Abstract
Background: As a type of breast cancer that has relatively strong invasiveness, triple negative breast cancer (TNBC) seriously affects the survival of patients. microRNAs (miRNAs) have been shown to exert a prominent regulatory effect on the disease, among which miR-133b is reported to be involved in the pathological mechanism of breast cancer, but its role in TNBC remains unclear. Methods: In this study, real-time quantitative PCR (RT-qPCR) and Western blotting (WB) were performed for detecting the expressions of miR-133b, fibroblast growth factor receptor 1 (FGFR1), and Wingless/Integrated (Wnt)-β-catenin pathway markers (Wnt1, β-catenin, nuclear-β-catenin, p-GSK-3β, GSK-3β, cyclinD1, and FOXQ1). With TNBC cells and DDP-resistant TNBC cells (TNBC/DDP cells) used as research objects, their proliferation and apoptosis were measured by Cell Counting Kit-8 (CCK-8) assays and Flow cytometry, respectively. Then, the targeted relationship between miR-133b and FGFR1 was verified by Dual luciferase reporter gene assay (DLRGA). Results: In our study, miR-133b was down-regulated while FGFR1 up-regulated in TNBC. The ectopic expression of miR-133b remarkably inhibited the proliferation and colony formation but induced apoptosis of TNBC cells, and inactivated the Wnt-β-catenin pathway. The knockdown of FGFR1 had similar effects. Additionally, miR-133b targeted and negatively regulated FGFR1. Up-regulating miR-133b or down-regulating FGFR1 could enhance the proliferation and DDP sensitivity of TNBC cells or TNBC/DDP cells. Up-regulating FGFR1 could offset the anti-TNBC cell survival and DDP sensitization shown by ectopic expression of miR-133b. Conclusion: To sum up, miR-133b can inhibit the growth and DDP resistance of TNBC cells by targeting FGFR1 and inactivating the Wnt-β-catenin pathway.
Keywords: Triple negative breast cancer, miR-133b, FGFR1, Wnt-β-catenin pathway
Introduction
As one of the main causes of female deaths, breast cancer (BC) is a heterogeneous tumor with different subtypes [1,2]. Triple negative breast cancer (TNBC) is the most invasive subtype with a relatively poor prognosis, because it lacks targeted therapies and has a low response to chemotherapy [3,4]. Moreover, the disease carried a higher risk of metastasis and recurrence, and was usually diagnosed in young women [5]. Therefore, it is of great significance to explore the molecular mechanism and chemical resistance of TNBC for formulating its molecular therapeutic strategies and enhancing its chemosensitivity.
microRNA (miRNA)-messenger RNA (mRNA) regulatory network has been widely used in the clinical treatment of tumors. As microRNA regulators, miRNAs modulate physiological and pathological processes through the targeted regulation of mRNAs, which process plays an important role in tumor growth and chemical sensitivity of TNBC [6-8]. Wang et al. indicated that miR-17 targets c-Jun activation domain binding protein 1 (JAB1) to promote tumor growth and cisplatin (DDP) resistance of TNBC [9]. As far as we know, chemotherapy is the standard therapeutic strategy for TNBC, and DDP is the most widely used chemotherapeutic drug for solid tumors. Although this drug has a certain curative effect on TNBC patients, it inevitably produces chemoresistance and results in unsatisfactory effects [10,11]. Based on the above reasons, this study mainly explored the molecular mechanism of DDP resistance in TNBC. We argue that the miR-133b-fibroblast growth factor receptor 1 (FGFR1) axis may mediate the tumor growth and DDP resistance of TNBC by regulating the Wnt-β-catenin pathway. miR-133b is related to the survival and metastatic behavior of BC cells, which can be inhibited by its ectopic expression. However, the role of miR-133b in TNBC has been rarely studied. Through an online target gene prediction website, we found that FGFR1 is a carcinogenic factor that has potential conservative binding sites with miR-133b, and its carcinogenicity has been revealed in esophageal squamous cell carcinoma, lung adenocarcinoma, and prostate cancer [12-14]. According to previous studies, miR-133b is correlated with the risk of BC, and involved in the molecular therapeutic mechanism of transducer of regulated CREB activity 1/2 (TORC1/2) inhibitors in TNBC [15,16]. Wingless/Integrated (Wnt)-β-catenin pathway, a classical regulatory pathway of cancer signal transduction, is regulated by FGFR1 to mediate the progression mechanism of BC, and can regulate the tumorigenesis and chemoresistance of TNBC [17,18].
In this study, we verified whether the miR-133b-FGFR1 axis could regulate the survival and DDP resistance of TNBC cells through the regulation of the Wnt-β-catenin pathway.
Materials and methods
Clinical samples
From November 2016 to November 2019, the breast cancer tissues and adjacent tissues were surgically obtained from 65 TNBC patients and then placed in liquid nitrogen containers for later use. All patients were initially diagnosed as TNBC by pathology [19]. Patients who had received radiotherapy and chemotherapy or who were complicated with other malignant tumors and severe organ dysfunction or infectious diseases were excluded. All patients and their family members signed the informed consent forms. Clinical Trial Ethics Committee, Affiliated Hospital of Southwest Medical University approved this experiment (Acceptance Number: KY2020232). The research process strictly followed the Declaration of Helsinki.
Cell culture, drug-resistant cell construction and transfection
Human normal mammary epithelial cells (MCF-10A) and TNBC cells (MDA-MB-468, HCC1395) were purchased from Otwo Biotech (Shenzhen) Inc., Shenzhen, China (HT-X1884, SAc0241, HTX2291C), and cultured (at 37°C) in a Roswell Park Memorial Institute (RPMI) 1640 medium (Beijing Greenherbs Science & Technology Development Co., Ltd., Beijing, China, 3026485) supplemeted with 10% fetal bovine serum (FBS; Lanso Biotechnology Co., Ltd., Shanghai, China, A3160801).
Construction of drug-resistant cells: The DDP-resistant TNBC cells (MDA-MB-468/DDP cells) were constructed through DDP (Chreagen, Beijing, China, 11866) interventions. DDP culture (1 μM) was used for DDP resistance maintenance. When the cells were in logarithmic growth phase, a suspension of 5×107 cells was prepared for the subsequent experiment. 10 mL of cell suspension was inoculated into the culture bottle for 24 hours, DDP (starting at 0.5 μM) was added to incubate for 48 hours, and the solution was then replaced with a fresh one. After digestion, DDP (2.5 μM) was added for further 48-hour treatment. Next, the DDP concentration was gradually increased (5 μM, 10 μM), and MDA-MB-468/DDP cells tolerant to 10 μM were obtained.
Transfection: miR negative control (miR-NC, 5’-UUCUCCGAACGUGUCACG-3’), miR-133b over-expression sequence (miR-133b, 5’-UUUGGU CCCCUUCAACCAGCU A-3’) and inhibitory sequence (inhibitor, 5’-UAGCUGGUUGAAGGGGAC CAAA-3’), negative control RNA (si-NC, 5’-UUCUCCGAACGUGUCACGUTT-3’), small interfering FGFR1 (si-FGFR1, 5’-GGAGGUGCUUCACUUAAGATT-3’), and FGFR1 over-expression vector (FGFR1) were respectively transfected into the cells using Lipofectamin 3000 kits (Lanso Biological Technology Co., Ltd., Shanghai, China, L3000-015), with the steps conducted in strict accordance with the kit instructions. Of them, FGFR1, confirmed to contain 3’UTR, was constructed by Shanghai Hanheng Biotechnology Co., Ltd., and a commercial confidentiality agreement was signed. Other transfection sequences were obtained from Shanghai Shiao Biomedical Technology Co., Ltd., and the si-NC used in this study was co-transfected with negative control plasmid. After 48-hour transfection, the cells were collected for subsequent experiments.
Real-time quantitative PCR (RT-qPCR)
Trizol reagents (Beijing Mairuibo Biotechnology Co., Ltd., Beijing, China; M NR0002) were adopted for extracting total RNA from the tissues and cells. After that, 5 μg of total RNA (each) was taken out to synthesize complementary DNA (cDNA) based on the instruction of reverse transcription kits (Biolab Science and Technology Co., Ltd., Beijing, China, BTN60906-PYW), and then 1 μL of cDNA was amplified. The amplification system contained cDNA (1 μL), upstream and downstream primers (0.4 μL each), 2X TransScript® Tip Green qPCR SuperMix (10 μL), passive reference dye (50X) (0.4 μL), and nuclease-free water that was finally added to supplement to 20 μL. β-Actin and U6 were used as internal references of FGFR1 and miR-133b, respectively, and 2-ΔΔct was used for data analysis. The forward sequence of FGFR1 was: 5’-GACGATCGATGCTAGCTACGTAGCT-3’, and the reverse sequence was: 5’-CGAGCTAGCTAGCTAGCTAGTCCAG-3’. The forward sequence of U6 was: 5’-GCTTCGGCAGCACATATACTAAAT-3’, and the reverse sequence was: 5’-CGCTTCACGAATTTGCGTGTCAT-3’. The forward sequence of FGFR1 was: 5’-CGCCAGGACCCGAACAG-3’, and the reverse sequence was: 5’-CAGTGAGCTCGATCCTCCTTT-3’. The forward sequence of β-Actin was: 5’-CAAAGGCCAACAGAGAGAAGAT-3’, and the reverse sequence was: 5’-TGAGACACACCATCACCAGAAT-3’.
Western blotting (WB)
Total protein was extracted using Radio-Immunoprecipitation Assay (RIPA) lysis buffer. The concentration was tested using bicinchoninic acid (BCA) protein assay kits (Future Biotech, Beijing, Beijing, China, FB327-01) and then adjusted to 4 μg/μL. After electrophoretic separation with 12% sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE) (Beijing Neobio Science & Technology Co., Ltd., Beijing, China, WB1103), the protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Biolab Science and Technology Co., Ltd., Beijing, China, YTB1201-NKC). After being stained by Ponceau S working solution (Biolab Science and Technology Co., Ltd., Beijing, China, RFT056-ZEX), the membrane was cleaned and sealed with 5% skimmed milk powder (Beijing Winter Song Boye Biotechnology Co. Ltd., Beijing, China, MP012) for 2 hours, and then added with antibodies against FGFR1, Wingless Type MMTV Integration Site Family Member 1 (Wnt1), β-catenin, nuclear-β-catenin (n-β-catenin), phospho-glycogen synthase kinase-3β (p-GSK-3β), GSK-3β (Beiyu Biotechnology Co., Ltd., Nanjing, China, BYFG-70R-37431, BYFG-70R-11918, BYCM-100029), cyclinD1, forkhead Q1 (FOXQ1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Kemin Biotechnology Co., Ltd, Shanghai, China, AF4196, ab5098, AF4196) with the dilution ratio of 1:500, and sealed overnight (at 4°C). After being cleaned to remove the primary antibodies, horseradish peroxidase-labeled goat anti-rabbit secondary antibody (1:1000) was added. All antibodies were purchased from Xinyu Biotechnology Co., Ltd., Shanghai, China, XY0650. The membrane was incubated (37°C, 1 hour) and then rinsed. Finally, the cells were luminesced with the electrochemiluminescence kit (Yanxi Biotechnology Co., Ltd., Shanghai, China, ECL-F-100) and developed, and the excess liquid was absorbed with filter papers. The gray values were analyzed.
Detection of cell proliferation by cell counting Kit-8 (CCK-8) assay
CCK-8 assay kits (Chreagen, Beijing, China, 120268) were used for detecting cell proliferation, with the steps performed strictly based on the instruction. The levels of cell proliferation at 0, 24, 48, and 72 hours were measured, respectively. After rinse with phosphate buffer solution (PBS; Taize Jiaye Technology Development Co., Ltd., Beijing, China, 20012027), the cells were cultured in a serum-free culture medium (90 μL; Taize Jiaye Technology Development Co., Ltd., Beijing, China, MEPI500CA) including 10 μL of CCK-8, for 2 hours (37°C, 5% CO2). The optical density (OD) values of cells were determined at 450 mm. DDP (0 μM, 0.5 μM, 2.5 μM, 5 μM, 10 μM) was used for interventions when the proliferation was compared between MDA-MB-468/DDP and MDA-MB-468 cells, and the other steps were conducted as previously described. After that, half maximal inhibitory concentration (IC50) was calculated based on the cell survival rate, and its value was defined as the DDP concentration when the OD value was reduced by 50%. The IC50 value was inversely proportional to DDP sensitivity.
Detection of apoptosis by flow cytometry (FCM)
After digestion with 0.25% trypsin (Chreagen, Beijing, China, 121520) and two rinses with PBS, the cells were added with 100 μL of binding buffer to prepare a 1×106 cells/mL suspension. After that, the suspension was sequentially added with AnnexinV-Fluorescein Isothiocyanate (FITC) and propidium iodide (PI) (10 μL each), and then incubated in the dark (room temperature, 5 minutes). The NovoCyte flow cytometer (Shanghai Ranger Apparatus Co., Ltd., Shanghai, China, AMG0002051) was used for detection. The apoptosis level was evaluated as the sum of early and late apoptosis levels, namely the sum of apoptosis percentage of AnnexinV+PI- and AnnexinV+PI+.
Dual luciferase reporter gene assay (DLRGA)
The downstream target genes of miR-133b were predicted by biological analysis with TargetScan 7.2 (http://www.targetscan.org/vert_72/). FGFR1-Wt (wild type) carrying miR-133b binding site was cloned into pMIR-REPORT luciferase reporter plasmid (Hewu Biotechnology Co., Ltd., Shanghai, China, P2155). Then FGFR1-Mut (mutant) was constructed using Quickchange II XL site-directed mutagenesis kit (Jinda Sun Technology Co., Ltd., Beijing, China, 200521). FGFR1-Wt (wild type), FGFR1-Mut (mutant), mimics, and miR-NC were transferred into the cells using Lipofectamin 3000 kits (Shanghai Lianshuo Biological Technology Co., Ltd., Shanghai, China, L3000-015). Luciferase activities were detected using DLRGA kits (Biolab Science and Technology Co., Ltd., Beijing, China, KFS303-LBV).
Statistical analysis
GraphPad 6 was used for data analysis and figure visualizing. The comparison between two groups was conducted by independent samples t test, and the comparison among multiple groups was conducted by one-way analysis of variance (ANOVA) and represented by F. Post hoc pairwise comparison was conducted by LSD-t test. The comparison of expression among multiple time points was analyzed by repeated measures ANOVA and represented by F. Bonferroni post hoc. Test was used to analyze the difference between groups. Receiver operating characteristic (ROC) curves were plotted to visualize the diagnostic values of miR-133b and FGFR1 in TNBC. Pearson test was performed to analyze the correlation of miR-133b with FGFR1 in the tissues. When P<0.05, the difference was statistically significant.
Results
miR-133b was down-regulated while FGFR1 up-regulated in TNBC
For exploring the role of miR-133b and FGFR1 in the development and progression of TNBC, we detected their expression in the TNBC tissues and cell lines. The expression or protein level of the former was down-regulated (P<0.01, Figure 1A and 1D), while the latter was up-regulated (P<0.01, Figure 1B and 1E). We also analyzed the correlation of their expression in the tissues, and found that there was a negative correlation between the two (r=-0.710, P<0.001, Figure 1C). These indicated that miR-133b and FGFR1 had abnormal expression in TNBC, suggesting that they may be involved in the pathogenesis of the disease (Figure 1).
Figure 1.
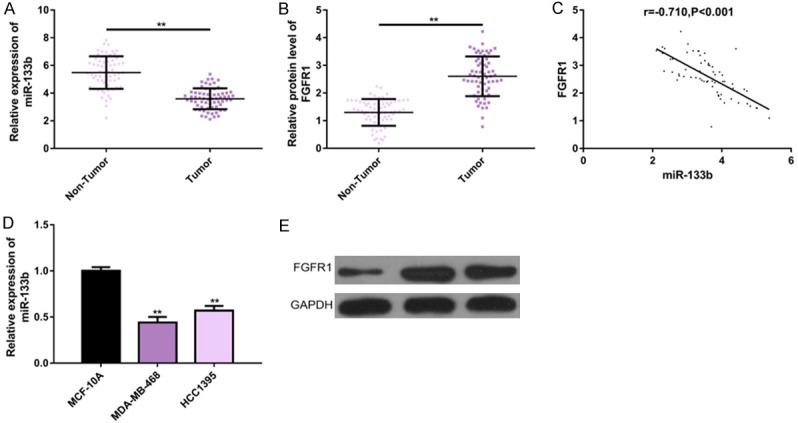
miR-133b and FGFR1 expression in TNBC. A, B. RT-PCR showed that miR-133b was down-regulated while FGFR1 was up-regulated in 65 TNBC tissues. C. miR-133b expression was negatively correlated with FGFR1 expression in TNBC tissues (r=-0.710, P<0.001). D, E. RT-PCR and protein imprinting analysis showed that miR-133b expression was upregulated while FGFR1 protein levels were downregulated in TNBC cell lines. Note: **P<0.01 between groups. Abbreviation: miR, microRNA; FGFR1, fibroblast growth factor receptor 1; TNBC, triple negative breast cancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Ectopic expression of miR-133b inhibited growth of TNBC cells
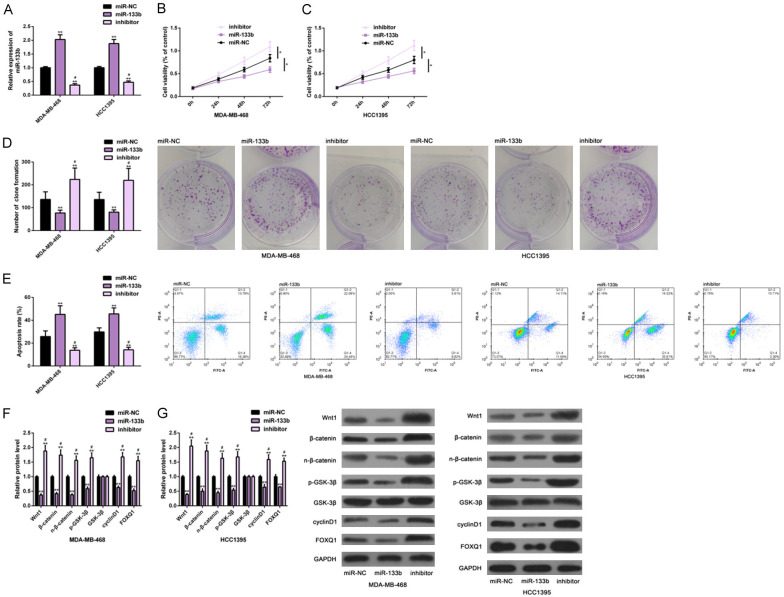
For further understanding the effects of miR-133b on TNBC cells, we conducted cell function experiments. At first, the ectopic expression and down-regulation of miR-133b in the cells were realized by the transfection of miR-133b and inhibitor (Figure 2A). Then, we found that the ectopic expression could remarkably inhibit the proliferation (Figure 2B, 2C) and colony formation (Figure 2D) of cells, induced the apoptosis (Figure 2E), and inactivated the Wnt-β-catenin pathway (Figure 2F, 2G). However, miR-133b down-regulation caused remarkably opposite results, that is, the proliferation (Figure 2B, 2C) and colony formation (Figure 2D) were remarkably enhanced, and the apoptotic rate (Figure 2E) was remarkably reduced, while the Wnt-β-catenin pathway was activated (Figure 2F, 2G). These indicated that the ectopic expression of miR-133b was conducive to inhibiting TNBC cells from growth, while its down-regulation aggravated TNBC, and that the mechanism of action may be associated with the regulation of this pathway (Figure 2).
Figure 2.
Effects of miR-133b on growth of TNBC cells. RT-PCR was used to test the expression of miR-133b in TNBC cells of each group (A); CCK-8 method was used to examine its influence on the proliferation level of TNBC cells (B, C); Colony formation test and flow cytometry were used to examine its influence on colony formation and apoptosis of TNBC cells (D, E); Western blot analysis was used to examine its influence on the Wnt-β-catenin pathway of TNBC cells (F, G). Note: *P<0.05, **P<0.01 vs. miR-NC or between groups; #P<0.05 vs. miR-133b. Abbreviation: miR, microRNA; NC, negative control; TNBC, triple negative breast cancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PI, propidium iodide; Wnt1, wingless-type MMTV integration site family, member1; FOXQ1, Forkhead box Q1.
FGFR1 down-regulation inhibited growth of TNBC cells
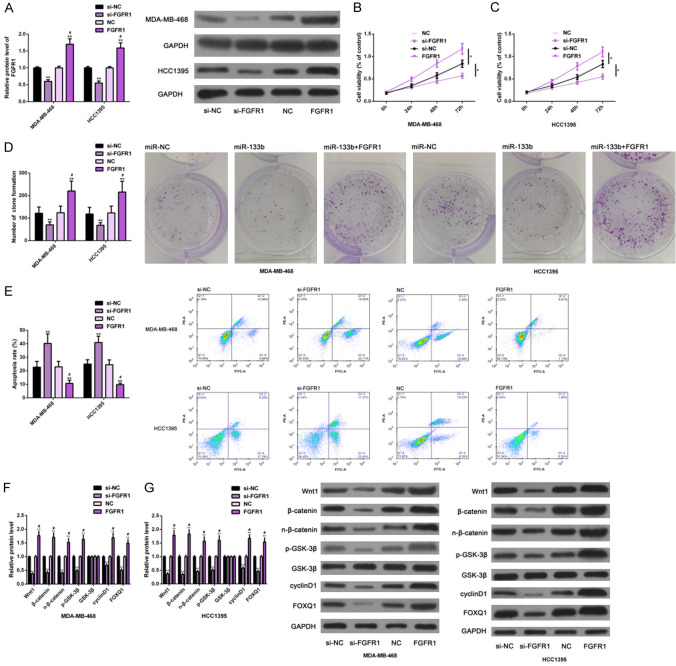
In the same way, we studied the effects of FGFR1 on TNBC cells. At first, FGFR1 was transfected with si-FGFR1 and FGFR1, respectively, to realize gene knockdown and over-expression (Figure 3A). In cell function tests, si-FGFR1 transfection had negative effects on the proliferation (Figure 3B, 3C) and colony formation (Figure 3D) of TNBC cells as well as on the activation of the Wnt-β-catenin pathway (Figure 3E), but it had positive effects on apoptosis (Figure 3F, 3G). However, FGFR1 transfection caused opposite results, namely, the cells’ proliferation (Figure 3B, 3C) and colony formation (Figure 3D) and the pathway’s activation (Figure 3E) were promoted, but the apoptosis (Figure 3F, 3G) was inhibited. All the results were statistically significant (P<0.05). The above results suggested that down-regulation of FGFR1 could inhibit the malignant growth of TNBC cells, while up-regulation of FGFR1 could promote the malignant growth of TNBC cells, which may also be related to its regulation of the Wnt-β-catenin pathway (Figure 3).
Figure 3.
Effects of FGFR1 on growth of TNBC cells. The expression of FGFR1 in TNBC cells of each group was detected by Western blot analysis (A); The effect of FGFR1 on proliferation of TNBC cells was detected by CCK-8 method (B, C); The effect of FGFR1 on colony formation and apoptosis of TNBC cells was detected by colony forming experiment and flow cytometry (D, E); The effect of FGFR1 on the Wnt-β-catenin pathway of TNBC cells was detected by Western blot analysis (F, G). Note: *P<0.05, **P<0.01 vs. si-NC; #P<0.05 vs. si-FGFR1. Abbreviation: FGFR1, fibroblast growth factor receptor 1; TNBC, triple negative breast cancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; si, short interfering; NC, negative control; PI, propidium iodide; Wnt1, wingless-type MMTV integration site family, member1; FOXQ1, Forkhead box Q1.
miR-133b negatively regulated FGFR1
To clarify the molecular mechanism of miR-133b, we verified its relationship with FGFR1. First, we found through the biological analysis that the two had conservative binding sites (Figure 4A). Second, according to the DLRGA, miR-133b only remarkably reduced FGFR1-Wt (P<0.01, Figure 4B, 4C), but had no strong effect on FGFR1-Mut in the two TNBC cell lines (P>0.05, Figure 4B, 4C). In addition, miR-133b could also negatively regulate FGFR1, which was mainly reflected in that up-regulating the former remarkably reduced the expression and protein levels of the latter (P<0.05, Figure 4D, 4E). These suggested that there was a targeted regulatory relationship between miR-133b and FGFR1, and the latter was the direct target of the former (Figure 4).
Figure 4.
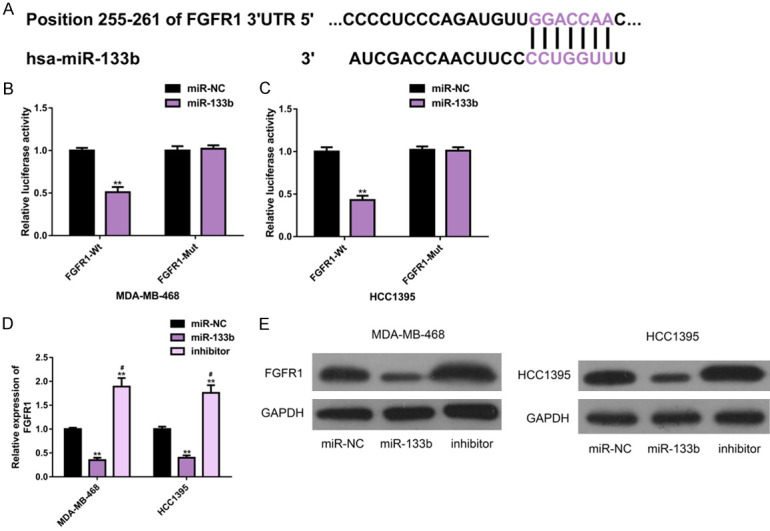
Regulatory effect of miR-133b on FGFR1. A. Targetscan7.2 analysis showed that miR-133b had a conservative binding site with FGFR1. B, C. Double luciferase gene report results. D, E. The effect of miR-133b on FGFR1 protein level and its protein map were analyzed by Western blot. Note: **P<0.01 vs. miR-NC; #P<0.05 vs. miR-133b. Abbreviation: miR, microRNA; NC, negative control; FGFR1, fibroblast growth factor receptor 1; WB, Western blotting; Wt, wild type; Mut, mutant.
Up-regulating miR-133b or down-regulating FGFR1 was conducive to inhibiting DDP resistance of TNBC cells
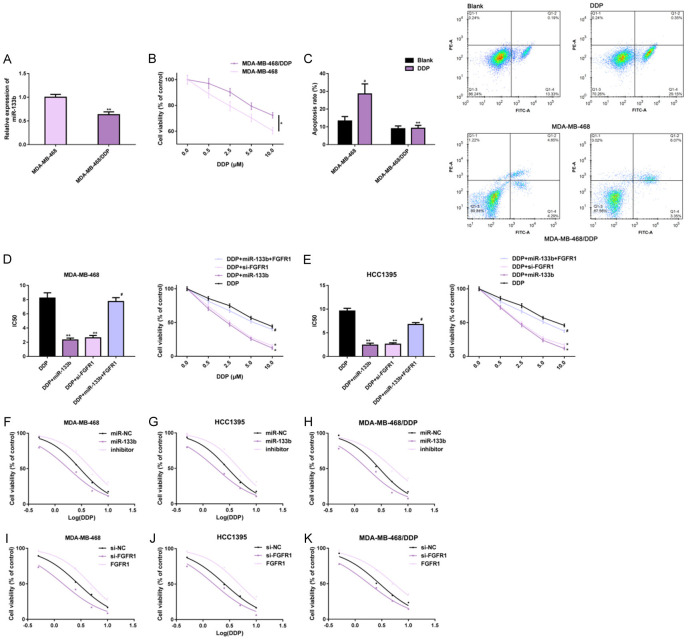
The effects of miR-133b and FGFR1 on DDP resistance in TNBC cells were investigated. miR-133b expression in MDA-MB-468/DDP cells was remarkably lower than that in MDA-MB-468 cells (Figure 5A). Under DDP exposure, the proliferation of both cells was reduced in a dose-dependent manner, while the proliferation levels were still higher in MDA-MB-468/DDP cells (Figure 5B). Under DDP stimulation, the apoptotic levels of MDA-MB-468 cells were remarkably elevated, while those of MDA-MB-468/DDP cells did not change remarkably (Figure 5C). This suggested that MDA-MB-468/DDP cells had better tumor malignancy and DDP resistance than those of MDA-MB-468 cells.
Figure 5.
Effects of miR-133b and FGFR1 on DDP resistance of TNBC cells. RT-PCR, CCK-8 method and flow cytometry were used to detect the expression, proliferation and apoptosis level of miR-133b in MDA-MB-468 and MDA-MB-468/DDP cells (A-C); CCK-8 method was used to analyze cell proliferation and IC50 under DDP intervention (D, E); Dose-response curve was visualized to analyze the effect of miR-133b on chemosensitivity (F-H) and FGFR1 on chemosensitivity (I-K). Note: *P<0.05, **P<0.01 vs. MDA-MB-468 or DDP or between groups; #P<0.05 vs. DDP+miR-133b; aP<0.01 vs. Blank. Abbreviation: miR, microRNA; NC, negative control; FGFR1, fibroblast growth factor receptor 1; TNBC, triple negative breast cancer; DDP, cisplatin; si, short interfering; PI, propidium iodide; IC50, half maximal inhibitory concentration.
Compared with those affected by DDP alone, DDP-exposed MDA-MB-468 and HCC1395 cells, which were affected by miR-133b up-regulation or FGFR1 down-regulation, had remarkably lower proliferation levels and IC50 values in a dose-dependent manner. In addition, the co-expression of miR-133b and FGFR1 remarkably weakened the effects of miR-133b up-regulation, that is, the co-expression of miR-133b and FGFR1 had no significant effect on IC50 (Figure 5D, 5E). These findings revealed that up-regulating miR-133b or down-regulating FGFR1 could interact with DDP to inhibit the cells’ malignant proliferation and enhance their sensitivity to DDP. We also plotted the dose-response curves, and found that DDP could inhibit the proliferation of MDA-MB-468, HCC1395, and MDA-MB-468/DDP cells in a dose-dependent manner. Furthermore, up-regulating miR-133b or down-regulating FGFR1 could inhibit the proliferation of these cells in different degrees, while down-regulating miR-133b or up-regulating FGFR1 could promote the proliferation (Figure 5F-K). This result further confirmed that miR-133b up-regulation or FGFR1 down-regulation is more beneficial to cells survival inhibition and DDP sensitization. All the results were statistically significant (P<0.05) (Figure 5).
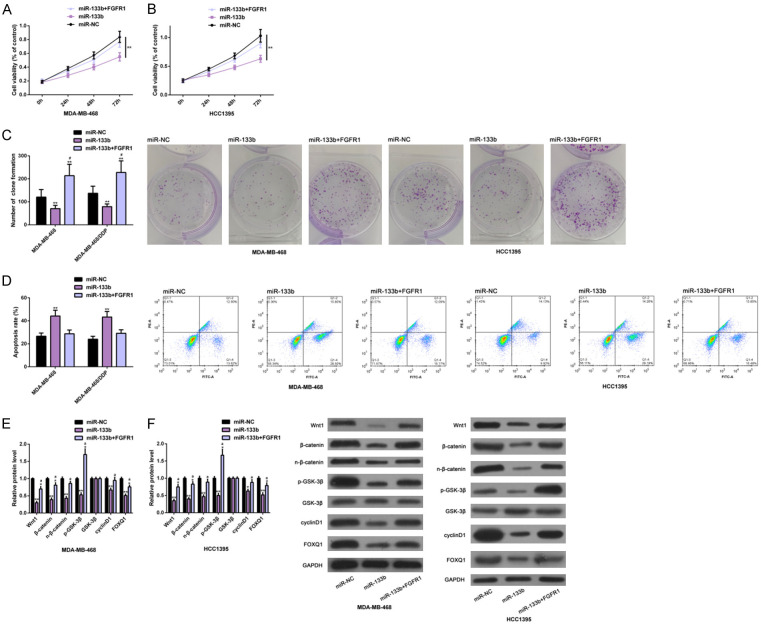
FGFR1 up-regulation could offset effects of ectopic miR-133b expression on growth of TNBC cells
The molecular mechanism of TNBC was further explored by the co-transfection of FGFR1 and miR-133b. The data showed that FGFR1 up-regulation could offset the anti-survival ability of TNBC cells induced by ectopic miR-133b expression. Specifically, the proliferation (Figure 6A, 6B) and colony formation (Figure 6C) of DA-MB-468 and MDA-MB-468/DDP cells were enhanced, while the apoptotic rate (Figure 6D) was remarkably reduced. And the above results were not significantly different from those of miR-NC transfection. In addition, similar effects were found on marker proteins of the Wnt-β-catenin pathway (Figure 6E, 6F). These results indicated that up-regulating FGFR1 could offset the inhibitory effect of ectopic expression of miR-133b on the growth of TNBC cells and the inactivation of the Wnt-β-catenin pathway. All the results were statistically significant (P<0.05) (Figure 6).
Figure 6.
Effects of upregulating FGFR1 on anti-TNBC cell growth induced by ectopic miR-133b expression. CCK-8, colony formation, flow cytometry and Western blot assays were carried out to analyze the proliferation and colony formation (A-C), apoptosis level (D) of MDA-MB-468 and MDA-MB-468/DDP cells and the Wnt-β-catenin pathway (E and F). Note: *P<0.05, **P<0.01 vs. miR-NC or comparison groups; aP<0.05, #P<0.05 vs. miR-133b. Abbreviation: miR, microRNA; NC, negative control; FGFR1, fibroblast growth factor receptor 1; TNBC, triple negative breast cancer; DDP, cisplatin; PI, propidium iodide; Wnt1, wingless-type MMTV integration site family, member1; FOXQ1, Forkhead box Q1.
Discussion
Accounting for 15-20% of BC, TNBC is typically characterized by abnormal deficiencies of estrogen and progesterone receptors [20]. Difficulties in treating this disease are that only 15% of patients have a good response to chemotherapy, while most are at higher risk of tumor recurrence or metastasis [21]. Therefore, it is critical to find out molecular targets for the tumor growth and chemosensitivity of TNBC, which is of great significance for improving the survival of TNBC patients.
In this study, we put forward that the miR-133b-FGFR1 axis has a great influence on the tumor growth and DDP resistance of TNBC. According to the results, both miR-133b and FGFR1 had abnormal expression in TNBC tissues and cells, and the former was abnormally down-regulated while the latter was abnormally up-regulated. Their expression in the tissues was closely and negatively correlated, suggesting that the two may have an antagonistic effect on the pathogenesis of TNBC. Increasing researchers get interested in the expression of miR-133b and FGFR1 in gynecological tumors. For instance, according to Chen et al., miR-133b is regulated by LINC02381 in cervical cancer, and it has an inhibitory effect on the proliferation and migration of cancer cells [22]. As reported by Li et al., miR-133b was remarkably reduced in the tissues and serum of patients with ovarian cancer, and could be regulated by miR-145 to affect the progression of tumor cells [23]. In a study by Ou et al., FGFR1 inhibitors could inhibit the survival of ovarian cancer cells [24]. Suh et al. reported that FGFR1 could stimulate the growth of breast cancer cells in the breast tumor microenvironment [25]. In our research, the ectopic expression of miR-133b or the knockdown of FGFR1 remarkably inhibited the proliferation and colony formation of TNBC cells and promoted apoptosis, accompanied by the inactivation of the Wnt-β-catenin pathway. The reversion of their expression caused opposite results. The cells’ malignant development was remarkably enhanced, and the pathway was remarkably activated. In this study, we evaluated the Wnt-β-catenin pathway by detecting protein levels of Wnt1, β-catenin, cyclinD1, and FOXQ1. It is known that Wnt1 and β-catenin are typical markers of this pathway, and the increase in their protein levels is related to the activation of this pathway and to stem-like characteristics of BC stem cells [26]. cyclinD1 is the downstream target gene of this pathway, and the inhibition of its expression is correlated with the inactivation of the pathway in TNBC cells [18]. Knocking down FOXQ1 helps to inhibit the activity of Wnt-β-catenin signal transduction pathway, whose regulation is mediated by FOXQ1 in the progression of colorectal cancer [27]. As reported by Xie et al., inhibition of the signal transduction of this pathway can inhibit the malignant process of TNBC cells [28]. Additionally, miR-133b regulates this pathway in gastric cancer and glioma [29,30].
AS a potential mechanism, miR-133b also has a targeted relationship with FGFR1, and the expression and protein levels of the latter are negatively regulated by the former. According to previous studies, the miR-133b-FGFR1 axis has a regulatory effect on osteosarcoma and skeletal muscle satellite cells, suggesting that miR-133b and FGFR1 have a certain relationship [31,32]. In our study, it was found that up-regulation of miR-133b or down-regulation of FGFR1 inhibited the proliferation of TNBC cells exposed to DDP and increased DDP sensitivity. In addition, up-regulating FGFR1 counteracted the anti-TNBC cell growth and DDP sensitization caused by ectopic expression of miR-133b. As reported by Lin et al., miR-133b could inhibit DDP resistance in non-small cell lung cancer via a targeted inhibition of glutathione-S-transferase P1 (GSTP1) [33]. According to Jang et al., down-regulating FGFR1 is helpful to enhance the DDP sensitivity of tumor heterogenic transplantation models in vivo [34]. All the above studies are similar to our research results.
Although this study confirmed that the miR-133b-FGFR1-Wnt-β-catenin molecular network can regulate the growth and DDP resistance of TNBC cells, there is still room for improvement. To begin with, it is necessary to search for more upstream target genes of miR-133b, which is of great significance to supplement the molecular regulation of this network. In addition, we can focus on studies of multi-drug resistance and supplement the molecular therapeutic significance of this regulatory network. What’s more, we can study the effect of miR-133b on metastasis and recurrence behavior of TNBC, which is of great value for increasing the potential pleiotropy of miR-133b. Therefore, we will gradually improve this research project based on the above contents.
Conclusion
To sum up, we propose for the first time that miR-133b can inhibit the growth and DDP resistance of TNBC cells by targeting FGFR1 and mediating the Wnt-β-catenin pathway, which may provide new insights for treating TNBC patients.
Acknowledgements
Sichuan Science and Technology Program 2019YFH0173. Major Project of Education Department in Sichuan Province 18ZA0517.
Disclosure of conflict of interest
None.
References
- 1.Muller KE, Marotti JD, Tafe LJ. Pathologic features and clinical implications of breast cancer with HER2 intratumoral genetic heterogeneity. Am J Clin Pathol. 2019;152:7–16. doi: 10.1093/ajcp/aqz010. [DOI] [PubMed] [Google Scholar]
- 2.Ryu JM, Yu J, Kim SI, Kim KS, Moon HG, Choi JE, Jeong J, Do Byun K, Nam SJ, Lee JE, Lee SK, Kim SW. Different prognosis of young breast cancer patients in their 20s and 30s depending on subtype: a nationwide study from the Korean Breast Cancer Society. Breast Cancer Res Treat. 2017;166:833–842. doi: 10.1007/s10549-017-4472-5. [DOI] [PubMed] [Google Scholar]
- 3.Jagadish N, Devi S, Gupta N, Suri V, Suri A. Knockdown of A-kinase anchor protein 4 inhibits proliferation of triple-negative breast cancer cells in vitro and in vivo. Tumour Biol. 2020;42:1010428320914477. doi: 10.1177/1010428320914477. [DOI] [PubMed] [Google Scholar]
- 4.Brady-West DC, McGrowder DA. Triple negative breast cancer: therapeutic and prognostic implications. Asian Pac J Cancer Prev. 2011;12:2139–2143. [PubMed] [Google Scholar]
- 5.Elsawaf Z, Sinn HP. Triple-negative breast cancer: clinical and histological correlations. Breast Care (Basel) 2011;6:273–278. doi: 10.1159/000331643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song J, Ye A, Jiang E, Yin X, Chen Z, Bai G, Zhou Y, Liu J. Reconstruction and analysis of the aberrant lncRNA-miRNA-mRNA network based on competitive endogenous RNA in CESC. J Cell Biochem. 2018;119:6665–6673. doi: 10.1002/jcb.26850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He K, Li WX, Guan D, Gong M, Ye S, Fang Z, Huang JF, Lu A. Regulatory network reconstruction of five essential microRNAs for survival analysis in breast cancer by integrating miRNA and mRNA expression datasets. Funct Integr Genomics. 2019;19:645–658. doi: 10.1007/s10142-019-00670-7. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Gong C, Xu R, Chen Y, Wang X. MicroRNA-5195-3p enhances the chemosensitivity of triple-negative breast cancer to paclitaxel by downregulating EIF4A2. Cell Mol Biol Lett. 2019;24:47. doi: 10.1186/s11658-019-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Oh DY, Leventaki V, Drakos E, Zhang R, Sahin AA, Resetkova E, Edgerton ME, Wu W, Claret FX. MicroRNA-17 acts as a tumor chemosensitizer by targeting JAB1/CSN5 in triple-negative breast cancer. Cancer Lett. 2019;465:12–23. doi: 10.1016/j.canlet.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Alam N, Koul M, Mintoo MJ, Khare V, Gupta R, Rawat N, Sharma PR, Singh SK, Mondhe DM, Gupta PN. Development and characterization of hyaluronic acid modified PLGA based nanoparticles for improved efficacy of cisplatin in solid tumor. Biomed Pharmacother. 2017;95:856–864. doi: 10.1016/j.biopha.2017.08.108. [DOI] [PubMed] [Google Scholar]
- 11.Lee JO, Kang MJ, Byun WS, Kim SA, Seo IH, Han JA, Moon JW, Kim JH, Kim SJ, Lee EJ, In Park S, Park SH, Kim HS. Metformin overcomes resistance to cisplatin in triple-negative breast cancer (TNBC) cells by targeting RAD51. Breast Cancer Res. 2019;21:115. doi: 10.1186/s13058-019-1204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, Liu S, Gan L, Wang J, Hu B, Xu H, Tong R, Yang H, Cristina I, Xue J, Hu X, Lu Y. FGFR1 signaling potentiates tumor growth and predicts poor prognosis in esophageal squamous cell carcinoma patients. Cancer Biol Ther. 2018;19:76–86. doi: 10.1080/15384047.2017.1394541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quintanal-Villalonga A, Molina-Pinelo S, Cirauqui C, Ojeda-Marquez L, Marrugal A, Suarez R, Conde E, Ponce-Aix S, Enguita AB, Carnero A, Ferrer I, Paz-Ares L. FGFR1 cooperates with EGFR in lung cancer oncogenesis, and their combined inhibition shows improved efficacy. J Thorac Oncol. 2019;14:641–655. doi: 10.1016/j.jtho.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Chen G, Liu Z, Liu S, Cai Z, You P, Ke Y, Lai L, Huang Y, Gao H, Zhao L, Pelicano H, Huang P, McKeehan WL, Wu CL, Wang C, Zhong W, Wang F. Aberrant FGFR tyrosine kinase signaling enhances the warburg effect by reprogramming LDH isoform expression and activity in prostate cancer. Cancer Res. 2018;78:4459–4470. doi: 10.1158/0008-5472.CAN-17-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Wang Y, Liu J, Chen Q, Pang D, Jiang Y. Effects of FGFR1 gene polymorphisms on the risk of breast cancer and FGFR1 protein expression. Cell Physiol Biochem. 2018;47:2569–2578. doi: 10.1159/000491653. [DOI] [PubMed] [Google Scholar]
- 16.Bhola NE, Jansen VM, Koch JP, Li H, Formisano L, Williams JA, Grandis JR, Arteaga CL. Treatment of triple-negative breast cancer with TORC1/2 inhibitors sustains a drug-resistant and notch-dependent cancer stem cell population. Cancer Res. 2016;76:440–452. doi: 10.1158/0008-5472.CAN-15-1640-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun WM, Tao W, Li JC, Zhu DM, Miao Y. MicroRNA-296 functions as a tumor suppressor in breast cancer by targeting FGFR1 and regulating the Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:10422–10432. doi: 10.26355/eurrev_201912_19681. [DOI] [PubMed] [Google Scholar]
- 18.Jitariu AA, Cimpean AM, Ribatti D, Raica M. Triple negative breast cancer: the kiss of death. Oncotarget. 2017;8:46652–46662. doi: 10.18632/oncotarget.16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Yan W, Yuan J, Wang Z, Wang C. Nek2B activates the wnt pathway and promotes triple-negative breast cancer chemothezrapy-resistance by stabilizing beta-catenin. J Exp Clin Cancer Res. 2019;38:243. doi: 10.1186/s13046-019-1231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin J, Chen D, Luo K, Lu M, Gu Y, Zeng S, Chen X, Song Y, Zhang Z, Zheng G, He Z, Liu H. Cip2a/miR-301a feedback loop promotes cell proliferation and invasion of triple-negative breast cancer. J Cancer. 2019;10:5964–5974. doi: 10.7150/jca.35704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Wang J, Zheng R, Song B, Huang L, Liu Y, Hao Y, Bai X. MiR-133 targets YES1 and inhibits the growth of triple-negative breast cancer cells. Technol Cancer Res Treat. 2020;19:1533033820927011. doi: 10.1177/1533033820927011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Zhang Z, Ma Y, Su H, Xie P, Ran J. LINC02381 promoted cell viability and migration via targeting miR-133b in cervical cancer cells. Cancer Manag Res. 2020;12:3971–3979. doi: 10.2147/CMAR.S237285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Zhang S, Zou Y, Wu L, Pei M, Jiang Y. miR-145 promotes miR-133b expression through c-myc and DNMT3A-mediated methylation in ovarian cancer cells. J Cell Physiol. 2020;235:4291–4301. doi: 10.1002/jcp.29306. [DOI] [PubMed] [Google Scholar]
- 24.Ou L, He X, Liu N, Song Y, Li J, Gao L, Huang X, Deng Z, Wang X, Lin S. Sialylation of FGFR1 by ST6GalI overexpression contributes to ovarian cancer cell migration and chemoresistance. Mol Med Rep. 2020;21:1449–1460. doi: 10.3892/mmr.2020.10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh J, Kim DH, Lee YH, Jang JH, Surh YJ. Fibroblast growth factor-2, derived from cancer-associated fibroblasts, stimulates growth and progression of human breast cancer cells via FGFR1 signaling. Mol Carcinog. 2020;59:1028–1040. doi: 10.1002/mc.23233. [DOI] [PubMed] [Google Scholar]
- 26.Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, Jin F. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/beta-catenin pathway. J Exp Clin Cancer Res. 2019;38:305. doi: 10.1186/s13046-019-1315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, Wang S, Wei Z. FOXQ1 mediates the crosstalk between TGF-beta and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther. 2015;16:1099–1109. doi: 10.1080/15384047.2015.1047568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie W, Zhang Y, He Y, Zhang K, Wan G, Huang Y, Zhou Z, Huang G, Wang J. A novel recombinant human Frizzled-7 protein exhibits anti-tumor activity against triple negative breast cancer via abating Wnt/beta-catenin pathway. Int J Biochem Cell Biol. 2018;103:45–55. doi: 10.1016/j.biocel.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Yang D, Zhao D, Chen X. MiR-133b inhibits proliferation and invasion of gastric cancer cells by up-regulating FBN1 expression. Cancer Biomark. 2017;19:425–436. doi: 10.3233/CBM-160421. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Zhao G, Zhang Y, Jiang H, Wang W, Zhao D, Hong J, Yu H, Qi L. Mesenchymal stem cell-derived exosomal microRNA-133b suppresses glioma progression via Wnt/beta-catenin signaling pathway by targeting EZH2. Stem Cell Res Ther. 2019;10:381. doi: 10.1186/s13287-019-1446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao G, Tian Z, Zhu HY, Ouyang XY. miRNA-133b targets FGFR1 and presents multiple tumor suppressor activities in osteosarcoma. Cancer Cell Int. 2018;18:210. doi: 10.1186/s12935-018-0696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin CF, Li Y, Ding XB, Li X, Zhang LL, Liu XF, Guo H. lnc133b, a novel, long non-coding RNA, regulates bovine skeletal muscle satellite cell proliferation and differentiation by mediating miR-133b. Gene. 2017;630:35–43. doi: 10.1016/j.gene.2017.07.066. [DOI] [PubMed] [Google Scholar]
- 33.Lin C, Xie L, Lu Y, Hu Z, Chang J. miR-133b reverses cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells. Int J Mol Med. 2018;41:2050–2058. doi: 10.3892/ijmm.2018.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang HS, Woo SR, Song KH, Cho H, Chay DB, Hong SO, Lee HJ, Oh SJ, Chung JY, Kim JH, Kim TW. API5 induces cisplatin resistance through FGFR signaling in human cancer cells. Exp Mol Med. 2017;49:e374. doi: 10.1038/emm.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]