Abstract
The endothelial cells from the microvasculature are key drivers and targets of inflammatory and thrombotic processes in microvascular diseases. The study of bioactive lipids in inflammatory processes has been largely based on two-dimensional endothelial cell cultures. Three-dimensional microvessels-on-a-chip provides an opportunity to monitor the inflammatory phenotype of human microvessels in a more physiological-relevant environment. This protocol describes the culture of endothelial cells as three-dimensional microvessels in the OrganoPlate. The microvessels are treated with tumor necrosis factor alpha to induce inflammation. The collection of samples from the microvessels is optimized for measuring bioactive lipids with liquid chromatography-mass spectrometry, providing a more informative metabolic readout as compared with functional assays.
Keywords: Microvessels, 3D cell culture, Oxidative stress, Metabolomics, Microfluidics, Inflammation, Organs-on-chips, Endothelial cells
Background
In recent decades, microvascular diseases have become a leading cause of mortality worldwide. Chronic activation of endothelial cells by cardiovascular risk factors can inflict the loss of pericytes, which play a critical role in microvascular stabilization. To diagnose and treat microvascular diseases, we aim to explore the association of circulating plasma factors with microvascular integrity.
In vitro mechanistic studies of bioactive lipids have been largely based on two-dimensional (2D) endothelial cell cultures that, due to lack of laminar flow and growth of cells on non-compliant stiff substrates, often display a proinflammatory phenotype. This complicates the assessment of inflammatory processes, such as tumor necrosis factor alpha (TNFα)-induced effects. Novel microfluidics-based perfused three-dimensional (3D) microvessels-on-a-chip models provide a unique opportunity to generate microvessels in a more physiological environment. The microvessels-on-a-chip are phaseguide-derived membrane-free microvessels grown from seeded endothelial cells on a 3D soft scaffold in the OrganoPlate 2-lane (van Duinen et al., 2017 ). To monitor the inflammatory phenotype of the 3D microvessels, we used an optimized targeted ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) measurement of a panel of pro- and anti-inflammatory bioactive lipids and generated expression profiles of TNFα-treated microvessels under flow ( Junaid et al., 2020a ).
Using the described protocol, we assessed the concentrations of prostaglandins, isoprostanes, lysophosphatidic acid (LPA) classes, sphingolipids, and platelet activating factor (PAF) secreted by primary human umbilical vein endothelial cells (HUVECs) seeded in the microvessels-on-a-chip. Conditioned medium perfused through TNFα-treated and control (untreated) microvessels was sampled, pooled, and measured by UPLC-MS/MS metabolomics ( Schoeman et al., 2018 ). A strong differential response was observed between untreated and TNFα-treated microvessels. Moreover, the relative concentrations of the bioactive lipids found in the microvessels-on-a-chip are similar to those found in normal human blood vessels.
Most assays performed with the OrganoPlate to study microvessels are related to immunostaining, permeability, and angiogenesis (van Duinen et al., 2019 , Junaid et al., 2020b ). These assays study morphological changes by imaging phenotypes, and although relevant, the metabolic readout of microvessels using metabolomics is more informative. In this protocol, coupling LC/MS-based metabolomics techniques with the OrganoPlate is novel and allows us to measure bioactive lipids, providing exceptional information regarding microvascular disease onset and progression and the discovery of new biomarkers of inflammation and oxidative stress in this context. The results are easily translatable to physiological values found in human microvessels. Moreover, metabolic readouts for inflammation and oxidative stress are highly valuable in the organ-on-a-chip field with respect to drug research. The following protocol using the OrganoPlate was developed for researchers to study microvessels with approaches that suit other microfluidic devices and organ models.
Materials and Reagents
1.5-ml Eppendorf tubes
15-ml centrifuge tubes
50-ml centrifuge tubes
2-ml pipettes (sterile)
5-ml pipettes (sterile)
10-ml pipettes (sterile)
P20 micropipette
P1000 micropipette
P20 tips (sterile)
P1000 tips (sterile)
0.2-ml Biopur repeater pipette tips (VWR, catalog number: 89232-968)
1.0-ml Biopur repeater pipette tips (VWR, catalog number: 89232-972)
2.5-ml Biopur repeater pipette tips (VWR, catalog number: 89232-974)
Cryovial of primary human umbilical vein endothelial cells (HUVECs; Lonza, catalog number: C2519A; stored in liquid nitrogen)
Cell counting slides (Bio-Rad, catalog number: 1450011)
T75 flasks
Ice
Dry ice
Trypan Blue
1% gelatin
-
Endothelial Cell Growth Medium 2 (EGM2; PromoCell, catalog number: C-39216; 4°C)
Basal Medium 2 (Promocell, catalog number: C-22211; 4°C)
Growth Medium 2 Supplement Mix (Promocell, catalog number: C-39216; -20°C)
Penicillin/Streptomycin (Sigma-Aldrich, catalog number: P4333; -20°C)
Phosphate-buffered saline (PBS)
TrypLE (Gibco, catalog number: 12604013)
OrganoPlate 2-lane (MIMETAS, catalog number: 9603-400-B)
Rat tail collagen type 1 (Trevigen, catalog number: 3440-005-01; 4°C)
37 g/L Na2CO3 (Sigma-Aldrich, catalog number: S5761; 4°C)
1 M HEPES buffer (Gibco, catalog number: 15630-056; 4°C)
Hank’s Balanced Salt Solution with calcium and magnesium (HBSS+; Life Technologies, catalog number: 24020117)
Tumor necrosis factor alpha (TNFα; Sigma-Aldrich, catalog number: H8916; -20°C)
Phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, catalog number: P8139; -80°C)
2-lane plate layout form (Supplementary Material 1)
70% ethanol
Equipment
Pipetboy
Eppendorf repeater M4 pipette (VWR, catalog number: 613-2890)
OrganoFlow (MIMETAS, catalog number: MI-OFPR-L)
Incubator (37°C, 5% CO2)
Water bath (37°C)
Eppendorf centrifuge 5810 (Sigma-Aldrich, catalog number: 022628188)
Cell culture hood
-80°C freezer
Liquid chromatography-tandem mass spectrometer (Shimadzu, catalog number: LCMS-8060)
Brightfield microscope
TC20 automated cell counter (Bio-Rad, catalog number: 1450102)
Software
Microsoft Excel
GraphPad Prism (GraphPad Software)
LabSolutions (Shimadzu)
MetaboAnalyst (https://www.metaboanalyst.ca/)
Procedure
This protocol details every step of culturing microvessels in the OrganoPlate 2-lane up to measuring and analyzing bioactive lipids. All procedures handling cells and the OrganoPlate are sterile and carried out in the cell culture hood. A general outline of the workflow is described below:
Day 1: Initiate the culture process and maintenance.
Day 2: Refresh the medium in the T75 flask.
Day 5:
Seed the collagen in the OrganoPlate 2-lane.
Harvest the cells from the T75 flask.
-
Seed the cells in the OrganoPlate 2-lane.
Day 8: Refresh the medium in the OrganoPlate 2-lane.
Day 9: Incubate the microvessels with TNFα and PMA overnight.
Day 10: Collect the medium from the microvessels, store the samples in a -80°C freezer and measure the bioactive lipids.
Day 1
-
Initiate the culture process and maintenance
Warm the 1% gelatin in a 37°C water bath.
Prepare EGM2 by adding Growth Medium 2 Supplement Mix and 5 ml Penicillin/Streptomycin to the Basal Medium 2.
Warm 50 ml EGM2 in a 37°C water bath.
Pipette 5 ml 1% gelatin to a T75 flask and place in the incubator for 30 min.
Pipette 11 ml 50 ml EGM2 into a 15-ml centrifuge tube.
-
Quickly thaw a cryovial of HUVECs in a 37°C water bath, being careful not to submerge the entire vial.
Important: Watch the cryovial closely; when the last sliver of ice melts, remove the cryovial.
Dispense the cells into the 15-ml centrifuge tube containing 11 ml EGM2.
Centrifuge the tube at 200 × g for 7 min to pellet the cells.
While the cells are centrifuging, take the T75 flask containing 1% gelatin from the incubator. Aspirate the gelatin and pipette 12 ml EGM2 into the flask.
Aspirate the supernatant from the centrifuged tube without disturbing the cell pellet.
Resuspend the cells in 500 µl EGM2 by gently pipetting to break up any clumps.
Pipette the cell suspension into the gelatin-coated T75 flask.
-
Gently rock the flask to evenly distribute the cells and place in the incubator.
Day 2
-
Refresh the medium in the T75 flask
Warm 12 ml EGM2 in a 37°C water bath.
Take out the T75 flask containing the HUVECs from the incubator (Figure 1).
-
Remove the medium, replace it with the warmed, fresh medium, and return the flask to the incubator.
Day 5
-
Seed the collagen in the OrganoPlate 2-lane
Warm the 1% gelatin in a 37°C water bath.
Place a 1.5-ml Eppendorf tube on ice.
Pipette 60 µl 1 M HEPES buffer and 60 µl 37 g/L Na2CO3 in the 1.5-ml Eppendorf tube and mix. Place the tube on ice.
Add 480 µl 5 mg/ml rat tail collagen type 1 to the HEPES/Na2CO3 mixture to a final concentration of 4 mg/ml collagen solution. Mix carefully to avoid the formation of bubbles. Place the tube back on ice.
Lay and hold the OrganoPlate flat with your non-pipetting hand in the cell culture hood (Figure 2).
Align the repeater pipette perpendicular to the plate and gently place the repeater pipette at the center of the gel inlet (Figure 3). Make sure that there is contact, but not pressure, between the tip and the gel inlet. Pipette 2 µl 4 mg/ml collagen solution into the gel inlet of each microfluidic unit.
-
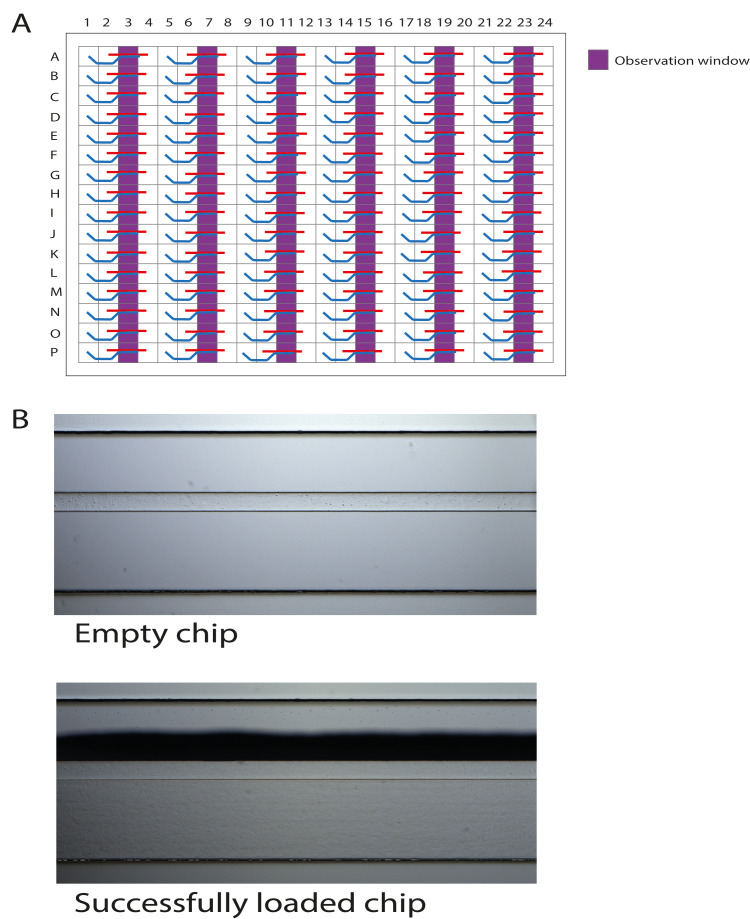
Utilize a brightfield microscope to check the filling patterns in the observation windows of the loaded chips (Figure 4A). The results of one loaded chip should be similar to those shown in Figure 4B.
Important: Meniscus formation as a shadow stretching along the entire observation window should be observed for a successfully loaded chip. If the collagen solution did not completely fill the channel, pipette an additional 2 µl.
-
Place the OrganoPlate in the incubator for 10 min to polymerize the collagen.
Important: Avoid incubating longer than 10 min to prevent the gel from collapsing or shrinking.
Pipette 25 µl HBSS+ into the gel inlet to prevent gel dehydration.
Pipette 50 µl HBSS+ into the observation window for optical clarity.
To coat the microchannels with gelatin, pipette 25 µl 1% gelatin into the medium inlet (Figure 5). Dispense the gelatin against the wall of the well to prevent air bubble formation.
Place the OrganoPlate in the incubator for at least 30 min.
-
Harvest the cells from the T75 flask and seed in the OrganoPlate 2-lane
Warm 50 ml EGM2 in a 37°C water bath.
Observe the HUVECs in the T75 flask under the brightfield microscope. They should be 80-100% confluent. If not, refresh the medium and leave them for an extra day in the incubator.
Aspirate the medium from the T75 flask.
Rinse the cells with 10 ml room-temperature PBS.
Aspirate the PBS from the flask.
Carry out Steps D4 and D5 again.
Cover the cells with 2 ml TrypLE solution.
Place the cells into the incubator for 3-5 min. Periodically examine the cell layer microscopically to check for cell detachment.
Allow the cell detachment to continue until approximately 90% of the cells are rounded up. At this point, tap the flask against the palm of your hand to release the majority of the cells from the cell culture surface.
After the cells are released, neutralize the TrypLE in the flask with 5 ml EGM2.
Quickly transfer the detached cells to a sterile 15-ml centrifuge tube.
Rinse the flask with 5 ml EGM2 to collect residual cells and add this rinse to the centrifuge tube.
Count the number of cells using the available cell counter device in your lab.
Centrifuge the harvested cells at 200 × g for 7 min to pellet.
While centrifuging the cells, transfer the OrganoPlate from the incubator to the cell culture hood.
Aspirate the gelatin from the medium inlet using an aspirator. Place the tip of the aspirator in the corner of the well and allow it to touch the bottom of the plate.
Pipette 25 µl EGM2 into the medium inlet.
Place the OrganoPlate back in the incubator.
After centrifuging, aspirate most of the supernatant.
Dilute the cells to a final concentration of 15 × 106 cells/ml with EGM2.
Transfer the cell suspension to a 1.5-ml Eppendorf tube.
Transfer the OrganoPlate from the incubator to the cell culture hood. Lay and hold the OrganoPlate flat with your non-pipetting hand.
Align the repeater pipette perpendicular to the plate and gently place the repeater pipette at the center of the medium outlet (Figure 5). Make sure that there is contact, but not pressure, between the tip and the medium outlet. Pipette 2 µl cell suspension into the medium outlet of each microfluidic unit. Regularly resuspend the cell suspension during seeding to ensure a homogenous cell density.
Utilize a brightfield microscope to observe the cells in the observation window. The cells should be homogeneous in the chip (Figure 6). If cells are not present, pipette an additional 2 µl.
Place the OrganoPlate in the incubator for 1 h to allow the cells to attach.
While the cells are attaching, clean and sterilize the OrganoFlow by spraying and wiping with 70% ethanol (Figure 7).
Place the OrganoFlow on a level surface in the incubator. Plug in the power adapter and turn on the device by pressing the ON/OFF switch located at the back of the instrument.
Press the CONFIRM button to calibrate the OrganoFlow.
Use the UP and DOWN buttons to choose the interval mode. In this mode, the OrganoFlow will move the platform to the desired rocking angle and will remain in this position for the selected interval duration.
Set the rocking angle to 7° and the interval to 8 min.
Press the CONFIRM button to start rocking.
After the cells have attached, add 50 µl EGM2 to the medium in- and outlets.
-
Place the plate parallel on the OrganoFlow in the incubator as shown in Figure 7 for the perfusion of medium from the medium inlets to the medium outlets and vice versa.
Day 8
-
Refresh the medium in the OrganoPlate 2-lane
Warm 15 ml EGM2 in a 37°C water bath.
Take out the OrganoPlate from the incubator.
-
Aspirate the medium from the medium in- and outlets. Place the tip of the aspirator in the corner of the wells and allow it to touch the bottom of the plate.
Important: Placing the tip of the aspirator directly on top of the medium in- and outlets may disturb the microvessels or cause the formation and trapping of air bubbles.
Pipette 50 µl warmed, fresh EGM2 into the medium in- and outlets.
-
Return the OrganoPlate to the OrganoFlow in the incubator.
Day 9
-
Incubate the microvessels with TNFα and PMA overnight
Take out the OrganoPlate from the incubator.
Observe the cells under a brightfield microscope. They should have formed fully confluent microvessels. An example of a microvessel that is perfectly arranged against collagen in the OrganoPlate 2-lane is shown in Figure 8.
Note in the 2-lane plate layout form, the microvessels that are perfectly arranged (Supplementary Material 1).
Partition the microvessels into groups, each containing four microvessels.
Select which groups will be the control, treated with 15 ng/ml TNFα, or treated with 20 ng/ml PMA. In Steps F9-F11, 50 µl medium will be pipetted into the medium in- and outlets of each microvessel.
Dilute TNFα in EGM2 to a final concentration of 15 ng/ml.
Dilute PMA in EGM2 to a final concentration of 20 ng/ml.
-
Aspirate the medium from the medium in- and outlets of the selected microvessels. Place the tip of the aspirator in the corner of the wells and allow it to touch the bottom of the plate.
Important: Placing the tip of the aspirator directly on top of the medium in- and outlets may disturb the microvessels or cause the formation and trapping of air bubbles.
Pipette 50 µl 15 ng/ml TNFα into the medium in- and outlets of the microvessels selected in Step F5.
Pipette 50 µl 20 ng/ml PMA into the medium in- and outlets of the microvessels selected in Step F5.
Pipette 50 µl EGM2 into the medium in- and outlets of the microvessels selected in Step F5. These are the controls.
-
Return the OrganoPlate to the OrganoFlow in the incubator and incubate for 18 h.
Day 10
-
Collect the medium from the microvessels and store samples in a -80°C freezer
Label 1.5-ml Eppendorf tubes with the experimental conditions.
Take out the OrganoPlate from the incubator.
-
Use a P200 micropipette to collect the medium from the medium in- and outlets.
Important: For each condition, the medium from four microvessels should be pooled to form one sample to allow analysis of metabolites at low concentrations due to low cell numbers.
Store the samples in a -80°C freezer.
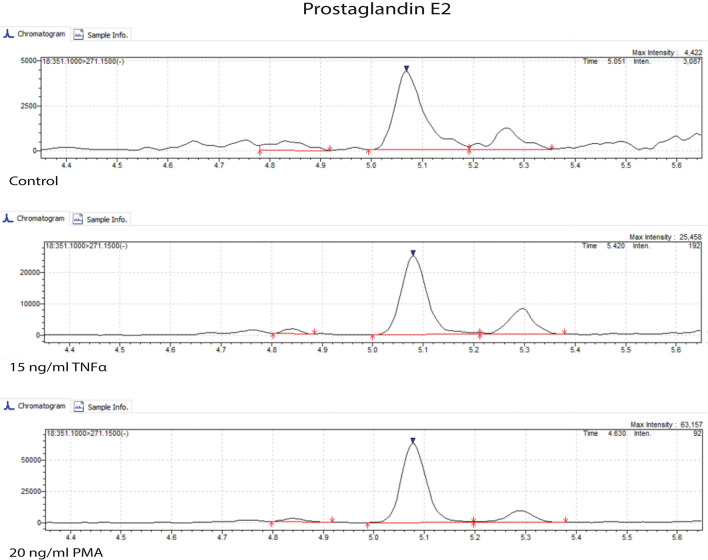
Carry out sample preparation according to the LC-MS platform available in your lab. For our work, we used the method described in the Materials and Methods section of Schoeman et al. (2018) to measure prostaglandins, isoprostanes, LPA classes, sphingolipids, and PAF. We used the serum extraction method. The samples were measured by ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) to obtain results such as those shown in Figure 9.
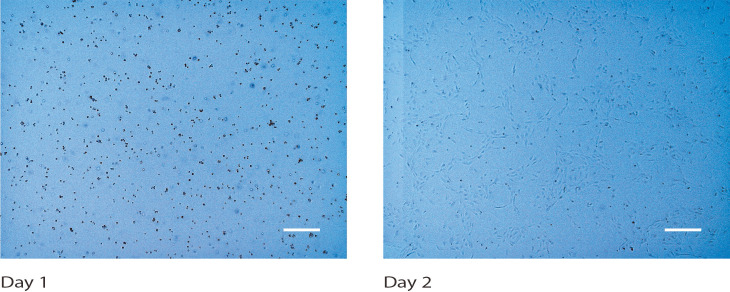
Figure 1. Human umbilical vein endothelial cells (HUVECs) cultured in a T75 flask.
Brightfield microscopy images were taken on days 1 and 2. Scale bars: 400 µm.
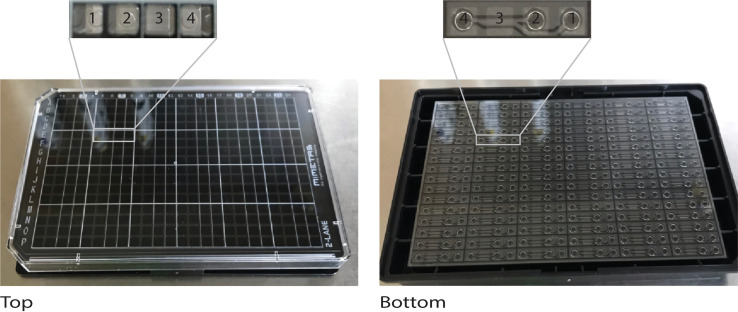
Figure 2. The OrganoPlate 2-lane for culturing microvessels.
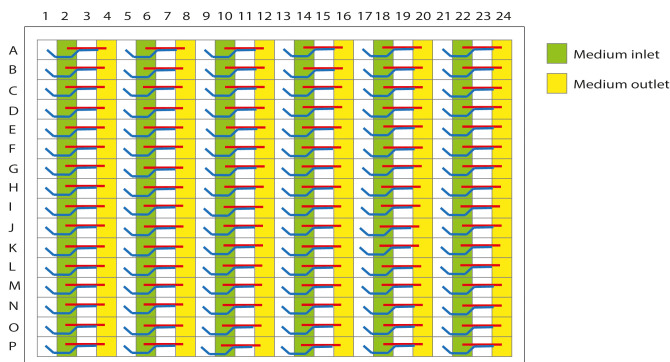
The microfluidic platform is based on a 384-well plate interface on top and 96 microfluidic devices integrated at the bottom. 1 = gel inlet; 2 = medium inlet; 3 = observation window to view the perfusion and gel channels; and 4 = medium outlet.
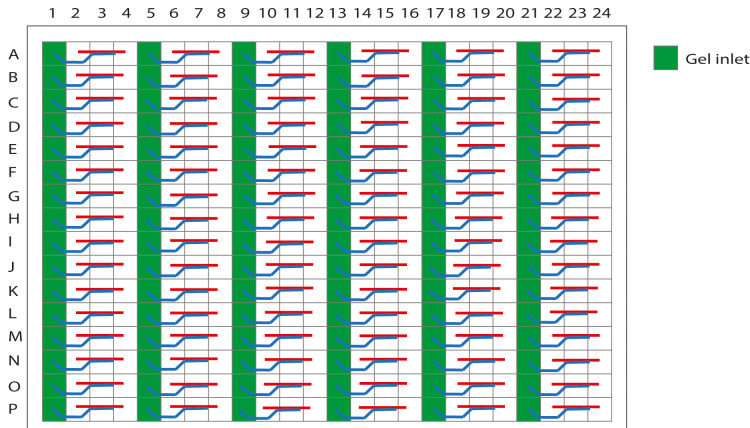
Figure 3. Schematic representation of the OrganoPlate 2-lane with 96 tissue chips indicating the gel inlets.
The wells in columns 1, 5, 9, 13, 17, and 21 are the gel inlets to load the collagen solution into the chips.
Figure 4. Schematic representation of the OrganoPlate 2-lane with 96 tissue chips indicating the observation windows.
A. The wells in columns 3, 7, 11, 15, 19, and 23 are the observation windows. B. Correct loading of the collagen solution is confirmed under a brightfield microscope by observing a shadow stretching along the entire observation window. This is a successfully formed meniscus.
Figure 5. Schematic representation of the OrganoPlate 2-lane with 96 tissue chips indicating medium in- and outlets.
The wells in columns 2, 6, 10, 14, 18, and 22 are medium inlets; the wells in columns 4, 8, 12, 16, 20, and 24 are medium outlets.
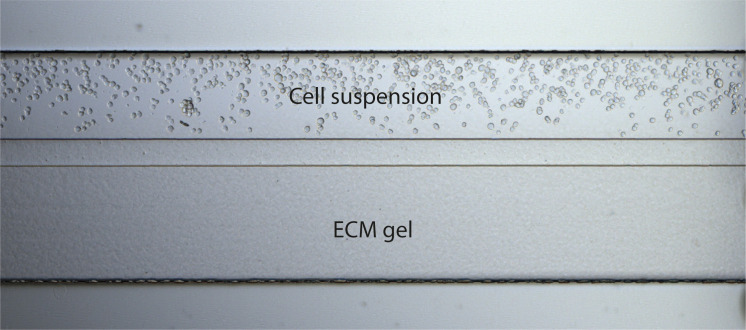
Figure 6. Brightfield microscopy image of the OrganoPlate 2-lane chip after seeding the cells.
Cells are in suspension next to the ECM gel.
Figure 7. The OrganoFlow for perfusing microvessels.
MIMETAS OrganoFlow with the OrganoPlate 2-lane placed on top.
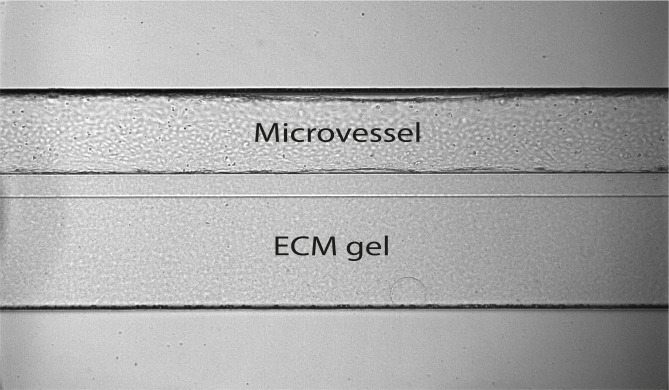
Figure 8. Microvessel in the OrganoPlate 2-lane.
The microvessel was cultured against collagen gel, forming a perfusable vessel.
Figure 9. Ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) chromatogram of prostaglandin E2.
Microvessels were untreated (control) or treated with 15 ng/ml TNFα and 20 ng/ml PMA for 18 h.
Data analysis
-
Use LabSolutions for peak determination and integration.
Note: Other software that are compatible with the available LC-MS platform in your lab can also be used for peak determination and integration.
Use MetaboAnalyst to determine the fold change by dividing the concentrations of bioactive lipids in TNFα- and PMA-treated samples with the concentrations of bioactive lipids in the control samples. Go to https://www.metaboanalyst.ca/ > Click Here to Start > Statistical Analysis > Upload your Data > Fold Change Analysis.
Export the data to Microsoft Excel and calculate the log2 of the fold change.
Copy and paste the calculated log2 from Microsoft Excel to GraphPad Prism and create a bar graph.
Acknowledgments
Our work was supported by the RECONNECT CVON Groot consortium, which is funded by the Dutch Heart Foundation. We acknowledge support from the TKI METABOCHIP project, which is co-funded by the PPP Allowance made available by Health-Holland, Top Sector Life Sciences & Health, to stimulate public–private partnerships. The measurement of bioactive lipids was derived from work by Schoeman et al. (2018). This protocol was derived from Junaid et al. (2020a) (doi: 10.7554/eLife.54754).
Competing interests
TH is the co-founder of the company that makes the OrganoPlates and OrganoFlow used in the protocol.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1. Junaid A., Schoeman J., Yang W., Stam W., Mashaghi A., van Zonneveld A. J. and Hankemeier T.(2020). Metabolic response of blood vessels to TNFalpha. Elife 9: e54754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Junaid A., Tang H., van Reeuwijk A., Abouleila Y., Wuelfroth P., van Duinen V., Stam W., van Zonneveld A. J., Hankemeier T. and Mashaghi A.(2020). Ebola Hemorrhagic Shock Syndrome-on-a-Chip. iScience 23(1): 100765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schoeman J. C., Harms A. C., van Weeghel M., Berger R., Vreeken R. J. and Hankemeier T.(2018). Development and application of a UHPLC-MS/MS metabolomics based comprehensive systemic and tissue-specific screening method for inflammatory, oxidative and nitrosative stress. Anal Bioanal Chem 410(10): 2551-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Duinen V., van den Heuvel A., Trietsch S. J., Lanz H. L., van Gils J. M., van Zonneveld A. J., Vulto P. and Hankemeier T.(2017). 96 perfusable blood vessels to study vascular permeability in vitro . Sci Rep 7(1): 18071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Duinen V., Zhu D., Ramakers C., van Zonneveld A. J., Vulto P. and Hankemeier T.(2019). Perfused 3D angiogenic sprouting in a high-throughput in vitro platform . Angiogenesis 22(1): 157-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.