Abstract
BACKGROUND
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is diagnosed via real time reverse transcriptase polymerase chain reaction (RT-PCR) and reported as a binary assessment of the test being positive or negative. High SARS-CoV-2 viral load is an independent predictor of disease severity and mortality. Quantitative RT-PCR may be useful in predicting the clinical course and prognosis of patients diagnosed with coronavirus disease 2019 (COVID-19).
AIM
To identify whether quantitative SARS-CoV-2 viral load assay correlates with clinical outcome in COVID-19 infections.
METHODS
A systematic literature search was undertaken for a period between December 30, 2019 to December 31, 2020 in PubMed/MEDLINE using combination of terms “COVID-19, SARS-CoV-2, Ct values, Log10 copies, quantitative viral load, viral dynamics, kinetics, association with severity, sepsis, mortality and infectiousness’’. After screening 990 manuscripts, a total of 60 manuscripts which met the inclusion criteria were identified. Data on age, number of patients, sample sites, RT-PCR targets, disease severity, intensive care unit admission, mortality and conclusions of the studies was extracted, organized and is analyzed.
RESULTS
At present there is no Food and Drug Administration Emergency Use Authorization for quantitative viral load assay in the current pandemic. The intent of this research is to identify whether quantitative SARS-CoV-2 viral load assay correlates with severity of infection and mortality? High SARS-CoV-2 viral load was found to be an independent predictor of disease severity and mortality in majority of studies, and may be useful in COVID-19 infection in susceptible individuals such as elderly, patients with co-existing medical illness such as diabetes, heart diseases and immunosuppressed. High viral load is also associated with elevated levels of TNF-α, IFN-γ, IL-2, IL-4, IL-6, IL-10 and C reactive protein contributing to a hyper-inflammatory state and severe infection. However there is a wide heterogeneity in fluid samples and different phases of the disease and these data should be interpreted with caution and considered only as trends.
CONCLUSION
Our observations support the hypothesis of reporting quantitative RT-PCR in SARS-CoV-2 infection. It may serve as a guiding principle for therapy and infection control policies for current and future pandemics.
Keywords: COVID-19, SARS-CoV-2, Viral load, Severe sepsis, Dynamics, Mortality
Core Tip: High viral load in Coronavirus-2 infections is an independent predictor of disease severity, mortality and prognosis. However there is a wide heterogeneity in fluid samples at different phases of the disease and data should be interpreted with caution. In aggregate, observations support the hypothesis of checking and reporting viral load by quantitative real time reverse transcriptase polymerase chain reaction, instead of binary assessment of a test being positive or negative. Longitudinal analysis with viral loads should be conducted for interpretation of outcome data. This may be the guiding principle for therapy and infection control policies for future pandemics.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic and associated mortality continues to rise and spread unabated in United States and worldwide. Coronavirus disease 2019 (COVID-19) infection is diagnosed via real time reverse transcriptase polymerase chain reaction (RT-PCR). However this assessment is qualitative and reported as a binary positive or a negative test. There is an urgent need to identify high risk patients early in the course of the illness, which includes rapid testing. Quantitative viral load may provide valuable assessment in risk stratification and may assist with early implementation of therapy in susceptible populations such as elderly, immunosuppressed patients with comorbidities.
Quantitative viral RNA load as determined by qRT-PCR assay and reported as cycle threshold (Ct < 38) value and/or log10 (viral copies/mL) from respiratory or blood specimens is a critical factor in diagnosing SARS-CoV-2 virus infection[1-60]. In addition, viral load dynamics in body fluids such as plasma, serum, urine, feces is emerging as a factor in determination of severe inflammation, infectiousness and transmissibility of COVID-19[1-60].
Similar association of high viral load along with age, comorbidities and elevated mortality were also demonstrated during the previous SARS-CoV, pandemic in Hong Kong in the year 2003 and MERS-CoV pandemic in middle east in 2012[61-64].
At present there is no Food and Drug Administration (FDA) Emergency Use Authorization issued for quantitative viral load assay in the current pandemic[59]. The intent of this research is to identify whether quantitative SARS-CoV-2 viral load assay correlates with clinical outcomes, particularly if there is any correlation with severity of infection and mortality? This a correlation study and does not imply causation. The author qualitatively examined the available data from different manuscripts to find patterns and generate a hypothesis for future research. These may assist clinicians; epidemiologist and health care policy makers develop strategies to improve care in COVID-19 sepsis.
MATERIALS AND METHODS
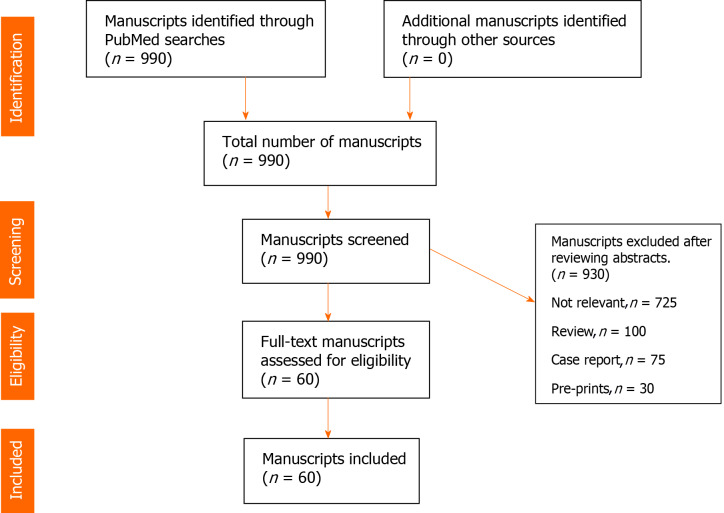
A systematic literature search was undertaken in PubMed/MEDLINE using combination of terms “COVID-19, SARS-CoV-2, Ct values, Log10 copies, quantitative viral load, viral dynamics, kinetics, severity of symptoms, sepsis, mortality’’ for a period between December 30, 2019 to December 31, 2020. Review of manuscripts was performed according to principles outlined in Cochrane handbook. Figure 1 (PRISMA flow diagram).
Figure 1.
PRISMA flow diagram.
Due to an explosion of COVID-19 related research and manuscripts, search was limited to adult (> 18 years) human subjects and published in English language journals. All data is retrospective, de-identified and conforms to the ethical principles in “Declaration of Helsinki”. Manuscripts from preprint non-peer reviewed servers, review articles and individual case reports were excluded. After screening 990 manuscripts, a total of 60 manuscripts which met the inclusion criteria were identified. Data on age, number of patients, sample sites, RT-PCR targets, disease severity, intensive care unit (ICU) admission, mortality and conclusions of the studies was extracted, organized and presented (Table 1). Other relevant articles with relevant information on viral load assessment and mortality, severity and infectiousness and transmission were also included for discussion purposes. During the course of the pandemic in the year 2020, the author followed the PubMed literature on the research question and carefully tracked and evaluated the consistency and quality of the published articles to ensure credibility, reliability, transferability and reduce the risk of bias. The full text of selected articles was fully read, and the key findings were extracted. To establish reliability the author recorded the data in a table and updated assessment of the results. The use of the tables for recording manuscripts provided this researcher with a chance to evaluate the results of the data provided in each manuscript and follow the trends in this topic. The table also helped in construction of concise conclusions of the data. The table is transparent and reproducible and may be useful for other researchers to follow upon.
Table 1.
Manuscript evaluating quantitative viral load assay and coronavirus disease 2019 outcomes. Sixty manuscripts meet the inclusion criteria
|
Ref./country
|
Number of patients
|
Age (yr)
|
Sampled sites
|
Quantitative viral load reported as Ct values or Log10 copies /mL /RTPCR gene target
|
Correlation with severity of sepsis
|
Correlation with mortality
|
P
value
|
Merits of the study/key points
|
| He et al[1], China | 94 | Median 47 yr | Nasopharynx | Ct values/; N gene | Not reported | Not reported | NR | Highest viral load at pre-symptomatic stage and infectiousness peaks before symptom onset. |
| Xu et al[2], China | 51 | Median 37 yr | Nasopharynx, BAL, Anal swab | Ct values/; ORF1ab and N gene | No | No | > 0.05 | The quantitative viral load and infectiousness may be the similar for primary (imported form epicenter) and secondary and tertiary exposed group of patients but decrease rapidly (in 14 d) in tertiary patients. |
| Lescure et al[3], France | 5 | Median 46 yr | Nasopharynx, Stool, Plasma | Log10copies/mL; RdRp-IP1 gene, E gene | No | Inadequate sample size | NR | Presymptomatic patients may have a high viral load and be highly infectious. |
| Liu et al[4], China | 76 | Median 50 yr | Nasopharynx | Ct values; Gene not reported | Yes | No | < 0.005 | Patients with severe COVID-19 have a higher mean viral load (60 times higher) and long shedding period. |
| To et al[5], China | 23 | Median 62 yr | Oropharynx | Log10copies/mL/; RdRp gene | Yes | Not reported | 0.56 | Peak viral load occurs at onset of symptoms and is correlated with increasing age and severity although not statistically significant. |
| Shen et al[6], China | 5 | Median 60 yr | Nasopharynx | Ct values; Gene not reported | Yes | No | NR | Patients with severe sepsis and high quantitative viral load benefit from convalescent plasma. The viral load became negative in all 5 patients in 12 d with clinical improvement. |
| Duan et al[7], China | 10 | Median 52.5 yr | Nasopharynx | Ct values; ORF1ab and N gene | Yes | No | < 0.001 | Resolution of severe sepsis and negative viral load with convalescent plasma infusion. |
| Chen et al[8], China | 48 | Median 63 yr | Oropharynx. serum | Ct values; ORF1ab and N gene | Yes | Yes | < 0.001 | Serum viremia and viral load associated with severity and poor prognosis. High RNAaemia is associated with elevated IL-6 levels. |
| Pan et al[9], China | 82 | Not reported | Oropharynx. Sputum, Stool | Log10copies/mL; N gene | Yes | Yes | NR | Viral load is high on presentation. Stool samples may turn positive later in the disease. |
| Cao et al[10], China | 199 | Median 58 yr | Oropharynx | Log10copies/mL; N and E gene | Not reported | No | NR | Lopinavir-Ritonavir did not aid with clinical improvement, reduce mortality or reduce the viral loads. |
| Wang et al[11], China | 237 | Median 65 yr | Oropharynx, Sputum | Log10copies/mL; Gene not reported | Not reported | Not reported | NR | Remdesivir group does not decrease viral load compared to control group, however it may have faster time to clinical improvement. |
| Zou et al[12], China | 18 | Median 59 yr | Nasopharynx, Oropharynx | Ct values; ORF1b | Not reported | Not reported | NR | High viral load begins in the presymptomatic period and may suggest high infectivity. |
| Wang et al[13], China | 23 | Median 56 yr | Nasopharynx, Oropharynx, sputum, fecal, urine, plasma | Ct values; RdRp and N gene | Yes | None | < 0.001 | High viral load and shedding from multiple tissues occurs for a prolonged period in severe cases. Feces remains positive for a prolonged time. |
| Wölfel et al[14], Germany | 9 | Not reported | Oropharynx, Sputum, stool, serum, urine | Log10copies/mL; RdRp and E gene | No | No | NR | High viral load begins in the presymptomatic period and may continue beyond 10 d after symptoms ensue suggest high infectivity. No positivity in stool, urine or serum. All cases were with mild symptoms. |
| Zheng et al[15], China | 96 | Median 55 yr | Nasopharynx, Oropharynx, sputum, fecal, urine, plasma | Ct values and Log10copies/mL; ORF1ab | Yes | Not reported | 0.03 | High respiratory viral load associated with disease severity and serum positivity and stool shedding occurs later and persists for a longer period. |
| Baggio et al[16], Swiss | 352 adults, 53 children | Mean 36.5 yr | Nasopharynx | Log10copies/mL; ORF1ab and E gene | Not reported | Not reported | NR | Children and adults can have same variation of viral loads, but risk of transmission and lower susceptibility in children may have other contributing factors. |
| Shi et al[17], China | 114 | Median 43.5yr | Oropharynx, serum | Log10copies/mL; N gene | Yes | Not reported | < 0.001 | High viral loads associated with severe sepsis in female patients. |
| Clementi et al[18], Italy | 200 | Mean 64 yr | Nasopharynx | Ct values; ORF1ab and E gene | Yes | Not reported | 0.08 | Higher viral loads associated with older age group and severity of sepsis. |
| Kwon et al[19], Korea | 31 | Mean 50 yr | Nasopharynx | Ct values; RdRp and N gene | Yes | None | 0.093 | High viral loads correlated with elevated cytokine profile and severity of sepsis. |
| Yu et al[20], China | 92 | Mean 55 yr | Sputum | Ct values/N and ORF1b | Yes | No | 0.017 | Higher baseline sputum viral load on admission is associated with severe disease. |
| Liu et al[21], China | 31 | Median 58 yr | Nasopharynx, sputum | Ct values; ORF1ab and N gene | Not reported | Not reported | NR | Viral load is higher in deep sputum samples and have a higher shedding and transmission capacity. |
| Zhou et al[22], China | 31 | Median 41 yr | Nasopharynx | Ct values; ORF1ab and N gene | No | No | NR | Asymptomatic patients have high viral loads and continue viral shedding and transmission. |
| Cheung et al[23],Hong Kong | 59 | Median 58.5 yr | Stool | Log10copies/mL; Gene not reported | No | No | = 0.019 | Stool viral loads are higher in patients with diarrhea and may persist after negative respiratory specimens. |
| Azzi et al[24], Italy | 25 | Mean 61.5 yr | Saliva | Ct values; 5’UTR | Yes | Not reported | = 0.04 | High salivary viral loads may be associated with severe disease and may persist after the negative respiratory specimens. High viral load associated with high serum LDH suggestive of tissue damage. |
| Chen et al[25], China | 22 | Median 36.5 yr | Saliva, feces, Oropharynx | Ct values; ORF1ab and N gene | No | No | NR | Sputum and stool viral load remains positive after pharyngeal samples turn negative. Indicating the infectivity may persist after negative pharyngeal samples. |
| Huang et al[26], China | 16 | Median 59.5 yr | Nasopharynx, sputum, tracheal aspirates, fecal, urine, plasma | Ct values; N gene | Yes | No | < 0.01 | In severe cases higher viral load is demonstrated in deep sputum and tracheal aspirates compared to upper respiratory tract specimens. |
| Pujadas et al[27], United States | 1145 | Mean 64.6 yr | Nasopharynx | Log10copies/mL; RdRp and N gene | Yes | Yes | = 0.003 | High viral load is an independent predictor of mortality. |
| Arons et al[28], United States | 57 | Mean 75 yr | Nasopharynx, Oropharynx | Ct values; N1 and N2 | No | Not reported | NR | High viral loads demonstrated in presymptomatic, asymptomatic cases, favoring high transmissibility in close knit nursing home population. |
| Huang et al[29], China | 308 | Median 63 yr | Nasopharynx, Oropharynx | Ct values; ORF1ab | Yes | Yes | < 0.001 | High viral load associated with critical disease and mortality. Sputum samples have higher viral loads. |
| Magleby et al[30], United States | 678 | Median 69 yr | Nasopharynx, | Ct values; ORF1b and E gene | Yes | Yes | < 0.001 | High viral load is an independent risk factor for severe sepsis, intubation and death. |
| Park et al[31], Korea | 46 | Median 26 yr | Nasopharynx, Oropharynx, sputum , Stool | Ct values; RdRp, N and E gene | No | No | NR | High fecal viral load and shedding, follows and persists after respiratory symptoms resolve for up to 50 d. |
| Yu et al[32], China | 76 | Median 40 yr | Nasopharynx, Oropharynx, sputum, urine, plasma | Ct values; ORF1b and N gene | Yes | None | < 0.001 | Digital droplet PCR is superior for patients with high suspicion but negative RTPCR. High viral load correlated with risk for progression and disease activity. |
| Blot et al[33], France | 14 | Median 67 yr | Broncho-alveolar fluid | Log10copies/mL; RdRp | Yes | Not reported | = 0.013 | Higher viral load associated with worse sepsis related organ failure (SOFA) scores. |
| Kim et al[34], Korea | 13 | Median 30 yr | Nasopharynx | Ct values; RdRp and E gene | No | No | NR | Patient with mild or asymptomatic infections are infectious before symptoms appear and 14 d of isolation may be sufficient in asymptomatic carriers. |
| Argyropoulos et al[35], United States | 205 | Median 60 yr | Nasopharynx | Log10copies/mL; RdRp and N gene | Decreased | Decreased | < 0.001 | Study shows inverse correlation of high viral load with duration, severity of sepsis and no correlation with survival. |
| Xu et al[36], China | 85 | Median 56 yr | Nasopharynx, Oropharynx, serum | Ct values; ORF1b and N gene | Yes | Yes | < 0.001 | Detection of high serum viral load in the serum increases the severity of organ damage, sepsis and mortality. |
| Veyer et al[37], France | 58 | Median 55.1 yr | Plasma | Log10copies/mL; ORF1b and N gene | Yes | Yes | = 0.036 | Detection of high Viral load in the serum increases the severity of sepsis and mortality. |
| Lin et al[38], China | 217 | Median 50 yr | Nasopharynx, Oropharynx, anal | Ct values; ORF1b and N gene | Yes | No | = 0.006 | Anal viral load remains positive longer and is correlated with severity of sepsis and ICU admission. |
| Wang et al[39], China | 275 | Median 49 yr | Oropharynx | Ct values; ORF1b and N gene | No | No | = 0.824 | Similar viral loads between severe and mild cases, no correlation of viral load to ICU admission, severity or mortality. |
| Kimball et al[40], United States | 23 | Mean 80.7 yr | Nasopharynx, Oropharynx | Ct values; N1, N2 genes | No | No | = 0.3 | High viral loads in unrecognized asymptomatic and presymptomatic patients may contribute to infectiousness and transmission. |
| Schwierzeck et al[41] ,Germany | 12 | Not reported | Nasopharynx | Ct values; E and RdRp genes | Yes | No | = 0.007 | High viral load, 200 times greater in symptomatic patients compared to asymptomatic patients. |
| Xia et al[42], China | 10 | Mean 56.5 yr | Nasopharynx | Ct values; ORF1ab and N gene | Yes | No | NR | Higher viral load associated with severe symptoms and increased neutrophil/lymphocyte ratio. |
| Huang et al[43], China | 41 | Median 49 yr | Nasopharynx, Oropharynx, sputum, BAL | Ct values; 5’UTR | No | No | No | Patients with high viral load with RNAeamia had severe infection, elevated cytokine levels, and mortality but not statistically significant. |
| Hagman et al[44], Sweden | 167 | Median 63 | Nasopharynx, Oropharynx, sputum, Blood | Ct values; E, RdRp, ORF1 genes | Yes | Yes | P < 0.05 | Viral RNAemia on admission was associated with eight fold increased risk of in hospital death. |
| Prebensen et al[45], Norway | 123 | Median 64 | Nasopharynx, Oropharynx, sputum, Blood | Ct values for respiratory specimens; Log10copies/mL for plasma samples; E gene | Yes | Yes | < 0.001 | Higher viral loads associated with ICU admission and death. |
| Hasanoglu et al[46], Turkey | 60 | Mean 32 | Nasopharynx, Oropharynx, sputum, urine, Blood, rectal | Ct values; RdRp gene | Decreased | Decreased | = 0.0141 | Viral loads in younger asymptomatic patients were significantly higher compared to elderly, symptomatic patients. |
| Kawasuji et al[47], Japan | 28 | Median 45 | Nasopharynx | Log10copies/mL; N gene | No | No | = 0.015 | High admission nasopharyngeal viral load associated with increased risk of transmission. |
| Bermejo-Martin et al[48], Spain | 250 | Median 66 | Nasopharynx, Oropharynx, sputum, urine, Blood, rectal | Log10copies/mL; N gene | Yes | Yes | < 0.001 | Increased serum viral load associated with increased severity, mortality and dysregulated host response. |
| Shlomai et al[49], Israel | 170 | Median 62 | Nasopharynx | Ct values; N gene | Yes | Yes | < 0.0001 | Increased hypoxemia, severity and eight fold increase in mortality. |
| Ra et al[50], Korea | 213 | Median 25 | Nasopharynx | CT value; E, N, RdRp gene | No | No | None | Comparable viral load in asymptomatic and symptomatic patients, asymptomatic patients contribute to ongoing transmission. |
| Faico-Filho et al[51], Brazil | 875 | Median 48 | Nasopharynx | Ct value; N gene | Yes | Yes | < 0.0001 | Admission nasopharyngeal viral load was independently associated with increased mortality. |
| Chen et al[52], China | 52 | Median 62 | Blood, oropharynx | Log10copies/mL; ORF1ab | Yes | Yes | < 0.001 | Increased RNAemia associated with severity, markers of inflammation and mortality. |
| Fajnzylber et al[53], United States | 88 | Median 57 | Nasopharynx, Oropharynx, sputum, Blood | Log10copies/mL; N gene | Yes | Yes | = 0.009 | Increased viremia associated with severity, progression and mortality. |
| Zhou et al[54], China | 195 | Median 66 | Oropharynx | Ct value; N gene, ORF1ab | Yes | Yes | < 0.005 | High viral load associated with multi organ failure and death. |
| Maltezou et al[55], Greece | 1122 | Mean 46 | Nasopharynx, Oropharynx | CT value; E, RdRp gene | Yes | Yes | < 0.05 | High viral load correlated with intubation and in hospital mortality. |
| Bitker et al[56], France | 129 | Median 69 | Nasopharynx, Oropharynx, sputum | Ct value; ORF1ab | Yes | Yes | < 0.05 | High viral load associated with increased mortality. |
| Carrasquer et al[57], Spain | 169 | Median 67 | Nasopharynx | Ct value; E, N gene, ORF1ab | No | No | = 0.029 | High viral load statistically not associated with in hospital mortality. |
| de la Calle et al[58], Spain | 455 | Mean 64 | Nasopharynx | Ct value; N gene | Yes | Yes | = 0.022 | High viral load associated with respiratory failure, and 30 d mortality. |
| Bryan et al[59], United States | 109 | Mean 65 | Nasopharynx | Ct value; N gene | Yes | Yes | = 0.01 | The high nasopharyngeal viral load on admission was independently associated with greater mortality. |
| Choudhuri et al[60], United States | 1044 | Mean 65 | Nasopharynx | Ct value; ORF1ab | No | Yes | < 0.001 | High viral load is an independent predictor of increased mortality. |
Data on country of origin, age, number of patients, sample sites, real time reverse transcriptase polymerase chain reaction targets, correlation with sepsis and mortality and key conclusions. NR: Not reported; ORF: Open reading frame; E: Envelope; N: Nucleocapsid; 5’UTR: 5 prime untranslated; RdRp: RNA dependent RNA polymerase; Ct: Cycle threshold; SOFA: Sequential organ failure assessment; ICU: Intensive care unit; COVID-19: Coronavirus disease 2019.
Due to a high heterogeneity in patient population, data from different countries, different methods in sampling, comorbidities, and different parameters used, the content was analyzed and is summarized using qualitative (descriptive) terms. Data with P value (< 0.05) was considered statistically significant.
RESULTS
Sixty manuscripts met the inclusion criteria with our research question, and are summarized[1-60]. Twenty eight manuscripts (46%) were reported from China[1,2,4-13,15,17,20-22,25,26,29,32,36,38,39,42,43,52,54], Eight (13%) studies from United States[27,28,30,35,40,53,59,60], Four (6%) were from France[3,33,37,56] and South Korea[19,31,34,50], Three (5%) from Spain[48,57,58], Two (3%) were from Italy[18,24] and Germany[14,41] and One manuscript (2%) was from Switzerland[16], Hong Kong[23], Sweden[44], Norway[45], Israel[49], Greece[55], Japan[47], Turkey[46], Brazil[51] (Table 1).
A total of 10514 patients were pooled from all reported studies. Quantitative RT-PCR and viral dynamics are reported in samples obtained from nasopharyngeal and oropharyngeal swabs, saliva, sputum, bronchial/tracheal lavage, feces, plasma/serum and urine samples. All studies had initial COVID-19 diagnosed on upper respiratory samples. Subsequent quantitative viral load was obtained and described from various other specimens and body fluids.
RT-PCR targets of SARS-CoV-2 virus included the following genes: ORF1 (open reading frame), N (Nucleocapsid), E (Envelope), RdRp (RNA dep RNA polymerase), 5’UTR (5’untranslated region). Forty-three studies (70%) reported viral kinetics in Ct values and 18 (30%) reported it as Log10 copies/mL values.
Association between viral load and disease severity
Thirty-six studies (7222 patients) demonstrated a significant association between pharyngeal viral load at onset of symptoms with severity of COVID -19 and ICU care[4-9,13,15,17-20,24,26,27,29,30,32,33,36-38,41,42,44,45,48,49,51-56,58,59]. The majority of these studies reported highest viral load at onset of symptoms.
Most studies consistently defined severity of illness and sepsis as: Respiratory rate ≥ 30 beats/min, resting-state oxygen saturation ≤ 93%, arterial partial pressure of oxygen/oxygen concentration ≤ 300 mm Hg or mechanical ventilation, shock, or multiple organ failure requiring care in ICU[4,8,29,65].
There is variation observed in kinetics, tissue distribution and antibody response between mild and severe infections. Wang et al[13] analyzed a cohort of 12 severe and 11 mildly ill patients and demonstrated a significant difference in the initial nasopharyngeal peak viral load (P < 0.001) between two groups. Subsequent prolonged viral shedding in other body fluids and stool occurred with detectable viral load for up to 40 d (days) in severely ill compared to 15 d in mildly ill group. Viral RNA was detected from respiratory tract, stool, plasma and urine samples in the severe group. Mildly ill patients had viral shedding restricted to respiratory tract and no virus was detected 10 d after onset of symptoms[13].
Yu et al[20] analyzed their cohort of 92 patients and observed that high viral load in baseline sputum samples was linearly associated with severity and risk of disease progression (P < 0.017).
Another cohort of 96 patients with mild and severe infections demonstrated similar viral kinetics. Respiratory viral load remained elevated in the severe group up to the third and fourth week after disease onset, compared to milder group where viral load peaked in the second week followed by a decline. Subsequent viral detection in serum samples was also higher in patients with severe disease than in patients with mild disease (45% vs 27%, P < 0.03)[15].
In general nasopharyngeal viral levels remained high in severe group and, begin to decrease after 14 d of symptom onset[4,15,65]. Subsequently, samples from other sites may also test positive for the virus. For example, viral load from stool samples were found to peak during the third and fourth weeks after disease onset and continue to remain positive during convalescence[9,13,15,19,25,31]. Some studies also reported presence of high viral load in stool up to 50 d after onset of COVID-19 symptoms[31,38].
Significance of viral load in stool remains unclear, whether it represents a true infection or residual viral nucleic acid and not transmissible live virus. Gastrointestinal epithelium also expresses angiotensin-converting enzyme II (ACE-2) receptors. Infection of gastrointestinal (GI) tract may occur primarily from swallowed nasopharyngeal secretions or due to dissemination to GI tract from viremia[23]. Eighteen studies (5479 patients) demonstrated a statistically significant (P value < 0.005) association between higher viral load in different samples and severity of disease[4,7,8,13,17,27,29,30,32,36,45,48,49,51,52,54-56].
Liu et al[4] analyzed their cohort of 46 mild and 30 severely ill patients with elevated nasopharyngeal viral load and demonstrated an association with severity. Viral load was 60 times higher in severe cases and with severe clinical outcomes (P < 0.005). Mild cases had viral clearance, with 90% of patients testing negative after 10 d. In contrast, all severe cases had persistently elevated viral load beyond 10 d of symptoms were elderly and required ICU care.
In a cohort of patients on dialysis, Schwierzeck et al[41] also demonstrated a similar association with severity. Ct values of symptomatic cases were significantly lower compared to asymptomatic cases (22.55, 29.94, respectively, P = 0.007), indicating approximately 200-fold higher viral load[41]. Similarly other authors from their cohorts from different countries Bermejo-Martin et al[48]; Spain, Shlomai et al[49]; Israel, Chen et al[52]; China, Zhou et al[54]; China, Maltezou et al[55]; Greece have demonstrated a statistically significant association between admission high viral load and intubation, ICU care and multi-organ dysfunction.
Collectively these data from different cohort of patients suggests that severe COVID-19 patients with a high viral load correlate with higher risk for severe infection with ICU admission and multi-organ dysfunction. Factors common to these cohorts was increased age, and active preexisting medical co-morbidities.
Association between viral load and inflammatory markers
Higher viral load on admission samples were also associated with elevated levels of IL-6, cytokines, lactate dehydrogenase (LDH), lymphopenia and elevated neutrophil/lymphocyte ratio; indicative of poor sequential organ failure assessment (SOFA) scores and associated with hyper-inflammatory state contributing to the severity of sepsis[8,19,24,29,33,36,37,42,48,49,52,65,66].
In a cohort of 48 patients, Chen et al[8] reported an association between high viral load in serum with elevated Il-6 Levels (≥ 100 pg/mL) and cytokine storm in critical compared to mildly ill patients (P < 0.001). These patients had a higher incidence of multi-organ failure and mortality.
Similarly Xia et al[42] in their cohort of 10 patients with severe illness and elevated nasopharyngeal viral load reported severe lymphopenia with CD4+ lymphocyte counts as low as 61 cells/uL (reference value: 355-1213 cells/µL). Neutrophil to lymphocyte ratio was also elevated in this group.
Liu et al[65] reported their cohort of 46 patients with severe illness and elevated nasopharyngeal viral load. CD4+ and CD8+ T lymphocyte count displayed a linear negative correlation (P < 0.001) with high viral count; and positively correlated with IL-2R, prothrombin time, lactate dehydrogenase, and hypersensitive troponin T (P = 0.002, P = 0.009, and P < 0.001, respectively). Also elevated, were levels of inflammatory factors, IL-2R, IL-6, IL-8 Levels in the severe compared to mild group (P = 0.022, 0.026, and 0.012, respectively)[65].
Blot et al[33] in their series of 14 patients demonstrated a positive correlation of high nasopharyngeal viral load on admission with risk of hypoxemia, increased oxygen requirements and SOFA score in respiratory distress syndrome patients (P = 0.013). Similar association with increase in severity of sepsis, organ damage and mortality was also reported by Xu et al[36].
Lucas et al[66] in their series of 113 patients with COVID-19 patients demonstrated an overall increase in cells of innate lineage and a reduction in T lymphocytic cell counts. High viral load correlated significantly with levels of IFNα, IFNγ, TNF and tumor necrosis factor-related apoptosis-inducing ligand. Chemokines responsible for monocyte recruitment correlated significantly with viral load in severe disease. Inflammasome associated cytokines were also elevated, including IL-1α, IL-1β, IL-6, IL-18 and TNF[66].
Similarly Han et al[67] in their series of 60 critical patients demonstrated high levels of cytokines TNF-α, IFN-γ, IL-2, IL-4, IL-6, IL-10 and C reactive protein (CRP). Serum IL-6 and IL-10 Levels were significantly higher in critically ill compared to moderately ill group. The levels of IL-10 positively correlated with CRP (r = 0.41, P < 0.01)[67].
Collectively these studies provide evidence that high viral load may be a surrogate marker for predicting inflammation and severity in COVID-19 infection.
Association between viral load and mortality
Subgroup analysis of 20 studies (7183 patients) demonstrated an association of admission viral load with in hospital mortality[8,9,27,29,30,36,37,45,46,48,49,51-56,58-60]. Majority of patients in this category were older (median > 65 years) and with medical comorbidities[8,9,29,30,33,36,37,45,46,48,49,58-60]. High admission viral load was an independent risk factor for in hospital mortality (P < 0.005)[8,27,29,30,36,46,48,49,51,52,54,59,60].
Pujadas et al[27] demonstrated an association of viral load as an independent predictor of mortality in a cohort of 1145 hospitalized patients. Mean log10 viral loads significantly differed between patients who survived [n = 807; mean log10 viral load 5.2 copies/mL (SD 3)] vs those who succumbed [n = 338; 6.4 copies/mL (SD2.7)]. Cox proportional hazards model was adjusted for age, sex, asthma, atrial fibrillation, coronary artery disease, chronic kidney disease, chronic obstructive pulmonary disease, diabetes, heart failure, hypertension, stroke, and race. The results demonstrate a significant independent association between viral load and mortality [hazard ratio 1.07 [95% confidence interval (CI): 1.03–1.11), P = 0.0014], and 7% increase in hazard for each log transformed copy/mL. Univariate survival analysis also demonstrated a significant difference in survival probability between high and with low viral load (P = 0.0003), with a mean follow-up of 13 d and a maximum follow-up of 67 d[27].
Magleby et al[30] in their cohort of 678 patients demonstrated that higher viral load was associated with increased age, comorbidities, smoking status, and recent chemotherapy. Mortality was highest, 35.0% in the high viral (Ct < 25; n = 220) followed by 17.6% in the medium viral (Ct 25-30; n = 216) and 6.2% with a low viral load (Ct > 30; n = 242; P < 0.001). The need for mechanical ventilation was also highest in the high viral (29.1%), compared to medium (20.8%) and low viral load (14.9%; P < 0.001) group. High viral load was independently associated with mortality [adjusted odds ratio (OR) 6.05; 95%CI: 2.92-12.52; P < 0.001] and intubation (adjusted OR 2.73; 95%CI: 1.68-4.44; P < 0.001) in multivariate models.
Similarly Huang et al[29] in their analysis of 308 patients demonstrated a high viral load associated with in-hospital mortality in (6/16) of critical patients, while no mortality was observed in the low viral load group (P < 0.0001). High viral load was associated with myocardial damage, elevated troponins, coagulopathy, abnormal liver and renal functions. Elevated IL-6, LDH, and elevated neutrophil counts and reduced CD4+, CD8+ lymphocytes were noted in deceased patients P < 0.0001)[29].
In a cohort of 109 patients Bryan et al[59] demonstrated high viral load on admission was associated with a significantly increased 30-d mortality (OR, 4.20; 95%CI, 1.62–10.86. Their data suggested that a CT value of 22 may serve as a useful discrete cutoff for significant viral replication that is associated with mortality[59].
In a cohort of 1044 patients, Choudhuri et al[60] demonstrated a statistical correlation of Ct value at admission was higher for survivors (28.6, SD = 5.8) compared to non-survivors (24.8, SD = 6.0, P < 0.001). After adjusting for age, gender, body mass index, hypertension and diabetes, increased cycle threshold was associated with decreased odds of in-hospital mortality (0.91, CI: 0.89-0.94, P < 0.001)[60].
Collectively these multiple cohort of patients from different studies shows a trend of the association of high viral load and mortality in hospitalized patients.
Association between viral load and infectivity, transmission and antibodies to SARS-CoV-2
Although not statistically significant, 20 studies (1857 patients) indicated the importance of high viral load dynamics with infectiousness and transmissibility (P > 0.05-0.53)[1-3,10-12,14,16,21,22,23,25,28,31,34,39,40,43,47,50].
Association between viral load and infectivity remains unclear, but earlier peak in viral load in SARS-CoV-2 infection suggests that infectivity may be higher earlier in the course than would be expected based on the SARS model[5,62,63].
Subgroup analysis suggests these patients are younger and had milder disease and may be highly infectious and transmit virus to the population given their asymptomatic or presymptomatic nature of illness. These studies shed light on high viral load and its association with infectivity and transmissibility. Highest respiratory viral load was noted at pre-symptomatic stage and infectiousness peaked before symptom onset[1,2,3,5,12,14,16,22,34,40,47,50].
He et al[1] demonstrated an infectiousness profile on 77 infector–infected transmission pairs. Highest viral load in oropharynx at the time of symptom onset correlated with infectiousness. Presymptomatic transmission was 44% (95%CI, 30%–57%) whereas infectiousness started at 12.3 d (95%CI, 5.9-17 d) before symptom onset and peaked at onset (95%CI: –0.9 to 0.9 d). They estimated that proportion of presymptomatic transmission was 37%-48%[1].
Xu et al[2] reported on 51 symptomatic patients, demonstrating transmission from primary (patients who visited the epicenter, Wuhan), to secondary (patients who came into contact with primary) and tertiary (patients who came into contact with only secondary cases). Their findings suggested incubation period in tertiary group was longer compared to primary and secondary groups (both P < 0.05). Ct values detected in tertiary were similar to those for the imported and secondary patients at the time of admission (both P > 0.05). For tertiary group, the viral load was undetectable in half of patients (52.63%) on day 7 and in all patients on day 14. One third of patients in imported and secondary groups remained positive on day 14 after admission. They concluded that infectivity of SARS-CoV-2 may gradually decrease in tertiary patients[2]. This study emphasizes that early quarantine and lock down measures may have mitigated the spread of disease in countries that enforced it strictly. The reason for decrease in infectivity from secondary to tertiary exposed patient remains unclear. Although speculative, this may be due to reduced quantitative viral load transmitted and other strict mask and quarantine measures[2,44].
Some reports demonstrated an association of high viral load and risk of transmission in a closed knit population[28,40]. In a cohort of 80 patients including both health care workers and nursing home residents from COVID-19 outbreak in Washington State, high viral load in unrecognized asymptomatic and presymptomatic patients contributed to infectiousness and transmission. Although the mortality was high in these patients, it did not correlate statistically with the viral load[28]. Similarly Kimball et al[40] analyzed their cohort of 23 patients from a long term care facility. Ten (43%) had symptoms on testing, and 13 (57%) were asymptomatic. Seven days after testing, 10 of these 13 previously asymptomatic residents had developed symptoms and were inferred as presymptomatic at time of testing. The Ct values indicated large quantities of viral RNA in asymptomatic, presymptomatic, and symptomatic residents, suggesting potential for transmission regardless of symptoms[40].
There are at present limits to our understanding and evidence in determining infectiousness and the risk of transmissibility. As described earlier, there is evidence of ongoing viral shedding in various body fluids after symptom resolution in COVID infection and may be prolonged, especially in stool samples compared to respiratory secretions (P < 0.001-0.5)[9,13,15,19,25,31,38,67]. Currently there is no reported evidence of fecal –oral transmission. Further the severity of illness also appears to extend the duration of viral shedding. However, based on current data, there is no convincing evidence that duration of shedding correlates with duration of infectivity. The viral nucleic acid detected in various body fluids later in the course of infection may represent non-viable fragments of virions.
Wölfel et al[14] demonstrated that live virus can be cultured from respiratory samples in patients with positive SARS-CoV-2 RT-PCR. However, the percentage of positive cultures declined and no live virus was successfully isolated after day 8 from symptom onset despite ongoing high quantitative viral load. Additionally, virus could not be isolated from samples less than 105 copies/mL. However a caveat with this cohort was that patients had mild symptoms and were young and middle aged adults. This emphasizes the point that elevated high viral load in convalescing patients may be suggestive but not a definitive factor in infectiousness and transmissibility[14].
There is evidence that children are susceptible to SARS-CoV-2 infection, but frequently do not have symptoms, raising possibility that children could be facilitators of viral transmission. Reports comparing viral kinetics in adults and pediatric patients have demonstrated that children, adolescents and adults can have same variation of viral load, but higher risk of transmission and asymptomatic illness in children may have other contributing factors[16,47,50].
The immune responses of the host to COVID-19 and its relation to infectivity and transmission remain unclear and data is emerging[5,13,59,68,69]. Most patients seroconvert by day 15 after symptom onset and Anti-SARS-CoV-2-NP or anti-SARS-CoV-2-RBD IgG levels correlate with virus neutralization[5]. While risk of transmission after symptom resolution and the presence of antibodies may be lower, it cannot be ruled out with available evidence[1-3,5]. Transmission by asymptomatic or minimally symptomatic individuals also appears likely and highlights the importance of contact tracing and isolation of exposed individuals, especially as transmission potential may be maximal early in course of infection as depicted in the nursing home cohort[28,40]. In their large series of 100 patients Li et al[68] demonstrated specific anti SARS-CoV-2 (IgM, IgG, IgA) antibodies to S-1, N, and RBD viral proteins in the serum within two weeks after onset and reached a peak in 17 d and maintained high levels up to 50 d post infection.
Fourati et al[69] demonstrated an inverse relationship of lower serum titer of neutralizing antibodies (anti-S1 Ig A and Ig G) with elevated nasopharyngeal viral load and severe COVID-19 sepsis. This may indicate an inability to clear infection and have a deleterious impact on survival. Patients who were alive at 28 d displayed higher titers of anti-S1 Ig A and Ig G on admission compared to those who succumbed[69]. Similar observation was demonstrated by Bryan et al[59]; this study demonstrated that detection of anti-SARS-CoV-2 nucleocapsid IgG is associated with lower viral loads in patients. They concluded that high viral loads almost never coexist with SARS-CoV-2 sera-positivity and suggest that persons with anti-SARS-CoV-2 antibodies on admission have reduced 30-d all-cause mortality[59]. Both these studies may suggest that presence of antibody titers on admission, coupled with molecular testing, may be particularly prognostic factor, helpful to assess the disease course for high risk patients who cannot provide a clinical history[59,69]. The mechanism may be due to lower host humoral immune response in the elderly patients with comorbidities.
The heterogeneity of the non-respiratory specimen’s limits its significance in explaining the risk of transmission and no correlation can be inferred. Further research is needed. In addition it is also important to determine viability of virus outside the respiratory and gastrointestinal tract at different stages of infection in both asymptomatic and symptomatic individuals. This will improve understanding of transmission risk and allow greater certainty around guidelines for appropriate contact tracing and quarantine periods[70].
DISCUSSION
SARS-CoV-2 is diagnosed based on nucleic acid test, detecting viral RNA. We briefly discuss the relevance of diagnostics in the context of our research question. Laboratories have set up their RT-PCR techniques with primers and probes and protocols, algorithms following guidelines from United States FDA and Center for Disease Control and Prevention (CDC) and World Health Organization[71]. A reference, limit of detection range is set by each laboratory based on reaction system and amplification conditions, specified according to manufacturer’s specifications[72]. These tests are high throughput and have high sensitivities and specificity. Bisoffi et al[73] demonstrated that nucleic acid tests have highest performance with 91.8% sensitivity, 100% specificity, 100% PPV (positive predictive value) and 97.4% negative predictive value). Some variation may exist in considering single gene targets. S and RdRp genes had highest sensitivity (94.1%) at their institution[73]. Factors that may affect sensitivity of tests are duration of illness, site of specimen collection, and viral load. Some authors have reported that false negative rates may occur in up to 30% tests[71]. However, at present there is no clear advantage of choosing one particular gene over another as long as the sample acquisition, preparation and device operations are performed by trained personnel and laboratories[70,71].
Viral load is the quantity of viral RNA in a given volume expressed as infectious particles per milliliter. This is also expressed as Log10 copies /mL or Ct value. Ct value represents the number of amplification cycles needed for a target gene to exceed a threshold detection level. It is inversely related to viral load; lower the value of Ct, higher the viral load[3,5,12,70,71]. For SARS-CoV-2 the test results are considered positive when multiple genes had a Ct value less than 38. If only one of target gene had a Ct value of < 38, it is reported as a single test positive[32]. Fung et al[74] compared the limit of detection for various assays and reported it to be between 85-499 copies/mL for CDC assays and 74 copies/mL with other commercial high-throughput laboratory analyzers. Digital droplet PCR is another technique useful in situations with a high suspicion of infection but a low viral load or a negative test. This test has an advantage of absolute quantification and higher sensitivity in viral RNA detection especially in low viral load samples[32,75].
Strengths and limitations of this manuscript
This study is a large pooled, qualitative content analysis of 60 manuscripts with a cohort of 10514 patients’ from different cohorts and countries evaluating patterns of quantitative viral load in predicting disease severity, mortality, risk of infectiousness, transmissibility, and prognosis in patients with COVID-19. The author presents the relative merits and discusses the objective data presented in these studies. This a correlation study and does not imply causation.
However, there are certain limitations in this study. Since there is a high heterogeneity of samples and data in the majority of these manuscripts, the content analysis is qualitative (narrative) and these data should be interpreted with caution and considered only as trends. Differences in distribution of age, sex, definition of disease severity, and other confounding variables such as medical comorbidities, different virologic tests and heterogeneous samples may contribute to different clinical outcomes. For instance very few studies adjusted their statistic models for the other medical morbidities which could have increased the risk for morbidity and mortality[4,6,7,15,19,27,30]. The majority of these studies are on hospitalized patients which has a potential bias of analyzing the more severely ill amongst the overall infected population. Further variations of ACE 2 receptors and expression in various tissues in different ethnic populations may play a role in virulence and transmissibility of this virus[76]. A viral nucleic acid load from a particular sample assay may not represent an exact systemic viral load in the body; further viral load may also not represent viable virions and may be falsely misleading. In addition there is no consistent trajectory of why certain samples test positive with high virus loads and others do not. Another important point to consider is that, majority of studies is from one country: China and from a few medical centers around the epicenter of outbreak, possibly leading to overlapping of population data in reported manuscripts. Other limiting factors may include the testing protocol and standards, set for RT-PCR targets vary between different laboratories[68-70]. Finally there is always a possibility of observer (author bias) which is to be considered.
Although majority of studies showed a positive association between a high viral load and mortality there were three studies with (434 patients) suggestive of an inverse correlation between the two. Argyropoulos et al[35] in their report on 205 patients demonstrated an inverse correlation of admission nasopharyngeal viral load with duration, severity of sepsis and no correlation with survival (P < 0.001). The reason for low mortality in this study is unclear. One possible explanation could be due to the fact that viral loads detected from nasopharyngeal samples were obtained at a later time point in the disease course. As we have described earlier, that SARS-CoV-2 viral load peaks earlier in the infection followed by cytokine storm and hyper-inflammation when the innate immune system is unable to control the initial viral replication[61]. At these later times points the viral replication may start to defervesce but the multi-organ dysfunction is secondary to systemic hyper-inflammatory response. Similarly Hasanoglu et al[46] on their cohort of 60 patients demonstrated an inverse relationship of high viral load with mortality; however their study had a mean age of 32 signifying a younger age group, where mortality is lower compared to older patients. Another group of 169 patients, reported from Spain by Carrasquer et al[57] demonstrated no statistical association of high viral load with in hospital mortality when adjusted to age, gender and serum cardiac troponin levels. The conclusions from this study suggested myocardial damage with medical comorbidities as the cause for increased mortality in susceptible population and not high viral loads.
Why is quantitative viral assay important?
Although infection and inflammation begins with the respiratory tract, it also involves extra pulmonary organs[77]. Isolation of viral nucleic acid in multiple tissues, blood and body secretions are indicative of systemic spread and are indicative of severe infection. Evidence from these manuscripts suggests that high viral load occurs in respiratory tract samples during presymptomatic period and peaks at the onset of symptoms and gradually declines over the next one to three weeks[1,2,3,5,9,12,14,16,22,34,40]. Increased viral load in respiratory tract represents active viral replication and a surrogate marker for predicting severity[28,32,37,61]. This is in contrast to previous SARS-CoV epidemic in 2003 where the peak viral load occurred during second week after symptoms appeared and was positively correlated with increased mortality[5,62,63]. This fact explains the increased infectivity and rapid transmission of SARS-CoV-2 compared to previous SARS-CoV epidemic[5]. Along with comorbidities, assessment of viral load from nasopharynx or sputum may determine the risk of severity of sepsis in symptomatic, hospitalized elderly patients[4,5,18]. High viral load is also associated with elevated cytokine, lymphopenia i.e., markers for inflammation and portends poor prognosis[8,24,33,36,37,42,52,65,66]. Early determination of viral load also has therapeutic benefits, such as administration of convalescent plasma, neutralizing antibodies, antiviral medicines and corticosteroids in susceptible elderly patients[6,7,11].
SARS-CoV-2 pandemic continues to spread unabated in United States and worldwide. This is particularly evident after the end of lock down and social distancing measures with increased mobility of the population. A report from a reference laboratory evaluated 29713 de-identified samples from respiratory tract. 14.9% of samples tested positive. Highest positivity rate was identified in males born between1964-1974. Patients between ages of 11-25 had highest viral load (> 10 Log10 copies/mL). The clinical symptoms or outcomes of these patients were not known. This study demonstrates that high viral load in younger group may be an important risk factor for infectivity and transmission in a community, regardless of their symptom status[78].
COVID -19 infections in younger asymptomatic patients, with high viral load may fare well due to their robust physiologic reserve. However, they are at highest risk for transmitting the disease and are called super spreaders. These infections generally appear asymptomatic or milder in younger population, but elderly patients bear the brunt of severe infection, hospitalization and mortality[61,62].
CONCLUSION
High SARS-CoV-2 viral load was found to be an independent predictor of disease severity and mortality in high proportion of studies, and may be useful in predicting the clinical course and prognosis of patients with COVID-19. However there is a wide heterogeneity in fluid samples and different phases of the disease and these data should be interpreted with caution and only considered as trends. In aggregate, these observations support the hypothesis of checking and reporting viral load by quantitative RT-PCR, instead of binary assessment of a test being positive or negative.
ARTICLE HIGHLIGHTS
Research background
High viral load has an implication in the clinical outcomes in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. At present there is no Food and Drug Administration Emergency Use Authorization for quantitative viral load assay in the current pandemic. Currently the coronavirus disease 2019 (COVID-19) tests are reported as a binary assessment of either positive or negative test.
Research motivation
The intent of this research is to identify whether quantitative SARS-CoV-2 viral load assay correlates with severity of infection and mortality?
Research objectives
To assess high viral load and its association with the severity, mortality, infectiousness in COVID-19 infections.
Research methods
A systematic literature search was undertaken for a period between December 30, 2019 to December 31, 2020 in PubMed/MEDLINE using combination of terms “COVID-19, SARS-CoV-2, Ct values, Log10 copies, quantitative viral load, viral dynamics, kinetics, association with severity, sepsis, mortality and infectiousness’’. Data on age, number of patients, sample sites, real time reverse transcriptase polymerase chain reaction (RT-PCR) targets, disease severity, intensive care unit admission, mortality and conclusions of the studies was extracted, organized and is analyzed.
Research results
High SARS-CoV-2 viral load was found to be an independent predictor of disease severity and mortality in high proportion of studies, and may be useful in predicting the clinical course and prognosis of patients with COVID-19.
Research conclusions
There is a wide heterogeneity in fluid samples and different phases of the disease and these data should be interpreted with caution and only considered as trends. In aggregate, these observations support the hypothesis of checking and reporting viral load by quantitative RT-PCR, instead of binary assessment of a test being positive or negative.
Research perspectives
In future, longitudinal studies with viral load should be monitored and analyzed, so it can be considered in interpretation of outcome data. It may also be a guiding principle for therapy and infection control policies for current and future pandemics.
Footnotes
Conflict-of-interest statement: The author declares that he has no competing interests.
PRISMA 2009 Checklist statement: The author has read the PRISMA 2009 checklist, and the manuscript was prepared and revised according to the PRISMA 2009 checklist.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: The Society for Surgery of the Alimentary Tract.
Peer-review started: February 5, 2021
First decision: March 17, 2021
Article in press: June 15, 2021
Specialty type: Virology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Carnevale S, Li JQ S-Editor: Liu M L-Editor: A P-Editor: Wang LL
References
- 1.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 2.Xu T, Chen C, Zhu Z, Cui M, Dai H, Xue Y. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis. 2020;94:68–71. doi: 10.1016/j.ijid.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, Gaymard A, Bouscambert-Duchamp M, Donati F, Le Hingrat Q, Enouf V, Houhou-Fidouh N, Valette M, Mailles A, Lucet JC, Mentre F, Duval X, Descamps D, Malvy D, Timsit JF, Lina B, van-der-Werf S, Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, Peiris M, Poon LLM, Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, Wei J, Xiao H, Qu J, Qing L, Chen L, Xu Z, Peng L, Li Y, Zheng H, Chen F, Huang K, Jiang Y, Liu D, Zhang Z, Liu Y, Liu L. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, Peng C, Yuan M, Huang J, Wang Z, Yu J, Gao X, Wang D, Yu X, Li L, Zhang J, Wu X, Li B, Xu Y, Chen W, Peng Y, Lin L, Liu X, Huang S, Zhou Z, Zhang L, Wang Y, Zhang Z, Deng K, Xia Z, Gong Q, Zhang W, Zheng X, Liu Y, Yang H, Zhou D, Yu D, Hou J, Shi Z, Chen S, Chen Z, Zhang X, Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, Men D, Huang Q, Liu Y, Yang B, Ding J, Li F. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, Liu X, Wang S, Wu X, Ge Q, He J, Zhan H, Qiu F, Guo L, Huang C, Jaki T, Hayden FG, Horby PW, Zhang D, Wang C. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, Li H, Wang S, Ruan S, Yang C, Mei C, Wang Y, Ding D, Wu F, Tang X, Ye X, Ye Y, Liu B, Yang J, Yin W, Wang A, Fan G, Zhou F, Liu Z, Gu X, Xu J, Shang L, Zhang Y, Cao L, Guo T, Wan Y, Qin H, Jiang Y, Jaki T, Hayden FG, Horby PW, Cao B, Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Zhang L, Sang L, Ye F, Ruan S, Zhong B, Song T, Alshukairi AN, Chen R, Zhang Z, Gan M, Zhu A, Huang Y, Luo L, Mok CKP, Al Gethamy MM, Tan H, Li Z, Huang X, Li F, Sun J, Zhang Y, Wen L, Li Y, Chen Z, Zhuang Z, Zhuo J, Chen C, Kuang L, Wang J, Lv H, Jiang Y, Li M, Lin Y, Deng Y, Tang L, Liang J, Huang J, Perlman S, Zhong N, Zhao J, Malik Peiris JS. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 15.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, Xie G, Lin S, Wang R, Yang X, Chen W, Wang Q, Zhang D, Liu Y, Gong R, Ma Z, Lu S, Xiao Y, Gu Y, Zhang J, Yao H, Xu K, Lu X, Wei G, Zhou J, Fang Q, Cai H, Qiu Y, Sheng J, Chen Y, Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baggio S, L'Huillier AG, Yerly S, Bellon M, Wagner N, Rohr M, Huttner A, Blanchard-Rohner G, Loevy N, Kaiser L, Jacquerioz F, Eckerle I. SARS-CoV-2 viral load in the upper respiratory tract of children and adults with early acute COVID-19. Clin Infect Dis. 2020:epub ahead of print. doi: 10.1093/cid/ciaa1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi F, Wu T, Zhu X, Ge Y, Zeng X, Chi Y, Du X, Zhu L, Zhu F, Zhu B, Cui L, Wu B. Association of viral load with serum biomakers among COVID-19 cases. Virology. 2020;546:122–126. doi: 10.1016/j.virol.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clementi N, Ferrarese R, Tonelli M, Amato V, Racca S, Locatelli M, Lippi G, Silvestri G, Clementi M, Mancini N. Lower nasopharyngeal viral load during the latest phase of COVID-19 pandemic in a Northern Italy University Hospital. Clin Chem Lab Med. 2020;58:1573–1577. doi: 10.1515/cclm-2020-0815. [DOI] [PubMed] [Google Scholar]
- 19.Kwon JS, Kim JY, Kim MC, Park SY, Kim BN, Bae S, Cha HH, Jung J, Kim MJ, Lee MJ, Choi SH, Chung JW, Shin EC, Kim SH. Factors of Severity in Patients with COVID-19: Cytokine/Chemokine Concentrations, Viral Load, and Antibody Responses. Am J Trop Med Hyg. 2020;103:2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Sun S, Shi Y, Wang H, Zhao R, Sheng J. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit Care. 2020;24:170. doi: 10.1186/s13054-020-02893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu R, Yi S, Zhang J, Lv Z, Zhu C, Zhang Y. Viral Load Dynamics in Sputum and Nasopharyngeal Swab in Patients with COVID-19. J Dent Res. 2020;99:1239–1244. doi: 10.1177/0022034520946251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou R, Li F, Chen F, Liu H, Zheng J, Lei C, Wu X. Viral dynamics in asymptomatic patients with COVID-19. Int J Infect Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung KH, Fung AY, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan KH, Yuen KY, Leung WK. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, Fasano M, Sessa F, Tettamanti L, Carinci F, Maurino V, Rossi A, Tagliabue A, Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Gao G, Xu Y, Pu L, Wang Q, Wang L, Wang W, Song Y, Chen M, Yu F, Yang S, Tang Y, Zhao L, Wang H, Wang Y, Zeng H, Zhang F. SARS-CoV-2-Positive Sputum and Feces After Conversion of Pharyngeal Samples in Patients With COVID-19. Ann Intern Med. 2020;172:832–834. doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, Zhang Y, Xu Z, Liu X, Li Y. SARS-CoV-2 Viral Load in Clinical Samples from Critically Ill Patients. Am J Respir Crit Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, Nadkarni G, Glicksberg BS, Houldsworth J, Cordon-Cardo C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA Public Health–Seattle and King County and CDC COVID-19 Investigation Team. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N Engl J Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang JT, Ran RX, Lv ZH, Feng LN, Ran CY, Tong YQ, Li D, Su HW, Zhu CL, Qiu SL, Yang J, Xiao MY, Liu MJ, Yang YT, Liu SM, Li Y. Chronological Changes of Viral Shedding in Adult Inpatients With COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:2158–2166. doi: 10.1093/cid/ciaa631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, Goyal P, Safford MM, Satlin MJ. Impact of SARS-CoV-2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients with Coronavirus Disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SK, Lee CW, Park DI, Woo HY, Cheong HS, Shin HC, Ahn K, Kwon MJ, Joo EJ. Detection of SARS-CoV-2 in Fecal Samples From Patients With Asymptomatic and Mild COVID-19 in Korea. Clin Gastroenterol Hepatol. 2020:epub ahead of print. doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, Gao G, Wang S, Ma C, Xie R, Wang F, Tan C, Zhu L, Guo Y, Zhang F. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin Infect Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blot M, Jacquier M, Manoha C, Piroth L, Charles PE Pneumochondrie study group. Alveolar SARS-CoV-2 Viral Load Is Tightly Correlated With Severity in COVID-19 ARDS. Clin Infect Dis. 2021;72:e446–e447. doi: 10.1093/cid/ciaa1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SE, Jeong HS, Yu Y, Shin SU, Kim S, Oh TH, Kim UJ, Kang SJ, Jang HC, Jung SI, Park KH. Viral kinetics of SARS-CoV-2 in asymptomatic carriers and presymptomatic patients. Int J Infect Dis. 2020;95:441–443. doi: 10.1016/j.ijid.2020.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argyropoulos KV, Serrano A, Hu J, Black M, Feng X, Shen G, Call M, Kim MJ, Lytle A, Belovarac B, Vougiouklakis T, Lin LH, Moran U, Heguy A, Troxel A, Snuderl M, Osman I, Cotzia P, Jour G. Association of Initial Viral Load in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Patients with Outcome and Symptoms. Am J Pathol. 2020;190:1881–1887. doi: 10.1016/j.ajpath.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu D, Zhou F, Sun W, Chen L, Lan L, Li H, Xiao F, Li Y, Kolachalama VB, Wang X, Xu H. Relationship Between serum SARS-CoV-2 nucleic acid(RNAemia) and Organ Damage in COVID-19 Patients: A Cohort Study. Clin Infect Dis. 2020:epub ahead of print. doi: 10.1093/cid/ciaa1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veyer D, Kernéis S, Poulet G, Wack M, Robillard N, Taly V, L'Honneur AS, Rozenberg F, Laurent-Puig P, Bélec L, Hadjadj J, Terrier B, Péré H. Highly sensitive quantification of plasma SARS-CoV-2 RNA shelds light on its potential clinical value. Clin Infect Dis. 2020:epub ahead of print. doi: 10.1093/cid/ciaa1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin W, Xie Z, Li Y, Li L, Wen C, Cao Y, Chen X, Ou X, Hu F, Li F, Tang X, Cai W. Association between detectable SARS-COV-2 RNA in anal swabs and disease severity in patients with coronavirus disease 2019. J Med Virol. 2021;93:794–802. doi: 10.1002/jmv.26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Liao B, Guo Y, Li F, Lei C, Zhang F, Cai W, Hong W, Zeng Y, Qiu S, Wang J, Li Y, Deng X, Li J, Xiao G, Guo F, Lai X, Liang Z, Wen X, Li P, Jiao Q, Xiang F, Wang Y, Ma C, Xie Z, Lin W, Wu Y, Tang X, Li L, Guan Y. Clinical Characteristics of Patients Infected With the Novel 2019 Coronavirus (SARS-Cov-2) in Guangzhou, China. Open Forum Infect Dis. 2020;7:ofaa187. doi: 10.1093/ofid/ofaa187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimball A, Hatfield KM, Arons M, James A, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Chisty Z, Bell JM, Methner M, Harney J, Jacobs JR, Carlson CM, McLaughlin HP, Stone N, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Russell D, Hiatt B, Gant J, Duchin JS, Clark TA, Honein MA, Reddy SC, Jernigan JA Public Health – Seattle & King County; CDC COVID-19 Investigation Team. Asymptomatic and Presymptomatic SARS-CoV-2 Infections in Residents of a Long-Term Care Skilled Nursing Facility - King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377–381. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwierzeck V, König JC, Kühn J, Mellmann A, Correa-Martínez CL, Omran H, Konrad M, Kaiser T, Kampmeier S. First Reported Nosocomial Outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 in a Pediatric Dialysis Unit. Clin Infect Dis. 2021;72:265–270. doi: 10.1093/cid/ciaa491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia XY, Wu J, Liu HL, Xia H, Jia B, Huang WX. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. J Clin Virol. 2020;127:104360. doi: 10.1016/j.jcv.2020.104360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagman K, Hedenstierna M, Gille-Johnson P, Hammas B, Grabbe M, Dillner J, Ursing J. SARS-CoV-2 RNA in serum as predictor of severe outcome in COVID-19: a retrospective cohort study. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prebensen C, Myhre PL, Jonassen C, Rangberg A, Blomfeldt A, Svensson M, Omland T, Berdal JE. SARS-CoV-2 RNA in plasma is associated with ICU admission and mortality in patients hospitalized with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasanoglu I, Korukluoglu G, Asilturk D, Cosgun Y, Kalem AK, Altas AB, Kayaaslan B, Eser F, Kuzucu EA, Guner R. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection. 2021;49:117–126. doi: 10.1007/s15010-020-01548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawasuji H, Takegoshi Y, Kaneda M, Ueno A, Miyajima Y, Kawago K, Fukui Y, Yoshida Y, Kimura M, Yamada H, Sakamaki I, Tani H, Morinaga Y, Yamamoto Y. Transmissibility of COVID-19 depends on the viral load around onset in adult and symptomatic patients. PLoS One. 2020;15:e0243597. doi: 10.1371/journal.pone.0243597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bermejo-Martin JF, González-Rivera M, Almansa R, Micheloud D, Tedim AP, Domínguez-Gil M, Resino S, Martín-Fernández M, Ryan Murua P, Pérez-García F, Tamayo L, Lopez-Izquierdo R, Bustamante E, Aldecoa C, Gómez JM, Rico-Feijoo J, Orduña A, Méndez R, Fernández Natal I, Megías G, González-Estecha M, Carriedo D, Doncel C, Jorge N, Ortega A, de la Fuente A, Del Campo F, Fernández-Ratero JA, Trapiello W, González-Jiménez P, Ruiz G, Kelvin AA, Ostadgavahi AT, Oneizat R, Ruiz LM, Miguéns I, Gargallo E, Muñoz I, Pelegrin S, Martín S, García Olivares P, Cedeño JA, Ruiz Albi T, Puertas C, Berezo JÁ, Renedo G, Herrán R, Bustamante-Munguira J, Enríquez P, Cicuendez R, Blanco J, Abadia J, Gómez Barquero J, Mamolar N, Blanca-López N, Valdivia LJ, Fernández Caso B, Mantecón MÁ, Motos A, Fernandez-Barat L, Ferrer R, Barbé F, Torres A, Menéndez R, Eiros JM, Kelvin DJ. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shlomai A, Ben-Zvi H, Glusman Bendersky A, Shafran N, Goldberg E, Sklan EH. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID-19 patients. Crit Care. 2020;24:539. doi: 10.1186/s13054-020-03244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ra SH, Lim JS, Kim GU, Kim MJ, Jung J, Kim SH. Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS-CoV-2 infection. Thorax. 2021;76:61–63. doi: 10.1136/thoraxjnl-2020-215042. [DOI] [PubMed] [Google Scholar]
- 51.Faíco-Filho KS, Passarelli VC, Bellei N. Is Higher Viral Load in SARS-CoV-2 Associated with Death? Am J Trop Med Hyg. 2020;103:2019–2021. doi: 10.4269/ajtmh.20-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, Wang G, Long X, Hou H, Wei J, Cao Y, Tan J, Liu W, Huang L, Meng F, Wang N, Zhao J, Huang G, Sun Z, Wang W, Zhou J. Dynamics of Blood Viral Load Is Strongly Associated with Clinical Outcomes in Coronavirus Disease 2019 (COVID-19) Patients: A Prospective Cohort Study. J Mol Diagn. 2021;23:10–18. doi: 10.1016/j.jmoldx.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fajnzylber J, Regan J, Coxen K, Corry H, Wong C, Rosenthal A, Worrall D, Giguel F, Piechocka-Trocha A, Atyeo C, Fischinger S, Chan A, Flaherty KT, Hall K, Dougan M, Ryan ET, Gillespie E, Chishti R, Li Y, Jilg N, Hanidziar D, Baron RM, Baden L, Tsibris AM, Armstrong KA, Kuritzkes DR, Alter G, Walker BD, Yu X, Li JZ Massachusetts Consortium for Pathogen Readiness. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou S, Yang Y, Zhang X, Li Z, Liu X, Hu C, Chen C, Wang D, Peng Z. Clinical Course of 195 Critically Ill COVID-19 Patients: A Retrospective Multicenter Study. Shock. 2020;54:644–651. doi: 10.1097/SHK.0000000000001629. [DOI] [PubMed] [Google Scholar]
- 55.Maltezou HC, Raftopoulos V, Vorou R, Papadima K, Mellou K, Spanakis N, Kossyvakis A, Gioula G, Exindari M, Froukala E, Martinez-Gonzalez B, Panayiotakopoulos G, Papa A, Mentis A, Tsakris A. Association Between Upper Respiratory Tract Viral Load, Comorbidities, Disease Severity, and Outcome of Patients With SARS-CoV-2 Infection. J Infect Dis. 2021;223:1132–1138. doi: 10.1093/infdis/jiaa804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bitker L, Dhelft F, Chauvelot L, Frobert E, Folliet L, Mezidi M, Trouillet-Assant S, Belot A, Lina B, Wallet F, Richard JC. Protracted viral shedding and viral load are associated with ICU mortality in Covid-19 patients with acute respiratory failure. Ann Intensive Care. 2020;10:167. doi: 10.1186/s13613-020-00783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrasquer A, Peiró ÓM, Sanchez-Gimenez R, Lal-Trehan N, Del-Moral-Ronda V, Bonet G, Gutierrez C, Fort-Gallifa I, Martin-Grau C, Benavent C, Vidal F, Bardají A. Lack of Association of Initial Viral Load in SARS-CoV-2 Patients with In-Hospital Mortality. Am J Trop Med Hyg. 2020;104:540–545. doi: 10.4269/ajtmh.20-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de la Calle C, Lalueza A, Mancheño-Losa M, Maestro-de la Calle G, Lora-Tamayo J, Arrieta E, García-Reyne A, Losada I, de Miguel B, Díaz-Simón R, López-Medrano F, Fernández-Ruiz M, Carretero O, San Juan R, Aguado JM, Lumbreras C. Impact of viral load at admission on the development of respiratory failure in hospitalized patients with SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021;40:1209–1216. doi: 10.1007/s10096-020-04150-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bryan A, Fink SL, Gattuso MA, Pepper G, Chaudhary A, Wener MH, Morishima C, Jerome KR, Mathias PC, Greninger AL. SARS-CoV-2 Viral Load on Admission Is Associated With 30-Day Mortality. Open Forum Infect Dis. 2020;7:ofaa535. doi: 10.1093/ofid/ofaa535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choudhuri J, Carter J, Nelson R, Skalina K, Osterbur-Badhey M, Johnston A, Goldstein D, Paroder M, Szymanski J. SARS-CoV-2 PCR cycle threshold at hospital admission associated with patient mortality. PLoS One. 2020;15:e0244777. doi: 10.1371/journal.pone.0244777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shenoy S. Coronavirus (Covid-19) sepsis: revisiting mitochondrial dysfunction in pathogenesis, aging, inflammation, and mortality. Inflamm Res. 2020;69:1077–1085. doi: 10.1007/s00011-020-01389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu CM, Poon LL, Cheng VC, Chan KS, Hung IF, Wong MM, Chan KH, Leung WS, Tang BS, Chan VL, Ng WL, Sim TC, Ng PW, Law KI, Tse DM, Peiris JS, Yuen KY. Initial viral load and the outcomes of SARS. CMAJ. 2004;171:1349–1352. doi: 10.1503/cmaj.1040398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY HKU/UCH SARS Study Group. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oh MD, Park WB, Choe PG, Choi SJ, Kim JI, Chae J, Park SS, Kim EC, Oh HS, Kim EJ, Nam EY, Na SH, Kim DK, Lee SM, Song KH, Bang JH, Kim ES, Kim HB, Park SW, Kim NJ. Viral Load Kinetics of MERS Coronavirus Infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation Between Relative Nasopharyngeal Virus RNA Load and Lymphocyte Count Disease Severity in Patients with COVID-19. Viral Immunol. 2020:epub ahead of print. doi: 10.1089/vim.2020.0062. [DOI] [PubMed] [Google Scholar]
- 66.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A Yale IMPACT Team. Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, Zhang P, Liu X, Gao G, Liu F, Jiang Y, Cheng X, Zhu C, Xia Y. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li L, Liang Y, Hu F, Yan H, Li Y, Xie Z, Huang L, Zhao J, Wan Z, Wang H, Shui J, Cai W, Tang S. Molecular and serological characterization of SARS-CoV-2 infection among COVID-19 patients. Virology. 2020;551:26–35. doi: 10.1016/j.virol.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fourati S, Hue S, Pawlotsky JM, Mekontso-Dessap A, de Prost N. SARS-CoV-2 viral loads and serum IgA/IgG immune responses in critically ill COVID-19 patients. Intensive Care Med. 2020;46:1781–1783. doi: 10.1007/s00134-020-06157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Widders A, Broom A, Broom J. SARS-CoV-2: The viral shedding vs infectivity dilemma. Infect Dis Health. 2020;25:210–215. doi: 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weissleder R, Lee H, Ko J, Pittet MJ. COVID-19 diagnostics in context. Sci Transl Med. 2020;12:eabc1931. doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 72.Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bisoffi Z, Pomari E, Deiana M, Piubelli C, Ronzoni N, Beltrame A, Bertoli G, Riccardi N, Perandin F, Formenti F, Gobbi F, Buonfrate D, Silva R. Sensitivity, Specificity and Predictive Values of Molecular and Serological Tests for COVID-19: A Longitudinal Study in Emergency Room. Diagnostics (Basel) 2020;10:669. doi: 10.3390/diagnostics10090669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fung B, Gopez A, Servellita V, Arevalo S, Ho C, Deucher A, Thornborrow E, Chiu C, Miller S. Direct Comparison of SARS-CoV-2 Analytical Limits of Detection across Seven Molecular Assays. J Clin Microbiol. 2020;58:e01535–20. doi: 10.1128/JCM.01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suo T, Liu X, Feng J, Guo M, Hu W, Guo D, Ullah H, Yang Y, Zhang Q, Wang X, Sajid M, Huang Z, Deng L, Chen T, Liu F, Xu K, Liu Y, Xiong Y, Chen G, Lan K, Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wen F, Huang X, Ning G, Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh SA, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Bilezikian JP, Landry DW. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kleiboeker S, Cowden S, Grantham J, Nutt J, Tyler A, Berg A, Altrich M. SARS-CoV-2 viral load assessment in respiratory samples. J Clin Virol. 2020;129:104439. doi: 10.1016/j.jcv.2020.104439. [DOI] [PMC free article] [PubMed] [Google Scholar]