Abstract
Circular RNAs (circRNA) are found to be closely associated with cancers as their possibility as “sponges” to miRNAs, thus liberating the downstream target mRNA. However, deep research is still needed to study the function of circRNA in papillary thyroid carcinoma (PTC). Here, we sought to explore new circRNA which could play an important role in the development of PTC. We filtered candidate circRNAs based on microarray data from public database and verified the result using qRT-PCR. We performed CCK8 assay, colony formation assay, apoptosis assay, transwell assays, and xenograft experiments to explore the function of selected circRNA on PTC. We predicted the miRNA targets of the circRNA and the target gene of miRNA through bioinformatic analysis and validated the target by mutant experiments. And by the use of overexpression experiments, knockdown experiments, and the functional assays mentioned above, we figured out the pathway behind the selected circRNA. Based on our data, we found that circNEURL4 was significantly decreased in the PTC samples and lower expression of circNEURL4 was closely associated with a poor prognosis of patients with PTC. Then, we proved that circNEURL4 could inhibit cell proliferation and invasion of PTC in vivo and in vitro. Furthermore, we demonstrated that circNEURL4 may binding to miR-1278 and thus indirectly improving the expression of LATS1. Our findings revealed that circNEURL4 may probably serve as a diagnostic marker to predict PTC patients’ prognosis and a possible therapeutic target to PTC via miR-1278/LATS1 axis.
Keywords: Papillary thyroid carcinoma, circNEURL4, miR-278, LATS1, cell invasion, cell proliferation
Introduction
Thyroid cancer is a kind of endocrine-related cancers, with a prevalence of about 2.1% in all cancer cases [1]. Papillary thyroid carcinoma (PTC) is the most common subtype of thyroid cancer, accounting for more than 90% of the new diagnoses [2]. Patients with PTC have a good prognosis based on traditional therapeutic management [3], and the 10-year mortality of PTC is less than 5% [4]. However, a small fraction of PTC cases present as large size, distant metastasis, and lymph node metastasis, and therefore, has a poor prognosis [5]. Recently, the incidence of high-risk PTC is increasing [1,6,7], calling for the importance of in-depth studies on the diagnosis and treatment of PTC, especially on its metastasis based on cell invasion.
Circular RNAs (circRNAs) are transcribed mostly from the exon region, form as loops, and belong to non-coding RNA (ncRNA) [8,9]. CircRNAs broadly participate in the bio-metabolism and often act as regulators to mRNAs or proteins [10,11]. How circRNAs regulate the expression of mRNAs is well studied. CircRNAs can perform as “sponges” to competitively bind to miRNAs and thus regulate the corresponding mRNAs in an indirect manner [12,13]. It has been widely reported that the circRNA-miRNA-mRNA axis plays a crucial role in different cancers [14,15]. Several studies have shown the relationship between circRNA and PTC, for example, circFOXM1 can upregulate the expression of HMGB1 by competitively inhibiting miR-1179 and thus promote the progress of PTC [16]; circBACH2 acts as an oncogenic RNA through binding to miR-139-5p and indirectly increasing the expression of LMO4 [17]; circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression [18]; hsa_circ_0007694 is down-regulated in PTC and can promoted apoptosis and inhibited proliferation, migration, and invasion in PTC cells [19]. Nevertheless, the mechanism of circRNA in regulating PTC progress needs further research, and new circRNAs await further exploration.
In this study, we found that circNEURL4 was significantly decreased in the PTC samples and cell lines. Furthermore, we demonstrated that circNEURL4 may perform as a PTC suppressor through competitively binding to miR-1278 and thus indirectly improving the expression of LATS1. Our findings revealed that circNEURL4 may probably serve as a diagnostic marker to predict PTC patients’ prognosis and a therapeutic target to cure PTC via miR-1278/LATS1 axis.
Materials and methods
Clinical specimens and ethics statement
Human samples involved in this study were managed using protocols approved by the Ethical Committee of The Second Hospital of Jilin University. Informed consents were obtained from all patients. 68 pairs of PTC tumor samples and matching contralateral normal samples were obtained from patients who were diagnosed with PTC and had undergone surgery at The Second Hospital of Jilin University. Collected samples were frozen in liquid nitrogen and stored at -80°C. Experiments on mice were performed with protocols approved by the Animal Research Committee of The Second Hospital of Jilin University.
Cell culture
The cell lines (KTC-1, IHH-4, BCPAP, TPC-1, and Nthy-ori 3-1) were purchased from Lonza Pharma & Biotech and cultured in RPMI-1640 medium (Lonza Pharma & Biotech) containing 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Invitrogen).
Transfection of plasmid, miRNA mimics, and miRNA inhibitors was performed using Lipo2000 according to the recommendations of the manufacturer (Thermofisher).
Construction of stable cell lines
Sequence expressed circNEURL4 was introduced into pLV-Puro vector (Hanbio Biotechnology) to construct pLV-circNEURL4. pLV-circNEURL4 or empty vector was transfected to BCPAP or TPC-1 and filtered with puromycin to construct stable cell lines as described previously [20].
Transfection
pLV-circNEURL4, and pLV were purchased from Hanbio Biotechnology. miR-1278 inhibitor, NC inhibitor, miR-1278 mimics, and miR-NC were purchased from Thermofisher Biotechnology. Transfection was performed using Lipo2000 (Thermofisher) according to the protocol of the manufacture.
Analyzing the circRNA microarray data
circRNA microarray data GSE93522 [21] was obtained from the GEO database. The data was analyzed using the Cluster and TreeView programs. The most down-regulated circRNA was pick up.
Prediction analysis
Prediction of the miRNA targets of circNEURL4 was done through the web tools CircBank (http://www.circbank.cn/), and Circular RNA Interactome (https://circinteractome.irp.nia.nih.gov/). Prediction of the binding site between circNEURL4 and miR-1278 was performed on Circular RNA Interactome (https://circinteractome.irp.nia.nih.gov/). Prediction of the binding site between LATS1’s 3’UTR and miR-1278 was performed on starbase (http://starbase.sysu.edu.cn/).
RNase R treatment
2 mg total RNA was incubated with or without 3 U/mg of RNase R (Epicenter Technologies) for 30 min at 37°C. Then, qRT-PCR was performed to detect the expression levels of circNEURL4 and NEURL4.
CCK8 assays
Cells (5000 cells/well in 96-well plates) were adjusted to different treatments for 24-72 h at 37°C. Then 10 μl CCK-8 reagent (Abcam) was added to each well and cultured for another 1 h at 37°C. The absorption values were measured by a microplate reader (Thermofisher) and were used to calculate the cell viability.
Colony-formation assays
Cells with different treatments were seeded at a density of 20 cells/cm2 in 12-wells plates and cultured at 37°C for 2 weeks. Then the cells were fixed, washed with PBS for 3 times, stained with Crystal Violet (Sangon), and pictured and counted using microscopy (Leica).
Apoptosis assays
Cells (5 × 105 cells/well in 6-well plates) were adjusted to different treatments for 24 h at 37°C. Then the cells were digested and stained with an Annexin V-FITC/propidine iodide (PI) double-staining kit (Abcam). The cells were then analyzed with FACS using CytoFlex (Beckman).
Cell migration and invasion transwell assays
Cell migration and invasion were measured with transwell assays. Cultured cells with 70-80% confluency were digested for the next experiments.
In brief, for cell migration, it was performed by cell culture insects invasion chambers which that carry membrane with 8 μm pores in 24-well plates (Corning). The cell suspensions were added into the upper chamber of a transwell filter. A serum-free medium was added to the upper compartment, and a medium containing 10% FBS was added into the lower compartment. After incubation at 37°C for 24 h, the cells in the lower upper chambers were fixed via methanol for 10 min and stained with 0.1% crystal violet solution. The cells on the upper-membranes were removed, and then the cells on the lower-membranes were counted under a microscope in five randomly chosen fields.
For cell invasion, the upper chambers were coated with Matrigel (BD Biosciences) before adding the cell suspensions.
Nuclear/cytosol fractionation
The separation of nuclear and cytosol RNA was performed using PARIS™ Kit (Thermofisher). Briefly, 1 × 107 cells were disrupted with 1mL cell fractionation buffer on ice for 10 min. Then samples were centrifuged for 5 min at 4°C with 500 g. The pellet was the nuclear part and the supernatant was the cytoplasmic lysate. The nuclear pellet was homogenized by 500 μl cell disruption buffer on ice. Next, 500 μl cytoplasmic and nuclear lysate were mixed with 500 μl 2 × lysis/binding solution and 500 μl ethanol respectively. Then, the samples were drawn through filter cartridges and washed 2 times. Finally, RNA was eluted with 40-60 μL of ~95°C elution solution.
RNA extraction and qRT-PCR
Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer’s protocol. miRNA Reverse Transcription Kit (Qiagen) was used for the synthesis of miRNA cDNA library and PrimeScript™ RT reagent Kit (Takara) was used for the synthesis of miRNA cDNA library. SYBR Green PCR Master Mix (Applied Biosystems) was used in a CFX96 Real-Time System thermocycler (Bio-Rad), and the RNA levels of the target genes were normalized to those of GAPDH (for mRNA) or U6 (for miRNA). All the primers’ sequences were listed in Table 1.
Table 1.
Primer sequences for qRT-PCR
| Gene | Primer sequence (5’-3’) |
|---|---|
| circNEURL4 | Forward primer: ATGCTCATGGCCTTGTCTTCA |
| Reverse primer: CTAGAGTGACGTTCTTGCCG | |
| miR-1278 | Forward primer: GGCTCTGGCTCCGTGTCTT |
| Reverse primer: CAGTGCAGGGTCCGAGGTATT | |
| miR-521 | Forward primer: ATGCCGCGCTCTTTCCTCGTC |
| Reverse primer: TCAGCGGGGACATCCTGAGC | |
| U6 | Forward primer: CAAATTCGTGAAGCGTTCCATA |
| Reverse primer: AGTGCAGGGTCCGAGGTATTC | |
| GAPDH | Forward primer: GCACCGTCAAGGCTGAGAAC |
| Reverse primer: ATGGTGGTGAAGACGCCAGT |
Western blot analysis
The procedure of western blot was the same as the previous paper [22]. All the primary and secondary antibodies were purchased from Abcam. An ECL detection kit (Thermofisher) was used to develop the bands. The pictures were captured by ChemiDoc XRS Plus (BioRad). The primary antibodies used are listed as follows: E-cadherin, N-cadherin, and GAPDH (Abcam).
Luciferase reporter assay
WT or mutant binding site sequences of circNEURL4 or LATS1’s 3’UTR (Figures 4D, 6A) was introduced to pmirGLO (Promega). Plasmids containing WT or mutant binding site were co-transfected with miR-1278 mimics (GenePharma) to BCPAP or TPC-1 using Lipofectamine 3000 (Invitrogen). 48 h post the transfection, cells were analyzed for luciferase activity using the Dual-Glo® Luciferase Assay System (Promega) and a MicroLumatPlus LB96V luminometer (Berthold). Relative luciferase activity was calculated as firefly luciferase activity/Renilla luciferase activity.
Figure 4.
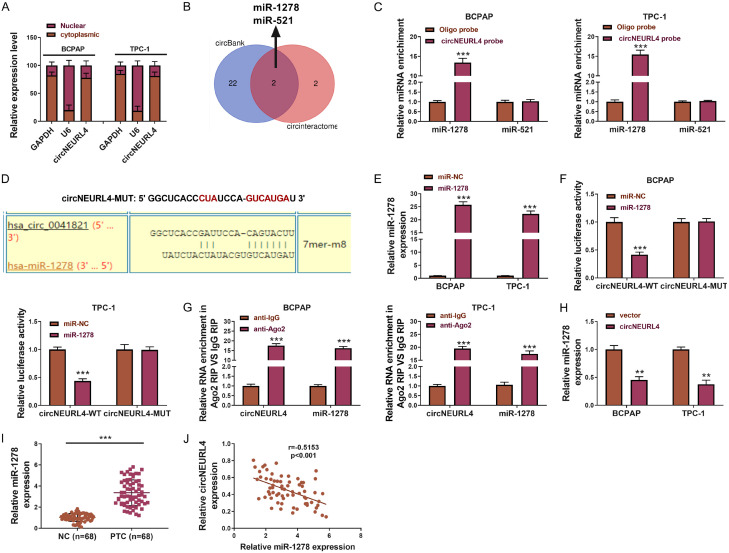
circNEURL4 binds to miR-1278 and they were antagonists to each other. A. The relative expression level of circNEURL4 in the nuclear and cytoplasm of BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector dectected by qRT-PCR. GAPDH was used as the reference of cytoplasm, U6 was used as the reference of nuclear. B. Vein graph showing the prediction of the miRNA targets of circNEURL4. C. The relative expression level of miR-1278 and miR-521 in BCPAP and TPC-1 using a circNEURL4-specific probe or oligo probe by RNA pull-down assays. D. Schematic representation of the predicted binding site of miR-1278 to circNEURL4 (circNEURL4-WT), and the mutant circNEURL4 designed by us (circNEURL4-MUT). E. The relative expression level of miR-1278 showing the efficiency of transfection dectected by qRT-PCR. F. The luciferase activity was determined using the Dual-Luciferase Reporter System. G. Relative RNA enrichment of circNEURL4 and miR-1278 in Ago2 RIP compared to IgG RIP. H. The relative expression level of miR-1278 in BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector dectected by qRT-PCR. I. The relative expression level of miR-1278 in PTC samples and their matching contralateral normal samples using qRT-PCR (n=68). J. Correlation test showing negative correlation of miR-1278 and circNEURL4 in PTC patients’ samples. Statistical results were presented as the mean ± SME. Two-tailed paired t-test, **P<0.01, ***P<0.001.
Figure 6.
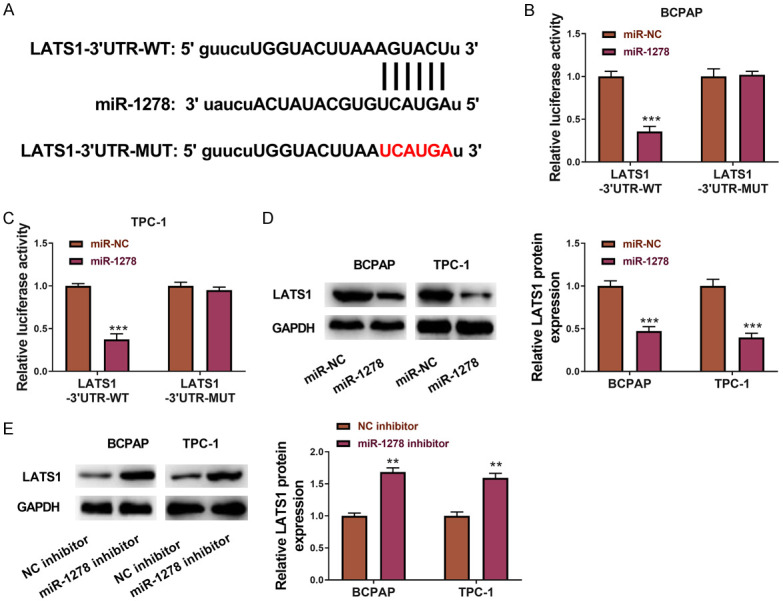
miR-1278 directly targets LATS1. A. Schematic representation of the predicted binding site of miR-1278 in the 3’UTR of LATS1 mRNA (LATS1-3’UTR-WT), and the mutant LATS1 3’UTR designed by us (LATS1-3’UTR-MUT). B. The luciferase activity was determined using the Dual-Luciferase Reporter System in BCPAP. C. The luciferase activity was determined using the Dual-Luciferase Reporter System in TPC-1. D. Western Blot and statistical results showing the expression of LATS1 in BCPAP and TPC-1 transfected with miR-1278 mimics or miR-NC. GAPDH was used as an internal reference. E. Western Blot and statistical results showing the expression of LATS1 in BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. GAPDH was used as an internal reference. Statistical results were presented as the mean ± SME. Two-tailed paired t-test, *P<0.05, **P<0.01, ***P<0.001.
RNA pulldown assay
The RNA pulldown assay was carried out as previously described [23]. In brief, BCPAP and TPC-1 cells were quantitated and treated with 1 ml of cell lysis buffer for 72 h. Before the cells were rotated overnight at 4°C, 1.5 μl of RNase inhibitor, 10 μl of streptavidin agarose beads, and 500 pM probe of circNEURL4 (Thermofisher) were added to BCPAP and TPC-1 cells. Total RNAs were then subjected to qRT-PCR analysis.
RNA binding protein immunoprecipitation (RIP)
The MagnaRIP RNA-Binding Protein Immunoprecipitation Kit was used to perform the Ago2 immunoprecipitation assay. Transfected cells were lysed with RIPA lysis buffer (150 mM NaCl, 20 nM Tris-HCl (PH 7.5), 1% NP-40, 2.5 mM sodium pyrophosphate, 1 mM Na2EDTA, 1 mM EGTA, 1% sodium deoxycholate, 1 μg/ml leupeptin, 1 mM Na3VO4, and 1 mM beta-glycerophosphate) for 30 min on ice. The cell suspension was then centrifuged at 14000 rpm for 15 min. After the addition of 2 μl of AGO2 antibody (Abcam) and 10 μl of beads (Thermofisher), the supernatant was rotated overnight at 4°C. The mixture was washed with lysis buffer. RNA extraction was performed using Trizol reagent and then subjected to qRT-PCR analysis.
Xenografts in mice
Male nude mice (6-8 weeks, purchased from Cyagen) were subcutaneously injected with 1 × 106 cells on the oxter. The length and width of tumor xenografts were measured weekly, and the tumor volume was calculated using the following formula: volume (cubic millimeters) = 0.5 × width2 × length. Five weeks post-injection, t the tumors were collected and weighed.
Statistical analysis
Data are shown as the mean ± S.E.M. Analyses of significant differences between groups were performed by Statistical Product and Service Solutions (SPSS) 21.0 using two-tailed Student’s t-tests or Tukey’s test. P<0.05 was considered statistically significant.
Results
Expression of circNEURL4 was decreased in PTC tissues and cell lines
We analyzed the circRNA microarray data of PTC samples from the NCBI database (GSE93522), which contains six PTC tumors and six matching contralateral normal samples. We found that hsa_circRNA_0041821 was the most downregulated circRNA in the PTC samples (Figure 1A). circNEURL4 (hsa_circRNA_0041821) was derived from the NEURL4 gene with the location on chr17:7225183-7225329, and the spliced mature sequence length of circNEURL4 is 146 bp. We verified the discovery in PTC tissues and cell lines. The relative expression of circNEURL4 was dramatically decreased in PTC tissues compared to their matching contralateral normal samples (n=68) (Figure 1B). We then analyzed the relationship between the expression level of circNEURL4 and the PTC patients’ clinical characters (detailed information of each patient is shown in Table 2). Interestingly, patients with advanced stages (III+IV) had a significantly lower circNEURL4 expression than patients with early stages (I+II) (Figure 1C). Moreover, patients with lymph node metastasis showed lower circNEURL4 expression (Figure 1D). We then separated the PTC tissue samples into two groups based on their circNEURL4 expression. One with a higher circNEURL4 expression than the median expression of the whole PTC tissue samples was separated into circNEURL4-high group and one with a lower circNEURL4 expression was separated into circNEURL4-low group. The circNEURL4-low group had a great worse survival curve (Figure 1E). These data manifested that the PTC patients had a lower expression of circNEURL4 and circNEURL4 expression level had a close relationship with the patients’ prognosis.
Figure 1.
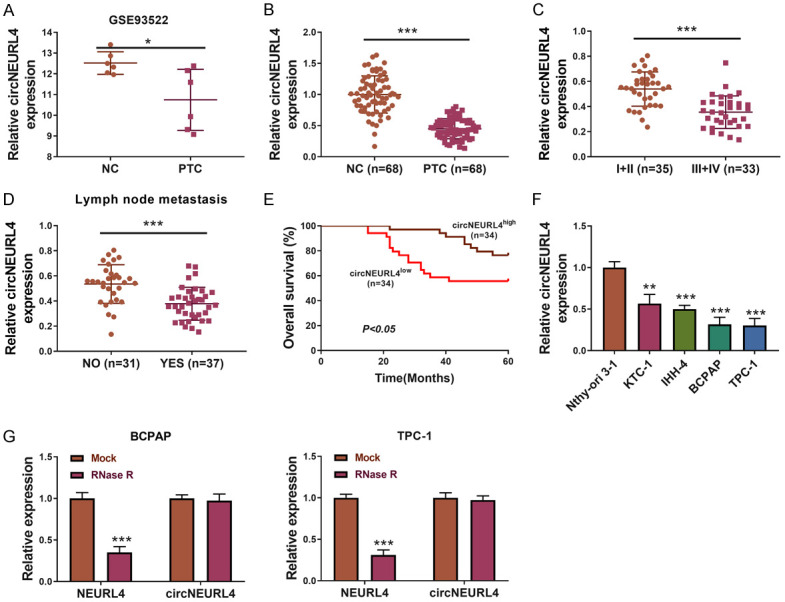
circNEURL4 is down-regulated in PTC samples and cell lines. A. circNEURL4 is down-regulated in the PTC samples from GSE93522. B. The relative expression level of circNEURL4 in PTC samples and their matching contralateral normal samples using qRT-PCR (n=68). C. The relative expression level of circNEURL4 in PTC patients with early stages (I+II) (n=35) or advanced stages (III+IV) (n=33) dectected by qRT-PCR. D. The relative expression level of circNEURL4 in PTC patients with metastasis (n=37) or without metastasis (n=31) dectected by qRT-PCR. E. PTC samples were separated into circNEURL4-high group and circNEURL4-low group with the median expression of circNEURL4 as a cut-off. The survival curve was drawn in the two groups. F. The relative expression level of circNEURL4 in KTC-1, IHH-4, BCPAP, TPC-1, and Nthy-ori 3-1 dectected by qRT-PCR. G. The relative expression level of circNEURL4 and NEURL4 in BCPAP or TPC-1 with or without the treatment of RNase R. Data were presented as the mean ± SME. Two-tailed paired t-test, *P<0.05; ***P<0.001.
Table 2.
Correlation between circNEURL4 expression and clinicopathological parameters of PTC patients
| Characteristics | Number | circNEURL4 expression | P | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| 34 | 34 | |||
| Age (years) | 0.625 | |||
| <45 | 38 | 20 | 18 | |
| ≥45 | 30 | 14 | 16 | |
| Gender | 0.622 | |||
| Male | 40 | 21 | 19 | |
| Female | 28 | 13 | 15 | |
| Tumor size (cm) | 0.331 | |||
| ≤3 | 30 | 18 | 14 | |
| >3 | 38 | 16 | 20 | |
| TNM grade | 0.002* | |||
| I-II | 35 | 24 | 11 | |
| III-IV | 33 | 10 | 23 | |
| Lymph Node Metastasis | 0.028* | |||
| NO | 31 | 20 | 11 | |
| YES | 37 | 14 | 23 | |
| Nodular Goiter | 0.145 | |||
| Negative | 36 | 21 | 15 | |
| Positive | 32 | 13 | 19 | |
P<0.05.
Similarly, compared to the normal thyroid cell line Nthy-ori 3-1, all the PTC cell lines (KTC-1, IHH-4, BCPAP, and TPC-1) showed a lower expression level of circNEURL4, of which BCPAP and TPC-1 were more obvious than the other two (Figure 1F). So we chose BCPAP and TPC-1 for our subsequent in vitro experiments. RNase R treatment also confirmed that circNEURL4 belonged to circRNA.
Based on the above data, we hypothesized that circNEURL4 may play a role in the tumorigenesis of PTC.
circNEURL4 inhibits the growth, migration, and invasion of PTC cells
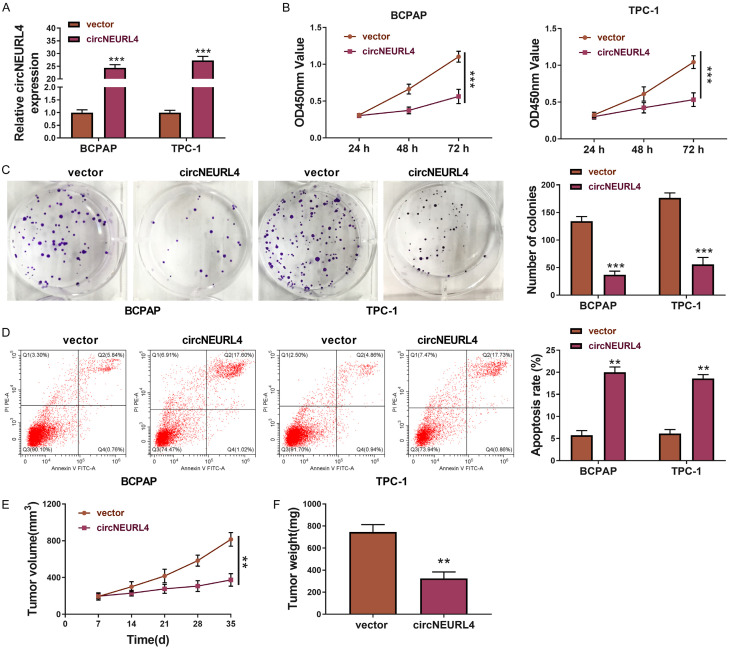
The cell lines (BCPAP-circNEURL4 and TPC-1-circNEURL4) that could stably over-express circNEURL4 were constructed (Figure 2A). The effects of circNEURL4 on PTC cells were determined. Firstly, overexpression of circNEURL4 decreased the cell proliferation (Figure 2C) and inhibited colony-formation (Figure 2C). Secondly, overexpression of circNEURL4 increased apoptosis in PTC cells (Figure 2D), revealing inhibition effects of circNEURL4 on tumor proliferation and survival in PTC cells.
Figure 2.
circNEURL4 inhibits the growth and promote the apoptosis of PTC cells. A. The relative expression level of circNEURL4 in BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector dectected by RT-qPCR. B. CCK8 assays of BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector. Absorbances were measured at 24 h, 48 h, and 72 h post cell seeding. C. Pictures and statistical results of colony-formation assays using BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector. D. Flow graph and statistical results of apoptosis assays using BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector. E. Tumor volumes of nude mice injected with TPC-1-circNEURL4 or TPC-1-vector 7, 14, 21, 28, 35 days post-injection. F. Tumor weights of nude micenjected with TPC-1-circNEURL4 or TPC-1-vector 35 days post-injection. Statistical results were presented as the mean ± SME. Two-tailed paired t-test, *P<0.05, **P<0.01, ***P<0.001.
Then, we explored the effects of circNEURL4 on PTC tumor formation in vivo. 1 × 106 TPC-1-circNEURL4 cells or TPC-1-vector cells were subcutaneously injected into nude mice, naming as circNEURL4 group or control group. The tumor volumes were measured every seven days, and the tumors were collected and weighed 35 days post the injection. Nude mice in the circNEURL4 group showed obviously slower growth rates and lighter tumor weights (Figure 2E, 2F).
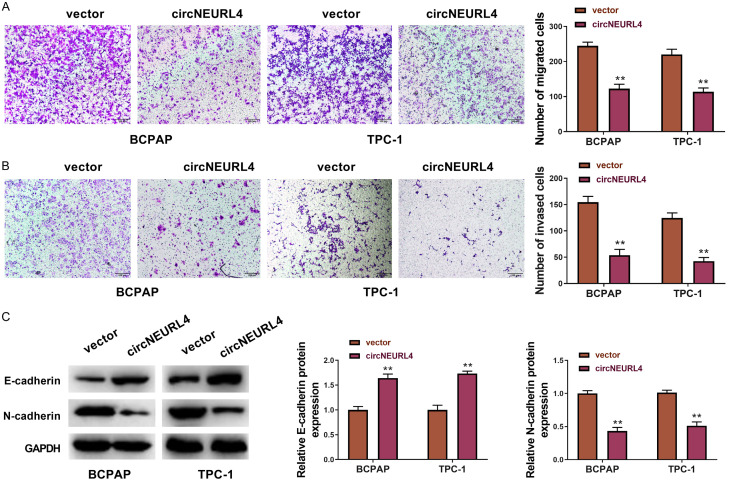
We further detected whether circNEURL4 affects the migration and invasion of PTC cells using transwell assays. The results indicated that circNEURL4 could distinctly reduce the migration and invasion of both BCPAP and TPC-1 cells (Figure 3A, 3B). It is well known that the migration or invasion of cancer cells is always accompanied by an increased expression of N-cadherin and a reduced expression of E-cadherin [24-26]. Our results showed that the expression of E-cadherin was increased and the expression of N-cadherin was decreased in cells with overexpression of circNEURL4 (Figure 3C), confirming the inhibition effects of circNEURL4 on the migration and invasion of PTC cells.
Figure 3.
circNEURL4 inhibits migration and invasion of PTC cells. A. Pictures and statistical results of migration assays using BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector. B. Pictures and statistical results of invasion assays using BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector. C. Western Blot and statistical results showing the expression of E-cadherin and N-cadherin in BCPAP-circNEURL4, TPC-1-circNEURL4, BCPAP-vector, and TPC-1-vector. GAPDH was used as an internal reference. Statistical results were presented as the mean ± SME. Two-tailed paired t-test, **P<0.01.
Thus, we proved that circNEURL4 had exciting effects on PTC cells’ growth, migration, and invasion, raising the problem of what’s the mechanism behind the phenomenon.
circNEURL4 binds to and regulated expression of miR-1278 negatively
circRNAs mostly function in the cytoplasm as “sponges” to miRNAs [12,13], and some circRNAs function in the nuclear by interacting with the genome DNA [27]. Here, we examined the location and mechanism of circNEURL4 on PTC cells. We detected circNEURL4 expression in the cytoplasm and nuclear respectively using qRT-PCR in BCPAP and TPC-1, representing nearly 80% circNEURL4 located at the cytoplasm (Figure 4A). So we proposed that circNEURL4 might probably function as “sponges” to miRNAs. And then, we filtered miR-1278 and miR-521 as the potential targets of circNEURL4 by circBank and Circular RNA Interactome (Figure 4B). We then performed circRIP assay and the results revealed that only miR-1278 could be remarkably enriched by a circNEURL4-specific probe (Figure 4C). To further explore the relationship between circNEURL4 and miR-1278, we detected the binding site between circNEURL4 and miR-1278 and designed a mutant sequence of circNEURL4 (Figure 4D). We used miR-1278 mimics and detected the efficiency of the transfection (Figure 4E). Cotransfection of circNEURL4-WT (reporter plasmids containing wild type binding site of circNEURL4 to miR-1278) and miR-1278 mimics showed an obvious reduction of relative luciferase activity, whereas cotransfection of circNEURL4-MUT (reporter plasmids containing mutant binding site of circNEURL4 to miR-1278) and miR-1278 mimics showed no change of luciferase activity to the control (Figure 4F), indicating that miR-1278 could directly bind to circNEURL4 and down-regulate its expression. Subsequent RIP assay illustrated that miR-1278 degraded circNEURL4 through an Ago-dependent manner (Figure 4G). Besides those above, we detected the expression of miR-1278 in BCPAP-circNEURL4 and TPC-1-circNEURL4 cell lines compared to their matching control cell lines and found that circNEURL4 could also down-regulate miR-1278’ expression (Figure 4H). Thus, we proved that circNEURL4 could directly bind to miR-1278 and they were antagonists to each other.
What’s more, we found that miR-1278 was highly expressed in our PTC samples (Figure 4I), with a negative correlation with the expression of circNEURL4 (Figure 4J).
Inhibition of miR-1278 decreases the growth, migration, and invasion of PTC cells
We had proved that circNEURL4 could bind to miR-1278 and their antagonism to each other in vitro and in vivo, so we wonder if inhibition of miR-1278 could have similar effects on PTC compared to the overexpression of circNEURL4.
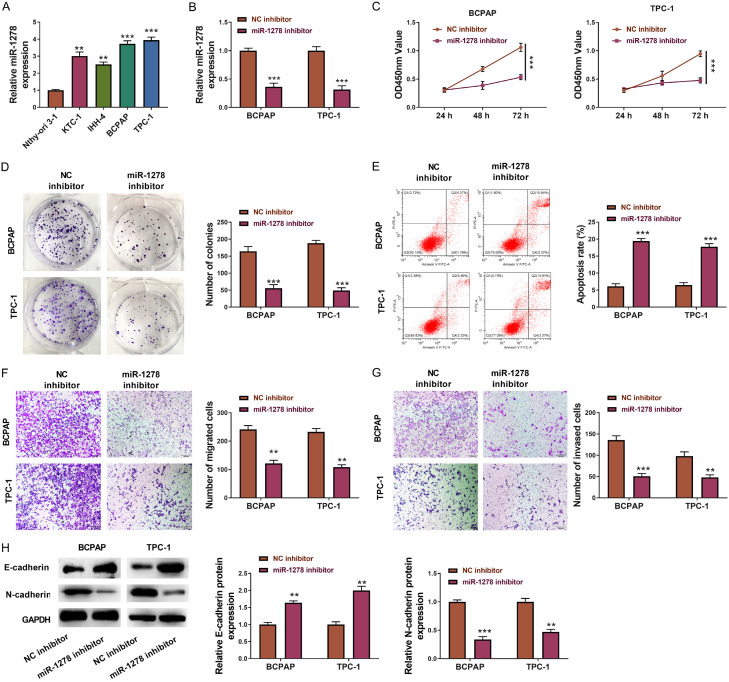
miR-1278 was highly expressed in the PTC cell lines (KTC-1, IHH-4, BCPAP, and TPC-1) compared to the normal thyroid cell line Nthy-ori 3-1, of which BCPAP and TPC-1 were most obvious (Figure 5A). Therefore, BCPAP and TPC-1 were chosen for subsequent experiments. After inhibiting miR-1278 expression in BCPAP and TPC-1 cells (Figure 5B), the cell viability and colony-formation were decreased (Figure 5C, 5D), and the apoptosis was increased (Figure 5E). Moreover, transwell assays manifested that inhibition of miR-1278 could notably reduce PTC cell lines’ migration and invasion (Figure 5F, 5G), accompanying with the increased expression of E-cadherin and reduced expression of N-cadherin (Figure 5H).
Figure 5.
Inhibition of miR-1278 decreases the growth, migration, and invasion of PTC cells. A. The relative expression level of miR-1278 in KTC-1, IHH-4, BCPAP, TPC-1, and Nthy-ori 3-1. B. The relative expression level of miR-1278 in BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. C. CCK8 assays of BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. Absorbances were measured at 24 h, 48 h, and 72 h post cell seeding. D. Pictures and statistical results of colony-formation assays using BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. E. Flow graph and statistical results of apoptosis assays using BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. F. Pictures and statistical results of migration assays using BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. G. Pictures and statistical results of invasion assays using BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. H. Western Blot and statistical results showing the expression of E-cadherin and N-cadherin in BCPAP and TPC-1 transfected with miR-1278 inhibitor or NC inhibitor. GAPDH was used as an internal reference. Statistical results were presented as the mean ± SME. Two-tailed paired t-test, *P<0.05, **P<0.01, ***P<0.001.
miR-1278 directly targets LAST1
To explore the downstream target gene that was liberated from miR-1278 by circNEURL4, we predicted the targets of miR-1278 through starbase and filtered out LATS1, a well-known cancer suppressor [28-30], as the potential candidate (Figure 6A). Cotransfection of LATS1-3’UTR-WT (reporter plasmids containing wild type binding site at LATS1’s 3’UTR to miR-1278) and miR-1278 mimics showed an obvious reduction of relative luciferase activity, whereas cotransfection of circNEURL4-MUT (reporter plasmids containing mutant binding site at LATS1’s 3’UTR to miR-1278) and miR-1278 mimics showed no change of luciferase activity to the control (Figure 6B, 6C). Transfection of miR-1278 mimics to BCPAP and TPC-1 significantly diminished the expression of LATS1 (Figure 6D), whereas transfection of miR-1278 mimics to BCPAP and TPC-1 significantly up-regulated the expression of LATS1 (Figure 6E). These data revealed that miR-1278 could directly bind to and suppress LATS1.
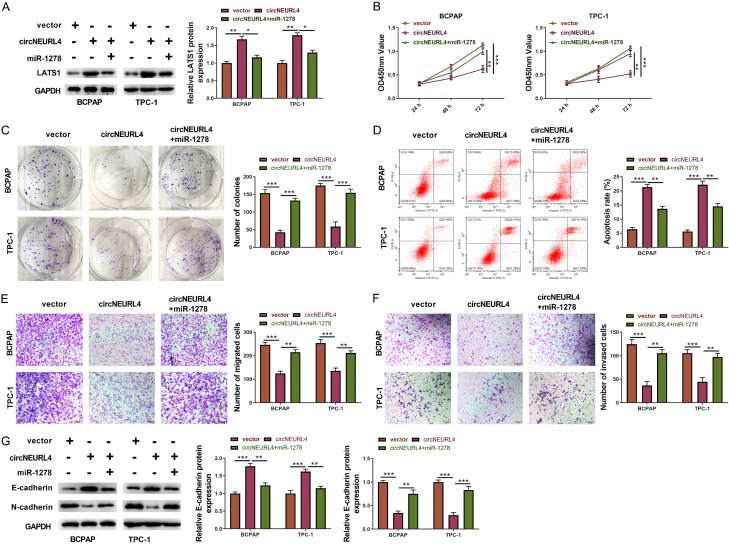
circNEURL4 regulates PTC through circNEURL4/miR-1278/LAST1 axis
With the validation of the direct binding of circNEURL4 to miR-1278 and miR-1278 to LATS1, we finally ought to prove the anti-tumor effect of circNEURL4/miR-1278/LATS1 axis in PTC cells.
We first detected the expression of LATS1 in three experimental groups: vector group (BCPAP or TPC-1 transfected with empty vector), circNEURL4 group (BCPAP or TPC-1 transfected with pLV-circNEURL4), and circNEURL4/miR-1278 group (BCPAP or TPC-1 transfected with pLV-circNEURL4 and miR-1278 mimics). Western Blot results showed circNEURL4 group had an increased expression of LATS1 relative to vector group, nevertheless, circNEURL4/miR-1278 group had a reduction in LATS1’s expression relative to circNEURL4 (Figure 7A), indicating that overexpression of circNEURL4 could up-regulate LATS1’s expression while additional miR-1278 could diminish the enhancement of circNEURL4 on LATS1.
Figure 7.
circNEURL4 regulates PTC through circNEURL4/miR-1278/LATS1 axis. A. Western Blot and statistical results showing the expression of LATS1 in BCPAP and TPC-1 with indicated conditions. GAPDH was used as an internal reference. B. CCK8 assays of BCPAP and TPC-1 with indicated conditions. Absorbances were measured at 24 h, 48 h, and 72 h post cell seeding. C. Pictures and statistical results of colony-formation assays using BCPAP and TPC-1 with indicated conditions. D. Flow graph and statistical results of apoptosis assays using BCPAP and TPC-1 with indicated conditions. E. Pictures and statistical results of migration assays using BCPAP and TPC-1 with indicated conditions. F. Pictures and statistical results of invasion assays using BCPAP and TPC-1 with indicated conditions. G. Western Blot and statistical results showing the expression of E-cadherin and N-cadherin in BCPAP and TPC-1 with indicated conditions. GAPDH was used as an internal reference. Statistical results were presented as the mean ± SME of three independent experiments. Dunnett’s multiple comparisons test, **P<0.01; ***P<0.001.
Moreover, compared to the circNEURL4 group, circNEURL4/miR-1278 group showed increased cell viability and colony formation (Figure 7B, 7C); littler apoptotic cells (Figure 7D); enhanced migration and invasion accompanying with reduced expression of E-cadherin and increased expression of N-cadherin (Figure 7E-G). These data confirmed the antagonistic function of miR-1278 to circNEURL4, and deeply confirmed the regulatory axis of circNEURL4/miR-1278/LATS1.
Discussion
Nowadays, increasing pieces of evidence suggest that circRNA plays a crucial role in the development of PTC [16-19]. However, most circRNAs that have been detected are oncogenic to PTC, indicating more researches are needed. Our research focused on the down-regulated circRNAs in the PTC samples, proposing to find a circRNA that could inhibit the development of PTC. Based on the public database, we filtered circNEURL4 as the most appropriate candidate for our subsequent research. We confirmed this finding in our PTC samples And then, proved its inhibitive effects on the growth, migration, and invasion of PTC cells. Furthermore, we reported that circNEURL4 could directly binding to miR-1278 and miR-1278 could directly interact with LATS1, illustrating the regulatory function of circNEURL4/miR-1278/LATS1 axis. Our data was the first one that showed the relationship between circNEURL4 and cancer. We found circNEURL4 could comprehensively affect the progress of PTC, including its growth, migration, and invasion. It will be very interesting to explore circNEURL4’s effect on other cancers. Whether its function is widely applicable or specific to PTC is unknown.
miR-1278 has been reported to participate in the development of other cancers, for example, it can interact with LINC00294 to promote glioma cells’ proliferation [31], and it is found in a circRNA/miRNA/mRNA regulatory network in renal cell carcinoma [32]. Our finding is the first report that miR-1278 performs as a PTC enhancer, expanding its research in cancers.
LATS1 is one of the core components belonging to the Hippo pathway and has been well studied to be a suppressor to different cancers, for example, lung cancer and hepatocellular carcinoma [28-30]. LATS1 is widely involved in cell metabolism, including cell proliferation and apoptosis [28]. Nevertheless, LATS1 has not been uncovered to be related to PTC. Our data revealed that LATS1 could negatively regulate the progress of PTC, confirming its role as a cancer suppressor.
Our finding showed that decreased expression of circNEURL4 was closely associated with a poor prognosis of patients with PTC and could inhibit the growth, migration, and invasion of PTC cells, shedding lights on the importance of circNEURL4/miR-1278/LATS1 axis in regulating PTC, and the potential as a target in clinical diagnosis and treatment.
Acknowledgements
This work was supported by the Science and Technology Conditions and Platform Construction Project of Jilin Science and Technology Department for Establishment of animal model of primary hyperthyroidism by immunizing homologous BALB/c mice with spleen cells activated by human thyroid stimulating hormone receptor cDNA (20180623020TC).
Disclosure of conflict of interest
None.
References
- 1.Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12:646–653. doi: 10.1038/nrendo.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang TS, Sosa JA. Thyroid surgery for differentiated thyroid cancer-recent advances and future directions. Nat Rev Endocrinol. 2018;14:670–683. doi: 10.1038/s41574-018-0080-7. [DOI] [PubMed] [Google Scholar]
- 3.Fröhlich E, Wahl R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat Rev. 2014;40:665–674. doi: 10.1016/j.ctrv.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Fagin JA, Wells SA. Biologic and clinical perspectives on thyroid cancer. N Engl J Med. 2016;375:1054–1067. doi: 10.1056/NEJMra1501993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomberg M, Feldt-Rasmussen U, Andersen KK, Kjaer SK. Thyroid cancer in Denmark 1943-2008, before and after iodine supplementation. Int J Cancer. 2012;131:2360–2366. doi: 10.1002/ijc.27497. [DOI] [PubMed] [Google Scholar]
- 6.Enewold L, Zhu K, Ron E, Marrogi AJ, Stojadinovic A, Peoples GE, Devesa SS. Rising thyroid cancer incidence in the United States by demographic and tumor characteristics, 1980-2005. Cancer Epidemiol Biomarkers Prev. 2009;18:784–791. doi: 10.1158/1055-9965.EPI-08-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent WDT, Hall SF, Isotalo PA, Houlden RL, George RL, Groome PA. Increased incidence of differentiated thyroid carcinoma and detection of subclinical disease. CMAJ. 2007;177:1357–1361. doi: 10.1503/cmaj.061730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saaoud F, Drummer I V C, Shao Y, Sun Y, Lu Y, Xu K, Ni D, Jiang X, Wang H, Yang X. Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers. Pharmacol Ther. 2020:107715. doi: 10.1016/j.pharmthera.2020.107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan L, Chen YG. Circular RNAs in immune response and viral infection. Trends Biochem Sci. 2020;45:1022–1034. doi: 10.1016/j.tibs.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang ZC, Guo XL, Li X. The novel roles of circular RNAs in metabolic organs. Genes Dis. 2017;5:16–23. doi: 10.1016/j.gendis.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zucko D, Boris-Lawrie K. Circular RNAs are regulators of diverse animal transcriptomes: one health perspective. Front Genet. 2020;11:999. doi: 10.3389/fgene.2020.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han D, Wang Y, Wang Y, Dai X, Zhou T, Chen J, Tao B, Zhang J, Cao F. The tumor-suppressive human circular RNA CircITCH sponges miR-330-5p to ameliorate doxorubicin-induced cardiotoxicity through upregulating SIRT6, survivin, and SERCA2a. Circ Res. 2020;127:e108–e125. doi: 10.1161/CIRCRESAHA.119.316061. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Z, Xia L, Fan S, Zheng J, Qin J, Fan X, Liu Y, Tao J, Liu Y, Li K, Ling Z, Bu Y, Martin KA, Hwa J, Liu R, Tang WH. Circular RNA circMAP3K5 acts as a microRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated SMC differentiation. Circulation. 2021;143:354–371. doi: 10.1161/CIRCULATIONAHA.120.049715. [DOI] [PubMed] [Google Scholar]
- 14.Ding B, Fan W, Lou W. hsa_circ_0001955 enhances proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS axis. Mol Ther Nucleic Acids. 2020;22:445–455. doi: 10.1016/j.omtn.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2020 doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 16.Ye M, Hou H, Shen M, Dong S, Zhang T. Circular RNA circFOXM1 plays a role in papillary thyroid carcinoma by sponging miR-1179 and regulating HMGB1 expression. Mol Ther Nucleic Acids. 2020;19:741–750. doi: 10.1016/j.omtn.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J, Shen Y, Kang J, Li J. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10:184. doi: 10.1038/s41419-019-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503:56–61. doi: 10.1016/j.bbrc.2018.05.174. [DOI] [PubMed] [Google Scholar]
- 19.Long MY, Chen JW, Zhu Y, Luo DY, Lin SJ, Peng XZ, Tan LP, Li HH. Comprehensive circular RNA profiling reveals the regulatory role of circRNA_0007694 in papillary thyroid carcinoma. Am J Transl Res. 2020;12:1362–1378. [PMC free article] [PubMed] [Google Scholar]
- 20.Quiñones-Pérez M, Cieza R, Ngo BKD, Grunlan MA, Domenech M. Amphiphilic silicones to reduce the absorption of small hydrophobic molecules. Acta Biomater. 2021;121:339–348. doi: 10.1016/j.actbio.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 21.Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS One. 2017;12:e0170287. doi: 10.1371/journal.pone.0170287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahmood T, Yang PC. Western blot: technique, theory, and trouble shooting. N Am J Med Sci. 2012;4:429–434. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Jin W, Song Y, Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol Cancer. 2017;16:166. doi: 10.1186/s12943-017-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amschler K, Beyazpinar I, Erpenbeck L, Kruss S, Spatz JP, Schön MP. Morphological plasticity of human melanoma cells is determined by nanoscopic patterns of E- and N-cadherin interactions. J Invest Dermatol. 2019;139:562–572. doi: 10.1016/j.jid.2018.09.027. [DOI] [PubMed] [Google Scholar]
- 25.Klymenko Y, Kim O, Loughran E, Yang J, Lombard R, Alber M, Stack MS. Cadherin composition and multicellular aggregate invasion in organotypic models of epithelial ovarian cancer intraperitoneal metastasis. Oncogene. 2017;36:5840–5851. doi: 10.1038/onc.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong D, Liu Q, Liu G, Xu J, Lan W, Jiang Y, Xiao H, Zhang D, Jiang J. Metformin inhibits castration-induced EMT in prostate cancer by repressing COX2/PGE2/STAT3 axis. Cancer Lett. 2017;389:23–32. doi: 10.1016/j.canlet.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Ratneswaran A, Kapoor M. Year in review: genetics, genomics, epigenetics. Osteoarthritis Cartilage. 2020 doi: 10.1016/j.joca.2020.11.003. S1063-4584(20)31171-7. [DOI] [PubMed] [Google Scholar]
- 28.Fallahi E, O’Driscoll NA, Matallanas D. The MST/hippo pathway and cell death: a non-canonical affair. Genes (Basel) 2016;7:28. doi: 10.3390/genes7060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song W, Zhang J, Zhang J, Sun M, Xia Q. Overexpression of lncRNA PIK3CD-AS1 promotes expression of LATS1 by competitive binding with microRNA-566 to inhibit the growth, invasion and metastasis of hepatocellular carcinoma cells. Cancer Cell Int. 2019;19:150. doi: 10.1186/s12935-019-0857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Yu X, Wei W, Liu Y. Long noncoding RNA MACC1-AS1 promotes the stemness of nonsmall cell lung cancer cells through promoting UPF1-mediated destabilization of LATS1/2. Environ Toxicol. 2020;35:998–1006. doi: 10.1002/tox.22936. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, Lv L, Zhang Z, Wei S, Zheng T. LINC00294 negatively modulates cell proliferation in glioma through a neurofilament medium-mediated pathway via interacting with miR-1278. J Gene Med. 2020;22:e3235. doi: 10.1002/jgm.3235. [DOI] [PubMed] [Google Scholar]
- 32.Bai S, Wu Y, Yan Y, Shao S, Zhang J, Liu J, Hui B, Liu R, Ma H, Zhang X, Ren J. Construct a circRNA/miRNA/mRNA regulatory network to explore potential pathogenesis and therapy options of clear cell renal cell carcinoma. Sci Rep. 2020;10:13659. doi: 10.1038/s41598-020-70484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]