Abstract
Diagnostics to accurately detect disease and monitor therapeutic response are essential for effective clinical management. Bioengineering, chemical biology, molecular biology, and computer science tools are converging to guide the design of diagnostics that leverage enzymatic activity to measure or produce biomarkers of disease. Here, we review recent advances in the development of these “activity-based diagnostics” and their application in infectious and noncommunicable diseases. We highlight efforts towards both molecular probes that respond to disease-specific catalytic activity to produce a diagnostic readout, as well as diagnostics that use enzymes as an engineered component of their sense-and-respond cascade. These technologies exemplify how integrating techniques from multiple disciplines with pre-clinical validation has enabled activity-based diagnostics that may realize the goals of precision medicine.
Keywords: diagnostics, enzymes, synthetic biomarkers, activity-based probes, protease activity, synthetic biology, CRISPR-Cas
Non-invasive diagnostics: methods, challenges, and possibilities
Accurate detection and diagnosis of disease are essential for effective clinical management and treatment. Rapid diagnostics for highly infectious diseases such as Ebola can enable early case detection and intervention, thereby informing impactful public health interventions [1]. Similarly, sensitive and specific diagnostics for noncommunicable diseases such as cancer offer the promise of improved therapeutic outcomes through early diagnosis of localized disease and accurate classification of high-risk patients [2–4]. Parallelizing therapeutic development with diagnostics that can measure treatment response could facilitate personalized treatment regimens tailored specifically to a patient’s disease state. Effective diagnostics improve patient outcomes by providing actionable information on the presence, prognosis, or progress of disease.
Traditional non-invasive diagnostic strategies rely on a combination of imaging tests and assays for endogenous biomarkers. Imaging tests, such as computed tomography for lung cancer and magnetic resonance imaging for brain scans, remain the clinical standard for non-invasive diagnostics and enable detection and localization of disease. However, they often suffer from poor specificity [5,6] and require investment in costly infrastructure and specialized personnel to interpret the findings. Recent advances in molecular diagnostics have yielded promising assays for endogenous disease biomarkers, such as nucleic acids for viral infections [7], stool-based tests for colon cancer screening [8], and circulating tumor DNA (ctDNA, see Glossary) [9–12] for cancer, that can be used in conjunction with or as an alternative to imaging.
An ideal molecular test for infectious diseases would be simple, rapid, inexpensive, and accurate; such a test would enable specific disease detection and isolation directly at the point-of-care and have tremendous implications for global health [13]. In oncology, ctDNA has emerged as a promising tool for noninvasive disease detection and evaluation of treatment response in patients with advanced cancers whose tumors shed ample cell-free DNA (cfDNA) into the bloodstream [12]. In this context, ctDNA has enabled comprehensive reconstruction of patient-specific mutation profiles via whole-exome sequencing [14] and can be used for longitudinal monitoring of treatment efficacy or disease relapse, as ctDNA levels are thought to scale with tumor burden [12]. However, this correlation with tumor burden presents fundamental sensitivity limits for early-stage, localized disease [12,15]. Multi-analyte blood tests that measure both ctDNA and protein biomarkers, such as CancerSEEK [10], have recently been developed as a means to improve detection rates. Though the median sensitivity of CancerSEEK was 70% for the eight common cancers tested, the median sensitivity for stage I cancers was only 43%. While multi-analyte tests can improve specificity and resolution, strategies that detect endogenous biomarkers in circulation face intrinsic sensitivity limitations for minimal residual or early-stage disease, due to low analyte concentration, high background signal, and biomarker clearance [12]. As a complementary strategy, engineered diagnostics that are selectively activated in disease states to generate amplified readouts may complement existing tests to help address these challenges and realize more accurate and accessible diagnostics.
Convergent efforts from bioengineering, chemical biology, and molecular biology have inspired a new class of smart diagnostics that leverage enzymatic activity to measure or produce biomarkers of disease. These engineered “activity-based diagnostics” (ABDx) offer the potential to overcome the limitations faced by current standard tests, as they harness the specialized substrate recognition and signal amplification properties of enzymes to achieve specific and sensitive disease detection (Figure 1). This Review broadly organizes ABDx into two classes, and focuses on recent efforts towards their pre-clinical development. First, we discuss molecular and chemical probes that monitor dysregulated enzyme activity as a functional biomarker of disease. Second, we highlight molecular and biological tools that use enzymatic activity as a means of sensing, measuring, or reporting on disease state. Throughout, we highlight the enabling technologies that have catalyzed the emergence of these diagnostics, the performance advantages afforded by ABDx, as well as strategies for enhancing ABDx specificity (Box 1) and supporting clinical translation through dialogue with regulatory agencies. Continued efforts to engineer and validate activity-based diagnostics will encourage their use as next-generation tests for precision medicine (Clinician’s Corner).
Figure 1. Principles of activity-based diagnostics (ABDx).
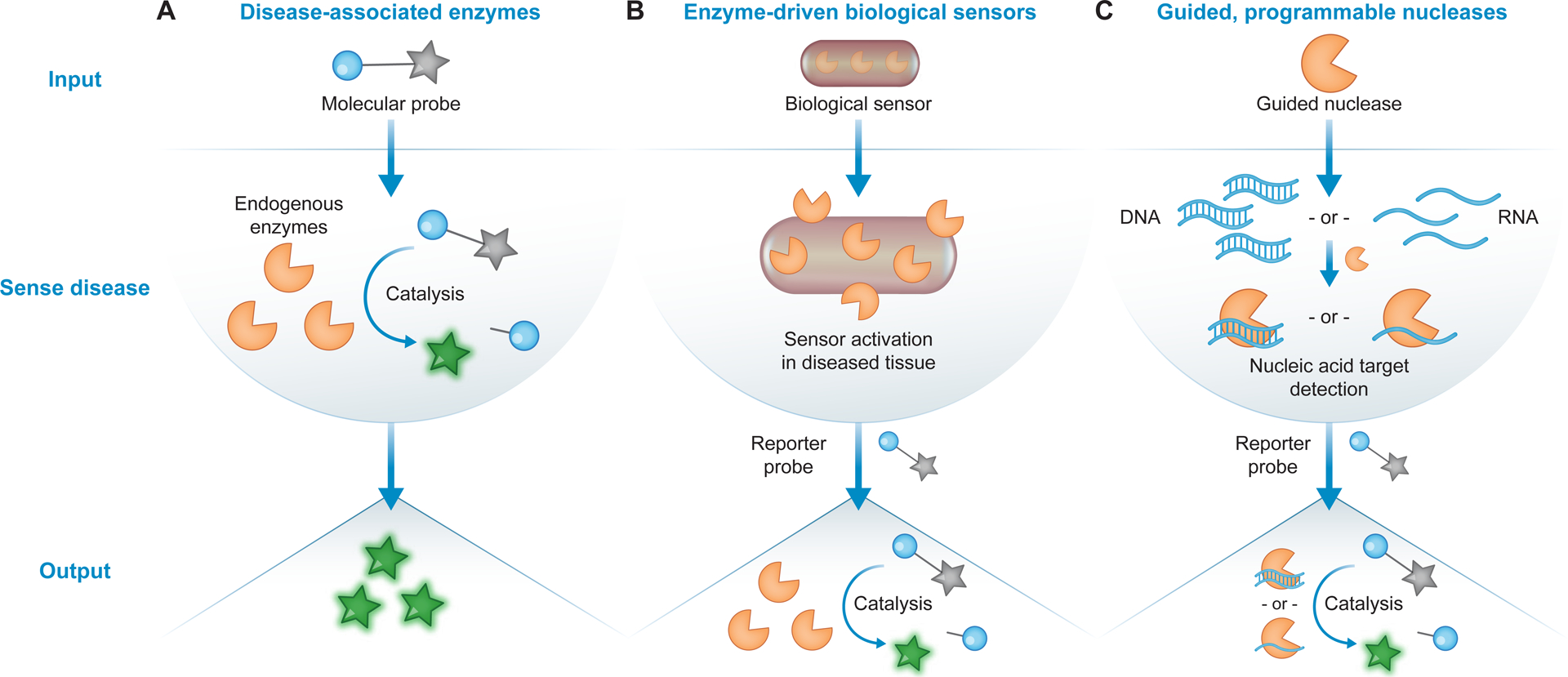
ABDx leverage enzymatic activity to measure or produce biomarkers of disease. (A) Molecular and chemical probes can be used to measure dysregulated enzyme (orange pacman) activity, either in vivo or ex vivo, as a functional biomarker of disease. Prior to catalysis, probes remain off (grey star). Enzyme-specific probe activation generates a measurable output as a diagnostic readout (green star). (B) Biological sensors, such as engineered bacteria or mammalian cells, carry enzyme-driven genetic circuits that enable them to sense and report on disease state. Synthetic reporter enzymes produced following sensor activation can generate amplified diagnostic readouts as output. (C) Guided, programmable nucleases, such as CRISPR-associated (Cas) effector enzymes, can be exploited for sequence-specific nucleic acid detection. Select nucleases can cleave synthetic reporter probes upon target binding to produce a signal-amplified readout of specific nucleic acid detection.
Box 1 |. Strategies for achieving ABDx specificity.
Enzyme target selection
Many ABDx measure enzyme activity as a functional biomarker of disease. However, enzyme regulation is complex, and putative targets may be highly expressed in other tissues besides the disease microenvironment, notably in benign comorbidities or healthy tissues.
‘Omics approaches and publicly available datasets can be used to nominate candidate enzymes dysregulated in a malignancy or disease of interest. Candidates that are promiscuously expressed or implicated in benign comorbidities can then be downselected.
Transcriptomic analysis of RNA-Seq and microarray data as well as proteomics approaches can be utilized to this end.
Probe and substrate engineering
ABDx probes can be engineered to be as specific as possible for a well-validated enzyme target of interest, reducing promiscuous activation.
High-throughput combinatorial screens incorporating unnatural amino acids [120] can inform the design of peptide-based probes.
Dual-targeting [76] and multivariate probes [46] that require processing by multiple enzymes can also improve selectivity.
ABP specificity can be enhanced by using specific enzyme inhibitors or substrates as the reactive warhead [38].
Targeting and localization of in vivo probes
Functionalizing in vivo probes with disease-targeting ligands [43] that engage active trafficking pathways may increase probe activation specifically in the disease microenvironment.
Cellular sensors that preferentially home to and expand in the site of disease, such as engineered probiotics [99] or macrophages [89], present an alternative strategy.
Signal multiplexing
Integrating multiple orthogonal signals can help drive diagnostic specificity.
Activity-based nanosensors (ABNs) detect and amplify protease activity in vivo to generate urinary reporters of disease [47]. ABNs can be multiplexed by using orthogonal mass-encoded reporters.
Large, combinatorial peptide substrate libraries [71,72] can be used for multiplexed protease activity profiling in patient-derived biospecimens.
Synthetic biology approaches integrating Boolean logic operations, such as DNA recombinase-based logic gates [101–103] and state machines [104] or protease-based circuits [106], can sense and respond to multiple environmental signals.
Nucleic acid detection with guided, programmable nucleases
Due to nucleotide base pairing dictated by an RNA guide, Cas effector enzymes can achieve highly specific recognition of nucleic acid targets. Following target detection, several CRISPR diagnostics leverage the collateral activity of select Cas enzymes to amplify signal for boosted sensitivity.
Introduction of synthetic mismatches in the guide RNA/target duplex has been shown to increase specificity [114].
For example, the SHERLOCK approach [114] can achieve specificity down to single-base mismatches, enabling pathogen identification, strain identification, and mutation identification in cfDNA.
Clinician’s Corner.
Traditional non-invasive diagnostic strategies, namely imaging (e.g., low-dose CT) and biomarker-based (e.g., PSA) tests, rely on peripherally detectable measures and can thus lack sensitivity and specificity. However, these limitations can be overcome with engineering, either by delivering probes that directly measure dysregulated biology at the site of disease, or by developing more sensitive, specific, and accessible in vitro tests. An emerging class of these engineered diagnostics, termed activity-based diagnostics (ABDx), includes probes that monitor dysregulated enzyme activity as a functional biomarker of disease as well as diagnostics that utilize enzymatic activity as an engineered component of their overall activation cascade (Figure 1).
Enzymes are dysregulated in many diseases and have several properties that motivate their use as biomarkers. In particular, dysregulated enzymes can play causal roles in disease progression, and a single enzyme molecule can catalyze multiple biochemical reactions, providing signal amplification and the opportunity for a highly sensitive diagnostic. Probes that measure disease-specific enzymatic activity can be used for noninvasive disease detection (e.g., by generating synthetic reporters of disease that filter into the urine), for intra-operative or ex vivo visualization of pathology (e.g., of tumor margins during resection surgeries; pathogen detection in clinical specimens), and for treatment response monitoring.
Diagnostics that utilize enzymatic activity as an engineered component of their activation cascade, such as engineered probiotics and CRISPR diagnostics, provide modular platforms for rapid and programmable detection of disease biomarkers. Probiotic diagnostics must be developed in clinically-relevant strains, tested in robust and relevant in vivo models, and rigorously evaluated for both performance and safety. Field evaluation and benchmarking of CRISPR diagnostics will demonstrate their potential for point-of-care deployment in resource-limited settings.
The involvement of clinicians throughout the development pipeline is essential for translation of ABDx. Given the engineered, synthetic, or biological nature of many ABDx devices (e.g., responsive nanomaterials, in vivo imaging probes, molecular or cellular recorders), chemical composition and dose should be selected in the context of clinical use case, validation in preclinical models, and consultation with regulatory agencies. Identification of appropriate patient populations for efficacy trials will be crucial to successful clinical validation of these emerging tools.
Enzyme activity as a functional biomarker of disease
Biomarkers, such as proteins or nucleic acids, are biological indicators of disease progression or therapeutic response [16]. Since enzymes, such as proteases, play critical roles in several biological processes that contribute to disease progression, many ABDx measure enzyme activity as a functional biomarker of disease [17] (Figure 1A). Furthermore, by leveraging the catalytic nature of enzymes for signal amplification, activity measurements may offer sensitivity advantages relative to endogenous blood biomarkers. Advances in chemistry and nanotechnology have enabled new molecular probes that are selectively activated by disease-associated enzymatic activity to generate a measurable diagnostic readout (Figure 1A).
Substrate and activity-based probes for molecular imaging in vivo
Several ABDx function by converting the activity of enzymes involved in disease progression into a imaging readout (Figure 2) [17,18]. For cancer in particular, accurate in vivo visualization of malignant tissues can aid in early diagnosis, surgical planning and resection, and monitoring of treatment response. As proteases play direct functional roles in all cancer hallmarks [17], imaging the proteases involved in cancer progression has emerged as a promising detection strategy.
Figure 2. Activatable probes for molecular imaging of disease-associated enzymes.
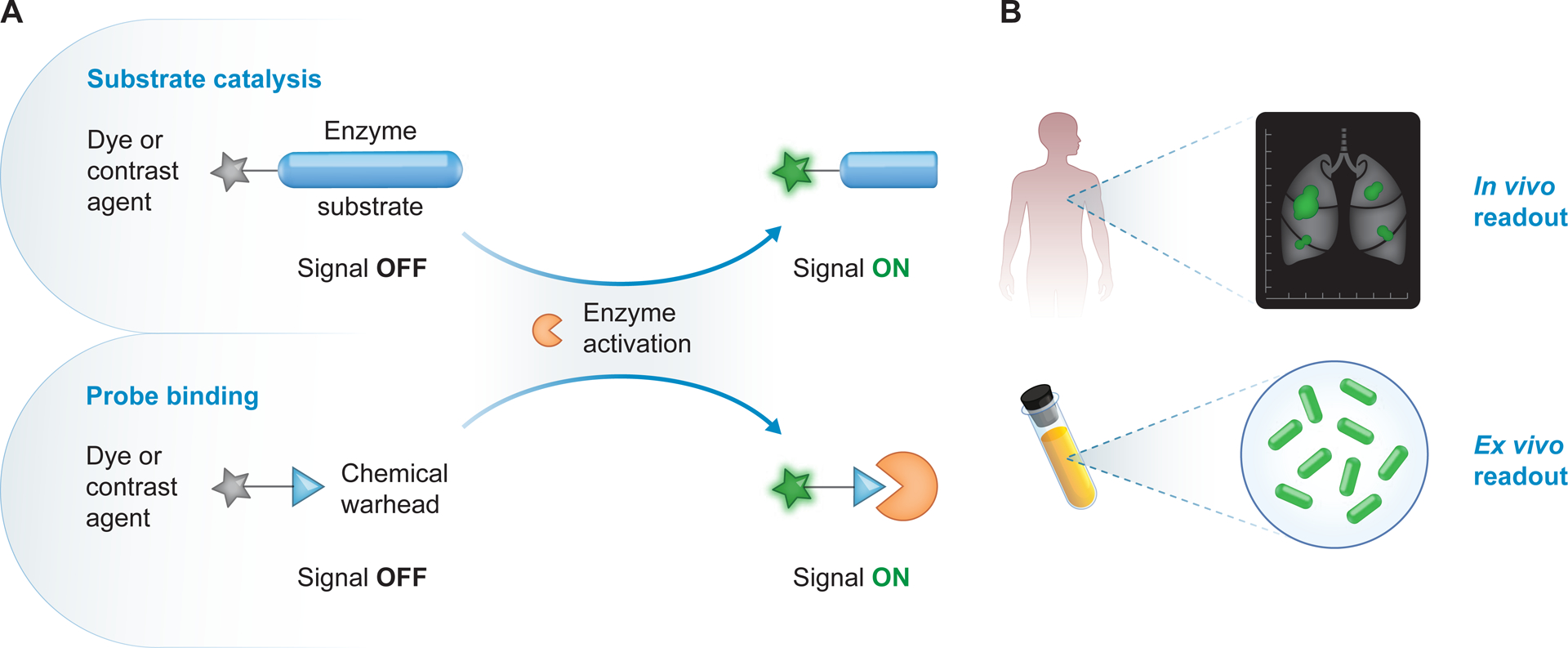
(A) Approaches that convert enzyme activity to an imaging readout include probes that assess substrate catalysis (top) or bind to the enzyme active site (bottom). Probes carry a dye or contrast agent and remain “OFF” prior to enzyme activation (grey star). Substrate-based probes carry an enzyme substrate and recognition motif (blue ellipse) that may be processed by active enzymes, while binding-based probes carry a chemical warhead (blue triangle) that can covalently bind to active enzymes. These probes turn “ON” to produce an imaging readout (green star) upon processing by enzymes such as proteases. (B) These probes can be used in vivo for diagnostic or intraoperative imaging (top), or ex vivo for visualization of tumor margins or for pathogen detection in clinical specimens (bottom).
Advances in chemistry and nanotechnology have spawned both substrate cleavage- and binding-based probes for molecular imaging of enzymes involved in cancer (Figure 2). Substrate-based probes that fluoresce in the near-infrared upon proteolytic cleavage have been extensively used for imaging-based tumor detection [19–24]. For example, the cathepsin-activated probe 6QC has shown pre-clinical promise in labeling lung, breast, and colon tumors [25], and has also been utilized for in vivo surgical guidance, where it can be detected intra-operatively with the daVinci Si Surgical System [25,26]. Cathepsin-responsive fluorescent probes [23,24] are currently under commercial development for in vivo tumor detection. These probes and similar technologies, such as topical probes activated by transpeptidases [27], could enable high-resolution intraoperative molecular imaging for guided tumor resection and debulking. In another strategy, a cell-penetrating peptide is released from a probe following proteolysis [28]; when functionalized with fluorescent acceptor-donor pairs [29,30] or magnetic resonance contrast agents [31], these activatable cell-penetrating peptides (ACPPs) have enabled visualization of primary tumors and metastases in several mouse models [30–32]. Improved ACPPs are currently under clinical development for intraoperative evaluation of lymph node metastases. Further, several enzyme-responsive nanoparticle systems have been developed for optical imaging of cancer in mouse models [31,33–37]. As an alternative to cleavable substrates, quenched activity-based probes (ABPs) [17,18,38] that covalently react with enzyme active sites through a chemical warhead have also been used to visualize tumors in vivo in mice [39–41]. While covalent labeling enables localization of proteolysis, the resulting enzyme inactivation prevents signal amplification, an important consideration for probe sensitivity.
In addition to optical approaches, analogous techniques utilize enzymatic activation for specific tumor detection with other imaging modalities, such as positron emission tomography (PET) [42] and magnetic resonance imaging (MRI) [35,37]. Dual readout probes for multi-modal assessment have also been developed [31]. One recent approach described a magnetism-based nanoscale phenomenon in which the distance between a paramagnetic enhancer and a superparamagnetic quencher tunes MRI signal [37]. Based on this method, an MMP-cleavable substrate can be used to link the enhancer and quencher, such that proteolysis by MMPs in the tumor microenvironment led to MRI contrast enhancement.
Despite these advances, there are a number of potential limitations to imaging-based ABDx. In regards to target specificity, cleavage promiscuity can hinder the discriminative power of single-substrate agents [17]. Further, several probes incorporate substrates responsive to enzymes, such as cathepsins [25,26], that are highly expressed in malignancy, benign diseases, inflammation, and even healthy tissues. Potential false positives could be attenuated through rational protease target nomination through ‘omic analyses and down-selection of highly expressed candidates (Box 1). Disease specificity could also be improved through active targeting (e.g., integrin [43] or receptor [44] docking) or multiplexing, though the multiplexing capacity of imaging is fundamentally limited by a paucity of orthogonal readouts [22] (Box 1). Thus, imaging probes that require multiple steps of enzymatic processing, such as those responsive to Boolean logic operations [45,46], may improve specificity. Furthermore, probe deployment in a focused clinical context, such as intraoperative visualization of tumor margins in reduction surgeries, can help eliminate false positives. Other significant challenges facing imaging-based ABDx include tissue penetrance, probe biocompatibility and clearance, and sensitivity, particularly for low levels of disease. As these are novel agents, the track through regulatory agencies is not yet established. Specifically, there is no defined path to FDA approval for optical contrast agents nor established precedent on desired outcomes for Phase II and Phase III trials, and product submissions continue to be considered on a case-by-case basis [18]. Completed and ongoing clinical trials for imaging-based ABDx will inform the design of future studies and help improve the efficiency and consistency of regulatory constructs.
Activity-based nanosensors for non-invasive detection of disease
As an alternative to imaging approaches that rely on contrast agents, Bhatia and colleagues have engineered a class of enzyme-responsive nanoparticles, termed activity-based nanosensors (ABNs), that respond to dysregulated protease activity in vivo to generate urinary reporters of disease (Figure 3) [47]. Following ABN administration, proteolytic cleavage of peptide substrates carried by ABNs liberates reporters that are small enough to be cleared by the kidney and detected in the urine. ABNs provide signal amplification advantages due to local enzymatic turnover and concentration of reporters in urine. Highly multiplexed detection can be achieved by leveraging orthogonal isotope-encoded reporters that can be read by mass spectrometry [47–49]. In a mouse model of colorectal cancer, a broadly MMP-responsive ABN panel with this encoding scheme exhibited superior diagnostic performance to the clinically approved blood biomarker carcinoembryonic antigen (CEA), as the urinary reporter signature enabled accurate detection (Area under the curve (AUC) = 0.94) of tumors ~60% smaller than those detectable with CEA (130 mm3 vs. 330 mm3 for ABNs and CEA, respectively) [47]. In addition to mass encoding, urinary detection can also be achieved with ligand-encoded reporters measurable by immunoassays [50–53].
Figure 3. Activity-based nanosensors (ABNs).
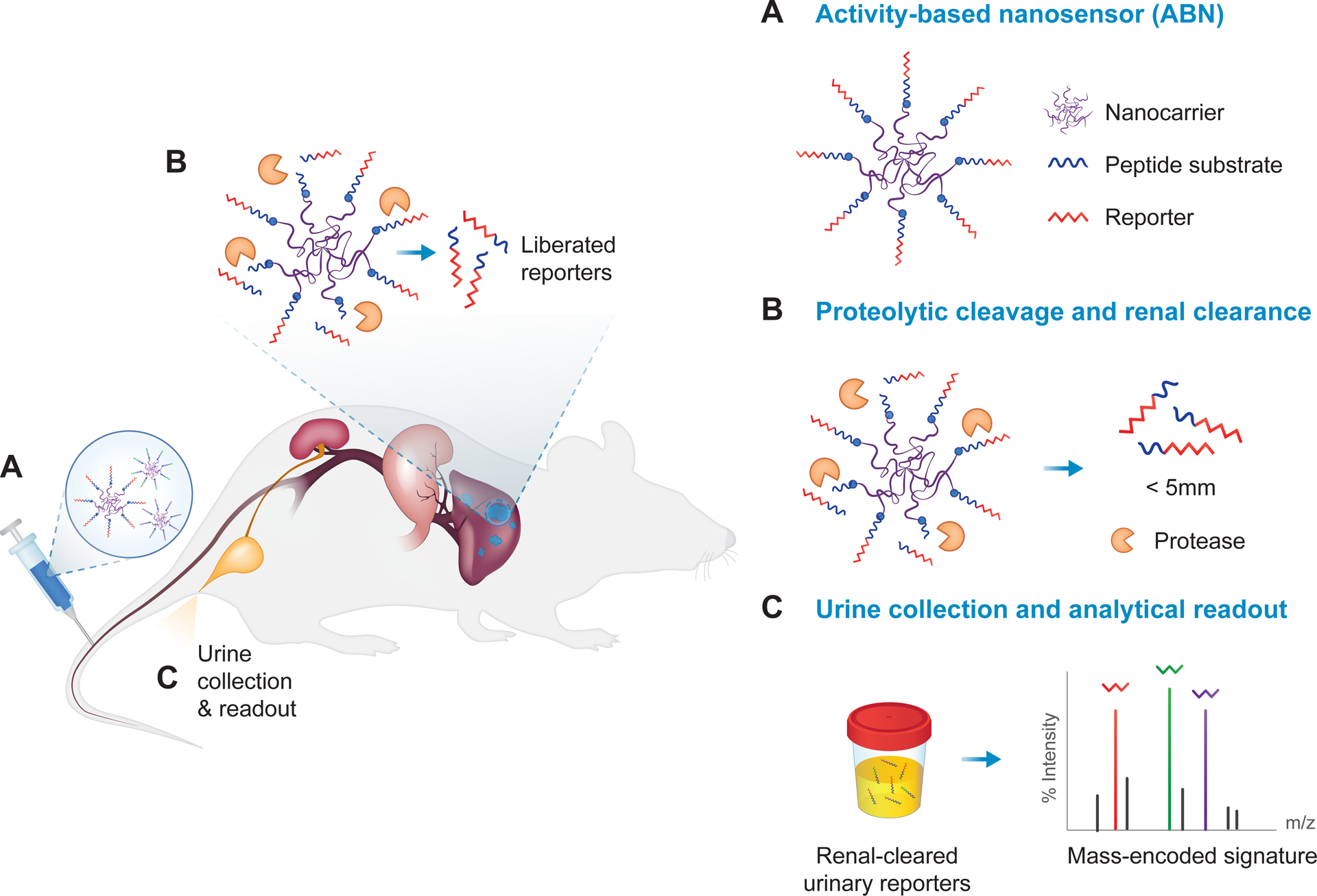
(A) Protease-cleavable peptide substrates appended with a reporter molecule, such as a mass-encoded barcode, are conjugated to a nanocarrier, such as a (poly)ethylene-glycol (PEG) scaffold. (B) The protease-sensitive ABNs are administered intravenously, subcutaneously, or intratracheally, and designed to specifically disassemble upon engagement with dysregulated proteases at the site of disease. After protease cleavage, liberated reporters, whose size is below the renal filtration limit of ~5 nm, are filtered into the urine. (C) Reporters can be detected in the cleared urine via a corresponding analytical method, such as liquid chromatography followed by tandem mass spectrometry (LC-MS/MS). Disease state is detected noninvasively based on urinary reporter signatures.
Both experimental and computational tools from seemingly disparate fields have yielded key advances in the evolution of the ABN platform. In work spanning nanoparticle engineering and transplant immunology, ABNs were recently engineered to measure the activity of granzyme B, a protease released by cytotoxic T cells as they engage and kill target cells, and were deployed as sensors for early diagnosis of acute transplant rejection in an allogenic skin graft model [54]. The demonstrated capacity of ABNs to measure immune-related protease activity could enable immunotherapy response monitoring and assessment of T cell-mediated autoimmune diseases. Recent work leveraged inorganic catalytic nanomaterials [55,56], specifically renal-clearable gold nanoclusters (AuNCs), to design ABNs wherein the peroxidase-mimicking catalytic activity of proteolytically liberated AuNCs could be monitored in urine to produce a visible color change as a diagnostic readout [57]. Paper-based [51,53] and colorimetric [57] readouts encourage point-of-care deployment of the ABN technology in resource-limited settings for both infectious [58] and non-communicable diseases.
The sensitivity of the ABN platform for in vivo tumor detection has been described through a multi-compartment pharmacokinetic model [59]. This in silico model informed the development of ultrasensitive ABNs that combined improved substrate presentation and active targeting for accurate detection of millimeter-sized tumors in mice [43]. In an orthotopic model of ovarian cancer, the accuracy of this urinary diagnostic exceeded that of the blood biomarker HE4, as ABNs achieved an AUC of 0.99 at an average tumor burden of 36 mm3 while HE4 was not elevated to a detectable level at this timepoint (AUCHE4 = 0.51). Further, transcriptomic analysis to identify proteases that are upregulated in disease and in vitro cleavage assays against peptide libraries to nominate substrates have been used to inform the design of multiplexed ABN panels [48,49]. Finally, preliminary work that couples ABN multiplexing with machine learning has yielded statistical algorithms for prospective diagnosis based on urinary reporter signatures [49].
While these results encourage the potential of ABNs, there are several limitations that must be considered. Namely, systemic (i.e., intravenous) delivery leaves ABNs susceptible to nonspecific activation in off-target organs, which can reduce signal-to-noise. Preliminary work in the context of lung cancer detection suggests that intrapulmonary ABN delivery may provide a means of maximizing delivery to the target organ to increase signal, while eliminating off-target activation to reduce noise [49]. However, even in the context of intrapulmonary delivery, ABNs are unable to precisely localize proteolysis and, by extension, the site of disease. Thus, ABNs may have clinical utility as companion tests to imaging approaches, for example as a rapid and cost-effective follow-up to diagnostic imaging to confirm the presence of malignancy or infection. Additionally, ABNs have largely been tested in cell transplant models of cancer. While intrapulmonary ABNs were recently deployed in a genetically-engineered mouse model of lung cancer [49], the model used only represents a subset of human disease. Clinical studies will be necessary to establish the utility of ABNs for detecting lung cancer in humans and for discriminating malignancy from benign comorbidities.
The specificity of ABNs for a disease of interest, particularly in the context of concomitant comorbidities, presents another significant challenge. Like imaging probes, ABNs are limited by the cleavage specificity of their substrates and the expression levels of protease targets. Since proteases are dysregulated not only in cancer but also in infection, inflammation, and fibrosis, ABN panels must be rationally designed with this in mind, and thorough in vivo validation in pre-clinical models and human trials will be necessary to ensure that ABNs are not confounded by benign disease etiologies or environmental insults such as alcohol consumption and tobacco smoke. Protease target nomination, peptide substrate engineering, and multiplexing may provide concrete means to improve specificity (Box 1). While mass-encoded ABNs enable multiplexing, the cost and equipment requirements of mass spectrometry may limit the accessibility of ABNs for routine use, particularly in resource-poor settings. Finally, the biocompatibility and clearance properties of nanosensor formulations must be established in clinical studies.
Continued efforts towards improved understanding of protease biology in both health and disease, methods for high-throughput substrate screening, and ex vivo assays for sensor validation in human specimens will be critical to design ABNs specific and responsive to human disease. Currently under clinical development for noninvasive monitoring of fatty liver disease, the ABN technology provides an example of how advances in nanomedicine, disease biology, and computational science can synergize to yield ABDx with clinical promise. Several of the described approaches, such as substrate optimization for specific enzymes [54], bioinformatics mining for candidate targets [48,49], and machine learning for predictive classification [49], are broadly applicable to the development of next-generation ABDx for human disease.
Ex vivo activity assays in clinical biospecimens
In addition to in vivo probes for diagnostic and intra-operative assessment, ex vivo measurements of disease-specific enzyme activity (Figure 1A) may enable differential diagnosis, relapse prediction, and treatment response monitoring. Active enzymes, such as proteases [60], have been found in a variety of human specimens, including tissue sections [61,62], blood [63], urine [60,64], and sputum [65]. Though several diagnostics are run on patient specimens, such as blood-based tests for cancer [9–11,66] or sputum-based tests for tuberculosis [67], assays and probes that rely on catalytic processing by disease-relevant enzymes could offer performance advantages driven by catalytic signal amplification.
Bridging molecular imaging and anatomic pathology, in situ zymography assays are a class of methods that measure and localize protease activities directly in tissue [43,44,61,62]. Chemical probes, such as ABPs [62] and substrate-functionalized nanoparticles [43], have shown potential for imaging proteolytic activity in frozen tissue sections. Similarly, topically-applied probes of dipeptidyl peptidase [68] and cathepsin [69] activity have been applied ex vivo on resected tissues to identify tumor margins. More broadly, in situ and topical enzyme activity measurements could be used to inform the development of ABDx or, with further validation, to serve as stand-alone diagnostics.
Approaches to bulk activity profiling have leveraged microfluidic [70] or mass spectrometry [71,72] techniques to measure protease activity in patient-derived biospecimens for diagnostic applications. In recent work, screening pancreatic cyst fluid samples against a multiplexed synthetic substrate library identified two aspartic proteases with increased activities in malignant, mucinous cyst fluid relative to benign specimens [72]. Activity of one identified protease, gastricsin, detected malignancy with 100% specificity and 93% sensitivity, significantly outperforming the widely-used molecular biomarker CEA, which exhibited 94% specificity and 65% sensitivity [72]. Emerging approaches that couple bulk activity profiling with mass tag [47,73] or DNA barcoding [74] schemes could enable quantitative substrate or ABP screens for diagnostic classification.
The convergence of discovery efforts in microbiology and of bio-orthogonal synthesis strategies from chemistry has led to the development of activatable molecular probes for pathogen detection and infectious disease diagnosis [75–78]. A striking example is in the design of enzyme-activatable fluorogenic probes for active tuberculosis (TB), a disease responsible for an estimated 1.6 million deaths in 2017 [79]. Standard diagnostics for active TB rely on sputum stains for the causative bug Mycobacterium tuberculosis (Mtb), which require extensive sample processing, have widely variable sensitivity (32 to 94%), and cannot distinguish live from dead bacteria, thereby precluding their ability to report on treatment efficacy or drug resistance [67,80]. Chemical probes whose signal is specifically activated and amplified by pathogen-specific enzymatic processing could overcome these limitations and enable next-generation ABDx for TB.
Biochemical investigations have yielded promising targets and pathways for new activity-based TB diagnostics. For example, trehalose mycolates, a class of glycolipids essential to Mtb viability, are the major constituents of the mycobacterial outer membrane. Accordingly, organic synthesis techniques can be used to design chemical probes that measure the activity of mycolyltransferases, which convert trehalose to trehalose mycolates to assemble this outer membrane [77,78]. In a recent diagnostic application, a fluorogenic trehalose analog, DMN-Tre, was developed to directly detect live Mtb in patient sputum [77]. Enzymatic conversion of DMN-Tre by mycobacterial acyl transferases enables its incorporation into the hydrophobic membrane, resulting in a dramatic fluorescence increase of an attached solvatochromic dye. While DMN-Tre can label live Mtb in human sputum following a single incubation step, it lacks specificity for Mtb compared to other bacterial strains that express mycolyltransferases.
Prior works have established Mtb’s resistance to β-lactam antibiotics due to expression of β-lactamase [81] and new enzymatic targets, such as DprE1, for anti-TB drugs [82]. A recent study leveraging these insights described an activatable fluorogenic probe, CDG-DNB3, that required two steps of enzymatic processing for specific detection of live Mtb [76]. CDG-DNB3 is first cleaved by a conserved Mtb β-lactamase to activate fluorescence [75,81], and then covalently modified by the Mtb essential enzyme DprE1 [82] to retain signal within the membrane. The combined use of two enzymes as diagnostic markers enabled detection of viable bacteria with specificity for Mtb over nontuberculosis mycobacteria [76]. Future efforts that continue to bridge microbiology, genetics, and organic chemistry could enable rapid target identification and probe design for new ABDx for TB and other infectious diseases.
Leveraging enzymatic activity to engineer molecular and cellular diagnostics
Advances in synthetic and molecular biology have spawned new tools, such as genetic memory circuits [83] and CRISPR-associated (CRISPR-Cas) systems [84], that have precipitated the emergence of diagnostic innovations that use enzymatic activity as a way to monitor for and report on disease state. In this section, we review recent work focusing on (1) activity-based probiotic and cellular biosensors (Figure 1B), and (2) CRISPR diagnostics (Figure 1C), and specifically highlight how integrative research can continue to accelerate development of these ABDx for precision health applications.
Engineering activity-based diagnostics with synthetic biology
Synthetic biology is a forward engineering approach that integrates experimental manipulation, computational modeling techniques, and principles of circuit design to produce specific, controlled behaviors in biological systems. The programmability, circuit design principles, and fast design-to-production cycles of synthetic biology afford the potential for diagnostics that can be rationally engineered and rapidly deployed to meet clinical needs. Recent advances in synthetic biology have promoted the design and use of bacteriophage [85], molecular recorders [86], mammalian cells [87–89], and programmable probiotics [90,91] for diagnostic applications.
Bacteria offer unique advantages as a chassis for programmable ABDx, as they can be readily manipulated to sense disease markers through enzyme-driven circuits or signaling cascades and to generate amplified, non-invasive diagnostic readouts. By bringing insights from microbiology and genetic circuit design together, researchers have engineered commensal bacteria to sense disease biomarkers and to report on their presence after in vivo interrogation. Diverse bacterial species possess sensing circuits responsive to disease-relevant signals, such as cytokines, metabolites, inflammatory markers, heme, hormones, pH, and temperature, as previously reviewed [91]. Two-component systems include a class of sensing circuits wherein signal transduction is mediated by the activity of sensory histidine kinases [92]. These circuits can be exploited to engineer probiotic diagnostics that respond to specific disease biomarkers via kinase-driven signal amplification, such as in sensing transient markers of inflammation in the gastrointestinal (GI) tract [93,94]. As a recent example, commensal mouse E. coli were engineered with a kinase-mediated sensing circuit synthetically linked to a memory element [95] and delivered orally to record exposure to the gut inflammatory marker tetrathionate [94]. Similar systems based on other enzyme-mediated signaling cascades could be used for the diagnosis of a diversity of disease states, such as fucose-sensing probiotics for detecting GI pathogens [96]. Furthermore, computational mining of genomic data [97] and library-based screening strategies [98] could be used to identify new bacterial biosensors for the design of more sensitive and specific circuits.
Owing to the programmability of genetic circuit design, in vivo bacterial diagnostics can be engineered to produce a synthetic reporter enzyme as an activity-based readout of disease (Figure 1B). This approach offers three potential modes of signal amplification: bacterial expansion in vivo, high levels of enzyme production from stably maintained plasmids, and enzymatic substrate turnover in generating the final readout. One study explored the capacity of orally-administered probiotic E. coli to preferentially expand in metastatic liver tumors, where the bacteria produced high levels of a reporter enzyme that could cleave a systemically-delivered substrate, which ultimately led to a color change detectable in the urine [99]. In addition to liver metastases, this technology may also enable detection of tumors in other organs exposed to bacterial loads from the GI tract, such as colorectal cancer.
While this system achieved high accuracy (AUC = 0.93) in a mouse model of liver metastases, it lacked any disease-specific sensing circuitry upstream of the enzymatic readout. Engineered sense-and-respond probiotics that link synthetic reporter enzymes downstream of biomarker sensing circuits have the potential for improved diagnostic performance. For example, a probiotic strain was recently engineered to detect quorum sensing molecules specific to the pathogen Vibrio cholerae via sensory histidine kinase-based signal transduction and, after in vivo interrogation in a mouse model of cholera, to generate a reporter enzyme whose activity could be readily queried in fecal samples (Figure 4) [100]. The signal amplification and substrate flexibility afforded by synthetic reporter enzymes encourage the deployment of these bacterial systems at the point-of-care for early detection of cancer and infectious diseases.
Figure 4. Enzyme-driven biological sensors.
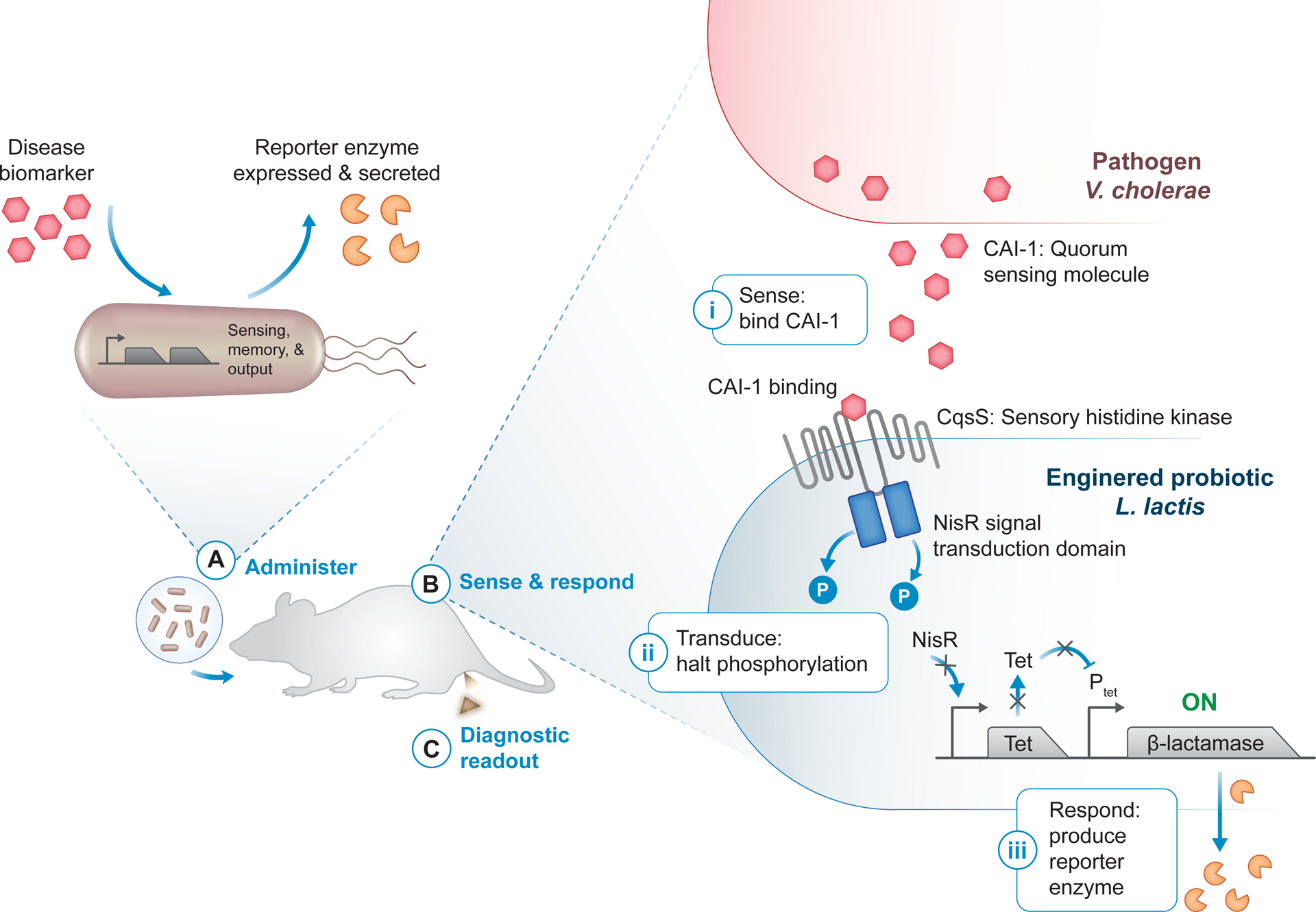
(A) Bacteria modified with sense-and-respond systems use disease-specific biomarkers (red hexagon) to sense a disease state and, in turn, produce a diagnostic readout. Synthetic reporter enzymes such as β-lactamase (orange pacman) can be expressed as a way to generate an activity-based readout of disease state. The engineered probiotic diagnostics can then be administered in vivo, for example orally for diagnostic applications in the gastrointestinal tract. (B) Quorum signaling through cholera autoinducer 1 (CAI-1, red hexagon) in Vibrio cholerae has been used to induce expression of the reporter enzyme β-lactamase in Lactococcus lactis engineered as a cholera diagnostic [100]. The engineered V. cholerae-sensing circuit consists of a hybrid two-component system, comprised of the sensory histidine kinase CqsS linked to the NisR signal transduction domain, and a TetR/Ptet reporter module. Upon CAI-1 binding to CqsS (i), phosphorylation through the two-component system is halted, preventing TetR expression and Ptet repression (ii). Without TetR, the β-lactamase reporter is freely expressed and secreted (iii). (C) An activity assay for the reporter enzyme can be used to produce a diagnostic readout, for example directly in collected fecal samples [100].
While engineered probiotics are perhaps the most widely used scaffold for applying synthetic biology to diagnostic design, genetically-encoded molecular recorders, such as DNA vectors that leverage a tumor-specific promoter to drive expression of a reporter enzyme [86], and mammalian cell biosensors [87–89] have also been applied as engineered ABDx. One example of the latter involved a subcutaneous cell implant that, in response to blood hypercalcemia associated with mouse models of colon and breast cancer, formed a visible “tattoo” via enzymatic production of the black pigment melanin [88].
An alternative strategy is the systemic delivery of engineered mammalian cells that are selectively activated in vivo to produce an activity-based readout. In a novel integration of ideas from synthetic biology and cell therapy, macrophages were engineered to express a synthetic reporter enzyme in response to adopting a tumor-associated (M2) metabolic profile [89]. In cell transplant models of colorectal cancer in immunodeficient or syngeneic mice, this system of “diagnostic adoptive cell transfer” achieved greater sensitivity and specificity than clinically-used cancer biomarkers. Namely, in a xenograft model that sheds the protein biomarker CEA, subcutaneous tumors of average volume 45 mm3 were discriminated more accurately by the macrophage sensor relative to CEA (AUCs of 0.914 and 0.829, respectively). The sensitivity of this cellular ABDx was also benchmarked to cfDNA in the syngeneic CT26 subcutaneous tumor model, where the macrophage sensor could detect 25 – 50 mm3 tumors while tumor-specific cfDNA mutations were only detectable for tumor volumes of 1,500 – 2,000 mm3. This 50-fold improvement in limit of detection highlights the potential for disease-activatable probes to circumvent the biological and mathematical limitations of endogenous biomarkers for early cancer detection [15].
The specificity of the macrophage sensor was also characterized in the context of co-occurring acute inflammation in a lung metastasis mouse model established via intravenous injection of the 4T1 breast cancer cell line, where no significant differences in accuracy were observed in the absence (AUC = 0.975) or presence (AUC = 1.00) of acute lung inflammation [89]. However, in models of lung and muscular inflammation, the macrophage sensors were activated during the wound healing and inflammation resolution phases, which are characterized by M2 macrophage influx. In light of these results, the specificity of this cellular ABDx for malignancy relative to confounding benign comorbidities must be validated. Greater signal multiplicity may enhance specificity, as the sensor in its current state measures a single M2-associated marker [89]. Further characterization will also be necessary to determine whether these M2-polarized macrophage sensors have an impact on tumor progression.
Tools from synthetic biology offer great promise for the future development of ABDx with improved functionality, clinical relevance, and performance. Enzyme-driven synthetic circuits, such as DNA recombinase-based logic gates [101–103] and state machines [104] or protease-based circuits [105,106], could provide the ability to logically integrate combinations or sequences of several signals for improved diagnostic specificity. For engineered probiotics, importing circuits optimized in model organisms into clinically-relevant strains is another significant consideration [107]. Future efforts to characterize the stability, safety, and performance of sense-and-respond circuits in commensal strains and in relevant disease models will be indispensable to the clinical translation of probiotic diagnostics. These studies will be critical to regulatory approval of engineered bacterial strains or cell sensors in anticipation of clinical implementation. Finally, automated circuit design with computational techniques such as machine learning could enable rapid development of more sensitive systems [108].
Another intriguing possibility is the integration of synthetic biology with other engineering disciplines to create hybrid diagnostics that interface more directly with the body. For example, an ingestible bacterial-electronic microcapsule was recently developed and deployed for accurate in situ diagnosis of GI bleeding in swine [109]. Bacteria engineered to sense heme and to produce a luciferase reporter enzyme in response were integrated with luminescence readout electronics that could transmit signal wirelessly to an external device. Alternatively, hybrid materials hosting genetically engineered bacteria could enable diagnostic wearables composed of both synthetic and living materials [110]. Advances in synthetic biology, low-power electronics, and soft materials may open new frontiers for novel activity-based diagnostic devices for noninvasive health monitoring.
Harnessing CRISPR-Cas biology for accurate, rapid, and accessible diagnostic tools
The discovery of microbial clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (CRISPR-Cas) adaptive immune systems has transformed the life sciences. These systems consist of sequence-specific RNA-guided nucleases that can hydrolyze nucleic acids with high enzymatic turnover. The programmable, efficient activity of CRISPR-Cas has been extensively exploited for gene editing in basic science and therapeutic applications [84]. Recent insights into the biology of CRISPR-Cas have led to new molecular diagnostics that leverage the activity of Cas effectors for sensitive, specific, and rapid nucleic acid detection (Figure 1C).
In a platform driven by engineered RNA biosensors, cell-free synthetic gene networks were coupled with a CRISPR-Cas9 module to yield a low-cost paper test for strain-specific Zika virus detection [111]. In this system, these engineered RNA sensors, termed toehold switches [112], detect “trigger” viral RNAs to ultimately produce a differential color change on paper. Strain-specific resolution is achieved by leveraging the requirement of an NGG protospacer adjacent motif (PAM) for Cas9 endonuclease activity (Figure 5A). Specifically, the presence or absence of a strain-specific PAM within the target RNA results in differential Cas9-mediated cleavage, such that active trigger molecules are only produced in the absence of Cas9 activity. This integration of guided Cas9 endonuclease activity with toehold switch RNA sensing enabled visual discrimination of American and African Zika viral strains with single-base resolution.
Figure 5. Guided, programmable nucleases for ABDx.
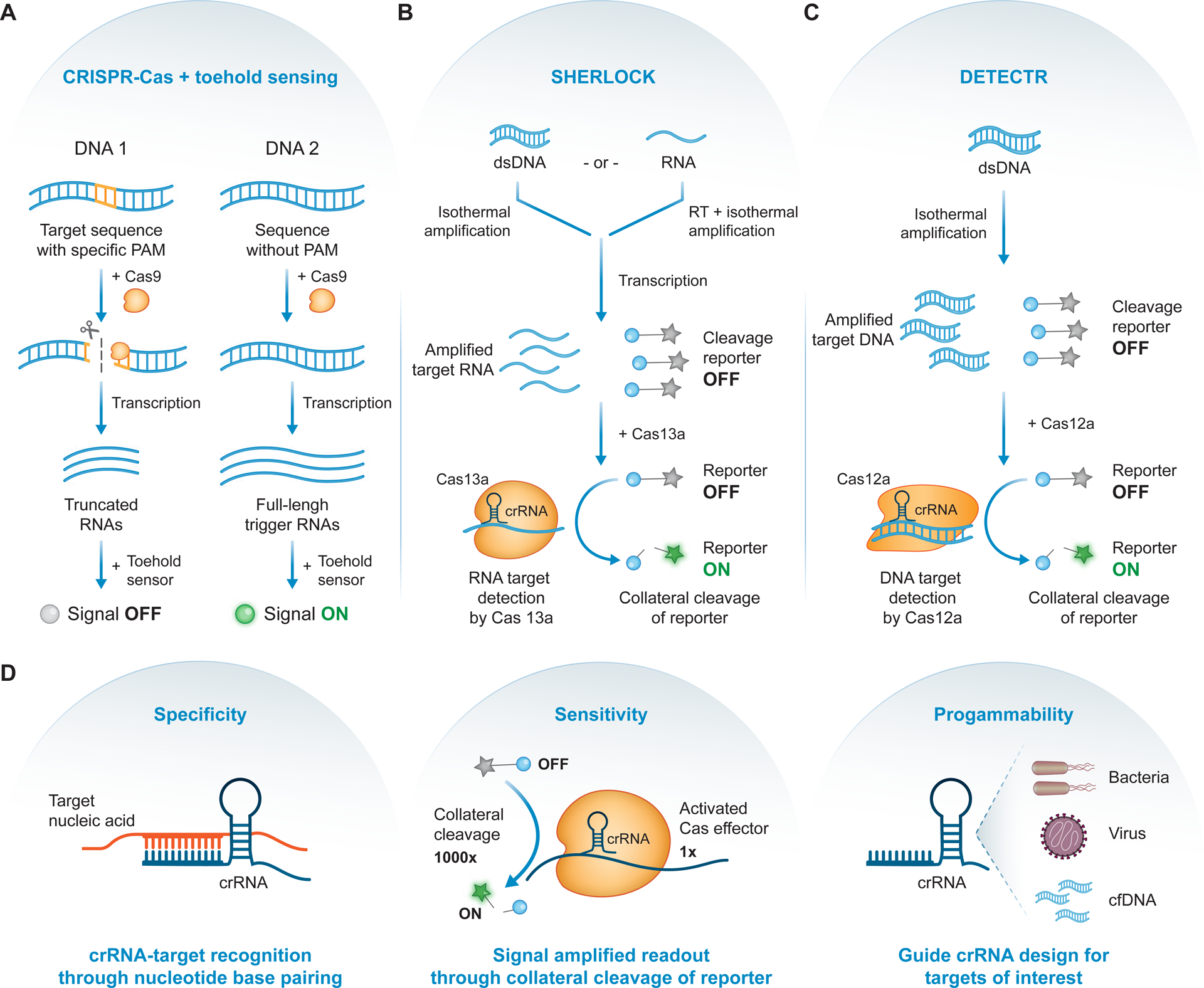
(A) Nucleic acid detection with CRISPR-Cas augmented toehold sensing. The presence of a specific protospacer adjacent motif (PAM; orange) in a DNA input results in cleavage by Cas9 with a PAM-specific guide RNA. Following transcription, this differential cleavage leads to either truncated or full-length trigger RNAs, which can be designed to differentially activate an engineered toehold sensor. Since only full-length triggers can activate the toehold sensor, a downstream signal can be used to discriminate the two inputs [111]. (B) SHERLOCK diagnostic test. Nucleic acids, either double-stranded DNA (dsDNA) or RNA, are amplified or reverse-transcribed (RT) and amplified, for DNA or RNA respectively. Following transcription, amplified target RNAs and fluorescently quenched cleavage reporters (grey star = fluorophore, blue circle = quencher) are incubated with Cas13a-crRNA complex. Detection and binding of the RNA target unleashes Cas13a’s collateral activity, resulting in reporter cleavage, fluorophore activation (green star), and signal amplification [114]. (C) DETECTR diagnostic test. Target dsDNA are amplified and, along with fluorescently quenched cleavage reporters, incubated with Cas12a-crRNA complex. Detection and binding of the dsDNA target unleashes Cas12a’s collateral activity, resulting in reporter cleavage, fluorophore activation, and signal amplification [115]. (D) CRISPR diagnostics achieve high specificity driven by crRNA guide-target base pairing (left), detection sensitivity from the highly efficient collateral cleavage of reporter molecules (middle), and diagnostic programmability due to facile guide RNA design for nucleic acid targets of interest (right).
Fundamental discoveries into the structure, function, and diversity of CRISPR-Cas systems have led to the emergence of next-generation CRISPR diagnostics. For example, Cas13a is an RNA-guided, RNA-targeting CRISPR effector that exhibits promiscuous ribonuclease activity upon target recognition [113]. One technique, termed SHERLOCK (Figure 5B), couples this collateral activity with isothermal amplification to achieve detection of target nucleic acids with attomolar sensitivity and single-base mismatch specificity [114]. Detection of a target sequence by a guided Cas13a unleashes its promiscuous ribonuclease activity, leading to activation of reporter molecules and robust signal amplification. This triggered collateral activity was exploited for detection of Zika virus isolated from human samples and for cancer mutation identification in cell-free DNA. An orthogonal CRISPR diagnostic system utilizes a distinct effector, Cas12a, which exhibits target-activated, non-specific single-stranded DNA endonuclease (ssDNase) activity [115]. Similar to SHERLOCK, this diagnostic platform, termed DETECTR (Figure 5C), leverages the target-specific “unlocking” of Cas12a’s highly efficient, indiscriminate ssDNase activity to produce an amplified detection signal only after Cas12a activation by its cognate DNA target, and achieved specific detection of cancer-associated human papilloma virus in human anal swab samples.
Both these platforms leverage the guided programmability of Cas enzymes for highly specific nucleic acid detection and exploit the promiscuous nuclease activity of two particular effectors for robust signal amplification. However, both SHERLOCK and DETECTR lack precise nucleic acid quantification, rely on a fluorescent readout, and are limited in their multiplicity. SHERLOCKv2 addresses these limitations by achieving improved quantification through optimization of the isothermal amplification step, point-of-care compatibility with a paper-based test, and increased multiplicity via use of orthogonal Cas effectors [116]. Finally, to support diagnostic deployment in any context, a protocol that enables direct detection of target molecules in clinical specimens was coupled with SHERLOCK for rapid visual diagnosis of viral infection in bodily fluids [117].
The CRISPR-Cas9 system has also been combined with next-generation sequencing (NGS) technologies to enrich low-abundance sequences in clinical samples [118]. In this approach, a sample of DNA or cDNA is subject to Cas9 digestion of target genes, producing cleavage products to which adapters are attached for subsequent amplification, enrichment, and sequencing of the target DNA. This approach was applied to clinical respiratory fluid samples to identify antimicrobial resistance-associated genes, with sub-attomolar sensitivity and up to 5 orders of magnitude greater enrichment relative to NGS alone. Future efforts that combine the enzymatic activity of CRISPR-Cas effectors with NGS may prove useful for detection of tumor mutations and for diagnostic molecular profiling in clinical samples.
The programmability of Cas enzymes offers tremendous potential for the development of responsive smart materials that can be actuated via nucleic acid sensing. Notably, DNA-embedded hydrogels were recently designed to respond to signal-triggered CRISPR-associated nuclease activity [119]. When integrated into a microfluidic device linked to an electronic circuit, one such actuated hydrogel modulated a conductive flow channel in response to Cas12a collateral activity, enabling detection of nucleic acid biomarkers, such as Ebola virus RNA and antibiotic resistance genes. Wireless electronic monitoring of this activity-based diagnostic readout was achieved by incorporating a microelectronic radio transmission module into the device. In exploiting the enzymatic properties of Cas12a for actuated smart materials that directly interface with electronic monitoring systems, this work highlights the promise of CRISPR-Cas for programmable ABDx that can be applied globally.
CRISPR-based technologies harness the unique enzymatic properties of Cas effector enzymes to achieve highly specific, sensitive, and programmable molecular diagnostics (Figure 5D). Strategies to combine the programmable activity of Cas enzymes with biomaterial, microfluidic, and bioelectronic interfaces may lead to new point-of-care diagnostic devices, similar to glucose monitors or home pregnancy kits, for cancer and infectious diseases. Future efforts that apply CRISPR diagnostics to clinical specimens from different diseases and rigorously benchmark performance against standard tests will support their clinical translation.
Concluding remarks
A variety of enabling technologies, including bio-orthogonal chemistry, CRISPR-Cas, and synthetic gene circuits, have armed researchers with the tools to design a powerful new class of diagnostic tests that leverage enzymatic activity to measure or produce biomarkers of disease. As an emerging class of technologies, these activity-based diagnostics (ABDx) must be evaluated in terms of both their relative advantages and disadvantages (Table 1). Looking forward, continued pre-clinical validation and technical development of activity-based diagnostics (ABDx) will be critical to reach the performance necessary for clinical use (see Outstanding Questions). A number of technical strategies, including probe and substrate optimization through high-throughput screening or targeted chemistry and signal multiplexing, may enable the design of more selective tests with the classification power necessary for high disease specificity (Box 1). Furthermore, applying machine learning to the chemical or biological signals generated by ABDx, alone or in conjunction with other modalities, will provide prospective statistical classifiers for validation in independent cohorts.
Table 1 |.
Summary and comparison of ABDx strategies
| Technology | Advantages | Disadvantages |
|---|---|---|
| In vivo imaging with cleavage- and binding-based probes |
|
|
| Activity-based nanosensors (ABNs) |
|
|
| Ex vivo probes and activity assays |
|
|
| Engineered probiotics and cellular sensors |
|
|
| CRISPR-Cas diagnostics |
|
|
Outstanding Questions.
What are the best strategies for optimizing the specificity, sensitivity, and predictive value of activity-based diagnostics (ABDx)? What role can machine learning play in the development of statistical classifiers based on data generated by ABDx (alone or in combination with other data)?
How can ABDx be pre-clinically and clinically benchmarked against other diagnostic modalities?
Can ABDx be used for noninvasive monitoring of treatment response, and could they provide sensitive prediction of responders vs. non-responders more rapidly than standard metrics of disease regression?
How can ABDx inform the design of multimodal technologies that integrate diagnostic modalities such as imaging, biofluid analytes, and “smart” devices such as wearable sensing technologies?
How can ABDx inform the design of “detect-and-treat” technologies that integrate both diagnostic and therapeutic modalities?
What are the most compelling clinical use cases for deployment of ABDx? Examples include early detection, intra-operative visualization, monitoring of therapeutic response, and detection of minimal residual disease.
What development pathways are most likely to support regulatory approval, reimbursement, and clinical adoption?
The emerging diagnostic tools described in this Review must be rigorously benchmarked, at both the pre-clinical and clinical levels, against standard tests to ensure sensitivity and specificity (Outstanding Questions). Head-to-head benchmarking against imaging and molecular diagnostics, including ctDNA and multi-analyte blood biomarkers, will help establish the concrete advantages that ABDx afford, identify specific clinical use cases for their deployment, and suggest opportunities for integration with other diagnostic modalities or medical devices. Further, studies that assess the ability of ABDx to monitor treatment response will clarify their potential as integrated diagnostics for precision health (Outstanding Questions).
There are several considerations to keep in mind on the path towards regulatory approval of ABDx (Outstanding Questions). First, projects should be designed with clear clinical motivation, components that are generally regarded as safe (GRAS), and proposed metrics for diagnostic success. For ABDx designed as in vivo sensors, such as imaging probes, nanoparticles, and engineered probiotics, in vivo efficacy experiments should be conducted in appropriate disease models and constructed with clinical use cases in mind. Additionally, the dosage regimens and safety of these in vivo sensors must be established both pre-clinically and clinically. The need to assess ABDx performance relative to standard and other emerging diagnostics cannot be emphasized enough, particularly in regards to establishing success and outcomes metrics for future clinical trials.
For many ABDx, such as optical imaging agents [18], there is no pre-established pathway to FDA approval. Similar to other emerging technologies, current FDA frameworks consider these product submissions on a case-by-case basis, often as combination products. As with any new modality, increased examples of performance and utility will establish more efficient regulatory frameworks for clinical translation. As these technologies grow in scope and number, sustained collaborations between researchers, clinicians, and regulatory officials will be critical to achieve clinical deployment (Clinician’s Corner). With further development, ABDx may provide an accurate way to intercept and manage disease and ultimately improve patient outcomes.
Highlights.
Effective diagnostics provide clinically-actionable information on the presence, prognosis, or progress of disease and are thus critical to improving patient outcomes.
Activity-based diagnostics (ABDx) leverage enzymatic activity to measure or produce biomarkers of disease. Because of the substrate recognition and catalytic signal amplification properties of enzymes, ABDx afford the potential for highly specific, sensitive, and programmable diagnostics.
Tools from an array of disciplines, such as bio-orthogonal chemistry, responsive nanomaterials, machine learning, synthetic gene circuits, and CRISPR-Cas systems, have enabled the development of engineered ABDx.
ABDx have shown promise in pre-clinical settings for the detection of both noncommunicable (e.g., cancer) and infectious diseases, and have been applied for molecular sensing in vivo and for in vitro diagnostics run on ex vivo biospecimens.
Acknowledgments
We thank Dr. H. Fleming (MIT), J. Kirkpatrick (MIT), and M. Anahtar (MIT) for critical reading and editing of the manuscript, and C. Malvar for assistance with figure illustration. We apologize for being unable to cite many pieces of primary literature due to length considerations. A.P.S. acknowledges support from the NIH Molecular Biophysics Training Grant and the National Science Foundation Graduate Research Fellowship. S.N.B is a Howard Hughes Medical Institute Investigator.
Disclosure statement
A.P.S. and S.N.B. are listed as inventors on patents and patent applications related to activity-based technologies. S.N.B. is a director at Vertex, co-founder and consultant at Glympse Bio, consultant for Cristal, Maverick, and Moderna, and receives sponsored research funds from Johnson & Johnson.
Glossary
- Activity-based diagnostics (ABDx)
Engineered diagnostics that leverage enzymatic activity to measure or produce biomarkers of disease
- Activity-based probes (ABPs)
Chemical probes that interact with catalytically active enzymatic targets and remain covalently bound to the active site
- Activity-based nanosensors (ABNs)
Enzyme-responsive nanoparticles that respond to dysregulated protease activity in vivo to generate urinary reporters of disease. ABNs have demonstrated utility in preclinical models of several disease conditions, including cancer, bacterial infection, and thrombosis
- Area under the curve (AUC)
A measure of the performance of a binary classifier, established by constructing receiver operating characteristic (ROC) curves and quantifying the area under the curve (AUC). AUC is used as a metric to characterize the predictive power of the candidate diagnostic in the tested condition, where a baseline AUC of 0.5 is equivalent to the predictive power of a random binary test
- Boolean logic operation
Boolean logic describes an algebra governed by the truth values “True” (1) and “False” (0). Boolean logic is dictated by the Boolean operators “AND”, “OR”, and “NOT”
- Cell-free DNA (cfDNA)
DNA in blood circulation that is not associated with cells
- Cell-penetrating peptide
Short peptides, usually positively charged, that facilitate the cellular uptake of associated cargo. The cargo can be covalently linked to the cell-penetrating peptide or associated with it through non-covalent interactions
- Chemical warhead
A reactive chemical group that can covalently bind to a target site on a biological target. In the case of activity-based probes, this warhead is usually an electrophile that can covalently link to a nucleophilic residue in the enzyme active site
- Circulating tumor DNA (ctDNA)
Cell-free DNA that is derived from tumor cells
- CRISPR-associated (Cas) effector
An enzyme that uses CRISPR guides to recognize and cleave nucleic acids
- DNA recombinase
An enzyme that catalyzes site-specific re-arrangement (excision, inversion, insertion, or translocation) of DNA
- Multi-compartment pharmacokinetic model
A mathematical model that describes the absorption, distribution, metabolism, and excretion of an administered molecule or particle in terms of transmission between physiological compartments (i.e., blood, tissue, or organ compartments)
- Paper-based test (also, paper test)
A simple paper-based (e.g., cellulose) device that can detect a target analyte in a sample without any specialized equipment beyond the test itself. Lateral flow assays are a prominent example, with home pregnancy tests being perhaps the most widely known example
- Predictive classification
The process of arranging samples into distinct classes or categories using a predictive statistical model
- Protease
An enzyme that hydrolyzes peptide bonds
- Reporter enzyme
An enzyme that is expressed and produced by an engineered diagnostic in response to activation. The activity of that enzyme can be queried in vivo or in vitro/ex vivo, depending on the nature of the sensor, to provide a diagnostic readout
- Sensory histidine kinase
An enzyme involved in signal transduction that modulates the phosphorylation of a response regulator in response to changes in an input signal
- Smart materials
Engineered materials that have properties that can be modulated in a controlled fashion by input stimuli such as small molecules, pH, or temperature
References
- 1.Broadhurst MJ et al. (2016) Diagnosis of ebola virus disease: past, present, and future. Clin. Microbiol. Rev 29, 773–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etzioni R et al. (2003) The case for early detection. Nat. Rev. Cancer 3, 243–252 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL et al. (2019) Cancer statistics, 2019. CA. Cancer J. Clin 69, 7–34 [DOI] [PubMed] [Google Scholar]
- 4.Prensner JR et al. (2012) Beyond PSA: the next generation of prostate cancer biomarkers. Sci. Transl. Med 4, 127rv3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Thoracic Society (2001) Guidelines for the management of adults with community-acquired pneumonia. Am. J. Respir. Crit. Care Med 163, 1730–1754 [DOI] [PubMed] [Google Scholar]
- 6.The National Lung Screening Trial Research Team (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med 365, 395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faye O et al. (2013) Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol. J 10, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imperiale TF et al. (2014) Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med 370, 1287–1297 [DOI] [PubMed] [Google Scholar]
- 9.Newman AM et al. (2014) An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med 20, 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen JD et al. (2018) Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan JCM et al. (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 [DOI] [PubMed] [Google Scholar]
- 12.Heitzer E et al. (2019) Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet 20, 71–88 [DOI] [PubMed] [Google Scholar]
- 13.Chertow DS (2018) Next-generation diagnostics with CRISPR. Science 360, 381–382 [DOI] [PubMed] [Google Scholar]
- 14.Adalsteinsson VA et al. (2017) Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun 8, 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque IS and Elemento O (2017) Challenges in using ctDNA to achieve early detection of cancer. bioRxiv DOI: doi: 10.1101/237578 [DOI] [Google Scholar]
- 16.Sawyers CL (2008) The cancer biomarker problem. Nature 452, 548–552 [DOI] [PubMed] [Google Scholar]
- 17.Dudani JS et al. (2018) Harnessing protease activity to improve cancer care. Annu. Rev. Cancer Biol 2, 353–376 [Google Scholar]
- 18.Garland M et al. (2016) A bright future for precision medicine: advances in fluorescent chemical probe design and their clinical application. Cell Chem. Biol 23, 122–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmood U and Weissleder R (2003) Near-infrared optical imaging of proteases in cancer. Mol. Cancer Ther 2, 489–496 [PubMed] [Google Scholar]
- 20.Weissleder R et al. (1999) In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat. Biotechnol 17, 375–378 [DOI] [PubMed] [Google Scholar]
- 21.Hilderbrand SA and Weissleder R (2010) Near-infrared fluorescence: application to in vivo molecular imaging. Curr. Opin. Chem. Biol 14, 71–79 [DOI] [PubMed] [Google Scholar]
- 22.Weissleder R (2006) Molecular imaging in cancer. Science 312, 1168–1172 [DOI] [PubMed] [Google Scholar]
- 23.Grimm J et al. (2005) Use of gene expression profiling to direct in vivo molecular imaging of lung cancer. Proc. Natl. Acad. Sci. U. S. A 102, 14404–14409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitley MJ et al. (2016) A mouse-human phase 1 co-clinical trial of a protease-activated fluorescent probe for imaging cancer. Sci. Transl. Med 8, 320ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ofori LO et al. (2015) Design of protease activated optical contrast agents that exploit a latent lysosomotropic effect for use in fluorescence-guided surgery. ACS Chem. Biol 10, 1977–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yim JJ et al. (2018) Optimization of a protease activated probe for optical surgical navigation. Mol. Pharm 15, 750–758 [DOI] [PubMed] [Google Scholar]
- 27.Urano Y et al. (2011) Rapid cancer detection by topically spraying a gamma-glutamyltranspeptidase-activated fluorescent probe. Sci. Transl. Med 3, 110ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang T et al. (2004) Tumor imaging by means of proteolytic activation of cell-penetrating peptides. Proc. Natl. Acad. Sci 101, 17867–17872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitney M et al. (2013) Ratiometric activatable cell-penetrating peptides provide rapid in vivo readout of thrombin activation. Angew. Chemie - Int. Ed 52, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savariar EN et al. (2013) Real-time in vivo molecular detection of primary tumors and metastases with ratiometric activatable cell-penetrating peptides. Cancer Res 73, 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson ES et al. (2010) Activatable cell penetrating peptides linked to nanoparticles as dual probes for in vivo fluorescence and MR imaging of proteases. Proc. Natl. Acad. Sci. U. S. A 107, 4311–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miampamba M et al. (2017) Sensitive in vivo visualization of breast cancer using ratiometric protease-activatable fluorescent cancer imaging agent, AVB-620. Theranostics 7, 3369–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welser K et al. (2011) Protease sensing with nanoparticle based platforms. Analyst 136, 29–41 [DOI] [PubMed] [Google Scholar]
- 34.Kwon EJ et al. (2015) Smart nanosystems: bio-inspired technologies that interact with the host environment. Proc. Natl. Acad. Sci 112, 14460–14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris TJ et al. (2006) Proteolytic actuation of nanoparticle self-assembly. Angew. Chemie - Int. Ed 45, 3161–3165 [DOI] [PubMed] [Google Scholar]
- 36.Harris TJ et al. (2008) Protease-triggered unveiling of bioactive nanoparticles. Small 4, 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi J et al. (2017) Distance-dependent magnetic resonance tuning as a versatile MRI sensing platform for biological targets. Nat. Mater 16, 537–542 [DOI] [PubMed] [Google Scholar]
- 38.Sanman LE and Bogyo M (2014) Activity-based profiling of proteases. Annu. Rev. Biochem 83, 249–273 [DOI] [PubMed] [Google Scholar]
- 39.Blum G et al. (2007) Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat. Chem. Biol 3, 668–677 [DOI] [PubMed] [Google Scholar]
- 40.Edgington LE et al. (2013) Functional imaging of legumain in cancer using a new quenched activity-based probe. J. Am. Chem. Soc 135, 174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdoes M et al. (2013) Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J. Am. Chem. Soc 135, 14726–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larimer BM et al. (2017) Granzyme B PET imaging as a predictive biomarker of immunotherapy response. Cancer Res 77, 2318–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon EJ et al. (2017) Ultrasensitive tumour-penetrating nanosensors of protease activity. Nat. Biomed. Eng 1, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desnoyers LR et al. (2013) Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med 5, 207ra144. [DOI] [PubMed] [Google Scholar]
- 45.Badeau BA et al. (2018) Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nat. Chem 10, 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widen JC et al. (2019) Multivariate “AND-gate” substrate probes as enhanced contrast agents for fluorescence-guided surgery. bioRxiv DOI: doi: 10.1101/695403 [DOI] [Google Scholar]
- 47.Kwong GA et al. (2013) Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat. Biotechnol 31, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudani JS et al. (2018) Classification of prostate cancer using a protease activity nanosensor library. Proc. Natl. Acad. Sci. U. S. A 115, 8954–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirkpatrick J et al. (2018) Noninvasive lung cancer detection via pulmonary protease profiling. bioRxiv DOI: doi: 10.1101/495259v1 [DOI] [Google Scholar]
- 50.Lin KY et al. (2013) Nanoparticles that sense thrombin activity as synthetic urinary biomarkers of thrombosis. ACS Nano 7, 9001–9009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren AD et al. (2014) Point-of-care diagnostics for noncommunicable diseases using synthetic urinary biomarkers and paper microfluidics. Proc. Natl. Acad. Sci. U. S. A 111, 3671–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warren AD et al. (2014) Disease detection by ultrasensitive quantification of microdosed synthetic urinary biomarkers. J. Am. Chem. Soc 136, 13709–13714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudani JS et al. (2016) Sustained-release synthetic biomarkers for monitoring thrombosis and inflammation using point-of-care compatible readouts. Adv. Funct. Mater 26, 2919–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mac QD et al. (2019) Non-invasive early detection of acute transplant rejection via nanosensors of granzyme B activity. Nat. Biomed. Eng 3, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao Y et al. (2015) Metal nanoclusters: novel probes for diagnostic and therapeutic applications. Chem. Soc. Rev. Chem. Soc. Rev 8636, 8636–8663 [DOI] [PubMed] [Google Scholar]
- 56.Loynachan CN et al. (2018) Platinum nanocatalyst amplification: redefining the gold standard for lateral flow immunoassays with ultrabroad dynamic range. ACS Nano 12, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loynachan CN et al. (2019) Renal clearable catalytic gold nanoclusters for in vivo disease monitoring. Nat. Nanotechnol 14, 883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buss CG et al. (2018) Protease activity sensors noninvasively classify bacterial infections and antibiotic responses. EBioMedicine 38, 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwong GA et al. (2015) Mathematical framework for activity-based cancer biomarkers. Proc. Natl. Acad. Sci 112, 12627–12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy R et al. (2009) Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol 27, 5287–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandooren J et al. (2013) Zymography methods for visualizing hydrolytic enzymes. Nat. Methods 10, 211–220 [DOI] [PubMed] [Google Scholar]
- 62.Withana NP et al. (2016) Labeling of active proteases in fresh-frozen tissues by topical application of quenched activity-based probes. Nat. Protoc 11, 184–191 [DOI] [PubMed] [Google Scholar]
- 63.Villanueva J et al. (2008) A sequence-specific exopeptidase activity test (SSEAT) for “functional” biomarker discovery. Mol. Cell. Proteomics 7, 509–518 [DOI] [PubMed] [Google Scholar]
- 64.Roy R et al. (2008) Tumor-specific urinary matrix metalloproteinase fingerprinting: identification of high molecular weight urinary matrix metalloproteinase species. Clin. Cancer Res 14, 6610–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simpson JL et al. (2005) Differential proteolytic enzyme activity in eosinophilic and neutrophilic asthma. Am. J. Respir. Crit. Care Med 172, 559–565 [DOI] [PubMed] [Google Scholar]
- 66.Bettegowda C et al. (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6, 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parsons LM et al. (2011) Laboratory diagnosis of tuberculosis in resource-poor Countries: Challenges and opportunities. Clin. Microbiol. Rev 24, 314–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Onoyama H et al. (2016) Rapid and sensitive detection of early esophageal squamous cell carcinoma with fluorescence probe targeting dipeptidylpeptidase IV. Sci. Rep 6, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walker E et al. (2017) Rapid visualization of nonmelanoma skin cancer. J. Am. Acad. Dermatol 76, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen CH et al. (2013) Multiplexed protease activity assay for low-volume clinical samples using droplet-based microfluidics and its application to endometriosis. J. Am. Chem. Soc 135, 1645–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Donoghue AJ et al. (2012) Global identification of peptidase specificity by multiplex substrate profiling. Nat. Methods 9, 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivry SL et al. (2017) Global protease activity profiling provides differential diagnosis of pancreatic cysts. Clin. Cancer Res 23, 4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lapek JD et al. (2019) Quantitative multiplex substrate profiling of peptidases by mass spectrometry. Mol. Cell. Proteomics 18, 968–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Navrátil V et al. (2017) DNA-linked Inhibitor Antibody Assay (DIANA) for sensitive and selective enzyme detection and inhibitor screening. Nucleic Acids Res. 45, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie H et al. (2012) Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nat. Chem 4, 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng Y et al. (2018) Rapid and specific labeling of single live Mycobacterium tuberculosis with a dual-targeting fluorogenic probe. Sci. Transl. Med 10, eaar4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamariza M et al. (2018) Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med 10, eaam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hodges HL et al. (2018) Imaging mycobacterial growth and division with a fluorogenic probe. Proc. Natl. Acad. Sci. U. S. A 115, 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.World Health Organization (2018) Global tuberculosis report 2018, (2018th edn) World Health Organization. [Google Scholar]
- 80.Pai M et al. (2016) Tuberculosis. Nat. Rev. Dis. Prim 2, 1–23 [Google Scholar]
- 81.Flores AR et al. (2005) Genetic analysis of the β-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to β-lactam antibiotics. Microbiology 151, 521–532 [DOI] [PubMed] [Google Scholar]
- 82.Makarov V et al. (2009) Benzothiazinones Kill Mycobacterium tuberculosis by blocking Arabinan synthesis. Science 324, 801–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brophy JAN and Voigt CA (2014) Principles of genetic circuit design. Nat. Methods 11, 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Komor AC et al. (2017) CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu TK et al. (2013) Advancing bacteriophage-based microbial diagnostics with synthetic biology. Trends Biotechnol 31, 325–327 [DOI] [PubMed] [Google Scholar]
- 86.Ronald JA et al. (2015) Detecting cancers through tumor-activatable minicircles that lead to a detectable blood biomarker. Proc. Natl. Acad. Sci. U. S. A 112, 3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ausländer D et al. (2014) A designer cell-based histamine-specific human allergy profiler. Nat. Commun 5, 4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tastanova A et al. (2018) Synthetic biology-based cellular biomedical tattoo for detection of hypercalcemia associated with cancer. Sci. Transl. Med 10, eaap8562. [DOI] [PubMed] [Google Scholar]
- 89.Aalipour A et al. (2019) Engineered immune cells as highly sensitive cancer diagnostics. Nat. Biotechnol 37, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Slomovic S et al. (2015) Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. U. S. A 112, 14429–14435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Riglar DT and Silver PA (2018) Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol 16, 214–225 [DOI] [PubMed] [Google Scholar]
- 92.Stock AM et al. (2000) Two-component signal transduction. Annu. Rev. Biochem 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 93.Daeffler KN et al. (2017) Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol 13, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riglar DT et al. (2017) Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol 35, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kotula JW et al. (2014) Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U. S. A 111, 4838–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pacheco AR et al. (2012) Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ziemert N et al. (2016) The evolution of genome mining in microbes -- a review. Nat. Prod. Rep 33, 988–1005 [DOI] [PubMed] [Google Scholar]
- 98.Naydich AD et al. (2019) Synthetic gene circuits enable systems-level biosensor trigger discovery at the host-microbe interface. mSystems 4, e00125–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Danino T et al. (2015) Programmable probiotics for detection of cancer in urine. Sci. Transl. Med 7, 289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mao N et al. (2018) Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med 10, eaao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Courbet A et al. (2015) Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med 7, 289ra83. [DOI] [PubMed] [Google Scholar]
- 102.Bonnet J et al. (2013) Amplifying genetic logic gates. Science 340, 599–603 [DOI] [PubMed] [Google Scholar]
- 103.Siuti P et al. (2013) Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol 31, 448–452 [DOI] [PubMed] [Google Scholar]
- 104.Roquet N et al. (2016) Synthetic recombinase-based state machines in living cells. Science 353, aad8559. [DOI] [PubMed] [Google Scholar]
- 105.Stein V and Alexandrov K (2014) Protease-based synthetic sensing and signal amplification. Proc. Natl. Acad. Sci. U. S. A 111, 15934–15939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gao XJ et al. (2018) Programmable protein circuits in living cells. Science 361, 1252–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mimee M et al. (2015) Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Camacho DM et al. (2018) Next-generation machine learning for biological networks. Cell 173, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 109.Mimee M et al. (2018) An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu X et al. (2017) Stretchable living materials and devices with hydrogel-elastomer hybrids hosting programmed cells. Proc. Natl. Acad. Sci. U. S. A 114, 2200–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pardee K et al. (2016) Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 [DOI] [PubMed] [Google Scholar]
- 112.Green AA et al. (2014) Toehold switches: De-novo-designed regulators of gene expression. Cell 159, 925–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abudayyeh OO et al. (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gootenberg JS et al. (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen JS et al. (2018) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gootenberg JS et al. (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science 360, 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Myhrvold C et al. (2018) Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Quan J et al. (2019) FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 47, e83–e83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.English MA et al. (2019) Programmable CRISPR-responsive smart materials. Science 365, 780–785 [DOI] [PubMed] [Google Scholar]
- 120.Kasperkiewicz P et al. (2014) Design of ultrasensitive probes for human neutrophil elastase through hybrid combinatorial substrate library profiling. Proc. Natl. Acad. Sci. U. S. A 111, 2518–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]


