Abstract
Background:
Dengue is a viral disease transmitted by female Aedes mosquitoes which are commonly found in tropical and subtropical areas. There is a dramatic increase in annual incidence rate of dengue attributed to urbanisation, poor environmental management as well as increased people mobility. Outbreak of dengue have been reported in Tanzania in recent years with Dar es salaam being the most affected region. Dengue is associated with haematological derangements and itindicates the severity of the disease. These changes have not been well elucidated in Tanzanian patients. The aim of this study was to determine these derangements among dengue patients admitted at Kairuki hospital in Dar es salaam, and compare these changes with non-dengue febrile patients.
Methods:
A retrospective cross sectional study was conducted among patients who were suspected to have dengue; tested for dengue IgM and their Complete Blood Count were tested during the index illness. This information was obtained from Kairuki hospital laboratory database. Haematological parameters were compared between dengue and non-dengue patients using SPSS Version 20.0. Binary logistic regression analysis was used to determine haematological predictors of dengue positive results.
Results:
A total of 255 patients were enrolled, whereby 188(73.7%) were dengue positive and 67 (26.3%) were negative. Dengue patients had relatively low mean total white blood cell counts compared to non-dengue patients (Students test= −2.7; p value=.007). Furthermore, Mean lymphocyte count was significantly low in dengue patients compared to non-dengue patients (Student's (t) test=−5.1; p<.001). Other haematological parameters were not significantly different. Lymphopenia was a significant predictor for dengue positive results (Adjusted Odd Ration =5.26 (95% CI=2.28–12.2; P value <.001).
Conclusion:
Patients with dengue had significantly low total white blood cell and lymphocyte count compared to non-dengue febrile patients. Lymphopenia is a significant haematological predictor for dengue positive results. Case defining signs and symptoms combined with these haematological changes may be used by clinicians as a guide to order confirmatory test for suspected dengue cases.
BACKGROUND
Dengue is a viral disease caused by the dengue virus that belongs into the flaviviridae family. There are 4 distinct dengue serotypes (DEN-1, DEN-2, DEN-3, and DEN-4). Despite their serotypical differences, they lead to the same clinical presentation. The infection by one serotype confers lifelong immunity to that particular serotype, but subsequent infection by other serotypes is associated with a more severe disease.1 Dengueis transmitted mainly by female Aedes aegypti and Aedes albopictus mosquitoes, which are mainly distributed in tropical and subtropical regions.2 There is a prevailing spread and increased cases of dengue fever. In 2017, it was estimated that 105 million people got infected with dengue, with 41,000 deaths, and an estimated incidence rate of 1,371 per 100,000 population.3 Variations in temperature, travel, rainfall, and degree of urbanisation are major influencers of transmission.4 Severe dengue is among the leading infectious cause of morbidity in Latin America and some Asian countries.5
Clinical presentation of dengue is not specific, thus, cannot be differentiated from other febrile illnesses on clinical grounds. In 2009, World Health Organization (WHO) developed clinical classification of dengue according to clinical signs and symptoms: dengue without warning signs, dengue with warning signs (abdominal pain, persistent vomiting, body fluid accumulation, mucosal bleeding, lethargy, hepatomegaly, elevated haematocrit, and rapid decrease in platelets count), and severe dengue.6 Severe dengue comprises of severe plasma leakage, severe bleeding and severe organ involvement.6 Most dengue infections are asymptomatic or mild disease. Only 10% develop severe disease. Case fatality rate of untreated or poorly treated severe cases is as high as 20%. However, with appropriate management and treatment, the mortality can be reduced up to 2%-5%.8 Tanzania has been experiencing sporadic dengue outbreaks. The most recent outbreaks occurred in 2010 followed by other outbreaks in 2012, 2013, 2014, 2018, and 2019. These occurred during the heavy rainy seasons that start from March through June. In the 2019 outbreak, the regions that were most affected included; Dar es salaam, Tanga, Pwani, Morogoro, Singida, and Kilimanjaro. In this episode, more than 3,500 cases and 3 deaths were reported9. Seroprevalence is also remarkable in other regions within Tanzania which signifies the wider spread of dengue in the country.10,11,12,13
Dengue is associated with several haematological changes such as leucopoenia, lymphopenia or lymphocytosis, thrombocytopenia and elevated haematocrit.14,15,16,17,18,19 These changes are immune mediated and may vary from one region or ethnic group to another due to heterogeneity and previous disease exposure.20,21,22 There is paucity of data describing these derangements in Tanzanian patients.
The aim of this study was to determine the haematological changes among patients with confirmed dengue infection compared to non-dengue patients with similar signs and symptoms attending Kairuki hospital in Dar es Salaam-Tanzania. If detected early, these changes may guide clinicians to perform specific confirmatory tests and provide appropriate management and counselling.
MATERIALS AND METHODS
Study Design and Study Site
We conducted a retrospective case-control study at Kairuki hospital which is a National referral level facility located in Kinondoni Municipal in Dar es salaam. It is also a teaching hospital for Hubert Kairuki Memorial University, providing both inpatient and outpatient care with capacity of up to 200 beds and an average of 600 outpatients per day.
Study Population and Participants Selection
The study involved patients who visited Kairuki hospital in the period between April to June 2019, were suspected to be having dengue and subjected to dengue serology test. Criteria for inclusion in the study were; availability of basic demographic information (age, sex, and residence), availability of dengue test results and Complete Blood Count (CBC). Those with indeterminate dengue test results and missing or invisible values in CBC were excluded from the study. We used the hospital database and laboratory logbooks to get basic demographic information, dengue test results as well as haematological findings from the Complete Blood Count (Beckman-Coulter, Model Act 10, Brea, CA, USA,). Dengue test was regarded as positive if patients tested positive either for non-structural protein-1 antigen (NS1-antigen), or immunoglobin M (IgM) for dengue by SD Bioline Duo Rapid Test (Standard Diagnostic, Inc., Gyeonggi-do, Korea) or both.
Sample Size Estimation
The minimum sample size was calculated using the formula for comparing 2 means from independent samples.23 The power of the study was set at 80% with type I(α) and II(β) error of 0.05 and 0.2 respectively. We used the total white blood cells as a reference value and assumed the Normal Standard Deviation of total white blood cells to be the same in both groups (dengue and non-dengue participants) at 1.5 × 109/l. We calculated the sample size that would enable us to detect the difference (variance) in mean total white blood cell count between the groups (dengue and non-dengue participants) from 1 × 109/L and above. The minimum sample size estimated in each group was calculated using the following formula:
where:
n= minimum sample size in each group
Zα= Critical value at α= 0.05 which is 1.96
Zβ=Critical value at β=0.2 which is 0.84
σ= standard deviation of the mean in both groups
d is the variance of the mean between the groups
Therefore,
Thus, the minimum sample size in each group was estimated to be 35 participants.
Data Analysis
Data was analysed using Statistical Package for Social Sciences (SPSS) version 20.0 (IBM Corp., Armonk, NY, USA). Proportions and categorical variables were compared using Chi square, Spearman's or Kendall's Tau Test whenever appropriate with their respective odd ratios and 95% confidence intervals. Continuous variables (absolute counts) between groups (dengue and non-dengue patients) were compared using Student's (t) test. Binary logistic regression analysis was used to determine demographic and haematological predictors of dengue positive results. Probability (p-value) of less or equal to 0.05 was considered statistically significant.
Ethical Considerations
Ethical clearance was obtained from the Ethical Review Committee of the Hubert Kairuki Memorial University teaching hospital, with approval number KH/RE/HKM/10/19. Strict confidentiality was maintained throughout data collection and analysis process. No personal identifiers were included in data mining and data analysis was performed anonymously.
RESULTS
A total of 255 participants were enrolled out of 662 patients who were subjected to dengue test. Participants who did not meet the selection criteria were excluded. Those were patients who had inadequate information, or patients who did not have complete blood count results (figure 1). Among the enrolled, 188(73.7%) tested positive for dengue while 67 (26.3%) tested negative. 215(84.3%) were between 41 and 60 year of age, while 8(3.5%) were below 20 years of age (Table 1)
FIGURE 1.

Screening and Enrolment Algorithm of Study Participants
TABLE 1:
Baseline Characteristics of Study Participants (N=255)
| Variable | Dengue Test Results | Chi-Square (X2) | p-Value | ||
|---|---|---|---|---|---|
| Positive N(%) | Negative N(%) | Total N(%) | |||
| Age of participants | |||||
| Below 20 years | 8 (4.3) | 1 (1.5) | 9 (3.5) | 1.46 | .7 |
| Between 21–40 years | 11 (5.9) | 3 (4.5) | 14 (5.5) | ||
| Between 41–60 years | 156 (83.0) | 59 (88.1) | 215 (83.3) | ||
| Above 60 years | 13 (6.9) | 4 (6) | 17 (6.7) | ||
| Total | 118 (100) | 67 (100) | 255 | ||
| Sex | |||||
| Male | 100 (53.2) | 30 (44.8) | 130 (51) | 1.4 | .24 |
| Female | 88 (46.8) | 37 (55.2) | 125 (49) | ||
| Total | 188 (100) | 67 (100) | 255 (100) | ||
| Attendance (Month) | |||||
| April | 70 (37.2) | 38 (56.7) | 108 (42.4) | 7.7 | .006*** |
| May | 118 (62.8) | 29 (43.3) | 147 (57.6) | ||
| Total | 188 (100) | 67 (100) | 255 (100) | ||
| Residence (Municipal) | |||||
| Kinondoni | 84 (44.7) | 25 (37.3) | 109 (42.7) | 4.6 | .31 |
| Ubungo | 34 (18.1) | 12 (17.9) | 46 (18) | ||
| Ilala | 24 (12.8) | 12 (17.9) | 36 (14.1) | ||
| Temeke | 18 (9.6) | 3 (3) | 21 (8.2) | ||
| Kigamboni | 28 (14.9) | 15 (22.4) | 43 (16.9) | ||
| Total | 188 (100) | 67 (100) | 255 (100) | ||
Most of the participants (42.7%) were residing in Kinondoni Municipal. Sexes were equally distributed among dengue positive and negative patients. Those with positive dengue results had relatively low mean total white blood cell counts compared do dengue negative participants [Student's test= −2.7; p value=.007 (figure 2)].
FIGURE 2.

Difference in Mean Total White Blood Cell Count between Dengue and Non-Dengue Febrile Patients at Kairuki Hospital, 2019
Mean total white blood cell count for dengue positive=4.7×109/L. Mean Total white blood cell count for dengue negative=5.6×109/L (Students (t) test= −2.7; p=.007)
In addition, mean lymphocyte count was significantly low in dengue patients compared to non-dengue patients [Students (t) test=−5.1; p<.001 (figure 3)].
FIGURE 3.

Difference in Mean Lymphocyte Count between Dengue and Non-Dengue Febrile Patients at Kairuki Hospital, 2019
Mean lymphocyte for dengue positive=1.01×109/L. Mean lymphocyte count for dengue negative=1.61×109/L Students (t) test=−5.1; p>.001
Patients with dengue had low mean platelet count compared to non-dengue patients, but the difference was not statistically significant [Student's test (t)=−0.86; p=.39 (figure 4)].
FIGURE 4.
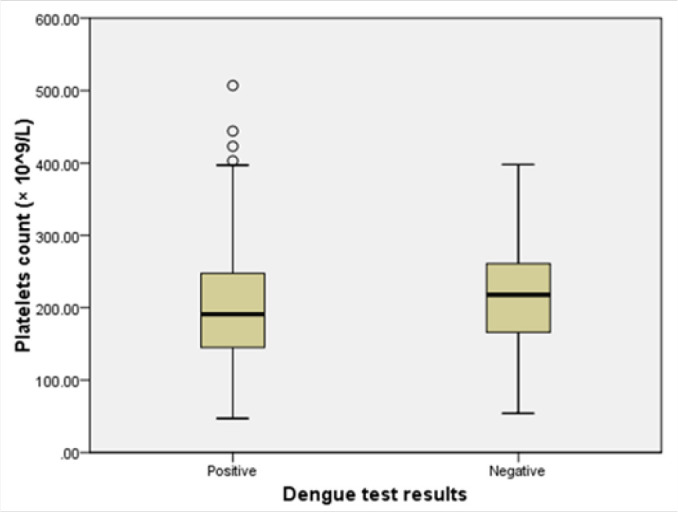
Difference in Mean Platelet Count between Dengue and Non-Dengue Febrile Patients At Kairuki Hospital, 2019
Mean platelets count for dengue positive patients=202.7×109/Mean platelets count for dengue negative patients=2 12.9×109/L (Student's test (t)=−0.86; p=0.39); p=.39)
After controlling the confounding variables by stratification of total white blood count, lymphocyte count, and platelets, lymphocyte count was found to be highly associated with dengue positive results (Adjusted Odd Ratio = 5.26 (95% CI=2.28–12.2; P value <.001) (table 2).
TABLE 2:
Social Demographic and Haematological Predictors of Dengue Positive Results among Febrile Patients At Kairuki Hospital, 2019
| Variables | Dengue serology test | COR | 95% Cl | P value | AOD | 95% Cl | P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive N (%) | Negative N (%) | TOTAL | Lower | Upper | Lower | Upper | |||||
| Residence(Municipal) | |||||||||||
| Kinondoni | 84(37.3) | 25(44.7) | 109(42.7) Ref | ||||||||
| Ubungo | 34(18.1) | 12(17.9) | 46(18.0) | 1.800 | 0.834 | 3.887 | 0.135 | ||||
| Ilala | 24(12.8) | 12(17.9) | 36(14.1) | 1.518 | 0.612 | 3.76 | 0.368 | ||||
| Temeke | 18(9.6) | 3(4.5) | 21(8.2) | 1.071 | 0.421 | 2.728 | 0.885 | ||||
| Others | 28(14.9) | 15(22.4) | 43(16.9) | 3.214 | 0.814 | 12.6 | 0.096 | 2.991 | 0.640 | 13.97 | 0.164 |
| TOTAL | 188(100) | 67(100) | 225(100) | ||||||||
| Age of participants | |||||||||||
| Below 20 years | 8(4.3) | 1(1.5) | 9(3.5) Ref | ||||||||
| Between 21–40 years | 11(4.9) | 3(4.5) | 14(5.5) | 2.462 | 0.232 | 26.1 | 0.455 | 3.727 | 0.267 | 51.93 | 0.328 |
| Between 41–60 years | 59(88.1) | 156(83) | 215(84.5) | 1.128 | 0.206 | 6.16 | 0.889 | ||||
| Above 60 years | 13(6.9) | 4(6.0) | 17(6.7) | 0.814 | 0.255 | 2.59 | 0.727 | ||||
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Sex | |||||||||||
| Male | 100(53.2) | 30(44.8) | 130(52) | 1.402 | 0.800 | 2.45 | 0.238 | 1.334 | .665 | 2.675 | 0.418 |
| Female | 88(46.8) | 37(55.2) | 125(49) Ref | ||||||||
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Platelet count(×l09/L) | |||||||||||
| 0.Normal (100–300) | 144(76.6) | 52(77.6) | 196(76.9) Ref | ||||||||
| Low (<100) | 19(10.1) | 7(10.4) | 26(10.2) | 0.886 | 0.376 | 2. OS | 0.782 | ||||
| Elevated>300 | 25(13.3) | 8(11.9) | 33(12.9) | 0.869 | 0.268 | 2.SI | 0.814 | ||||
| TOTAL | 188(100) | 67(100) | 225(100) | ||||||||
| Hematocrit (%) | |||||||||||
| Low (<35) | 52(27.7) | 17(25.4) | 69(27.1) Ref | ||||||||
| Normal (35–55) | 134(71.3) | 48(71.6) | 182(71.4) | 3.059 | 0.400 | 23.4 | 0.282 | 7.256 | .287 | 183.7 | 0.229 |
| Elevated (>55) | 2(1.1) | 2(3) | 4(1.6) | 2.792 | 0.383 | 20.3 | 0.311 | 1.785 | .099 | 32.26 | 0.695 |
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Haemoglobin(g/dl) | |||||||||||
| Low (<12) | 45(23.9)2 | 0(29.9) | 65(25.50 Ref | ||||||||
| Normal (12–16) | 140(74.5) | 45(67.2) | 185(72.5) | 1.500 | 0.232 | 9.6 | 0.670 | ||||
| Elevated (>16) | 3(1.6) | 2(3) | 5(2) | 2.074 | 0.336 | 12.8 | 0.432 | ||||
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Red blood cell count (×l012/L) | |||||||||||
| Low (<3.5) | 6(3.2) | 2(3) | 8(3.1) Ref | ||||||||
| Normal (3.5–5.5) | 170(90.4) | 58(86.6) | 228(89.4) | 1.750 | .275 | 11.1 | 0.554 | ||||
| Elevated (>5.5) | 12(6.4) | 7(10.4) | 19(7.5) | 1.710 | .643 | 4.5 | 0.283 | ||||
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Oesonophil absolute count (× 109/L) | |||||||||||
| Low (<0.02) | 5(2.7) | 1(1.5) | 6(2.4) Ref | ||||||||
| Normal (0.02–0.5) | 182(96.8) | 63(94) | 245(96.1) | 15. | 0.663 | 339.5 | 0.089 | 9.508 | .233 | 388.4 | 0.234 |
| Elevated (>0.5) | 1(0.5) | 3(4.5) | 4(1.6) | 8.667 | 0.885 | 84.8 | 0.064 | 6.659 | .510 | 86.8 | 0.148 |
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Total white Blood cells count (× 109/L) | |||||||||||
| Normal (4-10) | 102(54.3) | 52(77.6) | 154(60.4) Ref | ||||||||
| Low (<4) | 81(43.1) | 13(19.4) | 94(36.9) | 0.785 | 0.147 | 4.1 | .776 | 0.450 | 0.035 | 5.842 | .541 |
| Elevated (>10) | 5(2.7) | 2(3) | 7(2.7) | 2.492 | 0.437 | 14.2 | .304 | 1.862 | 0.115 | 30.22 | .662 |
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Neutrophil absolute count (× 109/L) | |||||||||||
| Low (<2) | 66(35.1) | 15(22.4) | 81(31.8) Ref | ||||||||
| Normal (2–7) | 111(59) | 48(71.6) | 159(62.4) | 1.6 | 0.447 | 5.7 | .470 | ||||
| Elevated (>7) | 11(5.9) | 4(6) | 15(5.9) | .841 | 0.255 | 2.7 | .776 | ||||
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Lymphocyte absolute count (×l09/L) | |||||||||||
| Low (<20) | 87(45.3) | 11(16.4) | 98(38.4) | ||||||||
| Normal (20–40) | 101(53.7) | 56(83.6) | 157(61.6) Ref | ||||||||
| TOTAL | 188(100) | 67(100) | 225(100) | ||||||||
| Basophil absolute count (× 109/L) | |||||||||||
| Low (<0.01) | 15(8) | 8(11.9) | 23(9) Ref | ||||||||
| Normal(0.01–0.1) | 173(92) | 59(88.1) | 232(91) | 0.639 | 0.258 | 1.5 | .334 | 0.617 | .208 | 1.825 | .383 |
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
| Monocyte Absolute count (× 109/L) | |||||||||||
| Low (<0.12) | 21(11.2) | 4(6) | 25(9.8) | 1.981 | .654 | 5.9 | 0.227 | 1.129 | .283 | 4.511 | .864 |
| Normal (0.12–1.2) | 167(88.8) | 63(94) | 230(90.2) Ref | ||||||||
| TOTAL | 188(100) | 67(100) | 255(100) | ||||||||
COR=Crude Odd Ratio, OR=Adjusted Odd Ratio, CI=Confidence Interval
DISCUSSION
The findings from this study suggest that patients with dengue have leucopoenia compared to non-dengue febrile patients. We have also demonstrated that lymphopenia is highly predictive of dengue positive results. This is similar to other studies conducted in other parts of the world.15,24,25,26 Elevated lymphocytes with atypical morphological features have been observed in some studies.27,14 However, we could not demonstrate the same in our study as we did not perform peripheral smears.
Elevated lymphocytes with atypical features have been associated with secondary dengue infection’ which indicates the role of immunological response’ and differences in cytokines produced during the first and subsequent disease exposure.28,29 This could be the result of augmented immune response due to secondary viral antigen exposure leading to a more severe form of the disease’a phenomenon also known as “the original antigenic sin”.30
Previous researches have demonstrated that cross reactive non neutralising antibodies from a previous dengue serotype bind to the new serotype and facilitate entry into the cells through Fc-receptors leading to activation of T-lymphocytes with subsequent lymphocytosis.31,32
Contrary to other studies, thrombocytopenia and elevated haematocrit were not significant in dengue positive patients compared to dengue negative patients. This could be due to the fact that these features are more common in patients with severe symptoms such and vascular leakage, haemorrhage and shock.18,33 This indicates that most of the patients in our study had a mild disease (classical dengue) with low likelihood of mortality.33,28
These findings (normal platelets, haematocrit and leucopoenia) with lymphopenia being predominant suggests that the current dengue infection could be the first exposure in most of the patients. Effective preventive measures are very important as further outbreaks with repeated infections will result into a more severe form of infection and possible increase in morbidity and case fatality.
CONCLUSION
Leukopenia and lymphopenia are the most common findings in dengue patients in non-endemic areas like Dar es salaam. Lymphopenia is the most reliable predictor for dengue positive results among febrile patients with similar signs and symptoms. Case defining signs and symptoms combined with these haematological changes may be used by clinicians as a guide to order confirmatory test for patients who are suspected to have dengue infection.
Acknowledgement:
We acknowledge the support from Kairuki Hospital administration for their permissionto use the patients’ data and hospital laboratory staff for their support in dataretrieval. We also acknowledge the support from Irene Maokola (HKMU MalariaResearch Office) for data entry.
Funding Statement
Funding: This study received did not receive any funding
Footnotes
Peer Reviewed
Competing Interests: None declared.
REFERENCES
- 1.Tuiskunen Bäck A, Lundkvist Å. Dengue viruses – an overview. InfectEcol Epidemiol. 2013;3(1):19839. 10.3402/iee.v3i0.19839. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gratz NG. Critical review of the vector status of Aedes albopictus. Med Vet Entomol. 2004;18(3):215–227. 10.1111/j.069-283X.2004.00513.x. Medline [DOI] [PubMed] [Google Scholar]
- 3.Zeng Z, Zhan J, Chen L, Chen H, Cheng S. Global, regional, and national dengue burden from 1990 to 2017: A systematic analysis based on the global burden of disease study 2017. E Clinical Medicine. 2021;32:100712. 10.1016/j.eclinm.2020.100712. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. 2013;496(7446):504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zambrano B, San Martin JL. Epidemiology of dengue in Latin America. Journal of the Pediatric Infectious Diseases Society. 2014. Sep 1;3(3):181–2. 10.1093/jpids/piu071. Medline [DOI] [PubMed] [Google Scholar]
- 6.WHO. New Edition 2009. Geneva; 2009. https://www.who.int/tdr/publications/documents/dengue-diagnosis.pdf. [Google Scholar]
- 7.Diagnosis G FOR. New edition 2009. 2009.
- 8.Kyle JL, Harris E. Global Spread and Persistence of Dengue. 2008;(April):71–92. 10.1146/annurev.micro.62.081307.163005 [DOI] [PubMed]
- 9.The Ministry of Health. Community Development, Gender E and, Children. National Clinical Management Guidelines for Patients with Dengue. Dar es Salaam; 2019.
- 10.Hertz JT, Saganda W, Munishi OM, et al. Chikungunya and dengue fever among hospitalized febrile patients in northern Tanzania. Am J Trop Med Hyg. 2012;86(1):171–177. 10.4269/ajtmh.2012.11-0393. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chipwaza B, Mugasa JP, Selemani M, et al. Dengue and Chikungunya fever among viral diseases in outpatient febrile children in Kilosa district hospital, Tanzania. PLoSNegl Trop Dis. 2014;8(11):e3335. 10.1371/journal.pntd.0003335. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vairo F, Nicastri E, Meschi S, et al. Seroprevalence of dengue infection: a cross-sectional survey in mainland Tanzania and on Pemba Island, Zanzibar. Int J Infect Dis. 2012;16(1):2011–2013. 10.1016/j.ijid.2011.09.018. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vairo F, Nicastri E, Yussuf SM, et al. IgG against dengue virus in healthy blood donors, Zanzibar, Tanzania. Emerg Infect Dis. 2014;20(3):465–468. 10.3201/eid2003.130150. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi AA, Gayathri BR, Kulkarni S. Correlation of thrombocytopenia with degree of atypical lymphocytosis as a prognostic indicator in dengue. Int J Res Med Sci. 2017;5(9):4041–4043. 10.18203/2320-6012.ijrms20173979. [DOI] [Google Scholar]
- 15.Choudhary S, Shivkumar BR, Shankar A, Manjunatha YA, Priyadarshini MM. Haematological changes in dengue fever. National Journal of Basic Medical Sciences. 2011;3(4):289–93. [Google Scholar]
- 16.Tahlan A, Bhattacharya A. Haematological profile of dengue fever. International Journal of Research in Medical Sciences. 2017;5(12):5367–71. 10.18203/2320-6012.ijrms20175456. [DOI] [Google Scholar]
- 17.Chaloemwong J, Tantiworawit A, Rattanathammethee T, et al. Useful clinical features and hematological parameters for the diagnosis of dengue infection in patients with acute febrile illness: a retrospective study. BMCHematol. 2018;18(1):1–10. 10.1186/s12878-018-0116-1. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azin FR, Gonçalves RP, Pitombeira MH, Lima DM, Castelo Branco I. Dengue: profile of hematological and biochemical dynamics. Revistabrasileira de hematologia e hemoterapia. 2012;34(1):36–41. 10.5581/1516-8484.20120012. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meena KC, Jelia S, Meena S, Arif M, Ajmera D, Jatav VS. A study of hematological profile in dengue fever at tertiary care center, Kota Rajasthan, India. Int J Adv Med. 2016. Jul;3(3):621–4. 10.18203/2349-3933.ijam20162506. [DOI] [Google Scholar]
- 20.Restrepo BN, Ramirez RE, Arboleda M, Alvarez G, Ospina M, Diaz FJ. Serum levels of cytokines in two ethnic groups with dengue virus infection. Am J Trop Med Hyg. 2008;79(5):673–677. 10.4269/ajtmh.2008.79.673. Medline [DOI] [PubMed] [Google Scholar]
- 21.Leslie TE. Dengue Fever and the Quandary of Race. Latin American and Caribbean Ethnic Studies. 2011. Nov 1;6(3):283–309. 10.1080/17442222.2011.617590. [DOI] [Google Scholar]
- 22.Cahill ME, Conley S, DeWan AT, Montgomery RR. Identification of genetic variants associated with dengue or West Nile virus disease: a systematic review and meta-analysis. BMC Infect Dis. 2018;18(1):1–18. 10.1186/s12879-018-3186-6. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espa S, Rodríguez MM, González-ramírez AR. Sample size calculation. AllergolImmunopathol (Madr). 2016;42(5):485–492. 10.1016/j.aller.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Musso D, Nhan TX, de Pina JJ, Marchi J, Texier G. The Use of Simple Laboratory Parameters in the Differential Diagnosis of Acute-Phase Zika and Dengue Viruses. Intervirology. 2019;62(2):51–56. 10.1159/000499567. Medline [DOI] [PubMed] [Google Scholar]
- 25.Herbinger KH, Hanus I, Schunk M, et al. Lymphocytosis and lymphopenia induced by imported infectious diseases: A controlled cross-sectional study of 17,229 diseased German travelers returning from the tropics and subtropics. Am J Trop Med Hyg. 2016;94(6):1385–1391. 10.4269/ajtmh.15-0920. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binh PT, Matheus S, Huong VTQ, Deparis X, Marechal V. Early clinical and biological features of severe clinical manifestations of dengue in Vietnamese adults. J Clin Virol. 2009;45(4):276–280. 10.1016/j.jcv.2009.04.004. Medline [DOI] [PubMed] [Google Scholar]
- 27.Khatri K, Rajani A, Khalla AR. Plasmacytoid lymphocytes: A diagnostic clue to dengue infection. Int J Sci Res. 2016. Mar;5(3):1002–5. 10.21275/v5i3.nov162022. [DOI] [Google Scholar]
- 28.Carlos CC, Mapua CA, Inoue S, et al. Comparison of clinical features and hematologic abnormalities between dengue fever and dengue hemorrhagic fever among children in the Philippines. Am J Trop Med Hyg. 2005;73(2):435–440. 10.4269/ajtmh.2005.73.435. Medline [DOI] [PubMed] [Google Scholar]
- 29.Chuansumrit A, Chaiyaratana W. Hemostatic derangement in dengue hemorrhagicfever. Thromb Res. 2014;133(1):10–16. 10.1016/j.thromres.2013.09.028. Medline [DOI] [PubMed] [Google Scholar]
- 30.Vaughn DW, Green S, Kalayanarooj S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181(1):2–9. 10.1086/315215. Medline [DOI] [PubMed] [Google Scholar]
- 31.Lei HY, Yeh TM, Liu HS, Lin YS, Chen SH, Liu CC. Immunopathogenesis of dengue virus infection. J Biomed Sci. 2001;8(5):377–388. 10.1007/BF02255946. Medline [DOI] [PubMed] [Google Scholar]
- 32.Guzman MG, Vazquez S. The complexity of antibody-dependent enhancement of dengue virus infection. Viruses. 2010;2(12):2649–2662. 10.3390/v2122649. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huy NT, Van Giang T, Thuy DHD, et al. Factors associated with dengue shock syndrome: a systematic review and meta-analysis. PLoSNegl Trop Dis. 2013;7(9):e2412. 10.1371/journal.pntd.0002412. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]


