:24:1) and chloroform, and then two volume of ethanol precipitated. The RNA analysis was performed according to the methods reported by Yang et al. (2014).
For Northern blotting, 30 µg total RNA was resolved by 19% PAGE electrophoresis in 1× TBE, and then transferred to a Hybond membrane (Amersham Biosciences, GE Healthcare, Little Chalfont, UK) at 200 mA for 2 h. The UV cross-linked membrane was hybridized in ULTRA hyb® Ultrasensitive Hybridization buffer (Ambion, Austin, TX, USA) using 3′ Biotin-labeled DNA oligo (TaKaRa, Otsu, Japan) antisense probes to the mature miRNA or U6 transcripts. Hybridization signals were detected using a Light Shift EMSA Kit (Thermo Scientific, Waltham, MA, USA) and imaged with an FLA-5000 Phosphor imager (FujiFilm, Tokyo, Japan). Quantifications were performed using ImageJ.
For RT-qPCR, total RNA was treated with DNase I (TaKaRa, Kyoto, Japan), followed by a phenol/chloroform extraction to remove DNA contamination. Approximately 4 μg of purified RNA was used for first-strand cDNA synthesis using PrimeScript® Reverse Transcriptase (TaKaRa) with oligo (dT) primers or specific primers. RT-qPCR was performed using the specific primer pairs in a MyiQ2 Two-color Real-time PCR Detection System (Bio-Rad, Richmond, CA, USA). Quantitative PCR for each gene was performed on at least three biological replicates. The relative transcript levels were determined for each sample by normalizing them to ACTIN cDNA.
For the detection of 3′ cleavage products from miRNA-targeted mRNAs, 5′ RACE was performed using the GeneRacer kit (Invitrogen). Total RNAs were directly ligated with the GeneRacer RNA oligonucleotide and reverse transcription reactions were carried out with gene-specific primers. PCR reactions were performed to quantify 3′ cleavage products. The same amount of RNAs was reverse transcribed with oligo (dT) to amplify UBQ5 or ACTIN as an internal loading control.
Before sRNA sequencing, sRNA libraries were constructed using the NEBNext Small RNA Library Prep Set kit. Then the libraries were sequenced on an Illumina Nova. The reads without 3′ adaptor and insert tag were filtered, and the clean reads were obtained. Adaptor-free reads that aligned to rRNA/tRNA/snRNA, though subjected to the BLAST search against Rfam version 10.1 (https://rfam.org/) and GenBank (http://www.ncbi.nlm.nih.gov/genbank/) database, were removed. The remaining reads were used for mature miRNA alignment as queries in a search against the miRBase version 21 database (http://www.mirbase.org/). The sRNA-seq data were uploaded into NCBI (PRJNA667212). The expression levels of miRNA were calculated based on RPM. To generate the heatmap, the average reads (RPMaverage) and standard deviation (SD) of each individual miRNA in different samples were first obtained, then the z-scores of this miRNA in each sample were calculated as (RPM − RPMaverage)/SD.
Protein analysis
Three-week-old plants were harvested and ground to a fine powder in liquid nitrogen before being mixed with PEB buffer (200 µL/0.1 g, 100 mM Tris–HCl pH6.8, 10% glycerol, 0.5% SDS, 0.1% Triton X-100, and 5 mM EDTA) at 4°C for 10 min. After centrifugation at 15,000 g for 10 min at 4°C, the supernatant was collected and mixed with 2 × SDS loading buffer (0.1 M Tris–HCl, pH 6.8, 10%SDS, 50% glycerin, and 0.2% Bromophenol blue) before 5 min boiling. The extracted proteins were resolved on 10% (w/v) SDS-PAGE and electroblotted on a nitrocellulose membrane (GE Healthcare, Chicago, IL, USA). The following antibodies were used in this study: anti-GFP (Rabbit, BBI Life Sciences; Lot.CC20AA0011, 1:5,000 dilution), anti-GFP (mouse, Proteintech, Lot.66002-I, 1:1,000 dilution for IP and 1:5,000 for immunoblot), anti-AGO1 (Agrisera, Vännäs, Sweden; Lot.1508, 1:3,000), anti-HYL1 (Agrisera; Lot.1602, 1:1,000), anti-Histone H3 (Agrisera; Lot.1904, 1:500), anti-PEPC (Agrisera; Lot.1805, 1:3,000), anti-ACTIN (Agrisera; Lot.1403, 1:5,000), IgG (ABClonal, Lot 9300014001, 1:10,000), anti-AP2 (Agrisera; Lot.1308, 1:2,000) anti-CSD2 (Agrisera; Lot.1502, 1:2,000), anti-REV(Abiocode, Agoura Hills, CA, USA; Lot.8641, 1:2,000), anti-SE (Agrisera; Lot.1501, 1:3,000), and anti-BiP2 (Agrisera; Lot.1310, 1:2,000) antibodies. TCP4 antibodies were used according to Liu et al. (2011). The secondary antibody was goat-developed anti-rabbit IgG (GE Healthcare; NA931V, 1:20,000 dilution). Quantifications were performed using ImageJ.
Nuclear-cytoplasmic fractionation
Three-week-old plants (0.5 g) were harvested and ground to a fine powder in liquid nitrogen and mixed with 2 mL/g of lysis buffer (20 mM Tris–HCl, pH 7.5, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 250 mM Suc, and 5 mM DTT) supplemented with protease inhibitor cocktail (50×; Roche, Basel, Switzerland). The homogenate was filtered through a double layer of Miracloth. The flow-through was spun at 1,500 g for 10 min, and the supernatant, consisting of the cytoplasmic fraction, was centrifuged at 10,000 g for 10 min at 4°C and collected. The pellet was washed 4 times with 5 mL of nuclear resuspension buffer NRBT (20 mM Tris–HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100) and then resuspended with 500 μL of NRB2 (20 mM Tris–HCl, pH 7.5, 0.25 M Suc, 10 mM MgCl2, 0.5% Triton X-100, and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail (Roche) and carefully overlaid on top of 500 μL NRB3 (20 mM Tris–HCl, pH 7.5, 1.7 M Suc, 10 mM MgCl2, 0.5% Triton X-100, and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail (50×; Roche). These samples were centrifuged at 16,000 g for 45 min at 4°C. The final nuclear pellet was resuspended in 400 μL lysis buffer. As quality controls for the fractionation, PEPC protein was detected and used as a cytoplasmic marker, and Histone H3 was used as a nuclear marker.
Co-IP
In the Co-IP assays, ∼2 g 10-d-old seedlings were ground into a fine powder using liquid nitrogen and then resolved in lysis buffer (25 mM Tris–HCl at pH 7.4, 25 mM KCl, 10 mM MgCl2, 0.1% NP-40, and 2× complete protease inhibitor cocktail; Roche). The protein A (protein G for GFP IP)-agarose beads (Bio-RAD, Hercules, CA, USA) were pre-cleared 3 times and incubated with antibodies for 30 min at 4°C and then added to the plant extraction for 4 h at 4°C. After three washes (20 mM Tris–HCl, pH 7.9, 0.5 M KCl, 10% glycerol, 1 mM EDTA, 5 mM DTT, 0.5% Nonidet P-40 and 0.2 mM phenylmethylsulfonyl fluoride), the proteins in the immunoprecipitates were subjected to immunoblot analyses.
RIP
Leaf tissue from 2-week-old transgenic Arabidopsis plants was ground in liquid nitrogen, then the cytoplasmic and nuclear samples of Col were mixed in 5 mL/g lysis buffer (50 mM Tris–HCl at pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 0.1% NP-40, and 2× complete protease inhibitor cocktail; Roche). Cell debris was pelleted by centrifugation using 9,500 × g for 15 min at 4°C. About 10% of the clarified lysate was collected for RNA extraction as input, and the rest was pre-cleared for 20 min at 4°C with 10-μL bed volume of protein A-agarose (30 μg protein A) per milliliter. Pre-cleared lysates were reacted with anti-HYL1 for 1 h at 4°C, and then with 50 μL bed volume of protein A-agarose (150 μg protein A) per mL for 3 h at 4°C. Precipitates were washed 3 times in lysis buffer and divided for protein and RNA analyses. RNA was recovered by a treatment with three volumes of proteinase K solution (100 mM Tris–HCl at pH 7.4, 10 mM EDTA, 150 mM NaCl, 2% SDS, and 0.2 μg/μL proteinase K) for 15 min at 65°C, extracted with saturated phenol and phenol:chloroform (1:1), and two volume of ethanol precipitated. Extracted RNA was then used for RT-qPCR analyses.
Data were analyzed according to Marmisolle et al. (2018), with some modifications. Essentially, the same volume of immunoprecipitated RNA from either HYL1-IP or negative control products was applied for reverse transcription followed by RT-qPCR. For each sample, the Ct number of gene A was normalized to its input (total RNA) Ct number, then the fold enrichment of HYL1-IP sample/negative control was calculated accordingly. Specifically, the calculation was done as: (1) △Ct [Normalized RIP] = (Ct [IP]−(Ct [Input]−Log2 (Input/IP dilution factor)); (2) △△Ct [RIP/control]=△Ct [Normalized RIP]−△Ct [Normalized negative control]; (3) Fold enrichment = 2(−△△Ct [RIP/control]).
Fluorescence microscopy
For the detection of HYL1 subcellular localization, HYL1 constructs and AGO1-mCherry were independently engineered into pCAMBIA1301. ER-mCherry was from Alexis Maizel lab, University of Heidelberg (Jouannet et al., 2012). Agrobacterium strains containing HYL1-GFP and AGO1-mCherry were used to co-transfect N. benthamiana. The fluorescence in N. benthamiana was observed using a Zeiss LSM 510 Meta confocal laser scanning microscope after transfection in 22.5°C growth chambers for 48 h. HYL1-GFP and ER-mCherry were used to co-transfect N. benthamiana and the fluorescence was observed using a Zeiss LSM 880 Meta confocal laser scanning microscope. The excitation/emission wavelengths were 488/500–550 nm for GFP, and 543/620–630 nm for mCherry. The images shown are representing at least 50 cells/protoplasts with same patterns.
Isolation and fractionation of polysomes
The isolation method was modified according to Lanet et al. (2009). Basically, 2 g of 20-d-old seedlings were ground in liquid nitrogen, and the powder was suspended in 6 mL chilled polysome buffer (100 mM Tris–HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, 5 mM EGTA, 15.4 units/mL heparin, 18 mM cycloheximide, 15.5 mM chloramphenicol and 0.5% (v/v) Nonidet P-40). Debris was removed by centrifugation at 1,200 × g for 5 min at 4°C twice. Approximately 2-mL aliquots of the resultant supernatant were loaded on the sucrose gradients [20%–25%–30%–40%–50% (w/w)] and centrifuged at 32,000 rpm for 150 min in a Beckman XL-70 rotor at 4°C. After centrifugation, 13 fractions were collected from the top (1) to the bottom (13) of the gradient based on the 254-nm absorbance, which monitored the ribosomal distribution. In total, 13 fractions were collected after centrifugation. Fractions were extracted in 20% TCA for protein analysis, and extracted using TRIzol for RNA analysis.
Statistical analyses
Statistical significance was calculated by a two-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001) and error bars indicate SD. One-way ANOVA followed by Tukey’s multiple comparisons test was used to determine the differences. Values of P < 0.05 were considered to be statistically significant. The results of statistical analyses are shown in Supplemental Data Set S2.
Accession numbers
HYL1 (AT1G09700), SE (AT2G27100), AMP1 (AT3G54720), REV (AT5G6069), AP2 (AT4G36920), AGO1 (AT1G48410), CSD2 (AT2G28190), TCP4 (AT3G15030), ARF10 (AT2G28350), SPL9 (AT2G42200), and UBQ5 (AT3G62250). The sRNA-seq data were uploaded into NCBI (PRJNA667212).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype and HYL1 expressions in transgenic plants.
Supplemental Figure S2. The distribution and expression of miRNAs in Col, hyl1-2, and NESHYL1 14 plants.
Supplemental Figure S3. The translational repression function of cytoplasmic HYL1 depends on miRNA.
Supplemental Figure S4. Nuclear HYL1 does not bind to miRNA-targeted mRNAs.
Supplemental Data Set S1. The primers used in this paper.
Supplemental Data Set S2. ANOVA and Student’s t test results for the data shown in the figures.
Supplementary Material
Acknowledgments
We thank Prof. Alexis Maizel (University of Heidelberg) for kindly sending the ER-mCherry construct and Prof. Hualing Mi (Shanghai Institute of Plant Physiology and Ecology) for help with the sucrose gradient analysis.
Funding
This work was supported by the National Programs for Science and Technology Development of China (Grant no. 2016YFD0101900) and the Natural Science Foundation of China (Grant Nos. 31771442, 31471883, and 31571261).
Conflict of interest statement: The authors declare no competing interests.
X.Y. and W.D. designed the research plan and performed the molecular and biochemistry experiments, and drafted the manuscript. W.R. performed the molecular and biochemistry experiments, and drafted the manuscript. Q.Z. provided the AMP1 results. F.W. revised the manuscript. Y.H. designed the research plan and wrote the manuscript. X.Y., W.D., and W.R contributed equally, and all have the right to put himself/herself first in bibliographic documents.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yuke He (heyk@sippe.ac.cn).
References
- Adam SA, Gerace L (1991) Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66: 837–847 [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP (2006) A family of microRNAs present in plants and animals. Plant Cell 18: 3355–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L, Yu A, Bouche N (2010) microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J 62: 454–462 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bologna NG, Iselin R, Abriata LA, Sarazin A, Pumplin N, Jay F, Grentzinger T, Dal Peraro M, Voinnet O (2018) Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant MicroRNA pathway. Mol Cell 69: 709–719 e705 [DOI] [PubMed] [Google Scholar]
- Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O, Brodersen P (2015) SKI2 mediates degradation of RISC 5'-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res 43: 10975–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW (2014) COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat Commun 5: 5867. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas DV, Bartel B (2008) Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol 67: 403–417 [DOI] [PubMed] [Google Scholar]
- Eamens AL, Kim KW, Curtin SJ, Waterhouse PM (2012) DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS One 7: e35933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483 [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M (2005) SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004a) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N (2004b) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T (2005) Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57: 173–188 [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y (2013) Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 52: 591–601 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A (2012) Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J 31: 1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE (1984a) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311: 33–38 [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE (1984b) A short amino acid sequence able to specify nuclear location. Cell 39: 499–509 [DOI] [PubMed] [Google Scholar]
- Karlsson P, Christie MD, Seymour DK, Wang H, Wang X, Hagmann J, Kulcheski F, Manavella PA (2015) KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc Natl Acad Sci USA 112: 14096–14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH (2008) Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol 49: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, Voinnet O, Robaglia C (2009) Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Doudna JA (2012) TRBP alters human precursor microRNA processing in vitro. RNA 18: 2012–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S. et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J (2013a) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B. et al. (2013b) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yang X, Wu F, He Y (2012) HYL1 controls the miR156-mediated juvenile phase of vegetative growth. J Exp Bot 63: 2787–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H, Li X, Liu Z, He Y (2013) HYL1 is required for establishment of stamen architecture with four microsporangia in Arabidopsis. J Exp Bot 64: 3397–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tang S, Jia G, Schnable JC, Su H, Tang C, Zhi H, Diao X (2016) The C-terminal motif of SiAGO1b is required for the regulation of growth, development and stress responses in foxtail millet (Setaria italica (L.) P. Beauv). J Exp Bot 67: 3237–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia L, Mao Y, He Y (2010) Classification and quantification of leaf curvature. J Exp Bot 61: 2757–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia L, Wang H, He Y (2011) HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J Exp Bot 62: 4367–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J (2006) SERRATE: a new player on the plant microRNA scene. EMBO Rep 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D (2012) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870 [DOI] [PubMed] [Google Scholar]
- Marmisolle FE, Garcia ML, Reyes CA (2018) RNA-binding protein immunoprecipitation as a tool to investigate plant miRNA processing interference by regulatory proteins of diverse origin. Plant Methods 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN (2011) Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature 475: 201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Raghuram B, Sheikh AH, Rustagi Y, Sinha AK (2015) MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J 282: 521–536 [DOI] [PubMed] [Google Scholar]
- Ran X, Zhao F, Wang Y, Liu J, Zhuang Y, Ye L, Qi M, Cheng J, Zhang Y (2020) Plant Regulomics: a data-driven interface for retrieving upstream regulators from plant multi-omics data. Plant J 101: 237–248 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM (2015) Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat Plants 1: 14027. [DOI] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B (2012) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA 109: 12817–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B (2014) Methylation protects microRNAs from an AGO1-associated activity that uridylates 5' RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA 111: 6365–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Wu F, Bai J, Li X, Yang X, Xue W, Liu H, He Y (2020) BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic Res 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacnun JM, Crespo R, Palatnik J, Rasia R, Gonzalez-Schain N (2020) Dual function of HYPONASTIC LEAVES 1 during early skotomorphogenic growth in Arabidopsis. Plant J 102: 977–991 [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA 104: 5437–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C, Willing EM, Rausch S, Schneeberger K, Laubinger S (2013) RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J 76: 433–445 [DOI] [PubMed] [Google Scholar]
- Su C, Li Z, Cheng J, Li L, Zhong S, Liu L, Zheng Y, Zheng B (2017) The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev Cell 41: 527–539 e525 [DOI] [PubMed] [Google Scholar]
- Szadeczky-Kardoss I, Csorba T, Auber A, Schamberger A, Nyiko T, Taller J, Orban TI, Burgyan J, Silhavy D (2018) The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res 46: 4632–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassi AH, Re DA, Romani F, Cambiagno DA, Gonzalo L, Moreno JE, Arce AL, Manavella PA (2020) The intrinsically disordered protein CARP9 bridges HYL1 to AGO1 in the nucleus to promote MicroRNA activity. Plant Physiol 184: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y (2007) The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shi Y, Li J, Xu L, Fang Y, Li X, Qi Y (2013) A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res 23: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang P, Wang B, Hsu CC, Tang K, Zhang H, Hou YJ, Zhao Y, Wang Q, Zhao C. et al. (2017) The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet 13: e1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H (2006a) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS (2012) Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc Natl Acad Sci USA 109: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA (2010) Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ren W, Zhao Q, Zhang P, Wu F, He Y (2014) Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res 42: 12224–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X (2006b) HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2' OH of the 3' terminal nucleotide. Nucleic Acids Res 34: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC. et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zhang S, Dou Y, Li S, Ren G, Chevalier D, Zhang C, Yu B (2018) DAWDLE interacts with DICER-LIKE proteins to mediate small RNA biogenesis. Plant Physiol 177: 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Guo X, Ge C, Ma Z, Jiang M, Li T, Koiwa H, Yang SW, Zhang X (2017a) KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc Natl Acad Sci USA 114: 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hu F, Sung MW, Shu C, Castillo-Gonzalez C, Koiwa H, Tang G, Dickman M, Li P, Zhang X (2017b) RISC-interacting clearing 3'- 5' exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.