Abstract
Enhancer of zeste homolog 2 (EZH2) is the catalytic core of polycomb repressive complex 2 (PRC2), which primarily methylates lysine 27 on histone H3 (H2K27me3), generating transcriptionally suppressed heterochromatin. Since EZH2 suppresses expression of genes involved in dentin formation, we examined the role of EZH2 in tooth development. Intriguingly, microCT analysis of teeth from mice with conditional Ezh2 knockout in uncommitted mesenchymal cells showed hyper-mineralization of enamel, which is produced by the epithelial-lineage cells, ameloblasts. Scanning electron microscopy analysis and nano-indentation of the incisor enamel from knockout mice revealed smaller inter-rod spaces and higher hardness compared to wild type enamel, respectively. Interestingly, expression of the calcium channel subunit gene, Orai2, was decreased compared to its competitor, Orai1, both in knockout mouse incisors and the ex vivo culture of ameloblasts with the surrounding tissues under EZH2 inhibition. Moreover, histological analysis of incisor from knockout mice showed decreased ameloblastin and expedited KLK4 expression in the ameloblasts. These observations suggest that EZH2 depletion in dental mesenchymal cells reduces enamel matrix formation and increases enamel protease activity from ameloblasts, resulting in enamel hyper-mineralization. This study demonstrates the significant role of the suppressive H3K27me3 mark for heterochromatin on enamel formation.
Keywords: Enamel hyper-mineralization, EZH2, histone methylation, dental follicle cell, ameloblast
1. Introduction
The tooth enamel formation process by ameloblasts is generally subdivided into three main functional stages: the presecretory, secretory, and maturation stages [1]. Ameloblasts become tall columnar cells in the secretory stage and secrete a large amount of enamel matrix proteins including ameloblastin. The secretory stage enamel is rich in proteins, shows low hardness, and contains thin hydroxyapatite crystals [2]. Upon reaching full thickness of matrix, ameloblasts transform into a shorter shape, and greatly decrease enamel matrix secretion. After this transition, ameloblasts secrete kallikrein-related peptidase-4 (KLK4) to remove the secreted matrix proteins from the enamel layer [1]. After removal of matrix proteins, the volume of remnant crystals expands within a mildly acidic environment [3]. This crystal expansion is associated with the supersaturation of enamel fluid, primarily with calcium and phosphate ions secreted by ameloblasts [4]. Disruption of these amelogenesis processes during tooth development leads to clinically visible enamel abnormalities.
The packaging of DNA by histones in chromatin is regulated by post-translational modifications of histone proteins, such as methylation, phosphorylation, acetylation, ubiquitylation, and sumoylation [5,6]. Polycomb group (PcG) proteins including two polycomb repressive complexes (PRCs) regulate cellular processes during embryonic development [7]. PRC1 catalyzes the monoubiquitination of histone H2A at lysine 119 (H2AK119ub1), whereas PRC2 primarily trimethylates lysine 27 on histone H3 (H3K27me3) through its core component, enhancer zeste homolog 2 (EZH2) [8]. EZH2 can catalyze the mono-, di-, and trimethylation of H3K27, thus reducing chromatin accessibility and promoting gene silencing [9]. Conditional knockout of Ezh2 in uncommitted mesenchymal cells causes multiple defects in skeletal patterning and bone formation [10,11]. Additionally, Ezh2 is expressed during tooth development in the tooth germ, enamel organ, dental papilla, and odontoblast layer [12]. EZH2 suppresses the differentiation and mineralization of human dental pulp cells (hDPCs) through the Wnt/β-catenin pathway [13], and supports their proliferation [14]. Based on these earlier findings, we postulated that epigenetic regulatory mechanisms mediated by EZH2 may regulate tooth development and maintenance of dental tissues.
To prove this hypothesis, in the present study we used conditional Ezh2 knockout mice (Ezh2fl/fl : Prrx1-Cre+), whose Ezh2 SET domain is deleted in the uncommitted mesenchymal cells. Since this Ezh2 knockout leads to the upregulation of bone marker genes expressed in odontoblasts, such as osteocalcin Bglap, bone sialoprotein Ibsp and alkaline phosphatase Alpl [10], the influence of the Ezh2 knockout on dentin formation was naturally predicted. However, interestingly the knockout mice showed an enamel hyper-mineralization (EHm), while showing a relatively subtle change in the dentin. This means that the activities of ameloblasts, an epithelial cell lineage, were affected by Ezh2 knockout in the adjacent mesenchymal cells. By analyzing this unanticipated phenomenon, we reveal the significant role of EZH2 in mesenchymal cells on amelogenesis and hypothesize a possible new function of dental follicle cells.
2. Materials and Methods
2.1. Animals
All mice were housed by 2~5 animals per cage with free access to food and water, in a facility with 12-h light-dark cycles. The wildtype (WT, Ezh2wt/wt : Prrx1-Cre+) and conditional knockout (Ezh2cKO, Ezh2fl/fl : Prrx1-Cre+) mice were obtained as shown before [10]. These animals were housed following the guidelines provided by the Mayo Clinic IACUC. C57BL/6J (B6) and Ai6(RCL-ZsGreen) [15] mice were purchased from Jackson Laboratories. Ai6 mice were crossed with Prrx1-Cre+ mice [16]. The mice harboring a conditional Ezh2fl/fl allele were genetically backcrossed with C57BL/6 [10,17]. The animal protocols were approved by the Rutgers University IACUC.
2.2. MicroCT analysis
Mandibles were dissected from 3-week-old WT and Ezh2cKO mice and stored in 70% ethanol at 4°C until processing. They were scanned by the microCT instrument, Skyscan 1172, at 70 kV, 7.45 μm per pixel, at 0.3-degree rotation. The machine was calibrated using phantoms with CaHA concentrations of 0.25 and 0.75 g/cm3. Reconstruction of raw images was performed using NRecon V1.4.0 software (Skyscan). Data were analyzed using CTan (CT-vol software). Slides from the mesial side to distal side of the first molar were used to measure the volume and density of dentin, enamel, and pulp [18]. For the incisor, slides from the apical end to mesial side of the first molar, and erupted tip were used for analysis.
2.3. Hardness test with nano indentation
The hardness and Young’s modulus of the enamel in mandibular incisor tips from WT and Ezh2cKO mice were measured by nanoindentation tests on the samples using a nanoindenter (iNano from Nanomechanics, Inc., Oakridge, TN). The incisor samples were fixed with 10% natural buffered formalin overnight and kept in 70% ethanol. A typical indentation test involves pressing a hard diamond (Berkovich) indenter into the enamel at a constant displacement rate until a peak force of 45 mN is reached, followed by unloading; the force-penetration (or displacement) response is measured continuously throughout the load-unload cycle. Young’s modulus and hardness properties for each indentation test were obtained by analyzing the force-displacement curves using a standard method described previously [19]; a Poisson’s ratio of 0.3 was assumed for all measurements. A minimum of seven and a maximum of twenty indentation tests with the Berkovich tip were carried out per sample.
2.4. The ex vivo culture of ameloblasts with the surrounding tissues
The samples were isolated by modifying Suzawa’s protocol [20] (Supplementary fig. S1). Mandibles from 7~10-days postnatal mice were collected, and their ascending rami, molars, molar-alveoli, and incisor tips were carefully removed. From the remained incisor with the alveolar bone, ameloblasts together with the surrounding tissues were isolated with microdissection by removing their pulp and dentin. The collected samples were incubated in DMEM/F12 media supplemented with 10% FBS and 1X Antibiotic-Antimycotic (Thermo Fisher).
2.5. Statistical Analysis
All statistical analyses in this study were performed using two-tailed Student’s t-tests.
Additional information regarding materials and methods is available in Appendix.
3. Results
3.1. Enamel hyper-mineralization (EHm) in mice with Ezh2 knockout in the mesenchymal cells
To investigate the effect of EZH2 on tooth development, we analyzed conditional Ezh2 knockout mice (Ezh2fl/fl : Prrx1-Cre+). The Ezh2 SET domain of these mice are deleted in uncommitted mesenchymal cells by Cre-recombination in cells expressing the mesenchymal stem cell marker Prrx1. Five to eight mandibular incisors from wild type (WT) and Ezh2 knockout (Ezh2cKO) mice (3 weeks old) were analyzed by microCT (Fig. 1). Intriguingly, significantly increased mineral densities were observed in enamels from the Ezh2cKO mice incisors and molars (Fig. 1, Left. Bright green parts), whereas dentin densities from the cKO mice were similar to those of the WTs (Table 1). The mineral densities of incisor enamel tips from cKO mice were 8.13% higher than those from WT mice, reaching approximately 2.97 g/cm3 (Table 1), which is close to the mineral density of pure hydroxyapatite (3.15 g/cm3). The mineral densities of the apical side incisor enamels, molar cones, tip side incisor dentins, and apical side incisor dentins from cKO mice were increased by 16.85%, 5.6%, 2.92%, and 3.72%, respectively (Table 1).
Fig. 1.
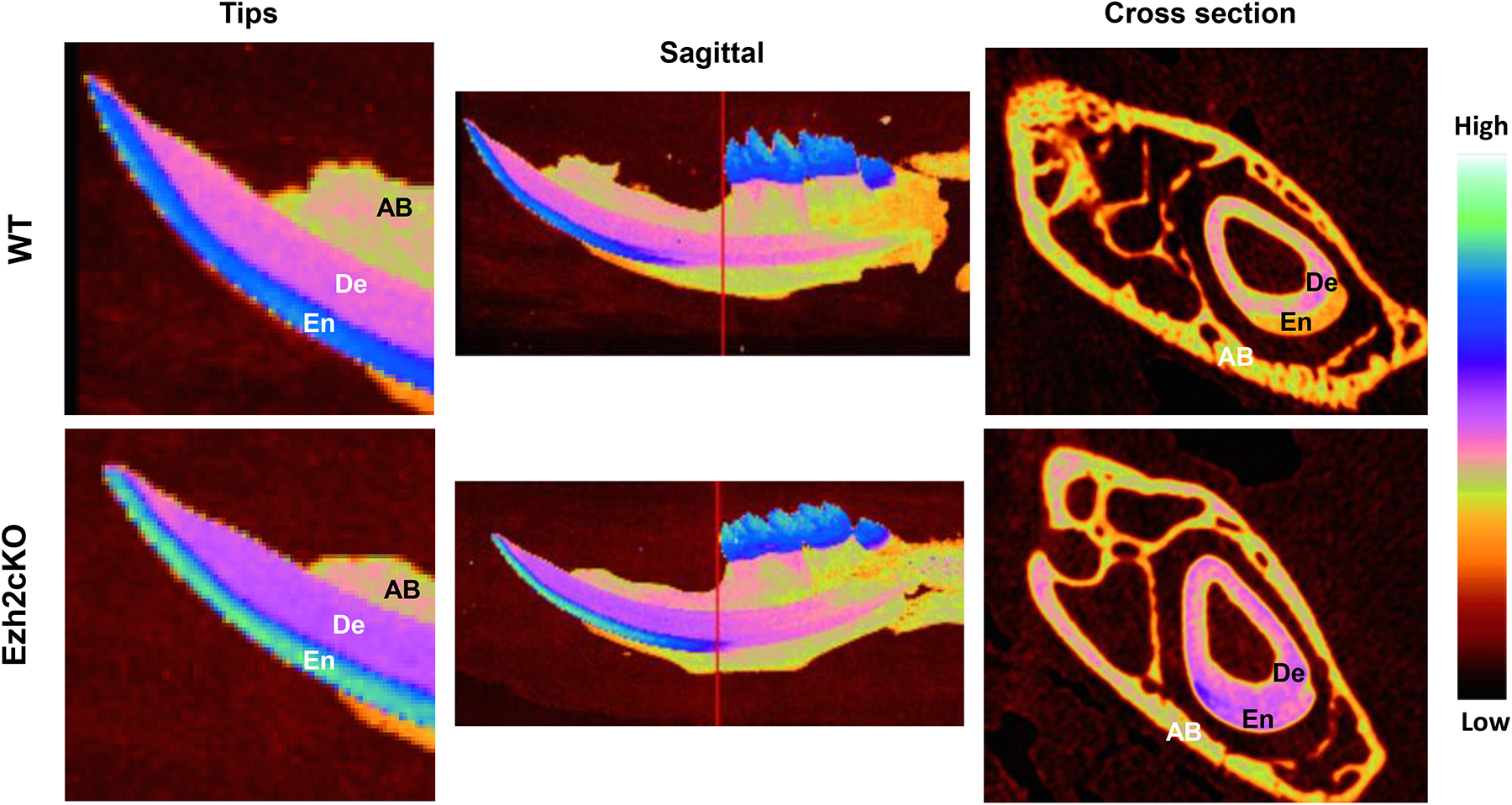
Typical microCT images of mandibles from the wild type (WT)(top) and conditional Ezh2 knockout (Ezh2cKO)(bottom) mice. Tip region (left), sagittal (middle) and cross section (right) images for those mice are shown. The mineral densities are displayed in pseudo-color, represented in the color bar at the far right. The red lines in sagittal images indicate the cross-section position. En, De, and AB indicate enamel, dentin, and alveolar bone, respectively.
Table 1.
Mineral density of incisors and molars from wild type or Ezh2 knock out mice.
| Wild typea | Ezh2 knock-outa | p-valueb | % increase | ||
|---|---|---|---|---|---|
| ENAMEL | Incisor tip | 2.75 (0.117) | 2.97 (0.097) | 0.017 | 8.13 |
| Incisor apical | 1.07 (0.099) | 1.24 (0.211) | 0.039 | 16.85 | |
| Molar | 2.68 (0.007) | 2.83 (0.107) | 0.033 | 5.60 | |
| DENTIN | Incisor tip | 1.46 (0.031) | 1.50 (0.025) | 0.025 | 2.92 |
| Incisor apical | 1.33 (0.023) | 1.38 (0.031) | 0.005 | 3.72 | |
| Molar | 1.34 (0.006) | 1.35 (0.007) | 0.351 | 0.66 |
Averaged mineral density (g/cm3) from five wild type and eight knock-out samples. Values in parentheses are standard deviations.
The p-values are calculated using two-tailed Student’s t-test.
To confirm tissue specificity of Cre-activity in the cKO mice, Prrx1-Cre+ mice were crossed with the corresponding line with the Ai6 (ZsGreen) reporter [15], in which the gfp variant, ZsGreen, is expressed only when recombination by Cre happens. As expected, ZsGreen was only expressed in the mesenchymal tissues among dental tissues in the Prrx1-Cre+:Ai6 mice (Supplementary fig. S2), and remarkably, no ZsGreen expression was observed in the ameloblasts or enamel organs. This suggests that Ezh2 knock out occurred only in the mesenchymal tissues, which affected ameloblasts or enamel organs, resulting in EHm.
3.2. Highly dense enamel rod structure in Ezh2 knockout mice incisors
The structure of hyper-mineralized enamel was analyzed with scanning electron microscopy (SEM) in the cross sections of the incisors from WT and Ezh2cKO mice (3 weeks old) (Fig. 2A). In the cKO mice enamel, each rod structure remained intact, however, the inter-rod spaces for remnant enamel matrix were remarkably smaller than that in WT mice (Fig. 2A). This observation suggests that hyper-mineralization is associated with a denser rod structure of enamel in cKO mice.
Fig. 2.
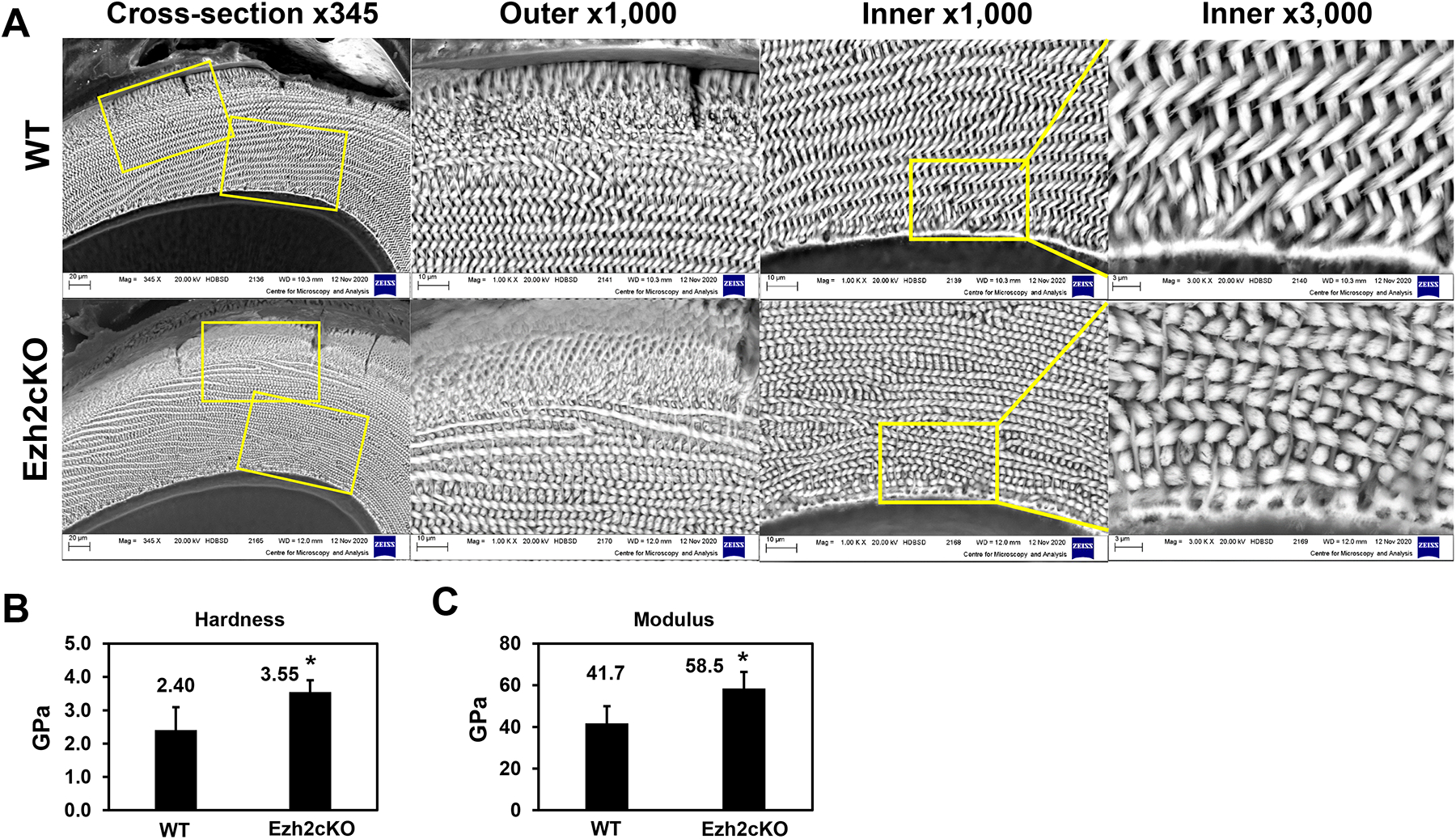
(A) The cross-section images at the eruption site of incisors from WT or Ezh2cKO mice, obtained by scanning electron microscopy. Yellow boxes indicate the outer or inner enamel areas shown magnified with indicated powers. The average values of (B) hardness and (C) Young’s modulus properties of the enamel from four WT and six cKO mandibular incisor tips were obtained using nanoindentation tests. A minimum of seven and a maximum of twenty indentation tests per sample were carried out on the enamel area. Error bars indicate standard deviations. *p < 0.05.
To determine if the dense rod structure affects the physical properties of enamel, the hardness and modulus were determined by nano-indentation (Supplementary fig. S3). The average hardness values of enamel in the incisor tips from cKO mice and WT mice were 3.55 GPa and 2.40 GPa, respectively (Fig. 2B). The enamel of cKO mice was 1.5-fold harder than that of WT and had a 1.4-fold higher Young’s modulus than that of the WT (Fig. 2B, and C). This result suggests that the denser rod structure results in Ezh2cKO EHm, making it significantly harder and stiffer.
To explore the machinery that mediates hyper-mineral deposition on the enamel matrix in Ezh2cKO mice, gene expression related to enamel calcium deposition was examined in the Ezh2cKO incisors using RT-qPCR (Fig. 3, Left). Among the results, Orai2 showed lower expression in Ezh2cKO incisors. ORAI2 is a subunit of the CRACC calcium channel and it competes with ORAI1, the homolog of ORAI2 with higher affinity [21]. It is possible that a decrease in ORAI2 made ORAI1 dominant and eventually increased the calcium transportation, as seen in previous reports [21,22]. Similar results were obtained from the ex vivo culture of ameloblasts with the surrounding tissues (Fig. 3, Right), which were isolated from WT mice mandibles and cultured for 24 h with or without the EZH2 inhibitor, GSK126. Importantly, all mesenchymal cells except dental follicles have been removed from this ex vivo culture (Supplementary fig. S1). Accordingly, if these changes of ameloblasts gene expression were triggered by the effect of any adjacent mesenchymal cell activity, the mesenchymal cells should be logically identified as dental follicles.
Fig. 3.
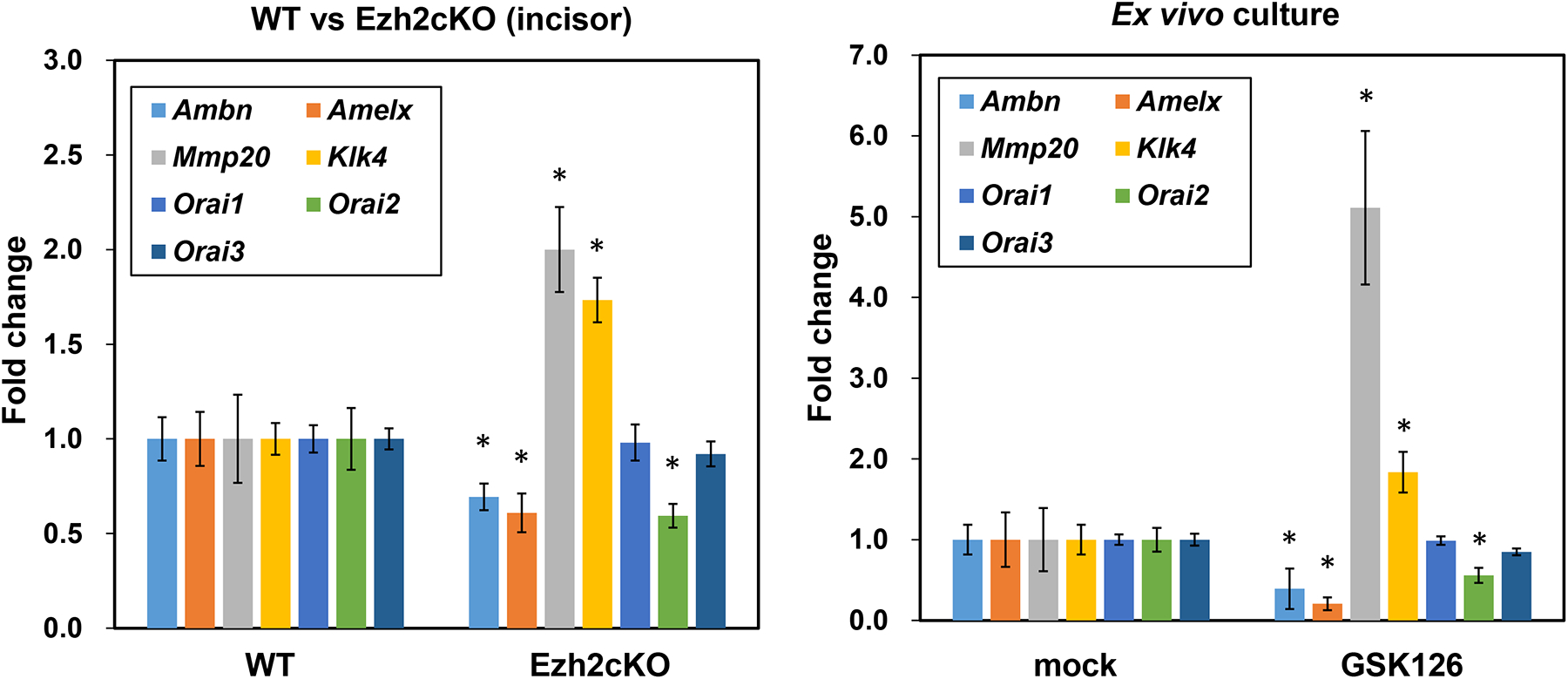
Profiling of ameloblast-related gene expression from WT or Ezh2cKO mice incisors (left) and from the ex vivo culture of ameloblasts with the surrounding tissues (right) using RT-qPCR. Each value was normalized to beta-actin level in the same sample. Each column shows the average from six samples. Error bars indicate standard errors of mean. *p < 0.05. Left: Total RNA from WT or Ezh2cKO mice incisors was isolated and the expression of indicated genes was analyzed. Right: Ameloblasts with the surrounding tissues from 7~10 days-old wild-type mice were cultured in DMEM/F12 media with or without 4 μM EZH2 inhibitor, GSK126. Total RNAs were isolated after 24 h.
3.3. Loss of EZH2 function represses enamel matrix genes but induces matrix protease genes
To elucidate the cause of EHm in Ezh2cKO mice, we measured ameloblast marker gene expression in cKO mice incisors (Fig. 3, Left). Interestingly, a significant increase in Klk4 and Mmp20 expression, as well as a decrease in ameloblastin and amelogenin expression were observed (Fig. 3, Left). Even clearer results were obtained from the ex vivo culture of ameloblasts with the surrounding tissues treated with GSK126 (Fig. 3, Right), which showed a substantial increase in Mmp20 and Klk4 and a decrease in ameloblastin and amelogenin. These findings suggest that the combination of reduced matrix protein and increased protease activity could contributes to the highly dense rod structure in the enamel of Ezh2cKO mice.
3.4. Decreased ameloblastin and accelerated KLK4 expression in ameloblasts from Ezh2 knockout mice
To corroborate the changes in expression of ameloblast marker genes, we analyzed enamel matrix protein ameloblastin (Fig. 4A) and protease KLK4 (Fig. 4B–E) in mice incisors by immunohistochemistry. Ameloblasts across the secretion stage in Ezh2cKO mice showed attenuated ameloblastin signals compared to WT mice as revealed by brown immunohistochemical staining in ameloblasts as indicated (Fig. 4A). In contrast, the KLK4 signal was detected at an earlier developmental stage of ameloblast differentiation in Ezh2cKO mice (Fig. 4E) and was stronger than that in WT mice as reflected by a line of brown staining (indicated with arrows; Fig. 4D, and E). These observations support the hypothesis that reduced matrix protein, by both increased protease activity and decreased matrix protein expression, contributes to the EHm in Ezh2cKO mice.
Fig. 4.
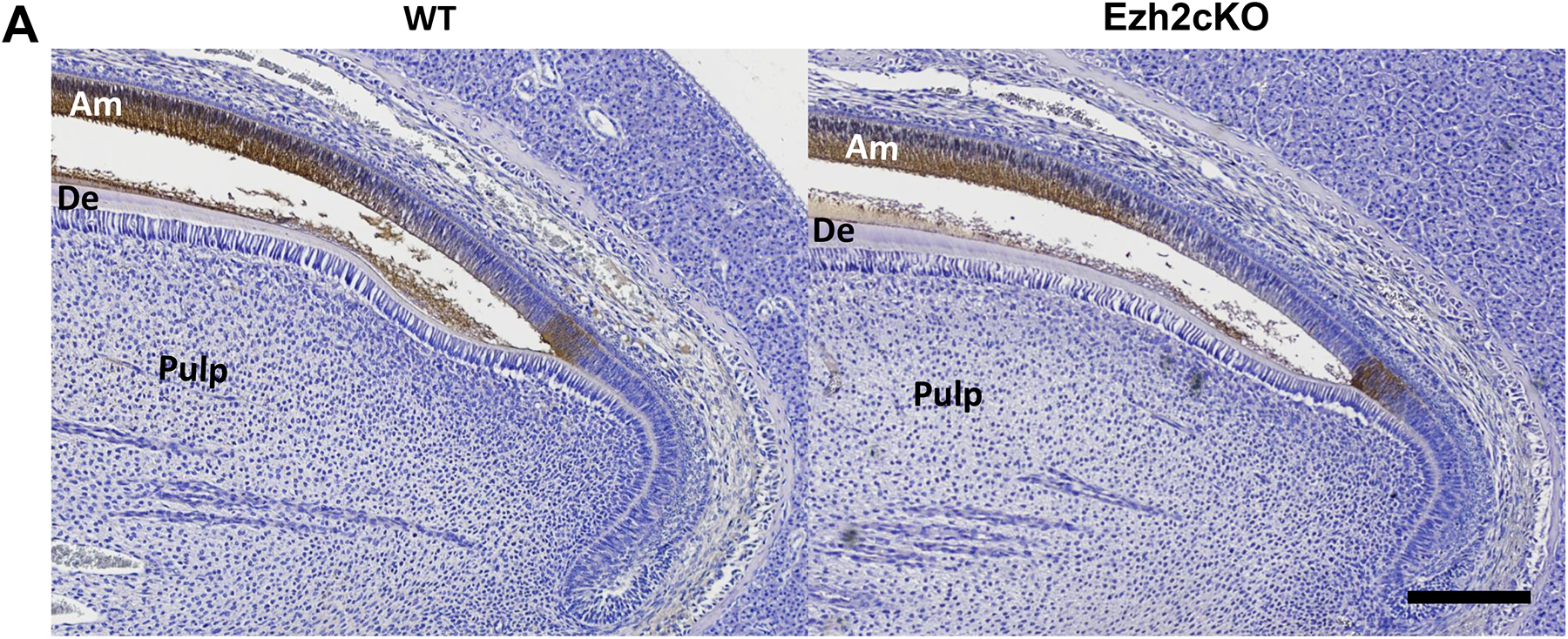
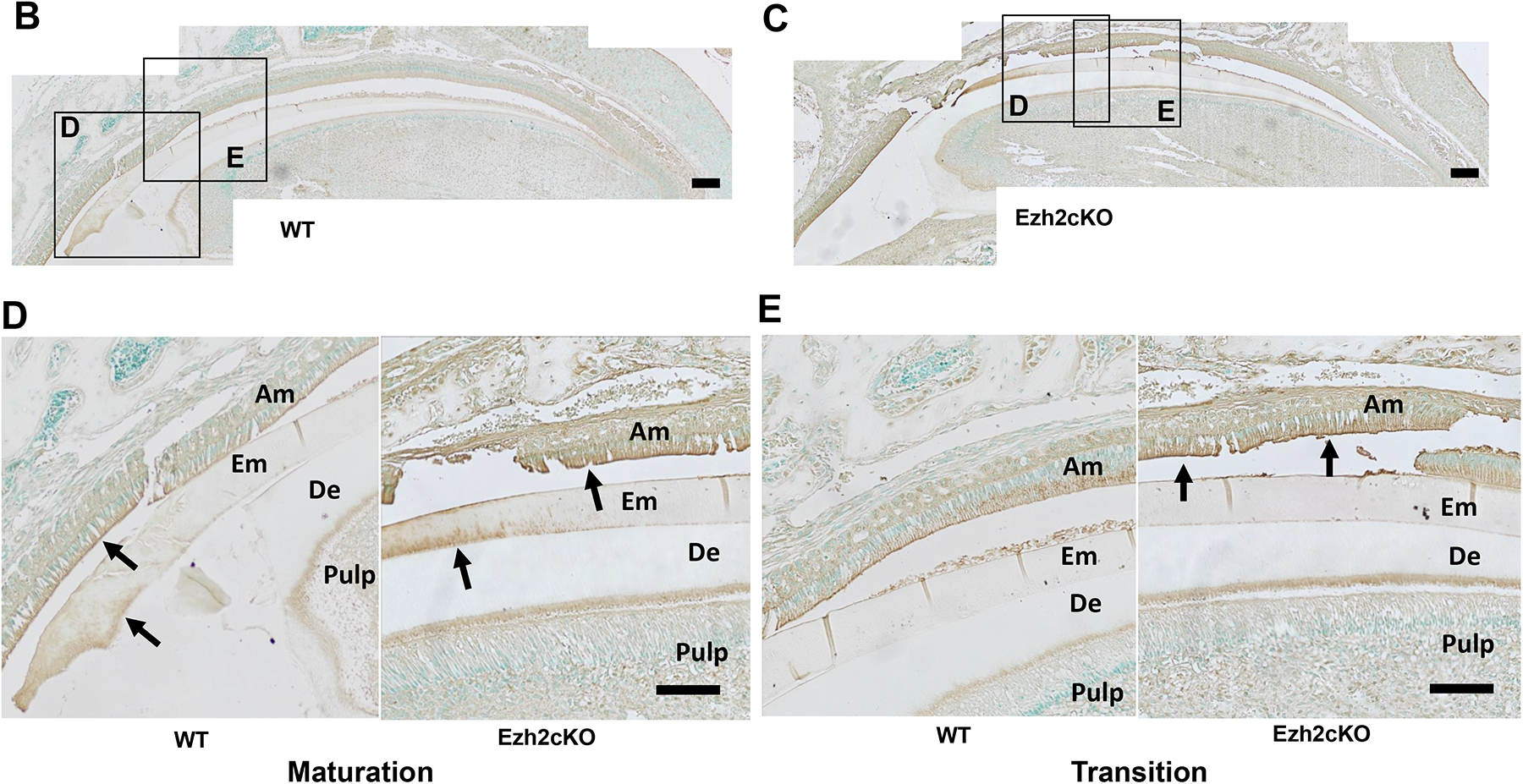
(A) Ameloblastin expression in the maxillary incisor from WT (left) or Ezh2cKO mouse (right). Sagittal sections of the incisor were stained with hematoxylin and DAB using HRP-conjugated anti-ameloblastin antibody (brown). Portions close to the cervical loop are shown. Ameloblastin signal is shown to be localized in ameloblasts and the enamel matrix surface. Scale bar = 100 μm. (B, C) KLK4 expression in maxillary incisor from (B) WT or (C) Ezh2cKO mouse. Sagittal sections of the incisor were stained with methyl green and DAB using HRP-conjugated anti-KLK4 antibody (brown). Scale bar = 200 μm. Magnified images (D) and (E) show maturation stage and transition stage ameloblasts from WT and knockout mouse, respectively. Arrows indicate residual KLK4 protein stained with DAB. Am: ameloblast, De: dentin, EM: enamel matrix. Scale bar = 100 μm.
4. Discussion
In this report, we performed a detailed examination of the EHm caused by conditional Ezh2 knockout in mesenchymal cells. So far, only a few reports have focused on the role of EZH2 in enamel formation or ameloblast development. For example, EZH2 is expressed in the enamel epithelium, stellate reticulum, and dental papilla in a spatio-temporal manner during murine tooth germ development [12]. Ezh2 knockout in dental mesenchymal cells affects root formation of mouse molars [23]. These emerging findings support our studies regarding the function of EZH2 in mesenchymal cells on enamel formation, and also reflect the complexity of epigenetic regulatory mechanism that control odontogenesis.
Unlike amelogenesis imperfecta (AI), scientific reports about EHm with genetic causes are quite rare. (1) EHm has been reported in vitamin D receptor (VDR) knockout mice [24,25]. Bone and dentin formation in these mice were impaired due to calcium metabolism deficiency. (2) Osteoprotegerin (OPG; Tnfrsf11b) knockout mice exhibit EHm as well as reduced alveolar bone mass [26]. (3) Bone sialoprotein (BSP; Ibsp) knockout have increased enamel mineralization, reduced bone mineral density, bone turnover, osteoclast activation, and impaired bone healing [27]. Interestingly, all three of the above gene knockouts causing EHm are related to bone formation and accordingly, all these mice exhibited deficiencies in osteogenesis.
To explain the EHm phenotypes of these mice with deficient osteogenesis, the above groups have proposed several theories. (1) Zhang et al. postulated that lacking a VDR may result in opening of tight junctions in ruffle-ended ameloblasts and enhance paracellular calcium transport as observed in the intestines [24,25]. (2) Soenjaya et al. proposed that periodontal dysfunction and the consequent lower rates of incisor eruption may underlie EHm [27]. In the present study, Ezh2cKO mice in the mesenchymal cells (Prrx1-Cre:Ezh2fl/fl) also show skeletal development deficiencies including shortened forelimbs, craniosynostosis, and clinodactyly due to precocious maturation of osteoblasts, as previously reported [10]. Hence, there can be some causality between EHm and bone formation defects. However, biological relationships may be even more complex, because the combination of AI and bone formation defects has been reported on several occasions [28–30].
The combination of accelerated protease expression and lower matrix protein expression observed in our study could explain EHm. Absence of functional KLK4 causes a hypomaturation enamel phenotype [31,32]. Ameloblastin overexpression results in AI-like problems [33]. Our study shows the opposite combination of above two cases, which would clarify the cause of increased mineralization. A reduced amount of enamel matrix may be easily degraded by augmented KLK4 and as a result, the originally small enamel matrix spaces could be quickly replaced with an increased amount of hydroxyapatite crystals.
Another biological mechanism which could result in EHm is decreased ORAI2 expression in ameloblasts from cKO mice (Fig. 3). Depletion of ORAI2 can result in dominance of ORAI1, an ORAI2 homolog with higher efficiency, on the cell surface and eventually increase the calcium transportation capacity of cells. This type of calcium transportation regulation depending on the ratio of ORAI1-ORAI2 has been found in chondrocyte cell lines and in T cells [21,22].
The pathways from Ezh2 in mesenchyme to the ameloblast genes involved in EHm remain to be fully clarified. Our observations from the ex vivo culture of ameloblasts with the surrounding tissues suggest the contribution of dental follicles to this phenomenon, since dental follicles were the only mesenchymal cells included in the culture (Supplementary fig. S1, S2). The effect of dental follicle on enamel formation is barely known except their apoptosis-inductivity on ameloblast-linage cells [34]. Our findings could have shown a possible new function of dental follicle on enamel formation, but its details are yet to be clarified. Specification of these pathways may provide an insight into potential strategies for the treatment of congenital AI. The local activation of the pathways in the developing teeth of AI patients could help in augmenting enamel mineralization, which will be a significant contribution to the future AI therapy.
Supplementary Material
Highlights.
The mice with conditional Ezh2 knockout in the uncommitted mesenchymal cells exhibited tooth enamel hyper-mineralization.
Smaller inter-rod spaces in the enamels were observed from the knockout mice using scanning electron microscopic analysis.
Incisors from the knockout mice showed decreased ameloblastin and expedited KLK4 expression in the ameloblasts.
Similar results were obtained from the ex vivo culture of ameloblasts with the surrounding tissues under EZH2 inhibition.
The combination of decreased enamel matrix protein and increased protease expression could explain the enamel hyper-mineralization.
Acknowledgements
This study was supported by National Institute of Dental and Craniofacial Research grant R01-DE025885 (to ES), National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01-AR049069 (to AJvW), and National Institute of Health instrument (micro-CT) grant S10OD010751-01A1 (to New York University College of Dentistry).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Bartlett JD, Dental enamel development: proteinases and their enamel matrix substrates, ISRN Dent 2013 (2013) 684607. 10.1155/2013/684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Smith CEL, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, Mighell AJ, Amelogenesis Imperfecta; Genes, Proteins, and Pathways, Front Physiol 8 (2017) 435. 10.3389/fphys.2017.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bronckers AL, Lyaruu D, Jalali R, Medina JF, Zandieh-Doulabi B, DenBesten PK, Ameloblast Modulation and Transport of Cl(−), Na(+), and K(+) during Amelogenesis, J Dent Res 94 (2015) 1740–1747. 10.1177/0022034515606900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lacruz RS, Smith CE, Kurtz I, Hubbard MJ, Paine ML, New paradigms on the transport functions of maturation-stage ameloblasts, J Dent Res 92 (2013) 122–129. 10.1177/0022034512470954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bannister AJ, Kouzarides T, Regulation of chromatin by histone modifications, Cell Res 21 (2011) 381–395. 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van Wijnen AJ, Westendorf JJ, Epigenetics as a New Frontier in Orthopedic Regenerative Medicine and Oncology, J Orthop Res 37 (2019) 1465–1474. 10.1002/jor.24305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang W, Qin JJ, Voruganti S, Nag S, Zhou J, Zhang R, Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications, Med Res Rev 35 (2015) 1220–1267. 10.1002/med.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang Y, Zhao W, Wang C, Zhu Y, Liu M, Tong H, Xia Y, Jiang Q, Qin J, Combinatorial Control of Recruitment of a Variant PRC1.6 Complex in Embryonic Stem Cells, Cell Rep 22 (2018) 3032–3043. 10.1016/j.celrep.2018.02.072. [DOI] [PubMed] [Google Scholar]
- [9].Chou RH, Chiu L, Yu YL, Shyu WC, The potential roles of EZH2 in regenerative medicine, Cell Transplant 24 (2015) 313–317. 10.3727/096368915X686823. [DOI] [PubMed] [Google Scholar]
- [10].Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ, van Wijnen AJ, Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2, J Biol Chem 290 (2015) 27604–27617. 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dudakovic A, Camilleri ET, Paradise CR, Samsonraj RM, Gluscevic M, Paggi CA, Begun DL, Khani F, Pichurin O, Ahmed FS, Elsayed R, Elsalanty M, McGee-Lawrence ME, Karperien M, Riester SM, Thaler R, Westendorf JJ, van Wijnen AJ, Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice, J Biol Chem 293 (2018) 12894–12907. 10.1074/jbc.RA118.002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zheng LW, Zhang BP, Xu RS, Xu X, Ye L, Zhou XD, Bivalent histone modifications during tooth development, Int J Oral Sci 6 (2014) 205–211. 10.1038/ijos.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li B, Yu F, Wu F, Hui T, A P, Liao X, Yin B, Wang C, Ye L, EZH2 Impairs Human Dental Pulp Cell Mineralization via the Wnt/beta-Catenin Pathway, J Dent Res 97 (2018) 571–579. 10.1177/0022034517746987. [DOI] [PubMed] [Google Scholar]
- [14].Hui T, A P, Zhao Y, Wang C, Gao B, Zhang P, Wang J, Zhou X, Ye L, EZH2, a potential regulator of dental pulp inflammation and regeneration, J Endod 40 (2014) 1132–1138. 10.1016/j.joen.2014.01.031. [DOI] [PubMed] [Google Scholar]
- [15].Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H, A robust and high-throughput Cre reporting and characterization system for the whole mouse brain, Nat Neurosci 13 (2010) 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ, Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer, Genesis 33 (2002) 77–80. 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- [17].Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A, Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement, Nat Immunol 4 (2003) 124–131. 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- [18].Matsumura S, Quispe-Salcedo A, Schiller CM, Shin JS, Locke BM, Yakar S, Shimizu E, IGF-1 Mediates EphrinB1 Activation in Regulating Tertiary Dentin Formation, J Dent Res 96 (2017) 1153–1161. 10.1177/0022034517708572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Oliver WC, Pharr GM, An Improved Technique for Determining Hardness and Elastic-Modulus Using Load and Displacement Sensing Indentation Experiments, J Mater Res 7 (1992) 1564–1583. Doi 10.1557/Jmr.1992.1564. [DOI] [Google Scholar]
- [20].Suzawa T, Itoh N, Takahashi N, Katagiri T, Morimura N, Kobayashi Y, Yamamoto T, Kamijo R, Establishment of primary cultures for mouse ameloblasts as a model of their lifetime, Biochem Biophys Res Commun 345 (2006) 1247–1253. 10.1016/j.bbrc.2006.04.122. [DOI] [PubMed] [Google Scholar]
- [21].Inayama M, Suzuki Y, Yamada S, Kurita T, Yamamura H, Ohya S, Giles WR, Imaizumi Y, Orai1-Orai2 complex is involved in store-operated calcium entry in chondrocyte cell lines, Cell Calcium 57 (2015) 337–347. 10.1016/j.ceca.2015.02.005. [DOI] [PubMed] [Google Scholar]
- [22].Vaeth M, Yang J, Yamashita M, Zee I, Eckstein M, Knosp C, Kaufmann U, Karoly Jani P, Lacruz RS, Flockerzi V, Kacskovics I, Prakriya M, Feske S, ORAI2 modulates store-operated calcium entry and T cell-mediated immunity, Nat Commun 8 (2017) 14714. 10.1038/ncomms14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jing J, Feng J, Li J, Han X, He J, Ho TV, Du J, Zhou X, Urata M, Chai Y, Antagonistic interaction between Ezh2 and Arid1a coordinates root patterning and development via Cdkn2a in mouse molars, Elife 8 (2019). 10.7554/eLife.46426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang X, Rahemtulla F, Zhang P, Beck P, Thomas HF, Different enamel and dentin mineralization observed in VDR deficient mouse model, Arch Oral Biol 54 (2009) 299–305. 10.1016/j.archoralbio.2009.01.002. [DOI] [PubMed] [Google Scholar]
- [25].Zhang X, Rahemtulla F, Zhang P, Li X, Beck P, Thomas HF, Normalisation of calcium status reverses the phenotype in dentin, but not in enamel of VDR-deficient mice, Arch Oral Biol 54 (2009) 1105–1110. 10.1016/j.archoralbio.2009.09.004. [DOI] [PubMed] [Google Scholar]
- [26].Sheng ZF, Ye W, Wang J, Li CH, Liu JH, Liang QC, Li S, Xu K, Liao EY, OPG knockout mouse teeth display reduced alveolar bone mass and hypermineralization in enamel and dentin, Arch Oral Biol 55 (2010) 288–293. 10.1016/j.archoralbio.2010.02.007. [DOI] [PubMed] [Google Scholar]
- [27].Soenjaya Y, Foster BL, Nociti FH Jr., Ao M, Holdsworth DW, Hunter GK, Somerman MJ, Goldberg HA, Mechanical Forces Exacerbate Periodontal Defects in Bsp-null Mice, J Dent Res 94 (2015) 1276–1285. 10.1177/0022034515592581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Malloy PJ, Feldman D, Genetic disorders and defects in vitamin d action, Endocrinol Metab Clin North Am 39 (2010) 333–346, table of contents. 10.1016/j.ecl.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Foster BL, Ramnitz MS, Gafni RI, Burke AB, Boyce AM, Lee JS, Wright JT, Akintoye SO, Somerman MJ, Collins MT, Rare bone diseases and their dental, oral, and craniofacial manifestations, J Dent Res 93 (2014) 7S–19S. 10.1177/0022034514529150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Irizarry AR, Yan G, Zeng Q, Lucchesi J, Hamang MJ, Ma YL, Rong JX, Defective enamel and bone development in sodium-dependent citrate transporter (NaCT) Slc13a5 deficient mice, PLoS One 12 (2017) e0175465. 10.1371/journal.pone.0175465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT, Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta, J Med Genet 41 (2004) 545–549. 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC, Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice, J Biol Chem 284 (2009) 19110–19121. 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML, A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression, J Biol Chem 278 (2003) 19447–19452. 10.1074/jbc.M300445200. [DOI] [PubMed] [Google Scholar]
- [34].Lee JH, Lee DS, Nam H, Lee G, Seo BM, Cho YS, Bae HS, Park JC, Dental follicle cells and cementoblasts induce apoptosis of ameloblast-lineage and Hertwig’s epithelial root sheath/epithelial rests of Malassez cells through the Fas-Fas ligand pathway, Eur J Oral Sci 120 (2012) 29–37. 10.1111/j.1600-0722.2011.00895.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


