Abstract
Pollen apertures are an interesting model for the formation of specialized plasma-membrane domains. The plant-specific protein INP1 serves as a key aperture factor in such distantly related species as Arabidopsis, rice, and maize. Although INP1 orthologs likely play similar roles throughout flowering plants, they show significant sequence divergence and often cannot substitute for each other, suggesting that INP1 might require species-specific partners. Here, we present a new aperture factor, INP2, which satisfies the criteria for being a species-specific partner for INP1. Both INP proteins display similar structural features, including the plant-specific DOG1 domain, similar patterns of expression and mutant phenotypes, as well as signs of co-evolution. These proteins interact with each other in a species-specific manner and can restore apertures in a heterologous system when both are expressed, but not when expressed individually. Our findings suggest that the INP proteins form a species-specific functional module that underlies formation of pollen apertures.
Pollen grains of flowering plants are surrounded by a robust wall, called exine. In most species, exine is deposited on the pollen surface non-uniformly, with certain regions of the surface receiving little to no exine material1. These regions develop into pollen apertures that help pollen to hydrate, change volume, and germinate2-6. Across species, apertures vary greatly in their number, positions, and morphology, contributing to diverse, species-specific patterns on the pollen surface1,2,7,8. Recently, we and others have demonstrated that prior to forming apertures, developing pollen forms distinct aperture domains in their plasma membrane, which accumulate specific combinations of proteins and lipids6,9-11. Apertures can thus be used to study how cells develop polarity and form membrane domains, as well as to understand how these mechanisms evolved to create the tremendous diversity of aperture patterns found in nature.
Aperture domains of the plasma membrane appear at the tetrad stage of pollen development, during which the four products of male meiosis (microspores) are transiently kept together under the common callose wall6,10. The positions, number, and morphology of the aperture membrane domains in microspores correspond to the aperture pattern of mature pollen. For example, in Arabidopsis pollen, apertures are shaped like three long and narrow meridional furrows (Fig. 1a). Accordingly, in Arabidopsis tetrads, each microspore develops three linear meridional domains of the plasma membrane, which attract the proteins D6 PROTEIN KINASE-LIKE3 (D6PKL3) and INAPERTURATE POLLEN1 (INP1)10,11. In contrast, in rice and other grasses, pollen has only one small round aperture, positioned at the distal pole. Correspondingly, the tetrad-stage microspores in rice develop at their distal poles a single aperture domain shaped like a tiny ring, which attracts the rice ortholog of INP1 (OsINP1)6.
Fig.1. INP2 is a new factor essential for the formation of pollen apertures.
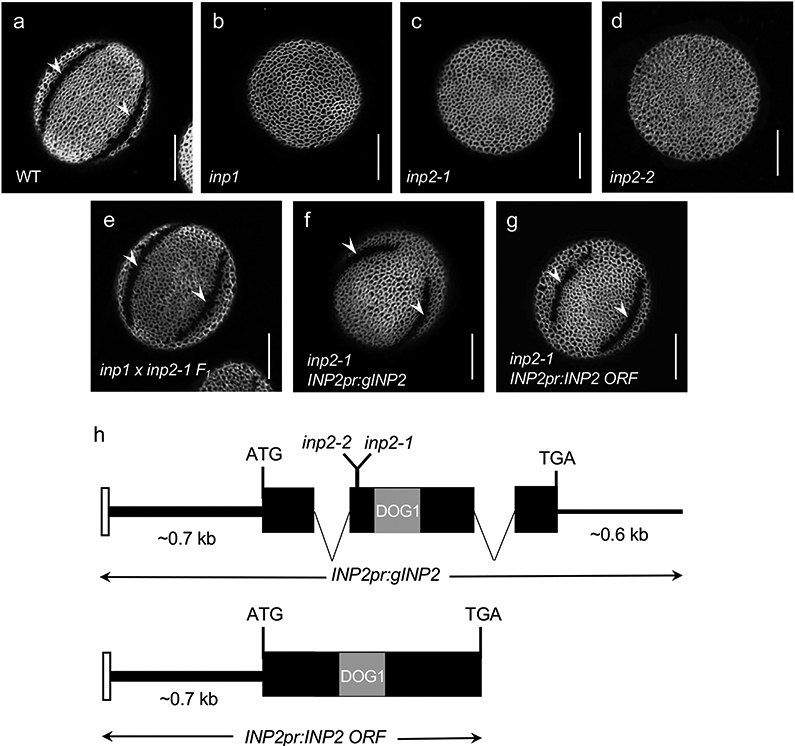
a-g, Confocal images of pollen grains stained with auramine O. Scale bars = 10 μm. a, Wild-type Arabidopsis pollen has three equidistant furrow-like apertures (two are visible here, arrowheads). b, inp1 pollen completely lacks apertures. c-d, Similar to inp1, inp2 pollen has normal exine, but completely lacks apertures (>100 pollen grains were imaged, with similar results; for inp2-2, two independent CRISPR plants were obtained, producing similar phenotypes). e, Pollen of the F1 progeny of the cross between inp1 and inp2 develops normal apertures (arrowheads), indicating that mutations disrupt different genes (eight plants (≥50 pollen grains per plant) were imaged, with similar results). f-g, INP2pr:gINP2 and INP2pr:INP2 ORF transgenes restore apertures (arrowheads) in inp2 (7/8 and 15/15 independent T1 lines, respectively; ≥ 50 pollen grains per line were imaged, with similar results). h, INP2 gene model and structure of the INP2pr:gINP2 and INP2pr:INP2 ORF complementation constructs. Black boxes indicate the protein-coding sequence of At1g15320. The region encoding the DOG1 domain is indicated by the grey box. The white box denotes a short region from the preceding gene, At1g15330, which was included in the constructs. Both the ~0.7 kb upstream region and the ~0.6 kb downstream region were included in the genomic construct. Positions of the inp2-1 and inp2-2 mutations are indicated on the gene model.
In both Arabidopsis and rice, as well as in maize, the INP1 protein was shown to be a major aperture factor whose loss causes a complete loss of apertures (Fig. 1b)6,9,12. INP1 is a plant-specific protein with a single recognizable domain – the DELAYED IN GERMINATION1 (DOG1) domain, whose function is unknown9. Although the biochemical function of INP1 remains to be identified, in both Arabidopsis and rice these proteins appear to play a role in keeping the aperture domains of the plasma membrane in close contact with the overlying callose wall6,10, which possibly protects these domains from the deposition of exine materials.
Since the role of INP1 as an essential aperture factor is conserved in such distantly related species as Arabidopsis, rice, and maize, it is reasonable to assume that INP1 orthologs across angiosperms are likely all involved in aperture formation. Intriguingly, though, many INP1 proteins show significant sequence divergence and cannot substitute for the loss of Arabidopsis INP19,12. This suggests that, despite their conserved involvement in aperture formation, INP1 proteins are likely functionally species-specific. We have previously proposed that such species specificity might be due to the presence of unknown aperture factors that have co-evolved with INP1 and help it to perform its function12.
Here, we present a novel aperture factor, INP2, that fulfills the role of a species-specific partner for INP1. INP2 resembles INP1 in its protein structure, patterns of expression, trends of evolutionary divergence, mutant phenotype, and genetic interactions. We provide evidence that INP2 is also functionally species-specific and that it physically interacts with INP1. Furthermore, we demonstrate that tomato orthologs of INP1 and INP2, which are unable to restore apertures in Arabidopsis mutants when only one of them is expressed, gain the ability to function in Arabidopsis when expressed together. The two INP proteins, therefore, behave as co-evolved species-specific partners that form a functional module required for the formation of pollen apertures.
Results
A new Arabidopsis mutant has the inaperturate pollen phenotype identical to the phenotype of the inp1 mutant
To discover genes involved in the formation of pollen apertures, we performed a forward genetic screen on an M2 population of Arabidopsis plants mutagenized with ethyl methanesulfonate (EMS). Since changes in pollen shape can serve as a proxy for aperture formation defects13,14, we screened these plants for unusual pollen shapes under dissecting microscopes. One mutant produced pollen that looked significantly rounder than the wild-type pollen, strongly resembling the phenotype of the inp1 mutants. An examination by confocal microscopy showed that, like inp1, pollen of this mutant completely lacks apertures (inaperturate phenotype) but had otherwise normal exine (Fig. 1c).
To test whether the mutation represented an allele of INP1 or disrupted another gene, we crossed the new mutant with the inp1-1 null mutant. In the F1 progeny of this cross, all pollen had normal apertures (Fig. 1e), demonstrating that the defect affected a gene other than INP1. This result also showed that, similar to inp1 and other previously discovered aperture mutants, the new mutation affected a gene with the sporophytic function. Because of the similarities with the inp1 mutant, we named the new gene INAPERTURATE POLLEN2 (INP2) and its mutant allele inp2-1.
The inp2-1 mutation disrupts the At1g15320 gene
Using positional cloning, we mapped the inp2-1 defect to a 146-kb region at the top of chromosome 1, containing 51 genes. To narrow down the list of gene candidates, we inspected their predicted identities as well as patterns of their mRNA expression reported in the TRAVA RNA-seq database15. We focused on the genes expressed in young flower buds (flowers 12-18 in the TRAVA nomenclature), as these buds include the tetrad stage of development associated with aperture formation. One gene, At1g15320, was prioritized as a particularly strong candidate as it is predicted to be expressed nearly exclusively in young buds (Extended Data Fig. 1) and encodes a protein with structural similarities to INP1 (see below). Sequencing of this gene from inp2-1 revealed a G-to-A substitution which created an early stop codon (Trp84Stop) (Fig. 1h). To independently confirm that INP2 is At1g15320, we targeted At1g15320 in the wild-type Col-0 background with CRISPR-Cas9 and generated an allele (inp2-2) with a 2-nt deletion that caused a frame shift after the amino acid 83 (Fig. 1h). The CRISPR mutant displayed the same inaperturate pollen phenotype as the original inp2-1 allele (Fig. 1d).
To further verify the identity of At1g15320 as INP2 and to define its regulatory regions, we created transgenic constructs containing either the genomic region of At1g15320 (including introns and the ~0.6-kb region downstream of the stop codon) or its open reading frame (ORF) (Fig. 1h). These constructs were placed under the control of the putative native promoter (a region of ~0.7-kb between the start codon of At1g15320 and the preceding gene At1g15330) and transformed into inp2-1. Both constructs successfully restored apertures in transgenic plants – 15/15 T1 plants with the ORF construct and 7/8 T1 plants with the genomic construct (Fig. 1f-g). Taken together, our results demonstrate that (1) At1g15320 encodes INP2 – a new factor essential for aperture formation; and (2) that the 0.7-kb upstream region is sufficient to drive functional expression of INP2. This promoter region was then used for all subsequent INP2 constructs transformed into Arabidopsis.
INP2 shares structural similarity with INP1
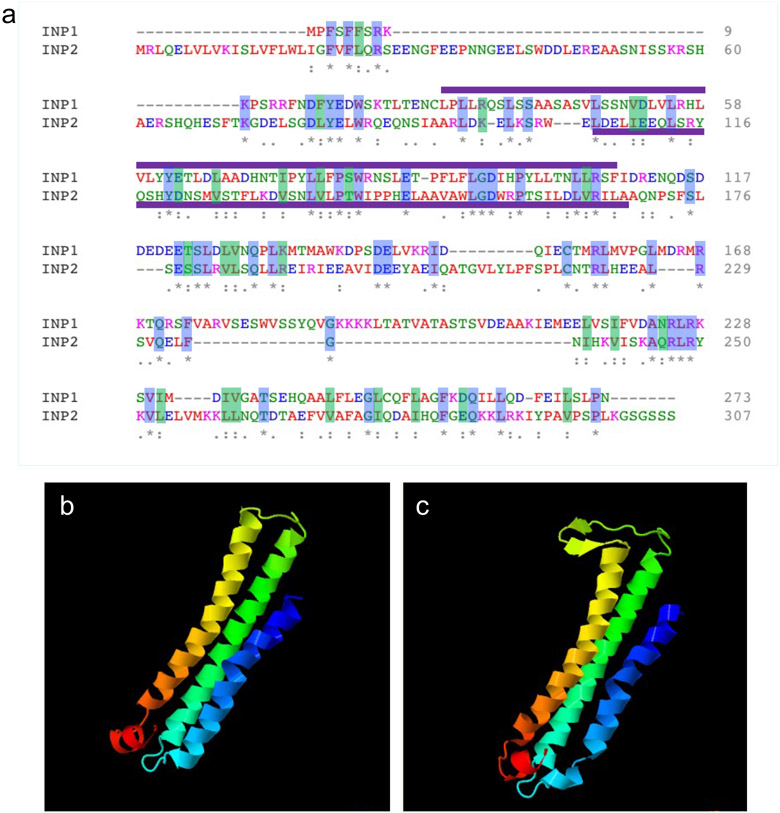
INP2 is a plant-specific protein of unknown biochemical function which shares certain similarities with INP1. Both proteins have similar size (273 aa for INP1 vs. 307 aa for INP2), are usually encoded in angiosperm genomes by single-copy genes, and contain the same domain – the plant-specific DELAYED IN GERMINATION1 (DOG1) domain (PFam14144) (Fig. 1, Extended Data Fig. 2a). This domain, typically associated with seed dormancy proteins and TGA bZIP transcription factors16, is the only recognizable domain in both INP proteins. Interestingly, although INP1 and INP2 share only limited homology with each other (23% sequence identity, Extended Data Fig. 2a), the protein-fold recognition software Phyre217 selected the same template for homology modeling of both proteins and predicted similar structures, with three alpha-helices, for their C-terminal regions (Extended Data Fig. 2b,c).
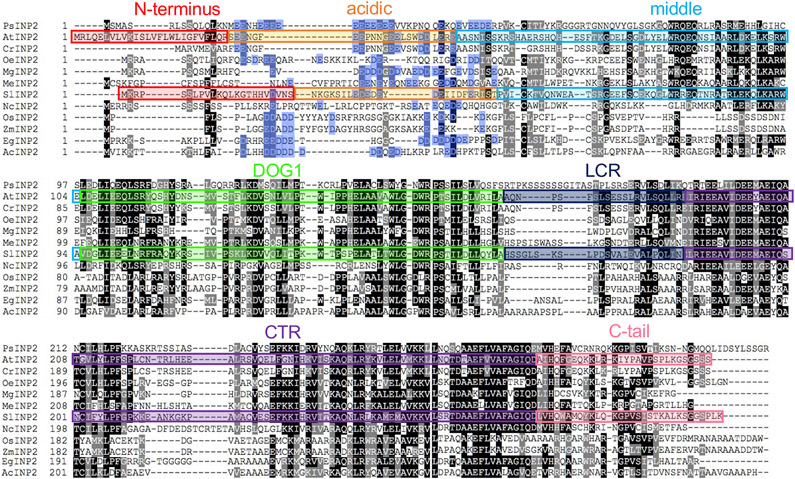
Protein alignments of INP1 and INP2 with their respective orthologs from other plants also revealed that, in eudicots, these proteins typically contain a region enriched in Asp and Glu residues. However, these acidic regions are positioned differently between INP1 and INP2. In the INP1 proteins, the acidic region follows the DOG1 domain9,12, whereas in the INP2 proteins it is located ahead of the DOG1 domain (Extended Data Fig. 3).
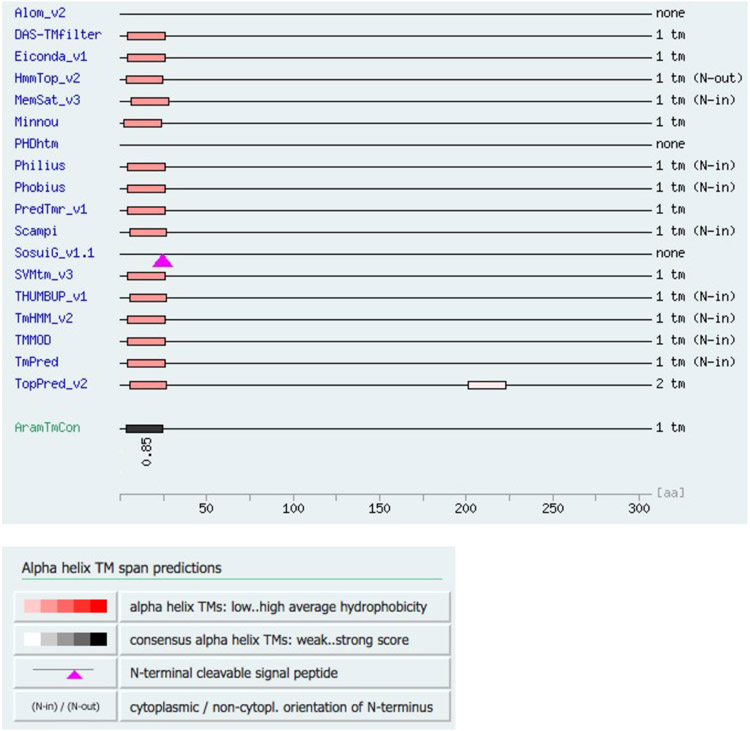
For Arabidopsis thaliana INP2 (AtINP2), multiple algorithms also predicted the existence of a transmembrane (TM) domain at its N-terminus (Extended Data Fig. 4), with most of the protein expected to be outside the cell, facing the extracellular space. Yet, the algorithms failed to identify a TM domain in many orthologs of AtINP2, including the highly related proteins from Arabidopsis lyrata and other members of the Brassicaceae family, suggesting that this is not a common feature of INP2 proteins. No lipid modifications are predicted for INP2.
INP2 is expressed in the developmental lineage of pollen at the time of aperture formation
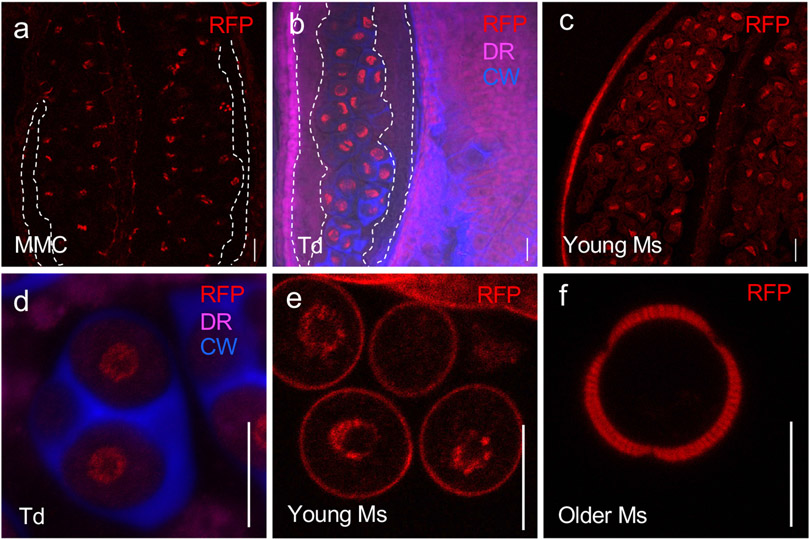
Publicly available RNA-seq data show that, like INP1, INP2 is expressed nearly exclusively in young buds containing pollen at or around the tetrad stage during which apertures form (Extended Data Fig. 1). To test whether in these buds INP2 is expressed in the male reproductive lineage, we expressed the nuclear marker histone H2B tagged with the Red Fluorescent Protein under the control of the INP2 promoter (INP2pr:H2B-RFP) in the wild-type Col-0 plants. This reporter, with its concentrated localization in the nucleus, was specifically chosen to help visualize the expression from the INP2 promoter, since, like INP1, INP2 is predicted to be expressed at low levels (Extended Data Fig. 1). The nuclear RFP signal was found in the dividing microspore mother cells, tetrad-stage microspores, and young free microspores (Fig. 2). The signal was absent in older microspores, the surrounding somatic tapetal cell layer and other anther layers (Fig. 2). This expression pattern matches that of INP19,10.
Fig. 2. INP2 is expressed in the male reproductive lineage at the time of aperture formation.
Images of anthers at different developmental stages (a-c) and magnified images of the cells from the male reproductive lineage at different developmental stages (d-f) expressing the transcriptional fusion construct INP2pr:H2B-RFP. Nuclear signal of H2B-RFP (red) is found in dividing microspore mother cells (MMC, a), tetrads of microspores (Td, b, d) and young free microspores (Ms, c, e). Older microspores (f) do not show nuclear H2B-RFP signal (peripheral red signal is due to the autofluorescence of the developing exine). No signal was observed in the tapetal layer of the anther (outlined by the white dashed lines in a, b). Besides RFP, the images in b, d show staining for callose wall (blue, CW, calcofluor white) and membranous structures (magenta, DR, CellMask Deep Red). Five independent T1 lines were imaged, with similar results. Scale bars = 10 μm.
To visualize the subcellular localization of the INP2 protein, we first created five constructs, in which INP2 was tagged with Yellow Fluorescent Protein (YFP) at four positions: at the N-terminus, at the C-terminus (either directly or following an 18-aa linker), after the predicted TM region, and internally – within the low-conservation region (see below). However, none of the YFP-tagged constructs rescued the inp2 mutant, suggesting that INP2 does not tolerate addition of sizable tags. This notion was supported by further experiments in which partial rescue of the mutant phenotype was achieved with constructs expressing INP2 tagged at the C-terminus with one or three copies of the small HA tag. Of these two types of constructs, the shorter HA1 construct produced better rescue (see Methods), yet no protein signal was detected in these lines with the anti-HA antibody in anther sections or whole-mount preparations, possibly owing to the low levels of the INP2 expression. This prevented us from determining whether INP2, like INP1, specifically localizes to the aperture domains in the plasma membrane of microspores.
Localization of INP1 and D6PKL3 to plasma-membrane aperture domains depends on the presence of INP2
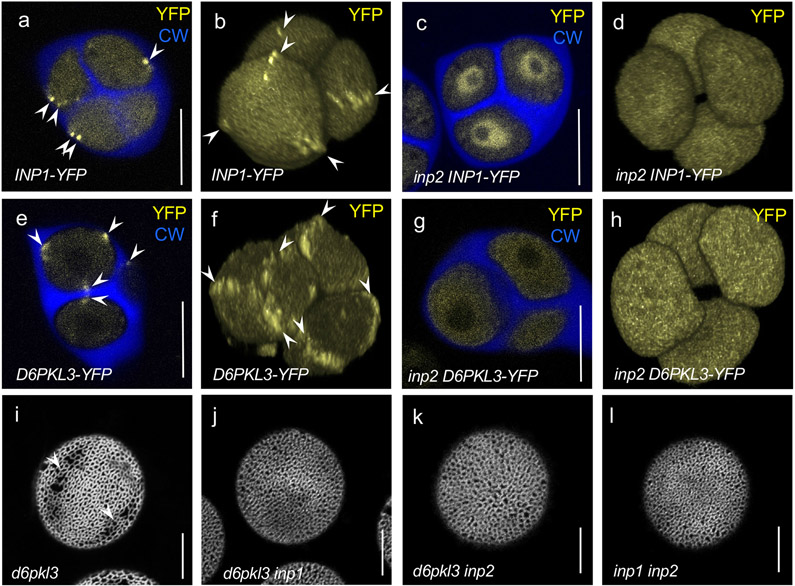
To test whether INP2 contributes to the distinct positioning of INP1 and another recently identified aperture factor, D6PKL3, both of which accumulate at the microspore aperture domains (Fig. 3a,b,e,f), we introgressed the previously characterized reporter constructs DMC1pr:INP1-YFP10 and D6PKL3pr:D6PKL3-YFP11 into the inp2 mutant background. In the absence of INP2, INP1-YFP failed to localize to the aperture domains of the plasma membrane, instead showing significant enrichment in the nucleoplasm (Fig. 3c,d). This result suggests that INP2 is involved either in targeting INP1 to the aperture domains or in keeping it there. Likewise, in the absence of INP2, the membrane-associated kinase D6PKL3-YFP lost its association with the aperture domains, instead displaying diffuse cytoplasmic localization (Fig. 3g,h). As D6PKL3 reacts the same way to the absence of INP111, both INP1 and INP2 are thus required to keep it at the aperture domains.
Fig. 3. INP2 is required for INP1 and D6PKL3 accumulation at the aperture domains and both inp1 and inp2 are epistatic to d6pkl3.
a-h, INP1-YFP and D6PKL3-YFP localization in tetrads of microspores in the presence and absence of INP2. Confocal optical sections (a, c, e, and g) and 3D reconstructions of tetrads of microspores (b, d, f, and h). YFP signal is shown in yellow and callose wall (CW, stained by calcofluor white) is shown in blue. Arrowheads point to the YFP signal at the aperture domains. INP1-YFP localizes to the aperture PM domains in the wild type (a, b) but loses this localization in the inp2 mutant (c, d), instead becoming enriched in the nucleoplasm. Experiments in (c, d) were repeated three times, with similar results. Likewise, D6PKL3-YFP localizes to the aperture domains in the wild type (e, f) but loses this localization in the inp2 mutant (g, h). Experiments in (g, h) were repeated two times, with similar results. i-l, inp1 and inp2 mutations are epistatic to d6pkl3, and do not cause additional phenotypic changes when combined. Confocal images of pollen grains stained with auramine O. d6pkl3 mutant pollen often develops apertures partially covered with exine (arrowheads) (i), whereas double mutants d6pkl3 inp1 (j), d6pkl3 inp2 (k), and inp1 inp2 (l) completely lack apertures. ≥ 3 plants (≥ 50 pollen grains per plant) were imaged in (i-l), with similar results. Scale bars = 10 μm.
To test for epistatic relationships between these aperture factors, we created double mutants of inp1 d6pkl3 and inp2 d6pkl3. Single mutations in D6PKL3 do not completely abolish aperture formation, instead producing ‘shadows of apertures’ that are partially covered with exine (Fig. 3i)11. Yet both double mutants produced completely inaperturate pollen (Fig. 3j,k), indicating that inp1 and inp2 are both epistatic to d6pkl3. To investigate the possibility of synergistic interactions between INP1 and INP2, we also created the inp1 inp2 double mutant. Its phenotype, however, was identical to those of single mutants (Fig. 3l), showing that the simultaneous loss of INP1 and INP2 does not cause any additional observable effects (e.g. in the exine deposition) and suggesting that these proteins behave as bona fide aperture factors. Taken together, the results presented so far are consistent with the notion that INP1 and INP2 occupy very similar positions in the aperture formation pathway and might coordinate their activities.
INP1 and INP2 are interacting proteins
Since INP1 and INP2 exhibit similarities in their protein structures, patterns of expression, mutant phenotypes, and genetic interactions, we hypothesized that they might physically interact. To explore this possibility, we used several approaches. An initial yeast two-hybrid (Y2H) assay with the full-length INP1 and INP2 did not result in yeast growth indicative of protein interaction. We reasoned, however, that lack of yeast growth would be expected if INP2 indeed had a TM domain at its N-terminus and most of the protein was extracellular.
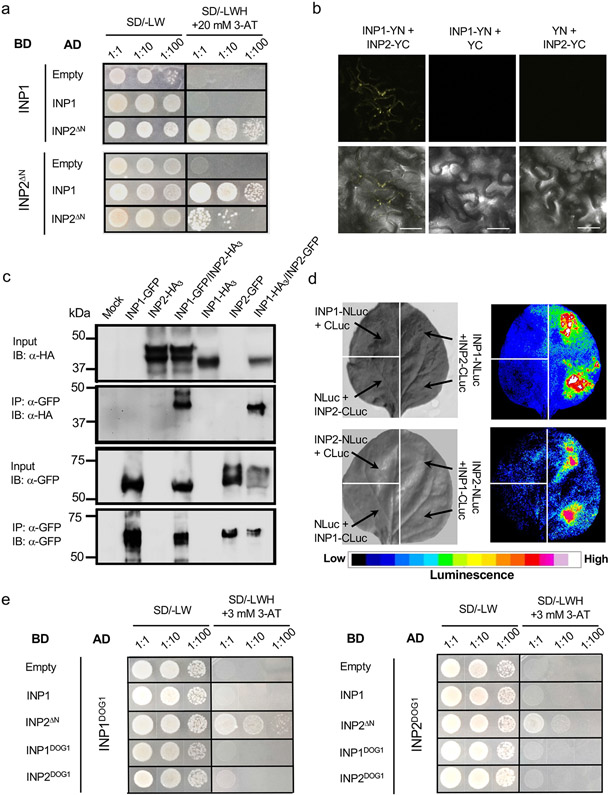
We, therefore, expressed INP2 in yeast without its first 24 amino acids, which contained the predicted TM domain. This truncated INP2 (INP2ΔN) showed strong interaction with INP1 in the Y2H system (Fig. 4a). In addition, this assay revealed that INP2 may be able to self-interact (Fig. 4a). We further verified the ability of INP1 and INP2 to interact in planta by expressing them in tobacco leaf cells and performing co-immunoprecipitation, bi-fluorescent molecular complementation (BiFC), and a split-luciferase assay (Fig. 4b-d).
Fig. 4. INP1 and INP2 physically interact.
a, Yeast two-hybrid assay of interaction between INP1 and INP2ΔN (lacking the N-terminal region). BD, DNA-binding domain; AD, activating domain; SD, synthetic defined medium. To test for the presence of both BD and AD constructs, leucine (L) and tryptophan (W) were excluded from the medium. To test for protein interaction, yeast were grown on media lacking L, W, and histidine (H) and containing 20 mM 3-aminotriazole (3-AT). b, BiFC experiments. INP1 and INP2 proteins fused, respectively, to the N- and C-terminal parts of YFP (YN and YC) were co-transformed into tobacco leaves to test for interaction. Co-transformation of INP1-YN with only YC and co-transformation of INP2-YC with only YN were used as negative controls. Top panels show YFP signal in leaf epidermis. Bottom panels show merged YFP and bright-field images. Scale bars = 50 μm. c, Co-immunoprecipitation experiments. INP1-HA3/INP2-GFP and INP1-GFP/INP2-HA3 pairs (or just single tagged proteins as negative controls) were co-expressed in tobacco leaves, precipitated with anti-GFP antibodies and visualized with anti-GFP or anti-HA antibodies. IP, immunoprecipitation; IB, immunoblot. ‘Mock’ indicates protein extract from leaves infiltrated only with buffer. d, Split-luciferase assay. Tobacco leaves were divided into sectors co-expressing indicated proteins containing the N-terminal (NLuc) and C-terminal (CLuc) parts of the firefly luciferase. Panels on the left show the bright-field images and panels on the right show the corresponding luminescence images. e, Yeast two-hybrid assay in which the DOG1 domains of INP1 (INP1DOG1) and INP2 (INP2DOG1) were tested for interaction with each other, self-interaction, and interaction with the full-length INP1 and with INP2ΔN. The description is the same as for a, except that 3 mM 3-AT was used here. Experiments in a-c and e were repeated three times and experiments in d were repeated two times (each time using multiple leaves from multiple plants), with similar results.
The DOG1 domain is the only recognizable protein domain present in these novel proteins. Although its function is unknown, it has been proposed that this domain might participate in protein-protein interactions18. We therefore used the Y2H assay to test the ability of the DOG1 domains from INP1 and INP2 to interact with each other and with the full-length (or nearly full-length in the case of INP2ΔN) proteins (Fig. 4e). INP1DOG1 was able to interact with INP2ΔN. In contrast, INP2DOG1 failed to interact with INP1 but showed some ability to interact with INP2ΔN, consistent with the finding that INP2 may self-interact. However, no interactions occurred when only the DOG1 domains were present (Fig. 4e), suggesting that these regions likely interact with other portions of INP2.
INP1 and INP2 exhibit similar trends of evolutionary sequence divergence
We previously reported that INP1 significantly diversified in angiosperm lineages9,12. Still, in several species these divergent orthologs were found to be involved in the formation of pollen apertures and able to localize to specific plasma-membrane aperture domains6,12, suggesting that, despite the significant difference in primary sequences, all INP1 proteins in angiosperms likely function as aperture factors. However, INP1 proteins appear to exhibit a significant degree of functional species specificity, since the divergent INP1 orthologs were not able to complement the aperture defects of the Arabidopsis inp1 mutant12. A possible interpretation of this result is that INP1 proteins might require the presence of co-evolved partners to perform their function.
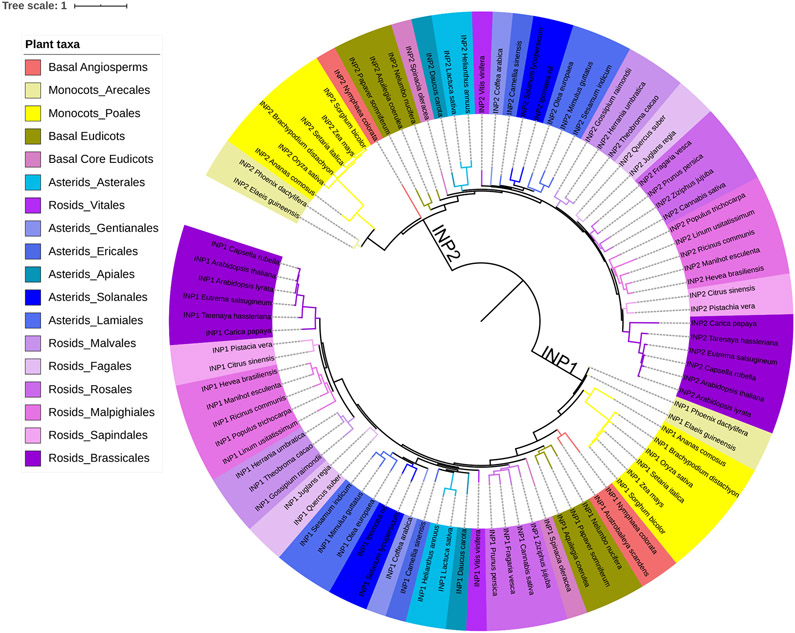
To see whether INP2 shows signs of co-evolution with INP1, we performed BLAST searches for INP2 homologs followed by phylogenetic analysis, revealing notable parallels between INP1 and INP2. Although proteins with the DOG1 domain appeared as early as green algae, we found distinct, well-supported INP1 and INP2 protein lineages only in gymnosperms and angiosperms. In angiosperms, they have greatly diversified and display similar trends of evolutionary divergence, resulting in phylogenetic trees of similar topology (Fig. 5). Orthologs of both INP1 and INP2 exist in various families of rosids, asterids, basal eudicots, monocots, and magnoliids. The INP1 sequence is also present among the transcripts from two ANA-grade basal angiosperms, Austrobaileya and Nymphaea. Failure to find an INP2 homolog in Austrobaileya, despite finding one in Nymphaea, could be due to the incompleteness of the database. Interestingly, both INP1 and INP2 are absent from the genome of Amborella, another basal angiosperm whose genome was published several years ago19.
Fig. 5. INP1 and INP2 exhibit similar trends of evolutionary sequence divergence.
Maximum likelihood phylogenetic tree of INP1 and INP2 sequences from a variety of angiosperm taxa (indicated by color coding). The INP1 and INP2 sequences cluster into two separate clades, which display similar topology.
Degrees of sequence divergence within the INP1 and INP2 angiosperm lineages are generally consistent with the phylogenetic relationships between species (Supplementary Table 1, Fig. 5). Both Arabidopsis INP1 and INP2 (AtINP1 and AtINP2) share between ~95% and ~70% protein sequence identity with their respective orthologs from closely related species in the Brassicaceae and Cleomaceae families. Sequence identity with orthologs from more distantly related eudicots drops to ~40-50%. In monocots, the similarities to AtINP1 and AtINP2 are further reduced: proteins from Arecaceae and Bromeliaceae families (e.g. palms and pineapple) exhibit ~45% to ~30% sequence identity with AtINP1 and AtINP2. In both INP1 and INP2 lineages, particularly distinct clades are formed by proteins from grasses (Poaceae) (Fig. 5, Supplementary Table 1): within each INP group, these proteins diverged greatly from the rest of their lineages (showing ~35% and ~20-25% identity, respectively, to AtINP1 and AtINP2) but retained >80% identity to their orthologs from other species of Poaceae despite the long evolutionary history of this monocot family20.
INP1 and INP2 are functionally species-specific
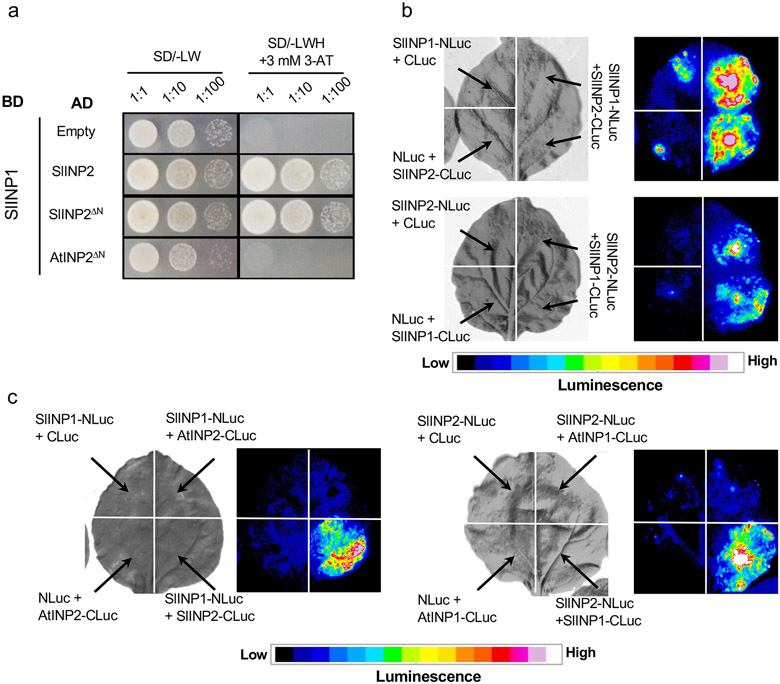
The similar evolutionary trends displayed by INP1 and INP2, as well as the ability of these proteins to interact, led us to hypothesize that INP2 might serve as a species-specific partner for INP1. We tested this hypothesis using the orthologs of INP1 and INP2 from tomato Solanum lycopersicum (SlINP1 and SlINP2) which both share ~45% amino acid identity with their Arabidopsis counterparts. Using the Y2H and split-luciferase assays, we confirmed the ability of SlINP1 and SlINP2 to interact (Fig. 6a,b). Furthermore, in both assays, the tomato INP proteins specifically interacted with each other and not with the Arabidopsis proteins (Fig. 6a,c).
Fig. 6. INP1 and INP2 interact in a species-specific manner.
a, Yeast two-hybrid assay testing SlINP1 interactions with SlINP2 (or SlINP2ΔN lacking the N-terminal region) and AtINP2DN. BD, DNA-binding domain; AD, activating domain; SD, synthetic defined medium. To test for the presence of both BD and AD constructs, leucine (L) and tryptophan (W) were excluded from the medium. To test for protein interaction, yeast were grown on media lacking L, W, and histidine (H) and containing 3 mM 3-aminotriazole (3-AT). b, Split-luciferase assay testing the ability of SlINP1 and SlINP2 to interact. Tobacco leaves were divided into sectors co-expressing indicated proteins containing the N-terminal (NLuc) and C-terminal (CLuc) parts of the firefly luciferase. Panels on the left show the bright-field images and panels on the right show the corresponding luminescence images. c, Split-luciferase assay testing the ability of INP1 and INP2 from Arabidopsis and tomato to interact with a protein from another species. Only the same-species interactions were observed. The description is the same as for b. All experiments were repeated at least twice, with similar results.
We demonstrated previously that SlINP1 was unable to localize to aperture domains and restore apertures when expressed in the Arabidopsis inp1 mutant12 (Fig. 7a). Here, we placed SlINP2 under the control of the AtINP2 promoter and transformed the AtINP2pr:SlINP2 construct into the Arabidopsis inp2-1 mutant. Similar to SlINP1, SlINP2 failed to restore apertures in the Arabidopsis inp2 mutant (12/12 T1 plants; Fig. 7b), suggesting that INP2 likely also exhibits functional species specificity.
Fig. 7. Tomato orthologs of INP1 and INP2 fail to function in Arabidopsis when expressed individually but gain this ability when co-expressed.
a-c, Neither SlINP1 (a) nor SlINP2 (b, c) are able to restore apertures in Arabidopsis pollen when expressed on their own. ≥ 10 T1 plants (≥ 50 pollen grains per plant) were analyzed, with similar results. d-e’, When both SlINP1 and SlINP2 are expressed in Arabidopsis, they restore short to medium apertures (arrowheads) in the inp1 (d, d’) and inp1 inp2 (e, e’) Arabidopsis mutants. Confocal images of pollen grains stained with auramine O. Both front and back views are shown for the same pollen grains in (d-d’) and (e-e’) to demonstrate positions of apertures. Experiments were repeated twice, with similar results (~90% of plants had short- to medium-size apertures, and the rest had no apertures). f-i, SlINP1-YFP localizes to the aperture domains in the presence of SlINP2 (h, i) but not when expressed on its own (f, g). Confocal optical sections (f, h) and 3D reconstructions of tetrads of microspores (g, i). YFP signal is shown in yellow and callose wall (CW, stained by calcofluor white) is shown in blue. Arrowheads point to the YFP signal at the aperture domains. Experiments were repeated twice, with similar results. Scale bars = 10 μm
Co-expression of SlINP1 and SlINP2 restores aperture formation in Arabidopsis mutants
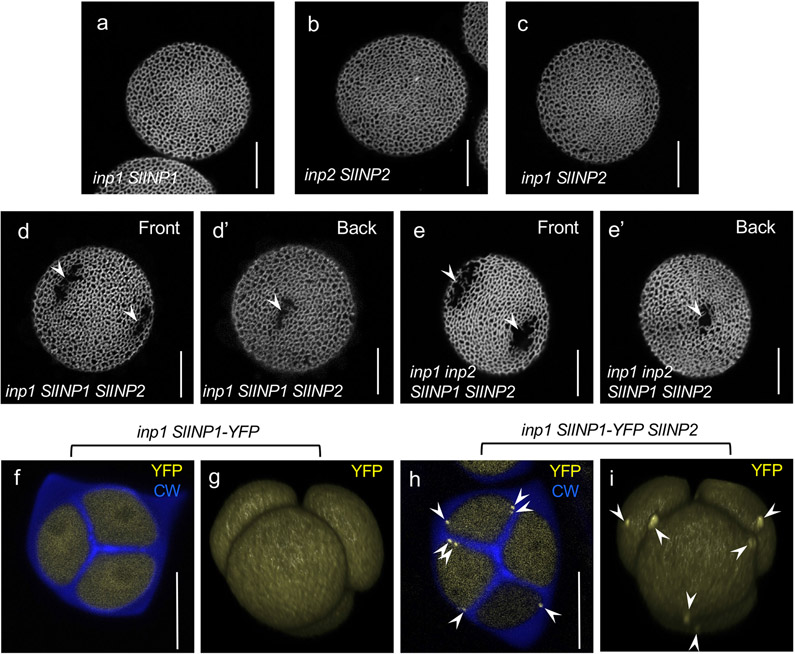
Since individually expressed SlINP1 and SlINP2 did not restore apertures in Arabidopsis (Fig. 7a,b), we tested if together they could gain functionality in that species. To this end, we transformed INP2pr:SlINP2 into inp1 (as expected, apertures were not restored in all 14 T1 plants; Fig. 7c)), and crossed these plants with the previously characterized DMC1pr:SlINP1-YFP inp1 transgenic lines12. In the resulting inp1 progeny that inherited just a single transgene, no apertures were restored. But, remarkably, among the inp1 progeny that inherited both transgenes, 91% of plants (64/70) produced pollen with short- to medium-size apertures (Fig. 7d,d’).
Because SlINP1 was tagged with YFP, we assessed its localization in Arabidopsis tetrad-stage microspores. As shown previously12, when expressed on its own, SlINP1-YFP fails to accumulate at the plasma-membrane aperture domains, instead localizing diffusely in the cytoplasm (Fig. 7f,g). However, in the presence of SlINP2, SlINP1-YFP gained the ability to successfully assemble into distinct puncta at the plasma-membrane aperture domains (Fig. 7h,i).
We further crossed the SlINP1 SlINP2 inp1 plants with inp2 plants to generate the double inp1 inp2 mutants carrying both transgenes. In the F2 and F3 generations, those double mutants that inherited both tomato transgenes also produced pollen with apertures (Fig. 7e,e’). Taken together, these results demonstrate that SlINP1 and SlINP2 act as species-specific partners in pollen aperture formation.
Certain regions of INP2 mediate its species specificity in Arabidopsis
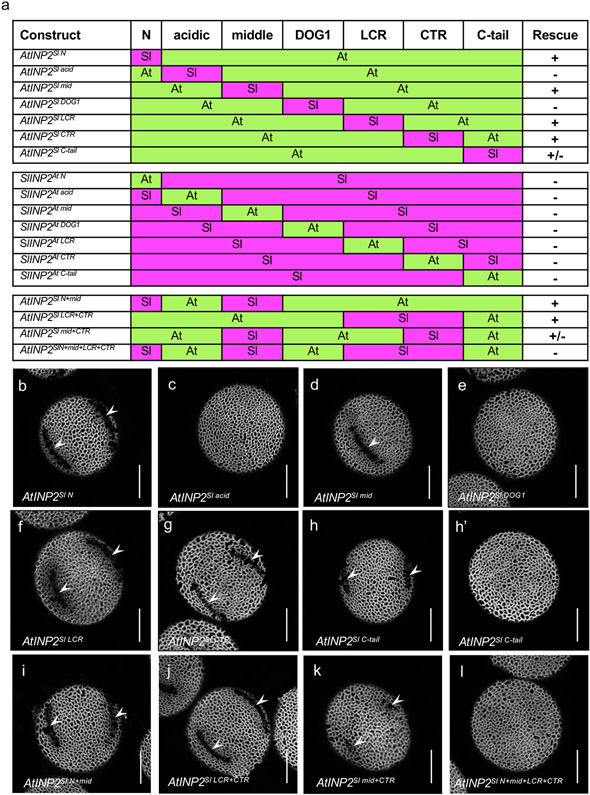
To identify sequences in INP2 responsible for the species specificity of this protein, we divided it into seven regions, based on position relative to the transmembrane and DOG1 domains and evolutionary conservation (Fig. 8a, Extended Data Fig. 3). Regions were chosen as follows: 1) the N-terminus (N), which in AtINP2 encompasses the predicted TM region; 2) the acidic region, which, besides its enrichment in Asp and Glu residues, is fairly divergent across species; 3) the middle region; 4) the DOG1 domain; 5) the low-conservation region (LCR), which shows high sequence divergence across species; 6) the conserved C-terminal region (CTR); and 7) the highly divergent C-tail. To test which of the AtINP2 regions were necessary for its function, we created seven chimeric transgenic constructs in which most of the sequence came from AtINP2, while one region at a time was replaced with the corresponding sequence from SlINP2 (Fig. 8a). In addition, to test if any of the AtINP2 regions were sufficient for its function in Arabidopsis, we created a complementary set of seven constructs; in those, most of the protein was from SlINP2 and a single region came from AtINP2 (Fig. 8a). These fourteen constructs were transformed into the Arabidopsis inp2 mutant, and their ability to complement the mutant phenotype was assessed.
Fig. 8. Certain regions of INP2 mediate its species specificity.
a, A diagram of 18 INP2 chimeric constructs containing regions from Arabidopsis (At, green) and tomato (Sl, magenta). The protein was divided into seven regions. The ability of a construct to restore apertures in the Arabidopsis inp2 mutant is indicated by ‘+’, the failure to restore apertures is indicated by ‘−’, and the ability to restore apertures in some but not all transgenic lines is indicated by ‘+/−’. b-m, Representative images of pollen grains produced by transgenic inp2 plants expressing different chimeric INP2 constructs. > 10 independent T1 lines were tested for each construct (≥ 50 pollen grains per line), with all or nearly all lines producing similar results, except in h, h’, and k where, as described, some plants produced short apertures and others – no apertures. Apertures are indicated by arrowheads. Scale bars = 10 μm
The AtINP2 constructs in which the N-terminus, the middle region, the LCR, or the CTR were replaced with the SlINP2 sequences all restored apertures in Arabidopsis (19/19, 10/12, 10/12, 19/20 T1 plants, respectively) (Fig. 8a, b, d, f, g), indicating that AtINP2 can tolerate the presence of tomato sequences in these four regions. The ability of the construct with the Sl N-terminus to function in Arabidopsis was surprising, as this region in SlINP2 is not predicted to contain a TM domain.
In contrast, the AtINP2 constructs with the SlINP2 acidic region or the DOG1 domain failed to rescue apertures in Arabidopsis (Fig. 8a,c,e) (11/11 T1 plants for both constructs), demonstrating that these regions are critical for species-specific interactions. In addition, although 5 out of 12 T1 plants expressing AtINP2 with the tomato C-tail had some ability to produce short apertures (Fig. 8h), overall, the AtINP2SlC-tail protein performed poorly: in 10 out of 12 T1 plants all or some pollen grains lacked apertures (Fig. 8h’). The divergent C-tail, therefore, likely also contributes to the protein’s species specificity.
The complementary set of the SlINP2 constructs with single AtINP2 regions showed that none of the AtINP2 regions was sufficient on its own to convert SlINP2 into a protein able to function in Arabidopsis (Fig. 8a, Extended Data Fig. 5; ≥ 10 T1 plants were analyzed for each construct). This suggests that sequences from more than one region contribute to the INP2 species specificity.
We then explored the extent to which AtINP2 can tolerate the simultaneous replacement of the regions which, individually, did not impact its functionality. The chimeric INP2s in which either the N-terminus and the middle region or the LCR and the CTR came from SlINP2 were still functional in Arabidopsis (Fig. 8a,i,j; 16/18 T1 plants for AtINP2SlN+mid and 18/18 T1 plants for AtINP2SlLCR+CTR). In contrast, the simultaneous replacement of the middle region and the CTR (AtINP2Slmid+CTR) resulted in a significant loss of protein activity: pollen developed either only very short apertures and ‘shadows of apertures’ (Fig. 8a,k; 10 /17 T1 plants) or no apertures at all (7/17 T1 plants). The replacement of all four of these regions (AtINP2SlN+mid+LCR+CTR) resulted in a completely non-functional protein (Fig. 8a,l; 13/13 T1 plants), indicating that while each of these four regions plays a less prominent role in the species-specific functionality of INP2 compared to the acidic region, the DOG1 domain, and the C-tail region, together they still provide important contributions.
Discussion
The diversity of pollen aperture patterns in nature likely reflects the diversity of mechanisms controlling formation of these structures. In this study, we identified and characterized the new pollen aperture factor INP2, which is essential for this process and acts as a species-specific partner for the previously discovered aperture factor INP1. While not closely related, INP1 and INP2 share multiple similarities, including their matching patterns of expression, identical mutant phenotypes, and the presence of the DOG1 domain (Figs. 1-3). Phylogenetic analysis suggests that the two INP genes are the result of an ancient gene duplication that occurred in the common ancestor of gymnosperms and angiosperms, with the diverging genes evolving non-redundant functions important for the formation of pollen apertures. INP1 and INP2 proteins interact with each other, display similar evolutionary trends, and show functional species specificity (Figs. 4-7), indicating that they have co-evolved to form a species-specific functional module that promotes aperture formation. The notion of species specificity of the components of this module is strongly supported by the ability of the tomato SlINP1 and SlINP2 proteins to restore apertures in Arabidopsis only when they are co-expressed, but not when expressed individually (Fig. 7). We have also demonstrated that SlINP1, which, on its own, does not assemble at the aperture domains in Arabidopsis tetrads12, gains the ability to do this in the presence of SlINP2 (Fig. 7h,i). Our data show that several regions of INP2, including the DOG1 domain, contribute to its species specificity (Fig. 8).
INP1 and INP2 are both proteins of unknown function. Accumulating evidence for the Arabidopsis INP1 indicates that it is a late-acting aperture factor that becomes attracted to the plasma membrane domains in microspores that are already pre-specified as aperture sites12,14,21. As such, even though INP1 and its partner INP2 are critical for aperture formation, they are unlikely to directly define positions and morphology of apertures and to contribute to the aperture diversity in that way. This idea is supported by the phenotype of the apertures restored in the Arabidopsis inp1 mutant by the expression of the tomato SlINP1/SlINP2 complex: the restored apertures did not resemble the colporate tomato apertures (PalDat: www.paldat.org)8,12, but were more like the colpate apertures of Arabidopsis (Fig. 7d-e’). It is, therefore, intriguing why these essential, yet late-acting aperture factors have diversified so significantly among angiosperms. This could indicate that different species have variations in the upstream mechanisms or differences in other interactors of the INP proteins. Relevant to this point, the apertures restored by the tomato complex in Arabidopsis were shorter than the normal Arabidopsis (or tomato) apertures (Fig. 7d-e’). While this could be due to reduced expression of the transgenes compared to that of endogenous genes, an alternative possibility is that, for optimal function, the tomato complex requires some additional species-specific component(s).
After aggregating at the aperture domains, the INP1 proteins in both Arabidopsis and rice participate in keeping the plasma membrane at these domains near the overlying callose wall and preventing the deposition of the exine precursor, primexine, at these sites6,10. How can the newly discovered INP2 protein contribute to aperture formation? There are several possibilities, and further investigations will be needed to distinguish between them. For instance, along with INP1, INP2 might directly be a part of the protein complex that assembles at the aperture domains and mediates their interaction with the callose wall. Alternatively, INP2 might be involved in the delivery of INP1 to its positions at the aperture domains. AtINP1 appears to aggregate on the extracellular side of the aperture domains, yet lacks a clear signal peptide or any obvious means to become anchored at the plasma membrane10. The proposed topology of AtINP2, with the TM domain at its N-terminus and most of the protein outside the cell, is consistent with the idea that the INP1/INP2 complex in Arabidopsis is extracellular. It is tempting to speculate that the interaction of AtINP1 with the extracellular portion of AtINP2 could provide a way to anchor AtINP1 at the aperture domains. Yet, further experimentation will be needed to validate the topologies of AtINP2 and its orthologs from other species, many of which, like SlINP2, lack predicted TM domains; to find a way to visualize the INP2 subcellular localization; and to establish whether the INP proteins from species with very different aperture patterns function in the same way. Intriguingly, OsINP1 from rice was recently shown to interact with the cytoplasmic portion of a lectin receptor-like kinase6, suggesting a possibility that in grasses, whose single pore-like apertures differ greatly from the three furrows in Arabidopsis and tomato, INP1 might have a role at the cytoplasmic side of the aperture domains. Since both INP lineages in grasses have significantly diverged from their counterparts in eudicots and some other monocots, it would be very interesting to determine whether in that plant family INP2 proteins are also involved in aperture formation and function in a complex with INP1 or whether they have evolved other functions.
In conclusion, our study uncovered a new essential player in the poorly understood mechanism underlying the formation of important patterning elements on the pollen surface and demonstrated that the two DOG1 domain-containing aperture factors form a protein complex whose components contribute to the species specificity of this molecular mechanism.
Methods
Plant materials and growth conditions
Plants were grown at 20-22°C with the 16-hour light:8-hour dark cycle in growth chambers or in a greenhouse at the Biotechnology facility at OSU. Besides the genotypes generated in this study, the following genotypes were used: Columbia (Col-0), Landsberg erecta (Ler), inp1-19, DMC1pr:INP1-YFP inp1-110,14; DMC1pr:SlINP1-YFP inp1-112; D6PKL3pr:D6PKL3-YFP d6pkl3-211.
Forward genetic screen
The genetic screen which led to the isolation of the inp2-1 mutant was recently described21. In brief, M2 plants (~10,000) from eight pools of ethylmethane sulfonate (EMS)-treated lines of Landsberg erecta background were screened for the presence of morphological abnormalities in their pollen (e.g. in size, shape, light reflection, ease of pollen release from anthers) identifiable with standard dissecting stereomicroscopes (Zeiss Stemi-2000C and Nikon SMZ745) at 75-80X magnification. Particular attention was paid to changes in pollen shape, known to be associated with aperture defects9,13. For primary screening, dry pollen did not undergo any treatment. At this level of magnification, pollen of the inp2-1 mutant looked rounder than the wild-type pollen. Pollen was then stained with auramine O as described14, and aperture defects were observed with confocal microscopy. inp2-1 was then backcrossed with Ler once. To test for complementation, inp2-1 mutant was crossed with inp1-1, and the pollen of their F1 progeny was observed with dissecting and confocal microscopes.
Mapping the inp2 defect
inp2-1 mutant with Ler background was crossed with Col-0, and the resulting F2 population was screened under a dissecting microscope for the presence of the round-pollen mutant phenotype. DNA was isolated from 189 mutants. First, the bulked segregant analysis22 placed the mutation to the top of chromosome 1. This was followed by map-based positional cloning using individual F2 mutants22. The INDEL-based PCR markers for this analysis were generated as previously described11, using combined information from the 1,001 Genomes Project database (http://signal.salk.edu/atg1001/index.php)23 and the Arabidopsis Mapping Platform (http://amp.genomics.org.cn/)24. The mutation was mapped to a 146-kb region between 5,151,424 bp and 5,297,411 bp.
To determine which of the 51 genes in this interval was responsible for the aperture defect in inp2, we used information from the Arabidopsis RNA-seq database TRAVA (travadb.org)15 to identify genes expressed in the young buds at or near the tetrad stage of pollen development (flowers 12-18). Although 37 of the 51 genes are expressed in buds at these stages, expression of only one of them, At1g15320, is specifically restricted to these tissues and stages. The finding that this gene, like INP1, encodes a novel protein with the DOG1 domain let us to further prioritize it as a strong candidate for INP2. Sequencing of this gene from the inp2-1 mutant revealed the presence of the point mutation which leads to a premature stop codon (Trp84Stop).
Sequence retrieval and phylogenetic analysis
Sequences of INP1 and INP2 homologs were retrieved by TBLASTN from NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), Phytozome v.12.1 (https://phytozome.jgi.doe.gov/pz/portal.html), the 1000 Plants project (OneKP-China National Gene Bank (https://db.cngb.org/onekp/)25 and PLAZA (https://bioinformatics.psb.ugent.be/plaza/)26. Accession numbers are provided in Supplementary Table 1. For multiple sequences alignment, MAFFT v7.017 (L-INS-i algorithm) was used. The alignment positions with more than 20% gaps were removed with trimAl27. ModelFinder28 accessed through IQ-TREE29 tested 546 protein models to find the best-fit model of evolution (INP1+INP2: JTT+R5). Lastly, the IQ-TREE program30 was used to construct phylogenetic trees, with the Maximum Likelihood (ML) method and 1,000 bootstrap replicates. The trees were visualized in iTOL v. 531.
Confocal microscopy
Preparation and imaging of mature pollen and tetrads were performed as previously described14. 3D reconstruction of tetrads was done using NIS Elements v.4.20 software (Nikon).
Generation of the CRISPR allele of INP2
The guide RNA for the At1g15320 gene was selected with the help of the CRISPR-PLANT platform (https://www.genome.arizona.edu/crispr/)32 and its sequence was cloned into the pHEE401E vector33 as described34 using primers Oligo-01-F-INP2-T1/Oligo-R-INP2-T1 (Supplementary Table 2). The resulting construct was transformed into the Agrobacterium tumefaciens strain GV3101, and Arabidopsis Col-0 plants were transformed using the floral dip method35. The T1 transformants were selected on hygromycin plates, and 18 seedlings were transferred to soil. Two of the 18 T1 plants displayed the inaperturate phenotype. Sequencing of the At1g15320 gene from these two plants revealed that both had the same homozygous single-nucleotide deletion which occurred one nucleotide before the PAM sequence and caused a shift in the open reading frame after the codon 83.
INP2 complementation and expression constructs
Primers used in this study are listed in Supplementary Table 2. All fragments for cloning were amplified with high-fidelity Phusion DNA polymerase (NEB, M0530 or Thermo Fisher, F530). To create the INP2pr:INP2 ORF construct, the INP2 promoter (a fragment of 701 bp from the end of the preceding gene to the start codon of INP2) and open reading frame (ORF) were amplified, respectively, with primer pairs INP2pr-IF-F/ INP2pr-IF-R and INP2 ORF-IF-F/ INP2-Stop-IF-R from the Col-0 genomic DNA and from the INP2 cDNA clone DQ446252 obtained from the Arabidopsis Biological Resource Center. The resulting two fragments were cloned into the SacI and SpeI sites in the pGR111 binary vector36 using In-Fusion cloning (Takara, #638950). A BamHI site was introduced in front of the INP2 start codon to facilitate subsequent cloning. To create the INP2pr:gINP2 construct, the genomic fragment, which included the coding sequence, introns, and the 567 bp region downstream of the INP2 stop codon, was amplified from Col-0 genomic DNA with primers INP2 ORF-IF-F/gINP2-SpeI-R and cloned downstream of the INP2 promoter between the BamHI and SpeI sites. Constructs were verified by sequencing and transformed into the A. tumefaciens strain GV3101. The inp2 plants were then transformed by floral dip. Transgenic T1 plants were selected with BASTA, and the presence of transgenes was confirmed with specific primers.
To generate the reporter construct INP2pr:H2B-RFP, the H2B-RFP fusion gene was cloned into the BamHI/SpeI sites downstream of the INP2 promoter in pGR111 and the construct was transformed into the Col-0 plants.
To express INP2 tagged at the C-terminus with one or three copies of HA (HA1 and HA3), we first used the In Vivo Assembly (IVA) cloning in E. coli37 to combine the INP2 ORF (without the stop codon), an 8-aa Gly-rich linker (G5PGS), and the corresponding HA tag in the pGEM-T-Easy vector (Promega). After confirming the sequences, the tagged INP2 sequences were placed into pGR111 under the control of the INP2 promoter and the resulting constructs were transformed into inp2. The shorter construct with HA1 produced better rescue. Out of the 14 INP2-HA1 T1 lines, 11 had long to medium-size apertures, two – short apertures, and one – no apertures. Out of the 13 INP2-HA3 T1 lines, four had medium-size apertures, four – short apertures, and five – no apertures. Note that the construct with HA3 inserted directly after INP2 (without the Gly-rich linker) essentially failed to rescue the mutant phenotype (2/9 T1 lines produced short apertures and the rest produced no apertures).
SlINP1, SlINP2, and Arabidopsis/tomato chimeric INP2 constructs
The tomato paralogs of INP1 and INP2 were identified in the tomato genome, respectively, as Solyc08g079050 and Solyc03g116770. The DMC1pr:SlINP1-YFP construct was previously described12 and the previously characterized transgenic lines of DMC1pr:SlINP1-YFP inp1 were used in this study. To create the INP2pr:SlINP2 construct, the tomato genomic DNA was amplified with primers BamHI-SlINP2-BF/SpeI-SlINP2-AR. The resulting fragment was digested with BamHI/SpeI and used to replace the AtINP2 gene in the INP2pr:gINP2 construct. The construct was transformed into the inp2 and inp1 mutants, transgenic T1 plants were selected with BASTA, and the presence of the transgene was confirmed with specific primers.
To genotype the F2 populations which segregated both the DMC1pr:SlINP1-YFP and INP2pr:SlINP2 transgenes in combination with the inp1 or inp1 inp2 mutations, the following primers and conditions were used: for SlINP1, 2-Sl-F/Sly INP1-R-NcoI primers; for SlINP2, AD23/AD8 primers; for inp1, a cleaved amplified polymorphism (CAPS) marker was used (22600-DF/22600-DR primers, SacI cuts the mutant allele, digestion products resolved on 1.5% agarose gels); for inp2, a derived CAPS (dCAPS) marker was used (At1g15320-BF/AD402 primers, AccI cuts the mutant band, digestion products resolved on 4% agarose gels). PCR reactions with 40 cycles (98°C for 15 sec, 55°C for 15 sec, 72°C for 30 sec) were performed in all cases.
To quickly generate multiple chimeras of AtINP2 and SlINP2, we used the ORF versions of the constructs and the IVA cloning method.37 The AtINP2 ORF was first amplified using primers pGEM-INP2-IF-F/INP2-pGEM-IF-R and cloned with the help of the In-Fusion procedure into the pGEM-T-Easy vector digested with SacI and NcoI. A BamHI site and a SpeI site were introduced, respectively, at the beginning and at the end of the AtINP2 sequence for ease of subsequent subcloning into the binary pGR111 vector. To obtain the SlINP2 cDNA, total mRNA was isolated from young tomato buds with Trizol (Thermo Fisher, #15596026) and converted into cDNA as previously described14. The SlINP2 ORF was then amplified with primers AD19/AD16 and used to replace AtINP2 between the BamHI and SpeI sites with the help of the IVA method in pGEM-T-Easy vector, which itself was amplified with AD17/AD14 primers. The resulting pGEM-T-Easy-based AtINP2 and SlINP2 constructs were used as backbone templates for chimeras, while pGR111-based constructs were used as the INP2-region templates in IVA cloning. 30 primers (AD1 through AD28, pGEM-INP2-IF-F, and INP2-pGEM-IF-R) were used in different combinations (as indicated in Supplementary Table S2) to obtain all 18 chimeras. IVA reactions were performed as described37: products of single PCR reactions amplified with sets of four primers were treated with DpnI (15 min at 37°C) to degrade templates and directly transformed into E. coli. Single-region chimeras were used as backbone templates to generate multi-region chimeras. All chimeric sequences were verified by sequencing, cut out with the BamHI/SpeI digestion, subcloned downstream of the INP2 promoter in the pGR111 vector, and re-sequenced again. The final constructs were transformed into the inp2 mutant; transgenic T1 plants were selected with BASTA, and the presence of the transgenes was confirmed with specific primers.
Yeast two-hybrid assay
The Y2H assays were performed as previously described11. The DNA-binding domain construct pB29-INP1 and the activation-domain construct pP6-INP1 were described previously11. Other constructs were created by cloning the coding sequences of AtINP2, SlINP1, SlINP2 or their truncated forms into the same vectors. Constructs were co-transformed in indicated combinations into the NMY51 yeast strain. Positive bait-prey co-transformants were selected on the synthetic dropout medium lacking Leu and Trp (-LW). To test for interaction, co-transformed yeast cells were grown on the medium that lacked Leu, Trp and His and contained either 3 mM or 20 mM 3-amino-1,2,4-triazole (-LWH+3-AT).
Bi-fluorescent molecular complementation assay
To create the 35Spr:nYFP-AtINP1 and 35Spr:cYFP-AtINP2 constructs, AtINP1 and AtINP2 ORFs were, respectively, amplified with AD17-BHL/AD18-BHL and AD469/470 primers, digested with PacI/XbaI, and inserted into 35Spr:nYFP(1–158) and 35Spr:cYFP(159-238) binary vectors38. Constructs were transformed into A. tumefaciens strain GV3101. Bacterial cultures containing these constructs were grown to OD600=0.4 and co-infiltrated into tobacco (Nicotiana benthamiana) leaves along with agrobacteria expressing the RNA silencing suppressor P1939 at the 1:1:2 ratio. Infiltrated leaves were grown for 5 days, after which samples were collected and imaged on a Nikon A1+ confocal microscope using identical settings for all imaging. Samples were excited with a 514-nm laser, and YFP emission was collected at 522 to 555 nm.
Co-immunoprecipitation
AtINP1 and AtINP2 ORFs were amplified through two sequential PCR reactions (1st PCR with primers AD326/488 for AtINP1 and primers AD370/489 for AtINP2; 2nd PCR with primers AD122/123 for both genes) and inserted into the pDONR207 vector through the Gateway BP recombination reaction (Invitrogen, #11789020). Gateway LR recombination reaction was then used to transfer these sequences into the pCsVMV:GFP-C-999 or pCsVMV:HA-C-1300 vectors40 (a gift from Prof. David Somers, OSU), producing four constructs: CsVMV-AtINP1-GFP, CsVMV-AtINP1-HA3, CsVMV-AtINP2-GFP, and CsVMV-AtINP2-HA3. All constructs were verified by sequencing and transformed into the A. tumefaciens strain GV3101 by electroporation. Tobacco leaves were infiltrated with different combinations of constructs (as described for BiFC above), harvested 5 days after infiltration, and stored at −80°C for at least one day. 1 μl of anti-GFP monoclonal antibody (A-11120, Molecular Probes by ThermoFisher Scientific) was incubated with 1 μl of protein A/G PLUS-agarose (SC-2003, Santa Cruz Biotechnology) and 8 μl IP buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 % NP-40, 1 mM EDTA, 3 mM DTT, 1 mM PMSF, 5 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin) overnight at 4°C with gentle agitation. Frozen tobacco leaves were ground in liquid nitrogen, and 500 μl of ground tissue were mixed with 500 μl of IP buffer. These samples were vortexed for 2-3 min and centrifuged at 18,400x g for 10 min at 4°C. Supernatant was added to the anti-GFP matrix agarose, and the mixture was incubated for 2 hours at 4°C with gentle agitation, followed by centrifugation at 845x g at 4°C for 3 min to collect immune complexes. Pellets were washed three times with ice-cold IP buffer, once with 1x PBS buffer and eluted with 50 μl of 2x SDS-PAGE sample buffer. Immunoblotting was performed as previously described (Lee et al., 2018). The following antibodies were used to detect the fusion proteins: rabbit anti-GFP (Abcam; ab6556), rat anti-HA (Sigma, 11867423001), anti-rabbit IgG peroxidase-conjugated antibodies (SeraCare/KPL; 5220-0283/04-15-06), and anti-rat IgG peroxidase-conjugated antibodies (SeraCare/KPL; 5220-0364/04-16-06). All antibodies were diluted 1:2000 in TBS-T buffer (1xTBS, 0.1% Tween 20) supplemented with 4% non-fat milk. After the final washes, the membranes were processed with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, 34577) and imaged with a MyECL imager (Thermo Scientific).
Split-luciferase assay
The coding sequences of INP1 and INP2 from Arabidopsis and tomato were PCR amplified, cloned into pDONR207 (Invitrogen), and transferred into pCAMBIA1300-GWNLuc and pCAMBIA1300-GWCLuc41 (a gift from Dr. David Mackey, OSU) using the Gateway technology. Resulting constructs were transformed individually into A. tumefaciens strain GV3101. Agrobacteria were collected and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, 150 μM acetosyringone) at a final concentration of OD600=0.8. Pairwise combinations of suspensions were infiltrated into young tobacco leaves, which were then allowed to grow for 3 days in light. 12-16 leaves were collected from 5-10 plants, the abaxial side of leaves was sprayed with 1 mM luciferin (Biosynth, L-8220), and kept in the dark at 4°C for 30 min. The bioluminescence images were captured using Azure Sapphire Biomolecule Imager (Azure Biosystems) and converted to heatmaps using the 16-color look-up table (LUT) from ImageJ v.1.53a.
Data Availability
All data supporting the findings of this study are available within the article, Supplementary Information files or from the corresponding author upon reasonable request. Source data are provided with this paper.
Extended Data
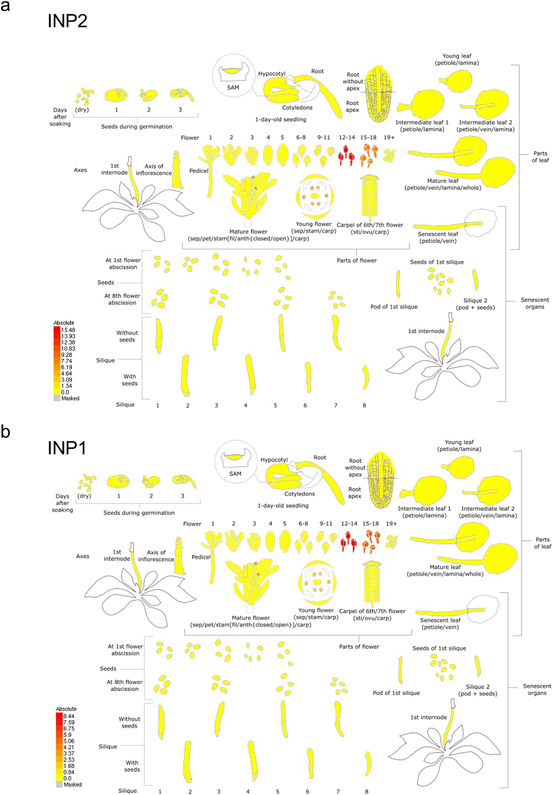
Extended Data Fig. 1. INP2 and INP1 display similar expression patterns, with both genes showing highest expression in young developing buds.
The RNA-seq data for INP2 (a) and INP1 (b) are from the dataset of Klepikova et al. (2016) and visualized with the BAR eFP Browser.
Extended Data Fig. 2. INP1 and INP2 proteins both contain the DOG1 domain and have similar structural organization predicted for their C-terminal parts.
a, Protein alignment between INP1 and INP2 proteins. Identical and similar (V/I/L, D/E, K/R, N/Q, and S/T) residues are shaded, respectively, in blue and green. The positions of the DOG1 domains predicted by Pfam are indicated by purple lines. b-c, Protein structures predicted by Phyre2 for C-terminal parts of INP1 (b) and INP2 (c) (confidence: >97% for both proteins). In both cases, the modeled regions cover 114 amino acids, which constitute, respectively, 42% of INP1 and 37% of INP2. The same template (c4clvB, nickel-cobalt-cadmium resistance protein NccX from Cupriavidus metallidurans 31a) was selected by the program in both cases.
Extended Data Fig. 3. Alignment of INP2 proteins from representatives of different angiosperm taxa.
The following species were used (from top to bottom): Papaver somniferum (basal eudicots, Papaveraceae), Arabidopsis thaliana (rosids, Brassicaceae) Capsella rubella (rosids, Brassicaceae), Olea europea (asterids, Oleaceae), Mimulus guttatus (asterids, Phrymacae), Manihot esculenta (rosids, Euphorbiaceae), Solanum lycopersicum (asterids, Solanaceae), Nymphaea colorata (basal angiosperms, ANA, Nympheaceae), Oryza sativa (monocots, Poaceae), Zea mays (monocots, Poaceae), Elaeis guineensis (monocots, Arecaceae), Ananas comosus (monocots, Bromeliaceae). The seven regions selected for creating AtINP2/SlINP2 chimeras are indicated by differently colored rectangles. Aspartate (D) and glutamate (E) residues in the acidic region are shaded in blue. Black shading indicates identical amino acids and grey shading indicates similar amino acids present at the same position in at least half of the aligned proteins.
Extended Data Fig. 4. Arabidopsis INP2 likely contains a transmembrane domain at its N-terminus.
Multiple TM discovery algorithms predict existence of the transmembrane domain at the N-terminus of INP2 from Arabidopsis thaliana (AtINP2), with the consensus score of 0.85 generated by the plant membrane protein database Aramemnon (AramTMCon).
Extended Data Fig. 5. None of the seven AtINP2 regions is sufficient on its own to convert SlINP2 into a protein able to function in Arabidopsis.
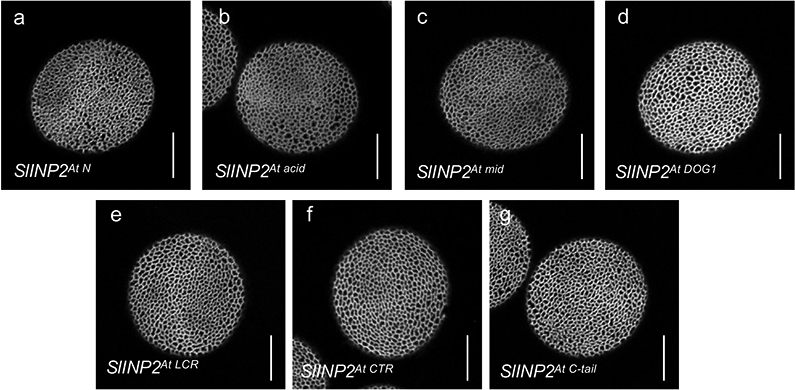
Confocal images of pollen grains produced by the transgenic inp2 plants expressing seven versions of chimeric SlINP2 constructs in which one region at a time was replaced with the corresponding regions from AtINP2. At least 10 independent T1 lines were tested for each construct (≥ 50 pollen grains per line), with similar results. Scale bars = 10 μm.
Supplementary Material
Acknowledgements
Funding for this project was provided to A.A.D. by the US National Science Foundation (MCB-1817835). We acknowledge the support from the US National Institutes of Health grant R35GM131760 (to I.B.Z.), the TRONDBUSS program (OSU and Norwegian Science and Technology University) (to I. M.M.), Herta Camerer Gross Postdoctoral Research Fellowship (to R.W.), University graduate fellowships (to P.C. and K.A.), OSU Mayers Undergraduate Summer Research Scholarship (to P.A.), NSF-REU supplement mechanism (to P.A. and A.H.), iCAPS internship from the Center for Applied Science (to A.H.), Undergraduate Research Scholarship from the OSU Arts and Sciences Honors Committee (to A.H.), and a Dr. Elizabeth Wagner Scholarship from the Department of Molecular Genetics at OSU (to A.H.). We thank members of the Dobritsa lab for discussions, Yuan Zhou for help with phylogenetic analysis, Vince Edwards, Noah Weyrick, and Stone Knapp for technical help, David Somers and David Mackey for vectors, Arabidopsis Biological Resource Center for DNA stocks, and the NCI-subsidized Genomics Facility at the OSU Comprehensive Cancer Center (CCSG:P30CA016058) for sequencing.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Furness CA & Rudall PJ Pollen aperture evolution – a crucial factor for eudicot success? Trends Plant Sci. 9, 154–158 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y & Dobritsa AA Formation of aperture sites on the pollen surface as a model for development of distinct cellular domains. Plant Science 288, 110222 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Heslop-Harrison J An interpretation of the hydrodynamics of pollen. Am. J. Bot 66, 737–743 (1979). [Google Scholar]
- 4.Katifori E, Alben S, Cerda E, Nelson DR & Dumais J Foldable structures and the natural design of pollen grains. PNAS 107, 7635–7639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vieira AM & Feijó JA Hydrogel control of water uptake by pectins during in vitro pollen hydration of Eucalyptus globulus. Am. J. Bot 103, 437–451 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Zhang X et al. Rice pollen aperture formation is regulated by the interplay between OsINP1 and OsDAF1. Nat. Plants 6, 394–403 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Wodehouse RP Pollen grains: Their structure, identification and significance in science and medicine. (McGraw-Hill, 1935). [Google Scholar]
- 8.PalDat. PalDat - a palynological database (www.paldat.org). (2018).
- 9.Dobritsa AA & Coerper D The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine. Plant Cell 24, 4452–4464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobritsa AA, Kirkpatrick AB, Reeder SH, Li P & Owen HA Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition. Plant Physiol. 176, 326–339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee BH et al. Arabidopsis protein kinase D6PKL3 is involved in formation of distinct plasma-membrane aperture domains on the pollen surface. Plant Cell 30, 2038–2056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P et al. INP1 involvement in pollen aperture formation is evolutionarily conserved and may require species-specific partners. J. Exp. Bot 69, 983–996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobritsa AA et al. A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 157, 947–970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeder SH, Lee BH, Fox R & Dobritsa AA A ploidy-sensitive mechanism regulates aperture formation on the Arabidopsis pollen surface and guides localization of the aperture factor INP1. PLOS Genet. 12, e1006060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD & Penin AA A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88, 1058–1070 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sall K et al. DELAY OF GERMINATION 1-LIKE 4 acts as an inducer of seed reserve accumulation. The Plant Journal 100, 7–19 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnani E et al. A comprehensive analysis of microProteins reveals their potentially widespread mechanism of transcriptional regulation. Plant Physiol. 165, 149–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amborella Genome Project. The Amborella Genome and the Evolution of Flowering Plants. Science 342, (2013). [DOI] [PubMed] [Google Scholar]
- 20.Kellogg EA Evolutionary History of the Grasses. Plant Physiology 125, 1198–1205 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plourde SM, Amom P, Tan M, Dawes AT & Dobritsa AA Changes in morphogen kinetics and pollen grain size are potential mechanisms of aberrant pollen aperture patterning in previously observed and novel mutants of Arabidopsis thaliana. PLOS Computational Biology 15, e1006800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukowitz W, Gillmor CS & Scheible W-R Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–806 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.1001 Genomes Consortium. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou X et al. A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant J. 63, 880–888 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Wickett NJ et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. PNAS 111, E4859–E4868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Bel M et al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46, D1190–D1196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capella-Gutiérrez S, Silla-Martínez JM & Gabaldón T trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A & Jermiin LS ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trifinopoulos J, Nguyen L-T, von Haeseler A & Minh BQ W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minh BQ et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letunic I & Bork P Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie K, Zhang J & Yang Y Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant 7, 923–926 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wang Z-P et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biology 16, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing H-L et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology 14, 327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clough SJ & Bent AF Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Dobritsa AA et al. LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis thaliana. Plant Physiol. 153, 937–955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Nafría J, Watson JF & Greger IH IVA cloning: A single-tube universal cloning system exploiting bacterial In Vivo Assembly. Sci Rep 6, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Kim J & Somers DE Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci U S A 110, 761–766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voinnet O, Pinto YM & Baulcombe DC Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. PNAS 96, 14147–14152 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Geng R, Gallenstein RA & Somers DE The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140, 4060–4069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen M et al. HOS15 is a transcriptional corepressor of NPR1-mediated gene activation of plant immunity. PNAS 117, 30805–30815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article, Supplementary Information files or from the corresponding author upon reasonable request. Source data are provided with this paper.