Abstract
Brain size and cognitive skills are the most dramatically changed traits in humans during evolution and yet the genetic mechanisms underlying these human-specific changes remain elusive. Here, we successfully generated 11 transgenic rhesus monkeys (8 first-generation and 3 second-generation) carrying human copies of MCPH1, an important gene for brain development and brain evolution. Brain-image and tissue-section analyses indicated an altered pattern of neural-cell differentiation, resulting in a delayed neuronal maturation and neural-fiber myelination of the transgenic monkeys, similar to the known evolutionary change of developmental delay (neoteny) in humans. Further brain-transcriptome and tissue-section analyses of major developmental stages showed a marked human-like expression delay of neuron differentiation and synaptic-signaling genes, providing a molecular explanation for the observed brain-developmental delay of the transgenic monkeys. More importantly, the transgenic monkeys exhibited better short-term memory and shorter reaction time compared with the wild-type controls in the delayed-matching-to-sample task. The presented data represent the first attempt to experimentally interrogate the genetic basis of human brain origin using a transgenic monkey model and it values the use of non-human primates in understanding unique human traits.
Keywords: human evolution, brain development, MCPH1, transgenic monkey, neoteny, cognition
INTRODUCTION
Expansion in brain size and improvement in cognitive skills are among the most fundamental evolutionary changes that set humans apart from other primates. Comparative genomic analyses between humans and non-human primates suggest that these dramatic phenotypic divergences may be due to several underlying genetic changes: rapid evolution of protein-coding genes [1,2] and non-coding RNA genes [3–5], emergence of human-specific segmental duplications [6–8], as well as alterations in gene expression [9–12] and epigenetic regulation [13–15]. Despite a great deal of effort in previous studies, we are still on the way in searching for the responsible genes and dissecting the genetic mechanisms that shape the human brain.
Among the reported genes that play important roles in human brain development, MCPH1 (also known as BRIT1) is one of the strong candidates that may contribute to human brain evolution [16]. It is one of the fast-evolving genes in primates [17]. In particular, MCPH1 has accumulated seven human-specific amino acid changes that are fixed in modern humans [17]. Our previous in vitro experiments showed that these human-specific protein-sequence changes could alter the regulation of MCPH1 on its downstream genes [18]. Importantly, at the transcriptional level, MCPH1 also has shown human-specific changes. During postnatal brain development, MCPH1 is abundantly expressed in humans, but less so in non-human primates (macaque and chimpanzee, Supplementary Fig. 1A) [11]. In addition, we have shown that the MCPH1 transcriptional activity was significantly higher in human than in rhesus monkey [18]. Collectively, current evidence suggests that not only the human-specific protein-sequence changes, but also gene-expression alteration of MCPH1 may contribute to human brain development and function.
MCPH1 encodes a pleiotrophic protein. It functions as a transcription factor by interacting with E2F1 (E2F transcription factor 1) to regulate cell cycle and cell apoptosis [19]. It also works as a DNA-damage response protein and is involved in chromatic remodeling to control DNA repair [16,20,21]. In the central nervous system, as a centrosome protein, MCPH1 plays a conserved role in neurogenesis by regulating the neuronal progenitor divison mode via the Chk1–Cdc25B pathway [22]. In humans, truncated mutations of MCPH1 cause primary microcephaly (MCPH, OMIM251200)—a rare human brain-developmental disorder, characterized by significantly reduced brain volume and mental retardation [23–25]. Consistently, the MCPH1 knockout animal models (mouse and monkey) reproduced the phenotypes of human microcephaly, notably the reduced brain size [22,26]. During human brain development, MCPH1 has the highest expression at the prenatal stage and the expression reduces after birth and remains at a constant level through adulthood (Supplementary Fig. 1B). At the prenatal stage, MCPH1 is highly expressed in all cell types in the cortex, including neural progenitor cells, inter-neurons, astrocytes and microglia cells [27] (Supplementary Fig. 1B). In a mouse study, it was demonstrated that MCPH1 controls precise mitotic spindle orientation and regulates the progenitor division mode to maintain brain size [22]. However, although MCPH1 loss of function causes abnormal brain development, resulting in a reduced brain size in human and animals, the functional consequence of the human-specific seqence and expression changes remains to be understood.
To interrogate the genetic basis of human brain evolution, the traditional mouse or rat models are less ideal due to the vast dissimilarities in brain size and structure between humans and rodents. Instead, a non-human primate transgenic model would be far more effective. The rhesus monkey (Macaca mulatta), an Old World Monkey species widely used for biomedical research, is an ideal choice, due to its high sequence similarity with humans (>93% for protein-coding genes) [28] and yet relatively large phylogenetic distance (about 25 million years of divergence from humans), which alleviates ethical concerns [29].
For MCPH1, the coding sequence similarity is 94.9% between human and rhesus monkey, while it is only 67.5% between human and mouse. Similarly, the 5’ non-coding sequence (∼5 kb) of MCPH1 likely contains regulatory elements for gene-expression regulation and it has 88.7% similarity between human and rhesus monkey, while it is only 40.4% between human and mouse. Additionally, we have shown that, during primate evolution, the MCPH1 promoter region has acquired a primate-specific E2F1-binding motif that is absent in rodents and other mammalian species [30]. Taken together, a rhesus monkey model is promising to study the functional impact of the human-specific changes (protein sequence and gene expression) on human brain evolution.
In this study, to mimic the human-specific genetic changes, using lentivirus transfection, we introduced the human MCPH1 copies (huMCPH1) into the rhesus monkey genome so that the transgenic (TG) monkeys have an overexpression of human MCPH1. We successfully generated eight first-generation (F0) and three second-generation (F1) TG monkeys carrying human MCPH1 copies. Brain-development tracking via magnetic resonance imaging (MRI) of a tissue section with cellular markers showed that the TG monkeys experienced delayed neuronal maturation and neural-fiber myelination, both of which are human-like features of brain-developmental neoteny. Accordingly, transcriptome analysis of prenatal and postnatal brain development revealed an altered gene-expression profile in neuro-progenitors and neurons with shifted expression time of synapse-related genes in the TG monkeys. Remarkably, our preliminary cognitive test detected an improved short-term memory in the TG monkeys.
RESULTS
Generation of transgenic monkeys carrying human MCPH1 copies
All animal procedures were conducted following the international standards, and were approved in advance by the Institutional Animal Care and Use Committee of Kunming Institute of Zoology, Chinese Academy of Sciences and Yunnan Key Laboratory of Primate Biomedical Research (Approval No: SYDW-2010002 and KBI_K001115033-01,01). Lentivirus delivery was used to introduce the human MCPH1 copy (huMCPH1) into the rhesus monkey genome. A high titer (>1 × 1010 infection particles per ml) simian immunodeficiency virus (SIV) vector containing lentivirus was produced for gene transfer (Supplementary Fig. 1C; see the ‘Methods’ section for more details). The human MCPH1 gene was cloned into the SIV vector containing an eGFP (enhanced green fluorescent protein) gene copy and a universal promoter (the CMV-enhanced chicken beta actin (CAG) promoter) (Supplementary Fig. 1D). The monkey oocytes were obtained by super-ovulation and fertilized in vitro (IVF). The early-cleavage-stage embryos were injected with 50–100 pl lentivirus.
In total, five pregnant surrogates produced eight F0 monkeys (T_01–T_08), among which six are twins (T_01/T_02, T_03/T_04 and T_06/T_07) (Fig. 1 and Table 1). Caesarean section was used to deliver baby monkeys at around 155 days’ gestation except for the twins (T_03 and T_04) with premature abortion at embryonic 136 days. Multiple tissues (blood, placenta, umbilical cord endothelial cells and skin) were sampled to test the transgenic status and all monkeys turned out to be positive (Supplementary Fig. 1E–P). We detected strong GFP signals in the nucleus of the TG monkeys (Supplementary Fig. 1P). Since MCPH1 is known to work in the nucleus [31], this result suggests that the huMCPH1 transgenes were correctly positioned in the cell.
Figure 1.
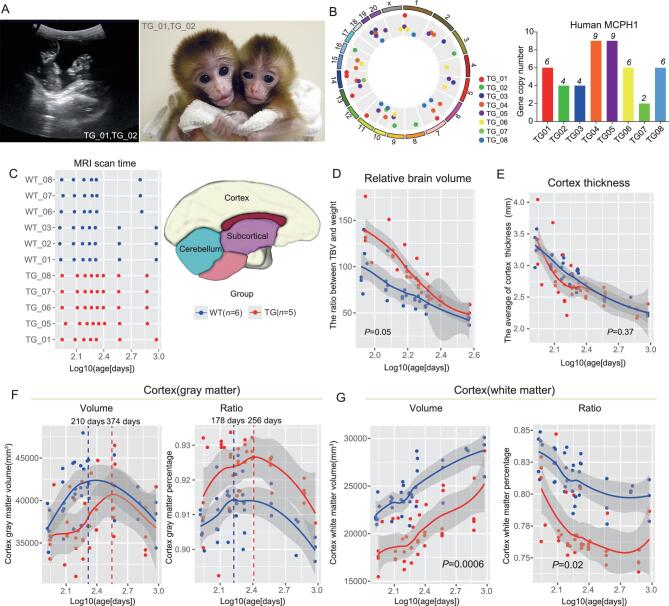
Brain-developmental tracking of the TG monkeys via structural MRI. (A) Left panel: the ultrasound image showing the twin monkeys (TG_01 and TG_02) at 58 days of gestation. Right panel: the newborn twin TG monkeys: TG_01 (male, left) and TG_02 (female, right). (B) Left panel: genomic distribution of the huMCPH1 transgene copies in the TG monkeys. The transgene insertion sites (dots) are randomly distributed on the chromosomes (outermost circle). Right panel: bar plot of the huMCPH1 copy numbers in the TG monkeys. (C) Left panel: the time points (log10(age-days)) of MRI scans of the five TG and six WT monkeys, with the first scan at about 2 months after birth and the last scan at 2–3 years old. Right panel: the schematic map of brain regions. (D) The change in relative brain volume (measured by the total brain volume divided by the body weight) during development. (E) The change in cortex thickness during development. (F) The change in cortex gray-matter volume and ratio during brain development. (G) The change in cortex white-matter volume and ratio during brain development. Group effect P-value was calculated based on LMM (linear mixed model) and P < 0.05 was taken as statistically significant. The dashed vertical lines indicate the peaks of cortex gray-matter volumes or ratios.
Table 1.
Information of the generated transgenic monkeys in this study.
| Monkey ID | Generation | Sex | Date of birth | Method of delivery | Status | huMCPH1 copy number |
|---|---|---|---|---|---|---|
| TG_01 | F0 | Male | 2011/6/15 | C-section | Live | 6 |
| TG_02 | F0 | Female | 2011/6/15 | C-section | Live, deceased at 76 days after birth | 4 |
| TG_03 | F0 | Male | – | C-section | Abortion at embryonic 136 days | 4 |
| TG_04 | F0 | Male | – | C-section | Abortion at embryonic 136 days | 9 |
| TG_05 | F0 | Male | 2015/6/18 | C-section | Live | 9 |
| TG_06 | F0 | Male | 2015/6/26 | C-section | Live | 6 |
| TG_07 | F0 | Female | 2015/6/26 | C-section | Live | 2 |
| TG_08 | F0 | Female | 2015/6/26 | C-section | Live | 6 |
| TG_09 | F1 | Male | – | C-section | Euthanized at embryonic E76 days | 5 |
| TG_10 | F1 | Male | – | C-section | Euthanized at embryonic E92 days | 6 |
| TG_11 | F1 | Male | – | C-section | Euthanized at embryonic E92 days | 5 |
To determine the integrated genomic locations and copy numbers, we conducted captured next-generation sequencing according to the reported method [32]. As expected for lentivirus, the huMCPH1 copies were randomly integrated into the monkey genomes. The eight TG monkeys have two to nine huMCPH1 copies (Table 1 and Fig. 1B). Importantly, all integration sites are located in either inter-genic or non-coding regions and presumably will not interfere with the function of the monkey endogenous genes (Supplementary Table 1).
For comparison, we recruited six wild-type (WT) monkeys (Fig. 1C and Supplementary Table 2) and initially they were divided into two groups. The first contained three age-matched monkeys (WT_01, WT_02 and WT_03) raised by their biological mothers. The second WT group contained three age-matched monkeys (WT_06, WT_07 and WT_08) who were separated from their biological mothers 6–25 days after birth and raised by humans under the same conditions as the TG monkeys (Supplementary Table 2). Unfortunately, TG_02 died of unknown cause 76 days after birth. A biopsy did not reveal any organ damage. As mentioned above, TG_03 and TG_04 were abortions at embryonic 136 days. For comparison of brain-tissue section and transcriptome analysis, six additional WT monkeys were sacrificed at the corresponding developmental stages (76 days after birth for WT_04 and WT_05, and embryonic 130–145 days for WT_09, WT_10, WT_11 and WT_12) (Supplementary Table 2). In total, we collected data from 8 TG monkeys and 12 WT monkeys.
Brain-development tracking using structural MRI suggests a delayed neural maturation
To test whether the integrated huMCPH1 copies influenced the brain development of the TG monkeys, we first performed a non-invasive analysis, namely structural MRI (Philips Achieva 3.0T TX). The tested monkeys included five TG monkeys (TG_01, TG_05, TG_06, TG_07 and TG_08) and six WT monkeys (WT_01, WT_02, WT_03, WT_06, WT_07 and WT_08). The MRI scans were conducted at scheduled intervals (once every month while 2–12 months old and then once every 6 months until 2–3 years old) (Fig. 1C). Both T1-weighted image and diffusion tensor imaging (DTI) data were collected. Using the T1 image data, we calculated the volumes of total brain (TB), cerebellum, lobes and subcortical regions based on the published method and monkey atlas [33] (Supplementary Fig. 2). To rule out the potential influence of feeding types, we first compared the two WT monkey groups (monkey feeding vs human feeding) and we did not detect any difference in volumes (Supplementary Table 3). Therefore, all WT monkeys were grouped together in the following analysis.
In general, the TB volume and body weight of the TG monkeys were smaller than the WT monkeys during early development, likely due to the C-section delivery of the TG monkeys at 155 days of pregnancy—about 1 week earlier than the natural delivery of the WT controls. Also, three of the five TG monkeys were twins, which usually weigh less than single-birth monkeys (all WT monkeys were single-birth). However, this difference became smaller when the monkeys grew older and the TG monkeys eventually caught up with the WT monkeys at about 3 years old (Supplementary Fig. 3A and B). Of note, the relative brain volume (TB volume adjusted by body weight) of the TG monkeys was larger than the WT monkeys during early postnatal development and this difference became invisible when the monkeys grew older (Fig. 1D, linear mixed model (LMM) model, group effect P = 0.05), while the cortex thickness was similar between them throughout development (Fig. 1E).
As the brain is mainly composed of cortex gray matter (GM, mostly neurons) and subcortical white matter (WM, mostly glial cells) [34], we next conducted segmentation analyses. Notably, during brain development, the cortex GM volume of the TG monkeys increased more slowly than that of the WT monkeys, and there was on average 164 days’ delay of peak time for the TG monkeys (Fig. 1F). When the cortex was divided into four lobes (frontal, parietal, occipital and temporal lobes), we saw the same pattern in all lobes (Supplementary Fig. 3C–F). Interestingly, the cortex GM ratios (the proportion of the cortex GM volume vs the TB volume) of the TG monkeys were larger than those of the WT monkeys with a similar ratio peak time delay (Fig. 1F, LMM model, group effect P = 0.06). By contrast, we did not detect such a delay in the cerebellum or subcortical region (Supplementary Fig. 3G and H).
The developmental pattern of WM was different from that of GM. There were no volume/ratio peaks for the cortex WM and the TG monkeys kept a significantly lower volume and cortex WM ratio than the WT monkeys during development (Fig. 1G, LMM model, group effect P = 0.0006) (Fig. 1G, LMM model, group effect P = 0.01). By contrast, the WM volume of the subcortical region did not show such a difference and an opposite pattern was seen for the cerebellum, although statistically not significant (Supplementary Fig. 3G and H, group effect P = 0.59). Given the observed patterns, we reasoned that the observed brain-developmental changes of the TG monkeys might reflect a delay in cortex development rather than a developmental impairment. Consistently with this view, the cortex GM volume curves of the TG and WT monkeys started to converge 1 year after birth (Fig. 1F) and the same pattern was observed when looking at the curves of the four lobes (Supplementary Fig. 3C–F). In particular, TG_01 and three WT monkeys (WT_01, WT_02 and WT_03) had MRI data at later stages (∼3 years old) and the cortex GM and WM volumes of TG_01 had already caught up with the WT monkeys (Supplementary Fig. 4), supporting the proposed brain-developmental delay.
To further explore the brain-developmental changes of the TG monkeys, we analysed the MRI-DTI data to evaluate the WM properties, growth of brain structures and fiber tracts that connect them [35]. The fractional anisotropy (FA) index was used in characterizing the degree of diffusion directionality and is sensitive to the axon size, density as well as the degree of myelination [36]. Consistently with the observation of brain-volume change, in three types of WM tracts (projection fibers, association fiber and commissural fibers), the TG monkeys exhibited relatively lower FA values compared with the WT monkeys, although statistically not significant (Supplementary Fig. 5A–C and Supplementary Fig. 6). The lower FA values suggest lower levels of myelination. A similar pattern was also observed when looking at the MD (mean diffusivity) values [37] (Supplementary Fig. 7). Hence, the MRI-DTI data indicated a lower myelination level in the TG monkeys, implying a delayed neural-fiber myelination and neural-network maturation, which seem to mimic the known brain-developmental delay (neoteny) of humans [38].
We also checked whether there was a correlation between the number of carried huMCPH1 copies and the brain structural measurements, and we did not find a significant correlation with any measurements including TB volume, cortex volume and cortex thickness, etc. (Supplementary Fig. 8), suggesting that a gene-dosage effect is not obvious.
Brain-tissue-section analysis indicates delayed neuronal differentiation
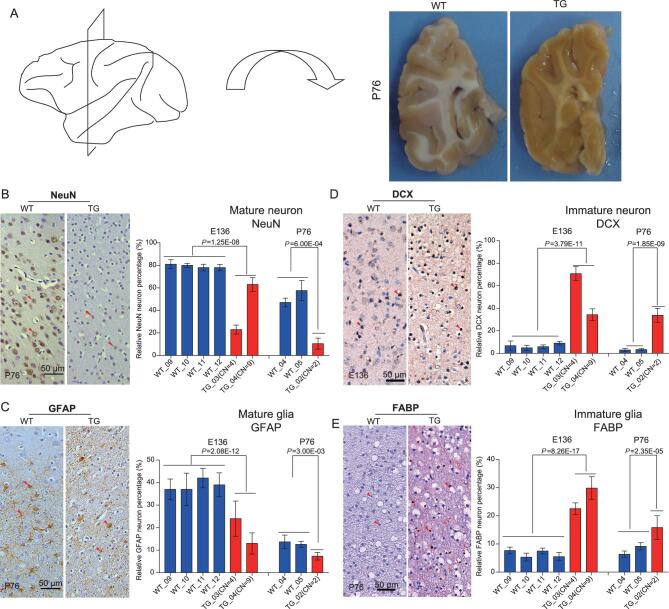
To detect brain-developmental changes at the cellular level, we conducted brain-tissue-section analysis of the frontal lobe at both the prenatal (two TG and four WT monkeys at embryonic day 136) and postnatal (one TG and two WT monkeys at 76 days after birth) stages (Fig. 2A). Two marker genes were used to examine the status of neural-cell proliferation and differentiation, including NeuN for matured neurons and glial fibrillary acidic protein (GFAP) for matured astrocytes. At the prenatal stage (E136), there were 80% NeuN-positive cells in the WT monkeys, contrasting with only 20% in TG_03 and 60% in TG_04 (P = 1.25E-08, two-tailed t-test; Fig. 2B). At the postnatal stage (P76), 40–60% of cells were NeuN-positive in the WT monkeys, but only 10% in TG_01 (P = 6.0E-04, two-tailed t-test; Fig. 2B). We detected similar ratio differences for astrocytes, with the WT monkeys having twice the ratio of GFAP-positive cells compared with that of the TG monkeys (P < 0.01, two-tailed t-test; Fig. 2C).
Figure 2.
Brain immunohistochemistry analysis with gene markers. (A) The schematic indication of the sampled frontal-lobe region of the P76 monkey brain. (B) Immunohistochemical staining of NeuN, the marker gene for matured neurons. Quantification of the NeuN-positive neurons indicates fewer matured neurons in the TG monkeys compared with the WT monkeys. (C) Immunohistochemical staining of GFAP, the marker gene for matured astrocytes. Quantification of the GFAP-positive astrocytes indicates decreased mature astrocytes in the TG monkeys. (D) Immunohistochemical staining of DCX, the marker gene for immature neurons. Quantification of the DCX-positive neurons indicates more immature neurons in the TG monkeys. (E) Immunohistochemical staining of FABP, the marker gene for immature astrocytes. Quantification of the FABP-positive astrocytes indicates more immature astrocytes in the TG monkeys. All histograms represent the mean ± SD of at least two sections and each section includes counts of four different visual fields. The red arrows indicate positively stained cells. The two-tailed unpaired t-test was used for statistical assessment and CN stands for copy number.
The reduced ratios of matured neurons and astrocytes in the TG monkeys would predict elevated ratios of immature cells. To test this, we used two additional makers, namely DCX (doublecortin) for immature neurons and fatty acid binding protein (FABP) for immature astrocytes. As expected, the TG monkeys possessed much higher ratios of immature neurons and glia cells compared with the WT monkeys (P < 0.001, two-tailed t-test; Fig. 2D and 2E). Of note, the total numbers of cells in the brain were similar between the TG monkeys and the WT monkeys (Supplementary Fig. 9). Collectively, this cell-level difference is consistent with the observed myelination delay of the MRI data, as the fiber tracts are mostly composed of glial cells and myelinated nerve cells (axons).
Brain-transcriptome analysis using bulk tissue
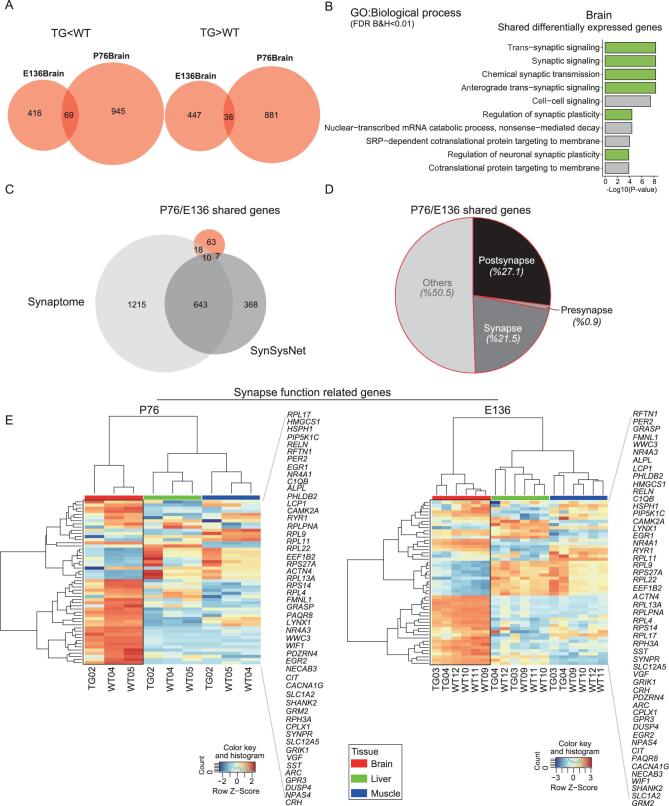
To gain insight into the molecular mechanism underling the speculated cortex-developmental delay, we conducted RNAseq of the prefrontal cortex of the prenatal (two TG vs four WT at E136) and postnatal monkeys (one TG vs two WT at P76), with liver and muscle as the references. As expected, the overall MCPH1 expression was much higher in the TG monkeys than in the WT monkeys for all tissue types and the integrated huMCPH1 copies had much higher expression than the endogenous monkey MCPH1 (Supplementary Figs 10A and 11A). In the brain, there was a large number of differentially expressed genes (DEGs) between the TG and the WT monkeys (970 genes at embryonic day 136 and 1933 genes at postnatal day 76) (Supplementary Figs 10B, C, 11B and C and Supplementary Table 4). The numbers of DEGs were comparable in muscles and much lower in livers. Only a small portion of DEGs overlapped among tissues (Supplementary Figs 10D and 11D and Supplementary Table 4), implying that the transgene huMCPH1 affects the gene expression of the TG monkeys in a tissue-type-dependent manner.
To see the functional enrichment of the DEGs in the brain, we performed gene ontology (GO) ontology analysis using the ToppGene Suite [39]. At the prenatal stage (E136), there were 350 significantly enriched functional categories for the TG-down-regulated genes (Supplementary Table 5, FDR B&H < 0.05) and the top four categories were all related to synaptic signaling (Supplementary Fig. 10E, left panel). In contrast, the enriched categories for the TG-up-regulated genes (60 categories, Supplementary Table 6) were mostly basic cellular functions such as translation and protein localization to the endoplasmic reticulum, not closely related to neural function (Supplementary Fig. 10E, right panel). Similarly, at the postnatal stage (P76), the top 10 enriched categories for the TG-down-regulated genes were all related to neuron differentiation and neuron development, and synaptic-signaling genes were also over-represented, while the top categories for the TG-up-regulated genes were mostly related to neuron projection (Supplementary Fig. 11E and Supplementary Table 7). The transcriptomic changes in the developing brains suggest that many neuron-maturation and differentiation-related genes were suppressed in the TG monkeys, consistently with the observed delay of neuron differentiation in the tissue-section analysis (Fig. 2).
Importantly, we found 107 brain DEGs shared between the prenatal E136 and postnatal P76 stages (Fig. 3A). GO ontology analysis with these 107 genes indicated that the highest enrichment category was synapse-related function, confirming the observed pattern when using all brain DEGs (Fig. 3B, 6/10). Remarkably, 35 of the 107 genes (32.7%) overlapped with the known synapse genes in the datasets of synaptome [40] and synsysnet [41] (Fig. 3C). Further analysis showed that about 50% of the 35 genes are either post-synapse- or synapse-related genes, and only 0.9% are pre-synapse-related genes (Fig. 3D). The hierarchical clustering analysis using the shared brain DEGs clearly distinguished the TG and the WT monkeys with the most prominent distinction between brain and muscle/liver (Fig. 3E). For example, NR4A1 is a downstream gene of MEF2A—a gene playing a critical role in activity-induced synaptic modification [42]. This gene showed 76% (at E136) and 26% (at P76) expression reduction in the TG monkeys. In the mouse model, overexpression of NR4A1 would eliminate dendritic spines while knock-down of NR4A1 could cause excessive number of spines and major postsynaptic density [43]. Together, these data suggest that, at the bulk-tissue level, the transgene huMCPH1 mainly suppresses the expression of neural differentiation and synapse-function-related genes.
Figure 3.
Brain-transcriptome analysis at prenatal E136 and postnatal P76. (A) Venn diagrams showing overlaps among differentially expressed genes (DEGs, TG vs WT) in the brain between E136 and P76. (B) Enriched gene clusters of DEGs in the brain. The green blocks indicate synapse-signaling-associated clusters. (C) Venn diagrams showing overlaps among the E136/P76 shared genes, the synaptome genes and the synsysnet genes. The hypergeometric tests indicate significant overlaps with the synaptome genes (P = 4.89E-08) and with the synsysnet genes (P = 7.74E-06). (D) Pie chart of the E136/P76 shared genes indicating that about 50% of the genes are synapse- and post-synapse-related genes. (E) Hierarchical clustering using the E136/P76 shared brain DEGs.
Generation of F1 TG monkeys and transcriptome analysis of fetal cortical lamina
To further dissect the impact of the huMCPH1 copies on brain development, we generated three F1 TG monkeys by IVF using the sperms of TG_01 that we proved were carrying the huMCPH1 copies in the germ line (Supplementary Fig. 12). The three F1 TG monkeys were sacrificed at embryonic day 76 (TG_09 at E76) and embryonic day 92 (TG_10 and TG_11 at E92)—the two developmental time points during the neurogenesis peak in rhesus monkeys [44]. Captured sequencing analysis indicated that the three F1 TG monkeys all carried the huMCPH1 copies at the same integrated sites as TG_01 (Supplementary Table 1). With the use of IVF, we also obtained five WT fetal monkeys at the corresponding developmental points (two WT at E76 and three WT at E92, Supplementary Table 2). To conduct more detailed developmental tracking, we sampled the frontal cortex and dissected (using laser micro-dissection) the brain tissue into four cortical laminae, as they reflect different stages of neural proliferation, differentiation and migration, including the cortical plate (CP), outer sub-ventricular zone (OSVZ), sub-ventricular zone (SVZ) and ventricular zone (VZ) (Fig. 4A). RNAseq was performed for each lamina.
Figure 4.
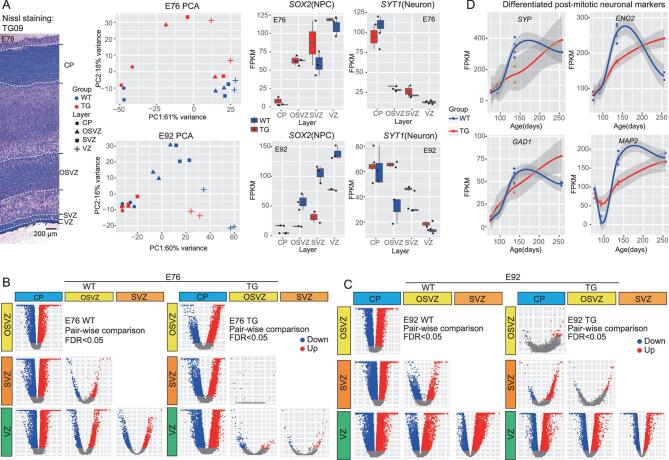
Transcriptome analysis of cortical laminae at E76 and E92. (A) Laser micro-dissection of cortical-laminae and RNAseq analysis. Left panel: Nissl staining of the E76 cortex (TG-09) showing the cortical laminae. Middle panel: the PCA maps of E76 and E92 based on expression levels of all genes. Right panel: cell-marker analysis with SOX2 for neuro-progenitor cell and SYT1 for neuron. (B) Volcano plots showing pairwise comparisons of gene expression between the indicated laminae of E76. (C) Volcano plots showing pairwise comparisons of gene expression between the indicated laminae of E92. (D) Four mature neuron gene makers showing expression delay in the TG monkeys.
First, we checked MCPH1 expression in WT monkey embryos and we found that, at E76, no difference existed between the CP and the germinal zone (OSVZ, SVZ and VZ), while, at E92, MCPH1 expression was higher in the germinal zone than in the CP, consistently with the reported pattern in mouse [24] (Supplementary Fig. 13A). As expected,
in the TG monkey embryos, the transgene huMCPH1 had a much higher expression than the endogenous MCPH1 in all laminae at both E76 and E92 (Supplementary Fig. 13B). The fold changes were different among different laminae. SVZ had the highest fold change at E76, while CP had the highest fold change at E92 (Supplementary Fig. 13B).
The principal component analysis (PCA) showed that the RNA profiles can distinguish different developmental cortical laminae. The VZ and CP were clearly separated, as they represent undifferentiated neuro-progenitors and differentiated neural cells, respectively, while the separation between the SVZ and OSVZ was incomplete because they are the intermediate stages in view of cell proliferation, migration and differentiation (Fig. 4A). Consistently, the marker genes for neuro-progenitors (SOX2) and neurons (SYT1) showed the expected expression pattern in these laminae (Fig. 4A). We then analysed DEGs of the four laminae between the TG monkeys and the WT monkeys. As expected, the transgene huMCPH1 caused expression changes of many genes and this pattern was more pronounced at E92 compared with E76 (140–350 genes for E76 and 3000–9000 genes for E92, Supplementary Table 8) (Supplementary Fig. 13C and D). The GO ontology analysis showed that, at E76, there were 25 enriched categories for CP, 26 for OSVZ, 41 for SVZ and 10 for VZ (Supplementary Tables 9–12, FDR B&H < 0.05). Among the top 10 categories, the enriched functional terms were cell development (CP, 5/10), synapse signaling (OSVZ, 5/10), cell differentiation and proliferation (SVZ, 5/10) and cilium function (VZ, 3/10) (Supplementary Fig. 13E, left panel, FDR B&H < 0.05). In contrast, there were a lot more enriched categories at E92 (453 for CP, 109 for OSVZ, 102 for SVZ and 786 for VZ) (Supplementary Tables 13–16, FDR B&H < 0.05), among which the top categories were mRNA catabolic process (CP, 3/10), neuron differentiation, neurogenesis and cell migration (OSVZ, 5/10), cell cycle and mRNA processing (SVZ, 10/10) and immune response (VZ, 8/10) (Supplementary Fig. 13E, right panel, FDR B&H < 0.05).
We next conducted lamina-to-lamina pairwise comparisons in the TG and the WT monkeys separately. Markedly, at E76, the TG monkeys exhibited much less between-lamina expression difference than the WT monkeys. In particular, there were no DEGs when comparing SVZ with OSVZ in the TG monkeys, contrasting with 557 DEGs in the WT monkeys (Fig. 4B and Supplementary Table 17). A similar pattern was seen at E92. For example, there were 61 CP vs OSVZ DEGs in the TG monkeys, while there were 9186 DEGs in the WT monkeys (Fig. 4C and Supplementary Table 18). This result suggests that cortical-lamina distinction is much weaker for the TG monkeys compared with the WT monkeys, supporting the proposed delay of neuronal maturation and differentiation. Consistently, we observed delayed expression peaks of four known neuron-differentiation markers, including SYP (Synaptic vesicle protein p38), ENO2 (Cytosolic protein t), GAD1 (Glutamic acid decarboxylase) and MAP2 (Cytoskeletal protein) (Fig. 4D).
Furthermore, in order to see the temporal pattern of gene-expression delay in the TG monkeys, we combined the RNAseq data of all four developmental stages, including E76, E92, E136 and P76 (6 TG vs 11 WT). We classified those genes showing expression delay in the TG monkeys into three types according to their expression peak times in the WT monkeys (Supplementary Fig. 14A and B). The Type-1 genes are those with an expression peak shift from E92 in the WT monkeys to E136 in the TG monkeys (e.g. the SLC44A2 gene). The Type-2 genes had a peak shift from E136 in the WT monkeys to P76 in the TG monkeys (e.g. the SYP gene), while the Type-3 genes had a peak shift from E92 in the WT monkeys to P76 in the TG monkeys (e.g. the CDK5 gene) (Supplementary Fig. 14A). In total, we identified 185, 347 and 50 genes for Type-1, Type-2 and Type-3 delays, respectively (Supplementary Fig. 14B). We then performed GO ontology analysis and only the Type-2 genes showed significant enrichment of functional categories, implying that the developmental stage close to birth was the most affected in the TG monkeys. Consistently with the results of bulk-tissue RNAseq, the Type-2 genes from the cortical laminae containing undifferentiated cells (VZ, SVZ and OSVZ) were mainly enriched for synapse-related functions such as trans-synapse signaling, chemical-synapse transmission and synaptic signaling (Supplementary Fig. 14C, P < 0.001) and the involved genes showed delayed expression peaks at P76 or later stages in the TG monkeys.
To test whether the observed gene-expression delay in the TG monkeys show human-like features, we obtained data from a previous study in which genes with human-specific expression delay were identified by comparing postnatal brain development in the prefrontal cortex of humans, chimpanzees and rhesus macaques [11]. We found that only the Type-2 genes were enriched in the reported Module-I gene set with human-specific expression delay (P < 0.0001; hypergeometric test; Supplementary Fig. 14D and Supplementary Table 19). For example, MEF2A is a Type-2 gene and also a Module-I gene, which not only plays a role in neuron differentiation [45], but also mediates a human-specific time shift of cortex synaptic development [11]. Hence, the patterns of gene-expression delay are consistent between the data from the bulk tissue and the data for the laminae, and many neural-differentiation-related genes were suppressed in the TG monkeys with human-like expression delays during brain development.
General behavior analysis and test of short-term memory
To test whether the observed brain-developmental delay at the molecular and cellular levels in the TG monkeys can be transformed into cognitive changes, we first performed an analysis of general behaviors (four TG vs four WT age-matched monkeys 24–36 months old; see the ‘Methods’ section for details). A total of nine indexes for general behaviors were measured [46], including self-injury behavior, stereotypical behavior, feeding, self-grooming, locomotion, resting, bouts of wake, wake and sleep. No difference was detected between the TG and the WT monkeys, suggesting that the transgene huMCPH1 did not cause abnormal behaviors in the TG monkeys (Supplementary Fig. 15).
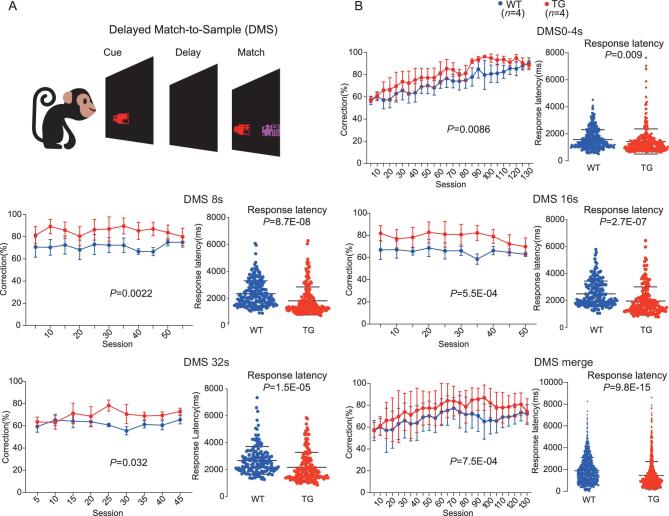
Next, we performed a test of short-term memory using the delayed-matching-to-sample (DMS) task, which was known to be correlated with prefrontal-cortex function [47–49]. The computerized touch-screen behavioral battery (Cambridge Neuropsychological Test Automated Batteries, CANTAB; Lafayette, USA) was used. The DMS task requires the monkeys to remember the color and the shape of a stimulus on the screen for a specified delayed time. The monkeys were initially habituated in a testing room for 5 days and then subject to touch training. Touch training was divided into two phases. In Phase-1, monkeys were subject to touch training for 15 days continuously. In Phase-2, monkeys were required to have a >85% correction rate for 3 continuous days in the training sessions and then were subject to the DMS task test (Fig. 5A, Supplementary Fig. 16 and Supplementary Video 1; more details about touch training is provided in the ‘Methods’ section). The results showed that, at 0 ∼ 4-sec delayed times, the TG monkeys performed significantly better than the WT monkeys (Fig. 5B, general linear model (GLM) model, group effect P = 0.0086) and this difference became more pronounced in the sessions with 8 and 16-sec delayed times (Fig. 5B; GLM model, 8 s, group effect P = 0.0022; 16 s, group effect P = 5.5E-04). When the delayed time extended to 32 sec, the difference remained (Fig. 5B, GLM model, group effect P = 0.032). When all sessions with different delayed times were combined together, the TG monkeys had a significantly better performance than the WT monkeys (Fig. 5B, GLM model, group effect P = 7.5E-04). Interestingly, we also observed significantly shorter reaction times (response latency) in the TG monkeys for all categories of delayed times (P < 0.001, two-tailed t-test, Fig. 5B). Collectively, the TG monkeys exhibited better performance in the DMS task than the WT monkeys, suggesting that the brain-developmental delay caused by the transgene huMCPH1 may have enhanced the short-term memory of the TG monkeys.
Figure 5.
Test of short-term memory using the DMS task. (A) Schematic diagram of the DMS task. (B) The results of the DMS trials indicating different performances between the TG and WT monkeys at different delay times including 0–4, 8, 16 and 32 sec. The bottom-right panel presents the merged DMS data of all delay times. The group effect P-values for correction percentage comparison were computed based on the general linear model (GLM) and corrections for multiple tests were conducted using Bonferroni. The P-values for reaction time were calculated using the two-tailed Student’s t-test.
DISCUSSION
MCPH1 is one of the strong candidates for human brain evolution, since it has accumulated a human-specific protein-sequence and gene-expression changes [17,18]. Ideally, a gene-replacement model would be preferred so that the influence of the endogenous monkey MCPH1 could be removed. However, due to the long generation time of monkeys (4–5 years), current gene-editing tools are still impractical in generating such a model in monkeys. We argue that a transgenic monkey model is practical and to a large extent can mimic the human-specific status. The transgenic monkeys carry the human MCPH1 copies so that the effect of the human-specific protein-sequence changes can be tested. At the same time, since the transgenic monkeys over-express the huMCPH1 transgene, this can mimic the human-specific increase in gene expression.
Because MCPH1 is a key gene for neurogenesis, one of the expected phenotypic outcomes in the transgenic monkeys would be a larger brain, which was not the case in this study. We showed that the TG monkeys carrying the huMCPH1 transgene did not manifest an enlarged brain size, implying that a single gene likely has a limited effect on neural progenitor pool proliferation during brain development. Alternatively, it is equally possible that the human-specific changes in MCPH1 may not enhance its known function in neuro-progenitor proliferation [22]; rather, they work on the unknown function of MCPH1 in neuronal maturation, neural plasticity and synapse signaling, which were supported by multiple lines of evidence presented in this study.
Our analyses found a developmental delay of GM in the brain of the TG monkeys, suggesting that the huMCPH1 transgene may delay neuron differentiation and maturation during brain development. Consistently with the cortex-developmental delay, there were much fewer mature neurons and glia cells in the TG monkeys compared with the WT monkeys during the early period of postnatal development. Consistently, the tissue-level transcriptome comparisons indicated a large amount of neuron differentiation and development, and synapse genes were suppressed in the TG monkeys, providing a possible molecular basis for the observed delay in cell maturation and fiber myelination in the brain. In fact, our previous in vitro experiments demonstrated that MCPH1 can act as a transcription repressor and repress telomerase activity [50]. Furthermore, the cortical-lamina transcriptome comparisons showed that the huMCPH1 transgene can influence gene expression at all laminae including the CP, OSVZ, SVZ and VZ as early as prenatal E76, indicating that the huMCPH1 transgene may alter neurogenesis by affecting the neuro-progenitor cell division and differentiation. In fact, previous mouse studies have already shown that MCPH1 is required for precise mitotic spindle orientation during neurogenesis [22]. Additionally, as expected, cortical-lamina pairwise comparisons suggested that the between-laminae differences in the TG monkeys were not as obvious as in the WT monkeys, consistently with the proposed delay in neural differentiation. Hence, the huMCPH1 transgene may have contributed to delaying cortical-lamina differentiation in the TG monkeys. Taken together, we propose that overexpression of the huMCPH1 transgene can cause neural-developmental delay due to the down-regulation of many neural-differentiation-related genes. Future experiments are warranted to reveal the detailed molecular pathways.
One hallmark difference between humans and non-human primates is that humans require a much longer time to shape their neuro-networks during development, greatly elongating childhood, namely the so-called ‘neoteny’. Myelination is the process of generating myelin sheaths around nerve fibers so that neural signals can be propagated more swiftly with less signal loss. This process is considered a key developmental aspect of the human brain and continues for at least 10–12 years after birth, providing an extended window of neural-network plasticity [51]. In fact, human neocortical myelination is developmentally protracted compared with that of chimpanzees [52]. We speculate that the observed neural-maturation delay in the TG monkeys may have extended their time window of neural-network plasticity, similar to the brain-developmental neoteny of humans. In support of our speculation, when we combined the RNAseq data of all developmental stages, we found that many of the delay genes were synapse-related genes, which are required for the experience-dependent process of neural-network plasticity [53]. More interestingly, most of the delay genes showed human-specific changes in the timing of synaptic development in the previous study [11]. For example, MEF2A and SYP were among the key genes showing human-specific delay of youth-like expression compared with chimpanzee and macaque [11]. Notably, synapse and spine density in the human-projection neurons is much higher than that in rhesus macaque, which is associated with the higher cognitive performance in humans [11,54].
The speculated extension of neural-network plasticity in the TG monkeys gained further support from our preliminary cognitive data. The TG monkeys showed an improved short-term memory, suggesting that the observed brain-developmental delay in the TG monkeys is beneficial, possibly through extending the time window of neural-network plasticity. More interestingly, the TG monkeys displayed a significantly shorter reaction time than the WT monkeys during the DMS task, which is another hint at cognitive improvement. More sophisticated cognitive tests are needed to understand the long-term effect of the huMCPH1 transgene in the TG monkeys.
Our findings demonstrated that transgenic non-human primates (excluding ape species) have the potential to provide important—and potentially unique—insights into the basic questions of what actually makes humans unique, as well as into disorders and clinically relevant phenotypes, such as neurodegenerative and social-behavior disorders that are difficult to study by other means [32,46,55]. But such gains must invariably be weighed against potential ethical concerns [29,56,57]. We noted that the transgenic monkey model also has limitations, including the influence of the endogenous monkey gene copy and the incapability to differentiate the effects of protein-sequence changes from gene-expression changes. Several recent technical improvements (e.g. CRISPR-Cas9) have shown the hope of conducting precision genome editing in monkeys [26,58–61], providing more powerful tools for future studies in understanding the genetic basis of human brain evolution.
METHODS
The detailed methods and materials are available as Supplementary Data at NSR online.
Supplementary Material
Acknowledgements
The wild-type MCPH1 plasmid was kindly provided by Prof. Xingzhi Xu (Capital Normal University, China). We are grateful to Prof. Xintian Hu and Drs. Dongdong Qin and Shihao Wu for their advice on behavioral and cognitive tests. The monkey icon in Fig. 5 was obtained using Freepick at www.flaticon.com. We thank Hui Zhang and Yan Guo for their technical assistance in this study.
FUNDING
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13000000 to B.S.), the National Natural Science Foundation of China (31730088 and 31621062 to B.S.) and the Youth Innovation Promotion Association of CAS (to L.S.).
AUTHOR CONTRIBUTIONS
L.S. and B.S. designed the study; L.S., X.L., J.J., T.L., M.L. and Q.L. performed the experiments; H.W. contributed to animal care and management; C.R.L. and J.H.W. assisted with the MRI image data analysis; Y.N., X.H., Y.C. and W.J. assisted in the monkey reproductive technique and virus-gene transfer; L.S., X.L., Y.L., Y.Y.M., X.S. and H.Y. conducted MRI scanning; Y.L. and T.L. assisted with the histological studies; L.S. and B.S. performed data analysis; L.S., X.L., J.J. and B.S. wrote the manuscript.
Conflict of interest statement . None declared.
REFERENCES
- 1. Clark AG, Glanowski S, Nielsen Ret al.. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 2003; 302: 1960–3. [DOI] [PubMed] [Google Scholar]
- 2. Bustamante CD, Fledel-Alon A, Williamson Set al.. Natural selection on protein-coding genes in the human genome. Nature 2005; 437: 1153–7. [DOI] [PubMed] [Google Scholar]
- 3. Necsulea A, Soumillon M, Warnefors Met al.. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014; 505: 635–40. [DOI] [PubMed] [Google Scholar]
- 4. Bentwich I, Avniel A, Karov Yet al.. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37: 766–70. [DOI] [PubMed] [Google Scholar]
- 5. Zhang R, Peng Y, Wang Wet al.. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res 2007; 17: 612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey JA, Gu ZP, Clark RAet al.. Recent segmental duplications in the human genome. Science 2002; 297: 1003–7. [DOI] [PubMed] [Google Scholar]
- 7. Dennis MY, Nuttle X, Sudmant PHet al.. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 2012; 149: 912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Florio M, Albert M, Taverna Eet al.. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 2015; 347: 1465–70. [DOI] [PubMed] [Google Scholar]
- 9. Khaitovich P, Hellmann I, Enard Wet al.. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 2005; 309: 1850–4. [DOI] [PubMed] [Google Scholar]
- 10. Enard W, Khaitovich P, Klose Jet al.. Intra- and interspecific variation in primate gene expression patterns. Science 2002; 296: 340–3. [DOI] [PubMed] [Google Scholar]
- 11. Liu X, Somel M, Tang Let al.. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res 2012; 22: 611–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He Z, Han D, Efimova Oet al.. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat Neurosci 2017; 20: 886–95. [DOI] [PubMed] [Google Scholar]
- 13. Zeng J, Konopka G, Hunt BGet al.. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am J Hum Genet 2012; 91: 455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shulha HP, Crisci JL, Reshetov Det al.. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol 2012; 10: e1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mendizabal I, Shi L, Keller TEet al.. Comparative methylome analyses identify epigenetic regulatory loci of human brain evolution. Mol Biol Evol 2016; 33: 2947–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pulyers JN, Journiac N, Arai Yet al.. MCPH1: a window into brain development and evolution. Front Cell Neurosci 2015; 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang YQ, Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum Mol Genet 2004; 13: 1131–7. [DOI] [PubMed] [Google Scholar]
- 18. Shi L, Li M, Lin Qet al.. Functional divergence of the brain-size regulating gene MCPH1 during primate evolution and the origin of humans. BMC Biol 2013; 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang S-Z, Lin F-T, Lin W-C. MCPH1/BRIT1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep 2008; 9: 907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X, Zhou ZW, Wang ZQ. The DNA damage response molecule MCPH1 in brain development and beyond. Acta Biochim Biophys Sin 2016; 48: 678–85. [DOI] [PubMed] [Google Scholar]
- 21. Peng G, Yim EK, Dai Het al.. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 2009; 11: 865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gruber R, Zhou Z, Sukchev Met al.. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol 2011; 13: 1325–34. [DOI] [PubMed] [Google Scholar]
- 23. Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet 2005; 76: 717–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson AP, Eastwood H, Bell SMet al.. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet 2002; 71: 136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trimborn M, Bell SM, Felix Cet al.. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet 2004; 75: 261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ke Q, Li W, Lai Xet al.. TALEN-based generation of a cynomolgus monkey disease model for human microcephaly. Cell Res 2016; 26: 1048–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhong S, Zhang S, Fan Xet al.. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 2018; 555: 524–8. [DOI] [PubMed] [Google Scholar]
- 28. Yan G, Zhang G, Fang Xet al.. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol 2011; 29: 1019–23. [DOI] [PubMed] [Google Scholar]
- 29. Coors ME, Glover JJ, Juengst ETet al.. The ethics of using transgenic non-human primates to study what makes us human. Nat Rev Genet 2010; 11: 658–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi L, Su B. Identification and functional characterization of a primate-specific E2F1 binding motif regulating MCPH1 expression. FEBS J 2012; 279: 491–503. [DOI] [PubMed] [Google Scholar]
- 31. Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem 2004; 279: 34091–4. [DOI] [PubMed] [Google Scholar]
- 32. Liu Z, Li X, Zhang JTet al.. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 2016; 530: 98–102. [DOI] [PubMed] [Google Scholar]
- 33. Liu C, Tian X, Liu Het al.. Rhesus monkey brain development during late infancy and the effect of phencyclidine: a longitudinal MRI and DTI study. Neuroimage 2015; 107: 65–75. [DOI] [PubMed] [Google Scholar]
- 34. Hofman MA. Size and shape of the cerebral cortex in mammals. Brain Behav Evol 1985; 27: 28–40. [DOI] [PubMed] [Google Scholar]
- 35. Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 2006; 51: 527–39. [DOI] [PubMed] [Google Scholar]
- 36. Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med 1996; 36: 893–906. [DOI] [PubMed] [Google Scholar]
- 37. Madden DJ, Bennett IJ, Burzynska Aet al.. Diffusion tensor imaging of cerebral white matter integrity in cognitive aging. BBA-Mol Basis Dis 2012; 1822: 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montagu MFA. Time, morphology, and neoteny in the evolution of Man. Am Anthropol 1955; 57: 13–27. [Google Scholar]
- 39. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pirooznia M, Wang T, Avramopoulos Det al.. SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics 2012; 28: 897–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. von Eichborn J, Dunkel M, Gohlke BOet al.. SynSysNet: integration of experimental data on synaptic protein-protein interactions with drug-target relations. Nucleic Acids Res 2013; 41: D834–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hawk JD, Abel T. The role of NR4A transcription factors in memory formation. Brain Res Bull 2011; 85: 21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Y, Wang Y, Erturk Aet al.. Activity-induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron 2014; 83: 431–43. [DOI] [PubMed] [Google Scholar]
- 44. Betizeau M, Cortay V, Patti Det al.. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 2013; 80: 442–57. [DOI] [PubMed] [Google Scholar]
- 45. Zhu B, Carmichael RE, Solabre Valois Let al.. The transcription factor MEF2A plays a key role in the differentiation/maturation of rat neural stem cells into neurons. Biochem Biophys Res Commun 2018; 500: 645–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y, Yu J, Niu Yet al.. Modeling rett syndrome using TALEN-Edited MECP2 mutant cynomolgus monkeys. Cell 2017; 169: 945–55. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuster JM, Alexander GE. Neuron activity related to short-term memory. Science 1971; 173: 652–4. [DOI] [PubMed] [Google Scholar]
- 48. Carlen M. What constitutes the prefrontal cortex? Science 2017; 358: 478–82. [DOI] [PubMed] [Google Scholar]
- 49. Goldman-Rakic PS. Cellular basis of working memory. Neuron 1995; 14: 477–85. [DOI] [PubMed] [Google Scholar]
- 50. Shi L, Li M, Su B. MCPH1/BRIT1 represses transcription of the human telomerase reverse transcriptase gene. Gene 2012; 495: 1–9. [DOI] [PubMed] [Google Scholar]
- 51. Paus T, Zijdenbos A, Worsley Ket al.. Structural maturation of neural pathways in children and adolescents: in vivo study. Science 1999; 283: 1908–11. [DOI] [PubMed] [Google Scholar]
- 52. Miller DJ, Duka T, Stimpson CDet al.. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci USA 2012; 109: 16480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. CSH Perspect Biol 2011; 3: a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duan H, Wearne SL, Rocher ABet al.. Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb Cortex 2003; 13: 950–61. [DOI] [PubMed] [Google Scholar]
- 55. Yang SH, Cheng PH, Banta Het al.. Towards a transgenic model of Huntington's disease in a non-human primate. Nature 2008; 453: 921–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jennings CG, Landman R, Zhou Yet al.. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci 2016; 19: 1123–30. [DOI] [PubMed] [Google Scholar]
- 57. Izpisua Belmonte JC, Callaway EM, Caddick SJet al.. Brains, genes, and primates. Neuron 2015; 86: 617–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Niu Y, Shen B, Cui Yet al.. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014; 156: 836–43. [DOI] [PubMed] [Google Scholar]
- 59. Liu H, Chen Y, Niu Yet al.. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 2014; 14: 323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Z, Cai Y, Liao Zet al.. Cloning of a gene-edited macaque monkey by somatic cell nuclear transfer. Natl Sci Rev 2019; 6: 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qiu P, Jiang J, Liu Zet al.. BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl Sci Rev 2019; 6: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.