ABSTRACT
The prognostic significance of miR-24 in tumors has not been determined. Therefore, we conducted a meta-analysis to systematically assess the correlation between miR-24 and its prognostic value in cancers PubMed, EMBASE, and Web of Science databases were used to search relevant articles (up to 1 October 2020). Studies that evaluated the prognostic value of miR-24 in tumors were included. The hazard ratio (HR) and odds ratio (OR) with 95% confidence intervals (CI) were used to evaluate survival outcomes and clinical characteristics. All data analyses were implemented using STATA 12.0 software. A total of 17 studies from 15 articles involving 1705 patients were collected for the meta-analysis. The pooled analysis revealed that elevated miR-24 expression was obviously associated with poor overall survival (OS) (HR = 1.66, 95% CI: 1.20–2.31). Furthermore, we also found that elevated miR-24 expression was positively correlated with tumor size (large or small) and tumor stage (III–IV vs I–II). Elevated miR-24 expression indicates poor prognosis and may be a promising prognostic marker in different cancers. Our findings needed to be verified through further investigations.
KEYWORDS: mir-24, prognosis, cancer, meta-analysis
Introduction
In recent years, cancer incidence has been on the rise, and studies investigating the molecular mechanisms involved have intensified. MicroRNA (miRNA) is considered to be closely associated with tumor progression. These small-molecule miRNAs are not directly involved in protein synthesis, but can affect mRNA-mediated gene expression by binding to the 3ʹ non-coding region of specific mRNA. Abnormal regulation of the expression of proto-oncogenes and tumor suppressor genes by miRNA plays an important part in tumorigenesis. Accumulated evidence discloses that miRNAs are indispensable in cancer progression and many different miRNAs have been applied to tumor diagnosis and prognosis [1,2].
MiR-24 is an important member of the miRNA family. It is located in two different gene clusters. One is the miR-23b/miR-27b/miR-24-1 gene cluster, located in the expression sequence region of chromosome 9 [3]. The other gene cluster is miR-23a/miR-27a/miR-24-2, located in the intergenic region of chromosome 19 [4]. Both share a common activated region miR-24-3p. MiR-24 is considered as a tumor-promoting factor and is abnormally expressed in many tumors. Studies have revealed that miR-24 involves in tumor cell proliferation, differentiation, invasion, and metastasis by regulating numerous genes, such as PTEN, TRIM11, SOX7, PRKCH, and P16 [5–9]. Recently, abnormal miR-24 expression has been reported to be correlated with the prognosis and clinical characteristics in tumors, including gastric cancer (GC), lung cancer (LC), colorectal cancer (CRC), acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), hepatocellular carcinoma (HCC), nasopharyngeal carcinoma (NPC), and tongue squamous cell carcinoma (TSCC) [10–24]. Most studies reported that high miR-24 expression predicted poor prognosis, while several other studies shown the opposite view. The results are controversial.
Nowadays, the prognostic value of miR-24 in tumors is not clear. Therefore, in this study, we have summarized the existing literatures and conducted a meta-analysis to comprehensively assess the prognostic significance of miR-24 in tumors.
Methods
Literature search
PubMed, Web of Science and EMBASE were used to conduct comprehensive search for eligible studies (up to 1 October 2020). The following terms were applied for the search strategy: ‘miR-24’ or ‘microRNA-24’ or ‘miRNA-24’ and ‘cancer’ or ‘carcinoma’ or ‘tumor’ or ‘tumour’ or ‘neoplasm’ and ‘prognosis’ or ‘survival’ or ‘prognostic’ or ‘outcome’. No limitation on language was applied to retrieve publications.
Study selection
The inclusion criteria for the studies were: 1) patients enrolled were pathologically diagnosed; 2) the expression of miR-24 and the prognosis outcomes of cancer patients were investigated; 3) the hazard ratio (HRs) with their 95% confidence interval (CI) of survival outcomes were directly reported or could be indirectly extracted from survival curves. The exclusion criteria were: 1) publications that were reviews, letters, meeting abstracts, animal studies, and non-comparative studies; 2) studies focusing on polymorphisms or methylation patterns of miR-24; 3) studies with insufficient data to retrieve HRs with CIs of survival outcomes; and 4) duplicated publications.
Data extraction and quality assessment
Two independent investigators extracted all data, and the third investigator resolved any disagreements. The following information was collected from the enrolled studies: the name of first author, publication country, publication year, tumor type, number of patients, miRNA type, sample source for miRNA detection, detection method, HRs with CIs of survival outcomes including overall survival (OS), disease-free survival (DFS), recurrence-free survival (RFS), and progression-free survival (PFS). For studies only providing survival curves, Tierney’s method was used to extract HRs and 95% CIs [25]. The quality of included studies was evaluated according to the Newcastle Ottawa scale (NOS) [26]. The NOS score ranges from 0 to 9, and studies with a NOS score >6 were defined as high quality.
Statistical analysis
Pooled HRs with 95% CIs were applied to assess the influence of miR-24 expression on cancer prognosis. The heterogeneity of pooled HRs was estimated by Cochran’s Q test and Higgins I2 statistic [27]. P-value <0.05 or I2 > 50% was defined as significant heterogeneity, and the random-effects model was used to pool HRs [28]. For all other instances, the fixed-effect model was used. The stability of the pooled HRs was evaluated by sensitivity analysis. In addition, subgroup analyses were also conducted. Begg’s and Egger’s tests were used to evaluate the publication bias, and P-value <0.05 was considered as significant publication bias [29,30]. The trim-and-fill method was used to assess the robustness of the meta-analysis results in the presence of publication bias [31]. P < 0.05 is considered statistically significance [30]. All the statistical analyses for this meta-analysis were executed in STATA version 12.0 (Stata Corporation, College Station, TX, United States).
Results
Brief introduction
The prognostic value of miR-24 in tumors has not been determined. Therefore, we present a meta-analysis to clarify the prognostic value of miR24 in various cancers. First, we searched the designated database to find the literatures on the prognostic value of miR-24 in tumors, and then extracted relevant data for meta-analysis. We discovered that elevated miR-24 expression was significantly associated with OS, tumor size (large or small) and tumor stage (III–IV vs I–II).
Literature selection
Articles investigating the prognostic value of miR-24 in tumors were searched and retrieved from the electronic databases. Six hundred and ninety-eight articles were initially collected. After excluding 302 duplicates, 396 articles remained. After trimming 366 irrelevant articles, 30 articles were examined. After screening the full text of these articles, 15 articles were excluded. Eventually, 15 articles published between 2012 and 2018 were included in the study [10–24]. The study flow chart is shown in Figure 1.
Figure 1.
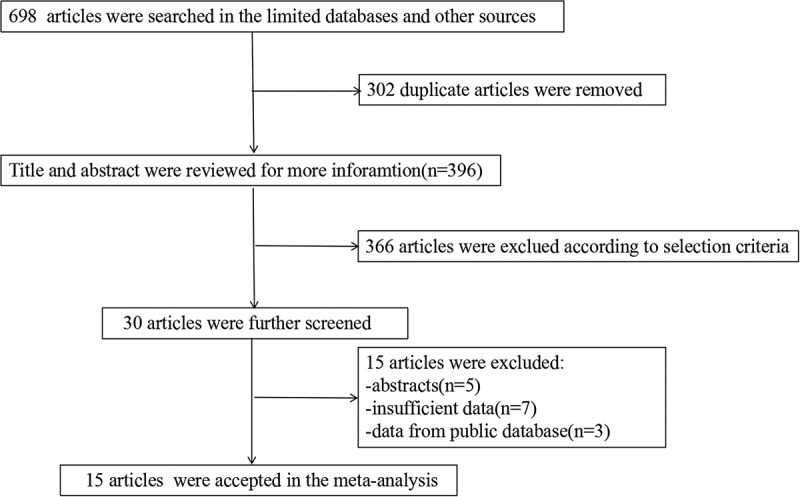
Flow chart of the literature search
Study characteristics
A total of 17 studies from 15 articles involving 1705 patients were enrolled in the study. Fourteen studies reported OS data, and seven studies reported DFS/PFS/RFS. One article was from Mexica, and all other articles originated from China. Four articles reported survival results in the multivariate analysis and 11 articles performed univariate analysis. All articles used RT-qPCR to detect the expression of miR-24. Two studies tested the expression of miR-24 in serum, and 15 studies tested the expression of miR-24 in tissues. The tumor types included were GC [10,19], CRC [11,13], leukemia [12,20], TSCC [22,23], HCC [15,17], LC [21,24], NPC [18], and osteosarcoma [16]. The average NOS score result was 6.5. Basic information about the included studies is displayed in Table 1.
Table 1.
The basic information of included studies
| Study | Country | Tumor type | No. of patients | microRNA type | Detected sample | Detected method | Analysis type | Survival analysis | Source of HR | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Dong 2018 | China | GC | 247 | miR‑24 | Tissue specimens | RT-qPCR | Multivariate | OS | Reported | 7 |
| Gao 2015 | China | CRC | 175 | miR‑24-3p | Tissue specimens | RT-qPCR | Multivariate | OS | Reported | 7 |
| Organista-Nava 2015A | Mexico | ALL | 111 | miR-24 | Tissue specimens | RT-qPCR | Univaritae | OS | K-M curves | 7 |
| Organista-Nava 2015B | Mexico | AML | 36 | miR-24 | Tissue specimens | RT-qPCR | Univaritae | OS | K-M curves | 7 |
| Kerimis 2017 | Greece | CRC | 182 | miR‑24-3p | Tissue specimens | RT-qPCR | Multivariate | OS, DFS | Reported | 7 |
| Le 2012 | China | LC | 82 | miR‑24 | Serum | RT-qPCR | Multivariate | OS | Reported | 7 |
| Liu 2014 | China | HCC | 207 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | OS, RFS | Reported | 7 |
| Liu 2018 | China | Osteosarcoma | 84 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | OS | K-M curves | 7 |
| Meng 2014 | China | HCC | 72 | miR‑24-3p | Tissue specimens | RT-qPCR | Multivariate | OS, DFS | Reported | 7 |
| Su 2018 | China | NPC | 23 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | PFS | K-M curves | 6 |
| Wu 2017 | China | GC | 28 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | OS | K-M curves | 7 |
| Yin 2015 | China | AML | 84 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | OS, RFS | K-M curves | 7 |
| Zhao 2015A | China | LC | 53 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | RFS | K-M curves | 6 |
| Zhao 2015B | China | LC | 67 | miR‑24 | Serum | RT-qPCR | Univaritae | RFS | K-M curves | 6 |
| Zhao 2016 | China | TSCC | 84 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | OS | K-M curves | 6 |
| Zhao 2017 | China | TSCC | 90 | miR‑24 | Tissue specimens | RT-qPCR | Multivariate | OS | K-M curves | 7 |
| Zhou 2018 | China | LC | 50 | miR‑24 | Tissue specimens | RT-qPCR | Univaritae | OS | Reported | 7 |
Abbreviation: GC: gastric cancer; LC: lung cancer; CRC: colorectal cancer; ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; HCC: hepatocellular carcinoma; NPC: nasopharyngeal carcinoma; TSCC: tongue squamous cell carcinoma.
Association between elevated miR-24 expression and OS
Due to the significant heterogeneity (I2 = 80.3%), the random-effects model was used. The results indicated that elevated miR-24 expression was closely related to OS (Figure 2). Elevated miR-24 expression revealed poor OS (HR = 1.66, 95% CI: 1.20–2.31).
Figure 2.
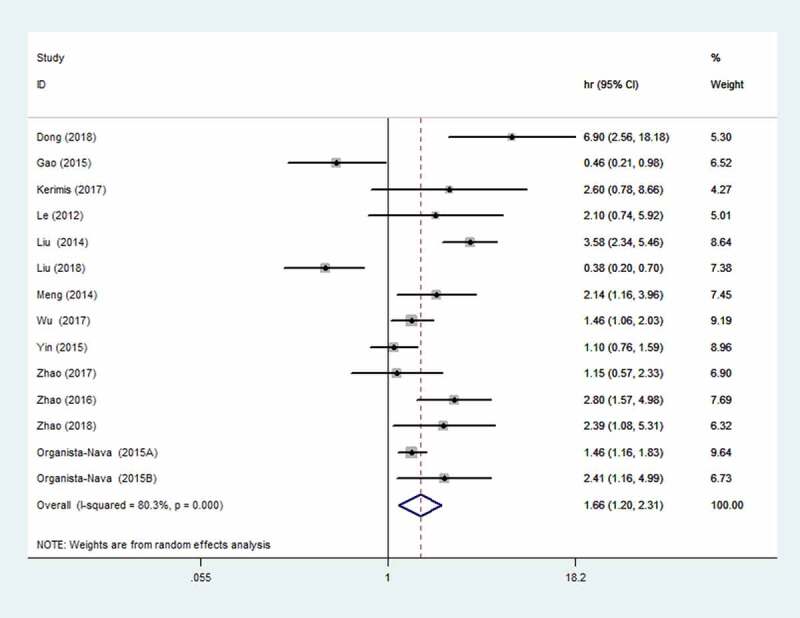
Forest plot of the association between elevated miR-24 expression and OS
Subgroup and regression analysis
In order to better detect the predictive value of miR-24, we performed subgroup analysis based on cancer type, analysis type, race, source of HR, sample size, detected sample, and miRNA type (Table 2). We observed a significant correlation between miR-24 and OS for the following subgroups: ALL, HCC, LC, univariate analysis, White race, reported, sample size <100, detection sample (tissue), and detection of miR-24. For other subgroups analyzed, no significant statistical differences were found.
Table 2.
Subgroup analysis for OS
| Stratifed analysis |
No. of studies |
Pooled HR (95%CI) |
P-value |
Heterogeneity |
||
|---|---|---|---|---|---|---|
| I2 (%) | P-value | Model | ||||
| Cancer type | ||||||
| GC | 2 | 2.96 (0.65–13.49) | 0.161 | 88.5 | 0.003 | Random |
| CRC | 2 | 1.02 (0.19–5.56) | 0.981 | 82.5 | 0.017 | Fixed |
| leukemia | 3 | 1.40 (1.17–1.69) | 0 | 48.8 | 0.142 | Fixed |
| TSCC | 2 | 1.84 (0.77–4.38) | 0.167 | 72.6 | 0.056 | Random |
| HCC | 2 | 3.03 (2.14–4.30) | 0 | 45.1 | 0.177 | Fixed |
| LC | 2 | 2.28 (1.21–4.29) | 0.011 | 0 | 0.843 | Fixed |
| Osteosarcoma | 1 | 0.38 (0.20–0.70) | ||||
| Analysis type | ||||||
| Univariate | 8 | 1.62 (1.10–2.39) | 0.015 | 84.3 | 0 | Random |
| Multivariate | 6 | 1.79 (0.87–3.65) | 0.112 | 76.7 | 0.001 | Random |
| Race | ||||||
| Caucasian | 3 | 1.55 (1.25–1.92) | 0 | 16.3 | 0.303 | Fixed |
| Asian | 11 | 1.61 (1.04–2.49) | 0.034 | 84.3 | 0 | Random |
| Source of HR | ||||||
| Reported | 7 | 2.01 (1.07–3.78) | 0.03 | 79.9 | 0 | Random |
| SC | 7 | 1.43 (0.99–2.05) | 0.056 | 78.5 | 0 | Fixed |
| Sample size | ||||||
| ≥100 | 5 | 2.04 (0.98–4.22) | 0.055 | 88 | 0 | Random |
| <100 | 9 | 1.50 (1.03–2.19) | 0.034 | 73.7 | 0 | Fixed |
| detected sample | ||||||
| Tissue | 13 | 1.64 (1.17–2.31) | 0.004 | 82.3 | 0 | Random |
| Serum | 1 | 2.095 (0.741–5.923) | ||||
| miRNA type | ||||||
| miR-24 | 11 | 1.76 (1.23–2.50) | 0.002 | 81.6 | 0 | Random |
| miR-24-3p | 3 | 1.32 (0.43–4.05) | 0.629 | 81.8 | 0 | Random |
Association between elevated miR-24 expression and DFS/PFS/RFS
Seven studies involving 688 patients reported DFS/PFS/RFS. The result revealed that the elevated miR-24 expression was significantly correlated with poor DFS/PFS/RFS (HR = 1.75, 95% CI: 1.17–2.62) (Figure 3). In addition, we also analyzed DFS and RFS data separately. We found that elevated miR-24 expression predicted poor DFS (HR = 2.31; 95% CI: 1.31–4.07) and RFS (HR = 2.02, 95% CI: 1.02–3.98).
Figure 3.
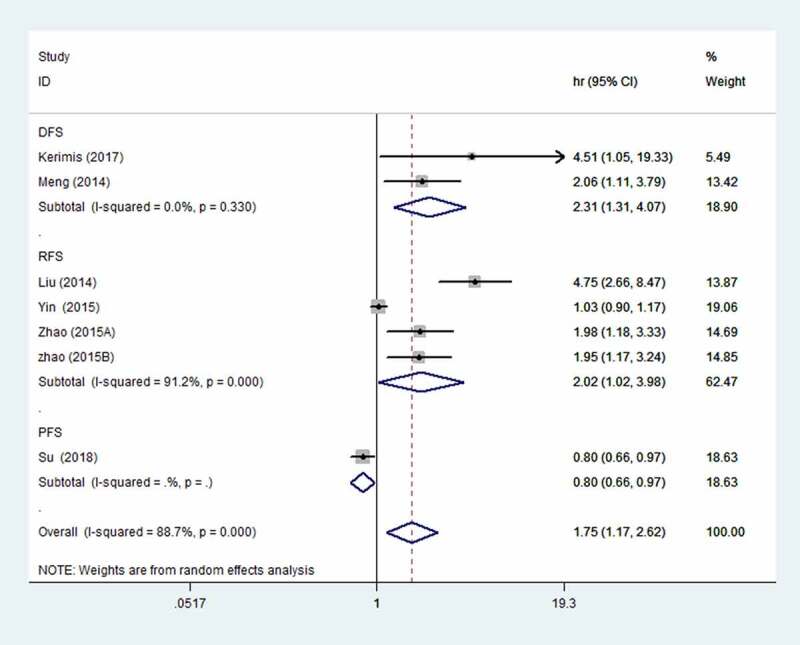
Forest plot of the association between elevated miR-24 expression and DFS/PFS/RFS
Elevated miR-24 expression and clinicopathological features
To investigate the relationship between elevated miR-24 expression and clinicopathological features, we extracted clinical data including gender, age, tumor diameter, tumor stage, and lymph node status from the published studies (Table 3). The pooled OR suggested that elevated miR-24 expression was positively correlated with tumor diameter (large or small)(OR: 1.25, 95 CI%: 1.04–2.15) and tumor stage (III–IV vs I–II) (OR: 2.41, 95 CI%: 1.53–2.90).
Table 3.
Relationship between elevated miR-24 expression and clinicopathological features
| Clinicopathologic features | No. of studies | Estimate OR (95%CI) | p-value | Heterogeneity | ||
|---|---|---|---|---|---|---|
| I2(%) | p-Value | Model | ||||
| Gender (Male vs Female) | 7 | 1.10 (0.73–1.65) | 0.658 | 0 | 0.603 | Fixed |
| Age (Old vs Young) | 7 | 0.84 (0.59–1.22) | 0.366 | 0 | 0.902 | Fixed |
| Tumor diamter (Big vs Small) | 5 | 1.25 (1.04–2.15) | 0 | 0 | 0.216 | Fixed |
| Tumor stage ((III–IV vs I–II) | 4 | 2.41 (1.53–2.90) | 0.002 | 30.8 | 0.014 | Fixed |
| Lymph node status (Yes vs No) | 2 | 0.89 (0.42–1.66) | 0.595 | 0 | 0.765 | Fixed |
Sensitivity analysis
Sensitivity analysis was conducted by excluding each study in turn to explore the stability of the results. The results did not differ significantly from the overall analysis (Figures 4 and Figures 5), revealing that the outcomes were stable.
Figure 4.
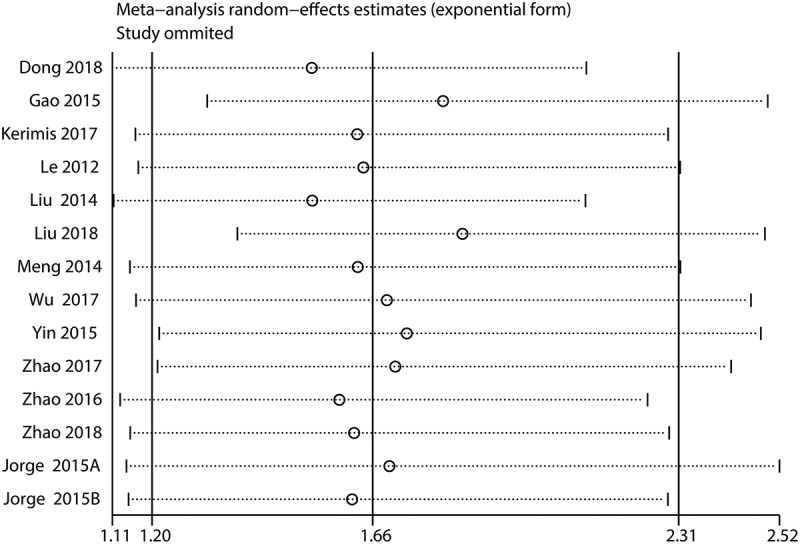
Sensitivity analysis for OS
Figure 5.
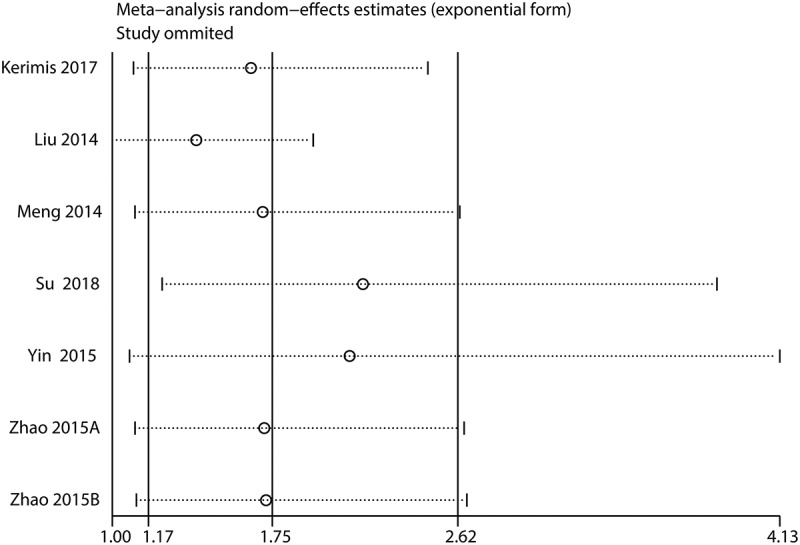
Sensitivity analysis for DFS/PFS/RFS
Publication bias
To determine whether there was a publication bias, funnel plots and Egger’s and Begg’s tests were used to test the publication bias. The funnel plots of OS and DFS/PFS/RFS were almost symmetrical. P-values for the Begg’s and Egger’s tests for OS were 0.743 and 0.609, respectively. The P-value was >0.05, which indicated there was no significant bias for OS (Figure 6). The P-values for the Begg’s and Egger’s Tests for DFS/PFS/RFS were 0.548 and 0.028, respectively, indicating that there was a certain bias (Figure 7(a)). Therefore, we adopted the trim-and-fill method for further analysis, and the results showed that the pooled HR for DFS/PFS/RFS was 1.357 (95%CI: 0.914–2.014), which confirmed the results were affected by publication bias (Figure 7(b)).
Figure 6.
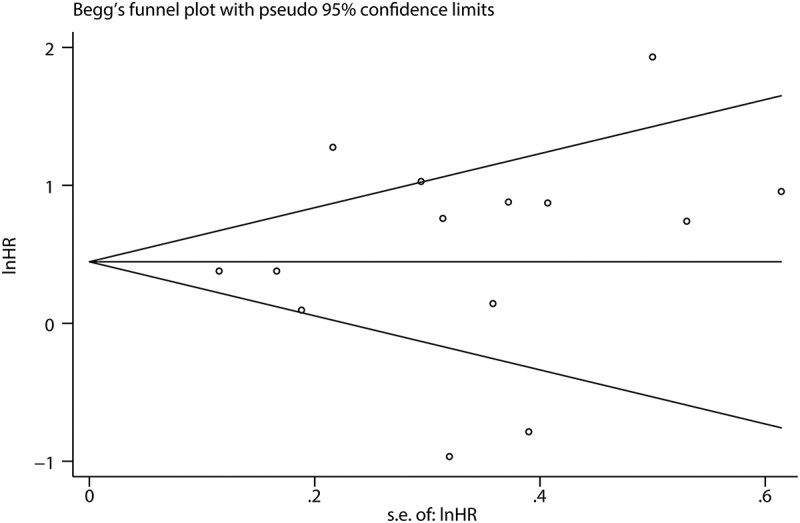
Funnel plots for publication bias for OS
Figure 7.
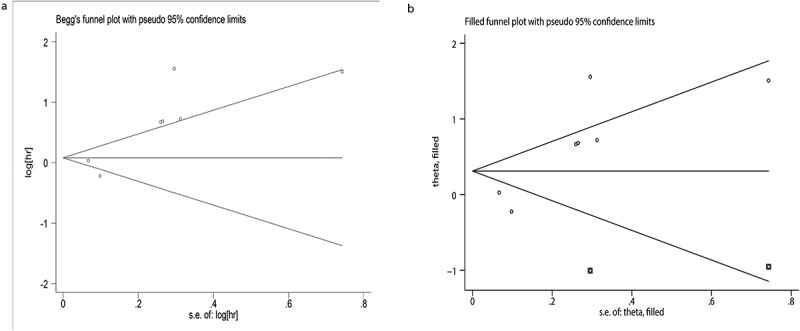
Funnel plots for publication bias for DFS/PFS/RFS. (a) Begg’s test to evaluate DFS/PFS/RFS data. (b) Trim and fill to evaluate DFS/PFS/RFS data
Discussion
Cancer is a serious global health problem, which not only imposes a heavy burden on the medical system, but also causes great distress to patients. According to statistics, there were more than 18.1 million new cancer patients and 9.6 million cancer deaths worldwide in 2018 [32]. Advanced tumors are often malignant, prone to relapse after treatment and have poor prognosis. Many diagnostic and prognostic markers have been applied to cancers, but the results may be not satisfactory [33–35]. Thus, it is important to identify more effective prognostic markers.
The relationship between miR-24 and cancer has always been a significant focus of research interest. MiR-24 can regulate tumor progression by degrading mRNA, destroying the stability of targeted mRNAs, or preventing their translation. MiR-24 is considered to act on an indispensable role in tumor development, tumor development, and it may be an effective prognostic marker for tumors. MiR-24 can regulate tumors in many ways. Du et al. found that miR-24 enhanced breast tumor cell invasion and metastasis through THE PTPN9/PTPRF/EGF signaling axis [36]. Roscigno et al. reported that miR-24 inhibits chemotherapy-induced apoptosis of breast cancer stem cells through the FIH1-HIFa signaling pathway, and enhances the resistance of tumor cells to hypoxia [37]. Lin et al. revealed that miR-24 can target P57 to promote the proliferation of oral squamous cell carcinoma cells [38]. Yu et al. disclosed that miR-24 promotes the proliferation, migration, and invasion of bladder cancer cells by inhibiting DEDD [39]. In lung cancer, miR-24 strengthened tumor proliferation and migration by directly targeting SOX7 in tumor cells and in a xenograft mouse model [7]. Dong et al. suggested that miR-24 could restrain liver cancer cell proliferation and accelerate tumor formation by targeting Metallothionein 1 M [40]. In addition, P53 has also been confirmed to be a regulatory gene targeted by miR-24 to promote liver cancer cell metastasis and invasion [41]. Chen et al. showed that miR-24 targeted ST7 through β-catenin/Tcf-4 signaling pathway to enhance the proliferation and invasion of glioma [42]. Liu et al. reported that miR-24 may achieve its biological function by regulating Bim [43]. Moreover, researchers discovered that miR-24 could promote colon cancer cell proliferation, invasion, and migration partially by repressing TRIM11 [5]. It can be seen from the above that miR-24 is responsible for different regulatory mechanisms in different tumors, and may be a potentially valuable target for tumor treatment.
Many studies have shown that miR-24 is abnormally expressed in a variety of tumors and has often been associated with prognosis, but a definitive conclusion has not been reached. Gao et al. explored 95 CRC patients who underwent radical surgery, and found that low miR-24 expression was significantly correlated with local invasion, lymph node metastasis and clinical stage and was a risk factor for poor prognosis [11]. Dong found that elevated miR-24 expression in GC tissue was correlated with the pathological differentiation degree, lymph node metastasis and depth of infiltration [10]. Further multivariate cox regression analysis showed that high miR-24 expression was a favorable prognostic factor in GC patients [10]. Zhao observed that lung cancer patients with high miR-24 expression were remarkably shorter OS [21], and miR-24 expression was correlated with tumor size and tumor node metastasis. In addition, Kerimis et al. revealed that high miR-24 expression predicted poor OS and DFS in patients with colorectal adenocarcinoma, independently of clinicopathological parameters [13]. These different results indicate that miR-24 plays different important roles in different tumors.
We implemented a meta-analysis to comprehensively clarify the prognostic role of miR-24 in different tumors. Our meta-analysis displayed that elevated miR-24 expression was closely correlated with adverse OS and DFS/PFS/RFS. In the subgroup analysis for OS, we found that elevated miR-24 expression was mainly associated with leukemia (1.40 [1.17–1.69]), HCC (3.03 [2.14–4.30]), and LC (2.28 [1.21–4.29]), which indicated that miR-24 may be a better predictor of outcome for these three types of tumors. Interestingly, we also found that regardless of the White or Yellow race, higher miR-24 expression was closely associated with poor prognosis. By comparing elevated miR-24 expression with clinicopathological data, we also found that elevated mir-24 expression was positively correlated with tumor diameter (large vs small) and tumor stage (III–IV vs I–II). Mir-24 may be involved in tumor development by influencing tumor proliferation, invasion, and metastasis. Thus, miR-24 may be a suitable and effective indicator of cancer prognosis.
Some unavoidable flaws exist in this study. Firstly, the analysis was from a handful of publications. Secondly, all enrolled studies were small-sample retrospective studies. Thirdly, most of the included studies were implemented in Asia, which affected the universality of the results. Fourth, some HRs and 95% CI were extracted from the survival curve, which may have led to an interpretation error. Fifth, the results need to be viewed with caution due to the heterogeneity. Finally, we did not compare the analysis with any public domain datasets or consortium with similar claims for miR-24.
Of course, our study has some advantages. Firstly, the sensitivity analysis showed that the meta-analyses were stable. Secondly, no publication bias was observed in the results of pooled OS. In addition, a previous meta-analysis explored the prognostic value of microRNA‑ 23A/24‑2 ‑ 27a, but the prognostic significance of miR-24 was not evaluated in depth because of the limited number of enrolled articles [44]. We further clarify the prognostic significance of miR-24 in tumors by compiling a large amount of literatures.
Conclusion
Our results indicated that elevated miR-24 expression predicts poor OS. We suggested that miR-24 could be a promising prognostic marker in cancers. Due to inevitable shortcomings, future prospective studies are necessary to verify our findings and further assess the relationship between miR-24 and cancers.
Funding Statement
This work was supported by grants from National Natural Science Foundation of China [No:81660158, 81460092, 81400372]; Natural Science Key Project of Jiangxi Province [No: 20161ACB21017]; Key Research Foundation of Jiangxi Province [No: 20151BBG70223, 20181BBG70004];
Article highlights
We revealed that elevated miR-24 expression was significantly associated with poor overall survival in patients with cancers.
we found that elevated miR-24 expression was positively correlated with tumor size (large or small) and tumor stage (III–IV vs I–II) in patients with cancers.
Elevated miR-24 expression indicates poor prognosis and can serve as an effective prognostic indicator in different cancers.
Abbreviation
HR, hazard ratio; OR, odds ratio; CI, confidence interval; OS, overall survival; DFS/PFS/RFS, disease-free survival/recurrence-free survival/progression-free survival; GC, gastric cancer; LC, lung cancer; CRC, colorectal cancer; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; HCC, hepatocellular carcinoma; NPC, nasopharyngeal carcinoma; TSCC, tongue squamous cell carcinoma; NOS, Newcastle Ottawa scale;
Authors’ contributions
Linsen Ye and Yi Shao designed this study. Rongqiang Liu, Weihao Kong and Shiyang Zheng participated in the literature search, formal analysis, investigation, methodology and writing of the manuscript. Other authors participated in the supervision. All authors approved the final manuscript.
Data availability statement
All data are in the manuscript and can be obtained from the corresponding author.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Bu J, Li H, Li XY, et al. Prognostic role of microRNA-126 for survival in malignant tumors: a systematic review and meta-analysis. Dis Markers. 2015;2015:739469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dai SL, Zhou J, Pan C, et al. Prognostic value of microRNA-145 in patients with various cancers: a meta-analysis. Cancer Biomark. 2015;15:507–513. [DOI] [PubMed] [Google Scholar]
- [3].Ding L, Ni J, Yang F, et al . Promising therapeutic role of miR-27b in tumor. Tumour Biol. 2017;39:1010428317691657. [DOI] [PubMed] [Google Scholar]
- [4].Chhabra R, Dubey R, Saini N.. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yin Y, Zhong J, Li SW, et al. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7:86755–86765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang MX, Zhang J, Zhang H, et al. miR-24-3p suppresses malignant behavior of lacrimal adenoid cystic carcinoma by targeting PRKCH to regulate p53/p21 pathway. PLoS One. 2016;11:e0158433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [7].Yan L, Ma J, Zhu Y, et al. miR-24-3p promotes cell migration and proliferation in lung cancer by targeting SOX7. J Cell Biochem. 2018;119:3989–3998. [DOI] [PubMed] [Google Scholar]
- [8].Lal A, Kim HH, Abdelmohsen K, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zheng X, Li J, Peng C, et al. MicroRNA-24 induces cisplatin resistance by targeting PTEN in human tongue squamous cell carcinoma. Oral Oncol. 2015;51:998–1003. [DOI] [PubMed] [Google Scholar]
- [10].Dong X, Liu Y. Expression and significance of miR-24 and miR-101 in patients with advanced gastric cancer. Oncol Lett. 2018;16:5769–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gao Y, Liu Y, Du L, et al. Down-regulation of miR-24-3p in colorectal cancer is associated with malignant behavior. Med Onco l. 2015;32:362. [DOI] [PubMed] [Google Scholar]
- [12].Organista-Nava J, Gómez-Gómez Y, Illades-Aguiar B, et al. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol Rep. 2015;33:1639–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kerimis D, Kontos CK, Christodoulou S, et al. Elevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinoma. Clin Biochem. 2017;50:285–292. [DOI] [PubMed] [Google Scholar]
- [14].Le HB, Zhu WY, Chen DD, et al. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190–3197. [DOI] [PubMed] [Google Scholar]
- [15].Liu YX, Long XD, Xi ZF, et al. MicroRNA-24 modulates aflatoxin B1-related hepatocellular carcinoma prognosis and tumorigenesis. Biomed Res Int. 2014;2014:482926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu L, Pan J, Wang H, et al. von Willebrand factor rescued by miR-24 inhibition facilitates the proliferation and migration of osteosarcoma cells in vitro. Biosci Rep. 2018;38. DOI: 10.1042/BSR20180372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meng FL, Wang W, Jia WD. Diagnostic and prognostic significance of serum miR-24-3p in HBV-related hepatocellular carcinoma. Med Oncol. 2014;31:177. [DOI] [PubMed] [Google Scholar]
- [18].Su B, Xu T, Bruce JP, et al. hsa-miR-24 suppresses metastasis in nasopharyngeal carcinoma by regulating the c-Myc/epithelial-mesenchymal transition axis. Oncol Rep. 2018;40:2536–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu XD, Liu Z, Long J, et al. Knockdown of miR-24 suppresses the proliferation, migration and invasion of gastric cancer cells and predicts a poor prognosis in gastric cancer. Int J Clin Exp Med. 2017;10:2911–2917. [Google Scholar]
- [20].Yin JY, Tang Q, Qian W, et al. Increased expression of miR-24 is associated with acute myeloid leukemia with t(8;21). Int J Clin Exp Pathol. 2014;7:8032–8038. [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao G, Liu L, Zhao T, et al. Upregulation of miR-24 promotes cell proliferation by targeting NAIF1 in non-small cell lung cancer. Tumour Biol. 2015;36:3693–3701. [DOI] [PubMed] [Google Scholar]
- [22].Zhao J, Hu C, Chi J, et al. miR-24 promotes the proliferation, migration and invasion in human tongue squamous cell carcinoma by targeting FBXW7. Oncol Rep. 2016;36:1143–1149. [DOI] [PubMed] [Google Scholar]
- [23].Zhao J, Chi J, Gao M, et al. Loss of PTEN expression is associated with high microRNA 24 level and poor prognosis in patients with tongue squamous cell carcinoma. J Oral Maxillofac Surg. 2017;75:1449.e1-1449.e8. [DOI] [PubMed] [Google Scholar]
- [24].Zhou N, Yan HL. MiR-24 promotes the proliferation and apoptosis of lung carcinoma via targeting MAPK7. Eur Rev Med Pharmacol Sci. 2018;22:6845–6852. [DOI] [PubMed] [Google Scholar]
- [25].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. [DOI] [PubMed] [Google Scholar]
- [27].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. [DOI] [PubMed] [Google Scholar]
- [28].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. [DOI] [PubMed] [Google Scholar]
- [29].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- [30].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed.). 1997;315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. [DOI] [PubMed] [Google Scholar]
- [32].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [33].Ding J, Cao J, Chen Z; Ding J, Cao J, Chen Z, et al . The role of long intergenic noncoding RNA 00511 in malignant tumors: a meta-analysis, database validation and review. Bioengineered. 2020;11:812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang C, Ren X, Zhang W, et al. Prognostic and clinical significance of long non-coding RNA SNHG12 expression in various cancers. Bioengineered. 2020;11:1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ma X, Bai J, Xie G, et al. Prognostic significance of microRNA-101 in solid tumor: a meta-analysis. PLoS One. 2017;12:e0180173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Du WW, Fang L, Li M, et al. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci. 2013;126:1440–1453. [DOI] [PubMed] [Google Scholar]
- [37].Roscigno G, Puoti I, Giordano I, et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget. 2017;8:19507–19521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lin SC, Liu CJ, Lin JA, et al. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010;46:204–208. [DOI] [PubMed] [Google Scholar]
- [39].Yu G, Jia Z, Dou Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol Rep. 2017;37:1123–1131. [DOI] [PubMed] [Google Scholar]
- [40].Dong X, Ding W, Ye J; Dong X, Ding W, Ye J,et al . MiR-24-3p enhances cell growth in hepatocellular carcinoma by targeting metallothionein 1M. Cell Biochem Funct. 2016;34:491–496. [DOI] [PubMed] [Google Scholar]
- [41].Chen L, Luo L, Chen W, et al. MicroRNA-24 increases hepatocellular carcinoma cell metastasis and invasion by targeting p53: miR-24 targeted p53. Biomed Pharmacother. 2016;84:1113–1118. [DOI] [PubMed] [Google Scholar]
- [42].Chen L, Zhang A, Li Y, et al. MiR-24 regulates the proliferation and invasion of glioma by ST7L via β-catenin/Tcf-4 signaling. Cancer Lett. 2013;329:174–180. [DOI] [PubMed] [Google Scholar]
- [43].Liu R, Zhang H, Wang X, et al. The miR-24-Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget. 2015;6:43831–43842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Quan J, Liu S, Dai K, et al. MicroRNA-23a/24-2/27a as a potential diagnostic biomarker for cancer: a systematic review and meta-analysis. Mol Clin Oncol. 2018;8:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are in the manuscript and can be obtained from the corresponding author.


