Abstract
Circular RNAs (circRNAs) in exosomes exhibit stable expression and are not easily degraded in plasma; a characteristic that makes them ideal as novel non-invasive tumor diagnostic markers. In this study, we examined different expression of circRNA in plasma exosomes of primary hepatocellular carcinoma patient and healthy volunteer by full transcriptome sequencing. Five circRNAs with up-regulated expression were selected, and large sample size verified their expression. Among them, it is further confirmed that exo_circ_0006602 is up-regulated in the large sample cohort. In addition, the expression level of exo_circ_0006602 was correlated with HBsAg (P<0.011), HBeAg (P=0.048), liver cirrhosis (P=0.001) and Edmondson-Steiner grade (P<0.001). The receiver operating characteristic (ROC) was used to evaluate the accuracy of exo_circ_0006602 as a diagnostic marker. The AUC value of exo_circ_0006602 was significantly highter than common serum tumor markers AFP and CEA. Exo_circ_0006602 combined with AFP can significantly improve the diagnostic accuracy. Cell function experiments show that exo_circ_0006602 can significantly improve the proliferation and invasion ability of liver cancer cell lines and also promoted the expression of tumor proliferation-related protein Snail. In conclusion, our results suggested that exo_circ_0006602 can be used as a potential non-invasive biomarker for the early diagnosis and screening of liver cancer, the sensitivity and specificity of diagnosis are higher than traditional tumor markers.
Keywords: Exosome, circRNA, hepatocellular carcinoma, diagnostic biomarker
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the fourth leading cause of cancer-related deaths worldwide. China has a high incidence of liver cancer, and more than half of all deaths from liver cancer globally occur in the country. Liver cancer patients usually do not show obvious clinical symptoms during the early stages of the disease, and this makes it difficult to diagnose early. Most patients are usually diagnosed when the condition is already at an advanced stage. Besides, after diagnosis, only 15% to 20% of patients do qualify for surgery [1]. Currently, the screening methods for early hepatocellular carcinoma are mainly medical imaging combined with tumor markers, such as alpha-fetoprotein (AFP) combined with either computed tomography (CT) or magnetic resonance imaging (MRI). However, these methods have shortcomings. For example, the positive rate of AFP expression in early-stage liver cancer is low, and about 30% of patients with liver cancer have negative AFP expression [2]. Therefore, there is a need to identify diagnostic markers with high sensitivity and specificity, which can be useful in timely diagnosis and treatment of liver cancer.
Exosomes are extracellular vesicles with a diameter of 30 to 150 nm. Most cells produce many exosomes, and therefore they are widely distributed in various body fluids, including plasma, urine, milk, and saliva [3]. Exosomes contain cell-specific lipids, antigens, and different nucleic acids such as messenger RNA (mRNA), microRNA (miRNA), long non-coding RNA (LncRNA), and circular RNA (circRNA). Exosomes have a lipid bilayer membrane structure, which can protect nucleic acids from degradation by extracellular RNases. The molecular composition of exosomes is stable and tissue-specific, which can reflect the state of the tissue of origin. And this stability makes them potentially useful as biomarkers [4].
Exosomes are the main medium of communication between cells, because exosomes can be captured by neighboring cells [5]. Studies have found that exosome-mediated transfer of specific small RNAs from stromal cells to epithelial cancer cells contributes to cancer progression [6]. Exosomes can be secreted by a variety of cells and can be detected in a variety of body fluids, which is considered an ideal source of biomarkers [7-9]. People are very concerned about the role of exosomes in the development of cancer, especially in the non-invasive diagnosis of cancer.
Circular RNAs (circRNAs) are recently identified novel kind of endogenous non-coding RNAs. They are produced by reverse splicing of a single-stranded RNA loop covalently closed at the 3’5’-phosphodiester linkage site of the precursor mRNA. They are more stable than linear RNA, which makes them potentially useful as diagnostic markers [10-12]. Some studies have found that circular RNA can be used as a diagnostic marker for tumors [13] and non-tumor tumors [14], which is more sensitive than common predictors and has a wide range of sources.
The current study aimed to assess the expression of Exo_circ_0006602 in plasma exosomes and evaluate its value as an early diagnostic marker.
Materials and methods
Sample collection
This study was conducted in accordance with the Helsinki Declaration and was approved by the Ethics Committee of Qilu Hospital of Shandong University (SDULCLL2019-1-17). Blood samples were collected from six patients with hepatocellular carcinoma from the Qilu Hospital of Shandong University in June 2018. All the patients met the characteristics of early-stage hepatocellular carcinoma (HCC) based on Barcelona Clinic Liver Cancer (BCLC) staging system. In addition, blood samples from 87 patients with HCC were collected from Qilu Hospital of Shandong University from July 2018 to July 2019. And none of these patients received chemotherapy and radiation therapy. From August 2020 to September 2020, blood samples of 13 patients with gastric cancer and 15 patients with cholangiocarcinoma were collected from Qilu Hospital of Shandong University. The pathological type was confirmed by histopathology. Also, controlled blood samples were collected from 30 healthy volunteers. All the blood samples were placed in EDTA anticoagulation tubes and centrifuged at 500 g for 5 min. Next, the supernatant was collected, and the remaining sample centrifuged again at 2000 g for 15 min to get more supernatant. The supernatant was placed in RNase-free tubes and store at -80°C.
Exosome extraction
ExoEasy Maxi Kit (Qiagen) was used to separate exosomes from plasma and cell culture medium following the manufacturers’ protocol. The XBP buffer was added to the sample in equal volumes and mixed thoroughly. The sample/XBP buffer mixture was applied onto exoEasy spin column and centrifuged at 500 g for 1 minute. Next, the flow-through was discarded and the spin column was placed in the same collection tube. A total of 10 ml of XWP buffer was added on the column and centrifuge at 5000 g for 5 minutes to remove the residual volume in the spin column. Subsequently, all the flow-through from the collection tube was discarded and the spin column was transferred to a new collection tube. Then, 400 μl-1 ml of XE buffer was added to the membrane and incubated for 1 minute. Finally, the column was centrifuged at 500 g for 5 minutes and the elute collected.
Exosomes identification
The transmission electron microscope was used to observe the morphological characteristics of exosomes extracted from plasma. Briefly, exosomes were fixed with osmic acid, washed with phosphate-buffered saline (PBS), dehydrated, and stained with uranyl acetate for 10 min, then tested on the machine. Detection of exosomal Nanoparticle Tracking Analysis (NTA) using Malvern nanoparticle size analyzer. Exosome specific markers CD63 and CD81 expression detected by Western blotting.
Circular RNA profiling analysis
Transcriptome high throughput sequencing was performed by CloudSeq Biotech (Shanghai, China). Ribosomal RNAs (rRNAs) were isolated from total RNAs using NEBNext® rRNA Depletion Kit (New England Biolabs, Inc., Massachusetts, USA) following the manufacturer’s instructions. The RNA libraries were constructed using rRNA-depleted RNAs with TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA) according to the manufacturer’s instructions. Libraries were screened for quality and quantified using the BioAnalyzer 2100 system (Agilent Technologies, USA). Library sequencing was performed using an Illumina Hiseq instrument with 150 bp paired-end reads. Paired-end reads were harvested from Illumina HiSeq 4000 sequencer and were quality controlled by Q30. After 3’ adaptor-trimming, low-quality reads were removed by cutadapt software (v1.9.3). The high-quality trimmed reads were used to analyse circRNAs. The high-quality reads were aligned to the reference genome/transcriptome using STAR software (v2.5.1b). And circRNAs were detected and identified using DCC software (v0.4.4). edgeR software (v3.16.5) was used to normalized the data and perform differentially expressed circRNA analysis. All raw data of the RNA sequencing have been uploaded to the GEO repository (GSE164953, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164953).
qPCR detection of exo-circ-RNA expression
ExoRNEasy kit (Qiagen) was used to isolate total RNA from exosomes according to the manufacturers’ instructions. Afterwards, the total RNA was quantified then transcribed to cDNA. Next, qPCR was used to detect the level of Exo-circRNA from the cDNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous control gene, whereas 2-ΔΔCt was used to indicate the relative expression level of Exo-circRNA.
Cell lines culture and cell proliferation, invasion assays
Human liver cancer cell line HepG2, MHCC97-L (97 L), HCC-LM3 (LM3) were purchased from the Cell Bank of the Chinese Academy of Sciences. The above cell lines were cultured in DMEM (Dulbecco’s Modified Eagle Medium) medium containing 10% FBS (fetal bovine serum) and 1% penicillin/streptomycin. The LM3 cell line has higher metastatic and proliferative capacity, and the 97 L cell line has lower metastatic and proliferative capacity [15].
To construct small interfering RNA (siRNA), knock down the expression of exo_circ_0006602 in HCC-LM3 (LM3) cell exosomes according to the instructions of the transfection reagent.
Incubate cells with exosomes (10 μg/ml) or PBS for 12 h. The cells (2 × 104 per well) were seeded into a 96-well plate, and the CCK8 (CK04, Dojido) reagent was added according to the instructions, and the OD values at 0, 24, 48, and 72 h were measured. For the cell invasiveness test, 1 × 104 cells were seeded in a Matrigel-coated (3422, Costar) cell. The medium in the chamber contains no serum, and the medium in the lower chamber contains 10% fetal bovine serum. After 48 h, stain with 0.5% crystal violet, observe and count under a microscope. Wound healing assays were used to analyze cell migration. The migrated cells at the edge of the wound were recorded at 0 and 24 h after wounding, and the results were calculated as follows: (average wound area at 24 h-average wound area at 0 h)/average wound area at 0 h.
Western blot analysis
The protein concentration in exosomes and cells was measured using the BCA protein detection kit (P0009, Beyotime). Separate the protein by 10% SDS-PAGE and transfer to polyvinylidene fluoride membrane (PVDF, Milipore). The membrane was blocked with 4% skim milk and incubated with primary antibody (1:1000 dilution) at 4°C overnight. After incubating the membrane with a secondary antibody (1:5000 dilution) for 1 h, the protein band was visualized using a fluorescent kit (P0018S, Beyotime) and a chemiluminescence imaging system (T-4600, Tanon). After quantifying protein bands using Image J, the samples were normalized to GAPDH protein levels.
Data analysis
Statistical analysis was performed using SPSS 22.0 and GraphPad Prism 7.0. Quantitative data were expressed as mean ± SD. Chi-square test was used to analyze the relationship between Exo-circRNA and clinicopathological characteristics of hepatocellular carcinoma. The ROC curve was used to evaluate the sensitivity and specificity of Exo-CircRNA in early HCC diagnosis. P<0.05 was considered statistically significant.
Results
Exosomes identification
The vesicles isolated from the plasma exhibited a unique bilayer membrane and goblet structure (Figure 1A). Based on nanoparticle tracking analysis, the diameter of the vesicles was 50-180 nm (Figure 1B), which was consistent with the typical size of exosomes. We detected the expression of exosome-specific markers CD81 and CD63 by Western blotting (Figure 1C).
Figure 1.
Identification of plasma-extracted exosomes. Identification of plasma-extracted exosomes. A. Transmission electron microscopy showed the morphological characteristics of exosomes. White arrows represent tipical exosomes in a single field of view under TEM, Scale bar =200 nm. B. Use a nanoparticle size analyzer to detect exosome size. C. Verification of exosomal surface marker protein by Western blotting, Exo represents the exosomal sample and negative control representing the supernatant when the exosomes are extracted.
Exosomal circRNA profile of the HCC patients
Based on the analysis results of the original circRNA expression profile data, 12,144 circRNAs were up-regulated (Figure 2A, 2B). A total of 143 of these up-regulated circRNAs were screened by fold change and p-value (fold change ≥2.0 and P<0.05). Significant differences in circRNAs expression were observed between HCC and NC groups. Of the 143 over-expressed circRNAs, 19 were identified as newly discovered circRNAs (Figure 2C). 121 of the identified circRNAs were less than 2,000 nucleotides (nt) in length (Figure 2D).
Figure 2.
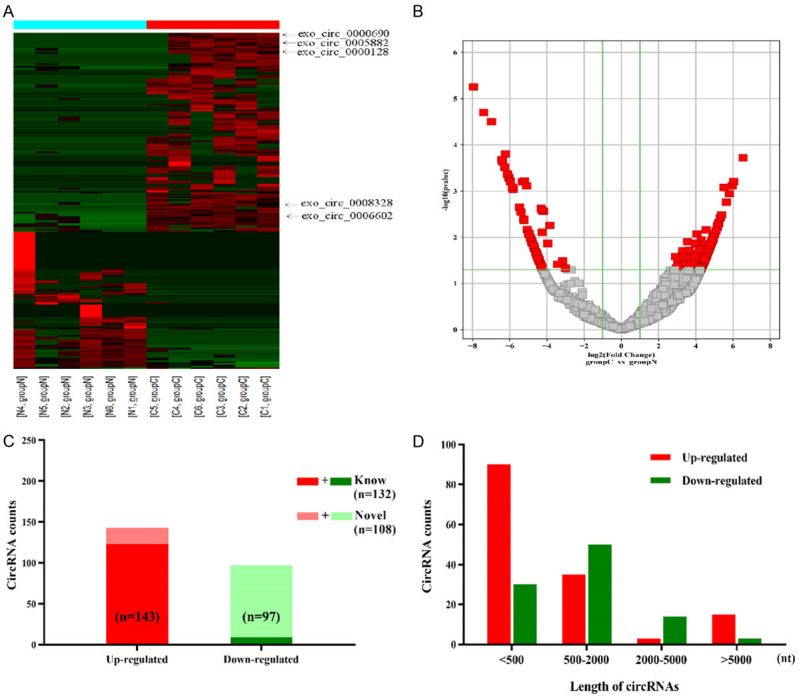
Expression profiles of circRNAs in plasma-extracted exosomes from six hepatocellular carcinoma patients compared to six healthy volunteers. A. Clustered heatmap with each column representing a Plasma sample and each row representing a circular RNA identified by circRNA sequencing. The red indicates upregulated circRNAs, and the green indicates downregulated circRNAs. The five circRNAs were validated by real-time qPCR (arrows). B. Volcano plots showed differential expression of circRNAs between Tumor plasma exosome and normal control (NC). Vertical line expressed as 2-fold (log2 scaled) up or down changes; the horizontal line represented a p value of 0.05 (-log10 scaled). Red spots indicated the differentially expressed circRNAs with statistical significance. C. 143 circRNAs were significantly upregulated (red). 19 novel circRNAs were identified. D. The distribution of the differentially expressed circRNAs based on the length of nuclear acids.
Circular RNA screening and validation
According to the inclusion criteria, we selected five circRNAs from 143 up-regulated circRNAs in HCC patients for qPCR verification (exo_circ_0000690, exo_circ_0005882, exo_circ_0000128, exo_circ_0008328, exo_circ_0006602). Four out of five circRNAs matched the sequencing results. Exo_circ_0006602 showed the highest fold change in the HCC group (Figure 3A).
Figure 3.

Exo_circ_0006602 expression level and diagnostic performance. A. Small samples detect expression of 5 circRNAs. B. Large samples detect expression of exo_circ_0006602 in plasma exosomes (Tumor group (n=87) and healthy control (n=30), P<0.001). C. The expression of exo_circ_0006602 in gastric cancer group cholangiocarcinoma group and HCC group. D. ROC curve was used to evaluate exo_circ_0006602, AFP and CEA diagnostic performances.
Samples from 87 HCC patients and 30 healthy volunteers were used to perform large-scale validation of exo_circ_0006602 by qPCR. Exo_circ_0006602 was significantly elevated in the HCC patients’ group (Figure 3B; Table 1). Compared with healthy volunteer group, the expression of exo_circ_0006602 was not increased in gastric cancer group and cholangiocarcinoma group (Figure 3C).
Table 1.
Patient’s hepatocellular carcinoma diagnosis index
| Tumor group | Healthy Control | t | p | |
|---|---|---|---|---|
| Number of Cases | 87 | 30 | ||
| Exo_circ_0006602 | 5.059±1.225 | 3.325±0.678 | 7.363 | <0.001 |
| AFP (ng/ml) | 146.667±217.608 | 4.800±3.875 | 12.349 | <0.001 |
| CEA (ng/ml) | 4.629±3.183 | 3.454±1.245 | 2.845 | 0.053 |
Diagnostic efficacy comparison
Furthermore, the receiver operating characteristic (ROC) was used to evaluate the accuracy of exo_circ_0006602 as a diagnostic marker. The Area Under the Curve (AUC) of exo_circ_0006602 to HCC patients was 0.907 (P<0.001). The optimal cut-off value was 4.03, with a sensitivity of 77.0% and a specificity of 93.3% (Figure 3D). The AUC value of exo_circ_0006602 was significantly highter than common serum tumor markers AFP (AUC=0.694, P<0.002) and CEA (AUC=0.589, P=0.146). Exo_circ_0006602 combined with AFP can significantly improve the diagnostic accuracy (AUC=0.942, P<0.0001, Table 2). In addition, we analyzed the TNM subgroups, and the AUC of phase I was the highest among all subgroups (AUC=0.9564, P<0.001, Figure 4). These results indicate that exo_circ_0006602 has a higher accuracy in the early diagnosis of HCC compared with the advanced stage.
Table 2.
ROC curve was used to evaluate tumor indicators and combined detection diagnostic performance
| Sensitivity | Specificity | AUC | p | |
|---|---|---|---|---|
| Exo_circ_0006602 | 0.770 | 0.933 | 0.907 | <0.001 |
| AFP | 0.345 | 0.867 | 0.694 | 0.002 |
| CEA | 0.402 | 0.933 | 0.589 | 0.146 |
| Combined Detection | 0.933 | 0.872 | 0.942 | <0.0001 |
Figure 4.
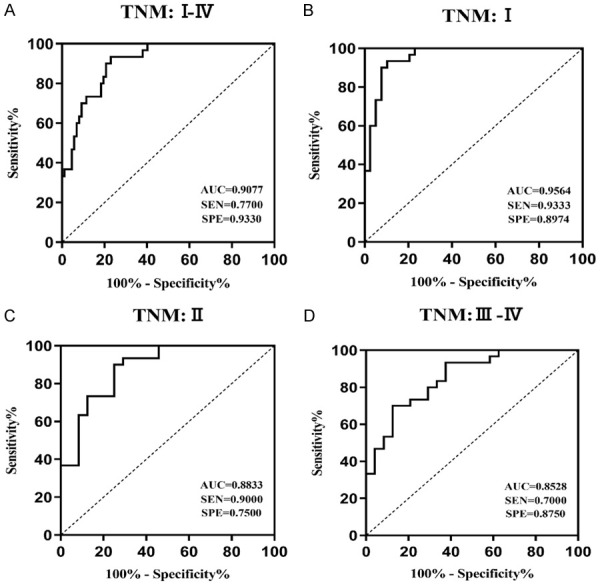
ROC curve analysis of exo_circ_0006602 expression level in the TNM subgroup. (A) 87 HCC cases stage I-IV; the AUC value was 0.9077. (B) in cases of TNM stage I, the AUC value was 0.9564. (C) in cases of TNM stage II, the cut-off value was 0.8833; and (D) in cases of TNM stage III-IV, the AUC value was 8528.
Association between Exo_circ_0006602 expression and clinicopathologic features
The correlation between Exo_circ_0006602 and clinicopathological parameters was analyzed based on the expression of circRNA and clinical data. As shown in Table 3, there was no correlation between exo_circ_0006602 expression level and age (P=0.154), gender (P=0.761), tumor nodule number (P=0.427) and tumor size (P=0.299). However, the expression level of exo_circ_0006602 was correlated with HBsAg (P=0.011), HBeAg (P=0.048), liver cirrhosis (P=0.001) and Edmondson-Steiner grade (P<0.001) (Table 3).
Table 3.
Correlation between exo_circ_0006602 expression and clinical pathologic characteristics
| Characteristics | Number of Cases | Expression of Exo_circ_0006602 | X2 | P value | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Gender | |||||
| Male | 54 | 28 | 26 | 0.093 | P=0.760 |
| Female | 33 | 16 | 17 | ||
| Age | |||||
| >61 | 27 | 13 | 14 | 0.092 | P=0.761 |
| ≤61 | 60 | 31 | 29 | ||
| Tumor size (cm) | |||||
| ≤5 | 56 | 30 | 26 | 0.565 | P=0.452 |
| >5 | 31 | 14 | 17 | ||
| Tumor nodule number | |||||
| Single | 64 | 31 | 33 | 0.442 | P=0.506 |
| Multiple | 23 | 13 | 10 | ||
| HBsAg | |||||
| (+) | 68 | 29 | 39 | 6.444 | P=0.011 |
| (-) | 19 | 15 | 4 | ||
| HBeAg | |||||
| (+) | 32 | 12 | 20 | 3.462 | P=0.048 |
| (-) | 55 | 32 | 23 | ||
| Liver cirrhosis | |||||
| (+) | 59 | 22 | 37 | 12.947 | P=0.001 |
| (-) | 28 | 22 | 6 | ||
| Edmondson-Steiner grade | |||||
| I-II | 63 | 25 | 38 | 10.839 | P<0.001 |
| III-IV | 24 | 19 | 5 | ||
Exo_circ_0006602 promotes proliferation and invasion of liver cancer cell lines
The expression of exo_circ_0006602 in hepatic cancer cell lines HepG2, MHCC97-L (97 L), HCC-LM3 (LM3) exosomes was detected by qPCR. The expression level of exo_circ_0006602 was the highest in HCC-LM3, and the expression level was the lowest in MHCC97-L cell line (Figure 5A). Co-culture of plasma exosomes from liver cancer patients with 97 L cell lines, plasma-derived exosomes increased the proliferation invasion and migration capacity of 97 L cell lines (Figure 5B-D).
Figure 5.

Exosomes from hepatocellular carcinoma patients’ plasma promote the function of liver cancer cells. A. Expression of exo_circ_0006602 in exosomes secreted by 97 L, HepG2 and LM3 cell lines. B. Plasma-derived exosomes from liver cancer increase the proliferation of 97 L cell lines. C. Plasma-derived exosomes from liver cancer patients increase invasive capacity of 97 L cell lines. D. Wound healing experiments showed that plasma-derived exosomes from liver cancer patients increased the migration capacity of 97 L cell line.
SiRNA (Small interfering RNA) significantly reduced exo_circ_0006602 expression in LM3-derived exosomes (Figure 6A). Silencing the expression of exo_circ_0006602 in LM3-derived exosomes, we found that the proliferation migration and invasion ability of LM3 cell line was significantly reduced (Figure 6B-D). Compared with the co-culture of normal LM3-derived exosomes, knocking out exo_circ_0006602 in LM3 exosomes significantly reduced the proliferation, invasion and migration capabilities of the 97 L cell line (Figure 6E-G).
Figure 6.
Exogenous exo_circ_0006602 promote the function of liver cancer cells. A. siRNA knocks down exo_circ_0006602 expression in LM3 exosomes. B-D. Silencing the expression of exo_circ_0006602 in LM3-derived exosomes, the proliferative, invasive and migration capacity of the LM3 cell line was significantly reduced. E-G. Knock out exo_circ_0006602 in LM3’s exosomes and co-culture with 97 L cell line, the proliferation, invasion and migration ability of 97 L cell line is reduced. H. Exogenous exo_circ_0006602 can promote the expression of epithelial-mesenchymal transition related protein Snail. Silencing the expression of exo_circ_0006602 in exosomes reduced the expression of epithelial-mesenchymal transition-associated protein Snail.
To uncover the mechanism driving the exo_circ_0006602 pro-tumorigenic effect on cancer cells, we analyzed the expression of Snail in cancer cell lines. Silencing the expression of exo_circ_0006602 in exosomes, Snail expression was reduced in the LM3 cell line. Plasma-derived and LM3-derived exosomes can increase the expression of Snail in 97 L cells (Figure 6H).
Discussion
At present, there is lack of an effective method for early diagnosis of liver cancer. Most patients still get diagnosed at an advanced stage and therefore miss the best time for surgery. The discovery of tumor markers AFP, CEA, CA199 significantly improved the diagnostic accuracy of early liver cancer. However, some benign tumors, benign lesions or other uncertain factors can affect the expression of these tumor markers, thus resulting in misdiagnosis. For example, alpha-fetoprotein is an ideal tumor marker for specific diagnosis of primary liver cancer; however, its expression also increases in patients with liver cirrhosis. Therefore, the diagnostic results are prone to false positives. Furthermore, some studies have indicated 57.14% as the sensitivity of AFP in early diagnosis of HCC [16-19]. Medical imaging is highly specific; however, its sensitivity is relatively low because it cannot distinguish very small tumors [20]. Therefore, there is a need to find a more effective tumor marker. And the advancement in exosomal research has been useful in the identification of these markers.
Exosomes are derived from late endocytic bodies (also known as multivesicular bodies, MVB), which deflate inwardly to form a multivesicular body containing multiple small vesicles. The source cells are different, and the specific components contained in the exosomes released by the secretion are also different. Exosomes are very stable in body fluids, and their lipid bilayer structure prevents them from being degraded. They transport their contents to specific target cells and therefore play an important regulatory role. The specific function of exosomes in physiological processes depends mainly on the cells from which they originate. Different cells produce exosomes that carry different biological information [21-23]. And the tumor-derived exosomes are different from the original exosomes in healthy cells. The level of expression of tumor-derived exosomes is several times higher/lower than normal exosomes. The environment inside exosomes is relatively simple and stable compared to the complex environment in tissues and cells [24-28]. Therefore, hepatocellular carcinoma-derived exosomes have the potential to serve as novel tumor diagnostic markers.
Exosomes contain mRNA, microRNAs, and proteins that can influence cell behavior and can, therefore, be used for the diagnosis of human diseases [29]. In 2015, Li et al. [30] revealed for the first time that exosomes are rich in circRNA. Wide-genome RNA-seq analysis also found that circRNA content was more abundant in exosomes compared with secreting cells. Unlike other exosomal contents, circRNAs are covalently closed single-stranded transcripts produced from exonic, intronic or intergenic regions, and lacks typical terminal structure. The lack of these structures makes them more stable and resistant to exonuclease R [31]. In 2015, Li et al. [32] also discovered that tumor exo-circRNA could enter the circulation and be isolated from the blood; a characteristic that makes it an ideal tumor diagnostic marker.
In the present study, various measures were adopted to reduce the error between cases as well as the influence of various factors on the results to improve the effectiveness of the tumor diagnostic markers. For example, studies have shown that platelets secrete several exosomes during the coagulation process, and nearly 50% of the exosomes in the serum come from the coagulation process [33-36]. Therefore, to screen tumor diagnostic markers more accurately and reduce the effect of platelet-derived exosomes on results, we selected plasma-derived exosomes as the research object. Also, we selected patients with early-stage hepatocellular carcinoma who did not receive radiation and chemotherapy and had pathological confirmation after surgery to prevent in-group errors. In the selection process of target circRNA, we also formulated strict screening criteria. For example, we selected circRNAs that were generally up-regulated in all cases to prevent the expression of circRNA in individual cases that could affect final Fold-Change and p-value. Finally, we chose circular RNAs that were between 300 bp and 3 kb for verification because circular RNAs that are smaller than 300 bp and larger than 3 kb might produce false positives.
Through screening of small samples and validation of large samples, it was revealed that exo_circ_0006602 was generally up-regulated in plasma exosomes of HCC. And exo_circ_0006602 is specifically expressed in the plasma of patients with hepatocellular carcinoma, but not in cholangiocarcinoma, gastric cancer and other tumors. The ROC curve evaluation also showed that the diagnostic efficiency of exo_circ_0006602 is higher than that of AFP, a traditional tumor marker for hepatocellular carcinoma. And exo_circ_0006602 combined with AFP can significantly improve the accuracy of early diagnosis of hepatocellular carcinoma. Moreover, we also found that exo_circ_0006602 can improve the invasion and proliferation of liver cancer cells. Exo_circ_0006602 also can promote the expression of tumor proliferation-related proteins in hepatoma cell lines. Overall, these findings showed that exo_circ_0006602 has great potential as a marker for early diagnosis of hepatocellular carcinoma and exo_circ_0006602 may have some impact on the progression of liver cancer.
The present study, however, had some limitations. First, the number of samples was insufficient. More samples were needed to determine the diagnostic efficacy of exo_circ_0006602. Secondly, this study did not involve follow-ups. Patients needed to be observed for a long time to obtain survival analysis data. Therefore further research involving larger sample size and follow-ups should be conducted to verify further the role and mechanism of exo_circ_0006602 in the occurrence and development of liver cancer.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lee WC, Lee CF, Cheng CH, Wu TJ, Chou HS, Wu TH, Soong RS, Chan KM, Yu MC, Chen MF. Outcomes of liver resection for hepatocellular carcinoma in liver transplantation era. Eur J Surg Oncol. 2015;41:1144–1152. doi: 10.1016/j.ejso.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 3.Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 4.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 5.Vanni I, Alama A, Grossi F, Dal Bello MG, Coco S. Exosomes: a new horizon in lung cancer. Drug Discov Today. 2017;22:927–936. doi: 10.1016/j.drudis.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Zeng AQ, Yu Y, Yao YQ, Yang FF, Liao M, Song LJ, Li YL, Yu Y, Li YJ, Deng YL, Yang SP, Zeng CJ, Liu P, Xie YM, Yang JL, Zhang YW, Ye TH, Wei YQ. Betulinic acid impairs metastasis and reduces immunosuppressive cells in breast cancer models. Oncotarget. 2018;9:3794–3804. doi: 10.18632/oncotarget.23376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Hernandez J, Olivares D, Forner MJ, Ortega A, Solaz E, Martinez F, Chaves FJ, Redon J, Cortes R. Urinary exosome miR-146a is a potential marker of albuminuria in essential hypertension. J Transl Med. 2018;16:228. doi: 10.1186/s12967-018-1604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Xin G, Sun D. Serum exosomal miR-328, miR-575, miR-134 and miR-671-5p as potential biomarkers for the diagnosis of Kawasaki disease and the prediction of therapeutic outcomes of intravenous immunoglobulin therapy. Exp Ther Med. 2018;16:2420–2432. doi: 10.3892/etm.2018.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JE, Eom JS, Kim WY, Jo EJ, Mok J, Lee K, Kim KU, Park HK, Lee MK, Kim MH. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: a pilot study. Thorac Cancer. 2018;9:911–915. doi: 10.1111/1759-7714.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Akhter R. Circular RNA and Alzheimer’s disease. Adv Exp Med Biol. 2018;1087:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y, Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadziyannis E, Sialevris K, Georgiou A, Koskinas J. Analysis of serum alpha-fetoprotein-L3% and des-gamma carboxyprothrombin markers in cases with misleading hepatocellular carcinoma total alpha-fetoprotein levels. Oncol Rep. 2013;29:835–839. doi: 10.3892/or.2012.2147. [DOI] [PubMed] [Google Scholar]
- 17.Choi JY, Jung SW, Kim HY, Kim M, Kim Y, Kim DG, Oh EJ. Diagnostic value of AFP-L3 and PIVKA-II in hepatocellular carcinoma according to total-AFP. World J Gastroenterol. 2013;19:339–346. doi: 10.3748/wjg.v19.i3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lampignano R, Neumann MHD, Weber S, Kloten V, Herdean A, Voss T, Groelz D, Babayan A, Tibbesma M, Schlumpberger M, Chemi F, Rothwell DG, Wikman H, Galizzi JP, Bergheim IR, Russnes H, Mussolin B, Bonin S, Voigt C, Musa H, Pinzani P, Lianidou E, Brady G, Speicher MR, Pantel K, Betsou F, Schuuring E, Kubista M, Ammerlaan W, Sprenger-Haussels M, Schlange T, Heitzer E. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (Pre)analytical work flows. Clin Chem. 2020;66:149–160. doi: 10.1373/clinchem.2019.306837. [DOI] [PubMed] [Google Scholar]
- 19.Shu H, Li W, Shang S, Qin X, Zhang S, Liu Y. Diagnosis of AFP-negative early-stage hepatocellular carcinoma using Fuc-PON1. Discov Med. 2017;23:163–168. [PubMed] [Google Scholar]
- 20.Schraml C, Kaufmann S, Rempp H, Syha R, Ketelsen D, Notohamiprodjo M, Nikolaou K. Imaging of HCC-current state of the art. Diagnostics (Basel) 2015;5:513–545. doi: 10.3390/diagnostics5040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynaston H, Mason MD, Tabi Z, Clayton A. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34:290–302. doi: 10.1038/onc.2013.560. [DOI] [PubMed] [Google Scholar]
- 22.Singh A, Fedele C, Lu H, Nevalainen MT, Keen JH, Languino LR. Exosome-mediated transfer of alphavbeta3 integrin from tumorigenic to nontumorigenic cells promotes a migratory phenotype. Mol Cancer Res. 2016;14:1136–1146. doi: 10.1158/1541-7786.MCR-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peinado H, Alec kovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar CM, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Corrigendum: melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2016;22:1502. doi: 10.1038/nm1216-1502b. [DOI] [PubMed] [Google Scholar]
- 24.Abudoureyimu M, Zhou H, Zhi Y, Wang T, Feng B, Wang R, Chu X. Recent progress in the emerging role of exosome in hepatocellular carcinoma. Cell Prolif. 2019;52:e12541. doi: 10.1111/cpr.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol. 2019;12:53. doi: 10.1186/s13045-019-0739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Li B. The functional role of exosome in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:2085–2095. doi: 10.1007/s00432-018-2712-7. [DOI] [PubMed] [Google Scholar]
- 27.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li LM, Liu ZX, Cheng QY. Exosome plays an important role in the development of hepatocellular carcinoma. Pathol Res Pract. 2019;215:152468. doi: 10.1016/j.prp.2019.152468. [DOI] [PubMed] [Google Scholar]
- 29.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lener T, Gimona M, Aigner L, Borger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, O’Driscoll L, Fais S, Falcon-Perez JM, Felderhoff-Mueser U, Fraile L, Gho YS, Gorgens A, Gupta RC, Hendrix A, Hermann DM, Hill AF, Hochberg F, Horn PA, de Kleijn D, Kordelas L, Kramer BW, Kramer-Albers EM, Laner-Plamberger S, Laitinen S, Leonardi T, Lorenowicz MJ, Lim SK, Lotvall J, Maguire CA, Marcilla A, Nazarenko I, Ochiya T, Patel T, Pedersen S, Pocsfalvi G, Pluchino S, Quesenberry P, Reischl IG, Rivera FJ, Sanzenbacher R, Schallmoser K, Slaper-Cortenbach I, Strunk D, Tonn T, Vader P, van Balkom BW, Wauben M, Andaloussi SE, Thery C, Rohde E, Giebel B. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, Janitz M. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109:401–407. doi: 10.1016/j.ygeno.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eitan E, Green J, Bodogai M, Mode NA, Baek R, Jorgensen MM, Freeman DW, Witwer KW, Zonderman AB, Biragyn A, Mattson MP, Noren Hooten N, Evans MK. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci Rep. 2017;7:1342. doi: 10.1038/s41598-017-01386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endzelins E, Berger A, Melne V, Bajo-Santos C, Sobolevska K, Abols A, Rodriguez M, Santare D, Rudnickiha A, Lietuvietis V, Llorente A, Line A. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer. 2017;17:730. doi: 10.1186/s12885-017-3737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuravi SJ, Yates CM, Foster M, Harrison P, Hazeldine J, Hampson P, Watson C, Belli A, Midwinter M, Nash GB. Changes in the pattern of plasma extracellular vesicles after severe trauma. PLoS One. 2017;12:e0183640. doi: 10.1371/journal.pone.0183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broggi MAS, Maillat L, Clement CC, Bordry N, Corthesy P, Auger A, Matter M, Hamelin R, Potin L, Demurtas D, Romano E, Harari A, Speiser DE, Santambrogio L, Swartz MA. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J Exp Med. 2019;216:1091–1107. doi: 10.1084/jem.20181618. [DOI] [PMC free article] [PubMed] [Google Scholar]




