Summary
There is an urgent need to identify which COVID-19 patients will develop life-threatening illness so that medical resources can be optimally allocated and rapid treatment can be administered early in the disease course, when clinical management is most effective. To aid in the prognostic classification of disease severity, we perform untargeted metabolomics on plasma from 339 patients, with samples collected at six longitudinal time points. Using the temporal metabolic profiles and machine learning, we build a predictive model of disease severity. We discover that a panel of metabolites measured at the time of study entry successfully determines disease severity. Through analysis of longitudinal samples, we confirm that most of these markers are directly related to disease progression and that their levels return to baseline upon disease recovery. Finally, we validate that these metabolites are also altered in a hamster model of COVID-19.
Keywords: COVID-19, SARS-CoV-2, metabolomics, longitudinal metabolite profiling, severity prediction, biomarker, machine learning, lipidomics, untargeted metabolomics
Graphical abstract

Highlights
Longitudinal profiling of plasma during COVID-19 reveals dynamic metabolic changes
Decreases in LPC and PC lipids early in the disease course predict severe COVID-19
Plasma LPCs and PCs are decreased in hamsters infected with SARS-CoV-2
Sindelar et al. combine untargeted metabolomics analysis of human plasma and machine learning to construct a model that predicts COVID-19 disease severity from the levels of 22 prognostic metabolites. The authors show that these metabolites change early in the disease course and are ultimately restored to control levels upon recovery.
Introduction
Coronavirus disease 2019 (COVID-19), which is caused by infection with the novel coronavirus SARS-CoV-2, has led to a global health crisis.1 To date, more than 150 million cases of COVID-19 have been reported worldwide and resulted in more than 3.2 million deaths.2 The infection-fatality rate of SARS-CoV-2 can be reduced with the appropriate care (e.g., intensive care unit beds, oxygen, staff, extracorporeal-life support, and therapeutics). Unfortunately, hospital resources can quickly become depleted in situations when cases spike.3 Although vaccination efforts are underway worldwide, SARS-CoV-2 infections continue to increase rapidly in a number of countries, such as India and Brazil, where medical facilities are being overwhelmed.4
Patients who develop critical illness from COVID-19 are best treated early in the disease course before the onset of severe symptoms.5,6 It is currently difficult to determine which subset of patients will develop life-threatening disease and, therefore, most benefit from receiving treatment when resources are limited.7 Early identification of these patients would allow optimal allocation of care. To that end, the objective of the current study was to identify metabolites in patient plasma that accurately predict life-threatening cases of COVID-19 before the onset of severe symptoms.
SARS-CoV-2 is an enveloped, single-stranded positive-sense RNA virus that gains entry into host cells through binding of the viral S protein to the angiotensin-converting enzyme 2 (ACE2) receptor.8,9 Multiple studies have established that patients infected with SARS-CoV-2 have metabolic dysregulation, possibly because of immune-triggered inflammation or other changes in host physiology.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 To date, however, unique alterations in metabolites upon SARS-CoV-2 infection have not been validated in large patient cohorts that have been profiled longitudinally from early after infection through recovery. Longitudinal assessment of metabolic trends is necessary to assess which changes are indicative of disease course.
In this study, we performed untargeted metabolomics profiling on the polar and non-polar fractions of 700 human plasma samples collected from 339 patients as part of the WU-350 cohort recruited during the first phase of the pandemic in St. Louis, Missouri. Untargeted metabolomics allows for the unbiased profiling of the human metabolome21 and has been successful at discovering metabolite biomarkers associated with disease pathology.22 Using machine learning (ML), we built a model of COVID-19 disease severity based on the metabolic profiles of samples collected from patients at study enrollment. The model led us to identify a panel of unique metabolite biomarkers that were highly indicative of disease severity. We confirmed that most of these metabolites were directly related to SARS-CoV-2 infections through comparison with patient demographics, comorbidities, clinical measurements, and longitudinal samples taken from individuals over the course of disease progression. Lastly, we validated that the same biomarkers appeared in an established hamster model of SARS-CoV-2 infection.23, 24, 25
Results
Clinical cohort WU-350: demographics
The clinical cohort presented in this study consisted of 155 female and 184 male participants. Of the 339 patients, 272 were considered SARS-CoV-2 positive (COV+) whereas 67 were considered SARS-CoV-2 negative (COV-). The COV+ and COV- assignment of each individual was confirmed by nasopharyngeal swab polymerase chain reaction (PCR). Significant differences were observed in several demographic factors for the COV+ cohort compared with the COV- cohort (Table 1). The COV+ group contained significantly older study participants (p < 0.0001; Figure S1A). The COV+ group also had significantly more African American, male, and non-smoking individuals. There was no significant difference in the body mass index (BMI) between the two groups (Figure S1B).
Table 1.
Demographics of all subjects
| Parameter | COV- | COV+ | p value |
|---|---|---|---|
| N | 67 | 272 | |
| Gender (M/F) | 28/39 | 156/116 | 0.0220 |
| Age (years) | 48 ± 16 | 60 ± 17 | <0.0001 |
| Race (African American/white/other) | 32/35/0 | 202/66/4 | <0.0001 |
| Body mass index | 30 ± 8 | 31 ± 9 | 0.3987 |
| COVID-19-like symptomsa | |||
| Any number of COVID19-like symptoms | 65 | 242 | 0.0437 |
| Fever | 29 | 128 | 0.5788 |
| Chills | 13 | 46 | 0.6300 |
| Conjunctival congestion | 1 | 1 | 0.2815 |
| Nasal congestion | 7 | 10 | 0.0229 |
| Headache | 18 | 23 | <0.0001 |
| Cough | 35 | 147 | 0.7907 |
| Sore throat | 14 | 23 | 0.0034 |
| Shortness of breath | 44 | 168 | 0.5540 |
| Nausea or vomiting | 15 | 41 | 0.1487 |
| Diarrhea | 11 | 38 | 0.6098 |
| Myalgia | 16 | 66 | 0.9476 |
| Fatigue | 21 | 53 | 0.0353 |
| Loss of taste or smellb | 0 | 4 | 0.3180 |
| Asymptomaticc | NA | 21 | |
| Acute respiratory failured | 9 | 100 | 0.0002 |
| Acute renal failured | 2 | 68 | <0.0001 |
| Comorbidities | |||
| Chronic kidney diseasee | 4 | 56 | 0.0050 |
| Diabetese | 15 | 120 | 0.0011 |
| Cancere | 8 | 24 | 0.4345 |
| COVID-19 drug treatmentf | |||
| Remdesivir | 2 | 43 | 0.0056 |
| Dexamethasone | 13 | 63 | 0.5087 |
| High/Low arterial pH | 5 | 83 | 0.0001 |
| Current smoker | 18 (27%) | 36 (13.2%) | 0.0063 |
| Hospital admissiong | 26 (38.8%) | 252 (92.6%) | <0.0001 |
| ICU admissiong | 10 (14.9%) | 129 (47.4%) | <0.0001 |
| Intubation and mechanical ventilation | 4 (6.0%) | 70 (25.7%) | 0.0005 |
| Deceased | 6 (8.9%) | 65 (23.9%) | 0.0071 |
| Deceased because of COVID-19h | |||
| 30 day mortality | N/A | 39 (76.5%) | |
| 60 day mortality | N/A | 49 (96.1%) | |
| 90 day mortality | N/A | 50 (98.0%) | |
| overall mortality | N/A | 51 (100%) | |
Table includes both training and test cohorts. Percentages are shown as the percentage of the group (COV- or COV+). Demographic data were last updated May 20, 2021. Data are presented as means ± SD, p values of numeric parameters were calculated by using a two-tailed Student’s t test with unequal variance, p values of categorical parameters were calculated by using a chi-square test. Abbreviations: M, male; F, female; N/A, not applicable.
Two COV- individuals were without symptoms but were exposed to a SARS-CoV-2-positive individual. Nine COV+ individuals had other symptoms (e.g., confusion, lethargy, an altered mental state, or breathing anomalies). Clinical metadata last updated October 16, 2020.
CDC guideline symptom was added to the symptom questionnaire late in the study; parameter is not available for most of the subjects.
A SARS-CoV-2 test was routinely administered at presentation to the hospital for reasons other than COVID-19 (e.g., accidents, pre-operation tests, regular checkups, cancer screening, injuries, or exposure to a SARS-CoV-2-positive individual).
During present hospitalization.
Recorded up to 1 year before the current admission or up to 1 year before the d0 sample for those who were not admitted to the hospital.
At any point during hospital stay.
Hospital and/or ICU admission of COV− group was for reasons other than COVID-19 (e.g., accidents, acute respiratory failure from bacterial pneumonia, intentional self-harm, possible heart failure, hypertension, trauma, and cancer).
Percentages shown as the percentage of total number of patients who died of COVID-19.
Of the 272 COV+ individuals, 252 were admitted to the hospital and 129 of those patients were admitted to the intensive care unit (ICU). As expected, the incidence of both factors (hospitalization and ICU admission) was significantly increased in the COV+ cohort. Treatment of severe COVID-19 cases often results in intubation and mechanical ventilation.26 In total, 70 of the COV+ patients required mechanical ventilation, whereas only four COV- individuals required mechanical ventilation. The therapeutics administered to treat COVID-19 in this cohort were dexamethasone and remdesivir. In total, 43 COV+ patients were treated with remdesivir and 63 with dexamethasone during their hospital stay (Table 1). The mortality rate in the COV+ group was 23.9%, which was significantly higher than the 8.9% mortality rate in the COV- group. A total of 65 COV- patients died, with 51 of the deaths being attributed to COVID-19 and 14 being attributed to other causes.
Of the 272 COV+ patients, 242 showed at least one COVID-19-related symptom mentioned by the Centers for Disease Control and Prevention (CDC), including fever, chills, conjunctival congestion, nasal congestion, headaches, cough, sore throat, shortness of breath, nausea or vomiting, diarrhea, myalgia, fatigue, and loss of taste or smell.27 Of the remaining COV+ cases, 21 showed no symptoms and were classified as asymptomatic. Nine subjects in the COV+ group displayed other symptoms, such as confusion, lethargy, an altered mental state, or breathing anomalies. Of the 67 COV- cases, 65 presented with at least one COVID-19-related symptom upon study entry, whereas two received a SARS-CoV-2 PCR test upon exposure to a SARS-CoV-2-positive individual. The frequency of COVID-19-related symptoms is shown in Table 1, and the distributions across the COV+ and COV- cohorts are depicted in Figure S1C. In both the COV- and COV+ groups, the number of COVID-19-related symptoms reported per individual was comparable. The breakdown of how many symptoms were experienced per individual in both the COV+ and COV- groups is shown in Figures S1D and S1E.
Next, we examined the distribution of comorbidities and self-reported medical history presented in the WU-350 cohort. Medical conditions were assigned based on ICD10 codes and the Elixhauser naming conventions.28 Chronic kidney disease (CKD) and diabetes (recorded up to 1 year before the current admission and up to 1 year before the day 0 (d0) sample for those who were not hospitalized) was significantly higher in the COV+ group compared with the COV- group (Table 1). The incidence of acute renal failure and acute respiratory failure during the present hospitalization was also significantly elevated in the COV+ group compared to the COV- group (Table 1). Furthermore, 31% of the COV+ individuals showed an abnormal arterial pH (acidosis or alkalosis) compared with 7.5% in the COV+ group. Of individuals in the COV+ group, 23.9% are now deceased. Of the COVID-19-related deaths, 76.5% occurred within 30 days, 96.1% within 60 days, and 98% within 90 days of study entry.
Study design
Blood was collected from study participants directly after enrollment in the WU-350 study (d0). Further longitudinal samples were collected 3 (d3), 7 (d7), 14 (d14), 28 (d28), and 84 (d84) days after the initial blood collection, when possible. The collection of longitudinal samples depended on survival of the study participants as well as the participants’ ability to complete study visits after being discharged from the hospital. A total of 703 human plasma samples from 341 patients were available for metabolomics profiling, including 324 d0 samples, 164 d3 samples, 111 d7 samples, 54 d14 samples, 31 d28 samples, and 19 d84 samples. All samples were divided into nine randomized sample batches and analyzed by liquid chromatography/mass spectrometry (LC/MS). An extract of the standard reference material (SRM) 1950 from NIST (National Institute of Standards and Technology, Metabolites in Frozen Human Plasma) was measured repeatedly as a quality control (QC), and blank samples were used to assess background signals. Polar and lipid metabolite fractions were extracted from each sample, and a global metabolomics profile was acquired in both positive and negative ion modes. Processing of the data led to the putative identification of 235 polar and 472 lipid metabolites based on accurate mass and MS/MS matching. Peak areas were extracted for those 707 metabolites to form the metabolic profile of each patient.
Given that the metabolic profiles were acquired over several months, the combined data showed strong batch effects as demonstrated by the principal component analysis (PCA) in Figure S2A. To remove the variance introduced by the individual batches, but not lose the differentiating biological variance within the research (WU-350) samples, we tested several normalization approaches (Figure S2B) and selected a combined batch correction (ComBat)29 approach that outperformed the other common normalization approaches tested (e.g., probabilistic quotient normalization, unit length, constant sum, quantile, etc.). After normalization, the metabolic profiles retained differences according to sample origin (WU-350, QC, and blank) as shown in Figure S2C but no longer clustered based on batch (Figure S2D).
The goal of this study was to find metabolic alterations that are predictive of disease severity in SARS-CoV-2-positive individuals. We used admission to the ICU during disease progression to classify patients as having severe or non-severe disease, as has been done previously.30,31 An ideal set of predictor metabolites would allow an individual presenting at the hospital and receiving a positive SARS-CoV-2 PCR-test result to be assessed for the likelihood of severe disease progression to best guide treatment at the earliest stage of hospitalization. Thus, we grouped the presented COV+ cohort into a non-severe (COV+ non-severe) group that did not require ICU admission and a severe group (COV+ severe) that did require ICU admission. For data interpretation purposes, three samples were excluded as the specimens originated from two patients who tested positive for other coronaviruses (NL63, HKu1). The final patient cohort consisted of 67 COV- cases, 143 COV+ non-severe cases, and 129 COV+ severe cases. Unsupervised analysis of the metabolic profiles for the 322 d0 samples available in our patient cohort demonstrated a clear trend in principal components space that separated COV+ severe, COV+ non-severe, and COV- patients (Figure 1A). Further, several significantly varying metabolites suggested that the metabolic profiles at d0 may indeed be predictive of disease severity. Hierarchical clustering analysis (HCA) of the 54 statistically significant metabolites (p < 0.05, Welch’s ANOVA) with an absolute fold change greater than two when compared with the COV- group revealed striking changes in multiple representatives of lipid classes, including lysophophatidylcholines (LPCs), phosphatidylcholines (PCs), and triglycerides (TGs). Further, several polar metabolites known to be related to COVID-19, including gluconate32 and dimethylguanosine,33 were also significantly altered (Figures 1B and S3).
Figure 1.
Study design
(A) Principal component analysis based on all polar (n = 235) and lipid (n = 472) metabolites in SARS-CoV-2-negative individuals (COV-, n = 59, green), SARS-CoV-2-positive individuals with non-severe disease (COV+ non-severe, n = 140, orange), and SARS-CoV-2-positive individuals with severe disease (COV+ severe, n = 123, red). Data shown are only from the sample provided during study entry(d0).
(B) Hierarchical clustering analysis of metabolic profiles of COV-, COV+ non-severe, and COV+ severe patients at d0. Represented are 54 significantly changing polar and lipid metabolites (p < 0.05, Welch’s ANOVA, Benjamini-Hochberg correction). Each column is a sample, and each row is a metabolite. Cell color corresponds to the normalized metabolite intensity in each sample.
(C) Human cohort of 67 SARS-CoV-2-negative and 272 SARS-CoV-2-positive participants. The cohort was divided into a training cohort and a test cohort. The study design incorporated six blood draws for SARS-CoV-2-positive individuals on days 0 (d0), 3 (d3), 7 (d7), 14 (d14), 28 (d28), and 84 (d84) after WU-350 study entry.
Predictive model of COVID-19 disease severity
The global trends in the d0 metabolic profiles visible in the PCA and HCA visualizations prompted us to develop an ML model of disease severity that would predict ICU admission caused by SARS-CoV-2 infection. Patients presented to the hospital and enrolled in the study at different times during their disease course, hence the d0 sample is variable with respect to days- post symptom onset. The time from patient-reported symptom onset to d0 sample collection ranged from 0 to 107 days, with 4 days being the median. In addition, 45 of the COV+ patients were on mechanical ventilation and 98 COV+ patients were admitted to the ICU when the d0 sample was collected (Figure S4A). Although this diversity added additional variance to the d0 metabolic signatures, it also enabled a more clinically relevant model to be constructed because patients will present to the hospital at various points in the disease course. We note that COV+ severe and non-severe patients were not significantly different in time from symptom onset to the d0 sample collection (Figure S4B). The 707 metabolites that composed the metabolic profiles served as the predictors for our ML model. To assess predictive power, we split our dataset into two distinct groups of COV+ patients: a training set (163 patients) that we used to select, optimize, and train our ML model and an independent test set (100 patients) that was only used to evaluate the model’s performance (Figure 1C; Tables S1 and S2). Using our training set, we evaluated the efficacy of five ML algorithms with 20-fold cross-validation and found that a linear ElasticNet34 regression model was the most effective (Figure S4C). After training the model, we applied it to the patients in the test set and assessed performance by using the area under the receiver operating characteristic curve (AUC). On the test set, we saw strong predictive performance (AUC = 0.72) that outperformed a simple model that only uses BMI and age to predict disease severity (Figure 2A) and is significantly more predictive than a random model (Figure 2B, see permutation test in method details). As further validation, when the trained model was applied to the COV- patients (no COV- patients were in the training set), the mean scores output by the model were lower than those for the COV+ non-severe and the COV+ severe patients in the test set (Figure S4D). This indicates that the model can differentiate disease severity and distinguish COV+ and COV- patients. We wish to emphasize that PCR is the gold standard to diagnose SARS-CoV-2 infection. As such, we present this result only as confirmation that our model correctly predicts disease severity and not as a potential diagnostic for viral infection.
Figure 2.
Predicting SARS-CoV-2 severity by machine learning
(A) Receiver operating characteristic (ROC) curves of prediction model on training set (green) and test set (blue). Random performance is shown in gray. ROC of BMI and age as predictors for severe COVID-19 (red) results in nearly random performance.
(B) Permutation test results from permuting training set labels and training the model on the permuted data. With every permutation, the area under the ROC curve (AUC) for the test set was computed. The histogram shows the distribution of these AUC values for 1,000 random permutations. The model performance on the test set when trained on the non-permuted training data results in an empirical p value of 0.002, as shown in blue.
(C) Variable importance in reduced ElasticNet prediction model for disease severity of SARS-CoV-2 infection in humans. Negative values are predictive of non-severe disease and positive values are predictive of severe disease. Variable importance is after the model is trained on the complete dataset.
(D) Profile plot of the normalized intensity (d0) of the prediction model metabolites grouped into COV- (control, n = 59), COV+ non-severe (n = 140), and COV+ severe (n = 123). The color of the lines reflects the mean intensity of each metabolite in the COV- patients.
(E) Boxplots showing predictor metabolite intensities (d0) in the COV-, COV+ severe, and COV+ non-severe groups. Box limits represent the quartiles of each sample group. Whiskers are drawn to 1.5× of the inter-quartile range.
We next sought to interpret which metabolites were most salient to the model’s predictions. First, we computed the variable importance of the model when trained on the complete dataset, which found 92 unique metabolites that contributed to the model’s predictions. Among that group of 92 compounds were metabolites that have been previously implicated in SARS-CoV-2 infection, such as kynurenate, nicotinamide, creatinine, LPCs, PCs, and others.11,12,14,32 The mean intensity of each metabolite in the COV-, COV+ non-severe, and COV+ severe groups can be seen in Figure S5. Next, we aimed to assess the robustness of the metabolites selected by the ML model. We used bootstrap resampling of our training dataset to construct confidence intervals for the variable importance of each of the 707 metabolites we profiled.35 The analysis led to the identification of 22 predictor metabolites that significantly contributed to the model’s fit. The structural identities of these metabolites were rigorously confirmed (see method details). Strikingly, manyof the predictor metabolites were LPCs. Using the reduced predictor set, we re-trained and re-optimized our ElasticNet model on the training set and assessed the predictive power on our test set. Using the reduced predictor set resulted in nearly an identical AUC to that of the full set of metabolites (AUC = 0.70) and still performed better than a random model or a model that used only BMI and age as predictors (Figures S6A and S6B). To ensure that our model is indeed capable of predicting future disease severity, we additionally evaluated our model on the portion of the test-set patients who were not admitted to the ICU on or before when the d0 sample was collected. We again found that our model’s performance remained high (AUC = 0.71) and continued to outperform a model based only on BMI and age (Figures S6C and S6D). The variable importance of the predictor metabolites when trained on the entire dataset is shown in Figure 2C. The mean intensity of the metabolites in the COV-, COV+ non-severe, and COV+ severe groups is shown in Figures 2D and S7. In addition, the intensities of these metabolites among test-set patients and patients not admitted to ICU on d0 are provided in Figure S8. To exclude the possibility that the metabolite alterations were due to the application of therapeutics, we additionally removed all d0 samples when dexamethasone or remdesivir were administered on the same day as sample collection. Using that subset, all predictor metabolites were still significantly altered. All LPCs and PCs that contributed to the model, as well as serine, presented a significant downward trend in signal abundance with disease severity. Conversely, the other polar metabolites (kynurenate and 1-methyladenosine), phosphatidylethanolamines (PEs), and ceramides exhibited a significant upward trend in signal intensity (Figure 2E).
Demographics, laboratory values, comorbidities, and COVID-19 severity
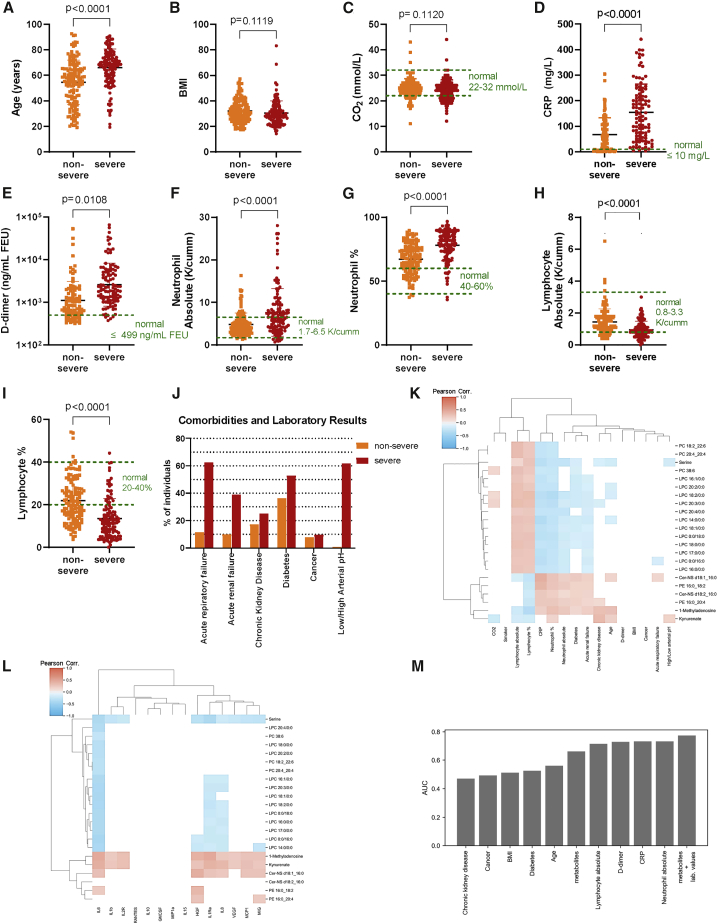
After evaluating the efficacy of the ML model, we wished to deduce the relationship of our metabolite predictors to COVID-19 disease severity. We examined whether these metabolites were reflective of an underlying condition/risk factor for severe disease or if they were related to the disease progression of COVID-19. We addressed the former by asking whether any of the predictor metabolites correlated with demographic factors, laboratory values, current severity predictors, or individual patient comorbidities available for the patient cohort. A comparison of the COV+ non-severe and severe groups identified several significantly different parameters (Table S3). The COV+ severe group was significantly older than the non-severe group (Figure 3A), but there was no significant difference in BMI (Figure 3B). CO2 levels were not significantly altered between groups (Figure 3C), with values mostly being in the normal range. In contrast, there were significantly increased levels of the inflammatory marker C-reactive protein (CRP; Figure 3D). D-dimer, absolute neutrophil count, and neutrophil percentage were also increased (Figures 3E–3G). Absolute lymphocyte count and lymphocyte percentage were decreased (Figures 3H and 3I). These data indicate more severe inflammation in the COV+ severe group compared with the non-severe group and are consistent with reports from previous studies.11,36,37 Neutrophil recruitment has also been shown to be dysregulated in severe cases of COVID-19.38, 39, 40, 41 Lymphopenia, abnormally low levels of blood lymphocytes, has been found to correlate with disease severity in COVID-19 and even to be predictive of disease severity.42, 43, 44, 45, 46
Figure 3.
COV+ patient parameters
(A–I) Demographics, comorbidities, and laboratory values of SARS-CoV-2-positive individuals grouped by disease severity (non-severe, severe) for age (A), BMI (B), CO2 (C), C-reactive protein (D), D-dimer (E), absolute neutrophil levels (F), neutrophil percentage (G), absolute lymphocyte levels (H), and lymphocyte percentage (I). Statistical significance (p < 0.05) was assessed by using a two-tailed Student’s t test with unequal variance for data shown in (A-I).
(J) Proportion of COV+ severe and non-severe patients with specific comorbidities and laboratory test results.
(K) Pearson correlation of listed demographic/laboratory results/comorbidities with abundances of the predictor metabolites.
(L) Pearson correlation of interleukin levels with abundances of predictor metabolites. Cells in the heatmaps (K and L) shaded white do not have a statistically significant Pearson correlation (p < 0.05, Benjamini-Hochberg correction).
(M) AUC values for patient comorbidities, demographics, laboratory values, the metabolite model, and a model that uses both the metabolites and laboratory values (absolute lymphocyte, absolute neutrophil, CRP, and D-dimer) when predicting disease severity on the test-set patients with complete information for all patient parameters.
Given that specific comorbidities increase the risk of having a severe case of COVID-19,31,47,48 we asked which co-morbidities were enriched in the COV+ severe group compared to the COV+ non-severe group (Figure 3J; Table S3). Diabetes, cancer, and chronic kidney diseases were recorded up to 1 year before hospital admission or up to 1 year before the d0 samples for non-hospitalized individuals. Although diabetes was significantly more prevalent in the COV+ group, the number of individuals with cancer and/or chronic kidney disease was not significantly different between the groups. Because acute respiratory failure and/or acute renal failure is a feature of severe COVID-19, the proportion of individuals suffering from either or both is significantly greater in the COV+ severe patients. Further, laboratory tests showed an increased proportion of individuals having an abnormal arterial pH (acidosis or alkalosis) in the COV+ severe group compared to the COV+ non-severe patients. The presented laboratory results (arterial pH, neutrophils, lymphocytes, D-dimer, CRP, and CO2) were obtained within 48 h of the first sample after study entry (d0).
Considering the number of significant associations in the patient parameters between COV+ severe and non-severe patients, we wanted to test whether any of our predictor metabolites significantly correlated with the clinical data after we controlled for disease severity (method details). To that end, we computed the Pearson correlation49 (for continuous parameters) or the point biserial correlation50 (for binary parameters) between each predictor metabolite and patient parameter (Figure 3K). The analysis revealed that many of the patient comorbidities were not significantly associated with the predictor metabolites. Smoking, BMI, and cancer were not significantly correlated with any of the predictor metabolites. Age and CKD had weak correlations with 1-methyladenosine (r = 0.32, 0.33), kynurenate (r = 0.20, 0.36), and serine (r = −0.19, −0.15). Kynurenate has been described previously51 as being associated with CKD. The most striking results are the significant positive correlations of the PEs (r = [0.20, 0.23], [0.26, 0.37]), ceramides (r = [0.14, 0.21], [0.26, 0.43]), and 1-methyladosine (r = 0.26, 0.20) with inflammatory markers (neutrophils and CRP) as well as the incidence of acute renal failure and diabetes. The PEs (r = [−0.07, −0.05]), ceramides (r = [−0.14, −0.12]), and 1-methyladenosine (r = −0.21) were also negatively correlated with lymphocyte levels, suggesting that these metabolite changes may be associated with inflammation. The LPCs, PCs, and serine showed the opposite trend as the PEs and ceramides. These molecules had positive associations with lymphocyte levels (r = [0.15, 0.34]) and negative associations with CRP (r = [−0.37, −0.23]) and neutrophil levels (r = [−0.28, −0.08]), suggesting that they too are associated with inflammation. We further correlated severity-associated interleukin levels that have been reported for a portion of our cohort in a prior publication (Figure 3L).52 Interleukin 6 (IL-6) levels, which have previously been shown to be a marker for disease severity,53, 54, 55 were inversely correlated with LPCs, PCs, and serine after controlling for severity (r = [−0.43, −0.26]). Similarly, 1-methyladenosine (r = 0.47), kynurenate (r = 0.44), Cer-NS d18:1_16:0 (r = 0.32), and PE 16:0_18:2 (r = 0.22) had significant positive correlations with IL-6, giving further evidence that many of the metabolite alterations are related to inflammation.
We then sought to assess the predictive power of our ML model relative to the predictive power of the patient comorbidities and laboratory values collected within 48 h of the d0 sample (CRP, D-dimer, neutrophil, and lymphocyte levels). For each patient parameter, we computed the AUC when predicting disease severity for each patient in the test set without missing values for any of those parameters (n = 68; Figure 3M). For all evaluated comorbidities, the model achieved a higher AUC. Conversely, the laboratory values (D-dimer, CRP, neutrophil, and lymphocyte levels) were all more predictive of disease severity than the metabolite model. However, when a new model was trained that used these laboratory values in combination with the biomarker metabolites, the performance of the combined model (AUC = 0.77) outperformed both the metabolite model (AUC = 0.66) and each laboratory value (AUC = [0.72, 0.73]), suggesting that the metabolite model is capturing complementary information to the laboratory values. In total, these results suggest that our predictor metabolites are indeed relevant to the pathogenesis of SARS-CoV-2 infection and not merely markers of other risk factors.
Longitudinal progression of predictor metabolites
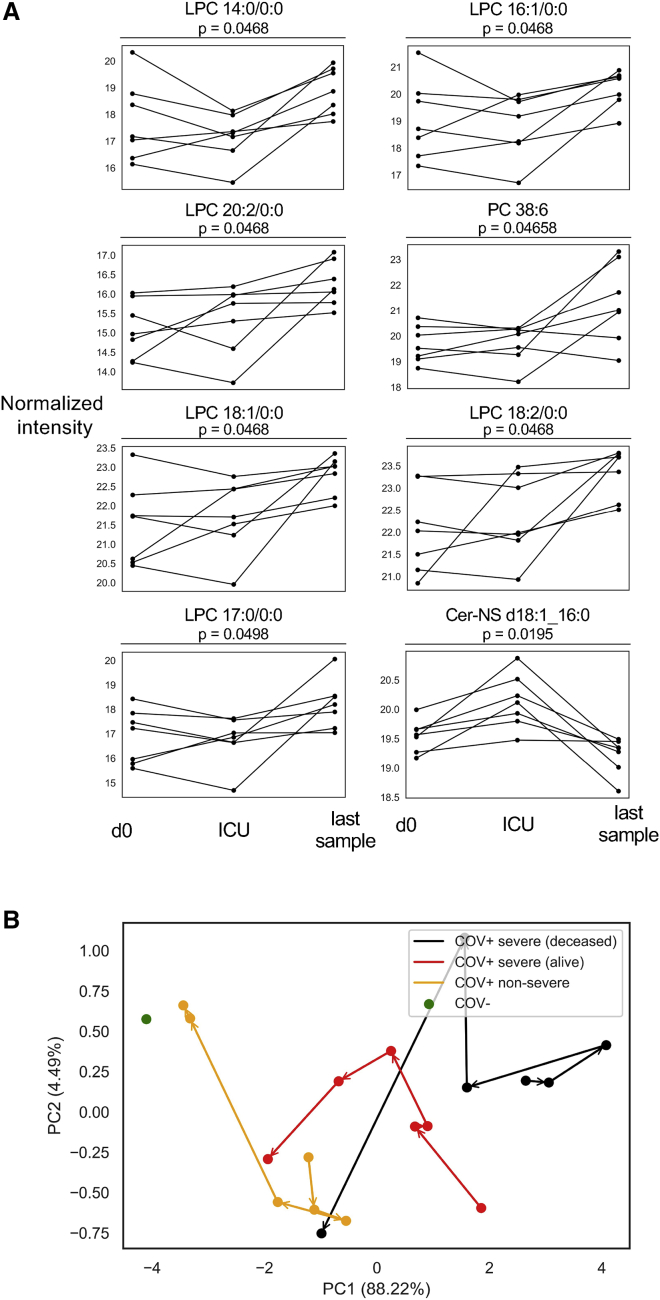
To give further confidence that our predictor metabolites are associated with COVID-19 pathogenesis, we aimed to determine how the levels of these metabolites changed over the course of the disease progression. First, we considered the portion of the COV+ severe cohort that survived SARS-CoV-2 infection. We sought to determine the temporal behavior of their metabolic profiles as patients reached peak disease severity and after recovery. Accordingly, we analyzed the longitudinal metabolite abundances from individuals who had severe disease but survived and were discharged from the hospital. We compared their initial d0 plasma sample to the sample taken closest to the day of ICU admission, when the disease had progressed to peak severity. We also compared their initial d0 plasma sample to the last sample provided by the patient at or after hospital discharge. For several LPCs and one PC, a V-shaped trend was observed (Figure 4A). After the initial sample (d0), the levels of these metabolites dropped further as the disease worsened, but then began to return to d0 levels during recovery. The reverse trend was observed for Cer-NS d18:1_16:0. Its levels significantly increased until the patients were admitted to the ICU, but its levels subsequently dropped to below the initial d0 values in the final sample obtained.
Figure 4.
Course of disease progression
(A) Prediction model metabolites that significantly vary in intensity as a function of disease progression for SARS-CoV-2-positive patients surviving severe disease (COV+ severe). d0 denotes the first sample after hospital admission, ICU denotes the sample collected closest to ICU admission, and the last sample is the final sample collected for the patient. Only patients for whom these time points were distinct samples were plotted. Statistical significance was assessed by a repeated-measures one-way ANOVA with a Benjamini-Hochberg correction.
(B) Principal component analysis showing the trajectory of the mean metabolic profile of the predictor metabolites in COV+ non-severe patients, surviving COV+ severe patients, and deceased COV+ severe patients. Deceased and surviving status was based on 90-day mortality. Patient samples were binned based on days post symptom onset (0–3, 4–6, 7–11, 12–18, 19–40, and 41–100). Samples taken when dexamethasone or remdesivir were administered were excluded. Asymptomatic patients were excluded. The mean d0 metabolic profile of COV- patients is shown in green.
These pronounced longitudinal trends in surviving COV+ severe patients raised the question of how the trajectory of disease progression (as marked by our predictor metabolites) differed among COV+ non-severe patients, surviving COV+ severe patients, and deceased COV+ severe patients. We also sought to compare the end points in these groups to the COV- d0 patients. We constructed representative metabolite profiles for the groups by using the levels of the predictor metabolites at different time windows relative to symptom onset (0–3, 4–6, 7–11, 12–18, 19–40, and 41–100 days post symptom onset) and performed a principal component analysis that enabled the trajectory of each group to be visualizedin two dimensions (Figure 4B). Samples from asymptomatic patients or patients administered the COVID-19-related therapies dexamethasone or remdesivir on the day of sample collection were excluded from this analysis. Notably, the analysis revealed three distinct trajectories with starting points that trended with disease severity. The groups then followed a common trajectory toward the COV- d0 sample. However, the COV+ non-severe patients recovered much quicker than the COV+ severe patients, reaching a similar metabolic profile to the d0 COV- patients in less than 40 days post symptom onset. The deceased COV+ severe patients had a similar metabolic profile during the 41–100 day window when compared to surviving COV+ severe patients. These results point toward distinct metabolic programs associated with the time course of infection and disease outcome. When examining the individual metabolite levels within the four groups at each time window, the deceased COV+ severe patients showed the same direction of dysregulation across the predictor metabolites as the surviving COV+ severe patients, but the magnitude of the perturbation was increased. In the 41–100 day window, less than 20% of the predictor metabolites were significantly altered, suggesting that metabolite levels return to control levels over time (Figure S9).
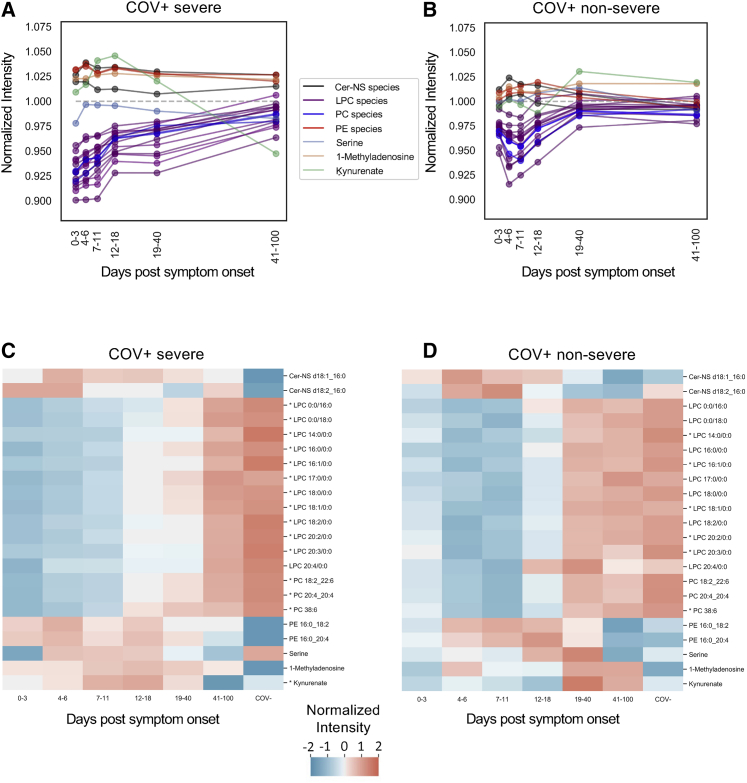
Next, we sought to compare the longitudinal progression of the predictor metabolites between the surviving COV+ patients. In the COV+ severe group, the LPC levels increased over the disease course to levels that were comparable to the COV- group (Figure 5A). In the COV+ non-severe group, the LPC levels recovered faster, reaching control levels in less than 40 days post symptom onset (Figure 5B). In total, 15 of the predictor metabolites showed a significant change (p < 0.05, Welch’s ANOVA) across the longitudinal time points in the COV+ severe group (Figure 5C). Of the LPCs, 11 were significantly increased over time. PC 38:6, PC 20:4_20:4, and PC 18:2_22:6 were also significantly increased with time. After initially being increased compared to the d0 sample of the COV- group, kynurenate showed a significant decreasing trend. Because of the lower sample numbers, the COV+ non-severe group had only six metabolites (five LPCs and one PC) that showed a significant trend (p < 0.05, Welch’s ANOVA; Figure 5D).
Figure 5.
Longitudinal trends in COV+ patients
(A) Profile plot of the mean predictor metabolite intensities relative to d0 COV- samples (n = 59, gray) in symptomatic SARS-CoV-2-positive individuals with severe COVID-19 (n = 123, COV+ severe).
(B) Profile plot of the mean predictor metabolite intensities in symptomatic SARS-CoV-2-positive individuals with non-severe disease (n = 140, COV+ non-severe).
(C and D) Heatmaps showing relative mean intensity of predictor metabolites in longitudinal profiles of symptomatic COV+ severe patients (C) or COV+ non-severe patients (D). The mean COV- d0 profiles are included as the control for reference. Patient samples were grouped based on symptom-onset windows. Patient samples where dexamethasone or remdesivir were administered were excluded from the analysis. Asymptomatic patients were excluded. ∗p < 0.05. Statistical significance was assessed by using a one-way Welch’s ANOVA with a Benjamini-Hochberg correction.
Syrian hamster model confirms metabolite changes in COVID-19
Lastly, we aimed to validate that the predictor metabolites we found in the human samples were directly related to COVID-19 and not to some unidentified correlate that could have confounded our analysis. As such, we used an established hamster model of SARS-CoV-2 infection.24,25 Syrian hamsters have been found to be susceptible to SARS-CoV-2 infection, with the virus mainly replicating in the upper and lower respiratory tract of intranasally challenged animals for approximately 6 days post infection. Diseased animals exhibit a loss of body weight and pathological lung inflammation. We obtained plasma samples from golden Syrian hamsters that were intranasally inoculated with SARS-CoV-2 according to Imai et al.,25 influenza virus according to Iwatsuki-Horimoto et al.,56 or nasally treated with saline solution as a mock infection. Viral infection of the lung and body-weight loss were confirmed in all SARS-CoV-2- and influenza-infected hamsters and were consistent with previous reports.25,56 All SARS-CoV-2-infected hamsters experienced greater body-weight loss (>5.1% of their initial body weight) compared to influenza-infected hamsters, and virus was detected in both the lungs and nasal cavity in both models (see method details; Figure S10).
After 2, 4, 6, and 14 days (d2, d4, d6, and d14) post infection, plasma was harvested from the SARS-CoV-2- and influenza-infected hamsters (Figure 6A) because those time points correlate with the well-established kinetics of pathology and infection in the hamster model of SARS-CoV-2.24,25,57 For the mock group, plasma was harvested on days 4 and 14 relative to the infection timeline. All plasma samples were subjected to the same LC/MS workflow as described above. Differences in the sample matrix prevented a direct comparison of metabolite intensities between rodent and human plasma, but we are able to make comparisons between the different experimental groups. We compared the predictor metabolite levels of samples from the three groups (SARS-CoV-2, influenza, and mock) harvested on d4 as a representative time point for early disease. Figure 6B shows the 11 significantly changing predictor metabolites (p < 0.05, Welch’s ANOVA) across the three groups. Of the LPCs, eight were significantly altered in animals infected with SARS-CoV-2. Although PC 20:4_20:4 was not detected in the hamster plasma, possibly because of matrix effects, the two detected PCs were significantly dysregulated, along with PE 16:0_20:4. All 11 of the significantly altered metabolites showed the same direction of dysregulation as what was observed in the d0 human samples. For all significantly varying metabolites except PE 16:2_20:4, infection with the influenza virus showed a similar trend of dysregulation relative to control samples but a very different magnitude compared to SARS-CoV-2 infection. This is consistent with milder disease in influenza-infected hamsters compared to hamsters infected with SARS-CoV-2 and suggests that the depression of LPCs and PCs may not be as specific to SARS-CoV-2 infection as changes in PEs.
Figure 6.
Syrian hamster model confirms SARS-CoV-2-dependent metabolite changes
(A) Experimental design of the Syrian hamster model. Hamsters (n = 3–6 per group) were infected through intranasal inoculation of SARS-CoV-2 (1e5 plaque-forming units [PFUs]), influenza virus (1e5 PFU), or nasally treated with a saline solution (mock) on day 0 (d0). After 2, 4, 6, and 14 days (d2, d4, d6, and d14) post infection, animals were sacrificed and exsanguinated.
(B) Comparing metabolite intensities among hamsters infected with influenza (n = 6), SARS-CoV-2 (n = 5), and mock (n = 6) on d4 shows many of the predictor metabolites are significantly altered in the hamster model (p < 0.05, Welch’s ANOVA). Box limits represent the quartiles of each sample group. Whiskers are drawn to 1.5× of the inter-quartile range.
(C and D) Metabolite changes during disease progression in SARS-CoV-2 (C) and influenza (D). Infected animals show a faster recovery during influenza. All groups are n = 6 with the exception of SARS-CoV-2 hamsters at d2 (n = 3) and d4 (n = 5). ∗p < 0.10, ∗∗p < 0.05. Statistical significance was assessed by using a two-tailed Student’s t test with unequal variance between d2 and d4 samples. All values were corrected with the Benjamini-Hochberg procedure.
Finally, we wanted to determine whether the predictor metabolites in hamsters showed longitudinal changes over the time course of infection. Indeed, when compared to the d4 control (mock infection) samples, there was a similar trend in hamsters infected with SARS-CoV-2 (Figures 6C and S11A) as in human COV+ samples. There was minimal time-dependent variation in the mock-infection samples, and none of the predictor metabolite levels were significantly different between d4 and d14 in those animals (Figure S11B). In SARS-CoV-2-infected hamsters, plasma LPC levels decreased on d4 compared to d2 and slowly recovered toward the control levels on d14. By comparison, for the influenza-infected animals, metabolite levels approached those of the control group more rapidly (Figures 6D and S11C). The similar patterns of alteration and recovery in LPC and PC levels across both hamsters and humans suggest that these lipid species are directly related to SARS-CoV-2 infection.
Discussion
The current study sought to predict COVID-19 disease severity based on the metabolic profiles of human plasma samples obtained early in the disease course, before the onset of critical illness. We applied untargeted metabolomics to profile a patient cohort of 339 individuals, which amounted to 700 study samples in addition to QC and method blanks. From the experiments, we putatively identified 235 polar metabolites and 472 lipid species. Using these compounds, we applied ML techniques to build a predictive model that accurately classified a patient’s disease severity from their d0 metabolic profile obtained at the time of initial study entry. Currently, risk assessment is primarily based on BMI and age.58 Even though we see a significant difference between the ages of patients in the non-severe and severe disease groups (p < 0.0001), the results of our study show that risk assessment based on a small panel of predictor metabolites is more reliable than age and BMI, thereby providing a better metric for resource allocation.
Although our linear ElasticNet model is relatively simple compared to other popular ML models that have been applied to metabolomics datasets previously (including artificial neural networks, support vector machines, or ensemble-based approaches, such as random forest), linear models can be easily interpreted and provide robust performance even when modeling innately non-linear biological systems.34,59 Notably, a previous study13 successfully used an ElasticNet model to predict disease severity from a multi-omic dataset. Other studies have used non-linear ML models, such as random forest, to predict disease severity from metabolomics, lipidomics, and/or proteomics profiles.10,12,19 While those studies found higher AUC scores than that of our model, they used considerably smaller patient cohorts than what our model was trained and evaluated on. Further, AUC scores are not directly comparable, unless they are calculated from the same dataset. When we tested non-linear models (support vector machines and random forest), we found worse cross-validated performance relative to ElasticNet (see Figure S4C).
Interpretation of our model led us to identify metabolites that predicted disease severity. Using a reduced predictor set, we were able to retrain our model and found similarly strong predictive ability. Of the predictor metabolites, most were LPCs and PCs that were more decreased in patients with severe COVID-19 when compared to patients with non-severe disease. The observation that LPC and PC levels are altered in patients with COVID-19 has been corroborated by multiple studies from around the world. Early studies of COVID-19 patients from China and Italy showed a decrease in PC levels and an increase in select LPCs,14,32,60 but recent studies from South America and Canada have shown that both circulating PC and LPC levels decrease in patients with COVID-19.10,19 Not only do our results support that PCs and LPCs are directly related to COVID-19 disease severity, they also build upon previous findings by showing that these metabolites are decreased early in the disease course and are, therefore, predictive of ICU admission. Additionally, upon disease recovery, we find that LPCs are restored to the levels of control individuals. We also confirmed that PCs and LPCs are decreased in a hamster model of SARS-CoV-2 infection and that the levels of those metabolites in animal plasma similarly recover over time.
The cellular mechanisms underlying the decreased LPC and PC levels in the plasma of patients with severe COVID-19 may be connected to uncontrolled inflammatory responses or viral pathogenesis. Multiple studies have shown that individuals with severe COVID-19 display a hyper-inflammatory state characterized by dysregulated production of cytokines, such as IL-1, IL-6, IL-10, and others.61, 62, 63 Fatty acids in plasma have also been correlated with sustained IL-6 release and renal dysfunction in a smaller cohort of patients with COVID-19,11 suggesting that altered inflammatory responses may affect levels of circulating metabolites. Consistent with prior studies, in our cohort, we found that PC and LPC levels in patients with COVID-19 correlated with inflammatory markers (e.g., IL-6, CRP, lymphocyte, and neutrophil levels). In addition, reductions in circulating LPC levels have also been found in patients with pneumonia-induced inflammation, infection of the lung,64 and sepsis in which a cytokine storm occurs.65 Alternatively, metabolic alterations may be directly related to the pathogenesis of SARS-CoV-2. Reprogramming of cellular metabolism is critical to sustain viral replication during coronavirus infection.66,67 SARS-CoV-2-infected monocytes upregulate genes involved in lipid biosynthesis and lipid remodeling, leading to the formation of lipid droplets.68 Additionally, a CRISPR-based screen for host factors required for the replication of coronaviruses identified genes involved in the sterol regulatory-element binding-protein (SREBP) pathway and the gene encoding the low-density lipoprotein (LDL) receptor. The SREBP pathway regulates cholesterol metabolism, and the LDL receptor influences circulating lipid levels.69 As such, the alterations in plasma metabolites found in our study may reflect systemic alterations in metabolism induced by sustained viral replication, especially in severe disease.
In summary, our large sample size that included longitudinal measurements of patient plasma, collection of patient metadata (laboratory values, comorbidities, and demographics), and use of an animal model allowed us to discover a small panel of metabolite predictors for COVID-19 severity and rigorously investigate the relationship of those metabolites to the pathology associated with SARS-CoV-2 infection. The model we present here to assess the risk of a severe case of COVID-19 does not require intensive computation or untargeted metabolomics, making it immediately applicable to most hospitals around the world. The metabolites presented could, for example, be quantitatively measured by triple quadrupole mass spectrometers, which are widely available in clinical laboratories.
Limitations of the study
Although the size of our patient cohort is a major strength of this study, sample collection started early in the pandemic (March 2020), when the appropriate treatment of SARS-CoV-2 infections was not yet established. Over the time of study enrollment, treatment plans changed (e.g., fewer individuals were treated by mechanical ventilation in the second half of the study). Additionally, although the goal was to collect longitudinal samples 3, 7, 14, 28, and 84 days after initial sample collection, we were unable to obtain samples from all patients at every time point. Subjects were less likely to provide plasma after being discharged from the hospital, and samples could not be collected after patients were deceased. Moreover, even though the d0 sample would ideally have been taken at or preceding hospital admission, this was not always possible. Some patients enrolled in the study several days after being admitted, or they were enrolled only after being transferred from another hospital. These practical challenges affected the statistical power of our temporal comparisons. Furthermore, it is important to note that the patients studied here were all from hospitals in the St. Louis region and were mostly collected before the emergence of variants, such as B.1.617.2 (delta). Further longitudinal studies of patients from other geographic regions who have been infected with variants are thus needed to confirm the broad applicability of our findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Viral vectors | ||
| H1N1 influenza A virus (A/California/04/2009) | NIH | N/A |
| SARS-CoV-2 (WA-1) | BEI Resources | Cat# NR-52281 |
| Biological samples | ||
| Human plasma samples | Washington University in St. Louis, Tissue Procurement Core (TPC), https://siteman.wustl.edu/research/shared-resources-cores/tissue-procurement-core-tpc/ | WU-350 |
| Reference human plasma | National Institue of Standards and Technolgoy | SRM1950 |
| Chemicals, peptides, and recombinant proteins | ||
| Acetonitrile LC/MS OPTIMA | Fisher Scientific | Cat#A955 4, CAS 75-05-8 |
| Euthanasia Solution SleepAway | Fort Dodge Animal Health | NAC No.: 10031902 |
| Formic Acid for LC-MS LiChropur | Sigma-Aldrich | Cat#5438040100, CAS 64-18-6 |
| Ketamine | Vedco, Inc., St Joseph, MO | NDC 50989-996-06 |
| Xylazine | Akorn, Inc., Lake Forest, IL | N/A |
| Mass Spectrometry Metabolite Library | IROA Technologies | Cat#MSMLS |
| Medronic Acid | Sigma-Aldrich | Cat#64255, CAS 1984-15-2 |
| Methanol LC/MS OPTIMA | Fisher Scientific | Cat#A456 4, CAS 67-56-1 |
| Methyl tert-butyl ether HPLC Plus | Fisher Scientific | Cat#650560, CAS 1634-04-4 |
| OPTIMA LC/MS 2-propanol (IPA) | Fisher Scientific | Cat#A461 4, CAS 67-63-0 |
| Water LC/MS OPTIMA | Fisher Scientific | Cat#W64, CAS 7732-18-5 |
| Deposited data | ||
| Raw and analyzed data WU-350 | NMDR, https://www.metabolomicsworkbench.org | NMRD:ST001849, https://doi.org/10.21228/M80981 |
| Metadata WU-350 | NMDR, https://www.metabolomicsworkbench.org | NMRD:ST001849, https://doi.org/10.21228/M80981 |
| Raw and analyzed data Syrian Hamster Model | NMDR, https://www.metabolomicsworkbench.org | NMRD:ST001853, https://doi.org/10.21228/M80981 |
| Metadata Syrian Hamster Model | NMDR, https://www.metabolomicsworkbench.org | NMRD:ST001853, https://doi.org/10.21228/M80981 |
| Experimental models: Organisms/strains | ||
| LVG golden Syrian hamsters | Charles River Laboratories (Kingston, NY) | LVG(SYR) |
| Software and algorithms | ||
| Biorender | BioRender.com | RRID:SCR_018316 |
| GraphPad Prism v3.8 | GraphPad | RRID:SCR_002798 |
| Lipid Annotator v1 B1.0.54.0 | Agilent Technologies | N/A |
| Mass Profiler Professional Software v15.5 | Agilent Technologies | N/A |
| MassHunter Workstation LC/MS Data Acquisition v10.1 B10.1.48 | Agilent Technologies | N/A |
| Python v3.7 | python | RRID:SCR_008394 |
| Scikit-learn v0.23.1 | N/A | https://scikit-learn.org |
| SciPy v1.4.1 | N/A | https://SciPy.org |
| Skyline 64-bit v20.2.0.343 | MacCoss Lab Software | RRID:SCR_014080 |
| statsmodels v0.11.1 | N/A | https://statsmodels.org |
| Thermo Scientific Xcalibur v4.2.28.14, AcquireX workflow | Thermo Fisher Scientific | RRID:SCR_014080 |
| Other | ||
| ACQUITY UPLC HSS T3 Column, 100Å, 1.8 μm, 2.1 mm X 150 mm | Waters | Cat#186003540 |
| VanGuard Pre-Column HSS T3, 1.7 μm, 2.1mm x 5mm | Waters | Cat#186003976 |
| SeQuant® ZIC®-pHILIC HPLC Column, 5μm polymer 100 × 2.1 mm | Merck Millipore | Cat#150462 |
| SeQuant® ZIC®-pHILIC Guard Kit, 5μm polymer 20 × 2.1 mm | Merck Millipore | Cat#150438 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Gary J. Patti (gjpattij@wustl.edu).
Materials availability
This study did not generate new unique reagents.
Resources used in this study are available to the scientific community upon request from Dr. Gary J. Patti (gjpattij@wustl.edu).
Experimental model and subject details
Human subjects and samples
Over the period of March to August of 2020, blood specimens of 341 individuals who presented at Barnes Jewish Hospital or Christian Hospital located in Saint Louis, Missouri, USA were collected. Inclusion criteria were a physician-ordered SARS-CoV-2 nasopharyngeal swab polymerase chain reaction (PCR) test with a positive or negative outcome, availability of gender and age information, and an age greater than 18. Informed consent was obtained from all study participants. The clinical cohort consisted of 155 female and 184 male participants. Out of the 339 patients, 272 were considered SARS-CoV-2 positive (COV+) and 67 were considered SARS-CoV-2 negative (COV-). Samples were collected at the time of enrollment (d0) to the study, and 3, 7, 14, 28, or 84 days post study entry. Clinically relevant medical information (e.g., patient-reported symptoms, date of symptom onset, age, race, and BMI) was collected at the time of enrollment from the subject or from the medical record. Data were retrieved for the current study on October 16th 2020.
Animal studies
All animal studies were performed at Mount Sinai School of Medicine. Outbred female LVG golden Syrian hamsters (6-8 weeks of age) were obtained from Charles River Laboratories (Kingston, NY). Experiments were performed as previously published.25,56 In short, the hamsters were anesthetized by intraperitoneal injection of a mixture of ketamine and xylazine prior to intranasal inoculation with 0.1 mL of 1e5 plaque-forming units (PFU) of SARS-CoV-2 (WA-1) or H1N1 influenza A virus (A/California/04/2009). On day 2, 4, 6, and 14 after infection, 3-6 anesthetized hamsters per infection group were euthanized by exsanguination followed by intracardiac injection of veterinary euthanasia solution (SleepAway; Fort Dodge). Plasma samples were exposed to germicidal UV-C light. A disease score was calculated based on virology and weight-loss data.
Study approval
Portions of the human study relevant to Barnes Jewish Hospital, Christian Hospital, and Washington University were reviewed and approved by the Washington University in Saint Louis Institutional Review Board (WU-350 study approval #202003085, and plasma metabolomics study approval #202004204). All animal studies were approved by the Institutional Care and Use Committee at Mount Sinai School of Medicine, following the humane care and use guidelines set by the institution.
Method details
Metabolomics sample preparation
Participant plasma, which had been stored at −80°C upon collection, was thawed on ice. A 50 μL aliquot was transferred onto a solid-phase-extraction (SPE)-system CAPTIVA-EMR Lipid 96-wellplate (Agilent Technologies) before addition of 250 μL of acetonitrile containing 1% formic acid (v/v) and 10 μM internal standard (consisting of uniformly 13C and 15N labeled amino acids from Cambridge Isotope Laboratories, Inc). The samples were mixed for 1 min at 360 rpm on an orbital shaker at room temperature prior to a 10 min incubation period at 4°C. Afterward, 200 μL 80% acetonitrile in water (v/v) was added to the samples. The samples were mixed on an orbital shaker (360 rpm) for an additional 10 min at room temperature. The samples were then eluted into a 96-deepwell collection plate by centrifugation (10 min, 57 x g, 4°C followed by 2 min, 1000 x g, 4°C). Polar eluates were stored at −80°C until the day of LC/MS analysis.
The SPE-plates were then washed twice with 500 μL 80% acetonitrile in water (v/v). Lipids still bound to the SPE-material were then released into a second elution plate, in two elution steps applying 2x 500 μL 1:1 methyl tert-butyl ether:methanol (v/v) onto the SPE cartridge and centrifuging for 2 min at 1000 g and 4°C. The combined eluates were dried under a stream of nitrogen (Biotage SPE Dry Evaporation System) at room temperature and reconstituted with 100 μL 1:1 2-propanol:methanol (v/v) prior to LC/MS analysis.
Hamster plasma samples were diluted 1:4 with methanol (v/v), vortexed for 30 s, and incubated at −20°C for 2 hours. Samples were centrifuged for 10 minutes at 13,500 x g at 4°C and supernatant was transferred to a new centrifuge tube, concentrated, and stored at −80°C until reconstitution as described above.
LC/MS analysis of polar metabolites
A 2 μLaliquot of polar metabolite extract was subjected to LC/MS analysis by using an Agilent 1290 Infinity II liquid-chromatography (LC) system coupled to an Agilent 6540 Quadrupole-Time-of-Flight (QTOF) mass spectrometer with a dual Agilent Jet Stream electrospray ionization source. Polar metabolites were separated on a SeQuant® ZIC®-pHILIC column (100 × 2.1 mm, 5 μm, polymer, Merck-Millipore) including a ZIC®-pHILIC guard column (2.1 mm x 20 mm, 5 μm). The column compartment temperature was maintained at 40°C and the flow rate was set to 250 μL·min-1. The mobile phases consisted of A: 95% water, 5% acetonitrile, 20 mM ammonium bicarbonate, 0.1% ammonium hydroxide solution (25% ammonia in water), 2.5 μM medronic acid, and B: 95% acetonitrile, 5% water, 2.5 μM medronic acid. The following linear gradient was applied: 0 to 1 min, 90% B; 12 min, 35% B; 12.5 to 14.5 min, 25% B; 15 min, 90% B followed by a re-equilibration phase of 4 min at 400 μL·min−1 and 2 min at 250 μL·min−1. Metabolites were detected in positive and negative ion mode with the following source parameters: gas temperature 200°C, drying gas flow 10 L·min−1, nebulizer pressure 44 psi, sheath gas temperature 300°C, sheath gas flow 12 L·min−1, VCap 3000 V, nozzle voltage 2000 V, Fragmentor 100 V, Skimmer 65 V, Oct 1 RF Vpp 750 V, and m/z range 50-1700. Data were acquired under continuous reference mass correction at m/z 121.0509 and 922.0890 for positive ion mode and m/z 119.0363 and 966.0007 for negative ion mode. Samples were randomized prior to analysis. In addition, a quality control sample was injected after every 12th sample to monitor signal stability of the instrument.
LC/MS analysis of lipid metabolites
A 2 μL aliquot of lipid metabolite extract was subjected to LC/MS analysis by using an Agilent 1290 Infinity II LC-system coupled to an Agilent 6545 QTOF mass spectrometer with a dual Agilent Jet Stream electrospray ionization source. Lipids were separated on an Acquity UPLC® HSS T3 column (2.1 × 150 mm, 1.8 μm) including an Acquity UPLC® HSS T3 VanGuard Pre-Column (2.1 × 5mm, 1.8 μm) at a temperature of 60°C and a flow rate of 250 μL·min−1. The mobile phases consisted of A: 60% acetonitrile, 40% water, 0.1% formic acid, 10 mM ammonium formate, 2.5 μM medronic acid, and B: 90% 2-propanol, 10% acetonitrile, 0.1% formic acid, 10 mM ammonium formate (dissolved in 1 mL water). The following linear gradient was used: 0-2 min, 30% B; 17 min, 75% B; 20 min, 85% B; 23-26 min, 100% B; 26 min, 30% B followed by a re-equilibration phase of 5 min.
Lipids were detected in positive and negative ion mode with the following source parameters: gas temperature 250°C, drying gas flow 11 L·min−1, nebulizer pressure 35 psi, sheath gas temperature 300°C, sheath gas flow 12 L·min−1, VCap 3000 V, nozzle voltage 500 V, Fragmentor 160 V, Skimmer 65 V, Oct 1 RF Vpp 750 V, and m/z range 50-1700. Data were acquired under continuous reference mass correction at m/z 121.0509 and 922.0890 in positive ion mode and m/z 119.0363 and 966.0007 in negative ion mode. Samples were randomized before analysis. In addition, a quality-control (QC) sample was injected after every 12th sample to monitor signal stability of the instrument.
Data preprocessing and normalization
Polar metabolite identifications were supported by matching the retention time, accurate mass, and MS/MS fragmentation data to our in-house retention time and MS/MS library created from authentic reference standards (Mass Spectrometry Metabolite Library supplied by IROA Technologies, Millipore Sigma, St. Louis, MO, USA) and online MS/MS libraries (Human Metabolome Database (HMDB, https://hmdb.ca 70), Mass Bank of North America (MoNA, https://mona.fiehnlab.ucdavis.edu/ 71), and mzCloud (https://www.mzcloud.org). Lipid iterative MS/MS data were annotated with the Agilent Lipid Annotator software. All data files were then analyzed in Skyline (Version 20.1.0.155) to obtain peak areas. m/z values of the metabolite and lipid target lists obtained from the metabolite identification workflow, which had at least an MS/MS match to an online library, were extracted under consideration of retention times.
Due to the risk of handling plasma samples from SARS-CoV-2-positive patients and not knowing how many batches of samples we would receive, we refrained from preparing a pooled sample and instead used the NIST SRM 1950 plasma reference material as a QC sample in each batch. The QC sample was injected after every 12th sample. After peak area extraction, batch effects were observed in the research samples (see Figure S2A). The research samples and QC data were used to test typical batch normalization methods (see Figure S2B), including constant sum, unit length, scale, percentile shift, minimum-maximum, probabilistic quotient normalization, quantile, and ComBat correction used in metabolomics.29,72, 73, 74, 75 In Figure S2B, the variance remaining in the research samples normalized to the variance in the QC samples is shown for each method. The higher this ratio, the more variance that remains in the research samples and the more batch-derived variance in the QC samples is reduced. ComBat correction outperformed the other batch-correction approaches tested by using this metric. After correction, samples are well clustered according to sample type (WU-350, QC, blank) as shown in Figure S2C. In addition, within the research samples, there is no clustering by batch (see Figure S2D).
Machine learning
Samples were split into two distinct cohorts for training and testing the ML model. d0 COV+ patient samples within batches 1-6 made up the training set and d0 COV+ patient samples from batches 7 through 9 made up the test set. Training and tests sets were treated independently except for batch normalization, which was carried out for all patients (including samples collected after d0 and COV- samples) together. Demographics of both training and tests sets are available in Tables S1 and S2.
Model selection was based on 20-fold cross validation of the training set. Five different ML models: logistic regression, ElasticNet linear regression, partial least-squares discriminant analysis (PLSDA), support vector machine (SVM), and random forest were selected for consideration based on interpretability and previous studies.10,12,59,76 Hyperparameters of all models and feature selection strategies were optimized by using 20-fold cross validation and a grid search. Two separate feature selection strategies were tested: a correlation-based approach and a statistic-based approach. In the correlation-based approach, the Pearson correlation was computed between each metabolite’s intensity and disease severity. Then, the top % of metabolites sorted by absolute correlation was taken as the predictors for the ML model. In the statistic-based approach, a Student’s t test was performed to assess the statistical significance of the differences in each metabolite’s intensity between COV+ severe and COV+ non-severe patients. Absolute fold-change and p value cutoffs were used to select metabolites. Performance was assessed with the area under the receiver operating characteristic curve (AUC). After optimization, ElasticNet regression achieved the highest AUC on the cross validated training dataset. The ElasticNet model is given below in Equation 1, where is the matrix of metabolic profiles (# of samples by # of metabolites), is the bias term, is the sample labels (0 = COV+ non-severe, 1 = COV+ severe), is the weight of each metabolite to the model prediction, is the weight of the regularization, and is the mixing parameter between the and norm regularization.
| (Equation 1) |
After optimization, the correlation-based feature selection was used by taking the top 33% most correlated metabolites and model hyperparameters were set to and . In the reduced predictor model, no feature selection was performed and model hyperparameters were set to and .
The variable importance of each metabolite in the ElasticNet model is easily computed from the optimized weights, . To normalize for the different abundances of the metabolites, each weight was normalized by the median abundance of the metabolite across all samples. The more positive the variable importance, the more predictive that metabolite is to severe disease. The more negative the variable importance, the more predictive the metabolite is to non-severe disease. To find the metabolites that significantly contribute to the model fit, the training dataset was resampled with replacement 10,000 times. At each iteration, the ElasticNet model was trained and the variable importance was calculated. After the iterations were complete, the 95% confidence interval of the variable importance was calculated for each metabolite by using the 2.5 and 97.5 percentiles. If this interval included zero, the metabolite did not significantly contribute to the model fit.
All ML analyses were carried out by using Python (v3.7), with extensive use of the packages SciPy (v1.4.1)77 and Scikit-learn (v0.23.1).78
Controlling for disease severity
To control for disease severity in the correlation analysis of the predictor metabolites, interleukin levels, and patient parameters (laboratory results, comorbidities, demographics), the mean value within each patient group (COV+ severe, COV+ non-severe, and COV-) was subtracted from each patient’s metabolite levels, interleukin levels, and patient parameters. This prevents weak correlations from occurring as a result of bulk changes associated with disease severity. After correction, the Pearson correlations between each corrected metabolite/interleukin measurement and patient parameter were calculated.
Confirming metabolite identities of predictor metabolites
The identities of the predictor metabolites were rigorously confirmed with authentic standards. For the polar compounds, authentic standards were purchased to not only match MS/MS spectra but also retention times. For lipids, one or two standards per lipid class were matched to an authentic standard to compare MS/MS spectra and retention times. PCs were identified based on m/z and the two characteristic fragments 184.0733 and 86.0964 in positive ionization mode. For PCs where no peaks for the acyl-chains were observed, only the sum composition can be given. PEs were identified based on the neutral loss of phosphorylethanolamine (141.0191) in positive mode. The fatty acyl composition could be derived from the spectra, but no differentiation of regioisomers was possible, as was the case for ceramides. To denote regiospecificity, metabolites whose regioisomers could be differentiated have their acyl-chains separated with a “/” while those that could not have a “_.” Cer-NS d18:1_16:0 was matched to its authentic standard. MS/MS data for Cer-NS d18:2_16:0 were matched to MS/MS library spectra. Cer-NS d18:3_16:0 eluted slightly before Cer-NS d18:1_16:0, as expected due to having one less double bond. LPCs were identified based on MS/MS spectral matches. Standards were available for LPC 14:0/0:0 and LPC 18:1/0:0. Their retention times were used as a reference for the other LPCs. The two regioisomers of LPCs (sn1 and sn2) were separated by liquid chromatography, with the sn1 isomer eluting later. They are also distinguished by their MS/MS spectra. 1-acyl-LPC (sn1) shows two main fragments (m/z 184.0733 and 104.1070), whereas the 2-acyl-LPC (sn2) has a more pronounced 184.0733 fragment. The 104.1070 fragment (choline) has been previously reported as being more abundant from sodiated LPCs.79 We note that sn2 LPCs can be converted to sn1 during sample preparation, and our sample preparation was not dedicated to preserve isomers.80,81
Acquiring MS/MS data
MS/MS spectra for polar metabolites and lipids were acquired by using an iterative approach in the MassHunter Acquisition Software (Version 10.1.48, Agilent Technologies) on an Agilent 6540 and 6545 QTOF, respectively. The same source settings as for MS1 data acquisition were used. MS/MS spectra were acquired at a scan rate of 3 spectra/s with different intensity thresholds and collision energies of 10, 20, and 40 V to increase identification rates.
To improve matching to Orbitrap spectral databases, MS/MS data for polar metabolites were acquired on an Orbitrap ID-X Tribrid mass spectrometer (Thermo Scientific). A Vanquish Horizon UHPLC system, with the same chromatographic conditions as described in the Method details, was interfaced with the mass spectrometer via electrospray ionization in both positive and negative ion mode with a spray voltage of 3.5 and 2.8 kV, respectively. The RF lens value was 35%. Data were acquired in data dependent acquisition (DDA) mode by using the built-in deep scan option (AcquireX) with a mass range of 67-900 m/z. MS/MS scans were acquired at 15K resolution on a NIST SRM 1950 plasma sample and from 4 individual samples (d0, d3, d7, and d14) in both positive and negative ion mode with different collision energies in the range of 20 NCE to 50 NCE for HCD and 30 NCE for CID to maximize identifications.
Quantification and statistical analysis
Statistical analysis
All statistical analyses were performed by using the SciPy (v1.4.1)77 and statsmodels (v0.11.1)82 Python packages and with the Mass Profiler Professional Software (Agilent Technologies, v15.5). All p values were corrected for multiple hypothesis testing by using the Benjamini-Hochberg procedure.83 All statistical tests used are described in the figure legends.
Permutation test
To assess the significance of the model fit and compare the predictive power to what is expected from random chance, we performed a permutation test. After the feature selection and model hyperparameters were optimized, the training dataset labels were permuted, and the model was retrained on the permuted data. Then, the performance of the model was assessed on the non-permuted test set and the AUC was computed. This process was repeated 1,000 times. The empirical p value was computed by calculating the percentage of the 1,000 permutations that achieved an AUC higher than that of the model’s performance when trained on non-permuted data.
Acknowledgments
This work was supported by funding from the National Institutes of Health grants R24OD024624 (G.J.P.) and R35ES2028365 (G.J.P.). This study used samples obtained from the Washington University School of Medicine’s COVID-19 biorepository, which is supported by the Barnes-Jewish Hospital Foundation; the Siteman Cancer Center grant P30 CA091842 from the National Cancer Institute of the National Institutes of Health; and the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the National Center for Advancing Translational Sciences of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the view of the NIH. This work was also partly supported by CRIP (Center for Research for Influenza Pathogenesis), a NIAID supported Center of Excellence for Influenza Research and Surveillance (CEIRS, contract HHSN272201400008C; W.-C.L., R.A.A., and A.G.-S.); by CRIPT (Center for Research for Influenza Pathogenesis and Transmission), a NIAID supported Center of Excellence for Influenza Research and Response (CEIRR, contract 75N93019R00028; R.A.A and A.G.-S.); by NIAID grant U19AI135972; by NCI grant U54CA260560; by supplements to NIAID grants U19AI135972, U19AI142733, and DOD grant W81XWH-20-1-0270; by the Defense Advanced Research Projects Agency (HR0011-19-2-0020); by the generous support of the JPB Foundation and the Open Philanthropy Project (research grant 2020-215611 (5384)); and by anonymous donors (A.G.-S.). The graphical abstract, Figure 1C, and Figure 6A were created with BioRender.com.
Author contributions
M.S. planned the metabolomics study, executed sample preparation, acquired and analyzed the LC/MS and MS/MS data, did statistical analyses, prepared figures, and wrote the manuscript. E.S. analyzed the data, did the statistical analyses, developed the prediction model, prepared figures, and wrote the manuscript. M.S.-H. executed sample preparation, acquired and analyzed the LC/MS and MS/MS data, prepared figures, and contributed to writing the manuscript. D.S.A. planned the metabolomics study and collected and managed demographic and clinical information. C.W.G., J.A.O, and P.A.M. designed and coordinated clinical studies and managed the COVID-19 biorepository. R.A., W.-C.L., K.A.-T., L.P.S., and A.G.-S. planned the hamster study, collected the samples, and prepared the hamster samples for metabolomics. G.J.P. planned the study, helped design experiments, contributed to data interpretation, and supervised all aspects of the research. All authors contributed to revising the manuscript.
Declaration of interests
The A.G.-S. laboratory has received research support from Pfizer, Pharmamar, Blade Therapeutics, Avimex, Dynavax, Kenall Manufacturing, ImmunityBio, Nanocomposix, Senhwa Biosciences, and 7Hills Pharma. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7 Hills Pharma, Avimex, Vaxalto, Accurius, and Esperovax. G.J.P. is a scientific advisor for Cambridge Isotope Laboratories. The Patti laboratory has a collaboration agreement with Agilent Technologies and Thermo Fisher Scientific. All other authors declare no competing interests.
Published: July 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100369.
Supplemental information
Data and code availability
The raw LC/MS data as well as the processed metabolic profiles and corresponding metadata for the human (deidentified) and animal samples is publicly available on the Metabolomics Workbench repository (NMRD:ST001849, ST001853; https://doi.org/10.21228/M80981). Custom code used to perform the ML analyses is available on GitHub (https://github.com/e-stan/covid_19_analysis)
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John Hopkins Coronavirus Resource Center . 2020. About us.https://coronavirus.jhu.edu/about [Google Scholar]
- 3.NIH . 2021. COVID-19 treatment guidelines: therapeutic management of hospitalized adults with COVID-19. Updated July 8, 2021.https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/ [Google Scholar]
- 4.Pal A. 2021. Wider image in COVID-hit India, a 26-year-old-doctor decides who lives and who dies. Reuters, May 5, 2021.https://www.reuters.com/world/asia-pacific/wider-image-covid-hit-india-26-year-old-doctor-decides-who-lives-who-dies-2021-05-05/ [Google Scholar]
- 5.Kim P.S., Read S.W., Fauci A.S. Therapy for early COVID-19: a critical need. JAMA. 2020;324:2149–2150. doi: 10.1001/jama.2020.22813. [DOI] [PubMed] [Google Scholar]
- 6.Goyal A., Cardozo-Ojeda E.F., Schiffer J.T. Potency and timing of antiviral therapy as determinants of duration of SARS-CoV-2 shedding and intensity of inflammatory response. Sci. Adv. 2020;6:eabc7112. doi: 10.1126/sciadv.abc7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kattan M.W., Ji X., Milinovich A., Adegboye J., Duggal A., Dweik R., Khouli H., Gordon S., Young J.B., Jehi L. An algorithm for classifying patients most likely to develop severe coronavirus disease 2019 illness. Crit. Care Explor. 2020;2:e0300. doi: 10.1097/CCE.0000000000000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser D.D., Slessarev M., Martin C.M., Daley M., Patel M.A., Miller M.R., Patterson E.K., O’Gorman D.B., Gill S.E., Wishart D.S. Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit. Care Explor. 2020;2:e0272. doi: 10.1097/CCE.0000000000000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5:140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overmyer K.A., Shishkova E., Miller I.J., Balnis J., Bernstein M.N., Peters-Clarke T.M., Meyer J.G., Quan Q., Muehlbauer L.K., Trujillo E.A. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2020;12:23–40.e7. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu D., Shu T., Yang X., Song J.-X., Zhang M., Yao C., Liu W., Huang M., Yu Y., Yang Q. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020;7:1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimhofer T., Lodge S., Whiley L., Gray N., Loo R.L., Lawler N.G., Nitschke P., Bong S.H., Morrison D.L., Begum S. Integrative modeling of quantitative plasma lipoprotein, metabolic, and amino acid data reveals a multiorgan pathological signature of SARS-CoV-2 infection. J. Proteome Res. 2020;19:4442–4454. doi: 10.1021/acs.jproteome.0c00519. [DOI] [PubMed] [Google Scholar]
- 16.Shi D., Yan R., Lv L., Jiang H., Lu Y., Sheng J., Xie J., Wu W., Xia J., Xu K. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism. 2021;118:154739. doi: 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doğan H.O., Şenol O., Bolat S., Yıldız S.N., Büyüktuna S.A., Sarıismailoğlu R., Doğan K., Hasbek M., Hekim S.N. Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 2021;93:2340–2349. doi: 10.1002/jmv.26716. [DOI] [PubMed] [Google Scholar]
- 18.Meoni G., Ghini V., Maggi L., Vignoli A., Mazzoni A., Salvati L., Capone M., Vanni A., Tenori L., Fontanari P. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 2021;17:e1009243. doi: 10.1371/journal.ppat.1009243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delafiori J., Navarro L.C., Siciliano R.F., de Melo G.C., Busanello E.N.B., Nicolau J.C., Sales G.M., de Oliveira A.N., Val F.F.A., de Oliveira D.N. Covid-19 automated diagnosis and risk assessment through metabolomics and machine learning. Anal. Chem. 2021;93:2471–2479. doi: 10.1021/acs.analchem.0c04497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen R. High-throughput approaches of diagnosis and therapies for COVID-19: antibody panels, proteomics and metabolomics. Future Drug Discov. 2021 doi: 10.4155/fdd-2020-0027. Published online November 10, 2020. [DOI] [Google Scholar]
- 21.Patti G.J., Yanes O., Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beger R.D., Dunn W., Schmidt M.A., Gross S.S., Kirwan J.A., Cascante M., Brennan L., Wishart D.S., Oresic M., Hankemeier T. Metabolomics enables precision medicine: “a white paper, community perspective.”. Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A., Satlin M.J., Campion T.R., Jr., Nahid M., Ringel J.B. Clinical characteristics of covid-19 in New York City. N. Engl. J. Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CDC . 2020. COVID-19: Symptoms of COVID-19. Updated February 22, 2021.https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html [Google Scholar]
- 28.Elixhauser A., Steiner C., Harris D.R., Coffey R.M. Comorbidity measures for use with administrative data. Med. Care. 1998;36:8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Fernández-Albert F., Llorach R., Garcia-Aloy M., Ziyatdinov A., Andres-Lacueva C., Perera A. Intensity drift removal in LC/MS metabolomics by common variance compensation. Bioinformatics. 2014;30:2899–2905. doi: 10.1093/bioinformatics/btu423. [DOI] [PubMed] [Google Scholar]
- 30.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrilli C.M., Jones S.A., Yang J., Rajagopalan H., O’Donnell L., Chernyak Y., Tobin K.A., Cerfolio R.J., Francois F., Horwitz L.I. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., Chua G.H., Zhang C., Meng F.P., Xu Z. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32:188–202.e5. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Migaud M., Gandotra S., Chand H.S., Gillespie M.N., Thannickal V.J., Langley R.J. Metabolomics to predict antiviral drug efficacy in COVID-19. Am. J. Respir. Cell Mol. Biol. 2020;63:396–398. doi: 10.1165/rcmb.2020-0206LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005;67:301–320. [Google Scholar]
- 35.Mendez K.M., Broadhurst D.I., Reinke S.N. Migrating from partial least squares discriminant analysis to artificial neural networks: a comparison of functionally equivalent visualisation and feature contribution tools using jupyter notebooks. Metabolomics. 2020;16:17. doi: 10.1007/s11306-020-1640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed Z., Zeeshan S., Foran D.J., Kleinman L.C., Wondisford F.E., Dong X. Integrative clinical, genomics and metabolomics data analysis for mainstream precision medicine to investigate COVID-19. BMJ Innov. 2021;7:6–10. [Google Scholar]
- 37.Luo X., Zhou W., Yan X., Guo T., Wang B., Xia H., Ye L., Xiong J., Jiang Z., Liu Y. Prognostic value of c-reactive protein in patients with coronavirus 2019. Clin. Infect. Dis. 2020;71:2174–2179. doi: 10.1093/cid/ciaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct. Target. Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J.H., Lee H.K. Re-analysis of single cell transcriptome reveals that the NR3C1-CXCL8-neutrophil axis determines the severity of COVID-19. Front. Immunol. 2020;11:2145. doi: 10.3389/fimmu.2020.02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 42.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020;95:834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target. Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., Li H., Luo M., Liu J., Wu L., Lin X., Li R., Wang Z., Zhong H., Zheng W. Lymphopenia predicted illness severity and recovery in patients with COVID-19: a single-center, retrospective study. PLoS ONE. 2020;15:e0241659. doi: 10.1371/journal.pone.0241659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jafarzadeh A., Jafarzadeh S., Nozari P., Mokhtari P., Nemati M. Lymphopenia an important immunological abnormality in patients with COVID-19: Possible mechanisms. Scand. J. Immunol. 2021;93:e12967. doi: 10.1111/sji.12967. [DOI] [PubMed] [Google Scholar]
- 46.Fathi N., Rezaei N. Lymphopenia in COVID-19: therapeutic opportunities. Cell Biol. Int. 2020;44:1792–1797. doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith S.M., Boppana A., Traupman J.A., Unson E., Maddock D.A., Chao K., Dobesh D.P., Brufsky A., Connor R.I. Impaired glucose metabolism in patients with diabetes, prediabetes, and obesity is associated with severe COVID-19. J. Med. Virol. 2021;93:409–415. doi: 10.1002/jmv.26227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain V., Yuan J.M. Predictive symptoms and comorbidities for severe COVID-19 and intensive care unit admission: a systematic review and meta-analysis. Int. J. Public Health. 2020;65:533–546. doi: 10.1007/s00038-020-01390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benesty J., Chen J., Huang Y., Cohen I. Springer-Verlag; 2009. Noise Reduction in Speech Processing. [Google Scholar]
- 50.Tate R.F. Correlation between a discrete and a continuous variable. point-biserial correlation. Ann. Math. Stat. 1954;25:603–607. [Google Scholar]
- 51.Gagnebin Y., Jaques D.A., Rudaz S., de Seigneux S., Boccard J., Ponte B. Exploring blood alterations in chronic kidney disease and haemodialysis using metabolomics. Sci. Rep. 2020;10:19502. doi: 10.1038/s41598-020-76524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mudd P.A., Crawford J.C., Turner J.S., Souquette A., Reynolds D., Bender D., Bosanquet J.P., Anand N.J., Striker D.A., Martin R.S. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci. Adv. 2020;6:eabe3024. doi: 10.1126/sciadv.abe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santa Cruz A., Mendes-Frias A., Oliveira A.I., Dias L., Matos A.R., Carvalho A., Capela C., Pedrosa J., Castro A.G., Silvestre R. Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 2021;12:613422. doi: 10.3389/fimmu.2021.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorham J., Moreau A., Corazza F., Peluso L., Ponthieux F., Talamonti M., Izzi A., Nagant C., Ndieugnou Djangang N., Garufi A. Interleukine-6 in critically ill COVID-19 patients: a retrospective analysis. PLoS ONE. 2020;15:e0244628. doi: 10.1371/journal.pone.0244628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwatsuki-Horimoto K., Nakajima N., Ichiko Y., Sakai-Tagawa Y., Noda T., Hasegawa H., Kawaoka Y. Syrian hamster as an animal model for the study of human influenza virus infection. J. Virol. 2018;92:e01693-17. doi: 10.1128/JVI.01693-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho J.S.Y., Mok B.W., Campisi L., Jordan T., Yildiz S., Parameswaran S., Wayman J.A., Gaudreault N.N., Meekins D.A., Indran S.V. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell. 2021;184:2618–2632.e17. doi: 10.1016/j.cell.2021.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.FDA . 2020. Fact sheet for health care providers emergency use authorization (EUA) of casirivimab and imdevimab.https://www.fda.gov/media/143892/download [Google Scholar]
- 59.Mendez K.M., Reinke S.N., Broadhurst D.I. A comparative evaluation of the generalised predictive ability of eight machine learning algorithms across ten clinical metabolomics data sets for binary classification. Metabolomics. 2019;15:150. doi: 10.1007/s11306-019-1612-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barberis E., Timo S., Amede E., Vanella V.V., Puricelli C., Cappellano G., Raineri D., Cittone M.G., Rizzi E., Pedrinelli A.R. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 2020;21:E8623. doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., Shi Y. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arshad H., Alfonso J.C.L., Franke R., Michaelis K., Araujo L., Habib A., Zboromyrska Y., Lücke E., Strungaru E., Akmatov M.K. Decreased plasma phospholipid concentrations and increased acid sphingomyelinase activity are accurate biomarkers for community-acquired pneumonia. J. Transl. Med. 2019;17:365. doi: 10.1186/s12967-019-2112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drobnik W., Liebisch G., Audebert F.X., Frohlich D., Gluck T., Vogel P., Rothe G., Schmitz G. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 2003;44:754–761. doi: 10.1194/jlr.M200401-JLR200. [DOI] [PubMed] [Google Scholar]
- 66.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mullen P.J., Garcia G., Jr., Purkayastha A., Matulionis N., Schmid E.W., Momcilovic M., Sen C., Langerman J., Ramaiah A., Shackelford D.B. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat. Commun. 2021;12:1876. doi: 10.1038/s41467-021-22166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Codo A.C., Davanzo G.G., Monteiro L.B., de Souza G.F., Muraro S.P., Virgilio-da-Silva J.V., Prodonoff J.S., Carregari V.C., de Biagi Junior C.A.O., Crunfli F. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab. 2020;32:437–446.e5. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., Sajed T., Johnson D., Li C., Karu N. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 72.Ghosh D. Statistical analysis of proteomics, metabolomics, and lipidomics data using mass spectrometry, Datta, Susmita and Mertens, Bart J.A. Springer Frontiers in Probability and the Statistical Sciences, 2017, 295 pages, £82.00, hardcover ISBN: 978-3-319-45807-6 [book review] Int. Stat. Rev. 2017;85:544–545. [Google Scholar]
- 73.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 74.Di Guida R., Engel J., Allwood J.W., Weber R.J., Jones M.R., Sommer U., Viant M.R., Dunn W.B. Non-targeted UHPLC-MS metabolomic data processing methods: a comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics. 2016;12:93. doi: 10.1007/s11306-016-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D.S., Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for COVID-19 (SARS-CoV-2) pandemic: a review. Chaos Solitons Fractals. 2020;139:110059. doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., SciPy 1.0 Contributors Author correction: SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:352. doi: 10.1038/s41592-020-0772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 79.Han X., Gross R.W. Structural determination of lysophospholipid regioisomers by electrospray ionization tandem mass spectrometry. J. Am. Chem. Soc. 1996;118:451–457. [Google Scholar]
- 80.Koistinen K.M., Suoniemi M., Simolin H., Ekroos K. Quantitative lysophospholipidomics in human plasma and skin by LC-MS/MS. Anal. Bioanal. Chem. 2015;407:5091–5099. doi: 10.1007/s00216-014-8453-9. [DOI] [PubMed] [Google Scholar]
- 81.Okudaira M., Inoue A., Shuto A., Nakanaga K., Kano K., Makide K., Saigusa D., Tomioka Y., Aoki J. Separation and quantification of 2-acyl-1-lysophospholipids and 1-acyl-2-lysophospholipids in biological samples by LC-MS/MS. J. Lipid Res. 2014;55:2178–2192. doi: 10.1194/jlr.D048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seabold S., Perktold J. In: van der Walt S., Millman J., editors. SciPy.org; 2010. Statsmodels: economic and statistical modeling with Python; pp. 92–96. (Proceedings of the 9th Python in Science Conference (SCIPY 2010)). [Google Scholar]
- 83.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw LC/MS data as well as the processed metabolic profiles and corresponding metadata for the human (deidentified) and animal samples is publicly available on the Metabolomics Workbench repository (NMRD:ST001849, ST001853; https://doi.org/10.21228/M80981). Custom code used to perform the ML analyses is available on GitHub (https://github.com/e-stan/covid_19_analysis)