ABSTRACT
The knowledge of genetic variation in Chinese patients with non–small-cell lung cancer (NSCLC) is still limited. We aimed to profile this genetic variation in 206 Chinese patients with NSCLC using next-generation sequencing. Tumor tissues or whole-blood samples were collected and subjected to whole-exome targeted next-generation sequencing, which included 565 tumor-associated genes, for somatic gene mutation screening and copy number variation (CNV) detection. Potential functions of most commonly mutated genes and genes with CNV were predicted by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Atotal of 18,749 mutations were identified using targeted next-generation sequencing, and 85.3% of them were missense mutations. Among the mutation, conversions between pyrimidine and purine were predominant, and C> T/G > A was the most common substitution type. High frequencies of mutations were noted in TP53 (47.6%), EGFR (41.7%), CREBBP (23.1%), KMT2C (16.9%), MUC2 (16.6%), DNMT3A (15.5%), LRP1B (15.5%), MUC4 (15.5%), CDC27 (15.2%), and KRAS (12.8%). EGFR and KRAS mutations were mutually exclusive. The tumor mutation load showed differences depending on gender and tumor type. CNV analysis showed that BCORL1 and ARAF have the highest copy number amplification, whereas KDM6A and RBM10 showed the highest copy number deletion. GO and KEGG analyses indicated that high-frequency mutations and CNV genes were concentrated in tumor-related PI3K-Akt, FoxO, and Ras signaling pathway. Cumulatively, we studied somatic gene mutations involved in NSCLC and predicted their clinical significance in Chinese population. These findings may provide clues for etiology and drug target of NSCLC.
KEYWORDS: sequencing, panel, lung, cancer, mutation
GRAPHICAL ABSTRACT
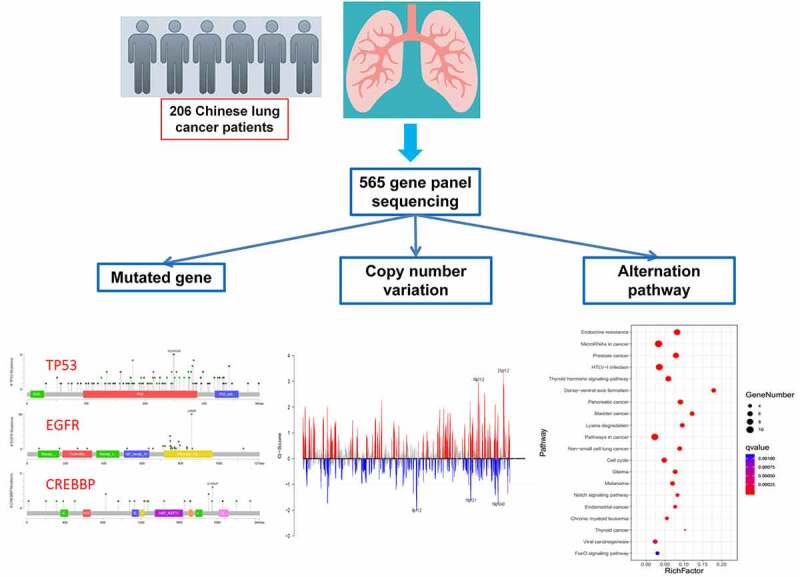
Introduction
Lung cancer has become the leading deadly malignancy in China and globally, in both men and women [1]. According to 2015 statistics, there were approximately 730,000 new cases of lung cancer in China and more than 430,000 people died from this disease. Lung cancer is divided into non–small-cell lung carcinoma (NSCLC) and small-cell lung carcinoma (SCLC) [2], with NSCLC accounting for more than 85% of cases [3]. Moreover, NSCLC has a high mortality rate. Despite extensive research on different treatment options, patients diagnosed with NSCLC (all stages) have a mortality rate of more than 50% within 1 year and an overall 5-year survival rate of less than 18% [4]. These data suggest that there is still a need for new targeted therapeutic drug research of NSCLC, and analyses of the underlying mechanism of NSCLC from a genetic level may provide clues for finding new therapeutic targets.
Next-generation sequencing (NGS) is an approach widely used for the characterization of genetic features. Using an NGS platform, whole-genome sequencing, whole-exome sequencing, whole-transcriptome sequencing, and targeted sequencing can be performed for multiple specific genomic regions. It is a high-throughput and economical method for detecting multiple genetic variations [5]. Many studies have used NGS to analyze genetic variation, tumor mutation burden, and microsatellite instability in solid tumors such as colorectal cancer, gastric cancer, and breast cancer [6,7]. Target sequencing is also used for the identification of variations in genes causing lung cancer. Based on these NGS data, several important genes related to lung cancer have been identified, for exampletumor protein P53 (TP53), phosphatase and tensin homolog (PTEN), epidermal growth factor receptor (EGFR), KRAS proto-oncogene, GTPase (KRAS), neurofibromin 1 (NF1), ATM serine/threonine kinase (ATM), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and fibroblast growth factor receptor 4 (FGFR4) [8–13]. However, the knowledge of genetic variation in NSCLC remains limited in the Chinese population. Existing studies have focused on a small range of genes. For example, Wen et al. performed NGS of 37 cancer-related genes and selected introns of eight genes [14]. Tsoulos et al. focused on a custom panel comprising 23 genes [13,15]. Therefore, a broader panel containing NSCLC-related genes of great significance for the diagnosis and precise treatment of NSCLC is still needed.
Here, we established a panel to detect somatic mutations in 206 samples from Chinese patients. To include as many NSCLC-related genes as possible, the panel comprised 565 genes that were associated with sensitivity and side effects of commonly used chemotherapeutic drugs in clinic and cancer risk. Our study expected to provide an overview of the characteristics of tumor genetic variation in Chinese patients with NSCLC, and provide clues for the clinical diagnosis to enable individualized therapy and find new therapeutic targets of NSCLC.
Materials and methods
Patient and DNA isolation
Surgically resected tumor tissues or venous blood samples were collected from 206 NSCLC patients. Genomic DNA was isolated from tissues or blood using the QIAGEN DNeasy Blood & Tissue Kit (#69504, Qiagen, Germany). All patients gave written informed consent to participate in this study.
Whole-exome next-generation and targeted gene sequencing
DNA libraries for whole-exome NGS were prepared using NEBNext® Ultra™ DNA Library Prep Kit (NEB #E7645, NEB, USA) for Illumina, in accordance with the manufacturer’s instructions. Whole-exome capture was performed using TruSeq Exome Enrichment kit (Illumina # 20020183, USA). For targeted gene sequencing, a panel comprising 565 tumor-related genes was prepared. Targeted genes were enriched with the TruSeq Custom Enrichment kits (Illumina). Samples were sequencing using the HiSeq X TEN platform (Illumina).
Bioinformatic analysis
The adapter sequence in the raw data was removed by cutadapt, after which high-quality reads were aligned to the human reference genome (hg19) using BWA [16] with the default parameters. Somatic mutations were detected by MuTect [17] based on the alignment. Somatic SNVs with high confidence were called if the following criteria were met: (I) both tumor and normal samples should have coverage of ≥10× at the genomic position; and (II) the variants should be supported by at least 5% of the total reads in the tumor. Copy number variation (CNV) for each tumor sample was determined by ADTEx [18]. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of mutated genes were performed using KOBAS [19]. Enriched terms were defined as those with FDR of <0.01.
Statistical analysis
The difference in Tumor mutation burden (TMB) between male and female and adenocarcinoma and squamous carcinoma were analyzed using Student’s t-test method. Correlation between TMB and age were analyzed using Pearson Correlation Coefficient method.
Results
Analyses of the underlying mechanism of NSCLC from a genetic level may provide clues for studying new therapeutic targets for drugs in NSCLC treatment; however, the knowledge of the genetic variation of NSCLC remains limited in Chinese population. Moreover, NGS is a widely used approach for the characterization of genetic characteristics. In this study, we established a panel containing 565 genes that were associated with sensitivity and side effects of commonly used chemotherapeutic drugs in clinic and cancer risk to detect somatic mutations in samples from 206 Chinese patients. A total of 18,749 mutations were identified using targeted NGS and 85.3% of them were missense mutations. Among the mutations, conversions between pyrimidine and purine were dominant, and C > T/G > A was the most common substitution type. High frequencies of mutations were noted in TP53 (47.6%), EGFR (41.7%), CREB binding protein (CREBBP) (23.1%), lysine methyltransferase 2 C (KMT2C) (16.9%), Mucin 2 (MUC2) (16.6%), DNA methyltransferase 3 alpha (DNMT3A) (15.5%), LDL receptor related protein 1B (LRP1B) (15.5%), Mucin 4 (MUC4) (15.5%), cell division cycle 27 (CDC27 (15.2%), and KRAS (12.8%). EGFR and KRAS mutations were mutually exclusive. The tumor mutation load showed BCL6 corepressor like 1 (BCORL1) and a-raf proto-oncogene (ARAF) have the highest copy number amplification, whereas lysine demethylase 6A (KDM6A) and RNA binding motif protein 10 (RBM10) showed the highest copy number deletion. GO and KEGG analyses indicated that high-frequency mutations and CNV genes were concentrated in the tumor-related PI3K-Akt, FoxO, and Ras signaling pathway.
Overview of somatic mutation in patients with NSCLC
To obtain an overview of somatic mutation in Chinese patients with NSCLC patients, we recruited 206 Chinese patients with NSCLC and performed targeted NGS. The mean age of the 206 enrolled patients with NSCLC was 65 years (range 54–86). Of these, 81 (39.3%) were male and 125 (60.7%) were female. Individual clinical information is listed in Table 1. To obtain the somatic mutation spectrum of the 206 patients, next-generation sequencing-based technology was used to capture 565 genes from tumor tissues and peripheral blood of patients with NSCLC. As shown in Figure 1(a), the coverage depth of the captured regions of most genes was at least 50×, with an average coverage depth of 914× (Table 1) (Figure 1(a)).
Table 1.
Clinical information of the 206 NSCLC patients
| SampleID | Gender | Age | Clinical diagnosis |
|---|---|---|---|
| P1 | Female | 63 | Non-small cell lung cancer |
| P2 | Male | 65 | Adenocarcinoma |
| P3 | Male | 79 | Adenocarcinoma |
| P4 | Male | 54 | Adenocarcinoma |
| P5 | Female | 68 | Adenocarcinoma |
| P6 | Male | 71 | Squamous |
| P7 | Female | 63 | Squamous |
| P8 | Male | 72 | Squamous |
| P9 | Male | 63 | Squamous |
| P10 | Male | 54 | Squamous |
| P11 | Female | 74 | Non-small-cell lung cancer |
| P12 | Male | 59 | Non-small-cell lung cancer |
| P13 | Male | 69 | Non-small-cell lung cancer |
| P14 | Male | 44 | Non-small-cell lung cancer |
| P15 | Male | 68 | Adenocarcinoma |
| P16 | Female | 49 | Non-small-cell lung cancer |
| P17 | Male | 57 | Adenocarcinoma |
| P18 | Female | 61 | Non-small-cell lung cancer |
| P19 | Male | 56 | Squamous |
| P20 | Male | 65 | Adenocarcinoma |
| P21 | Male | 63 | Squamous |
| P22 | Female | 64 | Non-small-cell lung cancer |
| P23 | Female | 57 | Non-small-cell lung cancer |
| P24 | Female | 45 | Adenocarcinoma |
| P25 | Female | 51 | Adenocarcinoma |
| P26 | Female | 50 | Adenocarcinoma |
| P27 | Male | 82 | Non-small-cell lung cancer |
| P28 | Male | 56 | Non-small-cell lung cancer |
| P29 | Female | 64 | Adenocarcinoma |
| P30 | Male | 71 | Non-small-cell lung cancer |
| P31 | Male | 46 | Adenocarcinoma |
| P32 | Male | 52 | Squamous |
| P33 | Female | 48 | Non-small-cell lung cancer |
| P34 | Male | 61 | Non-small-cell lung cancer |
| P35 | Male | 35 | Squamous |
| P36 | Male | 69 | Small cell lung cancer |
| P37 | Female | 69 | Non-small-cell lung cancer |
| P38 | Male | 64 | Non-small-cell lung cancer |
| P39 | Male | 65 | Non-small-cell lung cancer |
| P40 | Female | 75 | Non-small-cell lung cancer |
| P41 | Female | 58 | Adenocarcinoma |
| P42 | Female | 38 | Adenocarcinoma |
| P43 | Female | 63 | Non-small-cell lung cancer |
| P44 | Male | 62 | Non-small-cell lung cancer |
| P45 | Male | 79 | Non-small-cell lung cancer |
| P46 | Male | 51 | Non-small-cell lung cancer |
| P47 | Female | 60 | Adenocarcinoma |
| P48 | Male | 62 | Non-small-cell lung cancer |
| P49 | Male | 68 | Non-small-cell lung cancer |
| P50 | Female | 69 | Non-small-cell lung cancer |
| P51 | Female | 53 | Non-small-cell lung cancer |
| P52 | Female | 57 | Adenocarcinoma |
| P53 | Female | 61 | Adenocarcinoma |
| P54 | Male | 58 | Non-small-cell lung cancer |
| P55 | Male | 54 | Neuroendocrine |
| P56 | Female | 77 | Non-small-cell lung cancer |
| P57 | Female | 35 | Non-small-cell lung cancer |
| P58 | Female | 70 | Adenocarcinoma |
| P59 | Male | 79 | Non-small-cell lung cancer |
| P60 | Male | 66 | Non-small-cell lung cancer |
| P61 | Male | 68 | Non-small-cell lung cancer |
| P62 | Male | 68 | Non-small-cell lung cancer |
| P63 | Male | 61 | Non-small-cell lung cancer |
| P64 | Male | 80 | Non-small-cell lung cancer |
| P65 | Male | 70 | Non-small-cell lung cancer |
| P66 | Female | 39 | Adenocarcinoma |
| P67 | Female | 50 | Adenocarcinoma |
| P68 | Male | 67 | Non-small-cell lung cancer |
| P69 | Male | 49 | Non-small-cell lung cancer |
| P70 | Male | 72 | Adenocarcinoma |
| P71 | Male | 54 | Non-small-cell lung cancer |
| P72 | Male | 52 | Adenocarcinoma |
| P73 | Female | 68 | Non-small-cell lung cancer |
| P74 | Female | 73 | Non-small-cell lung cancer |
| P75 | Male | 69 | Adenocarcinoma |
| P76 | Female | 71 | Adenocarcinoma |
| P77 | Female | 66 | Non-small-cell lung cancer |
| P78 | Male | 69 | Adenocarcinoma |
| P79 | Male | 62 | Squamous |
| P80 | Male | 54 | Non-small-cell lung cancer |
| P81 | Female | 47 | Non-small-cell lung cancer |
| P82 | Male | 76 | Non-small-cell lung cancer |
| P83 | Male | 86 | Non-small-cell lung cancer |
| P84 | Male | 73 | Non-small-cell lung cancer |
| P85 | Male | 72 | Non-small-cell lung cancer |
| P86 | Male | 43 | Adenocarcinoma |
| P87 | Female | 67 | Adenocarcinoma |
| P88 | Male | 55 | Non-small-cell lung cancer |
| P89 | Male | 77 | Small cell lung cancer |
| P90 | Male | 57 | Non-small-cell lung cancer |
| P91 | Female | 54 | Adenocarcinoma |
| P92 | Male | 65 | Neuroendocrine |
| P93 | Female | 72 | Adenocarcinoma |
| P94 | Male | 62 | Squamous |
| P95 | Female | 45 | Non-small-cell lung cancer |
| P96 | Female | 45 | Non-small-cell lung cancer |
| P97 | Female | 51 | Adenocarcinoma |
| P98 | Female | 65 | Non-small-cell lung cancer |
| P99 | Male | 61 | Adenocarcinoma |
| P100 | Male | 79 | Squamous |
| P101 | Female | 64 | Adenocarcinoma |
| P102 | Male | 75 | Non-small-cell lung cancer |
| P103 | Male | 67 | Adenocarcinoma |
| P104 | Male | 72 | Non-small-cell lung cancer |
| P105 | Male | 79 | Adenocarcinoma |
| P106 | Female | 51 | Non-small-cell lung cancer |
| P107 | Female | 78 | Non-small-cell lung cancer |
| P108 | Male | 58 | Non-small-cell lung cancer |
| P109 | Female | 69 | Adenocarcinoma |
| P110 | Male | 82 | Non-small-cell lung cancer |
| P111 | Male | 76 | Non-small-cell lung cancer |
| P112 | Male | 61 | Adenocarcinoma |
| P113 | Female | 64 | Adenocarcinoma |
| P114 | Female | 69 | Non-small-cell lung cancer |
| P115 | Male | 85 | Adenocarcinoma |
| P116 | Male | 56 | Non-small-cell lung cancer |
| P117 | Female | 62 | Non-small-cell lung cancer |
| P118 | Male | 62 | Squamous |
| P119 | Male | 56 | Squamous |
| P120 | Male | 68 | Squamous |
| P121 | Male | 63 | Adenocarcinoma |
| P122 | Male | 58 | Non-small-cell lung cancer |
| P123 | Male | 64 | Adenocarcinoma |
| P124 | Male | 68 | Non-small-cell lung cancer |
| P125 | Male | 59 | Adenocarcinoma |
| P126 | Male | 67 | Non-small-cell lung cancer |
| P127 | Female | 78 | Non-small-cell lung cancer |
| P128 | Female | 66 | Non-small-cell lung cancer |
| P129 | Female | 67 | Non-small-cell lung cancer |
| P130 | Female | 57 | Non-small-cell lung cancer |
| P131 | Female | 74 | Non-small-cell lung cancer |
| P132 | Male | 55 | Non-small-cell lung cancer |
| P133 | Male | 62 | Squamous |
| P134 | Male | 66 | Squamous |
| P135 | Female | 56 | Non-small-cell lung cancer |
| P136 | Male | 60 | Non-small-cell lung cancer |
| P137 | Male | 81 | Non-small-cell lung cancer |
| P138 | Male | 63 | Non-small-cell lung cancer |
| P139 | Female | 49 | Adenocarcinoma |
| P140 | Male | 56 | Non-small-cell lung cancer |
| P141 | Male | 74 | Non-small-cell lung cancer |
| P142 | Female | 49 | Non-small-cell lung cancer |
| P143 | Male | 65 | Non-small-cell lung cancer |
| P144 | Female | 52 | Adenocarcinoma |
| P145 | Male | 40 | Non-small-cell lung cancer |
| P146 | Male | 66 | Adenocarcinoma |
| P147 | Female | 65 | Small cell lung cancer |
| P148 | Female | 68 | Large cell lung cancer |
| P149 | Male | 41 | Adenocarcinoma |
| P150 | Male | 54 | Adenocarcinoma |
| P151 | Female | 53 | Non-small-cell lung cancer |
| P152 | Male | 76 | Non-small-cell lung cancer |
| P153 | Female | 49 | Non-small-cell lung cancer |
| P154 | Female | 71 | Adenocarcinoma |
| P155 | Male | 69 | Non-small-cell lung cancer |
| P156 | Male | 60 | Adenocarcinoma |
| P157 | Male | 52 | Non-small-cell lung cancer |
| P158 | Female | 68 | Non-small-cell lung cancer |
| P159 | Male | 62 | Adenocarcinoma |
| P160 | Male | 75 | Non-small-cell lung cancer |
| P161 | Male | 65 | Non-small-cell lung cancer |
| P162 | Male | 65 | Non-small-cell lung cancer |
| P163 | Male | 55 | Non-small-cell lung cancer |
| P164 | Male | 68 | Non-small-cell lung cancer |
| P165 | Male | 57 | Adenocarcinoma |
| P166 | Female | 48 | Neuroendocrine |
| P167 | Male | 73 | Adenocarcinoma |
| P168 | Male | 62 | Adenocarcinoma |
| P169 | Female | 70 | Adenocarcinoma |
| P170 | Female | 61 | Non-small-cell lung cancer |
| P171 | Male | 65 | Adenocarcinoma |
| P172 | Male | 75 | Non-small-cell lung cancer |
| P173 | Male | 53 | Non-small-cell lung cancer |
| P174 | Female | 53 | Non-small-cell lung cancer |
| P175 | Male | 75 | Adenocarcinoma |
| P176 | Male | 40 | Non-small-cell lung cancer |
| P177 | Male | 65 | Non-small-cell lung cancer |
| P178 | Female | 67 | Non-small-cell lung cancer |
| P179 | Male | 70 | Non-small-cell lung cancer |
| P180 | Male | 55 | Non-small-cell lung cancer |
| P181 | Female | 68 | Small cell lung cancer |
| P182 | Male | 56 | Adenocarcinoma |
| P183 | Male | 66 | Non-small-cell lung cancer |
| P184 | Female | 70 | Non-small-cell lung cancer |
| P185 | Male | 62 | Squamous |
| P186 | Female | 55 | Adenocarcinoma |
| P187 | Female | 71 | Adenocarcinoma |
| P188 | Female | 63 | Adenocarcinoma |
| P189 | Female | 69 | Non-small-cell lung cancer |
| P190 | Female | 51 | Adenocarcinoma |
| P191 | Female | 46 | Adenocarcinoma |
| P192 | Female | 74 | Non-small-cell lung cancer |
| P193 | Female | 61 | Non-small-cell lung cancer |
| P194 | Male | 47 | Non-small-cell lung cancer |
| P195 | Male | 68 | Squamous |
| P196 | Male | 49 | Non-small-cell lung cancer |
| P197 | Female | 83 | Non-small-cell lung cancer |
| P198 | Female | 66 | Adenocarcinoma |
| P199 | Male | 56 | Squamous |
| P200 | Male | 54 | Non-small-cell lung cancer |
| P201 | Male | 51 | Non-small-cell lung cancer |
| P202 | Female | 70 | Adenocarcinoma |
| P203 | Female | 62 | Non-small-cell lung cancer |
| P204 | Male | 66 | Adenocarcinoma |
| P205 | Male | 56 | Non-small-cell lung cancer |
| P206 | Male | 63 | Adenocarcinoma |
Figure 1.
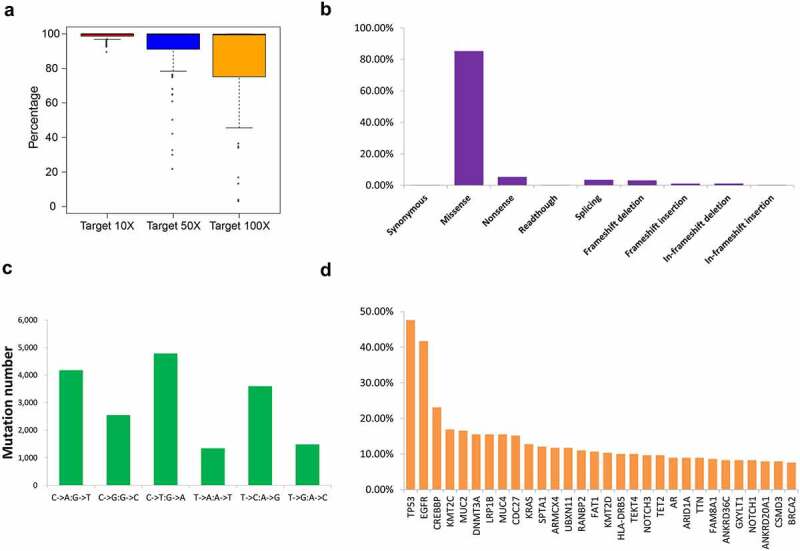
Overview of the mutation status of the 206 patients with NSCLC based on next-generation sequencing. (a) Coverage depth for gene regions. Distribution of gene mutation types (b) and single mutation types (c) in the 206 patients with NSCLC. (d) Schematic showing 30 genes with the highest mutation frequency
A total of 18,749 mutations were identified, and the dominant mutation type was missense mutation (85.3%) (Figure 1(b), Table 2). Single-mutation variation analysis revealed that the dominant base mutations predominantly involved purines (Figure 1(c)) and that C > T/G > A was the most common substitution type. Of the mutated genes, 79 had a mutation frequency of more than 5%. Among these, the top ten most frequently mutated genes were TP53 (47.6%), EGFR (41.7%), CREBBP (23.1%), KMT2C (16.9%), MUC2 (16.6%), DNMT3A (15.5%), LRP1B (15.5%), MUC4 (15.5%), CDC27 (15.2%), and KRAS (12.8%) (Figure 1(d)).
Table 2.
Mutation information of the 206 NSCLC patients
| Type | Number | Percentage |
|---|---|---|
| Synonymous | 32 | 0.17% |
| Missense | 15,985 | 85.26% |
| Nonsense | 996 | 5.31% |
| Readthough | 38 | 0.20% |
| Splicing | 653 | 3.48% |
| Frameshift deletion | 585 | 3.12% |
| Frameshift insertion | 199 | 1.06% |
| In-frameshift deletion | 214 | 1.14% |
| In-frameshift insertion | 47 | 0.25% |
| Total | 18,749 | 100.00% |
TMB analysis in patients with NSCLC
TMB has been proved to be an immunotherapy biomarker in clinical oncology, including NSCLC. To explore the association between TMB and NSCLC in Chinese patients, we performed comparative analysis of the sexes and different tumor subtypes showed that TMB in females was lower than that in males (Figure 2(a)). The median TMB for men is 6.6 Mutations/Mb, and the median TMB for women is 3.7 Mutations/Mb. The median TMB for men is 1.78 times that for women (Figure 2(a)).
Figure 2.
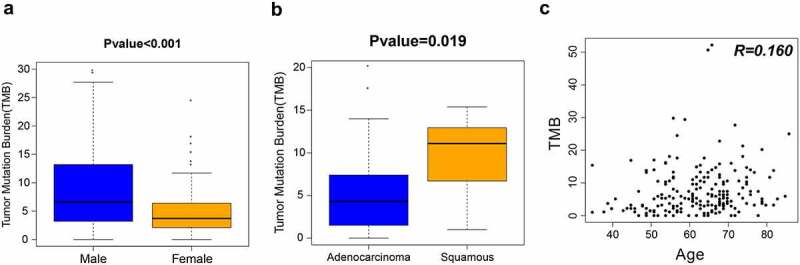
TMB analysis of 206 patients with NSCLC. (a) Differences in TMB by sex. (b) Differences in TMB by tumor type. (c) Correlation between TMB and age. TMB, tumor mutation burden
Significantly higher TMB was observed in squamous carcinoma than that in adenocarcinoma (Figure 2(b)). The median TMB of lung adenocarcinoma is 4.3 Mutations/Mb, and the median TMB of lung squamous is 11.1 Mutations/Mb, 2.58 times that of lung adenocarcinoma (Figure 2(b)).
To investigate the association between TMB and age, we compared TMB (range, 0–52.2 Mutations/Mb; median, 5.3 Mutations/Mb) and patient age (range, 35–86 years; median, 63 years). Correlation analysis showed that the correlation between the two was not significant (correlation coefficient R = 0.160, P = 0.074) (Figure 2(c)).
Analysis of most commonly mutated genes in patients with NSCLC
Gene mutation has been proved to be closely associated with tumor development, and identification of the isoform of gene mutation might benefit therapy. We analyzed the ten most frequently mutated genes in tumor tissues of patients with NSCLC and found that all patients had at least one high-frequency mutation. Of the 206 cases, no KRAS mutation was observed in patients with EGFR mutations (Figure 3). MutationMapper analysis showed that, in addition to DNMT3A, the mutation sites of the other nine high-frequency mutation genes were R249S/M, L858R, Q1950P, R886 C, T1488I/P, S2589, S2704P, C115R, G12 CN/D. Out of these nine, mutant hotspots of TP53 (R249S/M), EGFR (L858R), and KRAS (G12 CN/D) were located P53 DNA-binding domain, Protein tyrosine kinase domain and Ras family domain respectively (Figure 4).
Figure 3.
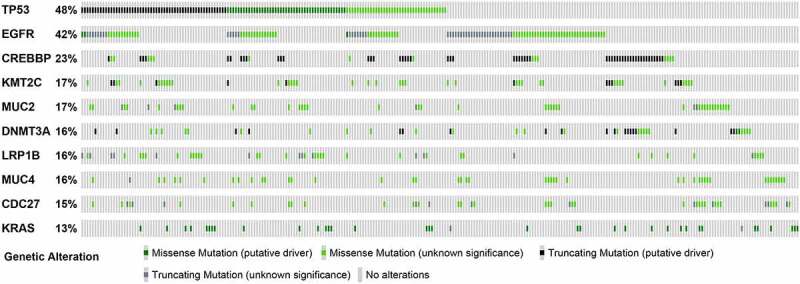
The top 10 high-frequency mutation genes of 206 patients were visualized by OncoPrinter. Each gray box from left to right represents the mutation of a sample
Figure 4.
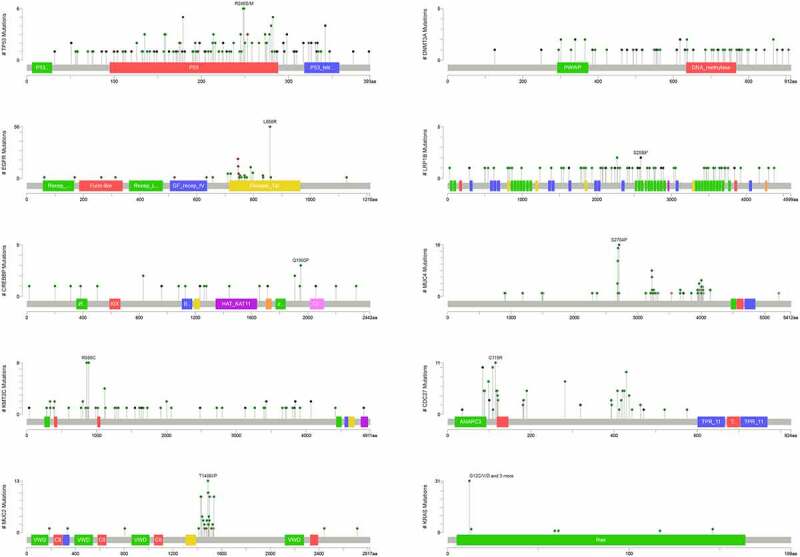
Diagram showing mutant sites and frequency of the top 10 genes harboring high-frequency mutations
GO and KEGG enrichment analyses showed that the top 10 high-frequency mutant genes were mainly enriched in terms of organelle lumen, membrane-enclosed lumen, intracellular organelle lumen, cellular macromolecule metabolic process, aromatic compound biosynthetic process (Figure 5(a)), and pathways including microRNAs in cancer, pathway in cancer, Notch signaling pathway, and FoxO signaling pathway (Figure 5(b)).
Figure 5.
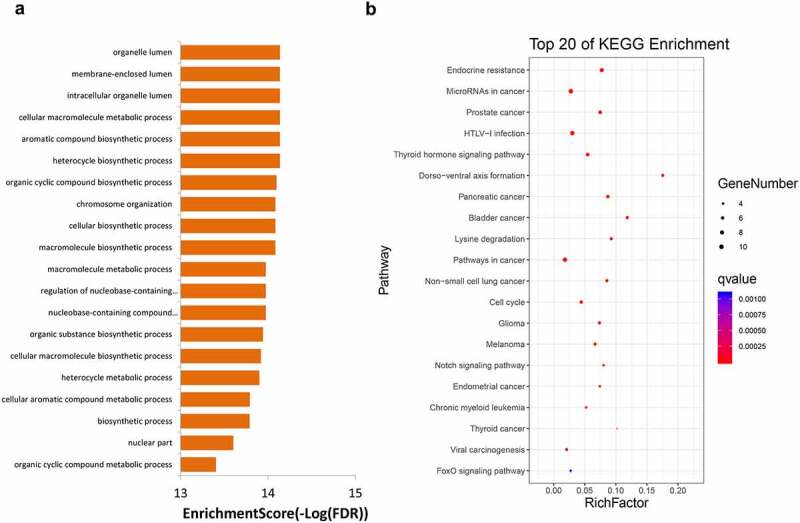
GO (a) and KEGG (b) enrichment analyses for genes with mutation frequency of >5%. GO, Gene Ontology, KEGG, Kyoto Encyclopedia of Genes and Genomes
Analysis of copy number variations in patients with NSCLC
Because CNV may indicate dysregulated gene and protein expression that may ultimately affect development and progression of NSCLC, we further explored gene CNV in Chinese patients with NSCLC. CNV analysis showed that 110 genes had copy number amplification. Among these, BCORL1, ARAF, GATA binding protein 1 (GATA1), bruton tyrosine kinase (BTK), and P21 (RAC1) activated kinase 3 (PAK3) were the genes with the highest copy number amplification (Figure 6(a)). These genes are mainly concentrated in the terms of protein binding, positive regulation of macromolecule metabolic process, regulation of cellular process, positive regulation of metabolic process, and regulation of macromolecule metabolic process (Figure 6(b)). KEGG analysis revealed that, for the genes with the highest copy number amplification, their predicted functions were enriched in transcriptional dysregulation in cancer, pathway in cancer, PI3K-Akt signaling pathway, and Ras signaling pathway (Figure 6(c)).
Figure 6.
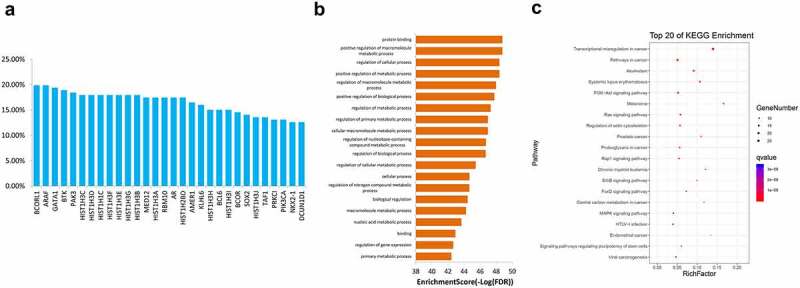
Of enrichment analysis of the 30 genes (a) and their GO (b) and KEGG enrichment analysis results (c) with the highest copy number increase in 206 samples. GO, Gene Ontology, KEGG, Kyoto Encyclopedia of Genes and Genomes
A total of 54 genes had copy number deletion. The genes with the highest copy number deletions were KDM6A, RBM10, TATA-box binding protein associated factor 1 (TAF1), ARAF, and stromal antigen 2 (STAG2) (Figure 7(a)). They were predicted to be enriched in terms of cellular macromolecule metabolic process, macromolecule modification, regulation of cellular process, macromolecule metabolic process, and cellular protein modification process (Figure 7(b)). The most enriched pathways were pathway in cancer, PI3K-Akt signaling pathway, and cell cycle (Figure 7(c)).
Figure 7.
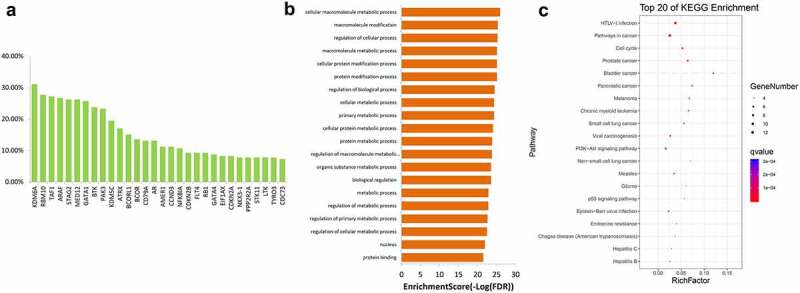
The top 30 genes with the highest copy number deletion in 206 samples (a) and their GO (b) and KEGG (c) enrichment analysis results. GO, Gene Ontology, KEGG, Kyoto Encyclopedia of Genes and Genomes
Discussion
The purpose of this study was to identify the mutational characteristics of 206 Chinese patients with NSCLC. We identified 18,749 mutations by using targeted NGS. Among these mutations, missense mutations were dominant. Base mutations were dominated by pyrimidine and purine conversions. The ten most frequently mutated genes were obtained. Notably, EGFR and KRAS mutations were mutually exclusive. There were differences in TMB between the sexes and pathological subtypes; however, TMB was not associated with age. Finally, 110 genes and 54 genes showed copy number amplification and copy number deletion, respectively. These genes were specifically enriched in the NSCLC-associated pathways.
Based on the targeted NGS, we determined the most frequently mutated genes in Chinese patients with NSCLC. These genes were TP53, EGFR, CREBBP, KMT2C, MUC2, DNMT3A, LRP1B, MUC4, CDC27, and KRAS. Mutations in these genes have been reported previously in NSCLC [20]. Interestingly, the genes with the highest mutation frequency differed in their rankings compared with the findings of a study on the American population. In the study, they showed that the most frequently mutated gene in this report is KRAS, followed by EGFR [10]. However, our results are also consistent with the results in some reports. For example, a study in Lebanon showed that mutations of TP53 are common molecular changes, occurring in over 50% of tumors [21,22]. In an NSCLC study with a small sample size, TP53 was also found to be the most frequently mutated gene in the Chinese population [15]. These indicate that TP53 mutation might be one of the genes affected in Chinese patients with NSCLC. In addition, our results also support the idea reported in a previous study that the mutant hotspot area of TP53 is located in the common R249 area [23]. It has been accepted that TP53 is an important tumor suppressor and the most commonly mutated gene in most cancers. As a prognostic factor in NSCLC, the presence of TP53 mutation suggested an aggressive feature and poor clinical outcome [24].
Our results show that EGFR ranks second in terms of the mutation frequency, at a rate of 41.7%. Based on previous studies, the mutation rate of EGFR in Chinese patients with NSCLC is approximately 30%–50% [23,25]. The frequency of EGFR mutations that we obtained is also in this . It is worth mentioning that we found the hotspot mutation L858R of the EGFR gene, which is also considered to be a high-frequency mutation in Asia [26,27]. There is evidence that patients harboring common EGFR mutations exhibit approximately 10 months progression free survival time after EGFR tyrosine kinase inhibitor (TKI) therapy, whereas those with uncommon EGFR mutations exhibit less response to EGFR TKI [28–30]. Therefore, our findings indicate that most Chinese patients with NSCLC might benefit from EGFR TKI treatment. However, in those NSCLC harboring dual TP53/EGFR mutations, especially missense mutations, low response is frequently observed [31]. In addition to TP53 and EGFR, KRAS is also a commonly mutated gene in NSCLC. In some reports, it is described that the frequency of conversion of KRAS in the Chinese is approximately 8% [25,32]. Here, we report a mutation rate of the KRAS gene of 12.8% [33].
In contrast to the widely reported high-frequency mutated genes mentioned above, CREBBP (23.1%), KMT2C (16.9%), MUC2 (16.6%), DNMT3A (15.5%), LRP1B (15.5%), MUC4 (15.5%), and CDC27 (15.2%) are currently reported less in the Chinese population, although mutations in DNMT3A and KMT2C have been identified in some studies [20,33–35]. Our results suggest some aspects of the mutational characteristics of these genes in Chinese NSCLC, suggesting functions of these genes in the etiology and treatment of NSCLC. It is worth mentioning that we observed that patients with NSCLC having EGFR mutations have no KRAS mutations, and vice versa. This is consistent with the previous assertion that EGFR and KRAS mutations are mutually exclusive in NSCLC, although some cases of EGFR and KRAS mutations being present together in some Asian populations, including in China, have been reported [25,36].
The genome in NSCLC is unstable and exhibits a wide range of gene CNVs. Because CNV is closely related to the expression of mRNA and protein, copy number amplification or deletion may affect the expression of tumor-related genes and the patient’s sensitivity to treatment and survival [37]. Analysis of the variation of copy number is helpful for learning underlying mechanisms and functions of related genes in patients with NSCLC. Our results show that the genes with the most increased copy number were BCORL1, ARAF, and GATA1, while those with the greatest deletion of copy number were KDM6A, RBM10, TAF1, ARAF, and STAG2. Among these genes, evidence suggests that patients with high expression of BCORL1 have a shorter 3-year survival than patients with its low expression [38]. In addition, RBM10 functions to inhibiting the proliferation of non-adenocarcinoma cells [39]. We speculate that the increase in BCORL1 copy number and deletion of RBM10 copy number may suggest their roles in the pathogenesis of NSCLC.
The results of GO and KEGG enrichment analyses of genes with frequent mutations and CNV suggest that the mutant genes are enriched in tumor-related terms and signaling pathways. These pathways include the PI3K-Akt signaling pathway, FoxO signaling pathway, and Ras signaling pathway. The correlation between activation of the Notch signaling pathway and poor prognosis of NSCLC has been confirmed [40,41]. PI3K-Akt is an important signaling pathway that regulates tumor formation, survival and metastasis [42,43]. One of its downstream factors is the FoxO signaling pathway. Akt promotes the phosphorylation of FoxO and inhibits the transcriptional function of FoxO, potentially resulting in the induction of apoptosis, which is involved in biological processes such as NSCLC radiosensitization and tumor growth inhibition [44–46]. Moreover, the Ras signaling pathway is a proto-cancer pathway. Multiple tumor-promoting factors and drugs have been found to modulate tumor progression through this pathway [47–49]. Based on KEGG analysis, we suggest that the high frequency of mutation genes and CNV genes are associated with these tumor-related pathways. Inhibitors targeting these pathways may thus have clinical significance.
It is interesting to find that TMB was higher in men than in women. Since we were unable to correlate the current data such as TMB with the treatment outcomes of men and women, the clinical prognostic value of genetic mutations could not be derived. Subsequent research on the links between the mutant genes and the clinical data of this patient population will further enrich the clinical value of the mutant gene.
Conclusion
The most common gene mutations in Chinese patients with NSCLC are missense mutations, and TP53, EGFR, CREBBP, KMT2C, MUC2 genes are the most frequently mutated genes. Several genes exhibited copy number amplification and copy number deletion. There were differences in TMB between the sexes and pathological subtypes; however, TMB was not associated with age. Our findings indicate that the panel is a good method for tumor molecular characterization In addition, our results are expected to provide clues for interpreting the etiology of NSCLC and performing drug target screening for this condition.
Funding Statement
The work was supported by Research and Development Projects in Key Areas of Guangdong Province (No. 2020B0404010002); Guangzhou science and technology plan project under grant number 201802020004.
Highlights
The TP53 gene occurs with the highest frequency in 206 Chinese patients with NSCLC.
TMB is higher in males than in females.
EGFR and KRAS mutations are mutually exclusive.
Genes with copy number variations are enriched in cancer-associated pathways.
Disclosure statement
The authors declare that they have no conflicts of interest.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- [1].Seigel R, Naishadham D, Jemal A.. Cancer statistics, 2014. Ca Cancer J Clin. 2014;64(1):9–29. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- [3].Wang L, Zhao D, Qin K, et al. Effect and biomarker of Nivolumab for non-small-cell lung cancer. Biomed Pharmacother. 2019;117:109199. [DOI] [PubMed] [Google Scholar]
- [4].Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. [DOI] [PubMed] [Google Scholar]
- [6].Han SW, Kim HP, Shin JY, et al. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS One. 2013;8(5):e64271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cai H, Jing C, Chang X, et al. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J Transl Med. 2019;17(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ng TS, Rajadurai P, Cheah YK. Molecular profiling of genetic alterations in selected non-small cell lung cancer. J Transdisciplin Biomed. 2017;1(1):47–52. [Google Scholar]
- [9].Lee W, Jiang Z, Liu J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465(7297):473–477. [DOI] [PubMed] [Google Scholar]
- [10].Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121(4):631–639. [DOI] [PubMed] [Google Scholar]
- [11].Araujo LH, Lammers PE, Matthews-Smith V, et al. Somatic mutation spectrum of non-small-cell lung cancer in African Americans: a pooled analysis. J Thorac Oncol. 2015;10(10):1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tsoulos N, Papadopoulou E, Metaxa-Mariatou V, et al. Tumor molecular profiling of NSCLC patients using next generation sequencing. Oncol Rep. 2017;38(6):3419–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wen S, Dai L, Wang L, et al. Genomic signature of driver genes identified by target next-generation sequencing in Chinese non-small cell lung cancer. The Oncologist. 2019;24(11):e1070–e1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiong D, Li G, Li K, et al. Exome sequencing identifies MXRA5 as a novel cancer gene frequently mutated in non-small cell lung carcinoma from Chinese patients. Carcinogenesis. 2012;33(9):1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Amarasinghe KC, Li J, Hunter SM, et al. Inferring copy number and genotype in tumour exome data. BMC Genomics. 2014;15(1):732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Frank R, Scheffler M, Michels SYF, et al. KEAP1-mutations in patients with non-small cell lung cancer (NSCLC). J Clin Oncol. 2015;33(15_suppl):8097. [Google Scholar]
- [21].Wong KM, Hudson TJ, McPherson JD. Unraveling the genetics of cancer: genome sequencing and beyond. Annu Rev Genomics Hum Genet. 2011;12(1):407–430. [DOI] [PubMed] [Google Scholar]
- [22].Shajani-Yi Z, de Abreu FB, Peterson JD, et al. Frequency of somatic TP53 mutations in combination with known pathogenic mutations in colon adenocarcinoma, non-small cell lung carcinoma, and gliomas as identified by next-generation sequencing. Neoplasia. 2018;20(3):256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vavala T, Monica V, Lo Iacono M, et al. Precision medicine in age-specific non-small-cell-lung-cancer patients: integrating biomolecular results into clinical practice-a new approach to improve personalized translational research. Lung Cancer. 2017;107:84–90. [DOI] [PubMed] [Google Scholar]
- [24].Hainaut P, Olivier M, Pfeifer GP. TP53 mutation spectrum in lung cancers and mutagenic signature of components of tobacco smoke: lessons from the IARC TP53 mutation database. Mutagenesis. 2001;16(6):551–553. author reply 555-556. [DOI] [PubMed] [Google Scholar]
- [25].Li S, Li L, Zhu Y, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110(11):2812–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. [DOI] [PubMed] [Google Scholar]
- [27].Xu J, He J, Yang H, et al. Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell lung cancer. Cancer Biomark. 2011;10(2):63–69. [DOI] [PubMed] [Google Scholar]
- [28].Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. [DOI] [PubMed] [Google Scholar]
- [29].Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. [DOI] [PubMed] [Google Scholar]
- [30].Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9(2):154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer. 2017;111:23–29. [DOI] [PubMed] [Google Scholar]
- [32].Wang R, Zhang Y, Pan Y, et al. Comprehensive investigation of oncogenic driver mutations in Chinese non-small cell lung cancer patients. Oncotarget. 2015;6(33):34300–34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang J, Li C, Wan F, et al. The rs1550117 A>G variant in DNMT3A gene promoter significantly increases non-small cell lung cancer susceptibility in a Han Chinese population. Oncotarget. 2017;8(14):23470–23478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yang B, Li J, Li F, et al. Comprehensive analysis of age-related somatic mutation profiles in Chinese young lung adenocarcinoma patients. Cancer Med. 2019;8(4):1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou Q, Peng X, Wang W, et al. P2. 04-20 TP53/KMT2C co-mutation as a novel biomarker for immunotherapy in non-small cell lung cancer patients. J Thoracic Oncol. 2019;14(10):S716. [Google Scholar]
- [36].Lee T, Lee B, Choi YL, et al. Non-small cell lung cancer with concomitant EGFR, KRAS, and ALK mutation: clinicopathologic features of 12 cases. J Pathol Transl Med. 2016;50(3):197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jabs V, Edlund K, Konig H, et al. Integrative analysis of genome-wide gene copy number changes and gene expression in non-small cell lung cancer. PLoS One. 2017;12(11):e0187246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhu XH, Tian J. The expression of BCL6 co-inhibitory factor-like protein 1 in the tissues of elderly patients with non-small cell lung cancer and its effect on long-term prognosis. Chin J Gerontol. 2018;12:27. [Google Scholar]
- [39].Bechara EG, Sebestyen E, Bernardis I, et al. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52(5):720–733. [DOI] [PubMed] [Google Scholar]
- [40].Yuan X, Wu H, Xu H, et al. Meta-analysis reveals the correlation of Notch signaling with non-small cell lung cancer progression and prognosis. Sci Rep. 2015;5(1):10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Galluzzo P, Bocchetta M. Notch signaling in lung cancer. Expert Rev Anticancer Ther. 2011;11(4):533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang C, Lan T, Hou J, et al. NOX4 promotes non-small cell lung cancer cell proliferation and metastasis through positive feedback regulation of PI3K/Akt signaling. Oncotarget. 2014;5(12):4392–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lai S, Wang G, Cao X, et al. EMP-1 promotes tumorigenesis of NSCLC through PI3K/AKT pathway. J Huazhong Univ Sci Technolog Med Sci. 2012;32(6):834–838. [DOI] [PubMed] [Google Scholar]
- [44].Zhang X, Tang N, Hadden TJ, et al. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta. 2011;1813(11):1978–1986. [DOI] [PubMed] [Google Scholar]
- [45].Chen G, Yu L, Dong H, et al. MiR‐182 enhances radioresistance in non‐small cell lung cancer cells by regulating FOXO 3. Clin Exp Pharmacol Physiol. 2019;46(2):137–143. [DOI] [PubMed] [Google Scholar]
- [46].Liu H, Zhou BH, Qiu X, et al. T63, a new 4-arylidene curcumin analogue, induces cell cycle arrest and apoptosis through activation of the reactive oxygen species-FOXO3a pathway in lung cancer cells. Free Radic Biol Med. 2012;53(12):2204–2217. [DOI] [PubMed] [Google Scholar]
- [47].Wu D, Zhao B, Qi X, et al. Nogo-B receptor promotes epithelial-mesenchymal transition in non-small cell lung cancer cells through the Ras/ERK/Snail1 pathway. Cancer Lett. 2018;418:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zheng G, Shen Z, Chen H, et al. Metapristone suppresses non-small cell lung cancer proliferation and metastasis via modulating RAS/RAF/MEK/MAPK signaling pathway. Biomed Pharmacother. 2017;90:437–445. [DOI] [PubMed] [Google Scholar]
- [49].Sun W, Ping W, Tian Y, et al. MiR-202 enhances the anti-tumor effect of cisplatin on non-small cell lung cancer by targeting the Ras/MAPK pathway. Cell Physiol Biochem. 2018;51(5):2160–2171. [DOI] [PubMed] [Google Scholar]


