Abstract
Purpose
Coronavirus disease 2019 (COVID-19) pandemic has spread worldwide rapidly and patients with cancer have been considered as a vulnerable group for this infection. This study aimed to examine the expressions of angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) in tumor tissues of six common cancer types.
Materials and Methods
The expression levels of ACE2 and TMPRSS2 in tumors and control samples were obtained from online databases. Survival prognosis and biological functions of these genes were investigated for each tumor type.
Results
There was the overexpression of ACE2 in colon and stomach adenocarcinomas compared to controls, meanwhile colon and prostate adenocarcinomas showed a significantly higher expression of TMPRSS2. Additionally, survival prognosis analysis has demonstrated that upregulation of ACE2 in liver hepatocellular carcinoma was associated with higher overall survival (hazard ratio, 0.65; p=0.016) and disease-free survival (hazard ratio, 0.66; p=0.007), while overexpression of TMPRSS2 was associated with a 26% reduced risk of death in lung adenocarcinoma (p=0.047) but 50% increased risk of death in breast invasive carcinoma (p=0.015).
Conclusion
There is a need to take extra precautions for COVID-19 in patients with colorectal cancer, stomach cancer, and lung cancer. Further information on other types of cancer at different stages should be investigated.
Keywords: COVID-19, Neoplasms, Gene expression, Prognosis, Pathway
Introduction
Since the first case of pneumonia with unknown causes was reported in Wuhan, Hubei Province, China in 12 December 2019, a novel coronavirus (coronavirus disease 2019 [COVID-19]) has been demonstrated and spread rapidly worldwide [1]. As of 4 July 2020, approximately 15 million cases have been confirmed and at least 600,000 people died due to this outbreak. Currently, only remdesivir and dexamethasone have been reported for their possible efficacy in the treatment of mild to moderate and severe patients with COVID-19, respectively [2,3]. Although recent results showed approximately 95% efficacy of BNT162b vaccine of Pfizer and BioNTech and mRNA-1273 vaccine of Russian Sputnik V and Moderna, the issues of little safety data, efficacy in high risk population and the duration of protection have remained unclear [4]. It is therefore essential to research strategies to prevent viral infection, especially factors associated with disease severity and mortality. Pooled analysis from our recent systematic review and meta-analysis showed that the history of cancer was associated with several critical conditions of COVID-19 (overall relative risk [RR], 2.91; 95% confidence interval [CI], 2.16 to 3.91), including severe infection (RR, 2.45; 95% CI, 1.67 to 3.58) and intensive care unit admission (RR, 2.26; 95% CI, 1.27 to 4.01) [5].
At the molecular level, spike protein sequence similarities between severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and SARS-CoV have been reported of approximately 76%–78% for the whole protein, 73%–76% for the receptor-binding domain, and 50%–53% for the receptor-binding motif [6], which strongly suggested that SARS-CoV-2 can use the SARS-CoV receptor angiotensin-converting enzyme 2 (ACE2) and employ a cellular transmembrane serine protease 2 (TMPRSS2) to enter human cells [7,8]. Differences in gene expression of ACE2 and TMPRSS2 among cancer types may therefore result in different impacts on the susceptibility to COVID-19 infection. Herein, we evaluated the genetic expression and prognostic of ACE2 and TMPRSS2 in six site-specific cancers (lung, breast, colorectum, prostate, stomach, and liver), which account for 50.9% of cancer incidence and 54.4% of cancer mortality globally [9]. Among them, lung, colorectum, and stomach have also remained three of top five common cancer sites and cancer deaths in Korea [10–14].
Materials and Methods
1. Gene expression analysis
The expression levels of ACE2 and TMPRSS2 in tumors and control samples were obtained from the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) database using RNA sequencing data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) [15]. The cutoff criterion was set at adjusted p-value < 0.01 and log2 fold change > 1 or < −1. Expression levels of ACE2 and RMPSS2 in lung adenocarcinoma (LUAD), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), and liver hepatocellular carcinoma (LIHC) and their subtypes (except STAD due to data unavailability) were compared with normal samples using a two-sample t-test and an analysis of variance (ANOVA).
2. Survival prognosis analysis
Expression of ACE2 and TMPRSS2 in the prognosis of six cancer types was examined utilizing the GEPIA2 database [15]. Median values of gene expression were selected as cut-off points in the prognostic analysis of overall survival (OS) and disease-free survival (DFS). Kaplan-Meier plot for the probability of survival throughout time, hazard ratio (HR) by a Cox proportional hazard model, and p-value by log-rank test for the OS and DFS between high and low expression groups was reported.
3. Biological functions of ACE2 and TMPRSS2
The list of 100 most co-expressed genes with ACE2 (S1 Table) and TMPRSS2 (S2 Table) in each tumor was obtained from the GEPIA2 database [15]. Then, Kyoto Encyclopedia of Genes and Genomes (KEGG) 2019 Human biological functions of ACE2 and TMPRSS2 were explored by applying the pathway enrichment analysis in the EnRichr database. p-values were selected at 0.05 for the cutoff [16].
Results
1. Expression analysis
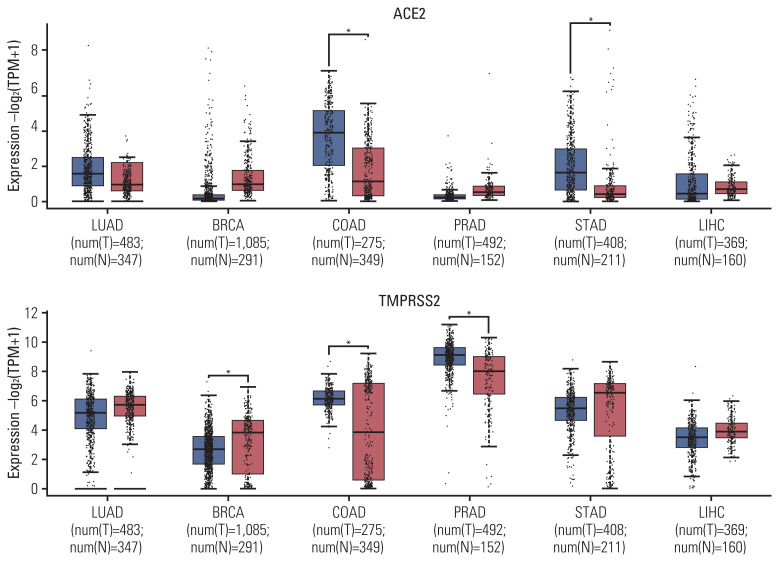
Data from the GEPIA2 database revealed that mRNA levels of ACE2 expression were significantly higher in COAD and STAD, whereas mRNA levels of TMPRSS2 expression were significantly lower in BRCA, but higher in COAD and PRAD, compared with the control group (Fig. 1).
Fig. 1.
Comparison of ACE2 and TMPRSS2 expression between tumors and control. ACE2, angiotensin-converting enzyme 2; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; TMPRSS2, transmembrane serine protease 2.
Analysis of subtype tumors from the GENT2 database also showed the significantly higher ACE2 expression of COAD subtypes including microsatellite instability–high (MSI-H), microsatellite instability–low (MSI-L), and microsatellite stable (MSS), in comparison with the control group (S3 Fig.). However, data on STAD subtypes were not available. In terms of TMPRSS2 expression, significantly lower mRNA levels were only observed for luminal A and luminal B subtypes of BRCA, but not for basal-like and human epidermal growth factor receptor 2 subtypes, compared with the control group (S4 Fig.). Significantly higher TMPRSS2 was also observed for i-Cluster 1 and i-Cluster 3, but not for i-Cluster 2 subtypes of PRAD, compared with the control group. Proximal inflammatory and proximal proliferative subtypes of LUAD additionally expressed lower mRNA levels of TMPRSS2 than the control group, but a significant association was not observed for the terminal respiratory unit subtype.
2. Prognosis analysis
The Kaplan-Meier plots for the effect of ACE2 and TMPRSS2 expression on OS and DFS in different tumor types are presented in S5–S8 Fig. and summarized in Table 1. Overall, higher levels of ACE2 in LIHC were associated with higher OS (HR, 0.65; p=0.016) and DFS (HR, 0.66; p=0.007). Additionally, higher levels of TMPRSS2 were associated with a 26% reduced risk of death in LUAD (p=0.047) but a 50% increased risk of death in BRCA (p=0.015).
Table 1.
Survival analysis of ACE2 and TMPRSS2 expression in different tumors
| Tumor | Outcome | ACE2 | TMPRSS2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| No. (high) | No. (low) | Hazard ratio | p-value | No. (high) | No. (low) | Hazard ratio | p-value | ||
| Lung adenocarcinoma | Overall survival | 239 | 237 | 1.20 | 0.19 | 239 | 239 | 0.74 | 0.047 |
|
| |||||||||
| Disease-free survival | 1.20 | 0.27 | 0.89 | 0.43 | |||||
|
| |||||||||
| Breast invasive carcinoma | Overall survival | 535 | 503 | 1.00 | 0.87 | 534 | 534 | 1.50 | 0.015 |
|
| |||||||||
| Disease-free survival | 0.89 | 0.54 | 1.10 | 0.72 | |||||
|
| |||||||||
| Colon adenocarcinoma | Overall survival | 135 | 135 | 0.73 | 0.20 | 135 | 135 | 0.92 | 0.73 |
|
| |||||||||
| Disease-free survival | 0.80 | 0.35 | 0.86 | 0.53 | |||||
|
| |||||||||
| Prostate adenocarcinoma | Overall survival | 242 | 236 | 0.66 | 0.51 | 246 | 246 | 0.54 | 0.38 |
|
| |||||||||
| Disease-free survival | 0.90 | 0.60 | 0.91 | 0.65 | |||||
|
| |||||||||
| Stomach adenocarcinoma | Overall survival | 192 | 192 | 0.88 | 0.41 | 192 | 192 | 1.10 | 0.68 |
|
| |||||||||
| Disease-free survival | 1.10 | 0.66 | 1.10 | 0.72 | |||||
|
| |||||||||
| Liver hepatocellular carcinoma | Overall survival | 182 | 182 | 0.65 | 0.016 | 182 | 182 | 0.94 | 0.71 |
|
| |||||||||
| Disease-free survival | 0.66 | 0.007 | 0.89 | 0.42 | |||||
ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane serine protease 2.
3. Pathway enrichment analysis
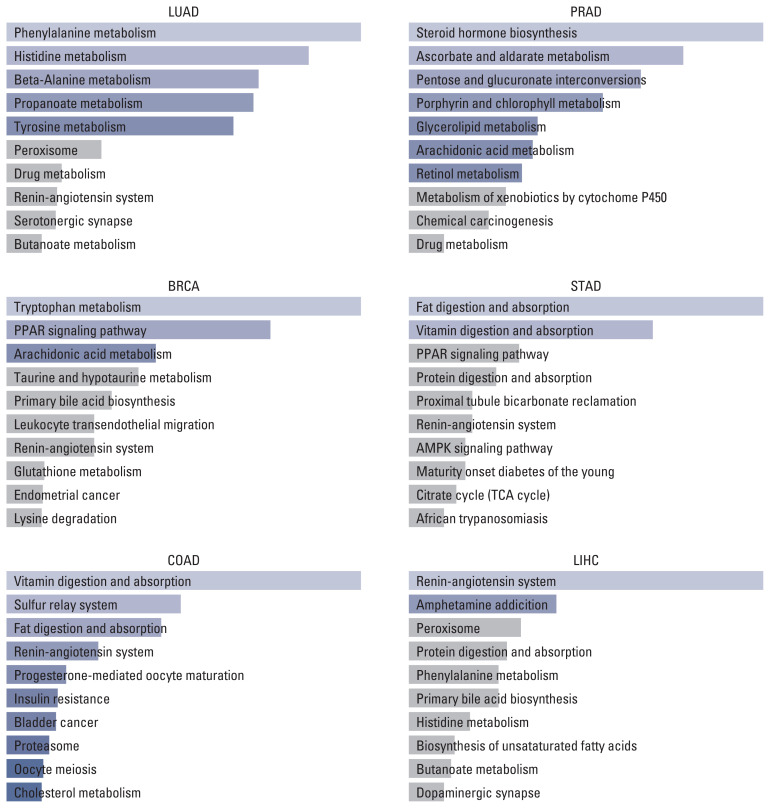
On the one hand, the result of the KEGG pathway enrichment analysis for ACE2 and their top 100 co-expressed genes is presented in Fig. 2 and S9 Table. The most significant pathways in LUAD, BRCA, COAD, PRAD, STAD, and LIHC were phenylalanine metabolism (p=0.003), tryptophan metabolism (p=0.001), vitamin digestion and absorption (p < 0.001), steroid hormone biosynthesis (p=0.003), fat digestion and absorption (p=0.001), and renin-angiotensin system (p=0.006).
Fig. 2.
Top 10 KEGG pathways of ACE2 and top 100 similar genes in different tumors. ACE2, angiotensin-converting enzyme 2; AMPK, AMP-activated protein kinase; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; PPAR, peroxisome proliferator-activated receptor; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma.
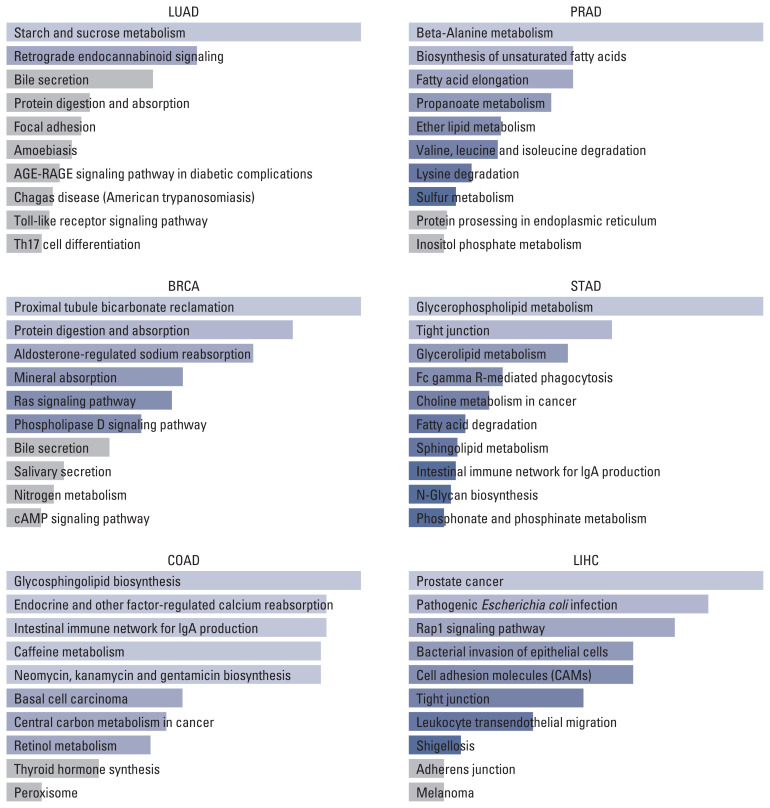
On the other hand, the result of the KEGG pathway enrichment analysis for TMPRSS2 and their top 100 co-expressed genes is presented in Fig. 3 and S10 Table. The most significant pathways in LUAD, BRCA, COAD, PRAD, STAD, and LIHC were starch and sucrose metabolism (p=0.014), proximal tubule bicarbonate reclamation (p=0.006), glycosphingolipid biosynthesis (p=0.022), beta-Alanine metabolism (p=0.001), glycerophospholipid metabolism (p=0.004), and prostate cancer (p=0.001).
Fig. 3.
Top 10 pathways of TMPRSS2 and top 100 similar genes in different tumors. BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; TMPRSS2, transmembrane serine protease 2.
Discussion
COVID-19 has become a global health emergency with a large number of confirmed cases and deaths that are much greater than any viral infection in the past. Severity condition and mortality have been noted to be significantly higher in patients with comorbidities, including cancer [17–19]. As cancer is heterogenous and cancer care still needs to continue during the pandemic, it is essential to determine which type of cancer will be at a high risk of getting severity condition with COVID-19 infection. Since SAR-CoV-2 employs ACE2 and TMPRSS2 for entering host cells and priming [8], we evaluated the expression of these genes that might be responsible for the severity of COVID-19 condition in patients with different types of cancer.
Our data showed the high expression of ACE2 and TMPRSS2 in COAD and STAD. For COAD, the coincidental result was confirmed at all subtypes regarding microsatellite status including MSI-H, MSI-L, and MSS. A similar finding has been reported in a study from the New York hospital system, which indicated that gastrointestinal cancer appeared to have a higher fatality rate than any type of solid tumor except lung cancer [19]. These might confirm the susceptibility of colorectal cancer and stomach patients to COVID-19 infection. Besides, at the mRNA level, we found that TMPRSS2 expressions were significantly lower in BRCA, but higher in COAD and PRAD, compared with control groups. Notably, it has been proved that TMPRSS2 was upregulated in prostate cancer under androgen stimulation [20]. Thus, it might suggest the risk of PRAD patients and possibly explain the higher proportion of infection prevalence of COVID-19 in males. Interestingly, for lung cancer, we did not see any overexpression of ACE2 but confirmed a significantly lower expression of TMPRSS2 in proximal inflammatory and proximal proliferative subtypes of LUAD than the control group. While it might be beneficial to have low ACE2 receptors for the entry of SARS-CoV-2, ACE2 has been proved to be responsible for anti-inflammatory, thus it might be considered as a protective factor during the infection process. Consequently, the downregulation of ACE2 in lung cancer patients might cause critical conditions by unstoppable inflammation. Given the fact that lung cancer patients account for a major proportion among cancer patients with COVID-19, further investigations to clarify specific mechanisms in lung cancer should be performed.
We then evaluated the association between the expressions of ACE2 and TMPRSS2 with the prognosis of six types of cancer. We noticed the overexpression of ACE2 in LIHC were associated with higher OS (HR, 0.65; p=0.016) and DFS (HR, 0.66; p=0.007), and upregulation of TMPRSS2 was associated with 26% reduced risk of mortality in LUAD (p=0.047) but 50% increased risk of death in BRCA (p=0.015). These expression statuses, however, have no correlation with OS and DFS in COAD, STAD, and PRAD. In return, there will be a need to assess the role of these genes in prognosis for COVID-19 patients with cancer.
By tracking the biological function of ACE2, TMPRSS2, and 100 similar genes, we can determine the top 10 pathways in six types of cancer. As the immune response is mainly driven in COVID-19 progression [21], we aimed to find the overlapping pathway which might contribute to the unfavorable prognosis in cancer with COVID-19. Nevertheless, no pathway has been considered a potential key. Approaches to extra center genes and pathways might lead to better findings.
In conclusion, our findings suggest the need for priority precautions for colorectal cancer, stomach cancer, and lung cancer during the COVID-19 pandemic. However, the risk of delaying appropriate treatment for cancer might be much greater than the risk of being exposed to COVID-19. Therefore, tailored cancer care with effective health delivery systems is crucial to bring final benefit for oncology patients. Further research on various cancer types/subtypes and cancer stages should be analyzed.
Footnotes
Ethical Statement
All data were obtained through free online databases and were therefore not subject to ethics approval.
Author Contributions
Conceived and designed the analysis: Hoang T, Nguyen TQ, Tran TTA.
Collected the data: Hoang T.
Performed the analysis: Hoang T.
Wrote the paper: Hoang T, Nguyen TQ, Tran TTA.
Conflicts of Interest
Conflict of interest relevant to this article was not reported.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
References
- 1.Kim SB, Huh K, Heo JY, Joo EJ, Kim YJ, Choi WS, et al. Interim guidelines on antiviral therapy for COVID-19. Infect Chemother. 2020;52:281–304. doi: 10.3947/ic.2020.52.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Recovery Collaborative Group. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, et al. Dexamethasone in hospitalized patients with COVID-19: preliminary report. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoang T, Anh TTT. Treatment options for severe acute respiratory syndrome, Middle East respiratory syndrome, and coronavirus disease 2019: a review of clinical evidence. Infect Chemother. 2020;52:317–34. doi: 10.3947/ic.2020.52.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Lancet. COVID-19 vaccines: no time for complacency. Lancet. 2020;396:1607. doi: 10.1016/S0140-6736(20)32472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang T, Tran TA, Kim J. Comparison of comorbidities in relation to critical conditions among COVID-19 patients: a network meta-analysis. Infect Chemother. 2020;53:13–28. doi: 10.3947/ic.2020.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M, Wang T, Zhou Y, Zhao Y, Zhang Y, Li J. Potential role of ACE2 in coronavirus disease 2019 (COVID-19) prevention and management. J Transl Int Med. 2020;8:9–19. doi: 10.2478/jtim-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and individual genetic susceptibility/ receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation: might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int J Mol Sci. 2020;21:3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 10.Jung KW, Won YJ, Kong HJ, Lee ES. Prediction of cancer incidence and mortality in Korea, 2018. Cancer Res Treat. 2018;50:317–23. doi: 10.4143/crt.2018.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019;51:417–30. doi: 10.4143/crt.2019.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung KW, Won YJ, Kong HJ, Lee ES. Prediction of cancer incidence and mortality in Korea, 2019. Cancer Res Treat. 2019;51:431–7. doi: 10.4143/crt.2019.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong S, Won YJ, Park YR, Jung KW, Kong HJ, Lee ES, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2017. Cancer Res Treat. 2020;52:335–50. doi: 10.4143/crt.2020.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung KW, Won YJ, Hong S, Kong HJ, Lee ES. Prediction of cancer incidence and mortality in Korea, 2020. Cancer Res Treat. 2020;52:351–8. doi: 10.4143/crt.2020.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–7. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee LY, Cazier JB, Angelis V, Arnold R, Bisht V, Campton NA, et al. COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: a prospective cohort study. Lancet. 2020;395:1919–26. doi: 10.1016/S0140-6736(20)31173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta V, Goel S, Kabarriti R, Cole D, Goldfinger M, Acuna-Villaorduna A, et al. Case fatality rate of cancer patients with COVID-19 in a New York hospital system. Cancer Discov. 2020;10:935–41. doi: 10.1158/2159-8290.CD-20-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4. [PubMed] [Google Scholar]
- 21.Ong EZ, Chan YF, Leong WY, Lee NM, Kalimuddin S, Haja Mohideen SM, et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:879–82. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.