Abstract
The RAG1-RAG2 recombinase (RAG) cleaves DNA to initiate V(D)J recombination. But RAG also belongs to the RNH-type transposase family. To learn how RAG-catalyzed transposition is inhibited in developing lymphocytes, we determined the structure of a DNA strand-transfer complex of mouse RAG at 3.1 Å resolution. The target DNA is a T form (T for transpositional target), which contains two >80° kinks towards the minor groove, only 3 bp apart. RAG2, a late evolutionary addition in V(D)J recombination, appears to enforce the sharp kinks and additional inter-segment twisting in target DNA and thus attenuate unwanted transposition. In contrast to strand-transfer complexes of genuine transposases, where severe kinks occur at the integration sites of target DNA and thus prevent the reverse reaction, the sharp kink with RAG is 1 bp away from the integration site. As a result, RAG efficiently catalyzes the disintegration reaction that restores the RSS (donor) and target DNA.
Keywords: T-form DNA, target DNA, major groove expansion, RAG2, CRISPR
Introduction
The RAG1-RAG2 recombinase (or RAG hereafter) shares its RNase H-like (RNH) catalytic core with many bacterial and eukaryotic transposases 1-3. The biological role of RAG is to cleave DNA in the immunoglobulin and T-cell receptor loci and initiate the process of V(D)J recombination that generates immune-system diversification in jawed vertebrates. Like DNA transposases, RAG cleaves both DNA strands at the end of the recombination signal sequences (RSS, equivalent to terminal inverted repeats of transposable elements, TIR) 1,4. After double-strand cleavage by RAG, the V, D and J coding segments with hairpin ends (Fig. 1) are processed and joined by the non-homologous end-joining pathway 5,6. In resemblance to the way DNA transposases excise mobile elements and insert them into new targets, RAG can integrate DNA with RSS ends into new loci, with duplication of target sequence, in vitro or ex vivo 7,8. But bona fide transposition by RAG, which would disrupt genome integrity, is very rare in cells and estimated to be 1 in 50,000 V(D)J recombination events in pre-B cell lines 9,10. Instead the RSS ends of DNA are joined and rendered harmless under normal circumstances 1,11. Given the structural and functional similarities between RAG and genuine transposases, it is unclear what prevents RAG from transposing cleaved RSS DNA to new targets in the genome. Specific regions in mouse RAG1 and RAG2 have recently been identified to cooperatively inhibit transposition by 100 fold 12. Interestingly, these regions are over 50 Å away from one another, and how they work together to inhibit transposition is unknown. Possible mechanisms of inhibition at the target capture or integration step 13, or by activating disintegration (reverse of integration) (Fig. 1) are to be resolved.
Fig. 1: Similarity of hairpin formation and disintegration catalyzed by RAG.
The 12/23RSS DNAs are shown in yellow and orange, and target DNA is drawn in purple. After hairpin formation, the coding ends are released from RAG. To be captured by RAG for transposition, target DNA has to undergo kinking and twisting (becoming T-form DNA). The strand-transfer complex (STC) can be reversed to target DNA and RSS (donor) DNAs by disintegration, the reverse of strand-transfer (or integration) reaction. Each RAG active site is marked by two divalent cations (green sphere). Open lilac circles indicate the scissile phosphates in target DNA.
RAG specifically binds two different recombination signal sequences, each composed of a conserved heptamer and nonamer but separated by a 12 or 23 bp non-conserved spacer and thus known as 12RSS and 23RSS 14-16, and cleaves at the borders of the 12/23RSS DNAs (Extended Data Fig. 1) 1. After resisting structural study for two decades, RAG proteins from mouse (mRAG) and zebrafish (zRAG) finally have yielded crystal and cryoEM structures of the entire DNA cleavage process, including apo RAG, pre-reaction RAG-DNA complex, and two DNA cleavage (nicking and hairpin formation) complexes 3,17,18 (Extended Data Fig. 1a). These structures reveal how a Y-shaped dimer of RAG1-RAG2 heterodimers pairs 12 and 23RSS DNA asymmetrically and undergoes large rearrangements of protein and DNA during reaction. In addition, our recent analysis of DNA nicking by mRAG has revealed that the active site undergoes re-configuration for sequential cleavage of two antiparallel DNA strands (manuscript under revision).
RNH-type transposases, including bacterial Tn5 and MuA and eukaryotic Mos1, retroviral integrase, and Hermes of the hAT family, have been extensively characterized 19,20. Transposition occurs when the 3′-OHs at transposon ends are inserted into a target site of 2-10 bp, forming the transposition intermediate called strand-transfer complex (STC) (Fig. 1). STCs are often more stable than transposase-DNA substrate complexes, as disintegration is disfavored by all known transposases 21,22. Structural analyses of STCs of the transposases and integrases 23-27 reveal similar binding of the RNase H-like catalytic core to the transposable DNA ends, with the target DNA usually kinked by 20° to 70° at the integration sites.
To find out what may prevent DNA transposition by RAG, we have determined a 3.1 Å-resolution cryoEM structure of mRAG complexed with RSS DNAs inserted into a new DNA target (STC). The STC can be further processed to full transposition with target duplication 28, or reversed to separate DNAs by disintegration (Fig. 1). In the mouse STC structure, the 5 bp target of DNA integration is forced by mRAG protein to make two sharp >80° kinks 3 bp apart towards the minor groove. The requirement of severe deformation of target DNA with stretched, flattened and inside-out major groove may present barriers to both target capture and strand-transfer reaction. Moreover, the product of strand transfer is prone to disintegration catalyzed by RAG, leading to a low probability of complete transposition.
Results
Structure of RAG strand-transfer complex (STC)
A preferred target site for transposition by mouse RAG is 5 bp of GC-rich sequence 7,8,29. We designed a 35 bp target DNA with CGGCG sequence in the center (5′-cgccg-3′ in the complementary strand) and synthetically linked it with the 12/23RSS DNA to mimic a DNA transposition intermediate (Fig. 1, Extended Data Table 1). An extended version of mouse RAG1 (aa 265-1040) and near full-length RAG2 (aa 1-520) were used in this study. To stop the disintegration reaction, we used an inactive mutant E962Q (in the DDE motif) for structural characterization. Using cryoEM single particle analysis, we initially determined the complete STC structure at 3.4 Å resolution. After excluding the asymmetric Y-stem portion, which contained the dissimilar RSS spacers, nonamer DNA and nonamer-binding domain of RAG1 (NBD), refinement without applying symmetry to the two Y-arms, so as to preserve the unique target (CGGCG), led to a 3.1 Å core STC structure of mRAG (Table 1, Extended Data Fig. 2, see Methods).
Table 1 ∣.
Cryo-EM data collection, refinement and validation statistics
| STC (EMD-20037, PDB 6OET) |
STCΔNBD (EMD-20036, PDB 6OES) |
|
|---|---|---|
| Data collection and processing | ||
| Magnification | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−/Å2) | 45 | 45 |
| Defocus range (μm) | −1.4 to −3.0 | −1.4 to −3.0 |
| Pixel size (Å) | 1.07 | 1.07 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 1,148,863 | 1,148,863 |
| Final particle images (no.) | 68,085 | 283,634 |
| Map resolution (Å) | 3.4 | 3.1 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 2.5-10 | 2.5-4.5 |
| Refinement | ||
| Initial model used (PDB code) | 5ZE0 | 5ZE0 |
| Model resolution (Å) | 3.5 | 3.1 |
| FSC threshold | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | −60 | −92 |
| Model composition | ||
| Nonhydrogen atoms | 19612 | 16832 |
| Protein residues | 1926 | 1784 |
| Ligands | 4 | 4 |
| B factors (Å2) | ||
| Protein | 77.35 | 44.98 |
| Ligand | 67.95 | 43.27 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.005 |
| Bond angles (°) | 0.917 | 0.751 |
| Validation | ||
| MolProbity score | 2.15 | 1.80 |
| Clashscore | 6.08 | 4.34 |
| Poor rotamers (%) | 3.44 | 2.87 |
| Ramachandran plot | ||
| Favored (%) | 93.82 | 96.39 |
| Allowed (%) | 6.18 | 3.33 |
| Disallowed (%) | 0 | 0.28 |
The RAG STC structure is superimposable with the hairpin-forming RAG-DNA complex (HFC) except for the integration sites, where the RSSs are covalently linked to target DNA (Fig. 2a). Between these two structures, the rmsd of RAG over 1437 pairs of Cα atoms is 0.6 Å. The similarity may appear surprising at first. In HFC, RSS DNAs are covalently linked to the coding flank DNA, which is replaced by flanks of the target DNA in STC (Extended Data Fig. 3). Transposition of the RSS DNAs would occur after the normal DNA cleavage and rapid release of the hairpin-end coding-flank DNAs 17,30,31, and requires the RAG-RSS complex to capture a target DNA (Fig. 1). However, the chemical nature of disintegration (reverse reaction of strand transfer) is the same type of transesterification as hairpin formation, both using a 3-OH′ on the flanking DNA to attack a scissile phosphate and replace one phosphodiester bond with another (Extended Data Fig. 3a,b). The main difference between the two is the linkage of the scissile phosphate, one belonging to the coding-flank and the other to the target DNA. In both STC and HFC, each RAG2 subunit interacts with 12 bp of flanking DNA, while RAG1 mainly interacts with the 12/23RSS DNAs (Fig. 2b,c). The target site CGGCG, which is unique in STC, interacts with an extended long loop of RAG2, LF2F3, that became traceable in HFC and STC.
Fig. 2: mRAG-DNA interactions in STC.
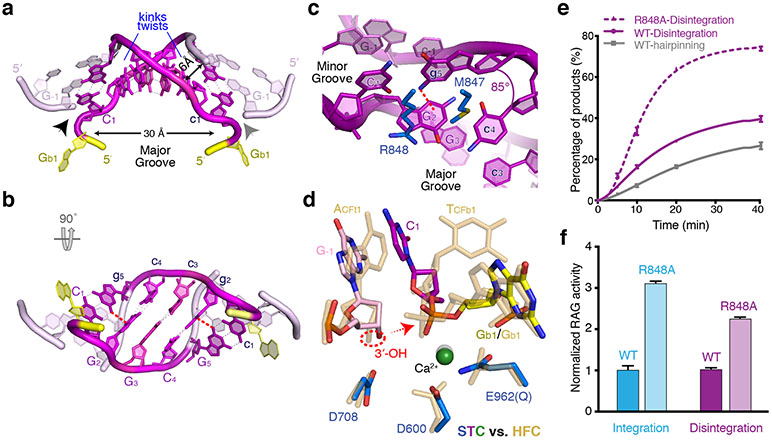
a, Superimposition of the HFC and STC structures. STC is shown in multiple colors, with 12- and 23- RSSs in yellow and target DNA in purple. The RSS DNAs in HFC (PDB: 6CG0) are shown in semi-transparent grey. The 85° bending angles are indicated. The region shown in panel d is boxed in dashed cyan. b, Protein-DNA interactions in STC. The T-form DNA undergoes segment-to-segment twist in addition to the sharp kink. The gray cylinder shows the target DNA without the twist, and the actual target DNA is shown in purple. Loop LF2F3 on RAG2 is marked and shown as a thick red tube. c, A rotated view of b. The target site is in a triangular shape when looking down the helical axis. R848 (RAG1) and C350 (RAG2) are 50 Å apart. d, LF2F3 of mAG2 interacts with the target-site DNA and also with αO (aa 823-841) of the RAG1 on the other Y-arm (trans). Molecular surfaces are shown for LF2F3 and the RAG1 in trans.
The T-form target DNA of mouse STC
The 5-bp target CGGCG retains Watson-Crick base pairing but undergoes dramatic conformational changes, departing drastically from B form. Two >80° kinks toward the minor groove between the first and second and between the fourth and fifth base pairs give the target DNA a U-shape appearance (Fig. 2a). Base-stacking at each kink site is completely lost, while surrounding each integration site (where the DNA strand is discontinuous) base-stacking is intact (Fig. 3a-c). The target DNA is segmented into three sections, two flanks and 3 bp between the two kink sites (C/GGC/G) (Fig. 2b). The central 3 bp are tilted ~45° relative to the helical axis and assume an inside-out structure with the major groove greatly expanded and exposed to solvent (Fig 3a-b). At each kink site, the flanking DNA helix is further twisted relative to the central 3 bp to open the major groove more widely (thus closing the minor groove) (Fig. 2b, Supplementary video 1). Accompanying the kinking and twisting, the Gs on opposite strands that frame each sharp kink form inter-strand cation-π (N2 of Gua to Gua base) interactions perpendicularly (Fig. 3b,c). With the two sharp kinks 3 bp apart, the DNA backbone of 2 nt before and 2 nt into the 5-bp target on the continuous strand (complementary to the DNA insertion site) forms two zig-zag turns reminiscent of a B- to Z-form DNA junction. The result is a triangular rather than circular appearance of the DNA when looking down the helical axis (Fig. 4a). The major groove of the 5-bp target site is expanded to 30 Å to receive the 12- and 23-RSS DNA ends for insertion (Fig. 2a and 3a,b). Meanwhile, the opposite minor groove is narrowed from 10 Å in B-DNA to 6 Å (ribose C4′ to C4′).
Fig. 3: The T-form DNA in STC.
a,b, Orthogonal views of the central 7 bps of target DNA and the very 3′ end of RSS (donor) DNA in RAG STC. The CGGCG target site is shown in magenta/purple, the flanking two basepairs on each side are in light pink, and the RSS DNA in yellow. c, The adjacent basepairs that frame the 85° kink form inter-strand cation-π interactions, as indicated by a red dashed line. d, Superimposition of the catalytic center in STC (multicolor) and HFC (semi-transparent sand color, PDB: 6CG0). The red dashed circle marks the 3′-OH, and the arrow shows the direction of nucleophilic attack. e, Activities of WT and R848A mutant mRAG in hairpin-formation and disintegration reactions. f, The R848A mutant RAG is more active than WT in both strand transfer (integration) and disintegration reactions. But the integration reaction is enhanced more than the disintegration, and the R848A mutant favors transposition more than WT. In e and f, mean and s.d. were calculated from three independent samples.
Fig. 4: Comparison of three different U-shaped DNAs.
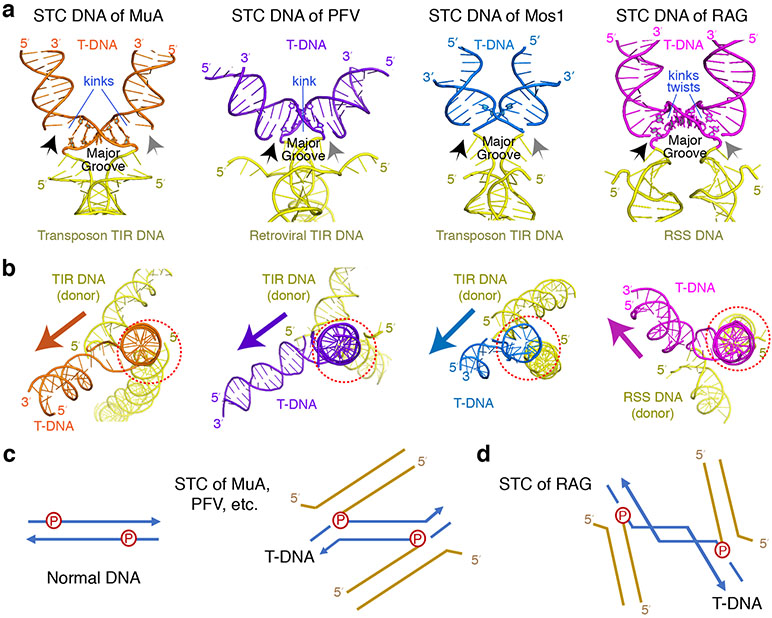
Two orthogonal views are shown with each protein-DNA complexes with protein structure omitted. The profile of DNA helix and the number of basepairs between two kink sites in each complex are marked.
An U-shaped DNA was first reported in an IHF-DNA complex 32 and later with topoisomerases II and IV 33,34, but in those cases the two severe bends are towards the major groove and more than 10 bp apart, and thus the circular helical nature remains unaltered (Fig. 4b,c). Two consecutive kinks towards the minor groove within 5 bp and additional inter-segment DNA twisting make the target DNA in this RAG complex the most severely distorted among transposase-DNA complexes. This sharply kinked DNA form with its greatly expanded major groove shares the general feature of bending towards the minor groove among target DNAs for transposition, but it differs from others in the severity and location of kinking, that is 1 bp inside of rather than at the integration site. Because it is a target in DNA transposition and bent toward the minor groove, we named it T-form DNA.
RAG performs efficient disintegration
The uniquely deformed T-form DNA is stabilized by RAG-DNA interactions, sparse at the DNA kinks per se and more abundant along the target flanks. At each sharp kink site, sidechains of R848 and M847 (belonging to the ZnH2 domain of RAG1) wedge between the two nearly perpendicular CG basepairs (Fig. 2c, 3c). Each target flank is surrounded by the ZnH2 domain of RAG1 adjacent to the integration site and by RAG2 over a stretch of 12 bp (Fig. 2b,c). In particular, the long loop LF2F3 of RAG2 (aa 333-342), contacts the zig-zag DNA backbone in the T-form target, where the minor groove is narrowest (Fig. 2). All other interactions between RAG and RSS and flanking DNA remain the same as in HFC.
LF2F3 of RAG2 is flexible in the apo, pre-reaction (PRC) and nick-forming complexes (NFC) , but it is involved in linking two Y-arms in HFC and STC by contacting RAG1 of the other RAG1-RAG2 heterodimer (Fig. 2d) 3,17,18,35. During hairpin formation, by linking the two coding flanks, LF2F3 complements NBD domains that bind the nonamer regions at the Y stem and probably secures the asymmetric pairing of 12 and 23RSS and their concerted cleavage. During transposition, by associating two Y arms and interacting with the narrowed minor groove of a target site, LF2F3 aids target DNA binding and contributes to the severe T-form DNA distortion. For a long time, RAG2 was thought to exist only in jawed vertebrates, where V(D)J recombination occurs. Very recently a RAG2-like protein (RAG2L) has been found in the invertebrate lancelet and forms a complex with RAG1 36, but the biological function of this RAG1-RAG2L, whether it performs DNA transposition or generates genome diversity as in V(D)J recombination, is unclear. Interestingly, LF2F3 is absent in the invertebrate RAG2L, which leads to diminished interactions with the flanking DNA and between two Y arms in lancelet HFC (Supplementary Fig. 1). These differences may underlie the non-concerted DNA cleavage and increased transposition by lancelet RAGL 12.
The catalytic RNH domain interacts with the scissile phosphate for hairpin formation in HFC and for disintegration in STC (reversal of strand transfer), and the two reactions are superimposable (Fig. 3d, Extended Data Fig. 3a, b). The freed 3′-OH nucleophile on the target flank and the scissile phosphate of the disintegration reaction are juxtaposed in STC, which suggests that disintegration (reversal of the strand transfer reaction) is imminent. Indeed, the disintegration reaction catalyzed by RAG is more efficient than hairpin formation under comparable reaction conditions (Fig. 3e). The highly efficient disintegration by RAG is in stark contrast to genuine transposases, in which strand transfer is overwhelmingly dominant 24,37.
Interestingly, R848A mutant RAG1 is twice as active in disintegration as the WT protein (Fig. 3e,f), indicating that the interaction of R848 with the T-form DNA kink site is not required for the reversal of transposition. Mutations of R848 to Met or Ala have been shown to increase transposition recently 12, which appears to be in discord with the observation that R848A mutant mRAG stimulates disintegration and thus reduces transposition. Because transposition is the sum of integration (strand transfer) and disintegration (reverse reaction) (Fig. 1), we checked the effect of R848A mutation on integration of RSS DNAs into a supercoiled target DNA (Methods) and found that it stimulated the strand-transfer reaction (3 fold) slightly more than disintegration (2 fold) (Fig. 3f). Therefore, our results support the finding that R848 inhibits transposition by mRAG.
Discussion
Comparison of target DNA distortion by transposases and by Cas1-Cas2
Prior to the STC of mouse RAG, structures of strand-transfer complexes have been reported for four retroviral integrases (Prototype Foamy Virus, Rous Sarcoma Virus, HIV, and Maedi-visna lentivirus, the eukaryotic mariner family member Mos1, and the bacteriophage transposase MuA 24-27,38-40 (Fig. 5a-b). The integration target varies from 2 bp (Mos1) to 4 (PFV), 5 (MuA, HIV and RAG) or 6 bp (RSV). With Mos1, the 2-bp target becomes completely unpaired, and one base is flipped out in the STC structure 25. For MuA transposition, the 5 bp target is severely kinked at each integration site (or transposon-target junction), resulting in a total bending of 150°, while for retroviral integration the overall DNA bending is milder, a single kink of ~40° in the center of the 4 to 6 bp target site and two gentle bends at the transposon-target junctions. In all cases, the target DNA is kinked toward the minor groove, and the transposon DNA ends always approach the expanded major groove of a target DNA (Fig. 5). Distinct from the STC of RAG, in which the severe DNA kinks occur within the 5 bp target and 1 bp inside of each integration site, kinks and distortions of T-form DNA with the genuine transposases occur at the integration sites and thus are likely to prevent the reverse reaction of transposition (Extended Data Fig. 3c-e). In accord with the biological role of these transposases, disintegration is rare 24,37.
Fig. 5: Distorted target DNA in transposition.
a, STC DNAs complexed with MuA, PFV, Mos1 and mRAG. The donor (TIR or RSS) and target DNA (T-DNA) joints are indicated by black (left) and grey (right) arrows. The major groove and kinks on target DNAs are marked. b, A view down the helical axis of one half of target DNA (right side in panel a) with one transposon end and integration site aligned (circled in dotted red), which reveals different orientations of the second transposon end (yellow) and target DNA (magenta) in RAG from other STC structures (marked by colored arrows). c,d, Diagrams of the DNA connection in the STCs of MuA, PFV and Mos1 (c) vs. RAG (d), which explain the different orientation in panel b and the additional inter-segment twist in target DNA with RAG.
The reason for target DNA bending towards the minor groove is that the two DNA ends (3′-OHs) for transposition or integration are no closer than 25 Å in all known STC structures, while target integration sites separated by 4-6 bp are only 16-20 Å apart in B-form DNA. Therefore, a target DNA has to be distorted into T-form with the major groove expanded to greater than 25 Å between two insertion sites. The different degree of kinking in target DNA reflects the nature of interaction of each transposase with flanking DNA. To compensate for the more severely kinked target DNA as observed in transposition by MuA and RAG, the protein-DNA interface is much more extensive than for the moderately kinked target found in retroviral integration. A pre-existing flexible and deformed site, such as a mismatched basepair or an insertion-deletion-loop, helps transposition by MuA as well as RAG 29,41.
One may wonder why targets of DNA transposition and integration are often 4-6 bp instead of 10 bp or longer, which would avoid the need for severe target DNA distortion. DNA targets longer than 20 bp do exist and are routinely found in DNA acquisition by CRISPR 42,43. Foreign DNAs of 21-72 bp in length (known as spacers in CRISPR and equivalent to transposon DNA) are acquired and inserted into a CRISPR locus in the host genome between “repeats” of 23-55 bps (equivalent to duplicated target sites). Indeed, the DNA spacer and repeat complexed with bacterial CRISPR Cas1-Cas2 are bent gently and smoothly in the equivalent STC 44 (Extended Data Fig. 4). Interestingly, the spacer DNA (transposon) ends still approach the major groove of the target site. With a long and smooth target in CRISPR, integration of two spacer ends, however, is uncoupled, and single-end integration occurs frequently 37. This sequential integration has been suggested to be necessary to correct mistakes by disintegration and thus enable the sequence- and location-specificity of DNA acquisition in CRISPR 37. In contrast, successful DNA transposition requires concerted two-end integration into a non-specific target site, during which each transposase subunit often forms cis and trans interactions with both DNA ends to keep them together. Distorted T-form target DNA may be a necessity born out of concerted integration.
RAG2 enforces T-form DNA distortion
The strand-transfer complex of RAG is unusual in the severity and location of the kinks of target DNA (Fig. 2, 3). Although a structure of target DNA captured by RAG before strand transfer is unavailable, the highly similar RAG STC and HFC structures, and nearly superimposable structures of target DNA before and after integration in a retroviral integration complex 38, lead us to expect that a target DNA has to adopt the sharply kinked T-form conformation to bind RAG for the strand-transfer reaction to occur (Fig. 1, 2b). Barriers to forming the kinked T-form conformation potentially make DNA transposition less likely, as an unwanted side reaction to V(D)J recombination. In addition, with the kinks in T-form DNA 1 bp away from the donor insertion sites in the STC of RAG, such distortion is no longer a barrier to the disintegration reaction.
The part of RAG that most extensively interacts with the T-form target DNA is RAG2 (Fig. 2). Although RAG2L has recently been found in the invertebrate lancelet and shares the six-bladed Kelch fold with mouse and zebrafish RAG2 12,36, four out of six blades including the loop LF2F3, which are involved in binding DNA flanks and linking two Y-arms, are dissimilar between RAG2 and RAG2L in sequence and structure (Supplementary Fig. 1). Interestingly, although essential for DNA cleavage in V(D)J recombination, RAG2 does not contribute to the active site formation nor sequence-specific binding of the RSS DNAs. The “acidic patch” of mRAG2 (aa 351-383), which is not essential for V(D)J recombination and disordered in all structures of mRAG determined to date, was found recently to inhibit transposition in the context of an R848M or R848A mutation in RAG1, but not with WT RAG112. Structurally, R848 of RAG1 and residue 350 of RAG2 are over 50Å apart (Fig. 2b,c) in all RAG structures. It must be the six-bladed Kelch structure of RAG2 that links the two together to inhibit transposition.
Comparing the known STC structures, the directions of transposon DNA ends approaching the integration target DNA in RAG STC are opposite to all others (Fig. 5b-d). This may be due to the fact that RAG cleaves the RSS DNA differently and makes a DNA hairpin on the coding flank DNA (Extended Data Fig. 1). Hermes transposase in the hAT family is closely related to RAG and cleaves DNA by forming hairpins on flanking DNAs rather than transposon ends. Although a Hermes STC structure is not yet available, comparison of Hermes HFC structures 45 with the HFC and STC of RAG by superimposition of the RNH catalytic domains shows that their active sites are well aligned (Fig. 6a,b), and the layout of the DNA substrates are similar. But the relative orientation of the coding or target flank DNAs differ between Hermes and RAG (Fig. 6a). The two DNA arms are much more “parallel” and thus the kinks in T-form DNA are more acute with RAG than with Hermes, which lacks a RAG2 equivalent, The relaxed coding flank DNA in Hermes would clash with RAG2, if present (Fig. 6c).
Fig. 6: RAG2 enforces the target DNA distortion.
a, Structure superposition of RAG STC and the hAT family transposase Hermes HFC (PDB: 6DWW) by one RNH domain (left) reveals that the two TIR and flank DNAs complexed with Hermes (colored green) are at a wider angle to each other than 12/23RSS DNAs (orange) and target DNA (purple) with RAG. The curved arrows indicate the narrowed crossing angle of DNA in the RAG STC. b, The RNH catalytic domain and the active site of RAG and Hermes are superimposable. c, With RNH domain superimposed, the flank DNA in Hermes would clash with RAG2 (hot pink).
Conclusions
Our analysis of DNA transposition by RAG has uncovered the special T-form DNA, which greatly deviates from the standard A or B forms and is more severely distorted than target DNA structures in active DNA transposases. The sharp kinks in the T-form DNA probably act as a barrier to strand transfer by RAG, and by being 1 bp away from the integration sites they also allow disintegration to occur readily (Fig. 3e). We suspect that the two consecutive sharp kinks and additional segmental twisting necessary for RAG transposition may be a result of evolutionary acquisition of the RAG2 subunits for V(D)J recombination 46. Our finding of the target DNA distortion imposed by the core of RAG2 (aa 1-350) provides a link between R848 of RAG1 at the kink site of target DNA and the acidic hinge of RAG2 50 Å away. We suggest that the acquisition of RAG2 may have been primarily to interfere with unwanted transposition. The roles of RAG2 in enhancement of V(D)J recombination and in DNA binding to coding flanks are means to that end.
Methods
Cell lines
HEK293T cells were originally obtained from Thermo Fisher Scientific and maintained as Yang laboratory stock. None of the cell lines used were authenticated.
Protein and DNA preparation
The WT and mutant mRAG proteins, which comprised WT, E962Q or R848A RAG1 (aa 265-1040) and T490A RAG2 (aa 1-520), were expressed as N-terminal His6-MBP fusions (on both RAG1 and RAG2) in HEK293T cells and purified as previously described 3,17. In addition to amylose affinity purification, a step of Mono Q anion exchange chromatography improved protein purity and eliminated a trace amount of DNA contamination. The buffer used in amylose affinity purification was 20 mM HEPES (pH 7.4), 500 mM KCl, 5% glycerol, 2 mM DTT, 0.5 mM EDTA. The salt concentration of protein samples coming off the amylose column was lowered to 100 mM before loading onto a Mono Q column (GE Healthcare) which was pre-equilibrated with 20 mM HEPES (pH 7.4), 100 mM KCl, 5% glycerol, 2 mM DTT, 0.5 mM EDTA. mRAG protein was eluted by a linear gradient of 100-500 mM KCl. The purified mRAG protein was buffer-exchanged into a storage buffer containing 20 mM HEPES (pH 7.4), 500 mM KCl, 20% glycerol, 0.1 mM EDTA, 2 mM DTT, concentrated to 6-8 mg/ml, and stored at −80°C. Human HMGB1 (amino acids 1–163) was prepared as reported previously 47.
Strand-transfer DNAs of 12- and 23-RSS used for structural analyses and biochemical assays (Table S1) were synthesized as ssDNA (Integrated DNA Technologies). Oligonucleotides longer than 20 nucleotides were purified by 8%–15% TBE-urea PAGE in a small gel cassette (Life Technologies). Gel purified oligonucleotides were then loaded onto a Glen Gel-Pak column (Glen Research) and eluted in deionized H2O. DNA was annealed in a Thermocycler in annealing buffer containing 20 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 50 mM NaCl.
DNA cleavage, disintegration and strand transfer assays
The hairpin formation and disintegration assays were performed in a reaction buffer containing 25 mM HEPES (pH 7.4), 100 mM KCl, 1 mM DTT, 0.1 mg/ml BSA, and 5 mM MgCl2. 50 nM each of Cy5- or FAM-labeled 12- and 23-RSS DNAs covalently linked to 20 bp coding flank (hairpin forming) or 35 bp target DNA (disintegration) (Table S1) were incubated with 50 nM of heterotetrameric WT or mutant (R848A) mRAG (tetramer), 100 nM HMGB1 and 200 nM H3K4Me3 peptide (Epicypher) at 37°C for 0 to 40 min. Reactions were stopped by adding an equal volume of formamide buffer (95% (v/v) formamide, 12 mM EDTA and 0.3% bromophenol blue) and heating at 95°C for 10 min. Cleavage products were separated by 15% TBE-urea PAGE, visualized and quantified using a Typhoon PhosphorImager (GE Healthcare). Plots of biochemical data show the mean ± SD from three independent experiments using Prism software (version 8.0).
The strand transfer (Integration) assay was carried out as previously reported7. Briefly, signal end complex (SEC) was first assembled by mixing WT or R848A mutant RAG, 12- and 23- RSS signal ends without coding flank and HMGB1 at 1:1:1:2 molar ration in a pre-reaction buffer (25 mM HEPES (pH 7.4), 100 mM KCl, 5 μM ZnCl2, 1 mM DTT, and 0.2 mM CaCl2) at 37 °C for 10 min. The strand transfer rection was carried out by mixing 300 ng supercoiled pUC19 plasmid, 100 nM SEC with 20 μM H3K4Me3 peptide in a reaction buffer (25 mM HEPES (pH 7.4), 100 mM KCl, 1 mM DTT, 0.1mg/ml BSA and 5 mM MgCl2) and incubating at 37 °C for 1 h. The reaction was stopped by adding 25 mM EDTA, and proteins were removed by treating with 0.4 mg/ml Proteinase K for 30 min at 37°C. DNA products were resuspended in 40 ul loading buffer after ethanol-precipitation and separated on a 1.5% agarose gel by electrophoresis. DNA bands were stained with ethidium bromide and quantified using a Typhoon PhosphorImager (GE Healthcare). Data from three independent experiments are averaged and shown with standard deviations using Prism software.
CryoEM sample preparation and data collection
To prevent reactions, we used the catalysis-deficient E962Q mutant mRAG. The purified E962Q mutant mRAG contained MBP-tags on both RAG1 and RAG2 subunits. MBP-mRAG protein and target DNA-linked 12- and 23-RSSs (Table S1), HMGB1 (aa 1-163) and H3K4Me3 peptide were mixed at 1:1.2:2.4:4 molar ratio in buffer containing 20 mM HEPES (pH 7.4), 100 mM KCl, 5 μM ZnCl2, 1 mM DTT, 5% glycerol and 5 mM CaCl2 and incubated at 37°C for 15 min. The mixture was further purified at 4°C by size exclusion chromatography on a Superdex 200 Increase 10/300 GL column (GE Healthcare) in buffer containing 20 mM HEPES (pH 7.3), 100 mM KCl, 1% glycerol, 1 mM DTT, 5 mM CaCl2. The elution peak fractions were pooled and used for cryoEM grid preparation. 3 μl of the purified STC (0.2 mg/ml) was spotted on freshly glow-discharged QUANTIFOIL R 1.2/1.3 (Cu, 300 mesh) grids at 22°C and blotted for 5 s. The frozen grids were stored in liquid nitrogen before use.
For structure determination, the frozen grids were loaded into a Titan Krios electron microscope operated at 300 kV for automated image acquisition with Leginon 3.1 48. Movies were recorded on a Gatan K2 Summit direct electron detector using the super-resolution mode at 130K nominal magnification (calibrated pixel size of 1.07 Å at the sample level, corresponding to 0.535 Å in super-resolution mode) and defocus values ranging from −1.4 to −3.0 μm. During data collection, the total dose was 45 e−/A2. The detailed collection statistics are shown in Table 1.
Structure analysis and model refinement
All frames in each collected movie were aligned and summed to generate both dose-weighted and dose-unweighted micrographs using Motioncorr2 49. The latter were only used for defocus determination. Particles on dose-weighted micrographs were picked using Gautomatch (developed by Dr. K. Zhang; https://www.mrc-lmb.cam.ac.uk/kzhang/ Gautomatch/) and extracted in RELION-2.1 using a box size of 280 * 280 pixels 50. Using the extracted particles, initial maps were obtained with cryoSPARC 51, and then served as the reference for template-based particle picking in Gautomatch and 3D classification in RELION 52. 2D classification and 3D classification were used to remove contamination and screen for the most homogeneous particles used for in-depth 3D structural analyses. The complete STC structure of mRAG was determined at 3.4 Å resolution and a 3.1 Å core STC structure was obtained by using a soft mask excluding the NBD-nanomer region (Extended Data Fig. 2). When calculating the STC∆NBD map, we used auto-sharpening in RELION_postprocess and obtained a B-factor of 92. When making the STC map, we manually lowered the B-factor generated by RELION to better show densities of the flexible NBD domain and the nonamer region. The anisotropy of the 3.1 Å STCΔNBD map was evaluated using 3D FSC 53 with a cutoff of 0.143.
All reported resolutions are based on the “gold standard” refinement procedure and the 0.143 Fourier Shell Correlation (FSC) criterion 54. Local resolution was estimated using Resmap 55. For model building, we used the 2.75-Å HFC crystal structure as an initial model to fit into the cryoEM STC map using Chimera 56, and then manually adjusted and rebuilt the model according to the cryoEM density in COOT 57. Phenix real-space refinement was used to refine the model. MolProbity and EMRinger 58 were used to validate the final model. The refinement statistics are shown in Table 1. The detailed classifications and map qualities of mRAG STC are shown in the Extended Data Fig. 2.
Reporting Summary
Further information on design of the research is available in the Nature Research Reporting Summary linked to this article.
Extended Data
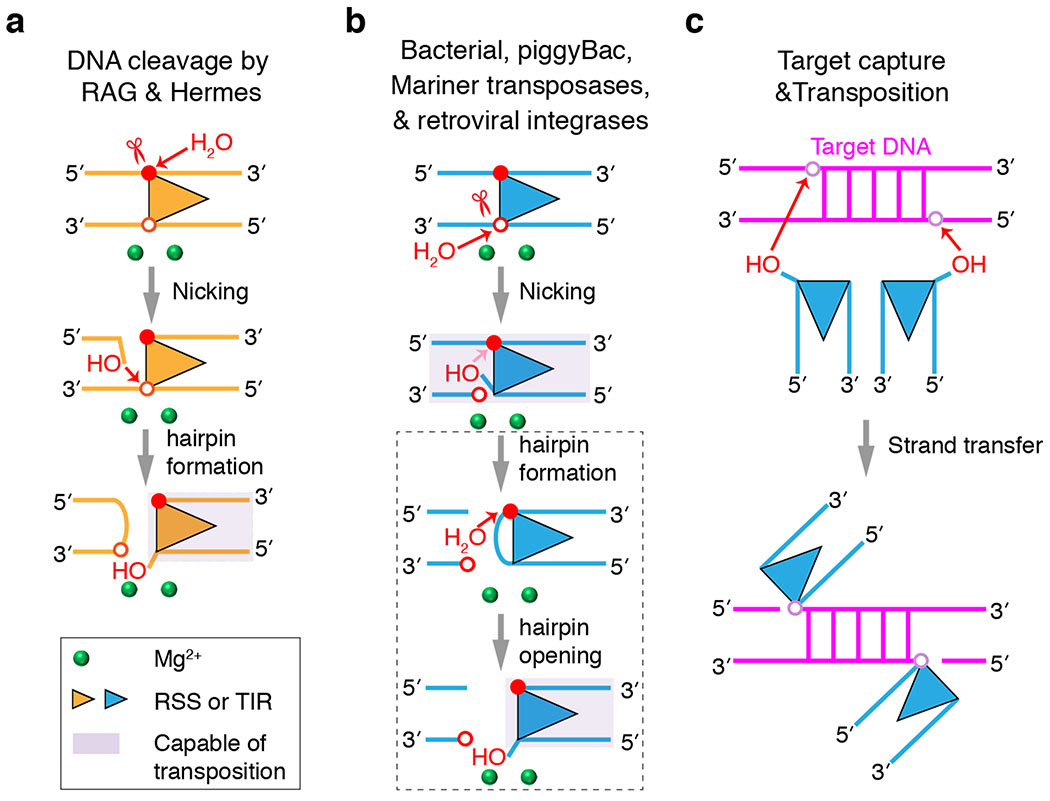
Extended Data Fig. 1. Two types of DNA cleavage mechanism used by RNase H-like transposases.
a, RAG and members of eukaryotic hAT transposase family, e.g. Hermes, cleave the top strand and generate a 5′ phosphate on the transposon end (terminal inverted repeat, TIR), or recombination signal sequence (RSS for RAG) first. Cleavage of the bottom strand occurs by hairpin formation on DNA flanking the TIR or RSS. The filled and open red circles indicate the scissile phosphates of the top and bottom strand, respectively. b, All bacterial and many eukaryotic transposases including retroviral integrases cleave the bottom strand first and generate a 3′-OH on the transposon end for transposition. The pink arrow before the hairpin formation step and the dashed grey box indicate that only a subset of transposases in this class undergo hairpin formation. The site of first nick is marked by a red scissor in a and b, and the transposition competent complexes are shaded. c, Target capture and strand transfer reaction. The target site in T-DNA, which is duplicated after transposition, is shown as a basepair ladder, and nucleophilic attack is indicated by red arrows.
Extended Data Fig. 2. Structure determination of RAG STC by cryoEM.
a, Flow chart for the cryoEM data processing. The maps with red bold letters are used for final model building of an intact STC and focused refinement without NBD and nonamer regions (STCΔNBD). b-c, A representative cryoEM micrograph (b) and 2D classes of different views (c). d, A surface presentation of the 3.06 Å STCΔNBD map (C1 symmetry). Colors are according to the local resolution estimated by ResMap, and the color scale bar is shown on its right. e, Angular distributions of all particles used for the final three-dimensional reconstruction shown in b. f, The FSC curves of STC map (C1). The "gold standard" FSC between two independent halves of the map (black line) indicates a resolution of 3.06 Å, and the blue line is the FSC between the final refined model and the final map. g, Directional FSC plots53 of the cryoEM reconstruction of STCΔNBD. h to k, Representative regions of the 3.06 Å STCΔNBD map (transparent grey surface). The maps of αX helix (h), heptamer plus one Ca2+ (i), L12 in RNH domain (j), and target DNA (k) are shown with the final structural models (cartoon or stick) superimposed.
Extended Data Fig. 3. Disintegration reaction is inhibited in RNH-type transposases.
a-b. Similarity between the hairpin formation in HFC (a) and disintegration in STC (b) catalyzed by RAG. The DNAs are colored in yellow (RSS), orange (the coding flank in HFC), and pink (the flank) and purple (the 5 bp target) of T-form DNA in STC. The RAG active site is marked by two divalent cations, shown as green spheres. The nucleophilic reaction is indicated by a red arrow. c-e. The reaction center for disintegration in RAG, PFV (PDB: 4BAC) and MuA (PDB: 4FCY). In the RAG STC (c), the 3′-OH nucleophile (in a dashed circle) is aligned for disintegration, but in the PFV STC (d), the entire nucleotide at the 3′-end is misaligned relative to the scissile phosphate. The direction of nucleophilic attack is marked by the dotted red arrow. In the MuA STC (e), the 75° kink at the integration site renders the 3′ end 15.1 Å away from the scissile phosphate.
Extended Data Fig. 4. Mild DNA distortion in complex with Cas1-Cas2.
The spacer is equivalent to the transposon DNA in transposition (TIR or RSS) and is colored in yellow. The repeat is equivalent to the target DNA in transposition and colored green. Because the target site is more than 20 bp, the repeat DNA is bent gently in the middle and far from the DNA integration sites.
Supplementary Material
Acknowledgements
W.Y. is grateful to Drs. W. Olson and S. Li for analyzing the T-form DNA structure. This research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases to M.G. (DK036167), W.Y. (DK036147 and DK036144), and ZHZ (GM071940). The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057, 1S10OD018111 and U24GM116792), NSF (DBI-1338135 and DMR-1548924) and CNSI at UCLA.
Footnotes
Competing interests
The authors declare no competing interest.
Data Availability
The accession codes for the cryoEM structures of STC are PDB 6OES and 6OET in the Protein Data Bank, and associated density maps are EMD-20036 and EMD-20037 in the Electron Microscopy Data Bank, as shown in Table 1.
References
- 1.Gellert M V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71, 101–32 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG & Swanson PC V(D)J recombination: mechanisms of initiation. Annu Rev Genet 45, 167–202 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Kim MS, Lapkouski M, Yang W & Gellert M Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature 518, 507–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuuchi K Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem 61, 1011–51 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Deriano L & Roth DB Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet 47, 433–55 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Boboila C, Alt FW & Schwer B Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol 116, 1–49 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Hiom K, Melek M & Gellert M DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell 94, 463–470 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Eastman QM & Schatz DG Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–51 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Chatterji M, Tsai CL & Schatz DG Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol 26, 1558–68 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy YV, Perkins EJ & Ramsden DA Genomic instability due to V(D)J recombination-associated transposition. Genes Dev 20, 1575–82 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alt FW & Baltimore D Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci U S A 79, 4118–22 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandt VL & Roth DB V(D)J recombination: how to tame a transposase. Immunol Rev 200, 249–60 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Sakano H, Huppi K, Heinrich G & Tonegawa S Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288–94 (1979). [DOI] [PubMed] [Google Scholar]
- 15.Lewis SM The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol 56, 27–150 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Lapkouski M, Chuenchor W, Kim MS, Gellert M & Yang W Assembly Pathway and Characterization of the RAG1/2-DNA Paired and Signal-end Complexes. J Biol Chem 290, 14618–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MS et al. Cracking the DNA Code for V(D)J Recombination. Mol Cell 70, 358–370 e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ru H et al. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell 163, 1138–1152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickman AB, Chandler M & Dyda F Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol 45, 50–69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkinson PW hAT Transposable Elements. Microbiol Spectr 3(2015). [DOI] [PubMed] [Google Scholar]
- 21.Steiniger-White M, Rayment I & Reznikoff WS Structure/function insights into Tn5 transposition. Curr Opin Struct Biol 14, 50–7 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Lesbats P, Engelman AN & Cherepanov P Retroviral DNA Integration. Chem Rev 116, 12730–12757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hare S, Gupta SS, Valkov E, Engelman A & Cherepanov P Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montano SP, Pigli YZ & Rice PA The mu transpososome structure sheds light on DDE recombinase evolution. Nature 491, 413–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris ER, Grey H, McKenzie G, Jones AC & Richardson JM A bend, flip and trap mechanism for transposon integration. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passos DO et al. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355, 89–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Z et al. Crystal structure of the Rous sarcoma virus intasome. Nature 530, 362–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahillon J & Chandler M Insertion sequences. Microbiol Mol Biol Rev 62, 725–74 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai CL, Chatterji M & Schatz DG DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res 31, 6180–90 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth DB, Nakajima PB, Menetski JP, Bosma MJ & Gellert M V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell 69, 41–53 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Ramsden DA & Gellert M Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev 9, 2409–20 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Rice PA, Yang S, Mizuuchi K & Nash HA Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87, 1295–306 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Dong KC & Berger JM Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450, 1201–5 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Laponogov I et al. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol 16, 667–9 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Ru H et al. DNA melting initiates the RAG catalytic pathway. Nat Struct Mol Biol 25, 732–742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang S et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell 166, 102–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright AV et al. Structures of the CRISPR genome integration complex. Science 357, 1113–1118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maertens GN, Hare S & Cherepanov P The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468, 326–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin Z, Lapkouski M, Yang W & Craigie R Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein Sci 21, 1849–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballandras-Colas A et al. A supramolecular assembly mediates lentiviral DNA integration. Science 355, 93–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanagihara K & Mizuuchi K Mismatch-targeted transposition of Mu: a new strategy to map genetic polymorphism. Proc Natl Acad Sci U S A 99, 11317–21 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN & Doudna JA Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527, 535–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nunez JK, Lee AS, Engelman A & Doudna JA Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Y, Ng S, Nam KH & Ke A How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550, 137–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hickman AB et al. Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase. Nucleic Acids Res 46, 10286–10301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carmona LM & Schatz DG New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J 284, 1590–1605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grundy GJ, Ramon-Maiques S, Dimitriadis EK, Kotova S, Biertumpfel C, Heymann JB, Steven AC, Gellert M, and Yang W . Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol Cell 35(2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Leiro R, and Scheres SHW A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct Biol 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14(2017). [DOI] [PubMed] [Google Scholar]
- 52.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai XC, Rajendra E, Yang G, Shi Y & Scheres SH Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swint-Kruse L, and Brown CS Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Kucukelbir A, Sigworth FJ, and Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, and Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, and Fraser JS EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession codes for the cryoEM structures of STC are PDB 6OES and 6OET in the Protein Data Bank, and associated density maps are EMD-20036 and EMD-20037 in the Electron Microscopy Data Bank, as shown in Table 1.