Abstract
Animal genomes are folded in topologically associating domains (TADs) that have been linked to the regulation of the genes they contain by constraining regulatory interactions between cis-regulatory elements and promoters. Therefore, TADs are proposed as structural scaffolds for the establishment of regulatory landscapes (RLs). In this review, we discuss recent advances in the connection between TADs and gene regulation, their relationship with gene RLs and their dynamics during development and differentiation. Moreover, we describe how restructuring TADs may lead to pathological conditions, which explains their high evolutionary conservation, but at the same time it provides a substrate for the emergence of evolutionary innovations that lay at the origin of vertebrates and other phylogenetic clades.
Keywords: topologically associating domains, evolution, chromatin structure, genetic diseases, evolutionary novelties, regulatory landscapes
Introduction
In the last years, the development of chromosome conformation capture (3C) techniques, together with remarkable advances in live cell imaging, have expanded our knowledge about the structural organization of animal genomes (Lieberman-Aiden et al., 2009; McCord et al., 2020; Jerkovic’ and Cavalli, 2021). 3C techniques consist of restriction enzyme digestion of crosslinked chromatin, followed by proximity ligation to generate chimeric molecules that are interpreted as interactions between two genomic regions (McCord et al., 2020). HiC is the genome-wide version of 3C techniques and its increasing resolution has allowed to discern different levels of 3D folding at different scales. At the megabases scale, gene-rich transcriptionally active regions tend to interact among them, while gene-poor heterochromatic regions also interact more frequently, leading to A and B compartments, respectively. At the sub-megabase scale, chromatin domains with high interaction frequency and relatively isolated from neighbor regions form topologically associating domains (TADs). Finally, below the scale of TADs, chromatin loops are formed by strong interactions between specific genomic regions, i.e., CCCTC-binding factor (CTCF) and enhancer-promoter loops (Bonev and Cavalli, 2016; Rada-Iglesias et al., 2018; Rowley and Corces, 2018).
Topologically associating domains are believed to facilitate interactions between cis-regulatory elements (CREs) and their target promoters, which otherwise would not occur at enough frequency to ensure a robust target gene expression (Galupa and Heard, 2017; Franke and Gomez-Skarmeta, 2018; Furlong and Levine, 2018). Therefore, TADs have been proposed as structural scaffolds for regulatory landscapes (RLs; Acemel et al., 2017), which are defined as “large genomic regions containing several long-range-acting regulatory sequences that control one or several target genes in a coordinated manner” (Spitz et al., 2003; Bolt and Duboule, 2020). However, whether TADs represent a privileged functional level in the chromosome folding hierarchy has been challenged by the increasing resolution of HiC assays that have uncovered nested structures at the subTAD level with relative insulation among them (Rao et al., 2014; Zhan et al., 2017; Hsieh et al., 2020; Krietenstein et al., 2020). In this review, we discuss the connections of the 3D genome with gene expression, the relationship between TADs and RLs, and their dynamics in the context of development, focusing on disease and evolution.
Formation of TADs by Loop Extrusion
TAD boundaries are enriched for the binding of CTCF (Phillips and Corces, 2009; Dixon et al., 2012; Merkenschlager and Nora, 2016), an 11-zinc-finger DNA binding protein that was previously known by its role in transcriptional insulation (Filippova et al., 1996; Bell et al., 1999). CTCF is distributed throughout the genome not only at TAD boundaries, but also at many other sites. However, its binding at boundaries has preference for a specific orientation, with its DNA binding motifs often positioned in convergent orientation between the two boundaries that define a TAD (Rao et al., 2014). CTCF co-localizes and determines the presence at TAD boundaries of the cohesin complex, which is also involved in establishing chromosomal interactions (Parelho et al., 2008; Hadjur et al., 2009). These latter observations led to the proposal of the so called “loop extrusion” model for TAD formation involving cohesin and CTCF (de Wit et al., 2015; Sanborn et al., 2015; Fudenberg et al., 2016). According to this model, TADs would arise by chromatin extrusion mediated by cohesin and counteracted by CTCF-mediated insulation, thus explaining both the increased interaction frequency within TADs and their relative insulation from neighbor TADs. Indeed, a recent study has shown that the N-terminal domain of CTCF is essential for blocking cohesin translocation from the interior of TADs, providing a molecular basis for the requirement of a specific binding polarity of CTCF for chromosome folding (Nora et al., 2020). The predictions of this model of TAD formation have been corroborated by the acute depletion of cohesin and CTCF in mammalian cells, which shows loss of chromatin loops and TAD insulation (Nora et al., 2017; Rao et al., 2017), as well as the depletion of factors regulating cohesin loading on chromatin, which leads to differences in the length of loops formed (Busslinger et al., 2017; Haarhuis et al., 2017; Schwarzer et al., 2017; Wutz et al., 2017).
Regulatory Landscapes and Chromatin Structure
Regulatory landscapes contain CREs that control the expression of their target genes (Bolt and Duboule, 2020). Different mechanisms have been proposed to explain the transcriptional control mediated by CREs: tracking, which implies that the RNA polymerase II would bind to enhancers and track along chromatin synthesizing RNA until it reaches the promoter; linking, where transcription factors (TFs) would oligomerize from the enhancer to the promoter; or looping, in which factors bound to both sides of the loop (enhancer and promoter) would interact with each other (Furlong and Levine, 2018). The latter mechanism is widely accepted and would be favored by CTCF- and cohesin-dependent chromatin folding and promoted by mediator and TFs as Yin Yang 1, a zinc-finger DNA binding protein that form dimers similarly to CTCF and anchors enhancer-promoter interactions (Kagey et al., 2010; Dowen et al., 2014; Weintraub et al., 2017). Direct evidence of the functionality of enhancer-promoter looping comes from studies in the β-globin locus, where forcing these interactions by artificial zinc fingers leads to gene activation (Deng et al., 2014).
An interesting feature of vertebrate RLs is that enhancers are broadly distributed throughout them and not in a gene-centric manner (Symmons et al., 2014). Indeed, the RLs of developmental genes, such as those encoding for lineage-specific TFs or signaling molecules, are characterized by their large sizes and the abundance of enhancers that confer tissue-specific expression to their target genes, thus explaining their common pleiotropy (Bolt and Duboule, 2020). How do enhancers regulate only their target genes? This may be explained in part by the coincidence of RLs coordinates with those of TADs (Symmons et al., 2014). According to this observation, RLs would be confined within TADs and TAD boundaries would correspond to transitions of regulatory domains, therefore ensuring the contact of enhancers with the appropriate target genes and avoiding promiscuous interference with genes located in neighbor domains (see below). However, restriction of RLs within TADs is not enough for gene activation and other mechanisms may influence the outcome of enhancer-promoter contacts (Serebreni and Stark, 2021), including phase separation, which refers to local microenvironments resulting from weak multivalent interactions that concentrate some factors and exclude others (Banani et al., 2017), and enhancer-promoter compatibility (see below).
The RL of the Shh gene, encoding an important morphogen involved in the patterning of the developing neural tube and limb buds, is one of the best studied cases in vertebrates. This gene is located within an evolutionary conserved TAD of around 1 Mb in size that contains other non-related genes and multiple tissue-specific enhancers (Lettice et al., 2003; Sagai et al., 2005, 2009; Jeong et al., 2006). In particular, the enhancer known as ZRS [zone of polarizing activity (ZPA) regulatory sequence] is responsible for Shh expression in the limb bud and is located around 900-kb away, in the intron 5 of the Lmbr1 gene (Lettice et al., 2003). Disruption of the ZRS sequence in mice causes loss of limbs, while in humans point mutations cause preaxial polydactyly (Sagai et al., 2005). Other paradigmatic examples of vertebrate RLs are the loci containing the HoxD and HoxA gene clusters, homeobox genes that are largely responsible for the patterning of several body structures including limbs. These clusters are located between two adjacent TADs that compartmentalize long-range regulatory interactions in two blocks at the spatial and temporal levels (Lonfat and Duboule, 2015). Thus, enhancers in the 3′ TAD preferentially contact “anterior” Hox genes, while enhancers in the 5′ TAD mostly interact with “posterior” Hox genes (Lonfat et al., 2014). In addition, a switch in the interactions of central genes explains a sequential transition between two regulatory phases (Andrey et al., 2013). The regulatory activity of these enhancers is therefore combined to generate the collinearity of Hox genes. Apart from these well-known examples, other cases have been studied and recently reviewed (Bolt and Duboule, 2020).
The Link Between Chromatin Structure and Function
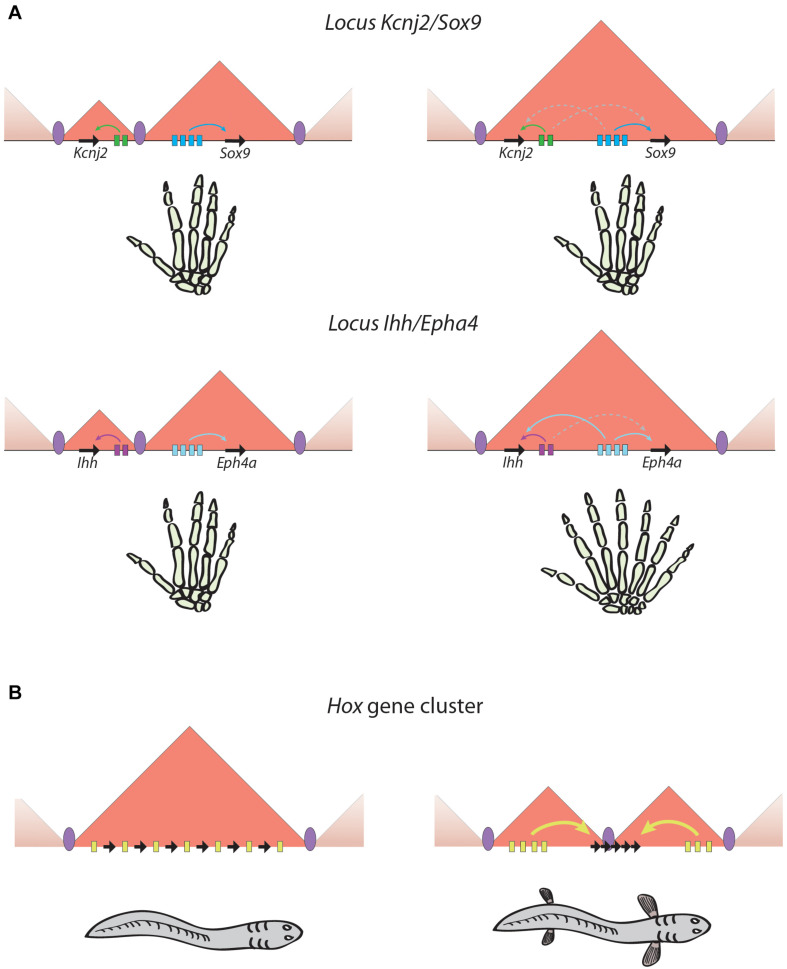
The relationship between TADs and gene expression remains an issue of open debate due to apparent discrepancies between different approaches. Studies analyzing structural variations encompassing TAD boundaries have provided strong links between the chromatin architecture of particular loci and the expression of the nearby developmental genes, which become miss-expressed causing developmental abnormalities (Spielmann et al., 2018; Ibrahim and Mundlos, 2020). Although the adoption of enhancers by genes that were not previously in contact with them explains some of these phenotypes (Lupiáñez et al., 2015; Franke et al., 2016), this is not always the case (Laugsch et al., 2019) and the effects of structural variations on gene expression are context-specific (Figure 1A), which suggests that additional mechanisms may be involved. Indeed, fusion of neighbor TADs by boundary removal at the Sox9-Kcnj2 locus does not result in major effects on gene expression (Despang et al., 2019). A study in Drosophila using highly rearranged balancer chromosomes concluded that TAD rearrangements did not result in altered expression of most genes (Ghavi-Helm et al., 2019). In agreement with this, the works describing alterations in chromatin structure and boundary deletions in the Shh locus show minor consequences on Shh expression not leading to developmental phenotypes (Paliou et al., 2019; Williamson et al., 2019).
FIGURE 1.
Phenotypical outputs of regulatory landscapes rearrangements. (A) Similar rearrangements may lead to very different situations. In the two examples illustrated in this panel, the fusion of two adjacent TADs produces no detectable phenotype in the case of the Kcnj2/Sox9 locus (Despang et al., 2019), and a striking polydactylia phenotype in the case of the Ihh/Eph4a locus (Lupiáñez et al., 2015). (B) TAD restructuring can generate evolutionary innovations by gaining new regulatory inputs or by increasing regulatory complexity. An example is the splitting of the HoxD and HoxA ancestral TAD found in non-vertebrate chordates in two TADs that are vertebrate-specific and could favor the emergence of vertebrate appendages (Acemel et al., 2016). Black arrows, genes; colored rectangles, enhancers; colored arrows, productive enhancer-promoter interactions; dashed arrows, unproductive enhancer-promoter interactions; purple ellipses, TAD boundaries.
The absence of transcriptional effects observed in some cases of TAD disruption may be explained by the lack of compatibility between enhancers and promoters. In this sense, it has been reported that transcriptional cofactors are able to activate only specific core promoters (Haberle et al., 2019), and that a particular group of highly conserved developmental enhancers, known as poised enhancers, contain CpG islands and are able to activate only developmental promoters also harboring CpG islands (Pachano et al., 2020). Moreover, it has been shown that enhancers within genomic regulatory blocks, which are regions containing a number of highly conserved non-coding elements, only activate particular target genes and not syntenic bystander genes (Akalin et al., 2009). While target genes commonly encode for developmental TFs and have promoters showing long CpG islands and multiple TF motifs, bystander genes often encode for proteins involved in unrelated functions, have different expression patterns and contain promoters with short CpG islands and few TF motifs. These observations indicate that enhancer-promoter compatibility mechanisms may determine the consequences of TAD restructuring at the transcriptional level.
On the other hand, several studies have attempted to remove architectural proteins and assess the effects of their loss genome wide. In this sense, the acute depletion of cohesin or CTCF in mammalian cells shows only a moderate effect in gene expression, affecting from some hundreds to few thousands of genes in different systems (Nora et al., 2017; Rao et al., 2017; Kubo et al., 2021). These studies have been limited in vivo due to the essential nature of cohesin and CTCF (Moore et al., 2012; Ju et al., 2013); however, this limitation was recently overcome by generating zebrafish ctcf knock-out embryos (Franke et al., 2020). In these embryos, a prolonged maternal contribution allows the survival of the mutant embryos until larval stages, when the absence of CTCF results in the miss-regulation of thousands of genes enriched in developmental functions (Franke et al., 2020). Although part of the effects seen in CTCF depletion or knock-out approaches may be indirect due to CTCF function as a TF, a subset of chromatin interactions involving lineage-specific genes change upon CTCF loss (Kubo et al., 2021). These data suggest that chromatin architecture might not be essential for the expression of most genes, but instead provide robustness to the expression of developmental genes that are frequently regulated by many CREs within complex RLs.
Chromatin Structure Dynamics During Development and Differentiation
An important question in the field of chromatin structure is how variable are TADs during dynamic processes, including embryonic development or cell differentiation. Studies of chromatin structure during animal early development have revealed a phase in which TADs become undetectable followed by a progressive reestablishment of chromatin folding (van der Weide and de Wit, 2019). However, there are some differences between species regarding when this phase happens in relation to the zygotic genome activation (ZGA). While in mammals the period without detectable chromatin structure takes place before the ZGA, with structure being progressively reestablished from there (Du et al., 2017; Gassler et al., 2017; Chen et al., 2019), similar to Drosophila (Hug et al., 2017), in zebrafish and medaka embryos it occurs during the ZGA and persist after it, being reestablished mostly during gastrulation (Kaaij et al., 2018; Nakamura et al., 2018), but there are observed discrepancies regarding the existence of chromatin structure before the ZGA in zebrafish (Kaaij et al., 2018; Wike et al., 2021). Although once formed, TADs seem to be very stable at later developmental stages, little is known about the correlation of these structures with progressive changes in gene expression.
Several studies have reported that TADs are largely stable when compared between embryonic stem cells and differentiated cells (Dixon et al., 2012, 2015; Nora et al., 2012). Consistent with this observation, a recent study has shown that chromatin structure in Drosophila embryos is preserved in different embryonic tissues despite lineage-specific gene expression (Ing-Simmons et al., 2021). However, another report showed a high variability of TAD boundaries across 37 human cell types (McArthur and Capra, 2021). It is worth noting that these apparent discrepancies might be explained either by the different biological systems used or by different resolutions in HiC experiments, as well as different computational methods to call TADs or TAD boundaries. At lower resolutions than TADs, it was shown that variations in intra-TAD contacts during mammalian differentiation corresponded with switches between active and inactive chromatin modifications and gene expression (Dixon et al., 2015). Indeed, enhancer-promoter interactions are highly dynamic and cell-type-specific during neural and erythroid differentiation, accompanying the activation of lineage-specific genes (Bonev et al., 2017; Oudelaar et al., 2020; Kubo et al., 2021). However, enhancer-promoter interactions are stably formed before gene activation in other contexts. Indeed, poised enhancers required for the activation of anterior neural genes are already engaged in contacts with their target genes in mESCs in a polycomb repressive complex 2-dependent manner (Cruz-Molina et al., 2017). However, enhancer-promoter loops in Drosophila precede target gene transcription and even TAD formation, being associated with paused RNA polymerase (Ghavi-Helm et al., 2014; Espinola et al., 2021). These observations suggest that the dynamics or stability of TADs and enhancer-promoter loops are highly context-specific, some of them being stable and others being dynamically regulated during development and differentiation.
Alterations of the 3D Genome Associated With Disease
Strong evidence for the importance of genome architecture for correct gene expression comes from studies showing that the disruption of 3D structure in particular loci, either by genomic rearrangements or alterations in TAD boundaries, lead to pathological situations, including developmental disorders or cancer (Ibrahim and Mundlos, 2020). A number of cases reported in the last years have provided a link between structural variations and disease. At the Epha4 locus, deletions, duplications and inversions disrupting TAD structure cause several limb malformations, including brachydactyly, polydactyly and F-syndrome, due to de novo enhancer-promoter interactions that lead to gene miss-expression of the surrounding genes Wnt6, Ihh and Pax3 (Figure 1A; Lupiáñez et al., 2015). At the Sox9 locus, duplications encompassing the neighbor Kcnj2 gene lead to the formation of a “neo-TAD” in which Kcnj2 is miss-regulated by interactions with new enhancers, leading to a limb malformation phenotype known as Cooks syndrome (Franke et al., 2016). A similar situation has been reported in the locus of GDPD1 gene in autosomal-dominant retinitis pigmentosa, caused by a rearrangement that place a TAD border and several retinal enhancers within this locus. This generates a “neo-TAD” and new contacts between the retinal enhancers and GDPD1 gene, which is overexpressed and likely contributes to the disease (de Bruijn et al., 2020). Finally, inversions at the TFAP2A locus in branchiooculofacial syndrome patients have been shown to disconnect this gene from its neural crest-specific enhancers, leading to its reduced expression and explaining the patient’s phenotype (Laugsch et al., 2019).
Genomic rearrangements can also lead to the fusion of genes that become overexpressed and function as oncogenes. This is the case of chromosomal translocations fusing the genes PAX3 and FOXO1 in alveolar rhabdomyosarcoma, which result in the fusion of both RLs and the activation of transcription from PAX3 promoter by enhancers from the FOXO1 RL (Vicente-Garcia et al., 2017). At larger scales, it has been found that higher order chromatin-folding structures can modulate interactions between different loci spanning several megabases in a highly aggressive type of squamous cell carcinoma (Alekseyenko et al., 2015; Rosencrance et al., 2020). The appearance of these interactions, called “megadomains,” responds to the formation of large regions of hyperacetylated chromatin due to the BRD4-NUT fusion oncoprotein. These fusions are generated by genomic translocations, usually between genes like BRD4 or BRD3 and NUT, which recruits the histone acetyltransferase p300 leading to hyperacetylated regions of up to 2 Mb. Both intra- and interdomain interactions are up-regulated, as well as the expression of the oncogenes SOX2, TP63 and MYC, which eventually contribute to tumorigenesis (Alekseyenko et al., 2015; Rosencrance et al., 2020; Eagen and French, 2021).
Apart from genomic rearrangements, alterations of TAD boundaries have also been linked to disease. The analyses of copy-number variants involving TAD boundary deletions revealed that a substantial proportion of cases could be explained by enhancer adoption (Ibn-Salem et al., 2014). In a recent work, CTCF binding sites surrounding the well-known ZRS enhancer of the Shh gene (see above) were removed by CRISPR-mediated genome editing in mice. Strikingly, the lack of these CTCF binding sites reduced the interaction between ZRS and Shh promoter, as well as the expression of the latter (Paliou et al., 2019). Nevertheless, this does not lead to a clear phenotype in these mice unless a hypomorphic allele of the ZRS is used, suggesting that chromatin structure provides robustness but does not determine enhancer-promoter communication. Indeed, a recent study from The Cancer Genome Atlas showed that only 14% of cancer-associated TAD boundary deletions resulted in significant changes in expression of the nearby genes (Akdemir et al., 2020). Moreover, mutations in CTCF binding sites have been frequently found in cancer (Katainen et al., 2015), which might lead to miss-expression of nearby genes causing tumorigenesis. Indeed, the disruption of boundaries demarcating insulated neighborhoods, which are chromatin domains smaller in size than TADs, leads to aberrant activation of proto-oncogenes, such as TAL1 and LMO2 associated with T-cell acute lymphoblastic leukemia (Hnisz et al., 2016). In the opposite situation, the overexpression of the Neurotensin gene NTS, a central nervous system neurotransmitter, has been recently related with melanomas due to a gained CTCF-mediated chromatin loop that establish contacts between the NTS promoter and a CRE in the intron of the LRRIQ1 gene located 800 Kb away (Chai et al., 2021).
It is worth to remark that mutations in the coding sequence or miss-expression of architectural proteins, i.e., cohesin and CTCF, can lead to a wide set of human pathological phenotypes (Singh and Gerton, 2015). For example, somatic mutations in cohesin have been associated with different forms of cancer (Mullenders et al., 2015; Fisher et al., 2017; Carico et al., 2021), which is also related to the important role of cohesin in the separation of sister chromatids during cell division (Nasmyth and Haering, 2009). In addition, CTCF has been proposed as a tumor suppressor gene, since mutations in its coding sequence have also been detected in different types of cancer (Filippova et al., 2002). Interestingly, different missense mutations affected specific zinc-finger domains, leading to defects in the binding to the promoters of a subset of genes involved in the regulation of proliferation, but not to others. Therefore, both cohesin and CTCF play an essential role in gene regulation that prevents pathological situations.
Conservation of TADs Across Genome Evolution
Several studies comparing chromatin structure in diverse species have reached the conclusion that TADs and their boundaries are largely conserved in animal genomes. The first HiC data comparing TADs in human and mouse cells found a general conservation of their boundaries (Dixon et al., 2012) and an in-depth analysis of conservation in mammals revealed that conserved TAD boundaries were associated with conserved CTCF sites, while divergent CTCF sites correlated with divergence of chromatin structure (Vietri Rudan et al., 2015). Indeed, CTCF binding at TAD boundaries is highly clustered and these sites are subjected to stronger selective constrains than other CTCF sites among closely related species (Kentepozidou et al., 2020; McArthur and Capra, 2021). Therefore, the strong selection against disruption of TAD boundaries in evolution is likely responsible for their enrichment in rearrangement breakpoints in vertebrates, being reshuffled as whole blocks during evolution (Krefting et al., 2018). A clear case of this conservation is the boundary splitting the RLs of the Six genes, which is conserved not only in vertebrates but also in echinoderms, illustrating the deep conservation of TADs involving important developmental genes (Gomez-Marin et al., 2015). Finally, studies analyzing highly conserved gene regulatory blocks, which are clusters of conserved non-coding elements around important developmental regulators, have revealed that a subset of TADs exhibit extreme non-coding conservation across metazoans (Harmston et al., 2017). Therefore, TADs involving developmental genes represent evolutionary conserved chromatin domains likely because they provide a scaffold for developmental RLs.
TAD Reshuffling Underlies the Emergence of Evolutionary Novelties
As commented above, genomic rearrangements involving TAD restructuring and the associated alterations in gene expression usually entail deleterious effects. Nevertheless, they also provide a substrate for evolution and changes in genome structure may result in the gain of new functions that underlie the appearance of evolutionary novelties during species evolution (Maeso et al., 2017; Figure 1B). This has probably been the case with the emergence of limbs in vertebrates, for which the restructuring of TADs at the HoxA and HoxD loci has been essential. As can be inferred from slow evolving, non-vertebrate animals like amphioxus, the ancestral Hox locus was organized in a single TAD encompassing all Hox genes and the enhancers regulating their expression. However, at the origin of vertebrates this TAD was split in two located at either sides of the cluster and separating some genes from the rest, while leaving some others in the hinge between both TADs (Figure 1B; Acemel et al., 2016). This organization allowed the spatial and temporal segregation of regulatory inputs that explains the collinearity of the HOX genes (see above; Andrey et al., 2013; Lonfat et al., 2014) and likely enabled the plasticity in the usage of the Hox patterning system that was essential for vertebrate evolutionary novelties such as the development of paired appendages. On the other hand, the gnathostome-specific expression of Shh in the limbs was shown to be originated between the two whole-genome duplications by a translocation that linked the RL of that gene with Lmbr1 (Irimia et al., 2012). Then, the ZRS enhancer could emerge in the intron 5 of Lmbr1, being critical for the emergence of paired and unpaired appendages (Letelier et al., 2018a).
Apart from TAD restructuring early in the evolution of vertebrates, other cases have been documented when comparing relatively close species. Thus, the regulatory cluster formed by rac3b, rfng and sgca genes emerged at the origin of the Ostariophysi fish superorder by a genomic rearrangement that brought in contact the RLs of rac3b/rfng and sgca, which are separated in Actinopterygii and tetrapods (Letelier et al., 2018b). Such rearrangement led to the formation of new regulatory contacts between rac3b and rfng promoters and the ancestral sgca RL, which was responsible to direct its expression to the hindbrain boundaries. These new regulatory interactions resulted in the co-option (which refers to the redeployment of pre-existing genetic or regulatory mechanisms for the acquisition of new functions or expression domains) of rac3b and rfng in the rhombomeres margins, thus promoting the formation of actomyosin cables characteristic of these structures (Letelier et al., 2018b). Moreover, it has been found that human brain tissue shows a subset of species-specific TADs compared with macaques that are associated with human-specific expression changes that are likely responsible of the higher complexity of the human brain (Luo et al., 2021). Similarly, interspecies differences in chromatin structure between human and chimpanzees are commonly associated with differences in gene expression (Eres et al., 2019). These differences between phylogenetically related species have also been discovered in two different species of Drosophila genus, D. melanogaster and D. triauraria. Only 25% of the TADs are orthologous between both species, and importantly these genomic rearrangements could be related with changes in gene expression (Torosin et al., 2020). These findings support the hypothesis that reorganization of genomic 3D structure may act as an important force in the rise of evolutionary novelties.
Concluding Remarks
Although the recent technological advances have allowed an increasingly detailed understanding of the 3D organization of the genome, many questions remain unanswered. How CREs operate within the context of TADs over their target genes in a mechanistic level remains incompletely understood, and to what extent the alterations of TADs lead to gene miss-expression is still an open debate. Moreover, our knowledge of how the restructuring of TADs leads to evolutionary innovations is limited to a few reported examples and the scarce availability of genome-wide chromosome conformation experiments in different species limits the comparative analyses of the 3D genome from an evolutionary perspective. Finally, the increasing applicability of the single cell technologies to chromosome conformation experiments will hopefully make possible to discern between chromatin structure in particular cell types and its association with cell type-specific gene expression during development and differentiation.
Author Contributions
JS-P and JT reviewed the articles and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize the authors whose work could not be discussed due to space limitations.
Footnotes
Funding. This research was funded by the European Research Council (ERC) under the European Union’s Horizon 2020 and Seventh Framework Program FP7 Research and Innovation Programs (ERC-AdG-LS8-740041). JT was funded by a 2019 Leonardo Grant for Researchers and Cultural Creators, BBVA Foundation (The BBVA Foundation accepts no responsibility for the opinions, statements and contents included in the project and/or the results thereof, which are entirely the responsibility of the authors). JS-P was funded by a postdoctoral fellowship from Junta de Andalucía (DOC_00512).
References
- Acemel R. D., Maeso I., Gómez-Skarmeta J. L. (2017). Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals. Wiley Interdiscip. Rev. Dev. Biol 6:e265. 10.1002/wdev.265 [DOI] [PubMed] [Google Scholar]
- Acemel R. D., Tena J. J., Irastorza-Azcarate I., Marlétaz F., Gómez-Marín C., De La Calle-Mustienes E., et al. (2016). A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 48 336–341. 10.1038/ng.3497 [DOI] [PubMed] [Google Scholar]
- Akalin A., Fredman D., Arner E., Dong X., Bryne J. C., Suzuki H., et al. (2009). Transcriptional features of genomic regulatory blocks. Genome Biol. 10:R38. 10.1186/gb-2009-10-4-r38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir K. C., Le V. T., Chandran S., Li Y., Verhaak R. G., Beroukhim R., et al. (2020). Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet. 52 294–305. 10.1038/s41588-019-0564-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko A. A., Walsh E. M., Wang X., Grayson A. R., Hsi P. T., Kharchenko P. V., et al. (2015). The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 29 1507–1523. 10.1101/gad.267583.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey G., Montavon T., Mascrez B., Gonzalez F., Noordermeer D., Leleu M., et al. (2013). A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340:1234167. 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]
- Banani S. F., Lee H. O., Hyman A. A., Rosen M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. C., West A. G., Felsenfeld G. (1999). The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98 387–396. 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- Bolt C. C., Duboule D. (2020). The regulatory landscapes of developmental genes. Development 147:dev171736. 10.1242/dev.171736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Cavalli G. (2016). Organization and function of the 3D genome. Nat. Rev. Genet. 17 661–678. 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G. L., Lubling Y., et al. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger G. A., Stocsits R. R., Van Der Lelij P., Axelsson E., Tedeschi A., Galjart N., et al. (2017). Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544 503–507. 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carico Z. M., Stefan H. C., Justice M., Yimit A., Dowen J. M. (2021). A cohesin cancer mutation reveals a role for the hinge domain in genome organization and gene expression. PLoS Genet. 17:e1009435. 10.1371/journal.pgen.1009435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai P., Yu J., Jia R., Wen X., Ding T., Zhang X., et al. (2021). Generation of onco-enhancer enhances chromosomal remodeling and accelerates tumorigenesis. Nucleic Acids Res. 48 12135–12150. 10.1093/nar/gkaa1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ke Y., Wu K., Zhao H., Sun Y., Gao L., et al. (2019). Key role for CTCF in establishing chromatin structure in human embryos. Nature 576 306–310. 10.1038/s41586-019-1812-0 [DOI] [PubMed] [Google Scholar]
- Cruz-Molina S., Respuela P., Tebartz C., Kolovos P., Nikolic M., Fueyo R., et al. (2017). PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell 20 689–705.e9. 10.1016/j.stem.2017.02.004 [DOI] [PubMed] [Google Scholar]
- de Bruijn S. E., Fiorentino A., Ottaviani D., Fanucchi S., Melo U. S., Corral-Serrano J. C., et al. (2020). Structural Variants Create New Topological-Associated Domains and Ectopic Retinal Enhancer-Gene Contact in Dominant Retinitis Pigmentosa. Am. J. Hum. Genet. 107 802–814. 10.1016/j.ajhg.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Vos E. S. M., Holwerda S. J. B., Valdes-Quezada C., Verstegen M. J. A. M., Teunissen H., et al. (2015). CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 60 676–684. 10.1016/j.molcel.2015.09.023 [DOI] [PubMed] [Google Scholar]
- Deng W., Rupon J. W., Krivega I., Breda L., Motta I., Jahn K. S., et al. (2014). Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158 849–860. 10.1016/j.cell.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despang A., Schöpflin R., Franke M., Ali S., Jerković I., Paliou C., et al. (2019). Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51 1263–1271. 10.1038/s41588-019-0466-z [DOI] [PubMed] [Google Scholar]
- Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E., Lee A. Y., et al. (2015). Chromatin architecture reorganization during stem cell differentiation. Nature 518 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen J. M., Fan Z. P., Hnisz D., Ren G., Abraham B. J., Zhang L. N., et al. (2014). Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159 374–387. 10.1016/j.cell.2014.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zheng H., Huang B., Ma R., Wu J., Zhang X., et al. (2017). Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Eagen K. P., French C. A. (2021). Supercharging BRD4 with NUT in carcinoma. Oncogene 40 1396–1408. 10.1038/s41388-020-01625-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eres I. E., Luo K., Hsiao C. J., Blake L. E., Gilad Y. (2019). Reorganization of 3D genome structure may contribute to gene regulatory evolution in primates. PLoS Genet. 15:e1008278. 10.1371/journal.pgen.1008278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinola S. M., Götz M., Bellec M., Messina O., Fiche J. B., Houbron C., et al. (2021). Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 53 477–486. 10.1038/s41588-021-00816-z [DOI] [PubMed] [Google Scholar]
- Filippova G. N., Fagerlie S., Klenova E. M., Myers C., Dehner Y., Goodwin G., et al. (1996). An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16 2802–2813. 10.1128/mcb.16.6.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G. N., Ulmer J. E., Moore J. M., Ward M. D., Hu Y. J., Neiman P. E., et al. (2002). Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 62 48–52. [PubMed] [Google Scholar]
- Fisher J. B., McNulty M., Burke M. J., Crispino J. D., Rao S. (2017). Cohesin Mutations in Myeloid Malignancies. Trends Cancer 3 282–293. 10.1016/j.trecan.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M., De la Calle-Mustienes E., Neto A., Acemel R. D., Santos-Pereira J. M., Gómez-Skarmeta J. L. (2020). CTCF knockout in zebrafish induces alterations in 1 regulatory landscapes and developmental gene expression 2 3. bioRxiv [Preprint]. 10.1101/2020.09.08.282707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M., Gomez-Skarmeta J. L. (2018). An evolutionary perspective of regulatory landscape dynamics in development and disease. Curr. Opin. Cell Biol. 55 24–29. 10.1016/j.ceb.2018.06.009 [DOI] [PubMed] [Google Scholar]
- Franke M., Ibrahim D. M., Andrey G., Schwarzer W., Heinrich V., Schopflin R., et al. (2016). Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538 265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L. A. (2016). Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 15 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong E. E. M., Levine M. (2018). Developmental enhancers and chromosome topology. Science 361 1341–1345. 10.1126/science.aau0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R., Heard E. (2017). Topologically Associating Domains in Chromosome Architecture and Gene Regulatory Landscapes during Development, Disease, and Evolution. Cold Spring Harb. Symp. Quant. Biol. 82 267–278. 10.1101/sqb.2017.82.035030 [DOI] [PubMed] [Google Scholar]
- Gassler J., Brandão H. B., Imakaev M., Flyamer I. M., Ladstätter S., Bickmore W. A., et al. (2017). A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36 3600–3618. 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Jankowski A., Meiers S., Viales R. R., Korbel J. O., Furlong E. E. M. (2019). Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51 1272–1282. 10.1038/s41588-019-0462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Klein F. A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., et al. (2014). Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512 96–100. 10.1038/nature13417 [DOI] [PubMed] [Google Scholar]
- Gomez-Marin C., Tena J. J., Acemel R. D., Lopez-Mayorga M., Naranjo S., de la Calle-Mustienes E., et al. (2015). Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. U. S. A. 112 7542–7547. 10.1073/pnas.1505463112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis J. H. I., van der Weide R. H., Blomen V. A., Yáñez-Cuna J. O., Amendola M., van Ruiten M. S., et al. (2017). The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169 693–707.e14. 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Arnold C. D., Pagani M., Rath M., Schernhuber K., Stark A. (2019). Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570 122–126. 10.1038/s41586-019-1210-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S., Williams L. M., Ryan N. K., Cobb B. S., Sexton T., Fraser P., et al. (2009). Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460 410–413. 10.1038/nature08079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmston N., Ing-Simmons E., Tan G., Perry M., Merkenschlager M., Lenhard B. (2017). Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 8:441. 10.1038/s41467-017-00524-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Weintraub A. S., Day D. S., Valton A. L., Bak R. O., Li C. H., et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351 1454–1458. 10.1126/science.aad9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. H. S., Cattoglio C., Slobodyanyuk E., Hansen A. S., Rando O. J., Tjian R., et al. (2020). Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 78 539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C. B., Grimaldi A. G., Kruse K., Vaquerizas J. M. (2017). Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 169 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Ibn-Salem J., Köhler S., Love M. I., Chung H. R., Huang N., Hurles M. E., et al. (2014). Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol. 15:423. 10.1186/s13059-014-0423-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim D. M., Mundlos S. (2020). Three-dimensional chromatin in disease: what holds us together and what drives us apart? Curr. Opin. Cell Biol. 64 1–9. 10.1016/j.ceb.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Ing-Simmons E., Vaid R., Bing X. Y., Levine M., Mannervik M., Vaquerizas J. M. (2021). Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat. Genet. 53 487–499. 10.1038/s41588-021-00799-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M., Royo J. L., Burguera D., Maeso I., Gomez-Skarmeta J. L., Garcia-Fernandez J. (2012). Comparative genomics of the Hedgehog loci in chordates and the origins of Shh regulatory novelties. Sci. Rep. 2:433. 10.1038/srep00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y., El-Jaick K., Roessler E., Muenke M., Epstein D. J. (2006). A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133 761–772. 10.1242/dev.02239 [DOI] [PubMed] [Google Scholar]
- Jerkovic’ I., Cavalli G. (2021). Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 10.1038/s41580-021-00362-w [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Ju L., Wing J., Taylor E., Brandt R., Slijepcevic P., Horsch M., et al. (2013). SMC6 is an essential gene in mice, but a hypomorphic mutant in the ATPase domain has a mild phenotype with a range of subtle abnormalities. DNA Repair 12 356–366. 10.1016/j.dnarep.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Kaaij L. J. T., van der Weide R. H., Ketting R. F., de Wit E. (2018). Systemic Loss and Gain of Chromatin Architecture throughout Zebrafish Development. Cell Rep. 24 1–10.e4. 10.1016/j.celrep.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467 430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R., Dave K., Pitkänen E., Palin K., Kivioja T., Välimäki N., et al. (2015). CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 47 818–821. 10.1038/ng.3335 [DOI] [PubMed] [Google Scholar]
- Kentepozidou E., Aitken S. J., Feig C., Stefflova K., Ibarra-Soria X., Odom D. T., et al. (2020). Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol. 21:5. 10.1186/s13059-019-1894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krefting J., Andrade-Navarro M. A., Ibn-Salem J. (2018). Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 16:87. 10.1186/s12915-018-0556-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N., Abraham S., Venev S. V., Abdennur N., Gibcus J., Hsieh T. H. S., et al. (2020). Ultrastructural Details of Mammalian Chromosome Architecture. Mol. Cell 78 554–565.e7. 10.1016/j.molcel.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N., Ishii H., Xiong X., Bianco S., Meitinger F., Hu R., et al. (2021). Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 28 152–161. 10.1038/s41594-020-00539-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugsch M., Bartusel M., Rehimi R., Alirzayeva H., Karaolidou A., Crispatzu G., et al. (2019). Modeling the Pathological Long-Range Regulatory Effects of Human Structural Variation with Patient-Specific hiPSCs. Cell Stem Cell 24 736–752.e12. 10.1016/j.stem.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Letelier J., de la Calle-Mustienes E., Pieretti J., Naranjo S., Maeso I., Nakamura T., et al. (2018a). A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 50 504–509. 10.1038/s41588-018-0080-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letelier J., Terriente J., Belzunce I., Voltes A., Undurraga C. A., Polvillo R., et al. (2018b). Evolutionary emergence of the rac3b/rfng/sgca regulatory cluster refined mechanisms for hindbrain boundaries formation. Proc. Natl. Acad. Sci. U. S. A. 115 E3731–E3740. 10.1073/pnas.1719885115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L. A., Heaney S. J., Purdie L. A., Li L., de Beer P., Oostra B. A., et al. (2003). A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12 1725–1735. 10.1093/hmg/ddg180 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E., Van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonfat N., Duboule D. (2015). Structure, function and evolution of topologically associating domains (TADs) at HOX loci. FEBS Lett. 589 2869–2876. 10.1016/j.febslet.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Lonfat N., Montavon T., Darbellay F., Gitto S., Duboule D. (2014). Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 346 1004–1006. 10.1126/science.1257493 [DOI] [PubMed] [Google Scholar]
- Luo X., Liu Y., Dang D., Hu T., Hou Y., Meng X., et al. (2021). 3D Genome of macaque fetal brain reveals evolutionary innovations during primate corticogenesis. Cell 184 723–740.e21. 10.1016/j.cell.2021.01.001 [DOI] [PubMed] [Google Scholar]
- Lupiáñez D. G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeso I., Acemel R. D., Gomez-Skarmeta J. L. (2017). Cis-regulatory landscapes in development and evolution. Curr. Opin. Genet. Dev. 43 17–22. 10.1016/j.gde.2016.10.004 [DOI] [PubMed] [Google Scholar]
- McArthur E., Capra J. A. (2021). Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am. J. Hum. Genet. 108 269–283. 10.1016/j.ajhg.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord R. P., Kaplan N., Giorgetti L. (2020). Chromosome Conformation Capture and Beyond: toward an Integrative View of Chromosome Structure and Function. Mol. Cell 77 688–708. 10.1016/j.molcel.2019.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., Nora E. P. (2016). CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genomics Hum. Genet. 17 17–43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- Moore J. M., Rabaia N. A., Smith L. E., Fagerlie S., Gurley K., Loukinov D., et al. (2012). Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS One 7:e34915. 10.1371/journal.pone.0034915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J., Aranda-Orgilles B., Lhoumaud P., Keller M., Pae J., Wang K., et al. (2015). Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 212 1833–1850. 10.1084/jem.20151323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura R., Motai Y., Kumagai M., Nishiyama H., Durand N. C., Kondo K., et al. (2018). CTCF looping is established during gastrulation in medaka embryos. bioRxiv [preprint]. 10.1101/454082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H. (2009). Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43 525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Nora E. P., Caccianini L., Fudenberg G., So K., Kameswaran V., Nagle A., et al. (2020). Molecular basis of CTCF binding polarity in genome folding. Nat. Commun. 11:5612. 10.1038/s41467-020-19283-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E. P., Goloborodko A., Valton A. L., Gibcus J. H., Uebersohn A., Abdennur N., et al. (2017). Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., Servant N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudelaar A. M., Beagrie R. A., Gosden M., de Ornellas S., Georgiades E., Kerry J., et al. (2020). Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat. Commun. 11:2722. 10.1038/s41467-020-16598-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachano T., Sánchez-Gaya V., Mariner-Faulí M., Ealo T., Asenjo H. G., Respuela P., et al. (2020). Orphan CpG islands boost the regulatory activity of poised enhancers and dictate the responsiveness of their target genes. bioRxiv [preprint]. 10.1101/2020.08.05.237768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliou C., Guckelberger P., Schöpflin R., Heinrich V., Esposito A., Chiariello A. M., et al. (2019). Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl. Acad. Sci. U. S. A. 116 12390–12399. 10.1073/pnas.1900672116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132 422–433. 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Phillips J. E., Corces V. G. (2009). CTCF: master weaver of the genome. Cell 137 1194–1211. 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Grosveld F. G., Papantonis A. (2018). Forces driving the three-dimensional folding of eukaryotic genomes. Mol. Syst. Biol. 14:e8214. 10.15252/msb.20188214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., et al. (2014). A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. P., Huang S. C., Glenn St Hilaire B., Engreitz J. M., Perez E. M., Kieffer-Kwon K. R., et al. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosencrance C. D., Ammouri H. N., Yu Q., Ge T., Rendleman E. J., Marshall S. A., et al. (2020). Chromatin Hyperacetylation Impacts Chromosome Folding by Forming a Nuclear Subcompartment. Mol. Cell 78 112–126.e12. 10.1016/j.molcel.2020.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M. J., Corces V. G. (2018). Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T., Amano T., Tamura M., Mizushina Y., Sumiyama K., Shiroishi T. (2009). A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development 136 1665–1674. 10.1242/dev.032714 [DOI] [PubMed] [Google Scholar]
- Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T. (2005). Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132 797–803. 10.1242/dev.01613 [DOI] [PubMed] [Google Scholar]
- Sanborn A. L., Rao S. S. P., Huang S. C., Durand N. C., Huntley M. H., Jewett A. I., et al. (2015). Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U. S. A. 112 E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., et al. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebreni L., Stark A. (2021). Insights into gene regulation: from regulatory genomic elements to DNA-protein and protein-protein interactions. Curr. Opin. Cell Biol. 70 58–66. 10.1016/j.ceb.2020.11.009 [DOI] [PubMed] [Google Scholar]
- Singh V. P., Gerton J. L. (2015). Cohesin and human disease: lessons from mouse models. Curr. Opin. Cell Biol. 37 9–17. 10.1016/j.ceb.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Spielmann M., Lupiáñez D. G., Mundlos S. (2018). Structural variation in the 3D genome. Nat. Rev. Genet. 19 453–467. 10.1038/s41576-018-0007-0 [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Duboule D. (2003). A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113 405–417. 10.1016/s0092-8674(03)00310-6 [DOI] [PubMed] [Google Scholar]
- Symmons O., Uslu V. V., Tsujimura T., Ruf S., Nassari S., Schwarzer W., et al. (2014). Functional and topological characteristics of mammalian regulatory domains. Genome Res. 24 390–400. 10.1101/gr.163519.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torosin N. S., Anand A., Golla T. R., Cao W., Ellison C. E. (2020). 3D genome evolution and reorganization in the Drosophila melanogaster species group. PLoS Genet. 16:e1009229. 10.1371/journal.pgen.1009229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weide R. H., de Wit E. (2019). Developing landscapes: genome architecture during early embryogenesis. Curr. Opin. Genet. Dev. 55 39–45. 10.1016/j.gde.2019.04.009 [DOI] [PubMed] [Google Scholar]
- Vicente-Garcia C., Villarejo-Balcells B., Irastorza-Azcarate I., Naranjo S., Acemel R. D., Tena J. J., et al. (2017). Regulatory landscape fusion in rhabdomyosarcoma through interactions between the PAX3 promoter and FOXO1 regulatory elements. Genome Biol. 18:106. 10.1186/s13059-017-1225-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D. T., Tanay A., et al. (2015). Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10 1297–1309. 10.1016/j.celrep.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A. S., Li C. H., Zamudio A. V., Sigova A. A., Hannett N. M., Day D. S., et al. (2017). YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171 1573–1588.e28. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wike C. L., Guo Y., Tan M., Nakamura R., Shaw D. K., Díaz N., et al. (2021). Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res. 31 981–994. 10.1101/gr.269860.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I., Kane L., Devenney P. S., Flyamer I. M., Anderson E., Kilanowski F., et al. (2019). Developmentally regulated Shh expression is robust to TAD perturbations. Development 146:dev179523. 10.1242/dev.179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G., Várnai C., Nagasaka K., Cisneros D. A., Stocsits R. R., Tang W., et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36 3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Mariani L., Barozzi I., Schulz E. G., Blüthgen N., Stadler M., et al. (2017). Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 27 479–490. 10.1101/gr.212803.116 [DOI] [PMC free article] [PubMed] [Google Scholar]