A MEE45–ANT pathway in maternal tissues modulates auxin biosynthesis and seed size in Arabidopsis.
Abstract
Seed size is a major factor determining crop yields that is controlled through the coordinated development of maternal and zygotic tissues. Here, we identified Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 (MEE45) as a B3 transcription factor that controls cell proliferation and maternally regulates seed size through its transcriptional activation of AINTEGUMENTA (ANT) and its downstream control of auxin biosynthesis in the ovule integument. After characterizing reduced seed and organ size phenotypes in mee45 mutants and finding that overexpression of MEE45 causes oversized seeds, we discovered that the MEE45 protein can bind to the promoter region of the ANT locus and positively regulate its transcription. ANT in-turn activates the expression of auxin biosynthetic genes (e.g. YUCCA4) in the ovule integument. Our results thus illustrate mechanisms underlying maternal tissue-mediated regulation of seed size and suggest that MEE45 and its downstream components can be harnessed to develop higher-yielding crop varieties.
Introduction
Seed size is a vital agronomic trait and determinant of crop yield (Moles et al., 2005; Adamski et al., 2009; Li and Li, 2016). Increasing seed size would provide an efficient method to raise grain yield and to meet the dietary caloric demands of the growing population worldwide (Khush, 2003; Song et al., 2007; Shomura et al., 2008; Fan et al., 2009; Zuo and Li, 2014). Accordingly, the regulation of seed size has been extensively studied in plant and crop sciences.
Seed size is determined by the coordinated development of maternal and zygotic tissues. In angiosperms, the zygotic tissues result from double fertilization, when one male sperm cell fuses with the two female polar nuclei to produce the triploid endosperm, while the other sperm cell combines with the egg to form the diploid embryo (Chaudhury et al., 2001; He et al., 2017). The seed coat is the maternal sporophytic tissue of the seed: it surrounds the embryo and endosperm, and is derived from the integument of the ovule (Haughn and Chaudhury, 2005). The integument forms the cavity within which the embryo and the endosperm will grow after fertilization, and become the seed coat that sets a physical constraint on final seed size (Adamski et al., 2009; Fang et al., 2012; Xia et al., 2013; Du et al., 2014). The diploid embryo, the triploid endosperm, and the maternal integument coordinate their development to determine the final size of the seed.
In Arabidopsis (Arabidopsis thaliana), several factors act through maternal tissues to control seed size (Jofuku et al., 2005; Ohto et al., 2005; Li et al., 2008; Adamski et al., 2009; Xia et al., 2013). The WRKY family transcription factor TRANSPARENT TESTA GLABRA2 promotes cell expansion in the ovule integument for seed growth (Garcia et al., 2005; Ohto et al., 2009). In contrast, the ubiquitin receptor DA1 (meaning large in Chinese) restricts cell proliferation in the integument of the developing ovule to physically limit seed size through the seed coat after fertilization (Li et al., 2008; Xia et al., 2013). The APETALA 2 (AP2) transcription factor controls seed size by limiting cell expansion in the ovule integument (Jofuku et al., 2005; Ohto et al., 2005, 2009). The AINTEGUMENTA (ANT) gene encodes an AP2-domain transcription factor (Weigel, 1995; Elliott et al., 1996; Klucher et al., 1996). Like the da1 and ap2 mutants, plants overexpressing ANT produce oversized organs and seeds (Mizukami and Fischer, 2000). Since ANT is a regulator of ovule integument initiation and ovule development (Elliott et al., 1996; Klucher et al., 1996), it acts maternally to control seed size (Li and Li, 2015).
The plant phytohormone auxin also regulates seed development. Among the 23 Arabidopsis AUXIN RESPONSE FACTOR (ARF) genes, which function in auxin-mediated gene expression, ARF2 is known to control seed size by restricting cell proliferation of the ovule integument (Schruff et al., 2006). Plants carrying mutations in the gene encoding the receptor kinase FERONIA (FER), which mediates auxin-regulated root hair growth (Duan et al., 2010; Westermann et al., 2019), produce seeds of larger size relative to the wild-type (WT) by increasing cell expansion in the ovule integuments (Yu et al., 2014). In rice (Oryza sativa), overexpression of BIG GRAIN1, which encodes a factor that participates in auxin transport, significantly increases grain size by promoting cell proliferation and expansion in spikelet hulls (Liu et al., 2015b) Thus, auxin signaling and transport appear to set a physical restriction for seed growth and seed size via the seed coat. A recent study showed that the ovule integuments affect seed development by supplying maternally produced auxin to the early embryo, suggesting that auxin from maternal tissues is apparently required for normal embryo development (Robert et al., 2018).
Loss-of-function mutations in the MATERNAL EFFECT EMBRYO ARREST (MEE) loci, identified based on defects in female gametophyte development by a Dissociation (DS) transposon insertion screen, exhibit defects in embryogenesis (Pagnussat et al., 2005). Here, we show that MEE45, a B3-type transcription factor, is required for ovule integument initiation and development. MEE45 is a seed size regulator that controls cell proliferation in the maternal integument and affects final seed size. Moreover, we determined that MEE45 directly binds to the promoter region of the ANT locus to activate ANT transcription. We also showed that the auxin biosynthesis capacity of maternal cells—as represented by the expression levels of the auxin biosynthetic gene YUCCA4 (YUC4)—is regulated by a MEE45–ANT regulatory module to support maternally controlled embryo development and seed size determination.
Results
MEE45 functions in controlling seed size
To genetically dissect the mechanisms of maternally controlled seed size, we collected Arabidopsis T-DNA insertion lines in predicted MEE genes with documented expression in ovules and/or embryos across several microarray studies. Phenotypic analysis of this collection of lines allowed the identification of three T-DNA insertion mutants in MEE45 (mee45-1, SALK_031530; mee45-2, SALK_038324; mee45-3, CS801014) with obvious seed growth defects. MEE45 encodes a B3-type transcription factor of unknown biological function (Supplemental Figure S1A). MEE45 transcript levels in mature ovules of mee45-1 mutant plants were significantly reduced compared to levels in the WT Columbia-0 (Col-0) (Supplemental Figure S1B). Seeds of the mee45-1 mutant were significantly smaller and lighter than those from the WT (Figure 1, A, B, I, and J; Supplemental Data Sets S1 and S2), but showed no significant difference in their seed germination rate or viability compared to the WT (Supplemental Figure S2, A, B, D, E, G, and H; Supplemental Data Sets S3 and S4). The embryo occupies the majority of the mature seed in Arabidopsis. To identify the cellular basis of the small seed phenotype of the mee45-1 mutant, we compared embryo sizes between mee45-1 and the WT. The mee45-1 mutant produced significantly smaller embryos relative to those of the WT, from the early globular embryo stage up to the mature embryo stage (Figure 2). However, embryo morphology was broadly comparable between the mutant and the WT at each developmental stage.
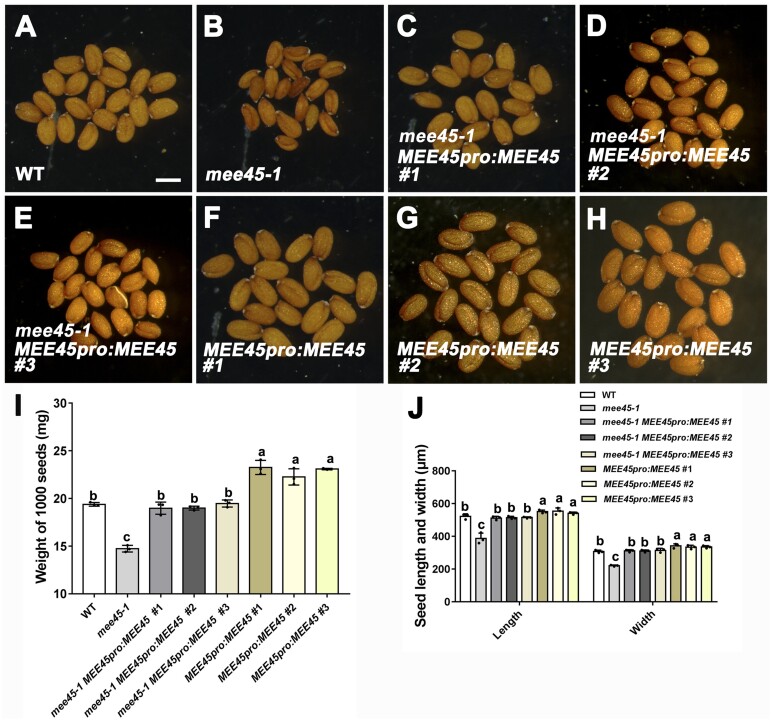
Figure 1.
MEE45 regulates seed size. A–H, Mature dry seeds of the WT (Col-0) (A), mee45-1 (B), mee45-1 MEE45pro:MEE45 line #1 (C), mee45-1 MEE45pro:MEE45 line #2 (D), mee45-1 MEE45pro:MEE45 #3 (E), MEE45pro:MEE45 line #1 (F), MEE45pro:MEE45 line #2 (G) and MEE45pro:MEE45 line #3 (H), Bar = 500 μm. I, Average seed mass per 1,000 seeds of the WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45 lines. J, Average seed length and width of the WT, mee45-1, mee45-1 MEE45pro:MEE45, and MEE45pro:MEE45 lines. The error bars in (I) and (J) represent the standard error of the mean, which were calculated from three sets of biological replicates. One-way analysis of variance (ANOVA) and Tukey’s multiple comparison tests were performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
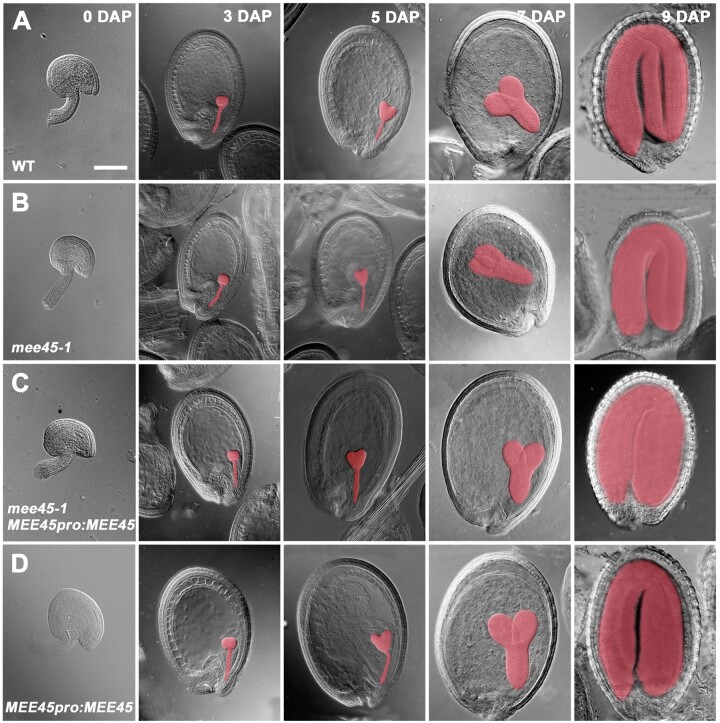
Figure 2.
Embryo development in WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45 plants. Whole-mount seeds observed by DIC microscopy at 0, 3, 5, 7, and 9 days after pollination (DAP) in the WT (A), mee45-1 (B), mee45-1 MEE45pro:MEE45 (C), and MEE45pro:MEE45 plants (D). Bar = 100 μm.
In contrast to the mee45-1 mutant, both mee45-2 and mee45-3 mutants accumulated extremely low levels of MEE45 transcripts, and were thus assumed to be knockout mutants (Supplemental Figure S1, A and B). Approximately 70%–80% mutant seeds of mee45-2 or mee45-3 were significantly smaller than those of the WT (Supplemental Figure S1, C–I; Supplemental Data Sets S5 and S6), although ∼75%–85% embryos in each mutant showed normal developmental patterns (Supplemental Figure S3, B and D). However, roughly 12%–20% embryos from each mutant displayed clear developmental defects as early as the globular stage (Supplemental Figure S3, C and E). Because of these additional developmental phenotypes affecting embryo morphology, we focused our subsequent investigations on the mee45-1 mutant for further analysis of seed size regulation.
The small seed phenotype observed in mee45-1 was rescued by the transgenic expression of MEE45 under the control of its own promoter (mee45-1 MEE45pro:MEE45; Figure 1, C–E, I, and J; Supplemental Figure S4; Supplemental Data Sets S1 and S2), confirming the role of MEE45 in the control of seed size. Moreover, we also found that transgenic lines with MEE45pro:MEE45 in the WT background produced seeds that were significantly larger and heavier relative to Col-0 seeds (Figure 1, F–J; Supplemental Figure S4; Supplemental Data Sets S1 and S2), and with oversized embryos from the early globular embryo stage up to the mature embryo stage (Figure 2). These results confirm that MEE45 exerts a function in mediating seed size and seed mass.
We measured the size and number of cotyledon epidermal cells of mature embryos in mee45-1 and the WT: we observed an increase in cell size that was accompanied by a significant decrease in cell number in the cotyledons of mee45-1 embryos as compared to the WT (Supplemental Figure S5; Supplemental Data Sets S7–S9). These findings suggest that a reduction in cell proliferation in embryos is responsible for the small seed size of mee45-1 plants.
Although the embryo was smaller in the mutant background, the endosperm in mee45-1 did not appear substantially different from that of the WT (Supplemental Figure S6, A–F). Moreover, there was no difference between mee45-1 mutant and WT seeds for the expression levels of genes known to mediate endosperm development (e.g. MINISEED3 [MINI3], HAIKU1 and 2 [IKU1/2], and SHORT HYPOCOTYL UNDER BLUE1 [SHB1]) (Zhou et al., 2009; Kang et al., 2013) (Supplemental Figure S6G).
To examine the effects of MEE45 function on organ growth in more detail, we measured the expression of MEE45 and found it to be much higher in leaves, flowers, and siliques than in roots (Supplemental Figure S7). We then evaluated the size of these organs in the mee45-1 mutant. Leaves, floral organs, and siliques were significantly smaller in the mee45-1 mutant compared to WT plants, without obvious alterations of their overall morphology at different developmental stages (Supplemental Figure S8; Supplemental Data Sets S10 and S11). These results support the notion that MEE45 contributes to the regulation of organ size in Arabidopsis.
MEE45 acts maternally to influence seed size
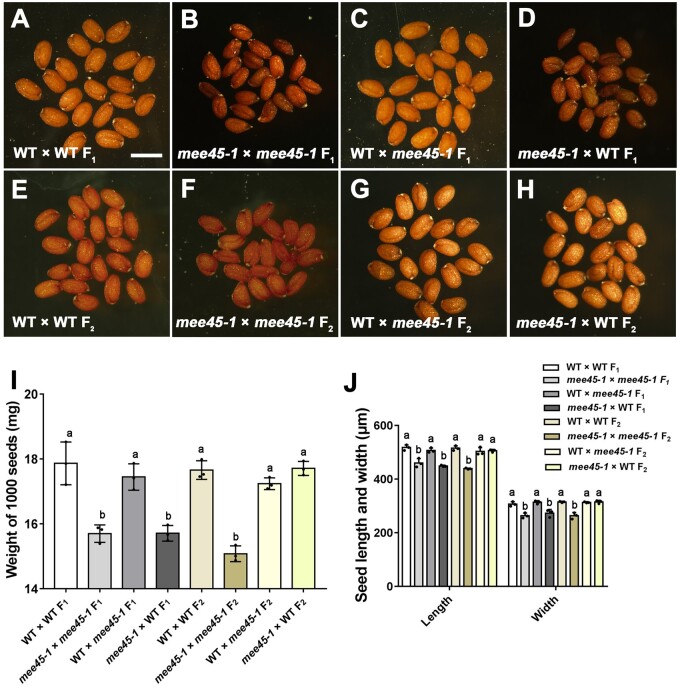
To determine whether the small seed phenotype of the mee45 mutant originated from maternal (the ovule) or zygotic tissues (the embryo), we performed reciprocal crossing experiments between mee45 and the WT Col-0. As shown in Figure 3 and Table 1, we only noticed the effects of the mee45 mutation on seed size when maternal plants were homozygous for the mee45-1 mutation. Seeds produced by homozygous mee45-1 maternal plants were consistently smaller than those produced by WT maternal plants, regardless of the genotype of the pollen donor (Figure 3, A–D, I, and J; Supplemental Data Sets S12 and S13). Furthermore, heterozygous F1 seeds derived from either a cross between a WT maternal parent and a mee45-1 pollen donor or between a mee45-1 maternal parent and a WT pollen donor produced F2 seeds with normal seed size, even for the 25% of the seeds that were homozygous for the mee45-1 mutation (Figure 3, E–J; Table 1; Supplemental Data Sets S12 and S13). Additionally, we observed the small seed phenotype and defects in seed development in heterozygous F1 seeds only when maternal plants were homozygous for the mee45-2 or mee45-3 mutations (Supplemental Figures S9 and S10; Supplemental Data Sets S14–S17). These results clearly indicate that the maternal genotype at the MEE45 locus determines the seed size in the next generation.
Figure 3.
MEE45 maternally controls seed size. A–H, Mature dry seeds of [WT × WT] F1 (A), [mee45-1 × mee45-1] F1 (B), [WT × mee45-1] F1 (C), [mee45-1 × WT] F1 (D), [WT × WT] F2 (E), [mee45-1 × mee45-1] F2 (F), [WT × mee45-1] F2 (G) and [mee45-1 × WT] F2 (H) plants. Bar = 500 μm. I, Average seed mass per 1,000 seeds of the indicated genotypes. J, Average seed length and width of the indicated genotypes. The error bars in (I) and (J) represent the standard error of the mean, calculated from three sets of biological replicates. One-way ANOVA and Tukey’s multiple comparison tests were performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
Table 1.
Maternal effects of MEE45 in controlling seed size
| Cross Group♀× ♂ | Cross Number | Zygote | Maternal Sporophyte | Seed Phenotype |
|---|---|---|---|---|
| Col-0 × mee45-1–/– F1 | 1 | +/– | +/+ | Normal, n = 51 |
| 2 | +/– | +/+ | Normal, n = 48 | |
| 3 | +/– | +/+ | Normal, n = 55 | |
| mee45-1 – / – × Col-0 F1 | 4 | +/– | –/– | Small, n = 55 |
| 5 | +/– | –/– | Small, n = 54 | |
| 6 | +/– | –/– | Small, n = 53 | |
| Col-0 × mee45-1–/– F2 | 7 | +/+ | +/– | Normal, n = 12 |
| +/– | +/– | Normal, n = 25 | ||
| –/– | +/– | Normal, n = 11 | ||
| 8 | +/+ | +/– | Normal, n = 13 | |
| +/– | +/– | Normal, n = 24 | ||
| –/– | +/– | Normal, n = 12 | ||
| 9 | +/+ | +/– | Normal, n = 13 | |
| +/– | +/– | Normal, n = 23 | ||
| –/– | +/– | Normal, n = 12 | ||
| mee45-1 – / – × Col-0 F2 | 10 | +/+ | +/– | Normal, n = 10 |
| +/– | +/– | Normal, n = 22 | ||
| –/– | +/– | Normal, n = 12 | ||
| 11 | +/+ | +/– | Normal, n = 12 | |
| +/– | +/– | Normal, n = 21 | ||
| –/– | +/– | Normal, n = 14 | ||
| 12 | +/+ | +/– | Normal, n = 11 | |
| +/– | +/– | Normal, n = 24 | ||
| –/– | +/– | Normal, n = 14 |
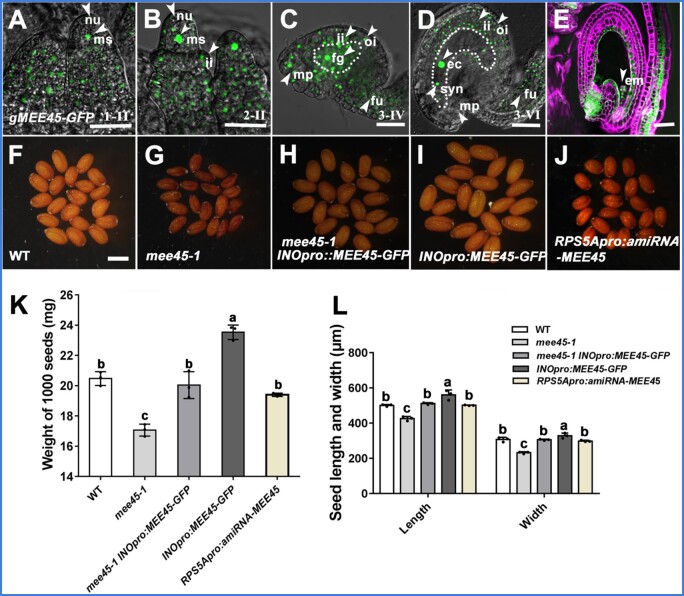
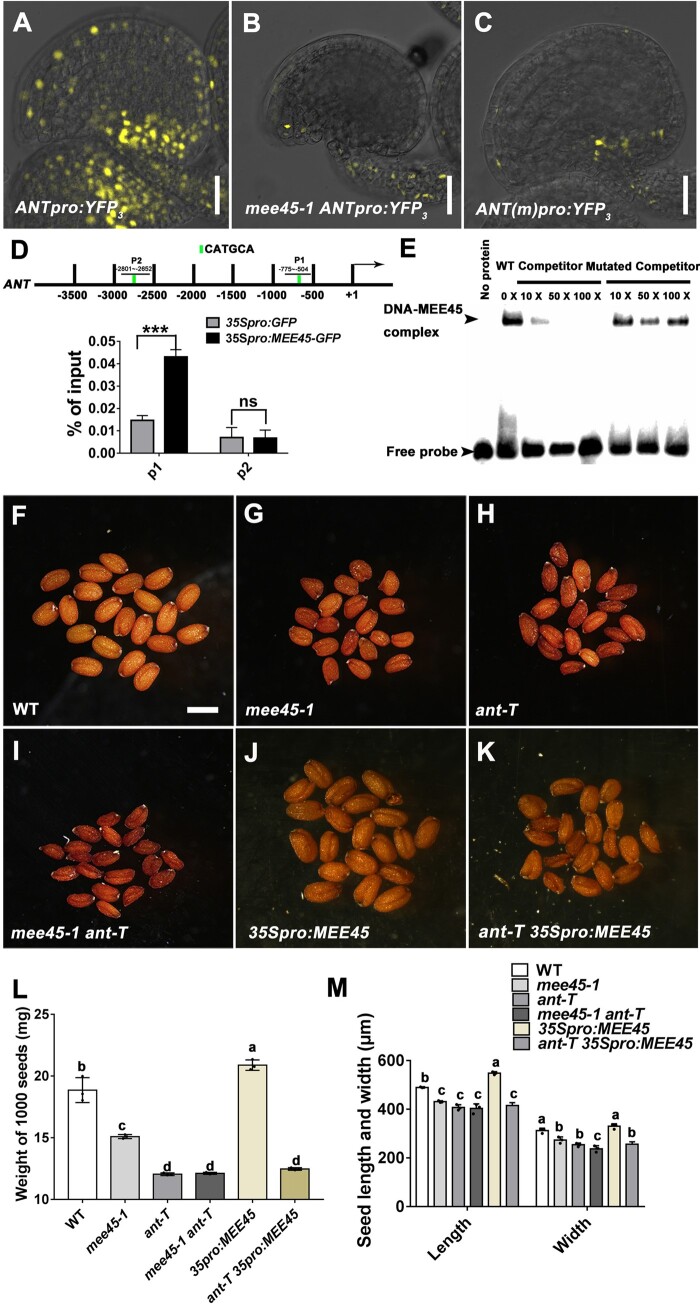
To investigate the accumulation patterns of the MEE45 protein, we introduced a transgene in WT plants consisting of the MEE45 coding sequence (CDS) cloned in frame with the green fluorescent protein (GFP) gene and placed under the control of the MEE45 promoter. We then detected the GFP signal in these gMEE45-GFP transgenic plants. As expected for a transcription factor, MEE45-GFP accumulated in the nucleus. The MEE45-GFP signal was strong during ovule development (Figure 4, A–D). We detected GFP fluorescence throughout the entire ovule primordium, as well as in the megasporocyte, the nucellus, and at the base of the nucellus where the inner and outer integument initiate (Figure 4, A and B). During the mature stages (3-IV and 3-VI) of ovule development, MEE45-GFP accumulated in the embryo sac, ovule integument, and the funiculus (Figure 4, C and D). After fertilization, we observed MEE45-GFP fluorescence in the seed coat, as well as in the embryo (Figure 4E).
Figure 4.
MEE45 functions in the ovule integument for the maternal control of seed size. A–E, Accumulation pattern of MEE45 in developing ovules (1-II (A), 2-II (B), 3-IV (C), and 3-VI (D) mature ovule stages) and seeds (2 DAP) (E) of gMEE45-GFP transgenic plants. Bars = 100 μm. nu, nucellus; ms, megasporocyte; ii, inner integument; oi, outer integument; fg, female gametophyte; ec, egg cell; syn, synergid; mp, micropyle; fu, funiculus; em, embryo . F–J, Mature dry seeds of the WT (F), mee45-1 (G), mee45-1 INOpro:MEE45-GFP (H), INOpro:MEE45-GFP (I), and RPS5Apro:amiRNA-MEE45 (J) plants. Bar = 750 μm. K, Average seed mass per 1,000 seeds of the indicated genotypes. L, Average seed length and width of the indicated genotypes. The error bars in (K) and (L) represent the standard error of the mean, calculated from three sets of biological replicates. One-way ANOVA and Tukey’s multiple comparison tests were performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
To investigate whether MEE45 expression in the integument of the ovule might be responsible for regulating seed size, we specifically expressed MEE45 in the integument of the mee45-1 mutant by using the INNER NO OUTER (INO) promoter (Supplemental Figure S11, A–C), which is strictly expressed in the ovule integument (Meister et al., 2002; Wang et al., 2016). INOpro:MEE45 expression completely rescued the small seed phenotype seen in the mee45-1 mutant, and resulted in normal seed sizes comparable to those of the WT (Figure 4, F–H, K, and L; Supplemental Data Set S19). Note that these findings are similar to the results obtained when MEE45 was driven by its own promoter (Figure 1, A–E). Moreover, INOpro:MEE45 transgenic lines in the WT background produced significantly oversized seeds (Figure 4, F, I, K, and L; Supplemental Data Set S19).
We also specifically reduced MEE45 transcript levels in seeds by using the RIBOSOMAL PROTEIN S5A (RPS5A) promoter (Supplemental Figure S11D), which is strictly expressed in the embryos and endosperm of seeds after fertilization (Weijers et al., 2001; Zhang et al., 2017), to drive the expression of an artificial microRNA (amiRNA) targeting MEE45. RPS5Apro:amiRNA-MEE45 transgenic lines produced normal-sized seeds showing no significant difference with those of the WT (Figure 4, F, J, K, and L; Supplemental Data Set S19), suggesting that the expression of MEE45 in the embryo does not contribute to its function in regulating seed size. Together, these results indicate that MEE45 functions in ovule integuments for the maternal control of seed size.
MEE45 induces cell proliferation in the ovule integument
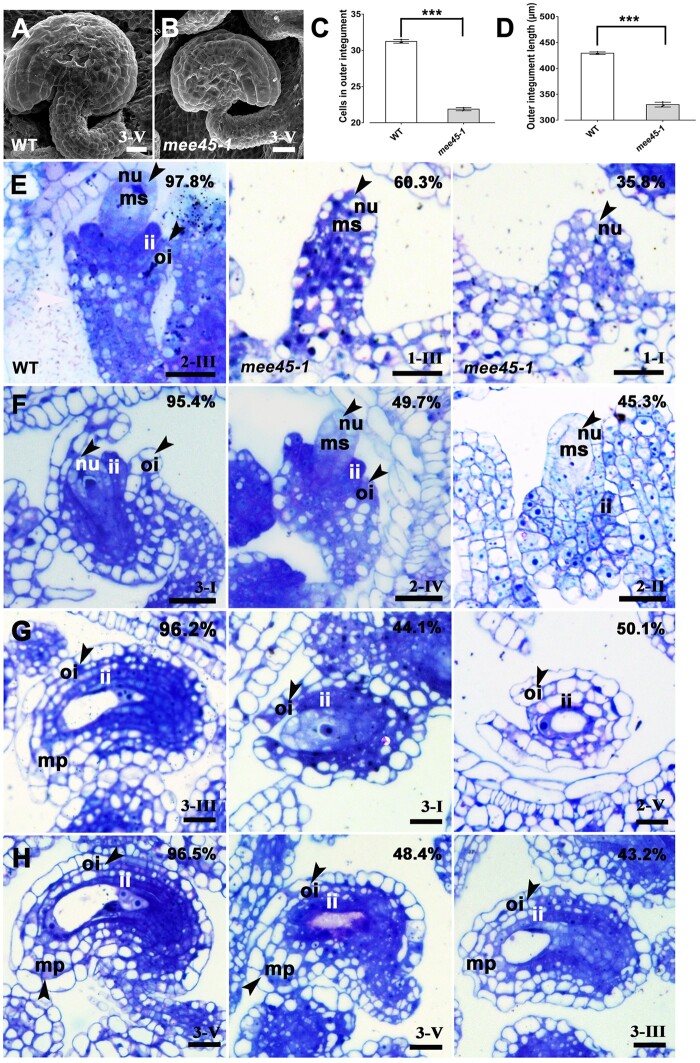
The ovule is a precursor of the seed and its maternal tissues affect both seed development and size (Xia et al., 2013; Du et al., 2014). Thus, we examined ovule size and discovered that it was significantly smaller in the mee45-1 mutant than in the WT (Figure 5, A and B). We also observed that the number of cells in the outer integument layer of mutant ovules was significantly lower compared to WT ovules. Consequently, the length of the outer integument in the mutant was much shorter than in the WT (Figure 5, C and D; Supplemental Data Sets S20 and S21). The CYCLIN B1;1 (CYCB1;1) gene is expressed at the G2/M phase of the cell cycle, and its expression has provided a useful marker to study cell proliferation during organ development (Schnittger and De Veylder, 2018). We compared the staining pattern of the CYCB1;1pro:β-GLUCURONIDASE (GUS) reporter in the ovules of mee45-1 and WT plants (Supplemental Figure S12). In mature ovules of the WT, there was strong GUS staining in the nucellus and in the integument cells enclosing it (Supplemental Figure S12A). In contrast, we detected hardly any GUS staining in ovule integument cells of the mee45-1 mutant, and these plants had only very weak GUS staining in the nucellus (Supplemental Figure S12B). Thus, our results demonstrate that MEE45 induces cell proliferation in the ovule, especially in the integument.
Figure 5.
Ovules in the WT and mee45-1 plants. A and B, Scanning electron microscopy of mature ovules (Stage 3-V) from WT (A) and mee45-1 (B). C, Number of cells in the outer integument of mature ovules (Stage 3-V) from WT and mee45-1 plants. D, Outer integument length of mature ovules from WT and mee45-1 plants. The error bars in (C) and (D) represent the standard error of the mean, calculated from three sets of biological replicates. ***P < 0.001, compared to the WT (Col-0) (Student’s t -test). E, Developing ovules from WT or mee45-1 carpels at Stage 10. F, Developing ovule from WT or mee45-1 carpels at stage 11. G, Developing ovule from WT or mee45-1 carpels at stage 12. H, Developing ovule from WT or mee45-1 carpels at Stage 13. Bars in (A), (B), and (E–H) = 100 μm. nu, nucellus; ms, megasporocyte; ii, inner integument; oi, outer integument; mp, micropyle.
Next, we compared ovule developmental progression between the mee45-1 mutant and the WT. The initiation and development of both the inner and the outer integuments in the mee45-1 mutant were significantly delayed relative to the WT (Figure 5, E–H). In stage 10 carpels of both WT and the mee45-1 mutant (Smyth et al., 1990; Guan et al., 2014), most WT ovules displayed the expanding inner and outer integuments identified as ovule Stage 2-III, ∼60% mutant ovules displayed their developing primordium at Stages 1-II, as well as roughly 35% mutant ovule primordia were at the earlier Stage 1-I (Figure 5E). Later in carpels at Stage 12, when WT ovules had fully developed inner and outer integuments enclosing the nucellus (ovule Stage 3-III), mutant ovules only displayed expanding inner and outer integuments at Stage 3-I or 2-V (Figure 5G). Although the mature integuments of mutant ovules did enclose the nucellus eventually (by carpel Stage 13) and did thus exhibit the same pattern as the WT, mutant ovules were significantly smaller than WT ovules (Figure 5H).
MEE45 directly activates ANT expression in the ovule integument
To investigate the potential regulatory roles of MEE45 during ovule development and in seed size control, we performed deep sequencing of the transcriptome (RNA-seq) to identify genes regulated by MEE45. We identified 1,578 differentially expressed genes (DEGs) in mee45-1 siliques compared to the WT (Supplemental Data Set S22), comprising 1,098 downregulated genes and 480 upregulated genes. The downregulated genes in mee45-1, which might be activated by MEE45, were enriched for functional annotations related to phytohormone metabolic processes, seed development, fatty acid biosynthetic processes, and environmental responses (Supplemental Figure S13A).
Our transcriptome analysis showed that the ANT gene is downregulated in mee45-1 siliques compared to the WT (Supplemental Figure S13, A and B). ANT was previously demonstrated to maternally control seed size by promoting cell proliferation in ovule integument (Elliott et al., 1996; Klucher et al., 1996; Li and Li, 2015). To determine whether ANT functions downstream of MEE45 in regulating the development of ovule integument, we examined ANT transcript levels in the mature ovules of WT and mee45-1 plants, as well as from mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45 (in Col-0) transgenic plants. ANT transcript levels were strongly diminished in the mature ovules of the mee45-1 mutant (Supplemental Figures S13C and S14), returned to WT levels in mee45-1 MEE45pro:MEE45 ovules, and significantly higher in MEE45pro:MEE45 ovules compared to those in the WT (Supplemental Figure S14). We further analyzed the expression pattern of ANT by determining the fluorescence accumulation pattern of a yellow fluorescent protein (YFP) construct driven by the ANT promoter (ANTpro:YFP) in mature ovules of WT and the mee45-1 mutant (Figure 6, A and B). Compared to the WT, YFP fluorescence was severely reduced in the mature ovule integuments of mee45-1 plants, suggesting that MEE45 promotes ANT expression in the ovule integument.
Figure 6.
MEE45 controls seed size by directly activating ANT expression. A and B, Expression pattern of ANT from ANTpro:YFP3 in mature ovules of WT (A) and mee45-1 (B). C, Expression pattern of ANT from ANTpro:YFP3 in mature WT ovules. Bars = 50 μm. D, ChIP-qPCR of the ANT promoter using an anti-GFP antibody in mature ovules of 35Spro:MEE45-GFP transgenic lines, using 35Spro:GFP ovules as negative control. MEE45 binds to region P1. The error bars represent the standard error of the mean, calculated from three sets of biological replicates. ***P < 0.001, ns, not significant, compared to the WT (Col-0) (Student’s t -test). E, Direct interaction between MEE45 and the ANT promoter, as determined by EMSA. The arrows indicate the position of the shifted bands and the free probe, respectively. F–K, Mature dry seeds of the WT (F), mee45-1 (G), ant-T (H), mee45-1 ant-T double mutants (I), 35Spro:MEE45 (J), and ant-T 35Spro:MEE45 (K) plants. Bar = 500 μm. L, Average seed mass per 1,000 seeds of the indicated genotypes. M, Average seed length and width of the indicated genotypes. The error bars in (L) and (M) represent the standard error of the mean, calculated from three sets of biological replicates. One-way ANOVA and Tukey’s multiple comparison tests were performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
To investigate whether MEE45 acts as a direct regulator of ANT transcription, we performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR; ChIP-qPCR) experiments using ovules from 35Spro:MEE45-GFP lines. We saw an enrichment for the P1 fragment of the ANT promoter (−775 bp to −504 bp relative to the transcription start site, Figure 6D), which contained the potential binding site for B3-type transcription factors CATGCA (–628 bp to –623 bp). Electrophoretic mobility shift assays (EMSAs) revealed that MEE45 binding to the P1 fragment of the ANT promoter was completely abolished when we mutated the CATGCA motif to ACCTAC (Figure 6E). Together, these data strongly suggest that MEE45 directly binds to the predicted CATGCA motif of the ANT promoter. Furthermore, YFP reporter constructs driven by the ANT promoter with a mutated version of the MEE45-binding site (CATGCA to ACCTAC) showed a strong reduction of YFP fluorescence in transgenic ovule integuments compared to a construct driven by the native ANT promoter (Figure 6, A and C). These results support the hypothesis that MEE45 functions as a direct regulator of ANT transcription in the ovule integument, specifically through its binding at the ANT promoter.
MEE45 controls seed size by activating the expression of its target gene ANT
Previous studies revealed that loss-of-function ant mutants have defective ovules that fail to form integuments or female gametophytes (Klucher et al., 1996). To investigate the genetic relationship between MEE45 and ANT in the control of ovule development and seed size, we identified a previously unexamined ant mutant with a T-DNA insertion in the 3′-untranslated region of the ANT locus (Supplemental Figure S15A). We refer to this new allele as ant-T. ANT transcript levels in mature ovules of the ant-T mutant were significantly reduced compared to those of the WT (Supplemental Figure S15B). Unlike published loss-of-function ant mutants, ant-T ovules produced functional female gametophytes and generated smaller seeds with normal seed germination and viability rates compared to the WT (Supplemental Figures S2 and S15, C, D, G, and H; Supplemental Data Sets S2, S4, S23, and S24), indicating that ant-T is a knockdown allele.
The small-seed phenotype seen in ant-T was completely rescued by the introduction of a 35Spro:ANT transgene. The same 35Spro:ANT transgene resulted in the production of significantly oversized seeds when introduced in the WT background (Supplemental Figure S15, E–H; Supplemental Data Sets S23 and S24). Compared to ovule development in the WT, ant-T ovule primordia appeared to form and enlarge normally, but integument initiation was delayed (Supplemental Figure S16, A–C). Although both the inner and outer integuments eventually enclosed the nucellus, mature ovules were smaller in ant-T compared to WT ovules (Supplemental Figure S16D), a phenotype shared with the mee45-1 mutant (Figure 5H). Therefore, ant-T can be classified as a new knock-down mutant with milder phenotypes than previously reported ant alleles.
We then generated mee45-1 ant-T double mutant plants through genetic crossing and discovered that their seed size and weight did not differ from those of the ant-T mutant, but were significantly smaller than those of the mee45-1 mutant (Figure 6, F–I, L, and M; Supplemental Data Sets S25 and S26). These results indicate that ant-T is epistatic to mee45-1 for the seed size phenotype. We also found that the ant-T mutant suppressed the seed size increase that we had observed earlier in MEE45-overexpressing transgenic lines (Figure 6, J–M; Supplemental Figure S17; Supplemental Data Sets S25 and S26). These genetic results demonstrate that ANT functions as a direct target of MEE45 in the maternal control of seed size.
MEE45 controls seed size by inducing YUC-mediated auxin biosynthesis in the ovule integument
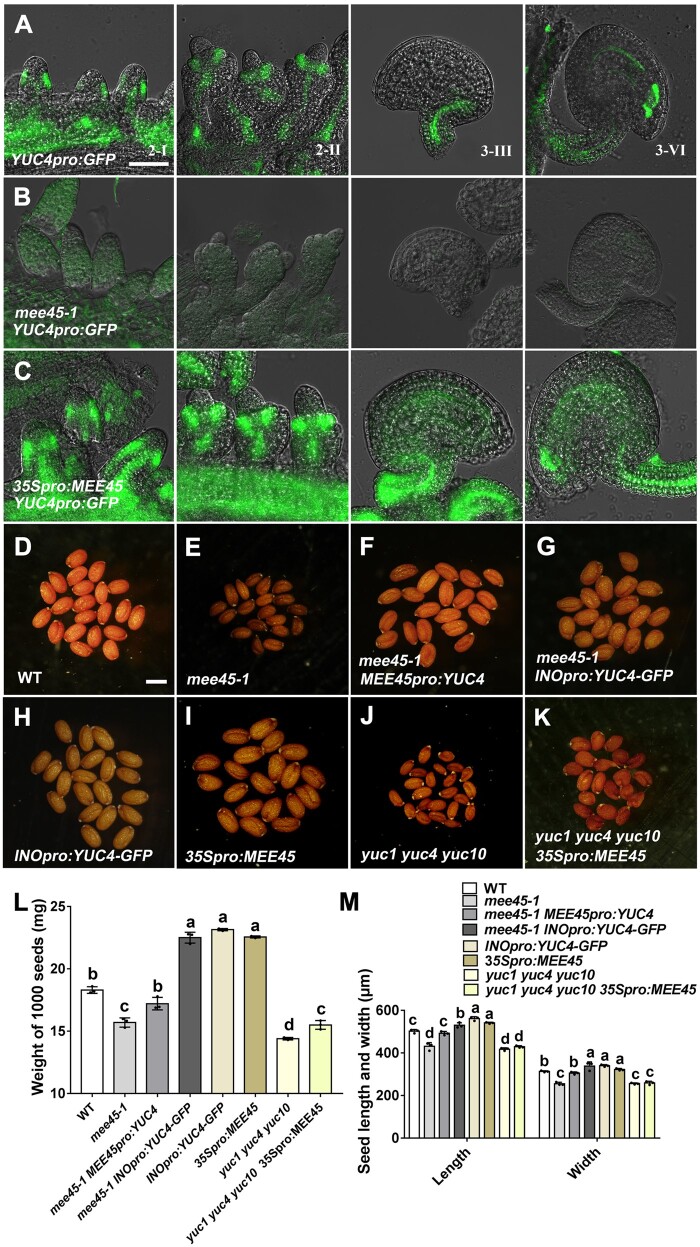
YUCs are auxin biosynthetic genes, and our transcriptomic analysis showed YUC1, YUC2, YUC4, YUC5, and YUC10 among significantly downregulated DEGs in the mee45-1 mutant compared to the WT (Supplemental Figure S13, A and B). We confirmed the significantly differential expression of YUC genes in the mature ovules of WT and mee45-1 mutant plants using qPCR (Supplemental Figure S13C). We further characterized the expression pattern of YUC4, whose differential expression was the most pronounced among differentially expressed YUCs, in the ovules of the mee45-1 mutant and in MEE45-overexpressing lines (Figure 7, A–C) by introducing a GFP reporter construct under the control of the YUC4 promoter. Compared to the WT, fluorescence from the YUC4pro:GFP reporter was severely reduced in the integuments of mee45-1 mutant ovules. In contrast, the GFP signal was significantly stronger in ovule integuments of MEE45-overexpressing lines relative to the WT. Moreover, the small-sized seed phenotype in the mee45-1 mutant was completely rescued when we expressed the YUC4 gene under the control of the MEE45 promoter (Figure 7, D–F, L, and M; Supplemental Data Sets S27 and S28). Thus, MEE45 promotes YUC-mediated auxin biosynthesis in the integument.
Figure 7.
Seed size in mee45-1 is reduced due to reduced auxin biosynthesis capacity in the ovule integument. A–C, Expression pattern of YUC4 in ovules at the 2-I, 2-II, 3-III, and 3-VI (mature ovule) stages of YUC4pro:GFP (A), mee45-1 YUC4pro:GFP (B) and 35Spro:MEE45 YUC4pro:GFP (C) transgenic plants. Bar = 50 μm. D–K, Mature dry seeds of the WT (D), mee45-1 (E), mee45-1 MEE45pro:YUC4 (F), mee45-1 INOpro:YUC4-GFP (G), INOpro:YUC4-GFP (H), 35Spro:MEE45 (I), yuc1 yuc4 yuc10 triple mutants (J), and yuc1 yuc4 yuc10 35Spro:MEE45 (K) plants. Bar = 500 μm. L, Average seed mass per 1,000 seeds of the indicated genotypes. M, Average seed length and width of the indicated genotypes. The error bars in (L) and (M) represent the standard error of the mean, calculated from three sets of biological replicates. One-way ANOVA and Tukey’s multiple comparison tests were performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
As the mee45-1 mutant displayed decreased expression of multiple auxin biosynthetic genes in ovules, we measured auxin content in the mutant and WT backgrounds. The total IAA content in mee45-1 siliques was significantly lower than in WT siliques (Supplemental Figure S18A). We further evaluated the fluorescence derived from the auxin reporter DR5:GFP in ovules of the mee45-1 mutant and MEE45-overexpressing lines. Similar to YUC4 expression, fluorescence from the DR5:GFP reporter was severely reduced in mee45-1 mutant ovules, but was significantly increased in ovules of MEE45-overexpressing lines compared to the WT (Supplemental Figure S18, C–E). Collectively, these results indicate that the mutation of MEE45 leads to decreased auxin levels and a localized downregulation of auxin response in ovules.
We hypothesized that a lack of auxin accumulation in ovule integument might cause the observed small seed size phenotype of the mee45-1 mutant. To test this hypothesis, we expressed YUC4 specifically in the integument of the mee45-1 mutant by using the aforementioned INO promoter (Supplemental Figure S18, B, F, and G). The INOpro:YUC4 transgene caused a strong and significant increase of seed size in the mee45-1 mutant, even producing oversized seeds as compared to the WT (Figure 7, D, E, G, L, and M; Supplemental Data Sets S27 and S28). Moreover, expression of the INOpro:YUC4 transgene in the WT background produced significantly oversized seeds compared to the WT control (Figure 7, D, H, L, and M; Supplemental Data Sets S27 and S28). These findings indicate that the small seed size in the mee45-1 mutant apparently results from an auxin deficiency in the integument and support the idea that auxin biosynthesis in ovule integument contributes to the control of seed size.
Overexpression of MEE45 resulted in oversized seeds (Figure 7, D, I, L, and M; Supplemental Data Sets S27 and S28). However, mutations of the YUC genes in the yuc1 yuc4 yuc10 triple mutant largely suppressed the seed size increase caused by MEE45 overexpression, displaying seeds with no significant difference in size or weight compared to the yuc1 yuc4 yuc10 triple mutant, which itself had significantly smaller seeds than the WT (Figure 7, I–M; Supplemental Figure S17; Supplemental Data Sets S27 and S28). Together, these observations provide strong evidence that MEE45 controls seed size by increasing auxin biosynthetic capacity in ovule integuments.
A MEE45–ANT pathway regulates auxin biosynthesis in the ovule integument
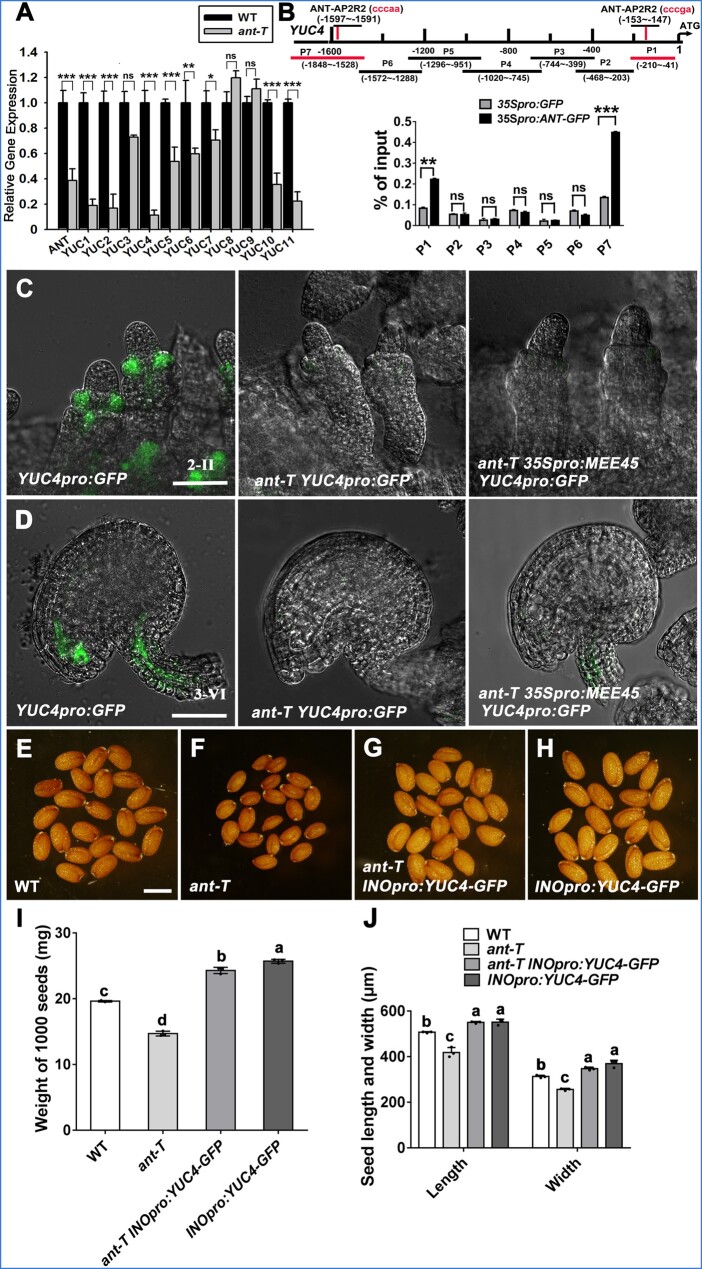
Given our findings that expression of both ANT and YUC genes requires MEE45, and our demonstration of ANT as a binding and transcriptional activation target of MEE45, we further tested whether YUC genes may act downstream of MEE45–ANT during integument development. Most YUC transcript levels were significantly lower in ant-T mutant ovules compared to the WT (Figure 8A). In agreement, ant-T mutant siliques had significantly reduced total IAA content compared to the WT (Supplemental Figure S18A). Furthermore, fluorescence of the YUC4pro:GFP reporter was significantly reduced in ovule integuments of the ant-T mutant (Figure 8, C and D).
Figure 8.
ANT activates auxin biosynthesis in the ovule integument. A, Relative expression levels of YUC genes in mature ovules of WT and ant-T. The error bars represent the standard error of the mean, calculated from three sets of biological replicates. ***P < 0.001, **P < 0.01, *P < 0.05, ns, not significant, compared to the WT (Col-0) (Student’s t-test). B, ChIP-qPCR of the YUC4 promoter using an anti-GFP antibody in the mature ovules of 35Spro:ANT-GFP transgenic plants, using ovules from 35Spro:GFP transgenic plants as negative control. ANT binds to regions P1 and P7. The error bars represent the standard error of the mean, calculated from three sets of biological replicates. **P < 0.01, ***P < 0.001, ns, not significant, compared to the WT (Col-0) (Student’s t-test). C, Expression pattern of YUC4 in stage 2-II ovules of YUC4pro:GFP, ant-T YUC4pro:GFP, and ant-T YUC4pro:GFP 35Spro:MEE45 transgenic plants. D, Expression pattern of YUC4 in stage 3-VI (mature) ovules of YUC4pro:GFP, ant-T YUC4pro:GFP and ant-T YUC4pro:GFP 35Spro:MEE45 transgenic plants. Bars = 50 μm. E–H, Mature dry seeds of the WT (E), ant-T (F), ant-T INOpro:YUC4-GFP (G) and INOpro:YUC4-GFP (H) plants. Bar = 500 μm. I, Average seed mass per 1,000 seeds of the indicated genotypes. J, Average seed length and width of the indicated genotypes. The error bars in (I) and (J) represent the standard error of the mean, calculated from three sets of biological replicates. One-way ANOVA and Tukey’s multiple comparison tests were performed. Statistically significant differences are indicated by different lowercase letters (P < 0.05).
To further investigate whether ANT directly interacts with the YUC4 promoter to regulate YUC4 expression, we performed a ChIP-qPCR assay using mature ovules from 35Spro:ANT-GFP lines (Figure 8B). Of the fragments (P1–P7) covering the entire presumptive 1,850 bp YUC4 promoter region, only the P1 and P7 fragments were strongly enriched by immunoprecipitation with the anti-GFP antibody, respectively, spanning the regions from –210 bp to –41 bp and from –1,848 bp to –1,528 bp. Each P1 and P7 fragments had one AP2 repeat (ANT-AP2R2) binding site, which is bound by ANT in vitro (Nole-Wilson and Krizek, 2000). These results suggest that ANT binds to the YUC4 promoter and activates its transcription.
Moreover, expressing YUC4 under the control of the INO promoter (INOpro:YUC4) rescued the small seed size seen in the ant-T mutant, even leading to seeds that were oversized compared to the WT, and showed no significant difference with the seeds of WT plants carrying the INOpro:YUC4 transgene (Figure 8, E–J; Supplemental Figure S18H; Supplemental Data Sets S29 and S30). Intriguingly, MEE45 overexpression in the ant-T background did not cause a similar extent of YUC4 expression induction as in the WT background: we detected the weak expression of YUC4 in ovule integuments from MEE45-overexpressing ant-T lines, but this level did not differ from the YUC4 level in the ant-T mutant (Figure 7, A and C; Figure 8, C and D). These results suggest that a MEE45–ANT pathway activates the expression of YUC4, thereby promoting the auxin biosynthesis capacity of cells in the ovule integument (Figure 9).
Figure 9.
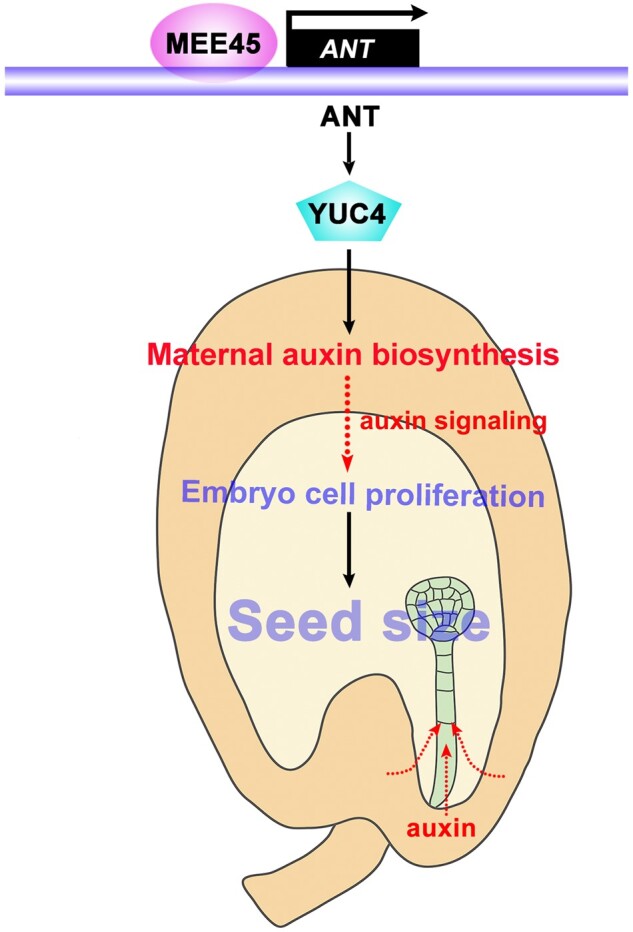
A working model for how a MEE45–ANT pathway in maternal tissues modulates auxin biosynthesis and seed size in Arabidopsis. The integuments are maternal tissues that surround the ovule, setting an upper limit for the embryo to grow and impacting the final seed size. In the ovule integument, MEE45 positively regulates ANT transcription by directly binding to the ANT promoter region, and ANT in turn directly interacts with the YUC4 promoter to activate YUC4 expression. MEE45 and ANT function in the same genetic pathway to regulate the expression of YUC4, thereby mediating maternal auxin biosynthesis. Auxin biosynthesized in the maternal ovule integument may generate a signal received by embryos or move directly into the embryos to affect their development, and further determines the seed size.
Discussion
Integuments are maternal tissues that surround the ovule and form the seed coat after fertilization. Integuments also provide the cavity for the growth of the embryo and the endosperm, and set an upper limit to seed size (Fang et al., 2012; Xia et al., 2013; Du et al., 2014). Here, we show that MEE45 is essential for the proliferation of integument cells and size control of mature ovules, which also controls embryo size in the limited space within seed coats. These findings indicate that MEE45 acts as a regulator in maternal tissues, and the regulation of ovule integument development by MEE45 clearly influences the determination of final seed size.
Several signaling pathways have been identified that control seed size in Arabidopsis and rice, including for example the IKU pathway, which integrates abscisic acid, brassinosteroids, and cytokinin signaling to control seed size by regulating endosperm development (Li et al., 2019). Seed size is also mediated by the growth of maternal tissues. Previous studies have suggested that maternal tissues contribute to seed size control: mutants with maternal tissue defects such as da1 and ap2 affect seed size through alternation of cell proliferation and/or expansion in ovule integuments (Li et al., 2008; Adamski et al., 2009). These studies have established that the effects on seed size of the da1 and ap2 mutants result from the genotypes of the maternal tissues, rather than the genotype of the zygotic tissues.
Transcriptional regulation is central throughout the plant life cycle, including during seed development. Several transcription factors have been identified as regulators of seed size. The Arabidopsis ANT gene encodes an AP2-like transcription factor and was identified as a regulator of integument initiation and ovule development, as well as organ growth (Elliott et al., 1996; Klucher et al., 1996; Mizukami and Fischer, 2000). ant mutants exhibit defects in ovule development with abnormal initiation of integuments, whereas overexpressing ANT produces oversized seeds resulting from a promotion of excessive cell proliferation in integuments. Thus, ANT is a transcription factor that maternally controls seed size. Another Arabidopsis regulator of cell proliferation and expansion is ARF2, which encodes a plant-specific B3 superfamily transcription factor involved in auxin signaling (Li et al., 2004). ARF2 acts maternally to control seed growth by restricting integument cell proliferation (Schruff et al., 2006). Mutation of ARF2 causes phenotypes including increased seed and organ size, abnormal flower morphology, and enhanced seed abortion (Schruff et al., 2006). A previous study showed that ARF2 binds to the ANT promoter and negatively regulates its transcription in seedling leaves and stems, as well as in developing siliques (Meng et al., 2015).
Similar to ARF2, another two Arabidopsis B3 domain transcription repressors, NGATHA-LIKE 2 (NGAL2; also named SUPPRESSOR OF DA1) and DEVELOPMENT-RELATED PcG TARGET IN THE APEX4 (also named NGAL3) were identified to act redundantly to maternally regulate seed size by limiting cell proliferation in integuments (Zhang et al., 2015). NGAL2 binds to the promoter of KLUH and negatively regulates its expression. In this study, we identified MEE45 as a B3 domain transcription factor that maternally regulates seed size. We show that MEE45 directly induces ANT expression in the ovule, which promotes integument cell proliferation and thereby regulates seed size. MEE45 and ARF2 both have ANT as their common target, but these two B3 domain transcription factors exert opposite transcriptional regulatory functions and therefore cause inverse effects on seed size. These findings highlight the multiple signaling pathways that function upstream of ANT for the maternal control of seed development. It will be interesting to further characterize how MEE45 and potentially several other B3 domain transcription factors differentially regulate maternal control of seed development via ANT, and such work may reveal the signaling components and perhaps environmental or cellular factors that function further upstream of these B3 domain transcription factors in specifying seed size.
Auxin controls cell division and cell expansion (Vandenbussche and Van Der Straeten, 2004; Leyser, 2005; Yao et al., 2019) Similar to ARF2 in Arabidopsis, BnARF18 in rapeseed (Brassica napus) represses auxin-responsive genes to maternally control seed size by restricting cell expansion (Liu et al., 2015a). Overexpression of ARF19 in purging nut (Jatropha curcas, an oilseed crop) results in an increased seed size (Sun et al., 2017). A recent study showed that ARF6 and ARF8 may mediate auxin responses in maternal tissues and contribute to seed development in Arabidopsis (Yao et al., 2019). Auxin signaling thus modulates maternal regulation of seed development and seed size. However, our current understanding of how auxin biosynthesis or accumulation controls seed size is limited. In this study, we show that the expression of auxin biosynthesis gene YUC4 in the ovule is positively regulated by ANT, whose expression is in turn directly induced by MONOPTEROS (also named ARF5) in floral primordium initiation (Yamaguchi et al., 2013). These data suggest a positive feedback regulation between ANT and auxin activity in the ovule, which may contribute to seed size control.
Previous studies have shown that maternally produced auxin in the integument after fertilization affects early embryo development (Robert et al., 2018). Our results provide new insights into the mechanisms by which auxin biosynthesis in integuments contributes to the regulation of seed development and final seed size. It will be intriguing to explore whether auxin biosynthesized in maternal integuments generates a signal received downstream by embryos; alternatively, auxin may move directly from maternal cells into embryos and thereby affect their development (Figure 9). Fertilization of the central cell is known to lead to the production of auxin in the endosperm, from which auxin is then exported to maternal tissues to drive seed coat development (Figueiredo et al., 2015). A defect in auxin export from the zygotic tissue to maternal tissues results in abnormal growth of maternal tissues and leads to further reduced seed size (Figueiredo et al., 2016). Together with these published findings, our results deepen our understanding of the mechanisms that regulate local auxin biosynthesis, signaling, and activity to coordinate the development of ovule integuments, embryos, and the endosperm, ultimately contributing to seed size control.
In conclusion, our results demonstrate that a MEE45–ANT pathway maternally regulates seed size. This pathway controls seed growth through the modulation of auxin biosynthesis genes in ovule integuments, tissues known to function as central hubs for the development of both maternal and zygotic tissues in plant reproduction (Figure 9) (Li and Li, 2016). These results improve our understanding of the mechanisms underlying maternal tissue-mediated regulation of seed size. Manipulating MEE45 and/or related pathways and components in crops may enable new technologies to increase seed yield in crop production.
Material and Methods
Plant materials and growth conditions
All mutants and marker lines used in this study were in the Arabidopsis (A. thaliana) Col-0 accession. The T-DNA insertion lines mee45-1 (SALK_031530), mee45-2 (SALK_038324), mee45-3 (CS801014), and ant-T (SALK_022770) were obtained from the Arabidopsis Biological Resource Center. The lines YUC4pro:GFP (Cheng et al., 2013), 35Spro:GFP (Su et al., 2020), and the yuc1 yuc4 yuc10 triple mutant (Cheng et al., 2006) have been described previously. We generated the mee45-1 ant-T double mutant by crossing mee45-1 with ant-T, and used F3 homozygous plants for imaging and measurements.
We surface sterilized all seeds before sowing them on 0.8% (w/v) agar solid medium (half-strength Murashige and Skoog (MS) medium, 1% sucrose, pH 5.7). After breaking dormancy at 4°C for 2 days in the dark, we allowed seeds to germinate and seedlings to grow under long-day conditions (16-h light/8-h dark) with white light 120 µmol/m2/s at 22°C for 10 days, before transplanting individual seedlings to soil for further growth.
Plasmid construction and plant transformation
We generated the MEE45pro:MEE45, MEE45pro:YUC4 constructs using the GreenGate system (Lampropoulos et al., 2013). We PCR-amplified a 1,970-bp fragment of the MEE45 promoter from Col-0 genomic DNA using 2× Phanta Max Master Mix (Vazyme, Nanjing, China ; P515-01), digested the resulting PCR products with BsaI, and then ligated the purified digested products into the BsaI site of the empty entry vector pGGA000 to yield a new promoter module. We amplified the MEE45 and YUC4 CDSs by PCR from Col-0 cDNAs, and ligated the PCR products at the BsaI site of the empty entry vector pGGC000 to generate new CDS modules. We then strung the four modules (pGGB003, pGGD002, pGGF005, and pGGZ001) with the promoter module and the CDS modules described above.
We generated the INOpro:MEE45-GFP, and INOpro:YUC4-GFP constructs using the GreenGate system. We PCR-amplified a 2,012-bp fragment of the INO promoter from Col-0 genomic DNA, digested the resulting PCR products with BsaI, and then ligated the purified digested products into the BsaI site of the empty entry vector pGGA000 to yield a new promoter module. We amplified the MEE45 and YUC4 CDSs by PCR from Col-0 cDNAs, and ligated the PCR products at the BsaI site of the empty entry vector pGGC000 to generate new CDS modules. We then strung the four modules (pGGB003, pGGD001, pGGF005, and pGGZ001) with the promoter module and the CDS modules described above.
Following the same strategy, to generate the RPS5Apro:amiRNA-MEE45 construct, the MEE45 amiRNA was generated by PCR amplification with the combinations of primers shown in Supplemental Table S1. The amiRNA-MEE45 construct was cloned into empty entry vector pGGC000 to generate a new module. We then strung the five modules (pGGA012, pGGB003, pGGD002, pGGF005, and pGGZ001) with the amiRNA module described above.
To generate the gMEE45-GFP construct, we PCR-amplified a 4,045-bp MEE45 genomic fragment and inserted it into the pENTR entry vector, before recombination into the pMDC107 vector (Gateway system, Invitrogen, Carlsbad, CA, USA) through Gateway LR recombination.
To generate the 35Spro:MEE45 and 35Spro:ANT constructs, we independently PCR-amplified the MEE45 and ANT CDS from Col-0 cDNAs, digested the PCR products with SacI and PstI, and ligated the purified and digested fragments into the pCAMBIA1300 vector.
To generate the 35Spro:ANT-GFP and 35Spro:MEE45-GFP constructs, we independently PCR-amplified the ANT and MEE45 CDSs from Col-0 cDNA, digested the PCR products with KpnI/SpeI and BamHI/KpnI, then ligated the purified and digested fragments into the pROK II-GFP vector.
To generate the ANTpro:YFP3 construct, we PCR-amplified a 2,019-bp DNA fragment of ANT genomic sequence upstream of the putative initiation codon and cloned in frame with NLS-3×YFP followed by a 35S terminator.
The primers used for plasmid construction are listed in Supplemental Table S1. The constructs were transformed into all genotypes using the floral dip method (Clough and Bent, 1998). Homozygous transgenic plants (T2 generation) were used in this study.
Analysis of seed size and seed mass
To determine seed mass, we weighed 1,000 mature dry seeds in each replicate. All measurements were repeated 3 times. After capturing photographs with a stereomicroscope connected to a computer, we used ImageJ to measure the length and width of seeds. Three replicates were used for statistical analysis, and in each replicate, ∼30 seeds were examined.
Total RNA isolation and real-time qPCR analysis
Total RNA was extracted from the indicated tissue using Ultrapure RNA Kit (CWbiotech, Beijing, China). First-strand cDNAs were then synthesized by FastQuant RT Kit (Tiangen Biotech, Beijing, China). SYBR Green Real-time PCR Master Mix (Tiangen Biotech) was used for qPCR, according to the manufacturer’s protocol. The SYBR Green Master mix was used to dilute each cDNA. For all samples, relative transcript levels were normalized to TUBULIN2, and all measurements were carried out in three biological replicates. To determine expression values, we used the comparative CT method. The primers used for qPCR analysis are listed in Supplemental Table S1.
Immunoblotting
To analyze MEE45 protein accumulation, total proteins were extracted from mature ovules of WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45 lines with extraction buffer (140 mM NaCl, 0.537 mM KCl, 0.2939 mM KH2PO4, 5 mM Na2HPO4, 0.01 mM EDTA, 0.05% NP-40, 5 mM DTT, 0.5 mM PMSF, 1× protease inhibitor cocktail). Samples were centrifuged at 4°C, at 12,000 r.p.m. for 60 min. The supernatant was then transferred to a new tube and used for gel electrophoresis. MEE45 protein was detected by immunoblot using anti-MEE45 antibody (1:1,000 dilution, polyclonal antibody produced by Abmart, using Arabidopsis MEE45 protein as antigen). ACTIN was detected by anti-ACTIN antibody (1:1,000 dilution, A01050; Abbkine Wuhan Shi, China) as loading control, followed by goat anti-rabbit (1:1,000 dilution, ZB-5301; ZSGB-BIO, Beijing, China) or goat anti-mouse (1:1,000 dilution, A0216; Beyotime, Shanghai, China) HRP-conjugated secondary antibody. Signal was visualized using a Universal Hood II (BIO-RAD, Hercules, CA, USA). Immunoblot experiments were repeated 3 times.
DIC microscopy and image processing
We observed embryo and endosperm development in cleared seeds using differential interference contrast (DIC) optics as previously described (Ohto et al., 2009). We cleared specimens with a chloralhydrate/glycerol/water (8:3:1) solution, and collected images on a DIC microscope connected to a computer for image capture.
Sectioning and cytological analysis
Siliques were cut at both ends and fixed in glutaraldehyde fixative under vacuum for 10 min. Siliques were then rinsed twice in 0.2 M phosphate buffer, pH = 7.0 before dehydration through a graded ethanol series: 10% (v/v with water), 30%, 50%, 70%, 95% (eosin added), and 100%, each time for 1 h. We incubated samples in propylene oxide for 1 h, followed by propylene oxide and epoxy (v/v 1:1) incubation overnight at room temperature. We then incubated the specimens with an epoxy resin accelerant for 2 days, changing the solution once a day. We finally embedded the specimens in the same solution at 42°C overnight, and then 60°C for 2 days. We used a Leica Microsystems RM2265 microtome to slice the specimens, followed by dyeing with 1% Toluidine Blue O. A stereomicroscope was used for image acquisition. Then, the sections were photographed with an Olympus BH-2 microscope equipped with an Olympus digital camera.
Scanning electron microscopy
Mature ovules were immersed in glutaraldehyde fixative for 20 h at room temperature and then rinsed 5 times for 20 min each in phosphate-buffered saline (PBS) buffer. Following immersion in osmium tetroxide for 5 h, we rinsed specimens in PBS buffer 3 times for 20 min each. The specimens were then dehydrated with a graded ethanol series (45%, 55%, 70%, 85%, 95%, and 100%). Critical-point drying was accomplished under liquid carbon dioxide. The specimens were mounted on stubs, sputter-coated with palladium gold, then examined with a scanning electron microscope (JSM-6610; JEOL, Tokyo, Japan).
Transcriptome analysis
We collected siliques 3 days after pollination from Col-0 and mee45-1 plants for RNA-seq analysis as three independent biological replicates per genotype. The libraries were constructed with NEBNext Ultra RNA Library Prep Kit for Illumina and checked on an Agilent 2100 Bioanalyzer. The libraries were then sequenced on a HiSeq 2500 platform (Illumina, San Diego, CA, USA). After removing adapters, reads with Ns, and low-quality reads, clean reads were mapped to the A. thaliana genome (version: Arabidopsis Information Resource [TAIR] 10) with TopHat2 with default parameters (Kim et al., 2013). Fragment per kilobase of exon per million reads mapped was used to estimate transcript abundance. Genes with at least two-fold change and a false discovery rate ˂0.1 were considered DEGs.
Observation of ovules development
Developing ovules were fixed in glutaraldehyde fixative under vacuum for 30 min, then incubated overnight at 4°C. The specimens were dehydrated in a graded ethanol series (30%, 40%, 50%, 60%, 70%, and 100%) for 30 min each. We cleared specimens with benzoate and benzyl alcohol (v/v 2:1) for 2 h, and acquired images with a Zeiss 880 laser scanning microscope with 488 nm laser excitation and 505–550 nm emission spectra.
GUS staining
Specimens (CYCB1;1pro:GUS, mee45-1 CYCB1;1pro:GUS) were stained in GUS staining solution (1 mM 5-bromo-4-chloro-3-indolyl-b-D-glucuronic acid, 100 mM Na3PO4 buffer, 3 mM each K3Fe(CN)6/K4Fe(CN)6, 10 mM EDTA, and 0.1% Nonidet P-40) and incubated at room temperature for 5 h. After GUS staining, chlorophyll was removed in 70% ethanol.
Fluorescence analysis
We acquired confocal laser-scanning micrographs of ovules and embryos with a Zeiss 880 laser scanning microscope. For GFP and YFP detection, we used 488 and 514 nm laser excitation, 505–550 nm, and 530–600 nm emission spectra for each respective excitation. We acquired, analyzed, and exported images with the help of the Zeiss ZEN software.
ChIP analysis
We performed ChIP on mature ovules collected from 35Spro:MEE45-GFP, 35Spro:ANT-GFP and 35Spro:GFP lines using an EZ ChIP Kit (Millipore, Burlington, MA, USA), according to previously described protocols (Su et al., 2020) with minor modifications. Bioruptor was used for sonication (250W, 20%, repeat 4–6 times; sonication buffer: 1 mL SDS Lysis buffer with 5 µL Protease inhibitor cocktail II). We used anti-GFP antibody (1:500 dilution, SAB5300167; Sigma St Louis, MO, USA) to precipitate chromatin. 35Spro:GFP lines were used as negative controls. For each ChIP assay, we performed three biological and three technical repeats. All primer sequences used for ChIP assays are listed in Supplemental Table S1.
EMSA
We performed EMSA as previously described (Su et al., 2020). We largely followed the instructions supplied with the Light Shift Chemiluminescent EMSA kit (Thermo Scientific, Waltham, MA, USA). To purify the MEE45-GST fusion protein, we cloned the MEE45 CDS into the pGEX-4T vector, and produced the protein in Escherichia coli BL21 (DE3), followed by purification with Glutathione Sepharose 4B beads (GE Healthcare, Chicago, IL, USA). Biotin-labeled probes were commercially synthesized. The WT sequence CATGCA corresponding to the –628 bp to –623 bp region of the ANT promoter, which contains the MEE45-binding site, was replaced with ACCTAC in the mutated probe.
Free IAA measurements
We harvested siliques 3 days after pollination from Col-0, mee45-1, and ant-T plants for free IAA measurements, using three independent biological replicates per genotype. Free IAA measurements were performed as previously described (Wang et al., 2015).
Seed germination and viability analysis
High-temperature and high-moisture conditions were used to evaluate Arabidopsis seed vigor. We tested seed germination or seed viability as previously described (Li et al., 2017). Seeds used to measure vigor were placed in a seed aging chamber at 83% relative humidity, 42°C for 72 h to accelerate aging, and then dried at room temperature for 24 h. Seeds treated with or without aging were surface-sterilized before sowing on 0.8% (w/v) agar solid half-strength MS medium for germination at 22°C. Germination was determined every 12 h. Three replicates were used for statistical analysis. In each replicate, 50 seeds were examined.
Accession numbers
Gene sequence data form this article can be found in the TAIR under the following accession number: MEE45 (At4g00260), ANT (At4g37750), MINI3 (At1g55600), IKU1 (At2g35230), IKU2 (At3g19700), SHB1 (At4g25350), YUC1 (At4g32540), YUC2 (At4g13260), YUC3 (At1g04610), YUC4 (At5g11320), YUC5 (At5g43890), YUC6 (At5g25620), YUC7 (At2g33230), YUC8 (At4g28720), YUC9 (At1g04180), YUC10 (At1g48910), and YUC11 (At1g21430). RNA-seq data discussed in this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE156491. All source data are provided with this article.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization and function of the MEE45 gene.
Supplemental Figure S2. Seed germination and viability of the WT, mee45-1 and ant-T.
Supplemental Figure S3. Mature ovules and embryo development in WT, mee45-2 and mee45-3 mutants.
Supplemental Figure S4. Relative expression of MEE45 in mature ovules of the indicated genotypes.
Supplemental Figure S5. The mature embryos of WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45 plants.
Supplemental Figure S6. Endosperm development in WT and mee45-1 seeds.
Supplemental Figure S7. Relative expression of MEE45 in different organs.
Supplemental Figure S8. The phenotypes of leaves, flowers, and siliques in mee45-1 mutants compared to WT plants.
Supplemental Figure S9. Reciprocal cross experiments between the WT and mee45-2.
Supplemental Figure S10. Reciprocal cross experiments between the WT and mee45-3.
Supplemental Figure S11. MEE45 Expression in the INOpro:MEE45-GFP and mee45-1 INOpro:MEE45-GFP transgenic lines.
Supplemental Figure S12. Cell proliferation indicated by CYCB1;1pro:GUS expression in the mature ovules of the WT and mee45-1 mutant.
Supplemental Figure S13. Functional enrichment and gene expression analysis of RNA-seq data in the WT and mee45-1 mutants.
Supplemental Figure S14. Relative expression of ANT in mature ovules of the indicated genotypes.
Supplemental Figure S15. Characterization and function of the ANT gene.
Supplemental Figure S16. Ovule development in the WT and ant-T plants.
Supplemental Figure S17. Relative expression of MEE45 in mature ovules of the indicated genotypes.
Supplemental Figure S18. IAA content and expression of YUC4 in mature ovules of the indicated genotypes.
Supplemental Table S1. Primers used for qPCR, ChIP-qPCR, and constructions of expression vectors
Supplemental Data Set S1. Figure 1I data: Seed mass per 1,000 seeds for the indicated genotypes.
Supplemental Data Set S2. Figure 1J data: Seed length and width for the indicated genotypes.
Supplemental Data Set S3. Supplemental Figure S2G data: Number of germinated seeds from WT, mee45-1 or ant-T mutants at the indicated hours after seed hydration without aging treatment.
Supplemental Data Set S4. Supplemental Figure S2H data: Number of germinated seeds from WT, mee45-1 or ant-T mutants at the indicated hours after seed hydration with aging treatment.
Supplemental Data Set S5. Supplemental Figure S1H data: Seed mass per 1,000 seeds of the WT, mee45-2 and mee45-3.
Supplemental Data Set S6. Supplemental Figure S1I data: Seed length and width of the WT, mee45-2 and mee45-3.
Supplemental Data Set S7. Supplemental Figure S5I data: Epidermal cell areas in the cotyledons of the WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45.
Supplemental Data Set S8. Supplemental Figure S5J data: Cotyledon areas of the WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45.
Supplemental Data Set S9. Supplemental Figure S5K data: Epidermal cell number in the cotyledons of the WT, mee45-1, mee45-1 MEE45pro:MEE45 and MEE45pro:MEE45.
Supplemental Data Set S10. Supplemental Figure S8F data: Leaf area of mee45-1 compared to WT (WT set to 100%).
Supplemental Data Set S11. Supplemental Figure S8G data: Silique length and width of mee45-1 compared to WT (WT set to 100%).
Supplemental Data Set S12. Figure 3I data: Seed mass per 1,000 seeds for the indicated F1 and F2 crosses.
Supplemental Data Set S13. Figure 3J data: Seed length and width for the indicated F1 and F2 crosses.
Supplemental Data Set S14. Supplemental Figure S9G data: Seed mass per 1,000 seeds for the indicated F1 crosses.
Supplemental Data Set S15. Supplemental Figure S9H data: Seed length and width for the indicated F1 crosses.
Supplemental Data Set S16. Supplemental Figure S10G data: Seed mass per 1,000 seeds for the indicated F1 crosses.
Supplemental Data Set S17. Supplemental Figure S10H data: Seed length and width for the indicated F1 crosses.
Supplemental Data Set S18. Figure 4K data: Seed mass per 1,000 seeds for the indicated transgenic lines.
Supplemental Data Set S19. Figure 4L data: Seed length and width for the indicated transgenic lines.
Supplemental Data Set S20. Figure 5C data: Number of cells in the outer integument of mature ovules from WT and mee45-1 plants.
Supplemental Data Set S21. Figure 5D data: Outer integument length of mature ovules from WT and mee45-1 plants.
Supplemental Data Set S22. Supplemental Figure S13 data: DEGs in mee45-1 siliques compared to the WT.
Supplemental Data Set S23. Supplemental Figure S15G data: Seed mass per 1,000 seeds for the indicated genotypes.
Supplemental Data Set S24. Supplemental Figure S15H data: Seed length and width for the indicated genotypes.
Supplemental Data Set S25. Figure 6L data: Seed mass per 1,000 seeds for the indicated genotypes.
Supplemental Data Set S26. Figure 6M data: Seed length and width for the indicated genotypes.
Supplemental Data Set S27. Figure 7L data: Seed mass per 1,000 seeds for the indicated genotypes.
Supplemental Data Set S28. Figure 7M data: Seed length and width for the indicated genotypes.
Supplemental Data Set S29. Figure 8I data: Seed mass per 1,000 seeds for the indicated genotypes.
Supplemental Data Set S30. Figure 8J data: Seed length and width for the indicated genotypes.
Supplemental Data Set S31. Statistical analysis.
Funding
This research was funded by the National Natural Science Foundation of China (31670320, 31730008, 31872669), the Natural Science Foundation of Shandong Province (ZR2017JL016), and the Program for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province (2019KJE011).
Conflict of interest
The authors declare no competing interest.
Supplementary Material
Y.H.S. and X.S.Z. designed the research and wrote the manuscript. Y.J.L. and Y.Y. performed the experiments. X.L. analyzed the RNA-seq data.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Ying Hua Su (suyh@sdau.edu.cn).
REFERENCES
- Adamski NM, Anastasiou E, Eriksson S, O’Neill CM, Lenhard M (2009) Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci USA 106: 20115–20120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Wang L, Sun W, Zhang Y, Zhou C, Su YH, Li W, Sun TT, Zhao XY, Li XG,. et al. (2013) Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol 161: 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Yu S, Wang C, Xing Y (2009) A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet 118: 465–472 [DOI] [PubMed] [Google Scholar]
- Fang W, Wang Z, Cui R, Li J, Li Y (2012) Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J 70: 929–939 [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C (2015) Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184. [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler C (2016) Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 5: e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10: e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 10: 472–477 [DOI] [PubMed] [Google Scholar]
- He S, Sun Y, Yang Q, Zhang X, Huang Q, Zhao P, Sun M, Liu J, Qian W, Qin G,. et al. (2017) A novel imprinted gene NUWA controls mitochondrial function in early seed development in Arabidopsis. PLoS Genet 13: e1006553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA 102: 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Li W, Zhou Y, Ni M (2013) A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet 9: e1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush G (2003) Productivity improvements in rice. Nutr Rev 61: S114–116 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate–-a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2005) The fall and rise of apical dominance. Curr Opin Genet Dev 15: 468–471 [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7: 193–204 [DOI] [PubMed] [Google Scholar]
- Li N, Li Y (2015) Maternal control of seed size in plants. J Exp Bot 66: 1087–1097 [DOI] [PubMed] [Google Scholar]
- Li N, Li Y (2016) Signaling pathways of seed size control in plants. Curr Opin Plant Biol 33: 23–32 [DOI] [PubMed] [Google Scholar]
- Li N, Xu R, Li Y (2019) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70: 435–463 [DOI] [PubMed] [Google Scholar]
- Li T, Zhang Y, Wang D, Liu Y, Dirk LMA, Goodman J, Downie AB, Wang J, Wang G, Zhao T (2017) Regulation of seed vigor by manipulation of raffinose family oligosaccharides in maize and Arabidopsis thaliana. Mol Plant 10: 1540–1555 [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Hu Z, Yang H, Zhang L, Li R, Deng L, Sun X, Wang X, Wang H (2015a) Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc Natl Acad Sci USA 112: E5123–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, Che R, Xu F, Hu B, Liang C, Chu J, Li J, Chu C (2015b) Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA 112: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister RJ, Kotow LM, Gasser CS (2002) SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Meng LS, Wang ZB, Yao SQ, Liu A (2015) The ARF2-ANT-COR15A gene cascade regulates ABA-signaling-mediated resistance of large seeds to drought in Arabidopsis. J Cell Sci 128: 3922–3932 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles AT, Ackerly DD, Webb CO, Tweddle JC, Dickie JB, Westoby M (2005) A brief history of seed size. Science 307: 576–580 [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA (2000) DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res 28: 4076–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Floyd SK, Fischer RL, Goldberg RB, Harada JJ (2009) Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod 22: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, Wójcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, et al. (2018) Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, De Veylder L (2018) The dual face of cyclin B1. Trends Plant Sci 23: 475–478 [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Su YH, Zhou C, Li YJ, Yu Y, Tang LP, Zhang WJ, Yao WJ, Huang R, Laux T, Zhang XS (2020) Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 117: 22561–22571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wang C, Wang N, Jiang X, Mao H, Zhu C, Wen F, Wang X, Lu Z., Yue G,. et al. (2017) Manipulation of auxin response factor 19 affects seed size in the woody perennial Jatropha curcas. Sci Rep 7: 40844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Van Der Straeten D (2004) Shaping the shoot: a circuitry that integrates multiple signals. Trends Plant Sci 9: 499–506 [DOI] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JG, Feng C, Liu HH, Ge FR, Li S, Li HJ, Zhang Y (2016) HAPLESS13-mediated trafficking of STRUBBELIG is critical for ovule development in Arabidopsis. PLoS Genet. 12: e1006269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D (1995) The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7: 388–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Geldner N, Offringa R, Jürgens G (2001) Seed development: early paternal gene activity in Arabidopsis. Nature 414: 709–710 [DOI] [PubMed] [Google Scholar]
- Westermann J, Streubel S, Franck CM, Lentz R, Dolan L, Boisson-Dernier A (2019) An evolutionarily conserved receptor-like kinases signaling module controls cell wall integrity during tip growth. Curr Biol 29: 3899–3908.e3893 [DOI] [PubMed] [Google Scholar]
- Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y (2013) The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D (2013) A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24: 271–282 [DOI] [PubMed] [Google Scholar]
- Yao X, Chen J, Zhou J, Yu H, Ge C, Zhang M, Gao X, Dai X, Yang ZN, Zhao Y (2019) An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol 180: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Li J, Huang Y, Liu L, Li D, Chen L, Luan S (2014) FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol Plant 7: 920–922 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Du L, Xu R, Cui R, Hao J, Sun C, Li Y (2015) Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 27: 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tucker E, Hermann M, Laux T (2017) A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev Cell 40: 264–277.e264 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Li J (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48: 99–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.