ABSTRACT
The permanent human settlement of the Tibetan Plateau (TP) has been suggested to have been facilitated by the introduction of barley agriculture ∼3.6 kilo-years ago (ka). However, how barley agriculture spread onto the TP remains unknown. Given that the lower altitudes in the northeastern TP were occupied by millet cultivators from 5.2 ka, who also adopted barley farming ∼4 ka, it is highly possible that it was millet farmers who brought barley agriculture onto the TP ∼3.6 ka. To test this hypothesis, we analyzed mitochondrial DNA (mtDNA) from 8277 Tibetans and 58 514 individuals from surrounding populations, including 682 newly sequenced whole mitogenomes. Multiple lines of evidence, together with radiocarbon dating of cereal remains at different elevations, supports the scenario that two haplogroups (M9a1a1c1b1a and A11a1a), which are common in contemporary Tibetans (20.9%) and were probably even more common (40–50%) in early Tibetans prior to historical immigrations to the TP, represent the genetic legacy of the Neolithic millet farmers. Both haplogroups originated in northern China between 10.0–6.0 ka and differentiated in the ancestors of modern Tibetans ∼5.2–4.0 ka, matching the dispersal history of millet farming. By showing that substantial genetic components in contemporary Tibetans can trace their ancestry back to the Neolithic millet farmers, our study reveals that millet farmers adopted and brought barley agriculture to the TP ∼3.6–3.3 ka, and made an important contribution to the Tibetan gene pool.
Keywords: Tibetans, millet farmers, barley agriculture, archaeology, mitochondrial genomes
INTRODUCTION
With an extreme and harsh environment that is generally considered inhospitable to humans, the Tibetan Plateau (TP) has been inhabited since the late upper Pleistocene [1–3]. In contrast to the earliest occupation by hunter gatherers during the Late Paleolithic era, whose contribution to the current Tibetan gene pool was rather limited [1], migration onto the plateau appears to have occurred during the Neolithic period [1,4–7]. Archaeological evidence further suggests that permanent human occupation began ∼3.6 kilo-years ago (ka), and was most likely facilitated by the introduction and utilization of cold-tolerant barley agriculture and sheep [8], which were first domesticated in west Asia around 10 ka [9,10]. However, it remains unknown who brought this exotic crop and livestock to the TP.
As revealed by archaeological evidence, modern humans settled extensively in the northeastern TP (below 2500 m above sea level (masl)) with millet cultivation around ∼5.2 ka, and further expanded to high-altitude plateau areas (above 2500 masl) ∼3.6 ka with barley and sheep from west Asia [8]. These observations have raised the question of whether barley agriculture was introduced to the TP by millet farmers from lower elevations or mediated by immigrations from the west. According to previous genetic studies, most Tibetan genetic components can be traced back to Neolithic immigrations from northern China [1,5,6], raising the possibility that it was millet farmers who brought barley agriculture to the TP around 3.6 ka. However, these studies, especially those on mitochondrial DNA (mtDNA), have also observed abundant lineages likely introduced to the Tibetans during the Holocene (which began ∼11.7 ka) [1,5,6,11]. Given the limited molecular resolution, and a lack of both genetic and archaeological evidence in previous studies, it remains unclear whether these lineages are associated with millet farmers or with the immigration of foragers during the early Holocene (∼11.7–8.3 ka) [12,13].
In this study, by exploring mitogenomes in Tibetans and surrounding populations (especially those with suggested Neolithic millet farmer ancestry) (Fig. 1 and Supplementary Tables S1-S3), as well as radiocarbon dating of cereal remains from archaeological sites on the TP and northern China (Supplementary Tables S4-S6), we aimed to address whether millet farmers contributed to the Tibetan gene pool at the time of barley dispersal to the TP (∼3.6 ka), thus providing deeper insight into the origin of Tibetans.
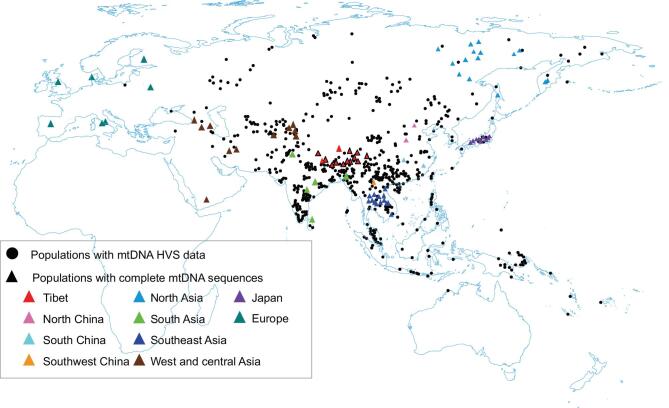
Figure 1.
Locations of samples in the present study. Black dots: populations with hypervariable segments (HVS) sequence data. Triangles: populations with complete mtDNA sequence data, including 671 newly sequenced samples from 11 Tibetan populations in this study (black outline) and 9789 from 109 populations from previous literature. Locations are indicated by different colors.
RESULTS
Haplogroups M9a1a1c1b1a and A11a1a are genetic legacies of Neolithic millet farmers
Comparing mtDNA variations between Tibetans and populations from surrounding regions (Supplementary Tables S1-S3), we found that 70.5% (473/671) of lineages—e.g. M9a1a1c1b1a, A11a1a, G3a1a, M13a1b, M13a2, M62b1 and M62b2—displayed de novo differentiation in contemporary Tibetans (Supplementary Fig. S1), with the remaining (29.5%) possibly introduced via recent gene flow (Supplementary Information (SI)-1; Supplementary Fig. S1, Supplementary Table S7). Among the de novo differentiation lineages, most (78.9%; 373/473) were <6 ka in age (Supplementary Table S8) when the Neolithic period on the TP began [13], consistent with previous suggestions of immigrations to the TP during the Neolithic period [1,5,6].
We analyzed the radiocarbon dates of cereal remains from different elevations along the TP and northern China, and found that millet was cultivated at Loess Plateau (<2500 masl and located to the northeast of TP) between 5.2 and 3.6 ka (Fig. 2a). Of note, barley and wheat remains began to appear in the Hexi Corridor, and the lower elevations of the northeastern TP, ∼4 ka [8,14], leading to a coexistence of indigenous millet and exotic barley–wheat cultivation in the area ∼4.0–3.6 ka (Fig. 2a). Thus, we shifted attention to haplogroups with ages within 5.2–4.0 ka, including A11a1a, A15a, A21, D4g2a1c1, D4j1a1f, D4s1, C4a1a1 and M9a1a1c1b1a (Fig. 2b, Supplementary Table S8).
Figure 2.
Paleoclimatological, archaeological and genetic chronologies on the Tibetan Plateau. (a) Radiocarbon dates of cereal remains (Supplementary Tables S4 and S5) and human bones with carbon isotope signals from different elevations in northern China (Supplementary Table S6). (b) Schematic tree of mtDNA lineages in Tibetans. Ages of independently differentiated nodes are shown. Coalescent ages estimated by coding regional variants, synonymous positions, and complete mitogenome variants are indicated by black, red and orange dots, respectively. Haplogroups within the 5.2–4.0 ka timeframe are in red. Paleolithic and other Neolithic haplogroups are in blue and green, respectively.
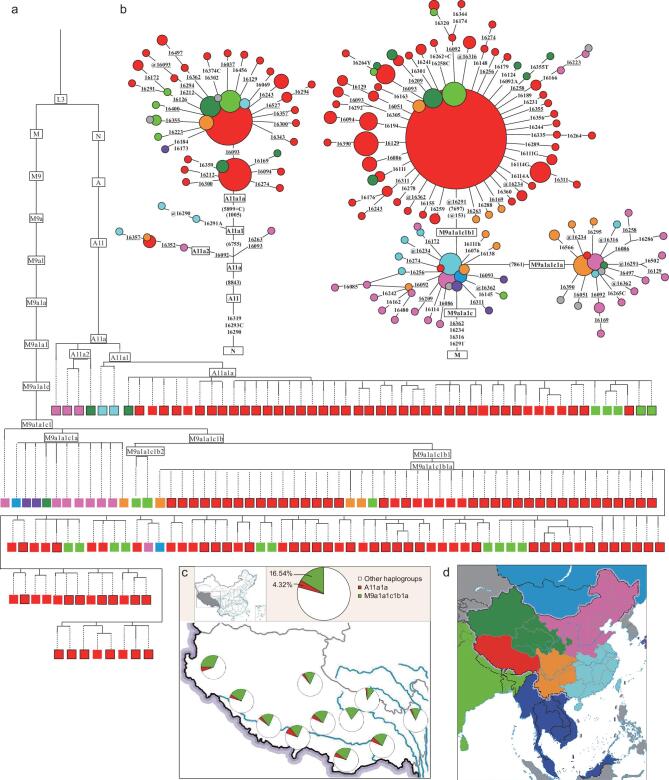
Phylogeographic analyses, very useful in identifying the origins and migrations of haplogroups [15], were performed to determine whether these haplogroups have roots in northern China (SI-2), where millet agriculture originated [16,17]. As a result, we identified two haplogroups (A11a1a and M9a1a1c1b1a), both with ancestor nodes (A11a and M9a1a1c1, respectively) distributed mainly in northern China (Fig. 3, Supplementary Fig. S2 and S3) and thus most likely originating from northern China. Coalescent ages of the two ancestor nodes were estimated to be 12.7–11.7 ka (A11a) and 10.1–6.4 ka (M9a1a1c1) (Table 1). These results matched the origin and development of millet agriculture, which originated about 11–9.5 ka [16] and subsequently became dominant in northern China from 10–6 ka [18,19], suggesting a close relationship between these two haplogroups and millet farming. Further support came from the observation that A11a2 (a subhaplogroup of A11a) and M9a1a1c2 (a sister haplogroup of M9a1a1c1) were also distributed in northern China (Fig. 3 and Supplementary Fig. S2) with ages estimated to be 7.0–5.0 ka (Table 1), thus showing a strong association with the intensive expansion of millet farming in the Yangshao period (7.0–5.0 ka) [18,19]. These findings strongly link A11a1a and M9a1a1c1b1a with the origin, development and westward spread of millet agriculture, and thus suggest that both haplogroups most likely represent the genetic legacy of the Neolithic millet farmers.
Figure 3.
Phylogeographic analysis of haplogroups A11a and M9a1a1c1. (a) Phylogenetic tree of haplogroups A11a and M9a1a1c1 based on whole mtDNA sequences from this study (black outline) and previous literature (no outline). (b) Median-joining networks of A11a and M9a1a1c1 based on HVS data set (Supplementary Table S3). Mutations outside of HVS-I and HVS-II are in parentheses. Recurrent mutations are underlined. `@' denotes a reverse mutation. (c) Frequencies of haplogroups M9a1a1c1b1a (green) and A11a1a (red) in Tibetan populations. Locations are indicated by different colors (d).
Table 1.
Age estimations of haplogroups M9a1a1c1b1a and A11a1a, as well as their ancestor and sister lineages.
| Coding region synonymous substitution† | Coding region (577–16 023)† | Complete mtDNA genome† | |||||
|---|---|---|---|---|---|---|---|
| Haplogroup | N | ρ ± σ | Age (ka) | ρ ± σ | Age (ka) | ρ ± σ | Age (ka) |
| A11a | 53 | 1.51 ± 0.93 | 11.88 ± 7.36 | 3.30 ± 1.30 | 11.67 ± 4.58 | 4.92 ± 1.49 | 12.73 ± 3.86 |
| A11a2 (Northern China) | 3 | 1.00 ± 1.00 | 7.87 ± 7.87 | 1.67 ± 1.00 | 5.89 ± 3.53 | 2.67 ± 1.25 | 6.89 ± 3.22 |
| A11a1a (Tibet) | 47 | 0.55 ± 0.13 | 4.36 ± 1.06 | 1.45 ± 0.21 | 5.11 ± 0.74 | 3.10 ± 0.85 | 8.03 ± 2.21 |
| M9a1a1c1 | 166 | 0.81 ± 0.13 | 6.40 ± 1.05 | 2.19 ± 0.95 | 7.73 ± 3.37 | 3.89 ± 1.34 | 10.06 ± 1.47 |
| M9a1a1c1a (Northern China) | 11 | 0.64 ± 0.24 | 5.01 ± 1.89 | 1.09 ± 0.30 | 3.85 ± 1.06 | 2.00 ± 0.43 | 5.17 ± 1.10 |
| M9a1a1c1b1a (Tibet) | 154 | 0.65 ± 0.09 | 5.11 ± 0.75 | 1.20 ± 0.15 | 4.24 ± 0.52 | 1.92 ± 0.19 | 4.97 ± 0.51 |
†Mutation rates from Soares et al. [36].
M9a1a1c1b1a and A11a1a were more frequent in Tibetans ∼3.6 ka
Of note, both haplogroups showed the highest frequencies (M9a1a1c1b1a: 16.5% and A11a1a: 4.3%) in Tibetans, accounting for 20.9% of the Tibetan gene pool (Supplementary Fig. S4). As this result does not necessarily represent their past distribution frequencies, we estimated their proportions through time and found that their percentages continued to increase since 5.14 ka, reaching 48.8% at 3.3 ka (Fig. 4 and Supplementary Table S9). After this, the proportion began to decrease and reached 19.1% by the present day (a result very close to the observed value of 20.9% in our Tibetan samples; Supplementary Table S9), which is probably due to immigrations to the TP during the historical period [20]. Thus, these results suggest a substantial genetic contribution of the Neolithic millet farmers from the Loess Plateau of northern China to the Tibetans ∼3.3 ka (Fig. 4; Supplementary Table S9), a time frame very close to that (3.6 ka) revealed by archaeological evidence when higher altitudes began to be permanently populated by groups who engaged in barley cultivation and sheep herding [8].
Figure 4.
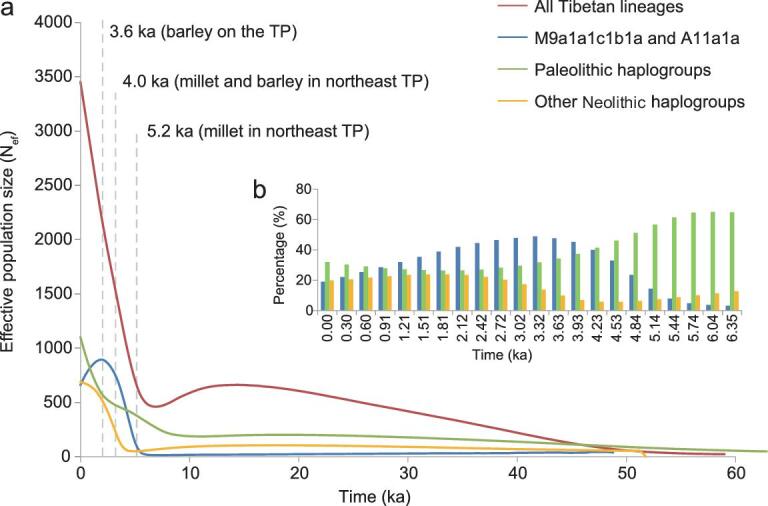
Proportional changes of two lineages of millet farmers (A11a1a and M9a1a1c1b1a) in Tibetans through time. (a) BSPs of all Tibetans (red), M9a1a1c1b1a and A11a1a (blue), Paleolithic lineages (green) and other Neolithic haplogroups (orange). (b) Proportions of M9a1a1c1b1a and A11a1a (blue), Paleolithic lineages (green) and other Neolithic haplogroups (orange) in Tibetans through time since the beginning of the Neolithic period.
M9a1a1c1b1a and A11a1a played important roles in shaping the genetic landscape of current Tibetans
Both M9a1a1c1b1a and A11a1a are distributed ubiquitously in contemporary Tibetans, with total frequencies ranging from 12.0% (Baqing County, Naqu) to 37.5% (Ngari) across all regional Tibetan populations studied (Fig. 3c). This suggests that the millet farmer lineages contributed to and existed in the proto-gene pool of Tibetans before their expansion to different regions on the TP. Indeed, much closer genetic affinity was observed among all regional Tibetan populations (Fig. 5a), with haplogroup M9a1a1c1b1a contributing most to this clustering pattern (Fig. 5b), implying that millet farmer components played an important role in shaping the genetic landscape of Tibetans.
Figure 5.
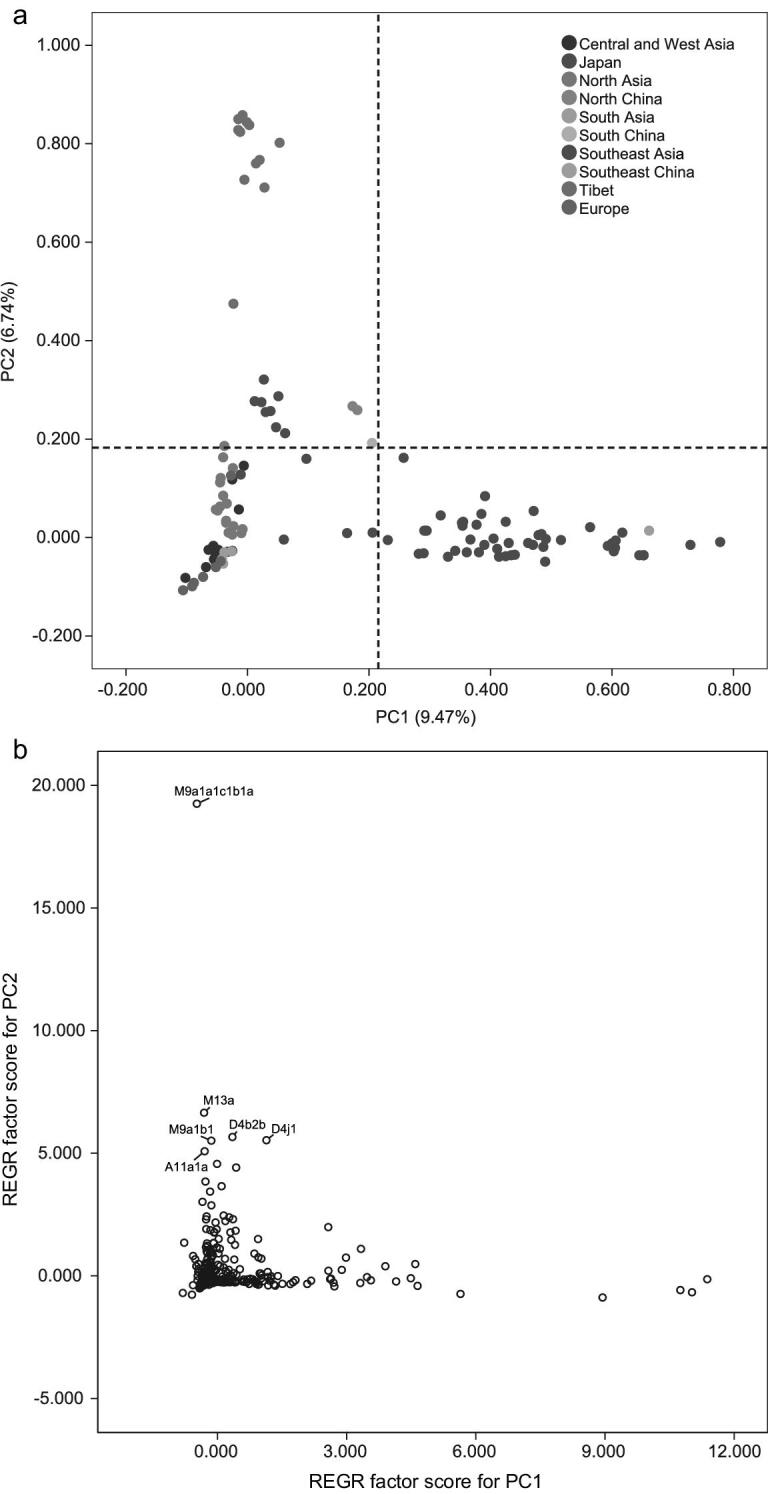
PCA of Tibetans and other Eurasian populations based on whole mitochondrial genome data. (a) PC map of 120 populations based on haplogroup frequencies. Populations from different regions are indicated by different colors. (b) Plot of haplogroup contribution of first and second PCs. Contribution of each haplogroup was calculated as the factor scores for PC1 and PC2 with REGR in SPSS.
DISCUSSION
In this study, we combined archaeological and genetic data to investigate the mode of spread of barley agriculture to the TP. Specifically, we identified that haplogroups A11a1a and M9a1a1c1b1a, which can trace their ancestry back to northern China ∼10 ka and underwent in situ differentiation in Tibetans 5.2–4.0 ka, likely represent the genetic legacy of Neolithic millet farmers in contemporary Tibetans. Interestingly, ancient samples excavated from different Neolithic sites in northern China, in which millet was the most important crop and thus most likely represent the remains of the Neolithic millet farmers, show close genetic affinity to our identified genetic legacy of millet farmers in the Tibetans. For example, haplogroup M9a1a1c (defined by HVS-I variants 16223–16291-16234–16316-16362), an ancestor type to M9a1a1c1b1a, has been observed at the Miaozigou (5.5–5.0 ka) [21,22] and Zhukaigou sites (4.2–3.5 ka) [23,24]. The above evidence argues for the existence of an ancestor lineage of haplogroup M9a1a1c1b1a in northern and northwestern China during the Neolithic period, thus strongly supporting their close relationship with Neolithic millet farmers.
The maternal genetic components of millet farmers, which accounted for 20.9% of current Tibetans and may have been more frequent prior to historical immigrations (40–50%), demonstrated substantial genetic contributions of Neolithic millet farmers from northern China into Tibetans. Moreover, this contribution, represented at least by the two haplogroups (A11a1a and M9a1a1c1b1a) identified in the present study, existed in Tibetans at relatively high frequencies for >3 ka, and even contributed to the genetic differentiation observed between the Tibetans and other ethnic groups. Therefore, by showing that the ancestry of the Tibetans on the TP can largely be traced back to millet farmers from northern China, our study indicates substantial migration of millet farmers onto the TP, which probably occurred during the Neolithic period. Although it was barley agriculture that finally promoted the permanent settlement of humans at high altitudes after 3.6 ka [8], the genetic origins of these settlers were primarily from Neolithic millet farmers in the Yellow River Basin, rather than immigrants from the west. Given that barley joined or displaced millet at some archaeological sites on the northeast TP during 4.0–3.6 ka [8], the most reasonable explanation for our observation is that millet farmers adopted barley agriculture after their arrival to the northeast TP, and further migrated to high altitudes after 3.6 ka with barley cultivation technology.
Of note, our observation based on mtDNA is somewhat similar to the Neolithic migration of males from northern China, in which a westward expansion of males initiated in the middle Yellow River Basin during 10.0–5.0 ka [4,25]. However, controversy still exists regarding whether these male lineages from northern China dispersed onto the TP [4], or just moved to the upper basin of the Yellow River and mixed with local people therein, who further occupied high altitudes after 3.6 ka [25]. To better understand the migration history of the Tibetans, investigations based on large-scale and high-resolution Y chromosome data are needed to explore whether the migration of millet farmers onto the TP also involved the expansion of males.
CONCLUSIONS
In summary, by combining archaeological and high-resolution genetic data, we have identified a substantial genetic contribution of Neolithic millet farmers, represented by haplogroups A11a1a and M9a1a1c1b1a, to contemporary Tibetans, which has even played an important role in shaping the Tibetan matrilineal landscape. Therefore, our study provides the first piece of evidence demonstrating that it was Neolithic millet farmers originating from northern China, rather than immigrants from the west, who brought barley agriculture to the TP ∼3.6 ka and contributed to the permanent human occupation of the TP.
METHODS
mtDNA sequencing
Blood samples from 671 Tibetans from 11 Tibetan populations were collected, covering all seven districts of Tibet (Fig. 1, Supplementary Table S1). Blood samples from an additional 11 Chinese individuals belonging to haplogroups of interest were also collected for mitogenome sequencing. The experimental protocol was approved by the Ethics Committee at the Kunming Institute of Zoology, Chinese Academy of Sciences. Informed consent was obtained from each individual before the study. Complete mtDNA genomes of the samples were enriched by capture-based strategies using a MyGenostics Human Mitochondria Capture Kit (MyGenostics Inc., Beijing, China). Sequencing was carried out using an Illumina HiSeq X Ten platform at MyGenostics. The average depth of sequencing was 4212×, ranging from 1003× to 40 278×.
mtDNA data collection
Additionally, 201 whole mitogenomes (Supplementary Table S2) and 7405 mtDNA hypervariable segments (HVS) sequences (Supplementary Table S3) from Tibetans were collected from previous literature and analyzed. For comparison, mtDNA data from surrounding areas, including 9588 mitogenomes (Supplementary Table S2) and 41 320 HVS data (Supplementary Table S3), were also included.
Archaeological data
Published radiocarbon dates of millet (n = 120, Supplementary Table S4), wheat and barley remains (n = 131, Supplementary Table S5) from archaeological sites at different elevations in northern China were collected. In addition, analysis of human bones with carbon isotope (δ13C) values (n = 81, Supplementary Table S6), which reflect the dietary consumption of C4 (millets) and C3 plants (rice, barley, wheat, and other natural vegetation) [26], were also collected from the literature.
Quality control and haplogroup allocations
Quality control was performed according to previous research [27]. Potential phantom mutations were checked and corrected. The sequencing outputs were edited and aligned by Lasergene (DNAStar Inc., Madison, Wisconsin, USA) and compared with the revised Cambridge Reference Sequence [28]. Haplogroup allocations of all mtDNAs (including fresh mitogenomes; Supplementary Table S10) were determined according to our previous work [29–31] and mtDNA tree Build 17 (http://phylotree.org/) [32].
Phylogeographic analysis
Phylogenetic trees of lineages of interest were reconstructed manually based on the complete sequences and confirmed by mtphyl software [33]. Rho (ρ) and standard errors (σ) were used to evaluate the coalescent ages of haplogroups [34,35]. The adopted mutation rates were obtained from previous research [36]. Contour maps of haplogroup spatial frequencies were constructed using the Kriging algorithm in Surfer 8.0 (Golden Software Inc. Golden, Colorado, USA).
Bayesian skyline plot analysis
Bayesian skyline plots (BSP) for effective population size (Nef) through time were reconstructed based on 13 protein coding regions using BEAST v1.7.5. [37], as described elsewhere [38,39]. In the BSP analysis, a general time-reversible substitution model with site-specific rates for the first, second and third codons was adopted to infer the ancestral gene trees. To estimate the timescale to the Nef change, a strict molecular clock with a fixed rate of 1.691e−8 substitutions per site per year [40] was chosen. Each Markov chain Monte Carlo simulation was run for 40 000 000 generations and sampled every 4000 generations, with the first 40 000 generations discarded as burn-in. The results were visualized with Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/). Population growth rate and time of population growth were calculated based on skyline plots, as per our previous study [41]. To avoid potential biases caused by sampling, we applied the pooled sampling strategy, which provides the best scheme for inferring demographic history [42], for BSP analysis. In detail, 20 samples were picked randomly from each of the 11 Tibetan populations and pooled as one data set to perform BSP analysis. Proportional changes of certain haplogroups in Tibetans through time were calculated based on Nef values of the haplogroups and all Tibetans in the BSP analyses.
Supplementary Material
Acknowledgements
We would like to thank Dr. Christine Watts for her help in honing the manuscript. We are grateful to all participants in this study.
FUNDING
This work was supported jointly and equally by the Strategic Priority Research Program (XDA20040102) and The Second Tibetan Plateau Scientific Expedition and Research (2019QZKK0607). Additional resources were provided by the Key Research Program of Frontier Sciences (QYZDB-SSW-SMC020), the National Key R&D Program of China (2018YFC2000400), the National Natural Science Foundation of China (31620103907, 31601017 and 41825001), Yunnan Applied Basic Research Project (2017FB044) and ‘Light of West China’ Program of Chinese Academy of Sciences (to Y.-C. L.).
Conflict of interest statement . None declared.
REFERENCES
- 1. Zhao M, Kong QP and Wang HWet al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci USA 2009; 106: 21230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu D, Lou HY and Yuan Ket al. Ancestral origins and genetic history of Tibetan Highlanders. Am J Hum Genet 2016; 99: 580–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang XL, Ha BB and Wang SJet al. The earliest human occupation of the high-altitude Tibetan Plateau 40 thousand to 30 thousand years ago. Science 2018; 362: 1049–51. [DOI] [PubMed] [Google Scholar]
- 4. Su B, Xiao CJ and Deka Ret al. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 2000; 107: 582–90. [DOI] [PubMed] [Google Scholar]
- 5. Qi XB, Cui CY and Peng Yet al. Genetic evidence of Paleolithic colonization and Neolithic expansion of modern humans on the Tibetan Plateau. Mol Biol Evol 2013; 30: 1761–78. [DOI] [PubMed] [Google Scholar]
- 6. Qin ZD, Yang YJ and Kang LLet al. A mitochondrial revelation of early human migrations to the Tibetan Plateau before and after the last glacial maximum. Am J Phys Anthropol 2010; 143: 555–69. [DOI] [PubMed] [Google Scholar]
- 7. Torroni A, Miller JA and Moore LGet al. Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol 1994; 93: 189–99. [DOI] [PubMed] [Google Scholar]
- 8. Chen FH, Dong GH and Zhang DJet al. Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 BP. Science 2015; 347: 248–50. [DOI] [PubMed] [Google Scholar]
- 9. Zeder MA. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc Natl Acad Sci USA 2008; 105: 11597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lev-Yadun S, Gopher A and Abbo S. The cradle of agriculture. Science 2000; 288: 1602–3. [DOI] [PubMed] [Google Scholar]
- 11. Wang LX, Lu Y and Zhang Cet al. Reconstruction of Y-chromosome phylogeny reveals two neolithic expansions of Tibeto-Burman populations. Mol Genet Genomics 2018; 293: 1293–300. [DOI] [PubMed] [Google Scholar]
- 12. Brantingham PJ and Xing G. Peopling of the northern Tibetan Plateau. World Archaeol 2006; 38: 387–414. [Google Scholar]
- 13. Zhang DJ, Dong GH and Wang Het al. History and possible mechanisms of prehistoric human migration to the Tibetan Plateau. Sci China Earth Sci 2016; 59: 1765–78. [Google Scholar]
- 14. Dong GH, Yang YS and Liu XYet al. Prehistoric trans-continental cultural exchange in the Hexi Corridor, Northwest China. Holocene 2018; 28: 621–8. [Google Scholar]
- 15. Olivieri A, Sidore C and Achilli Aet al. Mitogenome diversity in Sardinians: a genetic window onto an island's past. Mol Biol Evol 2017; 34: 1230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang XY, Wan ZW and Perry Let al. Early millet use in Northern China. Proc Natl Acad Sci USA 2012; 109: 3726–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao ZJ. New archaeobotanic data for the study of the origins of agriculture in China. Curr Anthropol 2011; 52: S295–306. [Google Scholar]
- 18. Dong GH, Zhang SJ and Yang YSet al. Agricultural intensification and its impact on environment during Neolithic Age in northern China. Chin Sci Bull 2016; 61: 2913–25. [Google Scholar]
- 19. Zhao ZJ. The process of origin of agriculture in China: archaeological evidence from flotation results. Quat Sci 2014; 34: 73–84. [Google Scholar]
- 20. Hazarika M. Tracing post-Pleistocene human movements and cultural connections of the eastern Himalayan region with the Tibetan plateau. Archaeol Res Asia 2016; 5: 44–53. [Google Scholar]
- 21. Zhang QC, Jacqueline TE and Wei Jet al. Paleodietary studies using stable carbon and nitrogen isotopes from human bone: an example from the Miaozigou Site, Qahar Youyi Qianqi, Inner Mongolia. Acta Anthropol Sin 2010; 29: 270–5. [Google Scholar]
- 22. Gao SZ, Zhang Y and Wei Det al. Ancient DNA reveals a migration of the ancient Di-qiang populations into Xinjiang as early as the early Bronze Age. Am J Phys Anthropol 2015; 157: 71–80. [DOI] [PubMed] [Google Scholar]
- 23. Wei J. Chronicle and related issues of primitive culture in Western Inner Mongolia. Chin Cult Relics 1985; 5: 77–8. [Google Scholar]
- 24. Wang HJ, Chang E and Cai DWet al. Mitochondrial DNA analysis of remains from Zhukaigou archaeological site in Inner Mongolia. J Jilin U (Med Ed) 2007; 33: 5–8. [Google Scholar]
- 25. Wang LX, Lu Y and Zhang Cet al. Reconstruction of Y-chromosome phylogeny reveals two Neolithic expansions of Tibeto-Burman populations. Mol Genet Genomics 2018; 293: 1293–300. [DOI] [PubMed] [Google Scholar]
- 26. Barton L, Newsome SD and Chen FHet al. Agricultural origins and the isotopic identity of domestication in northern China. Proc Natl Acad Sci USA 2009; 106: 5523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kong QP, Salas A and Sun Cet al. Distilling artificial recombinants from large sets of complete mtDNA genomes. PLoS One 2008; 3: e3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrews RM, Kubacka I and Chinnery PFet al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147. [DOI] [PubMed] [Google Scholar]
- 29. Kong QP, Yao YG and Sun Cet al. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet 2003; 73: 671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong QP, Bandelt HJ and Sun Cet al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet 2006;15: 2076–86. [DOI] [PubMed] [Google Scholar]
- 31. Kong QP, Sun C and Wang HWet al. Large-scale mtDNA screening reveals a surprising matrilineal complexity in East Asia and its implications to the peopling of the region. Mol Biol Evol 2011; 28: 513–22. [DOI] [PubMed] [Google Scholar]
- 32. van Oven M and Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30: E386–94. [DOI] [PubMed] [Google Scholar]
- 33. Eltsov N and Volodko N. mtPhyl-software tool for human mtDNA analysis and phylogeny reconstruction. http:/eltsov.org (11 November 2017, date last accessed). [Google Scholar]
- 34. Saillard J, Forster P and Lynnerup Net al. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 2000; 67: 718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forster P, Harding R and Torroni Aet al. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 1996; 59: 935. [PMC free article] [PubMed] [Google Scholar]
- 36. Soares P, Ermini L and Thomson Net al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet 2009; 84: 740–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drummond A and Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Atkinson QD, Gray RD and Drummond AJ. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc Biol Sci 2009; 276: 367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Atkinson QD, Gray RD and Drummond AJ. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol 2008; 25: 468–74. [DOI] [PubMed] [Google Scholar]
- 40. Ho SYW and Endicott P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am J Hum Genet 2008; 83: 142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gignoux CR, Henn BM and Mountain JL. Rapid, global demographic expansions after the origins of agriculture. Proc Natl Acad Sci USA 2011; 108: 6044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heller R, Chikhi L and Siegismund HR. The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS One 2013; 8: e62992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.