Abstract
Soybean accounts for more than half of the global production of oilseed and more than a quarter of the protein used globally for human food and animal feed. Soybean domestication involved parallel increases in seed size and oil content, and a concomitant decrease in protein content. However, science has not yet discovered whether these effects were due to selective pressure on a single gene or multiple genes. Here, re-sequencing data from >800 genotypes revealed a strong selection during soybean domestication on GmSWEET10a. The selection of GmSWEET10a conferred simultaneous increases in soybean-seed size and oil content as well as a reduction in the protein content. The result was validated using both near-isogenic lines carrying substitution of haplotype chromosomal segments and transgenic soybeans. Moreover, GmSWEET10b was found to be functionally redundant with its homologue GmSWEET10a and to be undergoing selection in current breeding, leading the the elite allele GmSWEET10b, a potential target for present-day soybean breeding. Both GmSWEET10a and GmSWEET10b were shown to transport sucrose and hexose, contributing to sugar allocation from seed coat to embryo, which consequently determines oil and protein contents and seed size in soybean. We conclude that past selection of optimal GmSWEET10a alleles drove the initial domestication of multiple soybean-seed traits and that targeted selection of the elite allele GmSWEET10b may further improve the yield and seed quality of modern soybean cultivars.
Keywords: soybean, domestication, yield, seed quality, SWEET
INTRODUCTION
Studies have suggested that global agricultural production needs to be doubled by 2050 to meet the rapidly growing population and diet shifts [1–3], translating into a need for increasing crop production by 2.4% per year [4]. Soybean is a major, multiuse crop that globally makes up 56% of the oilseed production and >25% of the protein used in human food and animal feed [5]. At the current average rate of annual-yield increase, only 55% of the necessary increase in soybean production can be reached by 2050. Thus, breeding soybeans with higher yields is urgently needed [4].
Cultivated soybean (Glycine max [L.] Merr.) was domesticated from wild soybean (G. soja Sieb. & Zucc.) in China over a period of 5000 years [6]. Seeds of wild soybeans are generally smaller and contain higher levels of protein. Cultivated soybeans produce larger seeds with higher oil content (Supplementary Fig. 1). Thus far, it has not been reported that a single gene can simultaneously alter seed size, oil content and protein content, although a number of quantitative trait loci (QTLs) that govern seed size, oil content and protein content in soybean were identified through previous genetic analyses (SoyBase, https://soybase.org/). Therefore, it remains unclear whether the improvement in these traits was achieved by selection of a gene with pleiotropic effects on these traits or by selection of individual genes that only affect each trait.
Sucrose is the main source of carbon energy delivered via the phloem to developing seeds [7] and sugars derived from sucrose metabolism play pivotal roles in seed development for many species [7–13]. Previous studies have demonstrated that SWEET proteins play important roles in sugar translocation to seeds and consequently affect seed setting, filling and composition [14–18]. For example, in Arabidopsis thaliana, mutation of AtSWEET11/12/15 impairs sucrose delivery from seed coat and endosperm to embryo and results in severe seed defects [16]. Similarly, knockout of OsSWEET11 and OsSWEET 15 in rice results in a complete loss of endosperm development [14,19]. Our previous study also illustrated that knockout of both GmSWEET15a and GmSWEET15b in soybean causes an extremely high rate of seed abortion [20].
In this study, we found that a pair of SWEET homologues, GmSWEET10a and GmSWEET10b, underwent stepwise selection that simultaneously altered seed size, oil content and protein content during soybean domestication. Our findings provide practical insights into how to improve soybean-seed traits, in particular seed size and oil content, by optimizing the combination of GmSWEET10a and/or GmSWEET10b alleles.
RESULTS
GmSWEET10a underwent selection during soybean domestication
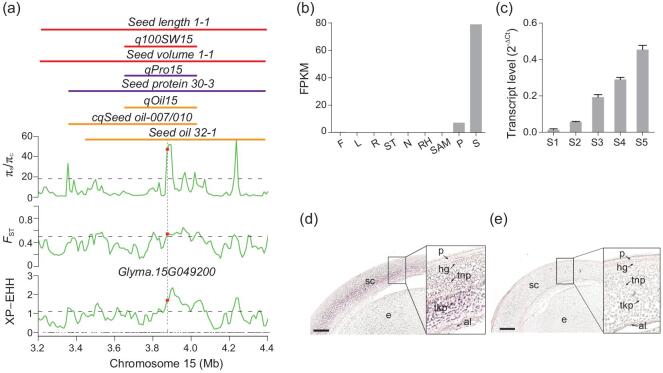
Using whole-genome re-sequencing data from >800 accessions with an average coverage depth of >13X for each accession [21,22], we identified a selective sweep on chromosome 15 from 3.87 to 4.0 Mb. The fixation of this enlargement was observed by different methods, including calculating the nucleotide diversity (π), the fixation index (FST) and the cross-population extended haplotype homozygosity (XP-EHH) (Fig. 1a). This selective sweep overlapped with several reported QTLs that related to seed size, oil content and protein content [23–28] (Fig. 1a and Supplementary Table 1). The results indicated that selected gene(s) in this region may be responsible for the simultaneous alternation of seed size, oil content and protein content in soybean domestication.
Figure 1.
GmSWEET10a was identified as a candidate pleiotropic gene that influences seed size, fatty-acid content and protein content. (a) Genetic variations (π, FST and XP-EHH values) were calculated between G. soja (S) and the cultivar (C) across the 1.2-Mb genomic region of the GmSWEET10a locus. The dashed horizontal lines indicate the genome-wide thresholds (top 5% of the genome) of the selection signals. The solid lines above the plot represent genomic locations of QTLs retrieved from SoyBase (https://soybase.org/; Supplementary Table 1). The red, orange and purple lines are QTLs for the seed size, seed oil and protein contents, respectively. The black dashed lines above the x-axis are annotated genes in this region. The red dots denote the GmSWEET10a gene, i.e. Glyma.15G049200. (b) Expression pattern of GmSWEET10a in different organs in Glycine max (Gm). Expression values were obtained from Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html#). F, flower; L, leaf; R, root; ST, stem; N, nodule; RH, root hair; SAM, shoot apical meristem; P, pod; S, seed; FPKM, fragments per kilobase of exon per million mapped. (c) Transcript abundance of GmSWEET10a in seed coats at different stages. The expression was detected by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). Transcript levels were calculated relative to soybean cyclophilin 2 (GmCYP2). (d) and (e) RNA in situ hybridization of GmSWEET10a showing specific expression in the seed coats. Cross-sections of developing seeds at S2–S3 hybridized with antisense (d) or sense (e) probes for GmSWEET10a. sc, seed coat; e, embryo; p, palisade layer; hg, hourglass; tnp, thin-walled parenchyma; tkp, thick-walled parenchyma; al, aleurone layer. Scale bars, 200 μm.
This selective sweep included 18 gene orders, among which Glyma.15G049200 (previously named GmSWEET10a [20]) encoded a member of the SWEET family of sugar transporters (Fig. 1a). SWEET proteins play important roles in seed development [14–16,19,20]. Transcriptional profiling data from Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html#) showed that GmSWEET10a was specifically expressed in seed and pod (Fig. 1b). Transcriptome data from Gene Networks in Seed Development (http://seedgenenetwork.net/soybean) indicated that GmSWEET10a was mainly expressed in the seed coat (Supplementary Table 2). Quantitative RT-PCR (qRT-PCR) showed that the expression of GmSWEET10a in the seed coats progressively increased during seed development and reached their peaks at the full-seed stage (S5 stage in Supplementary Fig. 2 and Fig. 1c). In situ RNA hybridization confirmed that GmSWEET10a was preferentially expressed in the thick-walled parenchyma of the seed coat (Fig. 1d and e), which are important for sucrose translocation to the embryo [29–31]. The known functions of SWEET proteins and the expression pattern of GmSWEET10a indicated that it might be the gene responsible for the simultaneous alternation of seed size, oil content and protein content during soybean domestication.
Association between seed traits and genetic variation of GmSWEET10a
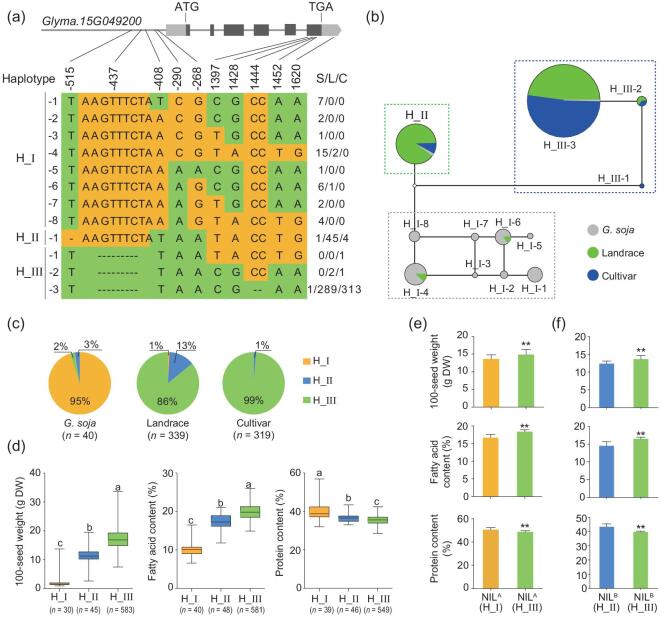
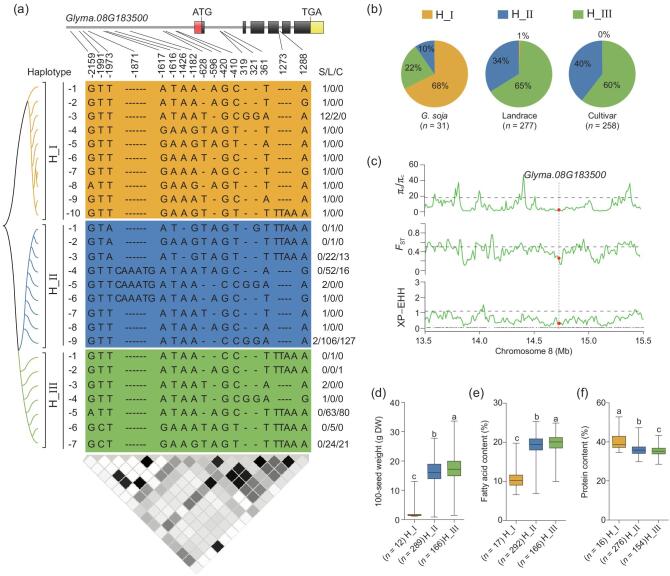
To verify our hypothesis, we first investigated the genetic variation of GmSWEET10a in wild and cultivated soybeans using our previously reported re-sequenced population [21,22]. After removing the polymorphisms with minor allele frequency <0.01 (MAF <0.01), 10 SNPs and In/Dels were found in GmSWEET10a in the re-sequenced population. These 10 genetic variants sorted the population into 12 haplotypes, which were represented by one to a few hundred accessions (Fig. 2a). Median-joining network analysis grouped the 12 haplotypes into three major groups, named H_I (including H_I-1 to H_I-8), H_II and H_III (including H_III-1 to H_III-3). H_I was mainly present in wild soybeans, H_II in landraces and H_III in cultivars (Fig. 2b). Allele-frequency investigation demonstrated that the proportion of H_I was significantly decreased in cultivated soybeans compared to that in wild soybeans, whereas the proportion of H_III was significantly increased in cultivated soybeans, indicating strong artificial selection of GmSWEET10a during soybean domestication (Fig. 2c).
Figure 2.
GmSWEET10a is a domestication gene that contributes to soybean seed size, fatty-acid content and protein content. (a) Haplotypes detected in the genomic region of GmSWEET10a. The SNP information of 871 re-sequenced accessions is derived from Zhou et al.'s data [21] and Fang et al.'s data [22]. The S/L/C indicates the accession number of soja/landrace/cultivar. (b) Median-joining network representing the relatedness of 12 GmSWEET10a haplotypes, each represented by a circle. Gray, green and blue circles represent wild soybeans, landraces and improved cultivars, respectively. (c) Frequency distribution of three haplotypes: H_I, orange; H_II, blue; H_III, green. (d) 100-seed weight, fatty-acid content and protein content of mature seeds in three haplotype populations (colors are the same as that in panel (c)). Box edges depict the interquartile range. The median is marked by a black line within the box. The number of samples in each haplotype (n) is shown under the haplotype label. The letters a, b and c indicate significant differences. P < 0.05 (Student's t-test). DW, dry weight. (e) and (f) Effect of two alleles of GmSWEET10a on seed traits. 100-seed weight, fatty-acid content and protein content of mature seeds from near-isogenic lines of GmSWEET10a with H_ I and H_ III haplotypes (e) or with H_II and H_ III haplotypes (f). NILsA derived from the hybrid combination between HJ117 (H_I) and JY101 (H_ III). NILsB derived from the hybrid combination between Suinong 14 (H_ III) and Enrei (H_II). Data are means ± s.d. ((e) NILA (H_I), n = 12; NILA (H_ III), n = 9; (f) n = 5). **P < 0.01 (Student's t-test).
Second, we looked for associations between the genetic variation of GmSWEET10a and seed-related traits, including seed size (indexed by 100-seed weight), protein content and oil content (indexed by total fatty acid) in the re-sequenced soybean accessions. The results showed that the seed size and the oil content of H_III were significantly higher than those of H_II and that these traits of H_II were significantly higher than those of H_I. In contrast, the protein content of H_III was significantly lower than that of H_II and that of H_II was significantly lower than that of H_I (Fig. 2d). The results indicated that selection at GmSWEET10a during soybean domestication pleiotropically affected the seed size, oil content and protein content. The decrease in protein content could also be a consequence of a rise in the seed size and oil content because the precursor supply may become limiting for protein synthesis when the GmSWEET10a-mediated sugar unloading from seed coats increases the carbohydrate state in developing embryos [32,33]. Since wild soybeans usually exhibit drastically smaller seeds, higher protein content and lower oil content than cultivated soybean (Supplementary Fig. 1), these three traits were further compared among different haplotypes only in the cultivated soybeans to eliminate the effect of genetic differences between wild and cultivated soybeans. Further, because only a few cultivated accessions had H_I haplotypes, only the differences between H_II and H_III cultivated soybeans were compared. In H_III haplotypes, the seed size and oil content were significantly higher but the protein content was lower than in H_II haplotypes (Supplementary Fig. 3).
GmSWEET10a simultaneously alters the seed size, oil content and protein content
To verify whether GmSWEET10a simultaneously affected the seed size, oil content and protein content, two pairs of near-isogenic lines (NILs) were developed: (i) NILsA carrying either H_I (NILA-H_I) or H_III (NILA-H_III) through a cross of HJ117 (carrying H_I) and JY101 (carrying H_III) (Fig. 2e); and (ii) NILsB carrying either H_II (NILB-H_II) or H_III (NILB-H_III) through a cross of Enrei (carrying H_II) and Suinong 14 (carrying H_III) (Fig. 2f). Phenotypic analysis showed that NILA-H_III or NILB-H_III lines had significantly higher 100-seed weight and oil content and lower protein content than did NILA-H_I or NILB-H_II, respectively (Fig. 2e and f).
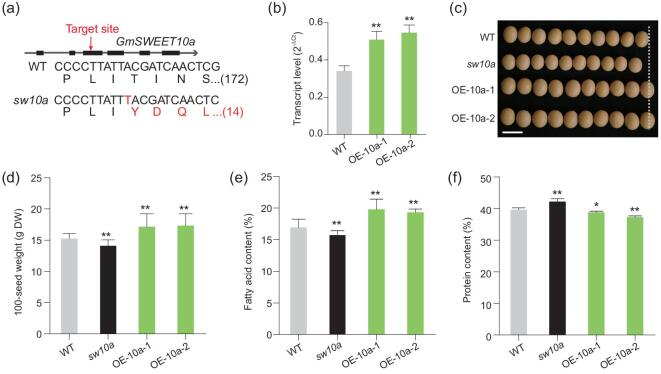
The functions of GmSWEET10a were further confirmed by genetic manipulation. A knockout line, named sw10a, was generated by an Agrobacterium-delivered CRISPR/Cas9 system in the soybean cultivar Williams 82 (Fig. 3a). Compared with Williams 82, sw10a seeds exhibited significantly decreased seed size, lower oil content and increased protein content (Fig. 3c–f). Two independent GmSWEET10a-overexpression lines, OE-10a-1 and OE-10a-2, were generated by introducing an additional copy of the GmSWEET10a genomic sequence into the Williams 82 genome, with a significantly increased transcript level of GmSWEET10a (Fig. 3b). Compared with Williams 82, the seed size and oil content were significantly increased and the protein content was significantly decreased in OE-10a-1 and OE-10a-2 (Fig. 3c–f). A recent study showed that a 9-base pair deletion in the promoter of GmSWEET10a upregulates the expression of GmSWEET10a, which potentially leads to increased oil content in cultivated soybeans [34]. Here, our results in transgenic soybean clarified that the larger seed size, higher oil content and lower protein content in H_III cultivars are indeed caused by the upregulation of GmSWEET10a.
Figure 3.
Effect of GmSWEET10a on seed size, fatty-acid content and protein content. (a) Genotype of the sw10a mutant edited by the CRISPR/Cas9 system. The arrow indicates the target site of the CRISPR/Cas9 editing in the region of exon 3 of GmSWEET10a. Changes in the DNA sequence in the targeted region and the amino-acid sequence of the sw10a mutant are highlighted in red. Numbers inside the brackets indicate the number of amino acids coded by the sequence. (b) Increased expression of GmSWEET10a was achieved in transgenic soybean lines OE-10a-1 and OE-10a-2 by introducing additional copies of the GmSWEET10a genomic sequence into the Williams 82 cultivar. (c) Seed appearance of the sw10a mutant, OE-10a-1 and OE-10a-2. Scale bars, 1 cm. (d)–(f) 100-seed weight (d), fatty-acid content (e) and protein content (f) of mature seeds from wild-type (WT), sw10a mutant, OE-10a-1 and OE-10a-2. DW, dry weight. Data are means ± s.d. ((d) n = 10; (e) and (f) n = 5). *P < 0.05; **P < 0.01 (Student's t-test).
Ongoing selection of GmSWEET10b, which is similar in function to GmSWEET10a
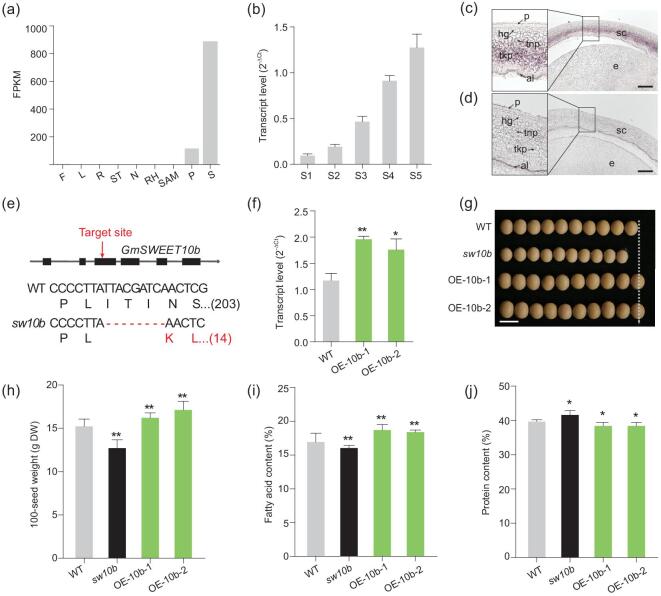
GmSWEET10b is a close homologue of GmSWEET10a. GmSWEET10b showed an expression pattern similar to, but at higher levels than, GmSWEET10a (Fig. 4a and b, and Supplementary Table 2). In situ RNA hybridization showed that GmSWEET10b also exhibited specific localization in the thick-walled parenchymatous layer of the seed coat, but not in the embryo (Fig. 4c and d). We investigated whether GmSWEET10b also played a role in controlling these three seed traits. GmSWEET10b-knockout and -overexpression lines were generated. The results demonstrated that GmSWEET10b had a similar function to that of GmSWEET10a (Fig. 4e–j). Moreover, a double knockout of both GmSWEET10a and GmSWEET10b in Williams 82 and Huachun 6 genetic backgrounds resulted in significantly smaller seed size, lower oil content and higher protein content than either of the single knockout lines or the WT (Williams 82) (Supplementary Fig. 4), indicating that GmSWEET10b and GmSWEET10a have functional redundancy in controlling seed development.
Figure 4.
Effect of GmSWEET10b on seed size, fatty-acid content and protein content. (a) Expression pattern of SWEET10b in different organs in Glycine max (Gm). Expression values were obtained from Phytozome 12 (https://phytozome.jgi.doe.gov/pz/-portal.html#). F, flower; L, leaf; R, root; ST, stem; N, nodule; RH, root hair; SAM, shoot apical meristem; P, pod; S, seed; FPKM, fragments per kilobase of exon per million mapped. (b) Transcript abundance of GmSWEET10b in seed coats at different stages. The expression was detected by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). Transcript levels were calculated relative to soybean cyclophilin 2 (GmCYP2). (c) and (d) RNA in situ hybridization of GmSWEET10b showing specific expression in the seed coats. Cross-sections of developing seeds at S2–S3 hybridized with antisense (c) or sense probes (d) for GmSWEET10b. sc, seed coat; e, embryo; p, palisade layer; hg, hourglass; tnp, thin-walled parenchyma; tkp, thick-walled parenchyma; al, aleurone layer. Scale bars, 200 μm. (e) Genotypes of the sw10b mutant edited by CRISPR/Cas9 system. The arrow indicates the target site in the region of exon 3 of GmSWEET10b. Changes in DNA sequence in the targeted region and amino-acid sequence of the sw10b mutant are highlighted in red. Numbers inside brackets indicate the number of amino acids coded by the sequence. (f) Increased expression of GmSWEET10b was achieved in transgenic soybean lines OE-10b-1 and OE-10b-2 by introducing additional copies of the genomic sequence into the Williams 82 cultivar. (g) Seed appearance of sw10b mutant and overexpression lines. Scale bars, 1 cm. (h)–(j), 100-seed weight (h), fatty-acid content (i) and protein content (j) of mature seeds from wild-type (WT), sw10b mutant, OE-10b-1 and OE-10b-2. DW, dry weight. Data are means ± s.d. ((h) n = 10; (i) and (j), n = 5). *P < 0.05; **P < 0.01 (Student's t-test).
Similarly, the genetic variation of GmSWEET10b was investigated in the re-sequenced population. The nucleotide polymorphisms of this gene classified the accessions into 26 haplotypes, which were then sorted into three major groups by further phylogenetic analysis (Fig. 5a). We found that, although the ratios of H_I to H_II and H_I to H_III were greatly decreased from wild soybeans to cultivated soybeans (Fig. 5b), GmSWEET10b did not show significantly artificial selection during soybean domestication at the genome-wide level (Fig. 5c). However, similarly to GmSWEET10a, the haplotypes mainly present in cultivated soybeans exhibited significantly higher seed size and oil content but lower protein content than the haplotype mainly present in wild soybeans (Fig. 5d–f), suggesting that GmSWEET10b may still be undergoing selection.
Figure 5.
GmSWEET10b is a potential domestication gene that contributes to soybean seed size, fatty-acid content and protein content. (a) Haplotypes detected in the genomic region of GmSWEET10b. The SNP information of 871 re-sequenced accessions is derived from Zhou et al.'s data [21] and Fang et al.'s data [22]. (b) Frequency distribution of three haplotypes of GmSWEET10b. (c) Genetic variations (π, FST and XP-EHH values) were calculated between G. soja (S) and the cultivar (C) across the 2.0-Mb genomic region of the GmSWEET10b locus. The dashed horizontal lines indicate the genome-wide thresholds (top 5% of the genome) of the selection signals. The black dashed lines above the x-axis are annotated genes in this region. The red dots denote the GmSWEET10b gene—Glyma.08G183500. (d)–(f) 100-seed weight (d), fatty-acid content (e) and protein content (f) of mature seeds in three haplotype populations. Box edges depict the interquartile range. The median is marked by a black line within the box. The number of samples in each haplotype (n) is shown under the haplotype label. The letters a, b and c indicate significant differences. P < 0.05 (Student's t-test).
GmSWEET10a and GmSWEET10b transport sucrose and hexose, likely from seed coat to embryo
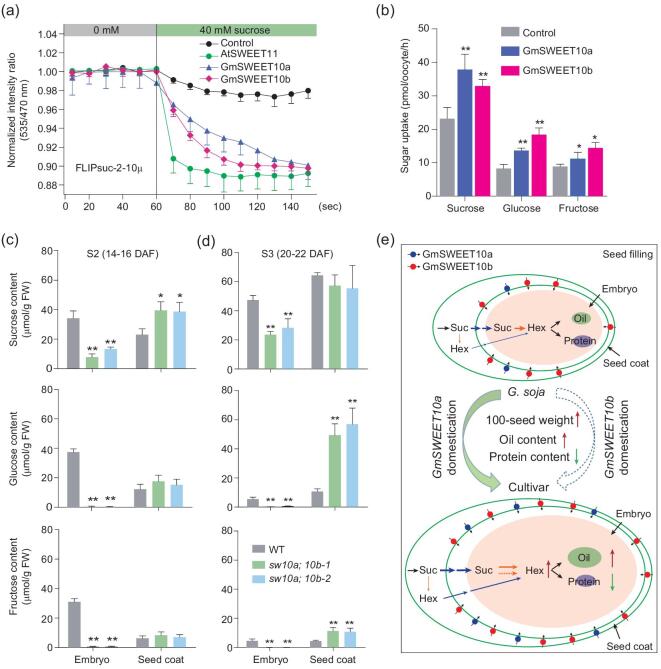
Previous studies have shown that SWEET proteins can transport either mono- or disaccharides or both [35–39]. To first test the sugar-transport activities of GmSWEET10a and GmSWEET10b, we used a newly improved, high-affinity sensor named FLIPsuc-2-10μ [40] with new N-terminal and C-terminal linkers and constructs with the 5’UTRs and codons optimized for humans. When this sensor was co-expressed with GmSWEET10a or GmSWEET10b in the human embryonic kidney line HEK293T, weak sucrose transport activity was detected (as a negative ratio change) when 40 mM sucrose was supplied (Fig. 6a). Sucrose transport by GmSWEET10a and GmSWEET10b was further confirmed by 14C-sucrose radiotracer uptake experiments in Xenopus oocytes (Fig. 6b). GmSWEET10a and GmSWEET10b can also uptake glucose and fructose in oocytes.
Figure 6.
Sugar-transporter activities of GmSWEET10a and GmSWEET10b. (a) Characterization of GmSWEET10a and GmSWEET10b sucrose-transport activity using FLIPsuc-2-10μ in HEK293T. Sensor only (black) and AtSWEET11 (green) were used as negative and positive controls, respectively. Data are means ± s.d. (n ≥ 8). (b) Sugar-uptake transport activities of GmSWEET10a and GmSWEET10b were tested in Xenopus oocytes. Oocytes were injected with water (negative control), GmSWEET10a or GmSWEET10b cRNA. Data are means ± s.d. (n = 3). *P < 0.05; **P < 0.01 (Student's t-test). (c) and (d) Sugar content in the developing seeds at S2 (14-16 DAF) (c) and S3 (20-22 DAF) (d) stages. sw10a;10b, double mutants at GmSWEET10a and GmSWEET10b. Data are means ± s.d. (n = 3). *P < 0.05; **P < 0.01 (Student's t-test). (e) A working model for the involvement of GmSWEET10a and GmSWEET10b in seed size, oil content and protein content during soybean domestication. The expression level of GmSWEET10a is significantly increased in cultivars at the seed-filling stage, which promotes more hexose accumulation in the embryo, resulting in a larger seed size, higher oil content and lower protein content due to the increased carbohydrate state. Selection of GmSWEET10b is ongoing and might use a mechanism similar to that of GmSWEET10a. Dark-blue arrows indicate the translocation of sugars from the seed coat to the embryo. Orange arrows indicate the breakdown of Suc into Hex by invertase or sucrose synthase. The red and green arrows represent ‘increase’ and ‘decrease’, respectively. Hex, hexose; Suc, sucrose.
It is possible that the reduced seed weight in the double sw10a;10b mutant is caused by the low availability of sugars in embryos, similar to what is observed in atsweet11;12;15. To investigate this, sugar levels were measured in isolated seed coats and embryos at the end of the transition phase (14–16 DAF) and storage phase I (20–22 DAF) [41] in WT (Williams 82) and sw10a;10b mutants. In the embryos of the sw10a;10b mutants, the glucose, fructose and sucrose levels were significantly lower at both stages compared to those of WT (Fig. 6c and d). In contrast, in the seed coat of the sw10a;10b mutants, the sucrose content was significantly higher at 14–16 DAF and the hexose content was significantly higher at 20–22 DAF compared with those of WT. Our results indicated that the transport of these three forms of sugar from the seed coat to the embryo are impaired in the sw10a;10b mutant (Fig. 6c and d). This suggests that GmSWEET10a and GmSWEET10b largely determine sugar partitioning between the seed coat and the embryo.
Previous studies showed that sugar allocation affects embryo development and regulates both fatty-acid biosynthesis and protein biosynthesis [41,42]. Thus, we speculated that the increased seed size and higher oil content that arose through soybean domestication might be caused by increasing the sugar content in the embryo through the selection of elite GmSWEET10a alleles.
DISCUSSION
Seed size, oil content and protein content are essential factors for soybean yield and quality. Each of these quantitative traits is controlled by multiple genetic loci. At least 267, 299 and 225 QTLs have been reported to be responsible for the seed size, oil content and protein content in soybean, respectively (retrieved from SoyBase, https://soybase.org/). In the study, we have shown that GmSWEET10a and GmSWEET10b are specifically expressed in the seed coat, likely transport sugars from seed coats to embryos and genetically regulate seed size and composition. GmSWEET10a is a QTL that genetically regulates seed size and composition simultaneously, and was subjected to strong artificial selection during soybean domestication. However, plots of the 100-seed weight against the fatty-acid content and the protein content from the natural population showed that there are cultivated soybeans combining the traits of large seed and high oil content or the traits of large seeds and high protein content (Supplementary Fig. 1d and e). Thus, selections on other genes that do not have pleiotropic effects on these three traits likely occur.
A working model of GmSWEET10a and GmSWEET10b function and their contribution to soybean domestication is proposed (Fig. 6e). At the seed-storage stage, sucrose, as the major carbon source, is delivered to the seed coat via the funicular phloem. Then sucrose, together with a few hexoses (including glucose and fructose) presumably hydrolysed from sucrose, is exported into the cell-wall space via GmSWEET10a and GmSWEET10b, and subsequently imported into the embryo by other sugar transporters [43]. Imported sugars are metabolized for energy generation and carbon skeleton supply for the synthesis of storage compounds including lipids, proteins and starch.
Sucrose concentrations in seed coats and embryos reach a high and steady level at the rapid-seed-growth stage [44]. Thus, sucrose flux across seed coats is particularly important to meet the increasing demand for a carbon source for a high rate of seed growth. Haplotype III of GmSWEET10a was selected during soybean domestication (from G. soja to G. max) because it confers a relatively higher expression of GmSWEET10a, which allows more sucrose to flux to the developing embryos at the rapid-seed-growth stage, and consequently leads to a higher seed-growth rate, larger seed and higher oil content. The selection of GmSWEET10b is currently ongoing and presumably leads to a selected function in contributing to seed-size-storage components, like GmSWEET10a.
The positive contribution of GmSWEET10a and 10b to seed size and oil content can be attributed to the following reasons. First, the elevated expression of the sugar transporter GmSWEET10a or GmSWEET10b can lead to the flux of more sugars into embryos from maternal tissues. It may trigger embryo cell division and expansion, and consequently larger seeds. Second, the increased transport of sugars into the embryos would result in an increase in the carbon resources for lipid synthesis. Some intermediates derived from glycolysis are directly or indirectly shared by lipid- and protein-synthesis pathways. Lipid synthesis may be enhanced due to more precursor of acetyl-CoA available from glycolysis and thus more lipid can be produced and accumulate. On the other hand, protein synthesis depends on both carbon and nitrogen availability. As GmSWEET10a or GmSWEET10b sugar-transporter activities increase, the nitrogen availability may become a limiting factor and thereby decrease the relative protein contents.
The knocking out of GmSWEET10a resulted in a 7.4% and 7.2% decrease in the 100-seed weight and fatty-acid content, but a 6.4% increase in the protein content (Fig. 3d–f). A similar effect was observed for its homologue GmSWEET10b (Fig. 4h–j). When both GmSWEET10a and GmSWEET10b were knocked out, the impact on these parameters increased to –40.2%, –40.7% and +32.1%, respectively (Supplemental Fig. 4b–d and f–h). These seed phenotypes supported that GmSWEET10a and GmSWEET10b are essential sugar transporters for sugar unloading from the soybean-seed coat to the embryos. It is worth noting that the knockout of GmSWEET10b has a stronger impact on the seed weight, as well as the oil and protein content, compared with GmSWEET10a (Figs 3d–f and 4h–j). This indeed is consistent with the transcript-level difference in their transcript abundance (Figs 1c and 4b), although not proportionate to the 10-fold difference at their transcript levels. It requires further study to determine whether their protein abundance and transporter activities correspond to their transcript abundance.
In addition to that, we retrieved the expression data of all the members of the SWEET and SUT/SUF family, which includes genes that have been implicated in exporting sucrose from the seed coat in pea and bean [45,46] in the seed coats from Gene Networks in Seed Development (Supplemental Table 2). Among the 27 SWEET and SUT/SUF genes analysed, only GmSWEET10s, GmSWEET13s and GmSWEET14s were expressed in seed coats. RT-qPCR showed that GmSWEET13s and GmSWEET14s were indeed expressed in seed coats, although at a lower level than in GmSWEET10a and GmSWEET10b (Supplemental Fig. 5a and b). Thus, whereas we speculate that, while GmSWEET10a and GmSWEET10b play essential roles in sugar unloading from the seed coats to the embryos, other uncharacterized sugar transporters, such as GmSWEET13s and GmSWEET14s, may also play roles, either due to their inherent function or as a compensation mechanism in the absence of GmSWEET10a and GmSWEET10b.
Introduction of an additional genomic copy of either GmSWEET10a or GmSWEET10b into soybean led to significant increases in yield, ranging from 11% to 20%, without the compromise of other agronomic traits (Supplementary Fig. 6). Genome editing is a powerful approach for targeted mutagenesis and has been successfully used for crop-trait improvement [47–50]. A recent study found that disruption of MIR396e and MIR396f by CRISPR/Cas9 significantly improves rice yield under nitrogen-deficient conditions [51]. We speculate that alteration of the expression of GmSWEET10a and GmSWEET10b by precise genome editing [52] may enhance seed and oil yield in soybean. Furthermore, since GmSWEET10b has not yet been fixed in cultivated soybeans, the further discovery and utilization of elite allele(s) of GmSWEET10b may provide a new avenue for future soybean breeding. Another member of the SWEET family, SWEET4, was found to be likely selected during the domestication of both maize and rice [15], indicating that a parallel selection of the SWEET family members may exist across different crop species during domestication. Identification of these genes would facilitate the improvement of current crops [53,54]. Thus, SWEET genes should be priority targets across a wide range of species for the improvement of crops and possibly even underused or undomesticated plants.
MATERIALS AND METHODS
For details, please see Supplementary data.
Supplementary Material
Acknowledgements
We thank Prof. Yong-Ling Ruan (University of Newcastle, Australia) and Prof. Heven Sze (University of Maryland) for their useful discussions; Dr. Y. Liu (Zhejiang University) for technical assistance; Mr. Long Yan (Hebei Academy of Agricultural and Forestry Sciences) for the help on the NILsB population construction and field trial; Prof. Nicholas P. Harberd (University of Oxford) and Ms. Anita K. Snyder for revising the manuscript; and B-H. Hou, a former member of the Wolf Frommer lab, for making the new FRET sensor, FLIPsuc-2-10μ.
Contributor Information
Shoudong Wang, State Key Laboratory of Plant Physiology and Biochemistry, College of Life sciences, Zhejiang University, Hangzhou 310058, China.
Shulin Liu, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jie Wang, College of Resources and Environment, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Kengo Yokosho, Institute of Plant Science and Resources, Okayama University, Kurashiki 710-0046, Japan.
Bin Zhou, Institute of Crop Science, Anhui Academy of Agricultural Sciences, Hefei 230031, China.
Ya-Chi Yu, Department of Plant Biology, School of Integrative Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Zhi Liu, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wolf B Frommer, Institute for Molecular Physiology and Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich Heine University of Düsseldorf, Düsseldorf, Germany.
Jian Feng Ma, Institute of Plant Science and Resources, Okayama University, Kurashiki 710-0046, Japan.
Li-Qing Chen, Department of Plant Biology, School of Integrative Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Yuefeng Guan, FAFU-UCR Joint Center for Horticultural Plant Biology and Metabolomics, Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Huixia Shou, State Key Laboratory of Plant Physiology and Biochemistry, College of Life sciences, Zhejiang University, Hangzhou 310058, China.
Zhixi Tian, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Funding
This work was supported by the National Natural Science Foundation of China (31771689) and the Ministry of Agriculture of China (2016ZX08004001) to H.S., the National Natural Science Foundation of China (31788103 and 31525018) to Z.T., Grant-in-Aid for Specially Promoted Research from the Japan Society for the Promotion of Science (KAKENHI grant 16H06296) to J.F.M., and a startup fund from the Department of Plant Biology, University of Illinois at Urbana-Champaign to L.C.
Author contributions
S.W., L.-Q.C., Y.G., H.S. and Z.T. designed the experiments and wrote the manuscript. S.W. performed the phenotyping assay of most soybean materials. S.L. and Z.L. performed the bioinformatics analyses. J.W. performed the phenotyping assay of sw10a;10b mutants (HC6) and in situ hybridization. K.Y. and J.F.M. designed and measured sugar transport in oocytes. B.Z. performed the management of soybean plants in field. W.B.F. provided sensor FLIPsuc-2-10μ. Y.-C.Y. detected the sucrose transport activity in HEK293T.
Conflict of interest statement. None declared.
REFERENCES
- 1. Godfray HCJ, Beddington JR, Crute IR et al. Food security: the challenge of feeding 9 billion people. Science 2010; 327: 812–8. [DOI] [PubMed] [Google Scholar]
- 2. Foley JA, Ramankutty N, Brauman KA et al. Solutions for a cultivated planet. Nature 2011; 478: 337–42. [DOI] [PubMed] [Google Scholar]
- 3. Tilman D, Balzer C, Hill J et al. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 2011; 108: 20260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ray DK, Mueller ND, West PC et al. Yield trends are insufficient to double global crop production by 2050. PLoS One 2013; 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilson RF. Soybean: market driven research needs. In: Stacey G (ed.). Genetics and Genomics of Soybean. New York: Springer, 2008. [Google Scholar]
- 6. Caldwell BE, Howell RW. Soybeans: Improvement, Production, and Uses. Madison: American Society of Agronomy, 1973. [Google Scholar]
- 7. Ruan YL. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol 2014; 65: 33–67. [DOI] [PubMed] [Google Scholar]
- 8. Jin Y, Ni DA, Ruan YL. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 2009; 21: 2072–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li B, Liu H, Zhang Y et al. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnol J 2013; 11: 1080–91. [DOI] [PubMed] [Google Scholar]
- 10. Wang L, Ruan YL. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol 2012; 160: 777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chourey PS, Taliercio EW, Carlson SJ et al. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet 1998; 259: 88–96. [DOI] [PubMed] [Google Scholar]
- 12. Fallahi H, Scofield GN, Badger MR et al. Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. J Exp Bot 2008; 59: 3283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weber H, Borisjuk L, Wobus U. Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 1996; 10: 823–34. [Google Scholar]
- 14. Yang JL, Luo DP, Yang B et al. SWEET11 and 15 as key players in seed filling in rice. New Phytol 2018; 218: 604–15. [DOI] [PubMed] [Google Scholar]
- 15. Sosso D, Luo D, Li Q-B et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 2015; 47: 1489–93. [DOI] [PubMed] [Google Scholar]
- 16. Chen LQ, Lin IWN, Qu XQ et al. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015; 27: 607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eom JS, Chen LQ, Sosso D et al. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol 2015; 25: 53–62. [DOI] [PubMed] [Google Scholar]
- 18. Chen LQ, Cheung LS, Feng L et al. Transport of sugars. Annu Rev Biochem 2015; 84: 865–94. [DOI] [PubMed] [Google Scholar]
- 19. Ma L, Zhang DC, Miao QS et al. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol 2017; 58: 863–73. [DOI] [PubMed] [Google Scholar]
- 20. Wang S, Yokosho K, Guo R et al. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol 2019; 180: 2133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou Z, Jiang Y, Wang Z et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 2015; 33: 408–14. [DOI] [PubMed] [Google Scholar]
- 22. Fang C, Ma Y, Wu S et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol 2017; 18: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diers BW, Keim P, Fehr WR et al. RFLP analysis of soybean seed protein and oil content. Theor Appl Genet 1992; 83: 608–12. [DOI] [PubMed] [Google Scholar]
- 24. Tajuddin T, Watanabe S, Yamanaka N et al. Analysis of quantitative trait loci for protein and lipid contents in soybean seeds using recombinant inbred lines. Breeding Sci 2003; 53: 133–40. [Google Scholar]
- 25. Salas P, Oyarzo-Llaipen JC, Wang D et al. Genetic mapping of seed shape in three populations of recombinant inbred lines of soybean (Glycine max L. Merr.). Theor Appl Genet 2006; 113: 1459–66. [DOI] [PubMed] [Google Scholar]
- 26. Shibata M, Takayama K, Ujiie A et al. Genetic relationship between lipid content and linolenic acid concentration in soybean seeds. Breeding Sci 2008; 58: 361–6. [Google Scholar]
- 27. Pathan SM, Vuong T, Clark K et al. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Sci 2013; 53: 765–74. [Google Scholar]
- 28. Yang HY, Wang WB, He QY et al. Identifying a wild allele conferring small seed size, high protein content and low oil content using chromosome segment substitution lines in soybean. Theor Appl Genet 2019; 132: 2793–807. [DOI] [PubMed] [Google Scholar]
- 29. Thorne JH. Morphology and ultrastructure of maternal seed tissues of soybean in relation to the import of photosynthate. Plant Physiol 1981; 67: 1016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patrick JW, Offler CE. Post-sieve element transport of sucrose in developing seeds. Funct Plant Biol 1995; 22: 681–702. [Google Scholar]
- 31. Wang X-D, Harrington G, Patrick JW et al. Cellular pathway of photosynthate transport in coats of developing seed of Vicia faba L. and Phaseolus vulgaris L. II. Principal cellular site(s) of efflux. J Exp Bot 1995; 46: 49–63. [Google Scholar]
- 32. Patil G, Mian R, Vuong T et al. Molecular mapping and genomics of soybean seed protein: a review and perspective for the future. Theor Appl Genet 2017; 130: 1975–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Warsame AO, O’Sullivan DM, Tosi P. Seed storage proteins of faba bean (Vicia faba L): current status and prospects for genetic improvement. J Agr Food Chem 2018; 66: 12617–26. [DOI] [PubMed] [Google Scholar]
- 34. Miao L, Yang S, Zhang K et al. Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol 2019; 225: 1651–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen LQ, Hou BH, Lalonde S et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010; 468: 527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tao YY, Cheung LS, Li S et al. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 2015; 527: 259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen LQ, Qu XQ, Hou BH et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012; 335: 207–11. [DOI] [PubMed] [Google Scholar]
- 38. Lin IW, Sosso D, Chen LQ et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014; 508: 546–9. [DOI] [PubMed] [Google Scholar]
- 39. Han L, Zhu YP, Liu M et al. Molecular mechanism of substrate recognition and transport by the AtSWEET13 sugar transporter. Proc Natl Acad Sci USA 2017; 114: 10089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lager I, Looger LL, Hilpert M et al. Conversion of a putative Agrobacterium sugar-binding protein into a FRET sensor with high selectivity for sucrose. J Biol Chem 2006; 281: 30875–83. [DOI] [PubMed] [Google Scholar]
- 41. Weber H, Borisjuk L, Wobus U. Molecular physiology of legume seed development. Annu Rev Plant Biol 2005; 56: 253–79. [DOI] [PubMed] [Google Scholar]
- 42. Hymowitz T, Collins FI, Walker WM et al. Relationship between content of oil, protein, and sugar in soybean seed. Agron J 1972; 64: 613–6. [Google Scholar]
- 43. Zhou YC, Chan K, Wang TL et al. Intracellular sucrose communicates metabolic demand to sucrose transporters in developing pea cotyledons. J Exp Bot 2009; 60: 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fader GM, Koller HR. Seed growth-rate and carbohydrate pool sizes of the soybean fruit. Plant Physiol 1985; 79: 663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou YC, Qu HX, Dibley KE et al. A suite of sucrose transporters expressed in coats of developing legume seeds includes novel pH-independent facilitators. Plant J 2007; 49: 750–64. [DOI] [PubMed] [Google Scholar]
- 46. Ritchie RJ, Fieuw-Makaroff S, Patrick JW. Sugar retrieval by coats of developing seeds of Phaseolus vulgaris L. and Vicia faba L. Plant Cell Physiol 2003; 44: 163–72. [DOI] [PubMed] [Google Scholar]
- 47. Chen K, Wang Y, Zhang R et al. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 2019; 70: 667–97. [DOI] [PubMed] [Google Scholar]
- 48. Mao YF, Botella JR, Liu YG et al. Gene editing in plants: progress and challenges. Natl Sci Rev 2019; 6: 421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li JS, Gao CX. Preface to the special topic on genome editing research in China. Natl Sci Rev 2019; 6: 389–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang JS, Zhang H, Botella JR et al. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol 2018; 60: 369–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang JS, Zhou ZY, Bai JJ et al. Disruption of MIR396e and MIR396f improves rice yield under nitrogen-deficient conditions. Natl Sci Rev 2020; 7: 102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hickey LT, Hafeez AN, Robinson H et al. Breeding crops to feed 10 billion. Nat Biotechnol 2019; 37: 744–54. [DOI] [PubMed] [Google Scholar]
- 53. Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell 2006; 127: 1309–21. [DOI] [PubMed] [Google Scholar]
- 54. Wang M, Li WZ, Fang C et al. Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 2018; 50: 1435–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.