Abstract
Artificial Intelligence (AI) methods have significant potential for diagnosis and prognosis of COVID-19 infections. Rapid identification of COVID-19 and its severity in individual patients is expected to enable better control of the disease individually and at-large. There has been remarkable interest by the scientific community in using imaging biomarkers to improve detection and management of COVID-19. Exploratory tools such as AI-based models may help explain the complex biological mechanisms and provide better understanding of the underlying pathophysiological processes. The present review focuses on AI-based COVID-19 studies as applies to chest x-ray (CXR) and computed tomography (CT) imaging modalities, and the associated challenges. Explicit radiomics, deep learning methods, and hybrid methods that combine both deep learning and explicit radiomics have the potential to enhance the ability and usefulness of radiological images to assist clinicians in the current COVID-19 pandemic. The aims of this review are: first, to outline COVID-19 AI-analysis workflows, including acquisition of data, feature selection, segmentation methods, feature extraction, and multi-variate model development and validation as appropriate for AI-based COVID-19 studies. Secondly, existing limitations of AI-based COVID-19 analyses are discussed, highlighting potential improvements that can be made. Finally, the impact of AI and radiomics methods and the associated clinical outcomes are summarized. In this review, pipelines that include the key steps for AI-based COVID-19 signatures identification are elaborated. Sample size, non-standard imaging protocols, segmentation, availability of public COVID-19 databases, combination of imaging and clinical information and full clinical validation remain major limitations and challenges. We conclude that AI-based assessment of CXR and CT images has significant potential as a viable pathway for the diagnosis, follow-up and prognosis of COVID-19.
Keywords: COVID-19, Computed tomography, Chest x-ray, Artificial intelligence, Radiomics, Deep learning, Deep radiomics
1. Introduction
Despite the rapid onset of the coronavirus disease 2019 (COVID-19), the ever-evolving knowledge of the disease's symptoms, and the medical consequences for both patients and clinicians, many radiologists remain unsure regarding whether or not to include COVID-19 pneumonia in the differential diagnosis [[1], [2], [3], [4]]. The rapid identification of COVID-19 and the extent of the infections will allow better control of the virus spread. In order to better manage the pandemic, early detection, severity scoring, and prediction are crucial.
Given the contagious nature of the SARS-CoV-2 virus, the timely recognition of patterns in COVID-19 images is highly important. Subject to availability of reverse transcription polymerase chain reaction (RT-PCR) kit for detecting COVID-19 infection, and according to the specific version of PCR, the results can be obtained in less than 1 h up to few days. In addition, while RT-PCR is highly specific, it can have low sensitivity, and studies have raised false-negatives in patients with abnormalities in chest Computed Tomography (CT) images confirmed with secondary follow-up RT-PCR to be positive for COVID-19 [5,6]. The potential for the detection of COVID-19 using minable quantitative data from chest x-ray (CXR)/CT images relies on development of adequate models for clinical use [[7], [8], [9]]. For instance, results of Deep Learning (DL) models on CXR from COVID-19 infected patients revealed that the elderly, comorbidities, as well as acuity of care are highly associated with the severity of the COVID-19 [10]. Therefore, utilizing artificial intelligence (AI) technology is has the potential to develop new approaches for prognosis, and follow up of COVID-19 infected cases.
AI-based analysis of medical imaging (including radiomics as a subset of it) are advanced automated image analysis approaches, with significant potential for precision medicine by means of data mining to provide insights into intra-regional heterogeneity of abnormal tissues [11,12]. Quantitative features extracted from standard of care medical images enables derivation of enhanced biomarkers of disease that could impact the clinical decision process. This so-called population-imaging approach may use either unstructured data from different modalities acquired for a specific purpose but possibly unrelated diagnostic purpose in broadly defined groups, or a single imaging test in a large cohort for multicentre longitudinal studies [13]. Imaging biomarkers established as such may provide key insights to disease processes as they describe lesions growth and tissue characteristics [14].
Conventional radiomics workflows involve extraction of so-called radiomics features (hand-crated or explicit features) from segmented regions of interest (ROI) [[15], [16], [17], [18], [19], [20]]. An alternative approach to this is deep leaning (DL), in which the features are implicitly derived utilizing neural networks [[21], [22], [23], [24], [25]]. Highly mature studies utilizing DL algorithms for detection and/or reporting the severity of COVID-19 infections are in Refs. [[26], [27], [28], [29], [30], [31], [32]]. Moreover, DL technically does not require segmented ROIs, and, if large enough datasets are provided, is able to focus on areas of importance. Each of these methods (radiomics and DL) has its own advantages (the former working better for small/medium datasets; the latter working better on large datasets). Other than that, hybrid methods integrating the two approaches (“deep radiomics”) have also been explored to utilize quantitative data extracted/derived from medical images [33]. Examples of this approach include initial generation of radiomics images at the voxel level fed into deep neural networks (DNNs), or alternatively, extraction of deep features as generated by DNNs (e.g., in the fully connected layer) combined with machine learning algorithms.
Radiomics (and the use of ML techniques to combine radiomics features into a model; i.e., radiomics signature) may be applied to the acquired datasets to enhance the assessment of diseases. However, linking radiomics (i.e., process of extracting quantitative image features) to biological or pathophysiological processes being investigated remains challenging, and has in the past hindered the translation of radiomics into clinical practice despite providing promising results into tissue characterization; this issue, however, is receiving further attention in recent years and is an active area of research [14]. Beyond the direct impact of COVID-19 AI-based methods for the diagnosis and prognosis of SARS-CoV-2 virus, an explicit research pathway may lead to establishment of a comprehensive prognostic approach to fight the spread of COVID-19.
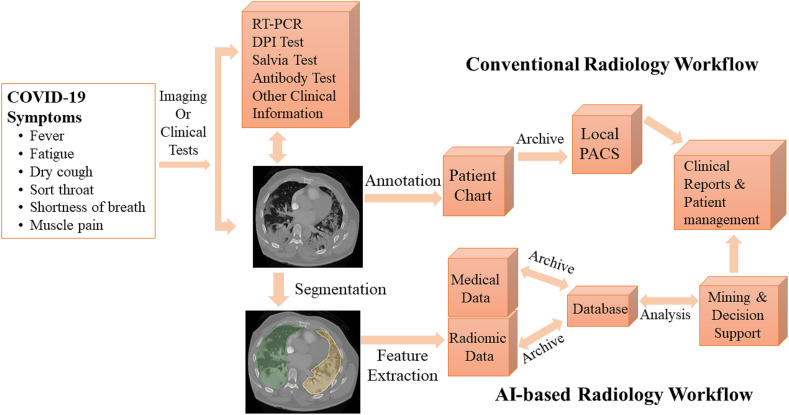
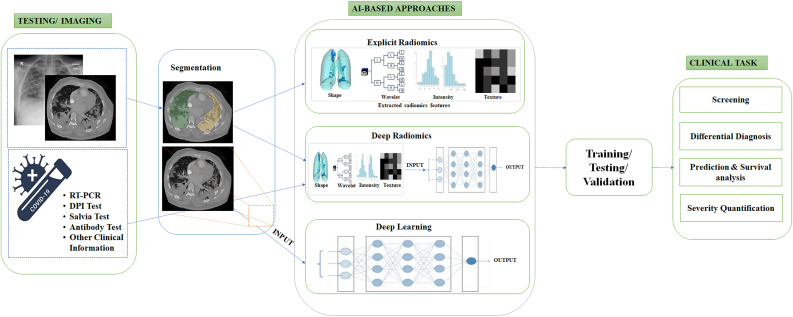
To this end, Born et al. [34] performed a systematic meta-analysis focusing on ML-based COVID-19 utilizing CXR, CT, and ultrasound images aiming to identify the most relevant articles. Bhattacharya et al. [35] focused on reviewing DL algorithms which were utilized in COVID-19 analysis and an overview of DL and its clinical impact over the last decade. Elsewhere, Shoeibi et al. [36] performed a complete review of DL techniques and tools for COVID-19 detection and lungs segmentation. Moreover, the challenges related to the automated detection of coronavirus infections using DL methods were reported. The authors summarized the COVID-19 prevalence in several parts of the world. Recently, Suri et al. [37] investigated a variety of comorbidities and their associated risks in acute respiratory distress syndrome and mortality. They elaborated on AI architectures and their extension from pre-COVID-19 to post-COVID-19 and the views of seven school-of-though were summarized. It is worth mentioning that non-imaging information obtained from genomics, proteomics, lipidomics, and transcriptomics, combined with AI-based approaches, can be valuable for the diagnosis and prognosis of COVID-19 [38,39]. To have a clearer picture on deploying AI-based approaches in clinical practice to help the fight against COVID-19, Fig. 1 illustrates the workflow of conventional methods (non-AI methods) and AI-based methods.
Fig. 1.
Conventional and AI-based radiology workflow in COVID-19.
Unlike the above-mentioned review articles, the aims of this work are to: (i) present a specific AI workflow utilizing CT and CXR images; and (ii) enhance the current knowledge towards improved future AI-based COVID-19 studies in terms of selecting appropriate segmentation, feature extraction, dimensionality reduction, and classification methods. Moreover, existing challenges in AI-based efforts are addressed, and recommendations on possible improvements are made; (iii) finally, the clinical impact of relevant AI-based COVID-19 studies are presented. This review focuses on AI-based COVID-19 studies utilizing CXR and CT imaging.
2. Utilization of AI to assess COVID-19 images
AI methods utilize machine learning (ML) approaches (as applied to radiomics features) vs. deep learning (DL) algorithms as directly applied to the images [4,5]. ML and DL have played significant roles to mine, interpret, and identify data patterns. ML models utilized for the diagnosis and/or prognosis of COVID-19 which were reported have included: Support Vector Machine (SVM) [[40], [41], [42]], Random Forest (RF) [43,44], Decision tree (DT) [45,46], and Logistic regression (LR) [47,48]. DL models frequently employed have been convolutional neural networks (CNNs) [49,50] and recurrent neural networks (RNNs) [51,52]. It was successfully demonstrated that AI-based model, combining CT/CXR modality and other clinical information, could be useful in screening COVID-19 that does not require radiologist input or physical tests. Xia et al. [53] performed a DL-based approach utilizing a classifier that combines clinical variables such as patient demographics, symptoms (cough, fever, sore throat, etc.), signs of infection (e.g., enlarged tonsils and lymph nodes), underlying diseases (e.g., hypertension, diabetes, etc.), and blood results with CXR data to distinguish COVID-19 from viral pneumonia in a simple, efficient, inexpensive, and accurate way. A recent study by Shiri et al. [54] revealed that integrating radiomics features with demographics and clinical data (gender, age, weight, height, BMI, medical history of comorbidities and vital signs), laboratory features (blood tests) and radiological data (scoring by radiologists) can help the prediction of overall survival in COVID-19 patients.
Overall, AI is a valuable technology for early detection of COVID-19 infections and proper health monitoring. CT and CXR have been identified and used as the imaging modalities of choice for the prognosis of COVID-19 infections. The following two sections (II.1 and II.2) describe AI-based signatures of COVID-19 studies utilizing CXR and CT scans.
2.1. Chest radiography (CXR)
Potential trends and information derived from the X-ray radiographs scans has proved to be useful in COVID-19 diagnosis since pulmonary infections have been detected through X-ray images [55]. CXR are commonly performed first and play role as an alternative viable choice when CT scanners are not available for fast diagnosis and monitoring the progression of COVID-19 cases [56,57]. In COVID-19 CXR findings are: pneumonia, namely bilateral peripheral and/or, subpleural ground glass opacity (GGO) and/or consolidation, unilateral non-segmental/lobar ground-glass or consolidative opacities or multifocal ground-glass/consolidative opacities without many particular distributions (Fig. 2 ). Moreover, using public CXR databases increased remarkably the number of AI-based COVID-19 studies that aimed to detect, follow, and predict outcome of SARS-CoV-2 infections [58]. DL-based approaches allowing automated analysis of CXR images significantly accelerated the analysis and processing time and helped speed-up the identification and control of the spread of COVID-19 [59].
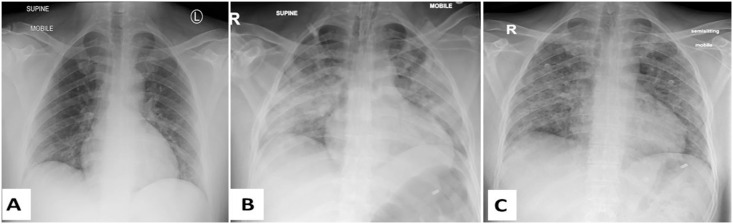
Fig. 2.
Three CXR images of a patient diagnosed with COVID-19 at days (A) 4, (B) 14, and (C) 29 of admission to the Royal Hospital, Muscat, Oman. The images in these 3 dates indicate: (A) bilateral peripheral lower lobe opacities, (B) bilateral mostly peripheral consolidations in the middle and lower lung zones, and (C) bilateral reticular opacities in the middle and lower lung zones.
Bukhari et al. [60] evaluated a total of 278 CXRs, applying ResNet-50 CNN architectures. These chest X-ray images were grouped into normal, pneumonia, and COVID-19. A ResNet-50 pre-trained architecture was chosen to diagnose COVID-19 pneumonia on related set of lung CXRs. The results illustrate that the diagnostic accuracy of the deep learning method was 98.2 %. Apostolopoulos et al. [61] investigated the possibility of extracting COVID-19 biomarker from 3905 CXRs. The randomly CXRs selected corresponded to: pulmonary edema, pleural effusion, chronic obstructive pulmonary disease, pulmonary fibrosis and COVID-19 infection. CNNs models were trained from scratch conventional diagnosis, to identify the CXRs between the 5 classes and COVID-19 and non-COVID-19 scans. The classification accuracy between the seven classes was 87.66 %. Moreover, accuracy, sensitivity and specificity of COVID-19 diagnosis were reported to be 99.2 %, 97.4 %, and 99.4 %, respectively.
Training a CNN model from scratch necessitates a large amount of training data and technical skills to determine the best model architecture for optimal convergence. Additionally, this requires time-consuming annotations by radiology experts. Due to computational requirements and memory constraints, CNN training takes a long time. Transfer learning has the advantage to decrease computational complexity and to speed-up the process. Basu et al. [62] successfully classified CXR images (accuracy of 90 %) into four classes: normal, pneumonia, other disease, and COVID-19, using a CNN pre-trained on normal and disease classes that were obtained from the National Institutes of Health (NIH) Chest X-ray freely-accessible database. The activation map was used to identify regions where the emphasis was classification of features. The average detection precision was found to be 95.2 %.
Hall et al. [63] obtained on overall accuracy of 91.2 % when they pertained 135 CXR of COVID-19 and 320 CXR of viral and bacterial pneumonia by a deep CNN (Resnet50 software). They suggested and recommended that CXR is an inexpensive, accurate and fast imaging modality for diagnosis of COVID-19. Wang et al. [64] proposed COVID-Net as a new deep learning architecture for prediction of COVID-19 disease using CXR. A dataset with a total of 5896 CXR images (358 COVID-19 and 5538 non-COVID-19) was studied. In total, four classes of cases were considered: (a) normal, (b) bacterial infection, (c) non-COVID infection, and (d) COVID-19 infection. Given the quantity of COVID-19 images collected, the distribution of inter-class images among their training sets and among their validation sets was highly unbalanced. They leveraged the principles of residual architecture design. A 93 % accuracy was obtained by the COVID-Net architecture emphasizing the ability to utilize new DNN architectures, and reflecting ever-evolving efforts on dedicated DL architectures.
Zhang et al. [13] performed advanced analysis using a DNN (CV19-Net) to differentiate COVID-19 from non-COVID-19 infections, on a total of 11105 CXR images (2060 COVID-19 cases and 3148 non-COVID-19 cases). State-of-art algorithms for the test set, CV19-Net achieved a sensitivity of 88 % and a specificity of 79 % by using a high-sensitivity operating threshold, and a sensitivity of 78 % and a specificity of 89 % by using a high-specificity operating threshold. They evaluated the performance of CV19-Net by choosing 500 CXR that were examined by the both CV19-Net and three radiologists. They reported an AUC of 0.90 for CV19-Net and an AUC of 0.85 obtained by radiologists.
2.2. Computed tomography (CT)
As COVID-19 continues to infect people all around the world, beside real-time reverse transcription polymerase chain reaction, CT plays an essential role in faster diagnosis. Various manifestations of COVID-19 on chest CT images (particularly, consolidation, GGO, or a mixture of GGO and consolidation) were important to distinguish between infections of the lungs [65]. Hence, early COVID-19 screening, differential diagnosis (see Fig. 3 ), and disease severity assessment/follow-up was achieved from reading chest CT scans (refer to Fig. 4 ). Moreover, CT images helped radiologists to visualize the effects of COVID-19 by means of 3D printing technology [66].
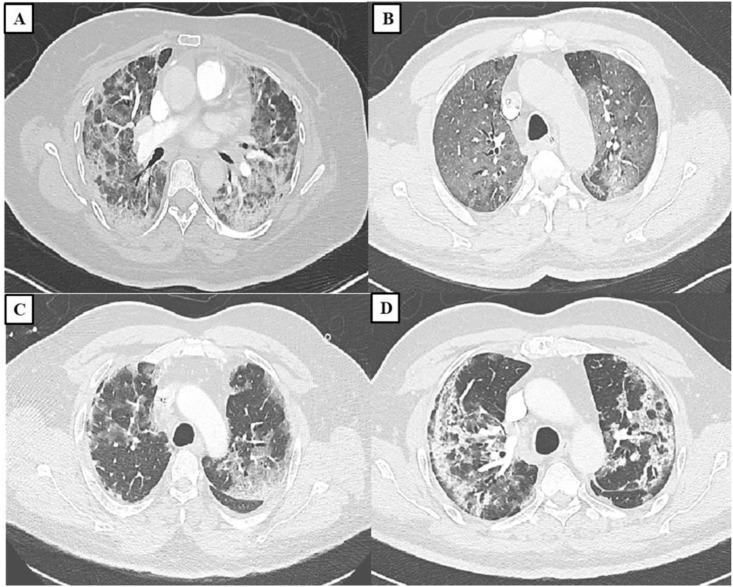
Fig. 3.
COVID-19 positive cases admitted at the Royal Hospital, Muscat, Oman; (A) A 57 year-old female admitted with COVID-19, admission day 24 with no wearable respiratory monitoring system. CT of the chest showing bilateral GGO with peripheral distribution; (B) A 63 year-old male patient admitted with COVID-19 with a history of desaturation. Images from a contrast enhanced CT of the thorax showing bilateral diffuse GGO; (C) A 78 year-old male admitted with severe bilateral COVID-19 pneumonia. CT shows bilateral peripheral GGO with prominent interstitial septae with crazy paving pattern; (D) A 59 year-old male patient with hypoxemia COVID-19. CT shows bilateral peripheral GGO with formation of peripheral bands sparing the subpleural area.
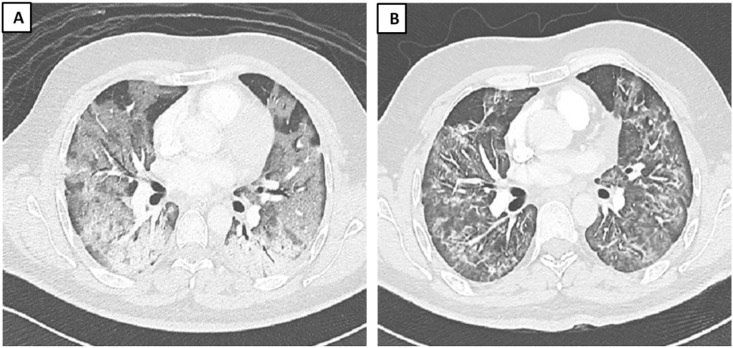
Fig. 4.
A 34 year-old male with COVID-19. (A) The first image was done on day15 from admission (day 19 from diagnosis). (B) The second image is done day 35 of admission (day 39 from diagnosis). The first image showing bilateral diffuse ground glass opacities. The follow-up showing improvement in the ground glass opacities but development of septal thickening and bands.
The most common CT findings, as lesion features, can be categorized into: (1) small peripheral/subpleural, bilateral, ground-glass opacities with/without consolidation; (2) crazy-paving pattern; (3) air bronchogram sign; (4) consolidation; (5) linear opacities; and (6) bronchial wall thickening [67]. In addition to detecting COVID-19 using deep learning methods, Shiri et al. [68] proposed a deep learning based algorithms for dose reduction in chest CT images. They implemented a residual DNN to generate high quality images from ultra-low dose CT images. Model were built on 970 chest CT images and evaluated on 170 external validation set of COVID-19 patients. They reported 89 % dose reduction in CT images with properly recovering most common lesion features in COVID-19 images. Shan et al. [69] suggested a method for reducing the prognosis time by automated methods of delineating CT images from 1 to 5 h to 4 to 3 min compared with manual method. They used a human-in-the-loop strategy to accelerate the delineation of CT images. The authors reported that this auto contoured regions could assist radiologists for their annotation refinements. Song et al. [70] conducted a deep leaning-based study on CT images of 88 COVID-19 and normal cases. They reported sensitivity of 93 % the model to distinguish COVID-19 from pneumonia. Ai et al. [71] emphasized that with RT-PCR as a reference, the sensitivity of chest CT imaging for COVID-19 was 97 %. Lin et al. [71] performed retrospective and multi-centre DL-based model study utilizing COVID-19 detection neural network (COVNet) for feature extraction from CT scans for COVID-19 diagnosis purpose. Community-acquired pneumonia and other non-pneumonia CT exams were included to study of the model. The pre-exam sensitivity and specificity in detecting community-acquired pneumonia in the test via COVNet were reported to be 87 % and 92 % respectively.
Although, CT has been reported as a most accurate tool to detect the COVID-19 [72], Elsewhere, Li and his colleagues Xia illustrated the dark side of CT scans on diagnosis of COVID-19. They found that CT findings of COVID-19 overlapped with the CT features of adenovirus infection due to this limitation in distinguishing between viruses and identification the infection patterns of other viruses [73]. In most recent study, Shiri et al. [54] implemented a holistic radiomics model with combining CT radiomics features (extracted from lung, and pneumonia lesion) with clinical features (demographic, laboratory and radiological score) for outcome prediction of COVID-19 patients. Model were build and evaluated on 106 and 46 patient data respectively and they reported the combining all information (AUC of 0.95, sensitivity of 0.88 and specificity of 0.89) outperformed radiomics only or clinical-only models.
3. AI-BASED workflows in the assessment of images for COVID-19
AI-based analysis has also been shown to have a role in detecting COVID-19. More specifically, radiomics investigations have demonstrated radiomics textural features capable of distinguishing COVID-19 form other type of pneumonia. Radiomics workflow in COVID studies, similar to others, includes image acquisition, lungs lesion segmentation and feature extraction, feature selection, signature construction, and the evaluation of developed model. By contrast, in the DL workflow, explicit feature extraction is omitted and the network used itself created the model based on implicitly derived features.
3.1. Image acquisition
Image acquisition is the first stage in the AI-based workflow. Currently available clinical imaging modalities allow wide variations in acquisition and image reconstruction protocols as illustrated in Table 1 . Image derived metrics such as radiomics features are sensitive to image acquisition settings, reconstruction algorithms and image processing methods [68].
Table 1.
Summary of image acquisition techniques in published articles.
| Reference | Modality- Device model | Tube Voltage (kV) | Tube current | Slice thickness (mm) | Collimation (mm) | Matrix size | Pitch |
|---|---|---|---|---|---|---|---|
| Fang et al. (2020) | CT- Philips Brilliance iCT; Dutch Philips | 120 | 100–400 mA | 0.9–5 | 0.625 | 512 × 512. | 0.914 |
| Guiot et al. (2020) | CT- Siemens Edge Plus, GE Revolution CT, GE Brightspeed | STANDARD reconstruction (no mention of acquisition and reconstruction parameters) | |||||
| Li et al. (2020) | CT- Discovery CT750HD, GE Healthcare | 120 | 250–400 mA | 5 | – | – | – |
| Fang et al. (2020) | CT- 64: Somatom, Siemens Healthcare; 256: Brilliance-16P, Philips Healthcare; 128: uCT 760, United Imaging Healthcare |
120 | – | 1 | 0.6 | 256 × 128 | – |
| Liu et al. (2020) | CT- Hitachi Medical, Japan | 120 | 180–400 mA | 5 | 0.625 | 512 × 512 | 1.5 |
| Wang et al. (2020) | CT- 16-MDCT, SOMATOM Emotion16, SIEMENS, Germany; 16-MDCT, Definition AS, SIEMENS, Germany; 64-MDCT, Optima CT680, GE, USA |
120 | 300 mAs | 5 | 0.625–1.25 | – | – |
| Zeng et al. (2020) | CT- Light-speed; GEHealthcare, Chicago, IL | 120 | 100–250 mAs | – | – | – | – |
| Shi et al. (2020) | CT- SOMATOM Perspective, SOMATOM Spirit, or SOMATOM Definition AS+ (Siemens Healthineers, Forchheim, Germany). |
120 | – | 1·5 or 1 and an interval of 1·5 or 1 | 0.6 | – | |
| Li et al. (2020) | CT | – | – | – | – | 512 × 512 | – |
| Zheng et al. (2020) | CT- GE Discovery CT750HD; GE Healthcare, Milwaukee, WI | 100 | 350 mA | 5 | 0.625 | 512 × 512 | 1.375 |
| Huang et al. (2020) | CT- Siemens, Germany; Philips, the Netherlands; and GE, USA |
120 | – | 1 or 1.5; and layer spacing, 1.5 | 0.6 | 128 × 128 | – |
| Juanjuan et al. (2020) | CT- 1212LightSpeed VCT (General Electric Medical Systems, USA), Somatom Sensation (Siemens Heathcare), Somatom Definition (Siemens Heathcare), and Somatom Definition AS+ (Siemens Heathcare).) |
STANDARD protocol (no mention of acquisition and reconstruction parameters) | |||||
| Wei et al. (2020) | CT- NeuViz 128 | 120 | 150 | 5 | – | 512 × 512 | 1.2 |
| Zheng et al. (2020) | CT- Ingenuity Core128, Philips Medical Systems, Best, the Netherlands; Somatom Definition AS, Siemens Healthineers, Erlangen, Germany |
120 | – | 1.5 | – | 512 × 512 | – |
| Li et al. (2020) | CT- uCT 780, United Imaging; or Somatom Force, Siemens Healthcare | Multiplanar reformatting (MPR) technique. | |||||
3.2. Segmentation
Image segmentation is an essential processing step that helps improve the accuracy image analysis and clinical reports [19]. By using image segmentation techniques, an image is divided into specific groups of pixels, assigned labels (lung regions, lesions, etc) and classified further according to these labels. The labels generated by image segmentation are then provided as an input to AI-based methods. Subsequently, radiomics features can be calculated from the segmented 2D/3D ROI in radiomics based analyses. In relation to the target ROIs, the segmentation method in AI-based COVID-19 are classified into, (a) the lung-region-oriented method, which is basically able to separate the entire lung and lung lobes in CT/CXR images; (b) the lung-lesion-oriented method; which tries to distinguish lesion in the whole lung from the regions. By segmenting lesions and healthy lungs in CT images, volume of infection and relative volume (lesion/lung) could be calculated, which further could be used as prognostic and severity scoring of COVID-19 patients. Automatic and semiautomatic segmentation approaches can either define features as COVID-19 or non-COVID-19.
Overall, segmentation techniques are specifically divided into four categories; (a) manual-based segmentation is defined as the delineation of the contours of anatomical regions that is performed by experts (e.g. radiologists, pathologists) [74]; (b) model-based segmentation is defined as the assignment of labels to pixels or voxels by matching the a priori known object model to the image data [75]; (c) DL-based segmentation (dominantly CNN-based) as used for automated feature extraction; and (d) hybrid segmentation methods which combines conventional and DL-based methods [76,77]. Segmentation of the lungs in COVID-19 infected cases consists of delineating the borders of the anatomical structures of lung or pneumonia lesions with computer-assisted contouring. It delineates regions of interest (ROIs) or volumes of interest (VOIs) in COVID-19 CT or CXR images; these are commonly: whole lungs, lung lobes, trachea, lung lesions, bronchus and pneumonia lesions. For segmentation, different methods were utilized including, thresholding, region-based, clustering-based, watershed-based algorithms [[78], [79], [80], [81], [82]]. Due to high variety of shape, size, boundary, type and manifestation of lesion in COVID-19 conventional algorithms failed to properly segment the lung and pneumonia lesions. In the COVID-19 pandemic, several ML-based algorithm were proposed for lung and lesion segmentation of COVID-19 radiological images [83,84]. Support Vector Machine (SVM) [85] is an ML method that has reported widely for supervised segmentation in radiomics-based COVID-19 studies [86,87].
Unlike conventional segmentation methods, unsupervised segmentation methods have typically relied on intensity or gradient analyses of the image via various strategies (i.e, using Inf-Net for COVID-19 CT images) to delineate the contours of the anatomical areas in the image. Such methods can as such dive deeper in considering several resolution levels in medical images. It is also possible to use unsupervised deep learning models for segmentation. Each and every layer will learn information from the CXR/CT images depending on the content of images or feature map. Deng-Ping et al. [88] proposed Inf-Net to determine coarse regions, which were followed by applying implicit models that boosted boundaries detection. They also used semi-supervised segmentation on COVID-19 SemiSeg and real CT images to render most of the unlabelled data. Surprisingly, they observed that the semi-supervised system enhanced volume learning capabilities compared to other cutting-edge programs.
To enhance the accuracy of predicted model utilizing two architecture methods (U-Net and Resnet-50) for segmentation of CT images of lung abnormalities was suggested. The proposed method enables the segmentation of ROIs and classify CT scans as COVID-19 and non-COVID-19 cases [[5], [6], [7], [8]]. As it was mentioned above, U-Net and its variant have been developed and have achieved fair segmentations in COVID-19 CT/CXR images. Çiçek et al. [35] recommended the 3D U-Net that utilizes the inter-slice info by replacing the layers in well-known U-Net method in 3D format. A VB-Net was used by Shan et al. [89] for more effective segmentation. Elsewhere, Tang et al. [86] adopted a VB-net [7] to perform accurate segmentation of the whole lungs and lung lesions from CT images. Using U-Net with the initial seeds provided by a radiologist, Qi et al. [37] presented segmentation of lesions in the lungs (see Table 2 ).
Table 2.
A listing of AI-based published articles.
| Reference | Modality/Subjects | Segmentation Method |
Feature Extraction | Type of Feature | Feature Selection/Derivation Methods | Model training | Model Validation | AI-based method | Task |
|---|---|---|---|---|---|---|---|---|---|
| Fang et al. (2020) | CT/46 COVID-19, and 29 other types of pneumonias | 2D/Manually/ITK-SNAP software (v. 3.6.0) | MATLAB (in-house developed tool-box) | Intensity-based statistical features, GLCM, GLRLM |
ML | SVM | 3-fold cross-validation | radiomics | Severity assessment |
| Guiot et al. (2020) | CT/181 COVID-19, 1200 non-COVID [110] | 2D and 3D/RadiomiX (OncoRadiomics SA, Liège, Belgium) |
RadiomiX(OncoRadiomics SA, Liège, Belgium) |
Statistics, texture, and shape | ML | Multivariable logistic regression with Elastic Net regularization | 10-fold cross validation | radiomics | Detection |
| Autee et al. (2020) | CXR/868 COVID-19, 9096 non-COVID [111] |
2D/U-NET | – | – | DL | Multi-layer perceptron stacked ensembling approach | 5-fold cross validation | DL | Diagnosis |
| Shuo et al. (2020) | CT/723 COVID-19 | 3D/Automated/U-Net, V-Net, and 3D U-Net++ | – | – | DL | ResNet-50, Inception networks, DPN-92, and Attention ResNet-50 18 | AUC curve | DL | – |
| Liping et al. (2020) | CT | 3D/Manually/ITK-SNAP software | QAK software | Histogram, shape factors, GLCM, RLM, GLZSM | ML | LASSO | AUC, accuracy, sensitivity, and specificity | radiomics | Diagnostic |
| Armando Ugo et al. (2020) | CXR [112] | Manually/MaZda 4.6) | MaZda 4.6 | Gray level histogram analysis, co-occurrence matrix, and Wavelet transform | ML | Partial Least Square Discriminant Analysis (PLS-DA), Naïve Bayes (NB), Generalized Linear Model (GLM), Logistic Regression (LR), Fast Large Margin (FML), Decision Tree (DT), RF, Gradient Boosted Trees (GBT), artificial Neural Network (aNN), SVM | radiomics | Diagnostic | |
| Fang et al. (2020) | CT/56 COVID-19, 34 other types of viral pneumonia [113] | 2D and 3D/Manually/uAI-Discover-NCP R001) | – | – | DL | variance analysis, spearman correlation analysis, and LASSO | – | DL | Prediction |
| Tang et al. (2020) | CT/176 COVID(non-severe or severe) | VB-net | uAI-Dicover-NCP | Infection volume, ratio of the whole lung, the volumes of GGO regions | ML | RF | 3-fold cross | radiomics | Severity assessment |
| Chen et al. (2020) | CT/51 COVID-19, 55 other disease | 3D/Automated/UNet++ | – | – | DL | UNet++ | – | DL | Detection |
| Li et al. (2020) | CT/400 COVID-19, 1396 Community acquired pneumonia, and 1173 non-pneumonia | 3D AND 2D/Automated/RestNet50 | – | DL | RestNet50 + Max pooling | AUC curve | DL | Diagnostic | |
| Hongmei et al. (2020) | CT/52 COVID-19 | 3D Slicer | 3D Slicer (version 4.10.0) | – | ML | LR,RF | 5-fold cross-validation | DL | Prediction |
| Zheng et al. (2020) | CT/313 COVID positive, 229 COVID negative | 3D/Automated/UNet | – | – | DL | DeCoVNet software | DeCoVNet | DL | Detection |
| Huang et al. (2020) | CT/89 COVID-19, 92 Non-COVID-19 | 3D/Manually/Lung Kit software (v. LK2.2) | PyRadiomics | Intensity statistics, shape features, GLCM, GLSZM, GLRLM, GLDM, NGTDM, wavelet features LoG Filtered features |
ML | LR | – | radiomics | Detection |
| Juanjuan et al. (2020) | CT/148 COVID-19 | – | – | Shape, histogram, GLCM, GLRLM, GLSZM, and GLDM | ML | LASSO | – | radiomics | Prediction |
| Liu et al. (2020) | CT/115 COVID-19 and 435 non-COVID-19 | 3D/Manually/itk-SNAP, (v. 3.4.0) | PyRadiomics | First order statistics, shape-based features (3D), GLCM, GLRLM, GLSZM, and GLDM | ML | LASSO | AUC | radiomics | Diagnosis |
| Wei et al. (2020) | CT/89 COVID-19 | 3D/Automate/LK2.1 software package | PyRadiomics | Histogram, GLCM, GLSZM, GLRLM |
ML | multivariate logistic regression method |
ROC analyses | radiomics | Severity assessment |
| Das et al. (2020) | CXR/COVID-19 (+), pneumonia (+) but COVID-19 (−) | – | – | – | DL | SVM, Random Back propagation network, Adaptive neuro-fuzzy inference system Convolutional neural networks VGGNet, Alexnet, Googlenet Inceptionnet V3 | Accuracy, f-measure, sensitivity, specificity, and kappa statistics | DL | Detection |
| Kabid et al. (2020) | CXR/non-COVID and COVID-19. | – | – | – | DL | Faster R–CNN | K-fold cross-validation | DL | |
| Singh et al. (2020) | CT/D1(233 COVID-19, 376 non-COVID-19) D2(53 COVID-19) D3(58 COVID-19) |
– | Principal Component Analysis, Autoencoder, Variance based Selector | – | DL | VGG16, Deep CNN, SVM, ELM, OS-ELM | AUC | Deep radiomics | Detection |
| Narin et al. (2020) | CXR/50 COVID-19, 50 normal | – | – | – | DL | InceptionV3, ResNet50, InceptionV2 | 5-fold cross validation | DL | Detection |
| Sethy et al. (2020) | CXR/25 COVID-19, 25 normal | – | – | DL | AlexNet, DenseNet201, GoogleNet, InceptionV3, ResNet18, ResNet50, ResNet101, VGG16, XceptionNet, InceptionNetV2 | Accuracy, Sensitivity, Specificity, False positive rate (FPR), F1 Score, MCC and Kappa | DL | Detection | |
| Hemdan et al. (2020) | CXR/25 COVID-19, 25 normal | – | – | – | DL | VGG, DenseNet201, ResNetV2, Inception, InceptionResNetV2, Xceptio, MobileNetV2 | ROC | DL | Diagnostic |
| Apostolopoulos et al. (2020) | CXR/224 COVID-19, 504 normal | – | – | – | DL | VGG19, MobileNet, Inception, Xception, InceptionResNet | 10-fold-cross-validation | DL | Detection |
| Tang et al. (2020) | CT/52 CoVID-19 | 3D/automatically/3DSlicer, U-net | PyRadiomics | Shape, wavelet features | DL | L R, RF | 5-fold cross-validation | radiomics | Severity assessment |
| Zhang et al. (2021) | CT/507 sets of Suspected COVID [32] | DL a built-in feature on InferScholar platform | PyRadiomics | First-order, shape, GLCM, GLRLM, GLSZM, NGTDM, GLDM | DL | SVM, multi-layer perceptron (MLP), logistic LR, XGBoost | 5-fold cross-validation | radiomics | Detection |
| Alqudah et al. (2019) | CXR/48 COVID-19 and 23 non-COVID-19 | ReLU layer | SVM | – | DL | AOCT-NET, SVM, RF | AUC | Deep radiomics | Detection |
| Basu et al. (2020) | CXR/302 COVID-19 and 108,948 normal | – | – | – | DL | AlexNet, VGGNet, RestNet | 5-fold cross-validation | DL | Severity assessment |
| Wenli et al. (2020) | CT/99 COVID-19 [114] | Automated/U-net | – | ML | RF | AUC | Deep radiomics | Severity assessment | |
| Joon et al. (2021) | CXR/338 COVID-19 | – | – | – | DL | DenseNet-121 | AUC | DL | Prediction |
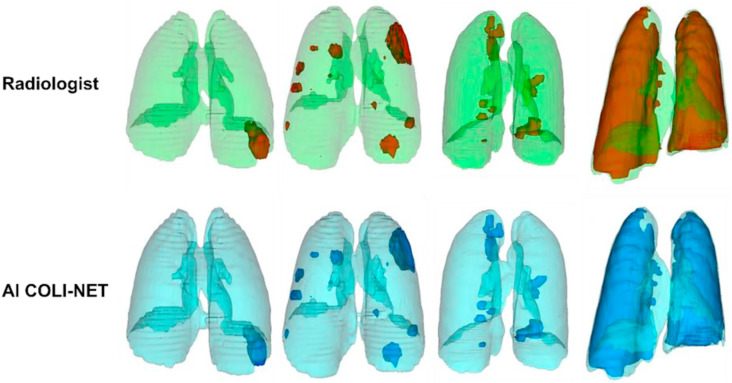
To evaluate segmentation accuracy, dice similarity coefficient (DSC) as a metric has been employed to determine the overlap between automatically segmented COVID-19 infection regions in the lungs and gold-standard manual delineations. Shan et al. [69] reported 92 % DSC for automated segmentation of COVID-19 infection using VB-Net. Elsewhere, Shan et al. [90] utilized DL-based method for determining the severity of COVID-19 using 549 CT scans of COVID-19 patients. They applied VB-Net, an automated segmentation tool, and reported 92 % DSC. In a more recent study, Shiri et al. [91] proposed a deep residual network based algorithm, COLI-NET, for whole lung and pneumonia lesion segmentation. The used 2358 clinical CT images (consisting of 347259 2D slices) for whole lung segmentation and 180 CT images (consisting of 17341 2D slices) for training of the networks. Evaluation was performed on 5 external validation datasets emanating from various centres in different countries (multi-scanner and multi-centre study). For lesion segmentation, they used transfer learning of networks from whole lung segmentations. The reported Dice coefficients were 0.03 ± 0.84 % (95 % CI, −0.12–0.18) and −0.18 ± 3.4 % (95 % CI, −0.8–0.44) for the lung and lesions, respectively. They also reported relative errors less than 5 % for first-order and shape radiomics features in both lung and lesions. Fig. 6 provides an example comparing manual segmentation performed by a radiologist and COLI-NET output for whole lung and lesion segmentation for COVID-19 patients at different stage (from mild to severe).
Fig. 6.
Radiologist and AI (COLI-NET) segmentation for whole lung and lesion segmentation for different stage of COVID-19 patients (from mild to severe).
3.3. Extraction of radiomics features
For the radiomics subset of AI-methods, effective feature extraction is a pillar of radiomics towards learning rich and informative representations from raw input data to provide accurate and robust results. Knowledge of the different types of radiomics features and core principals may facilitate interpretation of results and preselection of features for specific application. To extract the desired radiomics features of COVID-19, segmentation and processing of images should be performed accurately. In other words, feature extraction indicates the computation of features, where descriptors are used to determine attributes of the gray levels within the 2D/3D ROI [92]. Features have to be obtained so that they express the complexity of each volume as best possible, but cannot be excessively redundant or complex. To date, several techniques and algorithms have been applied to extract COVID-19 features, although no agreement exists about a standard method (see Fig. 5 and Table 2).
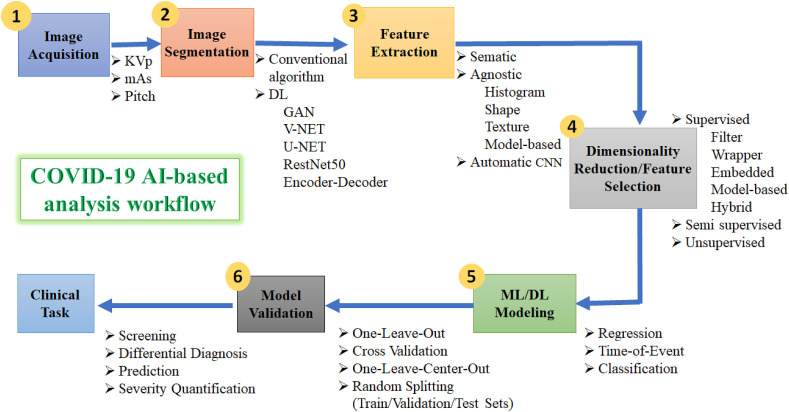
Fig. 5.
A schematic diagram of COVID-19 AI-based analysis workflow, involving different parameters and options for (1) image acquisition, (2) image segmentation, (3) extraction of features, (4) dimensionality reduction, (5) ML/DL modeling, and (6) model validation. Only few examples of parameters/features/algorithms are mentioned.
Different types of COVID-19 explicit radiomics features were identified, the most commonly found ones are shape, statistics, histogram, and texture features including Gray-Level Co-occurrence Matrix (GLCM), Gray Level Run Length Matrix (GLRLM), and Gray Level Size Zone Matrix (GLSZM). Moreover, the extraction depends on the amount of data, and different types of filters such as Wavelet decomposition or Gaussian filters that are carried out to identify the key points in the images during this step of the radiomics pipeline. Various sets of features can be studied and combined for developing the suitable model for diagnostic and prognostic purpose [40]. In DL approaches, features extraction is achieved implicitly via DNNs. Using these approaches, relevant features are extracted directly and automatically from the raw pixels of the original CT and CXR images [[93], [94], [95]] (see Table 2).
Alqudah et al. [96] utilizes SVM and RF for detection of COVID-19 at an early stage. They adopted a deep radiomics model for feature extraction. Specifically, they extracted features from the fully connected layer in a CNN, followed by use of machine learning to combine these deep features, to build a model to distinguish between COVID-19 vs. non-COVID-19 cases. Ghoshal and Tucker [97] used a Bayesian CNN (BCNN) method to distinguish the COVID-19 infection using CXRs. A classification accuracy of 90 % was reported. Salman et al. [98] employed a trained CNN for identifying the COVID-19 using CXRs. The sensitivity and specificity achieved from the model were 100 % and 100 %, respectively. Farooq and Hafeez [99] conducted a multi-stage fine-tuning scheme on pre-trained ResNet-50 architecture. The COVIDResNet model achieved on accuracy of 96.2 %. Asnaoui et al. [100] provided a comparative study on eight different learning techniques for the classification of COVID-19 using CXRs. MobileNet-V2 and Inception-V3 showed the highest accuracy of 96 % classification among the other models. Abbas et al. [101] used a specific deep CNN named as Decompose, Transfer, and Compose for diagnosis of COVID-19 utilizing CXRs. The accuracy and sensitivity of the suggested model were 95.12 % and 97.91 %, respectively.
3.4. Dimensionality reduction
Dimensionality reduction strategies, attempt to choose a small subset of the most relevant features from the all extracted features by removing irrelevant and noisy features. Once large number of features and well-curated datasets are available, they can then be used for data mining to identify radiomics signature. Feature selection is a significant step to avoid overfitting the model, removing non-informative or redundant predictors/features from the dataset. It hence reduces the computational complexity of the training, and can increase the model's performance. Overall, feature selection is important for proper applicability of the model in day-to-day clinical applications. In order to enhance the translation of radiomics into clinical practice, highly accurate and reliable ML approaches are required. Moreover, the performance of the model depends on how the various redundant and non-redundant features/variables are handled prior to testing the model.
The training data is defined as a collection of instances, represented by a set of patients, each with a collection of features and a desired category label. The classifier evaluates the training data and derives a model that can be used to predict the labels from the input features. For this process, AI approaches and statistical approaches were employed. At the other end, the data mining spectrum are hypothesis-driven methods that cluster features in line with information of content. By getting used of the advantages of these two approaches, the best models can be selected and further emphases in a particular medical context and, therefore, well-defined endpoint will be achieved. There are two categories of methods for dimensionality reduction: feature selection and feature extraction, which we discuss next. The former, extract a subset of features from a given set of (large) features. The latter, combines existing features into a reduced number of new features/dimensions; e.g. Principal Component Analysis (PCA) [102]. This term (“feature extraction”), as we use it here, should not be confused with extraction of hand-crafted radiomics features, which can also be referred to as feature extraction. In the present use, it is in the context of dimensionality reduction that it is used.
3.4.1. Feature selection
The feature selection step in Al-based radiomics workflow is the selection of the most relevant data for the respective task. Several steps are involved for feature selection based on machine learning algorithms and/or on the conventional statistical methods and data visualization. The selection process can be as follows: (a) select initial features; (b) visualize clusters of highly correlated radiomics metrics; (c) and allow selection of one representative feature per correlation cluster (patient cohort). The risk of model overfitting is high when the number of features in a model are high or the sample size for classification is low. Aiming to identify a proper sub-set of features from a given set of original features in data mining, feature selection is usually requiring pre-processing algorithms (PPA). PPA stages require (a) search strategy that utilized some methods to select a subset of features; (b) feature subsets utilizing classifiers, quality of the desired sub-set of features can be computed. Feature reduction methods are mainly grouped into two groups: Supervised and unsupervised. Two sub-categories are supervised technique are (1) filter- and (2) wrapper-based methods.
The filter-based method utilizes the data related specifications to assess the merits of the feature subset. The wrapper-based methods employ a specific classifier to estimate the significant features. Feature selection approaches using special group of filters tend to simultaneously select highly predictive but uncorrelated features. One of such filter is the Maximum Relevance Minimum Redundancy (MRMR) algorithm, which was first developed for feature selection of microarray data. Fu et al. [103] employed MRMR to find out the high correlation and low redundancy features from 2 sets of features (radiologists A and B) which were obtained to construct the COVID-19 radiomics signature.
Elsewhere, Li et al. [104] applied the Mann–Whitney U test to determine the correlation between features (extracted from 64 CT images of COVID-19 cases) and severity score. Then, the MRMR algorithm utilized to rank features according to their relevance to severity to select optimal feature subset. Rafid et al. [105] adopted filter-based and wrapper-based (hybrid method) to select the most relevant features. The MRMR and Double Input Symmetrical Relevance (in total 1144 features were obtained by discrete wavelet transform and CNN) feature selection methods along with recurrent feature elimination (RFE) techniques were adopted.
3.4.2. Feature extraction
Unsupervised techniques for feature reduction are divided into linear and nonlinear methods that aim to only keep low number of features. By adopting these methods, the new reduced set of features that was created from a combination of original features will be feed into the analysis. Hence, the original features will be discarded. In other words, the features that do not provide additional information will be removed. PCA is a multivariate statistical procedure that analyses dependent and inter-correlated features and extracts the important information by transforming it to a new set of orthogonal variables called principal components. A new set of principal components is obtained, each with certain variance, while the first principal component would have the highest contribution among others [106]. Subsequently, a number of important principal components are retained, while the rest are discarded, resulting in dimensionality reduction.
Rasheed et al. [107] employed PCA to further speed up the learning process as well as improve classification accuracy by selecting the highly relevant features. The overall reported accuracy without and with PCA was 98–100 % for Logistic regression and was 95–98 % for CNN for the detection of COVID-19 from CXR images. Khuzani et al. [108] utilized the kernel PCA method to reduce the dimensionality of original feature. By utilizing these methods, the obtained 64 new synthetic features out of 252 features. Attallah et al. [109] conducted a study to diagnose COVID-19 infections using CT images. They adopted PCA method on deep features. They reported 32 % computational coast reduction for analysis as well as accuracy of 94.7 % of accuracy, 95.6 % of sensitivity and of 93.7 % of specificity their COVID-19 detection mode.
4. Performance of radiomics-based methods in COVID-19 studies
The dominant approaches used in AI are ML and DL, which offer fast, automated, decisive strategies to enable improved assessment of COVID-19 infections. Using AI techniques, scientists succeeded in extracting features from COVID-19 CT and CXR datasets to extract findings that are specific and highly sensitive for early COVID-19 detection, prognosis and severity scoring. Moreover, ML and DL models have demonstrated comparable success to qualified radiologists and substantially increased the effectiveness of the radiology clinical practice. The related COVID-19 patterns differentiating COVID-19 infection from other lungs infections could be established if pertinent ML and/or DL analysis models are developed, trained and properly validated. Fig. 7 provides a comprehensive picture and summarises the AI-based methods in the context of COVID-19 reviewed in this paper.
Fig. 7.
Summary of AI-based methodology in COVID-19 studies.
4.1. Using CXR images
The SVM, as demonstrated by Sethy and Behera [115] is a classification method for COVID-19 CXR images compare to others deep learning-based methods. For the diagnosis of coronavirus infected patients, the technique proved to be useful for medical practitioners. For detecting COVID-19, ResNet50 + SVM, achieved better accuracy compare to FPR, F1 score, MCC, were 95.4 %, 95.5 %, 91.4 % and 90.8 %, respectively. Also, 115 COVID-19 and 435 non-COVID-19 cases were investigated by Liu et al. [116] and used to create a radiomics signature utilizing minimum LASSO, main radiomics features extracted from chest CT images were selected. The clinical model for the diagnosis of COVID-19 pneumonia ROC area of 0.98 and a successful cohort validation. The sensitivity and precision of the combined model were 85 % and 90 % respectively.
Different pre-trained CNN-based models (ResNet50, ResNet101, ResNet152, InceptionV3 and Inception-ResNetV2) were evaluated by Narin et al. [117] for the detection of COVID-19 using CXR. The pre-trained ResNet50 model provided the highest classification performance (96.1 % accuracy for Dataset-1, 99.5 % accuracy for Dataset-2 and 99.7 % accuracy for Dataset-3) among other four models used in the study. The ability of domain extension transfers learning (DETL) to detect COVID-19 was demonstrated by Basu et al. [62]. On a related large CXR dataset (normal, pneumonia, other illness, and COVID-19), they employed DETL, with a pre-trained CNN model. The overall accuracy of the model was found to be 90 %. In this study, Gradient Class Activation Map (Grad-CAM) for identifying and visualizing the areas where the model focused most during classification.
A weakly-supervised DL-based software system was tested by Zheng et al. [78] to detect COVID-19 using 3D CT images. A pre-trained UNet was used for lung region segmentation. The results showed 90 % sensitivity and 91 % specificity. The algorithm achieved an accuracy of 90 % to define COVID-19 and non-COVID groups. Ioannis and Tazani [118] extracted features from numerous images of CXR (COVID-19 disease, common and normal bacterial pneumonia). The findings indicate that DL using CXR can extract essential biomarkers related to COVID-19 disease. A 96.8 % accuracy, 98.7 % sensitivity, and 96.5 % specificity were reports in this study.
4.2. Using CT chest scans
On three separate batches of patients, including 148 patients (training sets), 264 patients (validation sets), Xu et al. [119] developed a radiomics-based model to create a prognosis prediction model and to check the predictive efficiency. They illustrated that the nomogram scoring system is a potential predictor for the short-term results of COVID-19 with a sensitivity of 81.3 % and specificity of 87.3 % in CT, C-reactive protein (CRP) and Radscore. In the independent validation datasets, the predictive performance of this model was also validated, giving a sensitivity of 88.8 % and specificity of 73 %. Huang et al. [120] investigated 181 cases of viral pneumonia grouped into COVID-19 and non-COVID-19. The combined models of CT signs and selected features showed that discrimination between COVID-19 and non-COVID-19 cases was better identified compared to the individual radiomics-based models. Combining CT signs and Radiomics characteristics achieved sensitivity was 91.9 % and specificity was 85.9 %; and accuracy was 88.9 %.
CT images of 176 COVID-19 patients were analysed by Tang et al. [86]. A RF model was trained and showed 87.5 % accuracy, 0.9 and 0.70 of true positive rate, and true negative rate, respectively for the detection of COVID-19. Chen et al. [79] studied CT images of 51 COVID-19 and 55 CT images of other types of diseases. The model achieved a per-patient sensitivity of 100 %, and accuracy of 95.3 %, a positive prediction value of 84.7 %. Moreover, they reported negative prediction value of 100 %; a per-image sensitivity of 94.4 %, with an accuracy of 98.9 % in retrospective datasets. Qi et al. [48] analysed radiomics-based models based on 6 s-order characteristics that were successful in distinguishing short-term and long-term hospitalization in patients using Logistic Regression-associated pneumonia and Random Forest-associated pneumonia in the research datasets, respectively. A sensitivity and specificity of 100 % and 89 % was achieved by the logistic regression model, and similar performance was shown by the RF model with a sensitivity and specificity of 75 % and 100 % in the test datasets.
Liu et al. [116] claimed that the decision curve analysis verified the clinical usefulness of the COVID-19 radiomics model for detection. Two tests were conducted on 115 patients COVID-positive and 435 COVID-negative ones. In order to distinguish COVID-19 by radiomics signature, a clinical model was developed and validated using specific data system such as CO-RADS. The model achieved 96 % sensitively compared to 75 % sensitivity obtained by the clinical model. Fang et al. [85] performed a study on 46 patients with COVID-19 and 29 other forms of pneumonia. Of the total lesions, 77 radiomics metrics were extracted. Multiple cross-validation was used to pick the primary characteristics after clustering by SVM to create the radiomics signature in the experiments. To test model efficiency, the AUC and calibration curve were used. The proposed model yielded the AUCs from the training set and the testing set of 86.2 % and 82.6 %, respectively. Wei et al. [121] retrospectively collected two groups conducted a study which included on CT images from 81 COVID-19 patients. Using LK2.1 software, the texture features were extracted. To find the characteristics with maximum correlation and minimum redundancy, the minimum redundancy and maximum relevance methods were carried out. To compare the efficiency of two models, ROC analysis were performed. The AUC values was 0.93 % and 0.95 % for textural features and clinical features, respectively.
A prognostic method for predicting poor results in COVID-19 based on CT imaging was suggested by Wu et al. [122]. A total of 492 patients were classified into (a) the early-phase group (CT scans one week after onset of symptoms); (b) the late-phase group (CT scans one week after onset of symptoms). The radiomics signature (RadScore software) was developed to build the low-pass Gaussian filter, and LASSO reduction methods/classification methods. Afterwards, the clinical model and the clinic-radiomics signature (CrrScore), was stablished by performing a Fine-Gray competing risk regression. They found that in group (a), the CrrScore estimated 85 % poorness, and predicted the probability of 28-day poor results of 86.2 %. In group (b), the RadScore alone achieved similar performance to the CrrScore in predicting poor outcome (88.5 %), and 28-day poor outcome probability (97.6 %).
5. Clinical impact of AI-based COVID-19 studies
The first significant challenge in dealing with patients with COVID-19 symptoms is to identify and prioritize cases so that the physician can isolate infected patients as soon as possible. For COVID-19 cases, a triage algorithm needs to prioritize those who require emergency medical care, according to the severity of infection. Radiomics models may assist radiologists and clinicians in making fast and accurate diagnosis and prognosis, to ensure appropriate clinical management and resources allocation. Moreover, radiomics models have the potential to distinguish COVID-19 from pneumonia caused by other etiologies. The major clinical impact highlighted by radiomics-based COVID-19 studies were in the areas of: screening of infection, identification and detection, prediction of disease progression, and analysis of survival rate. The following subsections summarize the most relevant studies for screening, diagnosis, prediction, and severity quantification of COVID-19 using AI-based methods.
5.1. Screening
Screening patients attending emergency departments for COVID-19 at time of overwhelming outbreak using RT-PCR is challenging process as that might take up to 24 h to obtain the results. Screening with Chest CT was adopted in many centres across the globe. The screening was augmented by using AI-based applications that played a major role in regions with acute shortage of imaging professionals’ and Radiologists. Fang et al. [85] used a SVM-based radiomics method to screen COVID-19 infection based on chest CT images from other types of pneumonia. This method achieved AUCs of 86 % and 83 % with the training and test sets, respectively. Rezaeijo et al. [123] successfully assessed the diagnostic value of several ML approaches for screening COVID-19. They reported an accuracy range of AUC between 30 % and 98 % for recursive feature elimination (RFE)+Multinomial Naive Bayes (MNB), Rebadging (BAG), and RFE + decision tree (DT) classifiers. They also reported an accuracy range of AUC between 30 % and 99 % for mutual information (MI)+MNB and RFE + k-nearest neighborhood (KNN) classifiers. In their study, the RFE + BAG and RFE + DT classifiers achieved the highest prediction accuracy of 98 %, followed by an accuracy of 97 % with MI Gaussian Naive Bayes (GNB) and logistic regression (LGR)+DT classifiers. The RFE + KNN classifier used for features selection achieved the highest AUC of 99 %, followed by RFE + BAG and RFE + gradient boosting decision tree classifiers. Chandra et al. [124] developed radiomics-based methods that uses a majority voting-based classifier to screen COVID-19 from chest X-ray images.
5.2. Differential diagnosis
Radiomics features were heavily investigated in an attempt to differentiate different types of viral pneumonia or to classify severity of COVID-19 pneumonia. A radiomics model combining 8 radiomics features and 5 selected clinical variables was constructed and used for the diagnosis of COVID-19 pneumonia [116] The combined radiomics model achieved a better diagnostic accuracy, compared to CO-RADS used by radiologists, with a 85 % sensitivity and 90 % specificity. Wang et al. [125] developed a radiomics-feature-based model that was significantly associated with the classification of COVID-19 pneumonia using a multi-classifier approach. The findings of this study extend the understanding of imaging characteristics of COVID-19 pneumonia. Moreover, Junior and co-authors [126] demonstrated that radiomics not only correlated with the etiologic agent of acute infections but also supported short-term risk stratification of COVID-19 patients. Elsewhere, Xie et al. [127] developed a non-invasive radiomics model using chest CT images for the detection of COVID-19 considering GGO lesions. The preliminary results demonstrated that the radiomic model could be used as supplementary tool for improving specificity for COVID-19 in a population confounded by GGO changes from other etiologies. Tabatabaei and co-authors [128] performed a preliminary study on CT scans using ML methods. The reported results were promising for differentiating COVID-19 and H1N1 influenza. Elsewhere, Guiot et al. [110] developed a AI framework to differentiate COVID-19 from other routine clinical conditions in a fully automated fashion, hence providing rapid accurate diagnosis of patients suspected of COVID-19 infections to enable early intervention.
5.3. Prediction
Providing an accurate prediction of the evolution of SARS-CoV-2 infections is expected to facilitate timely implementation of isolation procedures and early intervention as well as predicting patient's clinical outcome. Giraudo et al. [129] built a rapid CXR-based radiomics integrated model that incorporated demographics, first-line laboratory findings, and clinical findings of positive COVID-19 cases obtained upon admission. The model showed that a combination of radiomics and a basic inflammatory index obtained at admission can predict ICU admission. According to Gülbay et al. [130] COVID-19 and atypical pneumonia-associated GGO lesions and consolidation could be predicted with high accuracy (80 % in COVID-19 and 81 % in atypical pneumonia). Roundness and peripheral location were found to be the most effective characteristics for identifying a GGO lesion with COVID-19, but were both ineffective in predicting lesions in the consolidation stage. The findings of Chaddad et al. [131] demonstrated that deep CNNs with transfer learning can predict COVID-19 in CT and CXR images. The proposed model could aid radiologists in improving the accuracy of their diagnosis and the efficiency of managing COVID-19 patients. Sinha and Rati [132] employed an AI-based approach to predict survival in COVID-19-isolated individuals. The multi-autoencoders-based model was developed and tested on 5165 COVID-19 cases before it was validated on 1533 patients who were quarantined. The findings identified the key points in the outbreak spread, indicating that the models driven by machine intelligence and deep learning can be effective in providing a quantitative view of epidemical outbreak. The combined model was shown to have an accuracy of 99 %. Yuan et al. [133] reported a risk score-based approach that could predict the mortality of COVID-19 patients by more than 12 days with more than 90 % accuracy across all cohorts.
5.4. Severity quantification
Severity quantification is a common measure to assess patients’ health condition. A new AI-based method that automatically performs 3D segmentation and quantifies abnormal CT patterns in COVID-19 (e.g., GGO and consolidation) was introduced by Changanti et al. [134]. Based on DL and deep reinforcement learning, the method provided combined assessments of lung involvement to assess COVID-19 abnormalities (percentage of lungs involvement) and the existence of large opacities (severity scores). The method offers the potential of a better management of patients and clinical resources. Using a commercially available DL-based technology, Huang et al. [135] determined that quantification of lung opacification in COVID-19 from CT images was significantly different among groups with varying clinical severities. This method could potentially eliminate subjectivity in the evaluation process and follow-up of COVID-19 pulmonary findings. Shen et al. [81] performed a retrospective examination on COVID-19 cases based on severity score by radiologists. The computer-aided quantification showed a statistically significant percentage of lesions in lower lobes than the ones in the lower lobes assessed by the radiologists (R = 0.63, P < 0.05). On a total of 176 positive COVID-19 CT scans, Tang et al. [86] computed 63 features. Their findings revealed that the volume and ratio of GGO areas (in relation to the total lung volume) were strongly associated with the severity of COVID-19.
6. Limitations and recommendations for future studies
While in several studies, imaging-AI based approaches have demonstrated significant potential as non-invasive methods for diagnosis and prognosis of COVID-19, the field still faces several challenges. Technical choices can greatly influence the clinical applicability of AI-based methods. Sample size plays an important role in AI-based diagnosis and prognostic of COVID-19. Reliable and robust model derivation and validation efforts reduce risks of misrepresentation and false discoveries caused by small sample sizes. Increasing sample sizes through data collection from several clinical centres, coupled with robust AI derivation and validation efforts, may more reliable and generalizable outcomes. Some recent reports are less enthusiastic about AI research in the management of COVID-19 disease [136,137]. Future studies should address the concerns raised. Some potential avenues are discussed below.
6.1. Sample size and data collection
Translation of AI-based methods to routine clinical practice is hampered by the proper validation of existing research studies. Insufficient data due to small sample sizes and toy datasets for different types or stages of disease or infection (e.g. mycoplasma infections) can induce significant selection bias as well as imbalanced COVID-19 dataset [63,138]. A study on a subgroup of early COVID-19 paediatric patients was not successful due to lack of sufficient CT scans [28].
Five common categories of COVID-19 severity based on total lungs involvements (LI) (score 0: 0 % LI; score 1: 5 % LI; score 2: 5%–25 % LI; score 3: 26%–50 % LI; score 4: 51%–75 % LI; as well as score 5: 75 % lobar involvement) have been considered. The severity scores are classified as negative, moderate, non-severe, critical and extreme [139]. Elsewhere, given small sample size of COVID-19 paediatric patients were less in some of these categories, only two categories were examined [42]. A solution is to use pertained networks (transfer learning) followed by fine-tuning using COVID-19 datasets. However, employing pre-trained networks (commonly for non-medical applications) in real medical applications is still challenging.
For different types of pneumonia only one case was reported and therefore, the characteristics comparison between COVID-19 and other types of pneumonia failed [26,28,37]. Overall, for proper generalizability of models, multi-centric, large datasets are required. Further, using appropriate data augmentation, transfer learning based on other COVID-19 models, and federated learning frameworks, the accuracy of the models might be enhanced [25]. To attain adequate balance classes for AI analysis, some techniques such as modified loss functions or resampling utilizing Synthetic Minority Oversampling Technique (SMOT), down-sampling and up-sampling might be helpful.
6.2. Imaging protocols (non-standard)
Apart from the issue of sample size, imaging protocols are often not standardized for such studies. The effect of these settings on radiomics features was investigated to minimize their influence by eliminating features that are sensitive (i.e. not reproducible) to those parameters [140]. Different brands of imaging scanners are available in the market, rendering identical performances and standard protocols of scanning for COVID-19 patients highly problematic across different hospitals and imaging centres. Alternatively, harmonization of acquired imaging datasets might be helpful for data collection at different centres [141].
6.3. Imaging and non-imaging factors
It was reported that the impact of the virus on the lungs is highly related to the host factor [142]. The CT data on its own is not sufficient to identify the types of viral pneumonia. A study stated that clinical and clinic-radiomics combined model results in better diagnosis of COVID-19 pneumonia compare to COVID-19 reporting and data system (CO-RADS) only model [116]. A recent study revealed that AI-based prediction modeling using CXR radiographs was insufficient unless diagnostic test results such as RT-PCR are also available [143]. Hence, clinical features and laboratory examination data are required to be investigated in addition to imaging features [26]. In other words, AI-based models should not merely rely on images, and combination of imaging data with clinical information enhances model applicability.
6.4. Segmentation methods
Segmentation can significantly impact feature extraction and hence the classification and clinical outcomes. The expertise of the operator in semi-automated or manual methods is a key explicit feature and may dictate the occurrence of the outcomes. Chest imaging scans of COVID-19 infected patients that have small ground-glass lesions could be missed when the ROI is automatically delineated using existing methods. However, fully automated segmentation methods based on deep learning have the potential to replace less automated methods, if large, accurate reference truth datasets are generated for training. Overall, 3D segmentation networks and the adoption of precise ground-truth annotated by radiologists is desirable and more efficient for explicit radiomics feature extraction and analysis [25].
Meticulous segmentation of COVID-19 images makes extraction of the radiomics matrix a great challenge for accurate labelling and clustering of regions [91]. Nevertheless, AI-based analysis is used on a variety of datasets, including labelled, non-labelled, mixture or small labelled and huge number of non-labelled data. Depending on the objectives and expected outcomes of the studies, the lack of accurate segmentation models can be compensated for by employing deep learning methods. In particular, manual segmentation can be replaced utilizing large training datasets, neural networks and evaluation algorithms. The extracted features can be hence identified to facilitate rapid and more accurate diagnosis as well as timely management of COVID-19 patients.
6.5. Availability of COVID-19 databases
Sharing COVID-19 scan details in publications that focus on CT and CXR datasets has been critical for COVID-19 first-line diagnosis during the early stages of the pandemic [144]. Later, the availability of free online COVID-19 CT and CXR images, contributed to proliferation of AI approaches and high enthusiasm about fostering diagnosis and prognosis for COVID-19 patients [29]. Subsequently, number of AI-based COVID-19 publications has increased rapidly. Public datasets vary remarkably in terms of ancillary datasets that directly enhance the performance of the AI models. The majority of datasets are still private, and publications based on public datasets are neither comprehensive enough nor clinically useful [136]. To overcome this hurdle in the application of AI-based COVID-19 studies, there is significant need for ongoing construction and expansion of COVID-19 database from carefully curated imaging and non-imaging data.
7. Conclusion
Prompt assessment of COVID-19 can lead to improved management and control of potential transmission of the virus. Our review reveals the prevalence of AI techniques as applied to COVID-19 CT/CXR images to enhance the diagnosis and prognosis of COVID-19 pneumonia. Developing accurate and highly sensitive AI-based models for clinical assessment and follow-up of COVID-19 patients is critical. AI-based approaches consist of a chain of key steps towards proper identification of AI-based signatures. Opting for systematic development and assessment of techniques will be critical towards accurate, reliable and generalizable AI-based models. Our review also sheds light on the different aspects of AI-based analysis workflows in COVID-19 infections. The findings and recommendations can guide future studies to develop alternative methods and increase the accuracy and sensitivity of AI-based models. Moreover, there is prospect to convince clinical decision makers in establishing CXR/CT-based assessment of COVID-19 pneumonia as an alternative diagnostic pathway. Overall, we have also elaborated upon the full chain of AI-based workflows from image acquisition methods, through segmentation, feature extraction and classification methods as well as model development and validation. Moreover, the challenges and limitations of current COVID-19 AI-based studies were highlighted to help initiate further efforts to overcome current limitations.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This work was supported by the Omani Research Council Grant, grant number RC/COVID-MED/RADI/20/01.
References
- 1.Chi Q., Dai X., Jiang X., Zhu L., Du J., Chen Y., et al. Differential diagnosis for suspected cases of coronavirus disease. 2019: a retrospective study. BMC Infect. Dis. 2020;20(1):1–8. doi: 10.1186/s12879-020-05383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G., et al. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagnostic and interventional imaging. 2020;101(5):263–268. doi: 10.1016/j.diii.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiri I., Abdollahi H., Atashzar M.R., Rahmim A., Zaidi H. A theranostic approach based on radiolabeled antiviral drugs, antibodies and CRISPR-associated proteins for early detection and treatment of SARS-CoV-2 disease. Nucl. Med. Commun. 2020;41(9):837–8340. doi: 10.1097/MNM.0000000000001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu G., Zhou S., Wang Y., Li X. Machine learning: a predication model of outcome of SARS-CoV-2 pneumonia. 2020. [DOI] [PMC free article] [PubMed]
- 5.La Marca A., Capuzzo M., Paglia T., Roli L., Trenti T., Nelson S.M. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. Biomed. Online. 2020 doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dheyab M.A., Khaniabadi P.M., Aziz A.A., Jameel M.S., Mehrdel B., Oglat A.A., et al. Focused role of nanoparticles against COVID-19: diagnosis and treatment. Photodiagnosis Photodyn. Ther. 2021:102287. doi: 10.1016/j.pdpdt.2021.102287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giri B., Pandey S., Shrestha R., Pokharel K., Ligler F.S., Neupane B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2020:1–14. doi: 10.1007/s00216-020-02889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilyas M., Rehman H., Naït-Ali A. 2020. Detection of Covid-19 from Chest X-Ray Images Using Artificial Intelligence: an Early Review. arXiv preprint arXiv:200405436. [Google Scholar]
- 10.Blain M., Kassin M.T., Varble N., Wang X., Xu Z., Xu D., et al. Determination of disease severity in COVID-19 patients using deep learning in chest X-ray images. Diagn. Interventional Radiol. 2021;27(1) doi: 10.5152/dir.2020.20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arabi H., AkhavanAllaf A., Sanaat A., Shiri I., Zaidi H. The promise of artificial intelligence and deep learning in PET and SPECT imaging. Phys. Med. 2021;83:122–137. doi: 10.1016/j.ejmp.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Arabi H., Zaidi H. Applications of artificial intelligence and deep learning in molecular imaging and radiotherapy. European Journal of Hybrid Imaging. 2020;4(1):1–23. doi: 10.1186/s41824-020-00086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R., Tie X., Qi Z., Bevins N.B., Zhang C., Griner D., et al. Diagnosis of coronavirus disease 2019 pneumonia by using chest radiography: value of artificial intelligence. Radiology. 2021;298(2):E88. doi: 10.1148/radiol.2020202944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomaszewski M.R., Gillies R.J. The biological meaning of radiomic features. Radiology. 2021;202553 doi: 10.1148/radiol.2021202553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nazari M., Shiri I., Zaidi H. Radiomics-based machine learning model to predict risk of death within 5-years in clear cell renal cell carcinoma patients. Comput. Biol. Med. 2021;129:104135. doi: 10.1016/j.compbiomed.2020.104135. [DOI] [PubMed] [Google Scholar]
- 16.Shiri I., Hajianfar G., Sohrabi A., Abdollahi H., Ps S., Geramifar P., et al. Repeatability of radiomic features in magnetic resonance imaging of glioblastoma: test-retest and image registration analyses. Med. Phys. 2020;47(9):4265–4280. doi: 10.1002/mp.14368. [DOI] [PubMed] [Google Scholar]
- 17.Nazari M., Shiri I., Hajianfar G., Oveisi N., Abdollahi H., Deevband M.R., et al. Noninvasive Fuhrman grading of clear cell renal cell carcinoma using computed tomography radiomic features and machine learning. Radiol. Med. 2020;125(8):754–762. doi: 10.1007/s11547-020-01169-z. [DOI] [PubMed] [Google Scholar]
- 18.Du D., Gu J., Chen X., Lv W., Feng Q., Rahmim A., et al. Integration of PET/CT radiomics and semantic features for differentiation between active pulmonary tuberculosis and lung cancer. Mol. Imag. Biol. 2021;23(2):287–298. doi: 10.1007/s11307-020-01550-4. [DOI] [PubMed] [Google Scholar]
- 19.Du D., Feng H., Lv W., Ashrafinia S., Yuan Q., Wang Q., et al. Machine learning methods for optimal radiomics-based differentiation between recurrence and inflammation: application to nasopharyngeal carcinoma post-therapy PET/CT images. Mol. Imag. Biol. 2020;22(3):730–738. doi: 10.1007/s11307-019-01411-9. [DOI] [PubMed] [Google Scholar]
- 20.Edalat-Javid M., Shiri I., Hajianfar G., Abdollahi H., Arabi H., Oveisi N., et al. Cardiac SPECT radiomic features repeatability and reproducibility: a multi-scanner phantom study. J. Nucl. Cardiol. 2020 doi: 10.1007/s12350-020-02109-0. [DOI] [PubMed] [Google Scholar]
- 21.Shiri I., Arabi H., Geramifar P., Hajianfar G., Ghafarian P., Rahmim A., et al. Deep-JASC: joint attenuation and scatter correction in whole-body (18)F-FDG PET using a deep residual network. Eur. J. Nucl. Med. Mol. Imag. 2020;47(11):2533–2548. doi: 10.1007/s00259-020-04852-5. [DOI] [PubMed] [Google Scholar]
- 22.Leung K.H., Marashdeh W., Wray R., Ashrafinia S., Pomper M.G., Rahmim A., et al. A physics-guided modular deep-learning based automated framework for tumor segmentation in PET. Phys. Med. Biol. 2020;65(24):245032. doi: 10.1088/1361-6560/ab8535. [DOI] [PubMed] [Google Scholar]
- 23.Sanaat A., Shiri I., Arabi H., Mainta I., Nkoulou R., Zaidi H. Deep learning-assisted ultra-fast/low-dose whole-body PET/CT imaging. Eur. J. Nucl. Med. Mol. Imag. 2021 doi: 10.1007/s00259-020-05167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akhavanallaf A., Shiri I., Arabi H., Zaidi H. Whole-body voxel-based internal dosimetry using deep learning. Eur. J. Nucl. Med. Mol. Imag. 2020 doi: 10.1007/s00259-020-05013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiri I., AmirMozafari Sabet K., Arabi H., Pourkeshavarz M., Teimourian B., Ay M.R., et al. Standard SPECT myocardial perfusion estimation from half-time acquisitions using deep convolutional residual neural networks. J. Nucl. Cardiol. 2020 doi: 10.1007/s12350-020-02119-y. [DOI] [PubMed] [Google Scholar]
- 26.Xueyan M., Hao-Chih L., Kai-yue D., Mingqian H., Bin L., Chenyu L., et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;16:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imag. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Bao Y., Wen Y., Lu H., Luo H., Xiang Y., et al. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imag. 2020;39(8):2572–2583. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 29.Bai H.X., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296(3):E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L., et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020;30(12):6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessmann N., Sánchez C.I., Beenen L., Boulogne L.H., Brink M., Calli E., et al. Automated assessment of CO-RADS and chest CT severity scores in patients with suspected COVID-19 using artificial intelligence. Radiology. 2020 doi: 10.1148/radiol.2020202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Wang D., Shao J., Tian S., Tan W., Ma Y., et al. A deep learning integrated radiomics model for identification of coronavirus disease 2019 using computed tomography. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-83237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Born J., Beymer D., Rajan D., Coy A., Mukherjee V.V., Manica M., et al. On the role of artificial intelligence in medical imaging of COVID-19. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 35.Bhattacharya S., Maddikunta P.K.R., Pham Q.-V., Gadekallu T.R., Chowdhary C.L., Alazab M., et al. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: a survey. Sustainable cities and society. 2021;65:102589. doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoeibi A., Khodatars M., Alizadehsani R., Ghassemi N., Jafari M., Moridian P., et al. 2020. Automated Detection and Forecasting of Covid-19 Using Deep Learning Techniques: A Review. arXiv preprint arXiv:200710785. [Google Scholar]
- 37.Suri J.S., Agarwal S., Gupta S.K., Puvvula A., Biswas M., Saba L., et al. A narrative review on characterization of acute respiratory distress syndrome in COVID-19-infected lungs using artificial intelligence. Comput. Biol. Med. 2021:104210. doi: 10.1016/j.compbiomed.2021.104210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sardar R., Sharma A., Gupta D. Machine learning assisted prediction of prognostic biomarkers associated with COVID-19, using clinical and proteomics data. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.636441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S., Xu J., Luo P., Yang L., Song S., Tan X., et al. Plasma metabolomic profiles and clinical features in recovered COVID-19 patients without previous underlying diseases 3 Months after discharge. 2020. [DOI] [PMC free article] [PubMed]
- 40.Barstugan M., Ozkaya U., Ozturk S. 2020. Coronavirus (Covid-19) Classification Using Ct Images by Machine Learning Methods. arXiv preprint arXiv:200309424. [Google Scholar]
- 41.Shi F., Xia L., Shan F., Wu D., Wei Y., Yuan H., et al. Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. 2020. arXiv preprint arXiv:200309860. [DOI] [PubMed]
- 42.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., et al. A machine learning-based model for survival prediction in patients with severe COVID-19 infection. MedRxiv. 2020 doi: 10.1101/2020.02.27.20028027. [DOI] [Google Scholar]
- 43.Magar R., Yadav P., Farimani A.B. Potential neutralizing antibodies discovered for novel corona virus using machine learning. 2020. arXiv preprint arXiv:200308447. [DOI] [PMC free article] [PubMed]
- 44.Ong E., Wong M.U., Huffman A., He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front. Immunol. 2020;11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu H., Guo Y., Xiang Y., Sun C., Yang T. Data‐driven discovery of a clinical route for severity detection of COVID‐19 paediatric cases. IET Cyber‐Systems and Robotics. 2020;2(4):205–206. doi: 10.1101/2020.03.09.20032219. [DOI] [Google Scholar]
- 46.Fong S.J., Li G., Dey N., Crespo R.G., Herrera-Viedma E. Finding an accurate early forecasting model from small dataset: a case of 2019-ncov novel coronavirus outbreak. 2020. arXiv preprint arXiv:200310776. [DOI]
- 47.Meng Z., Wang M., Song H., Guo S., Zhou Y., Li W., et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv. 2020. [DOI]
- 48.Qi X., Jiang Z., Yu Q., Shao C., Zhang H., Yue H., et al. medRxiv; 2020. Machine Learning-Based CT Radiomics Model for Predicting Hospital Stay in Patients with Pneumonia Associated with SARS-CoV-2 Infection: A Multicenter Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae J., Kapse S., Singh G., Phatak T., Green J., Madan N., et al. 2020. Predicting Mechanical Ventilation Requirement and Mortality in COVID-19 Using Radiomics and Deep Learning on Chest Radiographs: A Multi-Institutional Study. arXiv preprint arXiv:200708028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Truhn D., Schrading S., Haarburger C., Schneider H., Merhof D., Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 2019;290(2):290–297. doi: 10.1148/radiol.2018181352. [DOI] [PubMed] [Google Scholar]
- 51.Yang N., Liu F., Li C., Xiao W., Xie S., Yuan S., et al. Diagnostic classification of coronavirus disease 2019 (COVID-19) and other pneumonias using radiomics features in CT chest images. 2020. [DOI] [PMC free article] [PubMed]
- 52.Feng Y., Liu S., Cheng Z., Quiroz J., Chen P., Lin Q., et al. medRxiv; 2020. Severity Assessment and Progression Prediction of COVID-19 Patients Based on the LesionEncoder Framework and Chest CT. [DOI] [Google Scholar]
- 53.Yang X., Weixiang C., Hongyi R., Jianping Z., Lihua W., Rui J., et al. A rapid screening classifier for diagnosing COVID-19. Int. J. Biol. Sci. 2021;17(5):539–548. doi: 10.7150/ijbs.53982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiri I., Sorouri M., Geramifar P., Nazari M., Abdollahi M., Salimi Y., et al. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients. Comput. Biol. Med. 2021;132:104304. doi: 10.1016/j.compbiomed.2021.104304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiaffino S., Tritella S., Cozzi A., Carriero S., Blandi L., Ferraris L., et al. Diagnostic performance of chest X-ray for COVID-19 pneumonia during the SARS-CoV-2 pandemic in Lombardy, Italy. J. Thorac. Imag. 2020;35(4):W105–W106. doi: 10.1097/RTI.0000000000000533. [DOI] [PubMed] [Google Scholar]
- 56.Borghesi A., Maroldi R. COVID-19 outbreak in Italy: experimental chest X-ray scoring system for quantifying and monitoring disease progression. La radiologia medica. 2020. [DOI] [PMC free article] [PubMed]
- 57.Jacobi A., Chung M., Bernheim A., Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin. Imag. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hammoudi K., Benhabiles H., Melkemi M., Dornaika F., Arganda-Carreras I., Collard D., et al. 2020. Deep Learning on Chest X-Ray Images to Detect and Evaluate Pneumonia Cases at the Era of COVID-19. arXiv preprint arXiv:200403399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das N.N., Kumar N., Kaur M., Kumar V., Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM. 2020 doi: 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bukhari S.U.K., Bukhari S.S.K., Syed A., Shah S.S.H. The diagnostic evaluation of Convolutional Neural Network (CNN) for the assessment of chest X-ray of patients infected with COVID-19. medRxiv. 2020. [DOI]
- 61.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative COVID-19 Biomarkers from X-Ray images with Deep Learning approach and image data related to Pulmonary Diseases. J. Med. Biol. Eng. 2020 doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basu S., Mitra S. 2020. Deep Learning for Screening COVID-19 Using Chest X-Ray Images. arXiv preprint arXiv:200410507. [Google Scholar]
- 63.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M. 2020. Finding Covid-19 from Chest X-Rays Using Deep Learning on a Small Dataset. arXiv preprint arXiv:200402060. [Google Scholar]
- 64.Wang L., Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. 2020. arXiv preprint arXiv:200309871. [DOI] [PMC free article] [PubMed]
- 65.Dai W.C., Zhang H.W., Yu J., Xu H.J., Chen H., Luo S.P., et al. CT imaging and differential diagnosis of COVID-19. Can. Assoc. Radiol. J. 2020;71(2):195–200. doi: 10.1177/0846537120913033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aydin A., Demirtas Z., Ok M., Erkus H., Cebi G., Uysal E., et al. 3D printing in the battle against COVID-19. Emergent materials. 2021;4:363–386. doi: 10.1007/s42247-021-00164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiri I., Akhavanallaf A., Sanaat A., Salimi Y., Askari D., Mansouri Z., et al. Ultra-low-dose chest CT imaging of COVID-19 patients using a deep residual neural network. Eur. Radiol. 2021;31(3):1420–1431. doi: 10.1007/s00330-020-07225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., et al. Lung infection quantification of covid-19 in ct images with deep learning. 2020. arXiv preprint arXiv:200304655. [DOI]
- 70.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. 2020 doi: 10.1101/2020.02.23.20026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020;296(2):E46–E54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 74.Qingsen Y., Bo W., Dong G., Chuan L., Wei Z., Jianhu S., et al. COVID-19 chest CT image segmentation network by multi-scale fusion and enhancement operations. IEEE transactions on big data. 2021;7(1):13–24. doi: 10.1109/TBDATA.2021.3056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Budak Ü., Çıbuk M., Cömert Z., Şengür A. Efficient COVID-19 segmentation from CT slices exploiting semantic segmentation with integrated attention mechanism. J. Digit. Imag. 2021 doi: 10.1007/s10278-021-00434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moradi S., Oghli M.G., Alizadehasl A., Shiri I., Oveisi N., Oveisi M., et al. MFP-Unet: a novel deep learning based approach for left ventricle segmentation in echocardiography. Phys. Med. 2019;67:58–69. doi: 10.1016/j.ejmp.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Zhao X., Zhang P., Song F., Fan G., Sun Y., Wang Y., et al. D2A U-net: automatic segmentation of COVID-19 CT slices based on dual attention and hybrid dilated convolution. Comput. Biol. Med. 2021;135:104526. doi: 10.1016/j.compbiomed.2021.104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., et al. medRxiv; 2020. Deep Learning-Based Detection for COVID-19 from Chest CT Using Weak Label. [DOI] [Google Scholar]
- 79.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv. 2020 doi: 10.1101/2020.02.25.20021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang L., Zhang X., Wang Y., Zeng X. Severe COVID-19 pneumonia: assessing inflammation burden with volume-rendered chest CT. Radiology: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200044. e200044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen C., Yu N., Cai S., Zhou J., Sheng J., Liu K., et al. Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. Journal of pharmaceutical analysis. 2020;10(2):123–129. doi: 10.1016/j.jpha.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elaziz M.A., Hosny K.M., Salah A., Darwish M.M., Lu S., Sahlol A.T. New machine learning method for image-based diagnosis of COVID-19. PloS One. 2020;15(6) doi: 10.1371/journal.pone.0235187. e0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 2020;126:104037. doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma J., Wang Y., An X., Ge C., Yu Z., Chen J., et al. Towards efficient covid-19 ct annotation: a benchmark for lung and infection segmentation. 2020. arXiv preprint arXiv:200412537. [DOI]
- 85.Fang M., He B., Li L., Dong D., Yang X., Li C., et al. CT radiomics can help screen the coronavirus disease 2019 (COVID-19): a preliminary study. Sci. China Inf. Sci. 2020;63(7) doi: 10.1007/s11432-020-2849-3. [DOI] [Google Scholar]
- 86.Tang Z., Zhao W., Xie X., Zhong Z., Shi F., Liu J., et al. 2020. Severity Assessment of Coronavirus Disease 2019 (COVID-19) Using Quantitative Features from Chest CT Images. arXiv preprint arXiv:200311988. [Google Scholar]
- 87.Keshani M., Azimifar Z., Tajeripour F., Boostani R. Lung nodule segmentation and recognition using SVM classifier and active contour modeling: a complete intelligent system. Comput. Biol. Med. 2013;43(4):287–300. doi: 10.1016/j.compbiomed.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 88.Deng-Ping F., Tao Z., Ge-Peng J., Yi Z., Geng C., Huazhu F., et al. Inf-net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imag. 2020;38(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 89.Fei S., Yaozong G., Jun W., Weiya S., Nannan S., Miaofei H., et al. Lung infection quantification of COVID-19 in CT images with deep learning. 2020. ArXiv preprint arXiv:200304655. [DOI]
- 90.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., et al. Abnormal lung quantification in chest CT images of COVID‐19 patients with deep learning and its application to severity prediction. Med. Phys. 2020;48(4):1633–1645. doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shiri I., Arabi H., Salimi Y., Sanaat A.H., Akhavanalaf A., Hajianfar G., et al. COLI-NET: fully automated COVID-19 lung and infection pneumonia lesion detection and segmentation from chest CT images. medRxiv. 2021:21255163. doi: 10.1101/2021.04.08.21255163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H.J., Andrearczyk V., Apte A., et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farid A.A., Selim G.I., Khater H.A.A. A novel approach of CT images feature analysis and prediction to screen for corona virus disease (COVID-19) 2020. [DOI]
- 94.Özkaya U., Öztürk Ş., Barstugan M. Big Data Analytics and Artificial Intelligence against COVID-19: Innovation Vision and Approach. Springer; 2020. Coronavirus (covid-19) classification using deep features fusion and ranking technique; pp. 281–295. [Google Scholar]
- 95.Islam M.Z., Islam M.M., Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Informatics in medicine unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alqudah A.M., Qazan S., Alquran H., Qasmieh I.A., Alqudah A. Covid-2019 detection using x-ray images and artificial intelligence hybrid systems. 2020. [DOI]
- 97.Ghoshal B., Tucker A. 2020. Estimating Uncertainty and Interpretability in Deep Learning for Coronavirus (COVID-19) Detection. arXiv preprint arXiv:200310769. [Google Scholar]
- 98.Salman F.M., Abu-Naser S.S., Alajrami E., Abu-Nasser B.S., Alashqar B.A. 2020. Covid-19 Detection Using Artificial Intelligence. [Google Scholar]
- 99.Farooq M., Hafeez A. 2020. COVID-ResNet: A Deep Learning Framework for Screening of COVID19 from Radiographs. arXiv preprint arXiv:200314395. [Google Scholar]
- 100.Asnaoui K.E., Chawki Y., Idri A. 2020. Automated Methods for Detection and Classification Pneumonia Based on X-Ray Images Using Deep Learning. arXiv preprint arXiv:200314363. [Google Scholar]
- 101.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021:854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carlotti D., Buss L., Leite G., Levin A., Sabino E., Nunes F., et al. Exploratory analysis of COVID-19 patients usingprincipal component analysis, feature selection and predictive algorithms. 2021. [DOI]
- 103.Fu L., Li Y., Cheng A., Pang P., Shu Z. A novel machine learning-derived radiomic signature of the whole lung differentiates stable from progressive COVID-19 infection: a retrospective cohort study. J. Thorac. Imag. 2020;35(6):361–368. doi: 10.1097/RTI.0000000000000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li C., Dong D., Li L., Gong W., Li X., Bai Y., et al. Classification of severe and critical covid-19 using deep learning and radiomics. IEEE Journal of Biomedical and Health Informatics. 2020;24(12):3585–3594. doi: 10.1109/JBHI.2020.3036722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mostafiz R., Uddin M.S., Alam N.-A., Reza M.M., Rahman M.M. Covid-19 detection in chest X-ray through random forest classifier using a hybridization of deep CNN and DWT optimized features. J. King Saud Univ. Comp. Inf. Sci. 2020 doi: 10.1016/j.jksuci.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh M., Bansal S., Ahuja S., Dubey R.K., Panigrahi B.K., Dey N. Transfer learning based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. Med. Biol. Eng. Comput. 2021;59:825–839. doi: 10.1007/s11517-020-02299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rasheed J., Hameed A.A., Djeddi C., Jamil A., Al-Turjman F. A machine learning-based framework for diagnosis of COVID-19 from chest X-ray images. Interdiscipl. Sci. Comput. Life Sci. 2021;13(1):103–117. doi: 10.1007/s12539-020-00403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khuzani A.Z., Heidari M., Shariati S.A. medRxiv; 2020. COVID-classifier: an Automated Machine Learning Model to Assist in the Diagnosis of COVID-19 Infection in Chest X-Ray Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Attallah O., Ragab D.A., Sharkas M. MULTI-DEEP: a novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ. 2020;8 doi: 10.7717/peerj.10086. e10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guiot J., Vaidyanathan A., Deprez L., Zerka F., Danthine D., Frix A.-N., et al. Development and validation of an automated radiomic CT signature for detecting COVID-19. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 111.Autee P., Bagwe S., Shah V., Srivastava K. StackNet-DenVIS: a multi-layer perceptron stacked ensembling approach for COVID-19 detection using X-ray images. Physical and Engineering Sciences in Medicine. 2020;43(4):1399–1414. doi: 10.1007/s13246-020-00952-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cavallo A.U., Troisi J., Forcina M., Mari P., Forte V., Sperandio M., et al. Texture analysis in the evaluation of covid-19 pneumonia in chest X-ray images: a proof of concept study. 2020. [DOI] [PubMed]
- 113.Fang X., Li X., Bian Y., Ji X., Lu J. Radiomics nomogram for the prediction of 2019 novel coronavirus pneumonia caused by SARS-CoV-2. Eur. Radiol. 2020;30(12):6888–6901. doi: 10.1007/s00330-020-07032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cai W., Liu T., Xue X., Luo G., Wang X., Shen Y., et al. CT quantification and machine-learning models for assessment of disease severity and prognosis of COVID-19 patients. Acad. Radiol. 2020;27(12):1665–1678. doi: 10.1016/j.acra.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sethy P.K., Behera S.K. Detection of coronavirus disease (covid-19) based on deep features. Preprints. 2020 doi: 10.20944/preprints202003.0300.v1. 2020030300. [DOI] [Google Scholar]
- 116.Liu H., Ren H., Wu Z., Xu H., Zhang S., Li J., et al. CT radiomics facilitates more accurate diagnosis of COVID-19 pneumonia: compared with CO-RADS. J. Transl. Med. 2020;19(1):29. doi: 10.1186/s12967-020-02692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. 2020. arXiv preprint arXiv:200310849. [DOI] [PMC free article] [PubMed]
- 118.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Juanjuan X., Mei Z., Zhilei L., Zhihui W., Tingting L., Yanliing M., et al. Development and Validation of a predictive model based on Radiomics topredict the short-term outcomes of patients with COVID-19. Research Square. 2020 doi: 10.21203/rs.3.rs-74208/v1. [DOI] [Google Scholar]
- 120.Huang Y., Zhang Z., Li X., Yang Y., Li Z., Zhou J., et al. 2021. CT-based radiomics combined with signs: a valuable tool to help physician discriminate COVID-19 and other viral pneumonia; p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei W., Xiao-wen H., Qi C., Ying-ming Z., Ya-going G. Identification of common and severe COVID-19: the value of CT texture analysis and correlation with clinical characteristics. Eur. Radiol. 2020;30:6788–6796. doi: 10.1007/s00330-020-07012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qingxia W., Shuo W., Liang L., Qingxia W., Wei Q., Yahua H., et al. Radiomics Analysis of Computed Tomography helps predict poor prognostic outcome in COVID-19. Theranostics. 2020;10(16):7231–7244. doi: 10.7150/thno.46428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Masoud R.S., Abedi-Firouzjah R., Ghorvei M., Sarnameh S. Screening of COVID-19 based on the extracted radiomics features from chest CT images. J. X Ray Sci. Technol. 2021:1–15. doi: 10.3233/XST-200831. [DOI] [PubMed] [Google Scholar]
- 124.Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in Chest X-Ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2021;165:113909. doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang L., Kelly B., Lee E.H., Wang H., Zheng J., Zhang W., et al. Multi-classifier-based identification of COVID-19 from chest computed tomography using generalizable and interpretable radiomics features. Eur. J. Radiol. 2021;136:109552. doi: 10.1016/j.ejrad.2021.109552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Junior J.R.F., Cardenas D.A.C., Moreno R.A., de Sá Rebelo M.D.F., Krieger J.E., Gutierrez M.A. Novel chest radiographic biomarkers for COVID-19 using radiomic features associated with diagnostics and outcomes. J. Digit. Imag. 2021:1–11. doi: 10.1007/s10278-021-00421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xie C., Ng M.-Y., Ding J., Leung S.T., Lo C.S.Y., Wong H.Y.F., et al. Discrimination of pulmonary ground-glass opacity changes in COVID‐19 and non-COVID-19 patients using CT radiomics analysis. European journal of radiology open. 2020;7:100271. doi: 10.1016/j.ejro.2020.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tabatabaei M., Tasorian B., Goyal M., Moini A., Sotoudeh H. Feasibility of radiomics to differentiate coronavirus disease 2019 (COVID-19) from H1N1 influenza pneumonia on chest computed tomography: a proof of concept. Iran. J. Med. Sci. 2021 doi: 10.30476/IJMS.2021.88036.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giraudo C., Frattin G., Fichera G., Motta R., Stramare R. A practical integrated radiomics model predicting intensive care hospitalization in COVID-19. Crit. Care. 2021;25:145. doi: 10.1186/s13054-021-03564-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gülbay M., Özbay B.O., Mendi B.A.R., Baştuğ A., Bodur H. A CT radiomics analysis of COVID-19-related ground-glass opacities and consolidation: is it valuable in a differential diagnosis with other atypical pneumonias? PloS One. 2021;16(3) doi: 10.1371/journal.pone.0246582. e0246582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chaddad A., Hassan L., Desrosiers C. Deep CNN models for predicting COVID-19 in CT and x-ray images. J. Med. Imag. 2021;8(S1) doi: 10.1117/1.JMI.8.S1.014502. 014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sinha A., Rathi M. Applied Intelligence; 2021. COVID-19 Prediction Using AI Analytics for South Korea; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuan Y., Sun C., Tang X., Cheng C., Mombaerts L., Wang M., et al. Development and validation of a prognostic risk score system for COVID-19 inpatients: a multi-center retrospective study in China. Engineering. 2020 doi: 10.1016/j.eng.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chaganti S., Balachandran A., Chabin G., Cohen S., Flohr T., Georgescu B., et al. 2020. Quantification of tomographic patterns associated with covid-19 from chest ct. arXiv preprint arXiv:200401279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., et al. Serial quantitative chest CT assessment of COVID-19: a deep learning approach. Radiology: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. e200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Summers R.M. Artificial intelligence of COVID-19 imaging: a Hammer in search of a nail. Radiology. 2021;298(3):E162–E164. doi: 10.1148/radiol.2020204226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tizhoosh H.R., Fratesi J. COVID-19, AI enthusiasts, and toy datasets: radiology without radiologists. Eur. Radiol. 2021;31(5):3553–3554. doi: 10.1007/s00330-020-07453-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos, Solit. Fractals. 2020;140:110170. doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Homayounieh F., Ebrahimian S., Babaei R., Karimi Mobin H., Zhang E., Bizzo B.C., et al. CT radiomics, radiologists and clinical information in predicting outcome of patients with COVID-19 pneumonia. Radiology: Cardiothoracic Imaging. 2020;2(4) doi: 10.1148/ryct.2020200322. e200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Janita E., Davide C., Stephanie T.L., Hatem A., Bettina B. Radiomics in medical imaging—“how-to” guide and critical reflection. insights into imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Varghese B.A., Cen S.Y., Hwang D.H., Duddalwar V.A. Texture analysis of imaging: what radiologists need to know. Am. J. Roentgenol. 2019;212(3):520–528. doi: 10.2214/AJR.18.20624. [DOI] [PubMed] [Google Scholar]
- 142.Yoo J.K., Kim T.S., Hufford M.M., Braciale T.J. Viral infection of the lung: host response and sequelae. J. Allergy Clin. Immunol. 2013;132(6):1263–1276. doi: 10.1016/j.jaci.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kwon Y.J., Toussie D., Finkelstein M., Cedillo M.A., Maron S.Z., Manna S., et al. Combining initial radiographs and clinical variables improves deep learning prognostication of patients with COVID-19 from the emergency department. Radiology: Artif. Intell. 2021;3(2) doi: 10.1148/ryai.2020200098. e200098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]