Abstract
Objective: To explore the significance of microRNA-155 (miR-155) and microRNA-46a (miR-46a) in the diagnosis of periodontitis (PD). Methods: A total of 41 patients with PD admitted to our hospital between September 2017 and August 2019 were enrolled into an observation group (obs group), and 35 healthy individuals during the same period were enrolled into a control group (con group). A qRT-PCR assay was carried out to quantify miR-155 and miR-146a expression, and the relationship between the expression of miR-155 and miR-146a and severity and clinical indicators of PD was analyzed. Results: The obs group showed significantly higher expression of saliva miR-155 and miR-146a than the con group (P<0.05), and the expression of them was positively correlated with the severity of their PD. It was also positively correlated with gingival index (GI), attachment loss (AL), plaque index (PLI), probing depth (PD), and bleeding on probing (BOP) of them. Additionally, according to the receiver operating characteristic (ROC) curves, the area under the curve (AUC), specificity, and sensitivity of miR-155 in diagnosing PD were 0.887, 78%, and 97.14%, respectively, and those of miR-146a in diagnosing PD were 0.745, 58.54%, and 88.57%, respectively. Conclusion: MiR-155 and miR-146a were both highly expressed in the saliva of patients with PD, and the expression of them was positively correlated with the severity and clinical indexes of PD, so miR-155 and miR-146a might be involved in the development and progression of PD.
Keywords: miR-155, miR-146a, periodontitis, clinical value, qRT-PCR
Introduction
Periodontitis (PD) is the most common chronic inflammatory disease in the oral cavity and is also one main cause of tooth loss among adults [1]. According to recent research results, PD can not only damage oral cavity, but also give rise to systemic microinflammation of patients. Studies have confirmed that PD is closely related to systemic diseases such as atherosclerosis, diabetes mellitus, lung diseases, digestive tract diseases, adverse pregnancy outcomes, cardiovascular and cerebrovascular diseases, chronic kidney diseases, and Alzheimer’s disease [2-4]. Currently, PD can be intervened with basic treatment, drug treatment, surgical treatment, relatively advanced laser treatment, and immunosuppressive treatment, but those treatments cannot achieve ideal bone tissue regeneration [5].
MiRNAs, as natural gene regulatory factors, regulate target genes at the post-transcriptional level and widely participate in various biological behaviors including cell metabolism, proliferation, differentiation, and apoptosis. They also play a regulatory role in the development of chronic inflammatory diseases [6-8]. MiR-155, as one of the members of the microRNA (miR) family, participates in various biological processes including cell proliferation, immunity, inflammation, as well as tumor and plays a role in inflammatory reactions and autoimmune diseases [9,10]. Recent studies have verified that abnormal expression of miR-155 takes a crucial part in the periodontal pathologic damage, and acts as an important part in the pathological mechanism of periodontal diseases [11]. As a miRNA closely related to inflammation, miRNA-146a has been verified to take a crucial part in immunity, tumor, and inflammation, and is highly expressed in various immune and inflammatory cells such as T lymphocytes, monocytes, and macrophages [12,13]. One study has found that miRNA-146a is highly expressed in synovioblasts and synovial tissues of cases with rheumatoid arthritis, which aggravates inflammatory response [14]. In contrast, miRNA-146a is lowly expressed in cases with systemic lupus erythematosus, which alleviates the immune inflammatory response [15]. There is little research on the expression of miR-155 and miRNA-146a in the saliva of patients with PD.
Therefore, this study determined and compared the expression of saliva miR-155 and miR-146a between patients with PD and healthy individuals, and analyzed the correlation between the expression of them and the severity and clinical indexes of PD, with the goal of exploring the clinical value of saliva miR-155 and miR-146a.
Materials and methods
Research objects
A total of 41 patients with PD in Beijing Stomatological Hospital, Capital Medical University from September 2017 to August 2019 were enrolled and assigned to the observation group (obs group), including 25 males and 16 females, with a mean age of 29.78±6.91 years. In addition, 35 healthy individuals in our hospital over the same period were enrolled and assigned to a control group (con group), including 18 males and 17 females, with a mean age of 28.59±6.81 years. The inclusion criteria of the patients: Patients meeting the diagnostic criteria of PD, and patients with complete clinical data. The study was approved by the Ethics Committee of Beijing Stomatological Hospital, Capital Medical University. All patients and their families agreed to participate in the study after being informed of the study, and signed informed consent forms. The exclusion criteria of the patients: Women during pregnancy, lactation or menstruation; patients with comorbid infectious diseases; patients with comorbid systemic diseases; patients who had taken antibacterial agents or immunomodulatory drugs within six months before being enrolled; patients who had received periodontal therapy within 3 months before being enrolled; patients with less than 15 retained teeth or missing full dentition; patients with a history of smoking or drinking; patients who had received periodontal treatment; patients who had undergone surgery or suffered from trauma in the past 3 months; and those with renal insufficiency, severe hypertension, respiratory tract disease, peripheral angiopathy or thrombotic diseases.
Quantification of miR-155 and miR-146a in saliva through qRT-PCR assay
Cotton swabs stained with citric acid were used to stimulate the oral cavity of each patient in the morning before treatment and at 3 months after treatment, and the patient was required to spit out saliva 2 min later. The saliva was stored in 10 ml centrifuge tubes without RNA enzyme to collect saliva specimens. Total RNA was obtained from the saliva in accordance with the procedure of a mirVana kit (Thermo Fisher Scientific), and miRNA was reversely transcribed into cDNA in accordance with procedures of PrimeScriptRTreagent reverse transcription kit (TakaRa), followed by an immediate fluorescence quantitative PCR in 7500 PCR instrument (Life Technologies). The conditions were as follows: 95°C for 2 min, followed by 35 cycles of 95°C for 5 s, 62°C for 10 s, and 72°C for 30 s, and then 95°C for 1 min, 62°C for 1 min, and 95°C for 15 s. Standardization was carried out using endogenous U6 small nuclear RNA (U6). Primers were all designed by Shanghai Sangon Biotech Co., Ltd. The experimental data were analyzed by 2-ΔΔCT method. The sequences of PCR primers are presented in Table 1.
Table 1.
Primer sequences of miR-155 and miR-146a for qRT-PCR assay
| Item | Forward primer | Reverse primer |
|---|---|---|
| MiR-155 | 5’-GGGCCTTCCCTGGAACAGGAGTCT-3’ | 5’-GGGAGATTCATGGTATCAAGCACCC-4’ |
| MiR-146a | 5’-GTGCAGGGTCCGAGGT-3’ | 5’-CAACACCAGTCGATGGGCTGT-4’ |
| U6 | 5’-CTCGCTTCGGCAGCACA-3’ | 5’-AACGCTTCACGAATTTGCGT-3’ |
Outcome measures
The PD severity of patients in the obs group was graded according to the indexes recommended by the Centers for Disease Control/American Academy of Periodontology (CDC/AAP): Mild: There were two adjacent sites with clinical attachment level (CAL) ≥3 mm or more on different teeth or two adjacent sites with probing depth (PD) ≥4 mm or more on different teeth, or there was one site on one tooth with PD ≥5 mm; Moderate: There were two adjacent sites with CAL ≥4 mm or more on different teeth or two adjacent sites with PD ≥5 mm or more on different teeth; Severe: There were two adjacent sites with CAL ≥6 mm or more on different teeth or there was one adjacent site with PD ≥5 mm or more. Patients with mild or moderate PD were enrolled into the same group. The same physician was arranged to examine and record the periodontal histological indexes of all remained teeth in each patient before intervention and at 3 months after intervention, including gingival index (GI), plaque index (PLI), probing depth (PD), as well as attachment loss (AL), and the readings of PD, AL, and bleeding on probing (BOP) were all obtained at six sites for every tooth, distobuccal (DB), midbuccal (B), mesiobuccal (MB), distolingual (DL), midlingual (L) as well as mesiolingual (ML) sites, and averaged as final results.
Statistical analyses
The obtained data were analyzed statistically with SPSS22.0. Measurement data were expressed as the mean ± standard deviation (x̅ ± sd), and compared with the t test. Enumeration data were expressed as rate, and analyzed using the χ2 test. In addition, Spearman’s rank correlation coefficient was adopted to evaluate the correlation between the expression of miR-155 and miR-146a and disease severity. P<0.05 indicates a significant difference.
Results
Comparison of general data
Comparison of general data between the obs group and the con group showed that there was no significant difference between them in general data such as red blood cell (RBC), body mass index (BMI), sex, age, primed lymphocyte typing (PLT), systolic blood pressure, diastolic blood pressure, Hb, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) (all P>0.05) (Table 2).
Table 2.
General data of the two groups
| Factor | The observation group (n=41) | The control group (n=35) | T/χ2 value | P-value |
|---|---|---|---|---|
| Sex | 0.701 | 0.403 | ||
| Male | 25 (86.21) | 18 (75) | ||
| Female | 16 (55.17) | 17 (70.83) | ||
| Age (Y) | 29.78±6.91 | 28.59±6.81 | 0.753 | 0.454 |
| BMI (Kg/m) | 23.87±2.81 | 23.69±2.72 | 0.283 | 0.778 |
| RBC (×1012/L) | 4.98±0.23 | 5.09±0.57 | 1.133 | 0.261 |
| PLT (×109/L) | 154.35±14.69 | 155.45±13.98 | 0.333 | 0.740 |
| Systolic blood pressure (mmHg) | 118.41±10.34 | 121.35±11.31 | 1.183 | 0.241 |
| Diastolic blood pressure (mmHg) | 76.89±12.43 | 77.78±12.14 | 0.312 | 0.754 |
| Hb (g/dl) | 14.46±0.87 | 14.72±0.82 | 1.333 | 0.187 |
| ALT (U/L) | 21.89±8.16 | 22.03±9.28 | 0.070 | 0.944 |
| AST (U/L) | 17.78±7.45 | 18.09±7.43 | 0.181 | 0.857 |
Comparison of clinical indexes of patients in the two groups
Comparison of clinical indexes between the two groups showed that the levels of PLI, GI, PD, and AL in patients with PD from the obs group were all significantly higher than those in healthy individuals from the con group (all P<0.001, Table 3).
Table 3.
Comparison of clinical indexes of patients in the two groups
| Item | PLI | PD (mm) | AL (mm) | GI |
|---|---|---|---|---|
| The observation group (n=41) | 0.93±0.05 | 6.41±0.15 | 6.46±0.55 | 3.35±0.51 |
| The control group (n=35) | 0.56±0.08 | 2.23±0.11 | 0.23±0.04 | 1.79±0.37 |
| t/χ2 value | 24.540 | 136.400 | 66.800 | 15.030 |
| P | <0.001 | <0.001 | <0.001 | <0.001 |
Comparison of the expression of saliva miR-155 and miR-146a
Comparison of the expression of saliva miR-155 and miR-146a between the obs group and the con group showed that the expression of them in the obs group was significantly higher than that in the con group (P<0.001, Table 4).
Table 4.
Comparison of the expression of saliva miR-155 and miR-146a
| Item | MiR-155 | MiR-146a |
|---|---|---|
| The observation group (n=41) | 1.73±0.54 | 2.24±0.53 |
| The control group (n=35) | 0.93±0.32 | 1.83±0.26 |
| t/χ2 value | 7.784 | 4.166 |
| P | <0.001 | <0.001 |
Correlation between the expression of saliva miR-155 and miR-146a and the severity of PD
According to the severity of PD in patients from the obs group, the mild PD, moderate PD, and severe PD were labeled as 1, 2, and 3, respectively, and then Spearman’s rank correlation coefficient was used for the analysis of the correlation between the PD severity and the expression of miR-155 and miR-146a. It came out that the expression of saliva miR-155 and miR-146a in patients with PD was positively correlated with the PD severity (r=0.657, 0.763, P<0.05) (Table 5 and Figure 1).
Table 5.
Correlation between the expression of miR-155 and miR-146a and the periodontitis severity
| r | 95% CI | P | |
|---|---|---|---|
| MiR-155 | 0.657 | 0.439-0.803 | <0.001 |
| MiR-146a | 0.763 | 0.595-0.867 | <0.001 |
Figure 1.
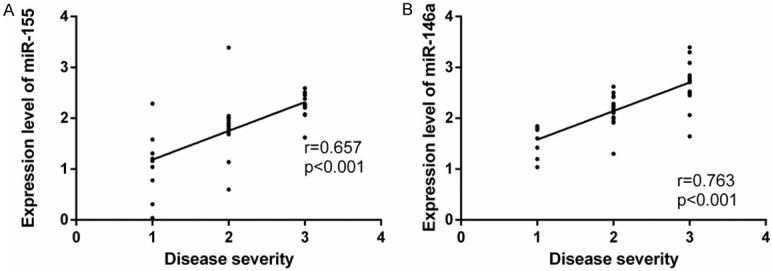
Correlation between the expression of miR-155 and miR-146a in the saliva of patients and the severity of their periodontitis. Analysis of the correlation between the expression of miR-155 in the saliva of patients and the severity of periodontitis. A: The expression of miR-155 in the saliva of patients was positively correlated with the severity of periodontitis (r=0.657, P<0.001). Analysis of the correlation between the expression of miR-146a in the saliva of patients and the severity of periodontitis. B: The expression of miR-146a in the saliva of patients was positively correlated with the severity of periodontitis (r=0.763, P<0.001).
Correlation between the expression of saliva miR-155 and miR-146a and clinical indexes of PD
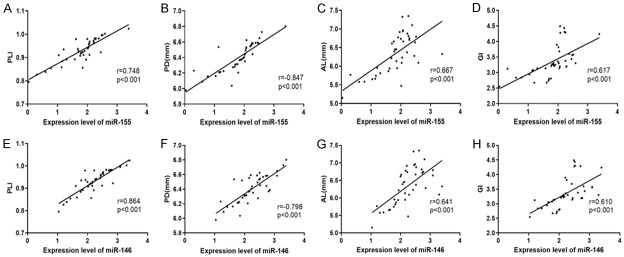
According to Spearman’s rank correlation coefficient on the relation between the expression of saliva miR-155 and miR-146a and clinical indexes of PD, the expression of them in patients with PD was positively correlated with PLI, GI, PD, and AL (Figure 2).
Figure 2.
Correlation between the expression of miR-155 and miR-146a in the saliva of patients and the clinical indexes of periodontitis. Analysis of the correlation between the expression of miR-155 in the saliva of patients with periodontitis and PLI. A: The expression of miR-155 in the saliva of patients with periodontitis was positively correlated with PLI (r=0.748, P<0.001). Analysis of the correlation between the expression of miR-155 in the saliva of patients with periodontitis and PD. B: The expression of miR-155 in the saliva of patients with periodontitis was positively correlated with PD (r=0.846, P<0.001). Analysis of the correlation between the expression of miR-155 in the saliva of patients with periodontitis and AL. C: The expression of miR-155 in the saliva of patients with periodontitis was positively correlated with AL (r=0.667, P<0.001). Analysis of the correlation between the expression of miR-155 in the saliva of patients with periodontitis and GI. D: The expression of miR-155 in the saliva of patients with periodontitis was positively correlated with GI (r=0.617, P<0.001). Analysis of the correlation between the expression of miR-146a in the saliva of patients with periodontitis and PLI. E: The expression of miR-146a in the saliva of patients with periodontitis was positively correlated with PLI (r=0.864, P<0.001). Analysis of the correlation between the expression of miR-146a in the saliva of patients with periodontitis and PD. F: The expression of miR-146a in the saliva of patients with periodontitis was positively correlated with PD (r=0.798, P<0.001). Analysis of the correlation between the expression of miR-146a in the saliva of patients with periodontitis and AL. G: The expression ofmiR-146a in the saliva of patients with periodontitis was positively correlated with AL (r=0.641, P<0.001). Analysis of correlation between the expression of miR-146a in the saliva of patients with periodontitis and GI. H: The expression of miR-146a in the saliva of patients with periodontitis was positively correlated with GI (r=0.610, P<0.001).
Diagnostic value of saliva miR-155 and miR-146a for PD
The receiver operating characteristic (ROC) curves of the expression of miR-155 and miR-146a in diagnosing PD showed that the area under the curve (AUC) of miR-155 in saliva in the diagnosis was 0.887 (95% CI: 0.804-0.970), and the specificity, sensitivity, and cut-off value of it were 78%, 97.14%, and 1.602, respectively, while the AUC of miR-146a in saliva in the diagnosis was 0.745 (95% CI: 0.633-0.858), and the specificity, sensitivity, and cut-off value of it were 58.54%, 88.57%, and 2.151, respectively (Table 6 and Figure 3).
Table 6.
Diagnostic value of saliva miR-155 and miR-146a for PD
| Index | AUC | 95% CI | Specificity (%) | Sensitivity (%) | Cut-off |
|---|---|---|---|---|---|
| MiR-155 | 0.887 | 0.804-0.970 | 78.00 | 97.14 | 1.602 |
| miR-146a | 0.745 | 0.633-0.858 | 58.54 | 88.57 | 2.151 |
Figure 3.
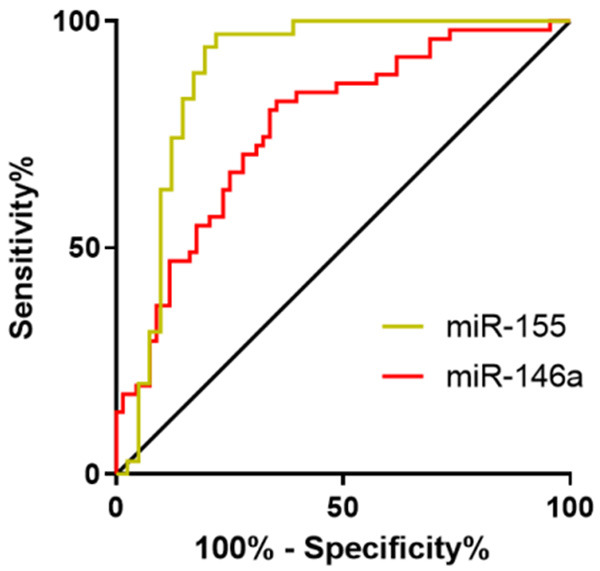
Diagnostic value of miR-155 and miR-146a in the saliva for periodontitis. The AUC of miR-155 for diagnosing periodontitis was 0.887 (95% CI: 0.804-0.970), and the specificity, sensitivity, and cut-off value of it were 78%, 97.14%, and 1.602, respectively, while the AUC of miR-146a in saliva in diagnosing periodontitis was 0.745 (95% CI: 0.633-0.858), and the specificity, sensitivity, and cut-off value of it were 58.54%, 88.57%, and 2.151, respectively.
Discussion
PD is a specific chronic inflammatory disease. Plaque and tartar in the mouth of patients with gingivitis or PD press and stimulate gingival tissue for a long time, which can give rise to inflammatory edema in different degrees in periodontal supporting tissue and compromise the health of periodontal tissue in patients [16,17].
Although the predisposing factor of PD is the destruction of periodontal tissue by pathogenic microorganisms, the immune mechanism of inflammation in patients plays an essential role in the development of PD. Under the long-term repeated stimulation of pathogenic microorganisms, periodontal tissue and cells will generate excessive reactive oxygen species, thus forming oxidative stress. Such a mechanism can eventually damage periodontal tissue through release of massive inflammatory factors and hydrolases and cause direct destruction to tissue biomacromolecules [18,19]. In addition, inflammation affects the function of stem cells in bone regeneration, which is the main difficulty in the treatment of periodontal diseases. MiRNA not only regulates cell metabolism, proliferation, differentiation, and apoptosis, but also regulates inflammatory response [20], so research on miRNA provides a good direction for understanding this pathological process [21]. In the present study, we quantified miR-155 and miR-146a in the saliva of patients with PD and healthy individuals, finding that the expression of them in the obs group was significantly higher than that in the con group. Earlier studies have also shown that miR-155 and miR-146a are abnormally expressed in patients with PD. For instance, Xie et al. [22] detected miRNA in patients with PD and healthy individuals through RT-PCR, and found that miR-146a expression in the two kinds of individuals was significantly different, and it was up-regulated in patients with PD. Moreover, Sipert et al. [23] have quantified miR-146a and miR-155 in the dental pulp, gingiva, and periodontal membrane fibers of the same individual, and revealed that under the stimulation of lipopolysaccharide, miR146a and miR155 in gingival fibers increased significantly compared with periodontal membrane fibers. However, the number of samples they studied was small and the expression of saliva miR-146a and miR-155 was not analyzed.
In the present study, we analyzed the correlation between the expression of saliva miR-155 and miR-146a in patients with PD and the severity of their PD, finding that the expression of saliva miR-155 in the patients was positively correlated with the severity of their PD to a certain extent (r=0.657), but the correlation was weaker than that between the expression of saliva miR-146a and the severity of PD (r=0.763), which implies that the expression of saliva miR-155 and miR-146a in patients with PD may be closely related to the severity of PD. In recent years, there is little research on the relationship between the expression of miR-155 and miR-146a and PD. Motedayyen et al. [24] studied 20 patients with PD and 10 healthy individuals, and found that the expression of miR-146a in gingival tissue specimens from patients with PD was significantly higher than that in gingival tissue specimens from healthy individuals, and the expression was related to clinical indexes such as PD and AL and inflammatory factors. According to our Pearson correlation analysis on the relation between the expression of saliva miR-155 and miR-146a and clinical indexes of PD, the expression of them in patients with PD was positively correlated with PLI, PI, PD, and AL, which suggests that up-regulated miR-155 and miR-146a in oral local secretions may also be one of the mechanisms of PD progression.
The diagnosis level of early PD is relatively poor, because the majority of patients do not have obvious clinical symptoms in the early clinical stage, and the diagnosis of it is relatively difficult due to costly examination including oral endoscopy [25,26]. Detection of saliva indexes takes an important part in the diagnosis of infectious diseases. The detection is relatively convenient, economical, and highly operable, so it is easy to popularize. At the end of the study, according to the ROC curves of miR-155 and miR-146a expression in the diagnosis of PD, the AUC, specificity, and sensitivity of miR-155 in diagnosing PD were 0.887, 78%, and 97.14%, respectively, and those of miR-146a in diagnosing PD were 0.745, 58.54%, and 88.57%, respectively, which suggests that both of them have certain value in the diagnosis of PD.
This study has provided clues for the roles of miR-155 and miR-146a in PD lesions by comparatively analyzing the expression of saliva miR-155 and miR-146a in PD patients and healthy individuals and exploring its correlation with severity and clinical indexes of PD, as well as the diagnostic value of them for PD. However, the specific mechanism is still unclear, and miR-155 and miR-146a in other types of specimens from PD patients have not been studied. It is hoped that these deficiencies can be addressed in future research.
In conclusion, miR-155 and miR-146 were both highly expressed in the saliva of cases with PD, and the expression of them was positively correlated with the severity and clinical indexes of PD, somiR-155 and miR-146 might participate in the development and progression of PD.
Disclosure of conflict of interest
None.
References
- 1.Wang Q, Kang J, Cai X, Wu Y, Zhao L. The association between chronic periodontitis and vasculogenic erectile dysfunction: a systematic review and meta-analysis. J Clin Periodontol. 2016;43:206–215. doi: 10.1111/jcpe.12512. [DOI] [PubMed] [Google Scholar]
- 2.Alfakry H, Malle E, Koyani CN, Pussinen PJ, Sorsa T. Neutrophil proteolytic activation cascades: a possible mechanistic link between chronic periodontitis and coronary heart disease. Innate Immun. 2016;22:85–99. doi: 10.1177/1753425915617521. [DOI] [PubMed] [Google Scholar]
- 3.Gaur S, Agnihotri R. Alzheimer’s disease and chronic periodontitis: is there an association? Geriatr Gerontol Int. 2015;15:391–404. doi: 10.1111/ggi.12425. [DOI] [PubMed] [Google Scholar]
- 4.Ma L, Chu WM, Zhu J, Wu YN, Wang ZL. Interleukin-1beta (3953/4) C→T polymorphism increases the risk of chronic periodontitis in Asians: evidence from a meta-analysis of 20 case-control studies. Arch Med Sci. 2015;11:267–273. doi: 10.5114/aoms.2015.50961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dosseva-Panova VT, Popova CL, Panov VE. Subgingival microbial profile and production of proinflammatory cytokines in chronic periodontitis. Folia Med (Plovdiv) 2014;56:152–160. doi: 10.2478/folmed-2014-0022. [DOI] [PubMed] [Google Scholar]
- 6.Segal M, Slack FJ. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov. 2020;15:987–992. doi: 10.1080/17460441.2020.1765770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang N, Li Y, Wang G, Ding Y, Jin Y, Xu Y. Tumor necrosis factor-alpha suppresses adipogenic and osteogenic differentiation of human periodontal ligament stem cell by inhibiting miR-21/Spry1 functional axis. Differentiation. 2017;97:33–43. doi: 10.1016/j.diff.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suo T, Chen GZ, Huang Y, Zhao KC, Wang T, Hu K. miRNA-1246 suppresses acute lung injury-induced inflammation and apoptosis via the NF-kappaB and Wnt/beta-catenin signal pathways. Biomed Pharmacother. 2018;108:783–791. doi: 10.1016/j.biopha.2018.09.046. [DOI] [PubMed] [Google Scholar]
- 10.Song J, Hu Y, Jiang X, Zhu W, Wu Z, Dong S. Profiling of novel microRNAs elicited by EV71 and CA16 infection in human bronchial epithelial cells using high-throughput sequencing. Virus Res. 2018;247:111–119. doi: 10.1016/j.virusres.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Al-Rawi NH, Al-Marzooq F, Al-Nuaimi AS, Hachim MY, Hamoudi R. Salivary microRNA 155, 146a/b and 203: a pilot study for potentially non-invasive diagnostic biomarkers of periodontitis and diabetes mellitus. PLoS One. 2020;15:e0237004. doi: 10.1371/journal.pone.0237004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. 2008;36:1211–1215. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- 13.Li YY, Cui JG, Dua P, Pogue AI, Bhattacharjee S, Lukiw WJ. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci Lett. 2011;499:109–113. doi: 10.1016/j.neulet.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavasolian F, Hosseini AZ, Soudi S, Naderi M. miRNA-146a improves immunomodulatory effects of MSC-derived exosomes in rheumatoid arthritis. Curr Gene Ther. 2020;20:297–312. doi: 10.2174/1566523220666200916120708. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 16.Miller K, Treloar T, Guelmann M, Rody WJ Jr, Shaddox LM. Clinical characteristics of localized aggressive periodontitis in primary dentition. J Clin Pediatr Dent. 2018;42:95–102. doi: 10.17796/1053-4628-42.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munz M, Chen H, Jockel-Schneider Y, Adam K, Hoffman P, Berger K, Kocher T, Meyle J, Eickholz P, Doerfer C, Laudes M, Uitterlinden A, Lieb W, Franke A, Schreiber S, Offenbacher S, Divaris K, Bruckmann C, Loos BG, Jepsen S, Dommisch H, Schaefer AS. A haplotype block downstream of plasminogen is associated with chronic and aggressive periodontitis. J Clin Periodontol. 2017;44:962–970. doi: 10.1111/jcpe.12749. [DOI] [PubMed] [Google Scholar]
- 18.Nazir MA. Prevalence of periodontal disease, its association with systemic diseases and prevention. Int J Health Sci (Qassim) 2017;11:72–80. [PMC free article] [PubMed] [Google Scholar]
- 19.Holick MF. The vitamin D deficiency pandemic: approaches for diagnosis, treatment and prevention. Rev Endocr Metab Disord. 2017;18:153–165. doi: 10.1007/s11154-017-9424-1. [DOI] [PubMed] [Google Scholar]
- 20.Bartold PM, Van Dyke TE. Host modulation: controlling the inflammation to control the infection. Periodontol 2000. 2017;75:317–329. doi: 10.1111/prd.12169. [DOI] [PubMed] [Google Scholar]
- 21.Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie YF, Shu R, Jiang SY, Liu DL, Zhang XL. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int J Oral Sci. 2011;3:125–134. doi: 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sipert CR, Morandini AC, Dionisio TJ, Trachtenberg AJ, Kuo WP, Santos CF. MicroRNA-146a and microRNA-155 show tissue-dependent expression in dental pulp, gingival and periodontal ligament fibroblasts in vitro. J Oral Sci. 2014;56:157–164. doi: 10.2334/josnusd.56.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motedayyen H, Ghotloo S, Saffari M, Sattari M, Amid R. Evaluation of microRNA-146a and its targets in gingival tissues of patients with chronic periodontitis. J Periodontol. 2015;86:1380–1385. doi: 10.1902/jop.2015.150319. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S. Evidence-based update on diagnosis and management of gingivitis and periodontitis. Dent Clin North Am. 2019;63:69–81. doi: 10.1016/j.cden.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Wu YC, Ning L, Tu YK, Huang CP, Huang NT, Chen YF, Chang PC. Salivary biomarker combination prediction model for the diagnosis of periodontitis in a Taiwanese population. J Formos Med Assoc. 2018;117:841–848. doi: 10.1016/j.jfma.2017.10.004. [DOI] [PubMed] [Google Scholar]