Abstract
By shaping where individuals move, habitat configuration can fundamentally structure animal populations. Yet, we currently lack a framework for generating quantitative predictions about the role of habitat configuration in modulating population outcomes. To address this gap, we propose a modelling framework inspired by studies using networks to characterize habitat connectivity. We first define animal habitat networks, explain how they can integrate information about the different configurational features of animal habitats, and highlight the need for a bottom–up generative model that can depict realistic variations in habitat potential connectivity. Second, we describe a model for simulating animal habitat networks (available in the R package AnimalHabitatNetwork), and demonstrate its ability to generate alternative habitat configurations based on empirical data, which forms the basis for exploring the consequences of alternative habitat structures. Finally, we lay out three key research questions and demonstrate how our framework can address them. By simulating the spread of a pathogen within a population, we show how transmission properties can be impacted by both local potential connectivity and landscape-level characteristics of habitats. Our study highlights the importance of considering the underlying habitat configuration in studies linking social structure with population-level outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00442-021-04967-y.
Keywords: Habitat configuration, Habitat networks, Landscape connectivity, Movement networks, Social networks
Introduction
Animals rarely move unrestrictedly, as the physical habitat environments they depend on are often heterogeneous and uneven (Fahrig 2007; Kovalenko et al. 2012; Lovett et al. 2005). The physical configuration of habitats, such as the spatial arrangement of habitat components and their physical attributes (e.g. heterogeneity, size, and quality), can fundamentally determine the patterns of habitat potential connectivity (i.e. where animals of a species can go), which eventually determine how populations of given species are functionally connected (e.g. socially or genetically). Thus, habitat configuration can have broad implications for population and community dynamics across spatial and temporal scales, including ecological interactions (Jordano 2016; Plitzko and Drossel 2015; Ryser et al. 2019), community structure (Altermatt and Holyoak 2012; Henriques-Silva et al. 2013; Wilson et al. 2016), and speciation (Naka and Brumfield 2018). Habitat configuration can also determine the rates of social interactions among conspecifics, thus shaping the social structure of populations (Emlen and Oring 1977; Farine and Sheldon 2019; Gosling 1991; He et al. 2019; Leu et al. 2016). Ultimately, the physical configuration of habitats shapes the distributions of genes (Armansin et al. 2020; Beninde et al. 2016; Phillipsen and Lytle 2013), pathogens (Altizer et al. 2003; Loehle 1995; Silk et al. 2019), and information (Aplin et al. 2015; Laiolo and Tella 2005, 2006) in populations. Understanding the effects of habitat physical configuration on animal population and community dynamics is particularly important in a rapidly changing world, where natural populations increasingly face anthropogenic habitat changes.
How individual animals are socially structured has many consequences for populations (Allen et al. 2017; Aplin et al. 2012; Keeling 1999; Montiglio et al. 2018). The best example for this perhaps comes from studies on pathogen transmission (Cantor et al. 2020; Prado et al. 2009; Sah et al. 2018; Silk et al. 2019) describing how patterns of social or physical connections among individuals at local and global scales can impact the speed of transmission and the magnitude of disease outbreaks. Specifically, more clustered connections—where the number of shared social connections between individuals, or triads , , and , are more represented in the population—can increase the local spread (among immediate contacts) but decrease the speed and global reach of pathogen transmission (Keeling 2005; Read and Keeling 2003; Sah et al. 2018). However, to unravel the role of social structure in shaping ecological and evolutionary dynamics, we need to also understand the mechanisms that shape animal social structure. Alongside social decisions, features of the physical habitat environments can play a major role in shaping where animals move, who they (re-)encounter, and how often they interact with one-another (He et al. 2019). For example, a study in sleepy lizards (Tiliqua rugosa) found that habitats with more barriers increased the rates of encounters among individuals, increasing the density and clustering of the social networks (Leu et al. 2016), which may have implications for the spread of infectious pathogens (Tildesley et al. 2010; White et al. 2018). Early socioecological models have linked the spatiotemporal distribution of resources and risks to social behaviour (van Schaik 1989; Wilson 1975), while more recent models have focussed the behavioural mechanisms underlying social structure (Cantor and Farine 2018; Farine 2019; Ilany and Akçay 2016; Kappeler 2017; Spiegel et al. 2016). However, we also require quantitative tools that explicitly link configurational properties of habitats to social structures to enable us to generate testable hypotheses on the role of the physical habitat environments on socially mediated population outcomes.
The features of animal habitats are typically multi-faceted—they can be described by the heterogeneity, sizes, abundance and spatial arrangements of habitat components (Tokeshi and Arakaki 2012). For a given animal species, these features determine habitat potential connectivity, indicating where individuals can move, thereby, the behaviours that they express and the subsequent consequences for populations (Gilarranz et al. 2017). For example, Doherty et al. (2019) found that the shape of habitats, specifically whether habitats were wider (i.e. forming a rectangle) or thinner (forming a narrow strip), structured the movements of radio-tracked agamid lizards (Pogona barbata); specifically, activity area and daily movement rates were lower among individuals inhabiting thinner habitats. The actual movements of animals are then the outcomes of a range of drivers (Nathan et al. 2008), including habitat potential connectivity and individuals’ actual behavioural decisions (e.g. where to move for resources and/or mates), which over time determine how populations of a species are functionally connected (Calabrese and Fagan 2004; Tischendorf and Fahring 2000). Among these drivers, the spatial components inherent to many emergent ecological patterns have received increasing attention in ecology (Fletcher et al. 2013; Gilarranz et al. 2017).
Spatial networks (Barthélemy 2011) have been used to characterize metapopulation spatial structures and the spatial configuration of habitats (Dale and Fortin 2010; Fall et al. 2007; Urban and Keitt 2001; Urban et al. 2009), and the spatial patterns of connectivity of animal habitats (Alther and Altermatt 2018; Bodin and Norberg 2007; Fall et al. 2007; Galpern et al. 2011; Lookingbill et al. 2010; Marini et al. 2019; Minor and Urban 2008; Poli et al. 2020; Urban and Keitt 2001; Urban et al. 2009). For example, Robertson et al. (2018) used long-term mark–resight data to construct networks that characterize the functional connectivity among habitat patches of snail kite (Rostrhamus sociabilis plumbeus) to evaluate the relative roles of among-patch movement and reproduction in modulating the effective connectivity of the species’ distribution range. In such networks, nodes often represent habitat or resource patches (e.g. nesting sites, Galpern et al. 2011; Urban et al. 2009), that is, areas crucial for survival and reproduction (Fahrig and Merriam 1985) as opposed to the landscape matrix (Ziolkowska et al. 2014). How these connections are defined determines what these networks are depicting. Typically, the connections are inferred from movements of individuals, gene flow, the species’ biological attributes, or from the characteristics of the environment itself (see Calabrese and Fagan 2004 for the definitions of connectivity metrics; but see review on the use of these connectivity metrics in Galpern et al. 2011).
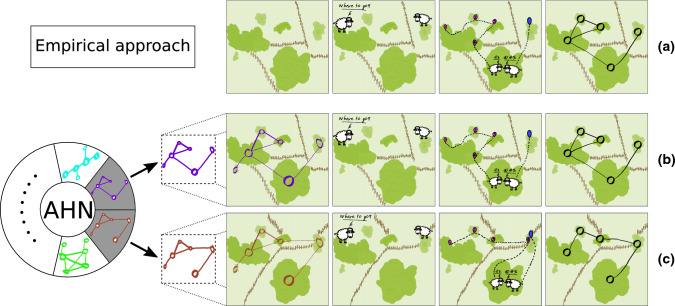
Spatial networks constructed following the approaches outlined above have been instrumental in studies of animal movements (Gilarranz et al. 2017; Robertson et al. 2018) and community structure (Altermatt and Holyoak 2012). However, they typically do not allow us to make broader, or more general, predictions on the linkages between spatial network structures and processes in animal populations and communities. This is because the structures of empirically constructed spatial networks are usually derived from, and thus inherently limited in scope by, the specific habitats and species under study (Baguette et al. 2013; Baranyi et al. 2011; e.g. Fig. 1a). Thus, they limit our understanding on how various configurational features of habitats (e.g. landscape linearity and spatial scale) might consistently shape population or community outcomes. For example, networks explicitly built to characterize the potential connectivity of the Yangtze and the Rhine rivers for freshwater organisms can be used to explore the relationships between these specific network structures and processes in populations or communities. Yet, the specificity of such networks, as originated from the specific spatial configurational features (e.g. landscape geometry, elevation) of those habitat systems for targeted species, may limit our ability to explore a broader spectrum of plausible habitat networks, including those that might not yet exist but could emerge from present habitat networks (e.g. through rerouting of river flow or the construction of a dam). Moreover, empirical networks provide limited ability to explore how specific dimensions of network properties (e.g. patterns of connectivity) affect populations while holding other properties constant (e.g. network size, spatial scales at which these networks are defined; e.g. Fig. 1b, c). For example, empirical networks have suggested that highly modular social structures (i.e. multilevel social structures) play an important role in promoting cultural evolution (Migliano et al. 2020); yet, it has been subsequently shown that such effects of modularity contribute relatively little in promoting cultural evolution by simulated networks, which allow testing the effects of network connectivity itself while controlling other network properties such as the number of links (Cantor et al. 2021). Such limitations may make it hard to generalize insights and predictions on the relationship between the physical habitat environment and biological processes.
Fig. 1.
Two distinct approaches for understanding the role of habitat configuration in shaping animal population (or community) structures. a In most studies, animals are observed living and moving (e.g. via GPS tracking) within given time windows in specific habitats, from which characteristics of the connectivity of the focal habitat area are inferred or modelled (e.g. by resistance surface modelling, network-based landscape connectivity modelling, or circuit theory). By contrast, (b) with a bottom–up approach, we can simulate networks to depict the physical configurations of specific habitats, and then model individual movements (or more complex behaviours) in these habitats, from which we can gain sights on how observed structures (e.g. patterns of movements and social interactions) emerged. With this approach, we can also (c) simulate habitat networks controlling for key parameters (e.g. network connectivity), thus producing alternative scenarios that can control (or not) for features that are hypothesized to play a major role in shaping biological processes in populations. Here, we illustrate two simulated networks, one of which (b) can exactly depict the configuration of the given habitat for the focal species (a), while the other depicts a habitat that maintains some characteristics (e.g. the same distributions and sizes of habitat patches, represented by nodes) as the given habitat (a), but provides alternative patterns of potential connectivity (by randomizing the spatial distribution of movement barriers that determine which patches are connected)
One way to overcome the limitations of empirical networks is to simulate networks using generative models (Granovetter 1973; Watts and Strogatz 1998). While many generative models exist (e.g. Barabási et al. 2001; Erdős and Rényi 1960), none considers the inherent spatial dependency of animal habitat networks, and thus generate networks might not capture their fundamental properties (but see Carraro et al. 2020 for a recent solution for riverine habitats). One generative model of random networks that can incorporate spatial components is the random geometric network model (Dall and Christensen 2002; Penrose 2003). In geometric networks, nodes are anchored in space and are connected whenever their Euclidean distance is below a given threshold. By producing distance-based spatial networks (Barthélemy 2011), the random geometric network model provides a starting point for generating habitat networks. However, random geometric networks remain limited by a fixed spatial extent (i.e. a consistent square landscape) and a fixed threshold for determining the presences of links. In nature, the geometry of habitats and landscapes for a species, and the topological properties of their potential connectivity, can vary widely. For example, the distance between two patches does not exclusively dictate their potential connectivity—barriers such as waterscapes can restrict movement of a terrestrial animal between two patches in close proximity, while patches that are far apart can be connected by movements (as demonstrated by empirical evidence that primates can use roads to efficiently move between distant areas of their home ranges, Green et al. 2020; Strandburg-Peshkin et al. 2017).
Here, we address the need for a generative model of animal habitat networks by extending the random geometric network model to generate more plausible spatial networks. We first define animal habitat networks and outline the key configurational features of animal habitats that can be captured by such networks. Next, we describe a modelling framework—available in the accompanying R package AnimalHabitatNetwork, for simulating animal habitat networks explicitly tailored to depict the diverse physical configurations of animal habitats. We show that our network simulation algorithm can be tuned to capture the patterns of potential connectivity of real habitats efficiently, thereby providing the basis for explorations of alternative scenarios. Doing so is important, as making predictions requires producing realistic alternative scenarios. We propose three key research questions related to our modelling framework. Finally, we illustrate how our framework can be used to simulate animal habitat networks with varying patterns of connectivity to investigate the implications of habitat configuration for populations by embedding a Susceptible-Infected-Recovered (SIR) epidemic model in our modelling framework. Taken together, our findings provide new insights on the linkages between habitat configuration and population-level outcomes and highlight how the application of an explicit and quantitative framework to simulate habitat networks can help us gain a better mechanistic understanding of the role of habitat configuration in shaping the dynamics of ecological, evolutionary processes and their conservation implications.
A multi-dimensional framework for modelling animal habitat configuration
Defining animal habitat networks
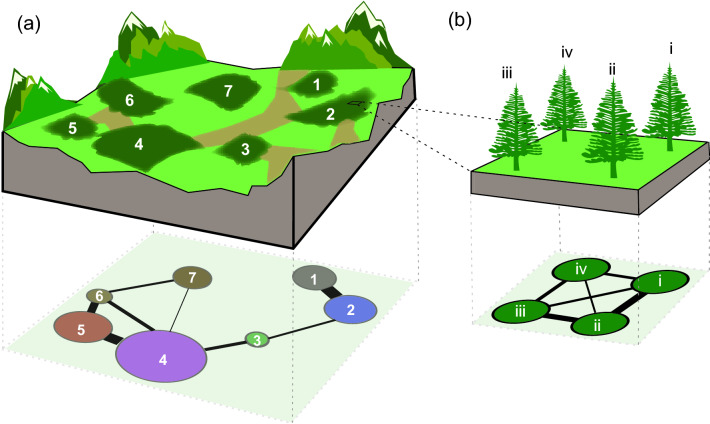
Animal habitats are defined by taking both the species-level properties (such as locomotion and space use characteristics of a focal species) and the environmental features into account. Here, we highlight the five fundamental dimensions proposed by Tokeshi and Arakaki (2012) for characterizing habitat physical configuration as a means of defining components in animal habitat networks. These dimensions are (1) spatial scale (spatial resolution and extent) at which the landscape and its elements are defined, (2) composition diversity (heterogeneity), (3) size (area), (4) abundance or density (number of discrete habitat patches/units per area), and (5) spatial arrangement (distribution) of habitat components. With these dimensions and following the definitions of connectivity metrics (i.e. structural, potential, and realized connectivity; Calabrese and Fagan 2004; Taylor et al. 1993; Tischendorf and Fahring 2000; Urban and Keitt 2001), we define animal habitat networks (Fig. 2) as network-based explicit depictions of (1) the spatial organization and (2) the physical attributes (e.g. heterogeneity and area) of given numbers of habitat components at given spatial scales, and (3) the potential connectivity indicating where animals of a given species can move.
Fig. 2.
Networks explicitly characterizing the physical configurations of animal habitats. We illustrate how five dimensions for assessing habitat configuration proposed by Tokeshi and Arakaki (2012) can be integrated and applied to construct animal habitat networks. These dimensions are (1) spatial scale (spatial resolution and extent), (2) composition diversity (heterogeneity), (3) size (area), (4) abundance or density (number of discrete habitat units per area), and (5) spatial arrangement (distribution) of habitat components. (a) A hypothetical landscape composed by forest fragments (numbered components) within a heterogeneous matrix with potential movement corridors (light green, which account for the presences of links between nodes) and physical barriers (light brown, which account for the absence of links between nodes). The physical features and spatial organization of the habitat components can be represented by a connected network at a large spatial scale, with a high composition diversity (fragments of different tree species), different habitat sizes (small and large fragments), high abundance (7 fragments), and heterogeneous spatial arrangement (fragments unequally distributed and connected by movement corridors across the landscape). (b) The physical features and spatial arrangement of habitat components can be characterized at different spatial scales. Here, part of the forest (fragment 2) can be represented by a connected network at a finer spatial scale (e.g. trees as habitat components), with a low composition diversity (the same tree species), small habitat size (single trees), low abundance of components (4 trees), and uniform spatial arrangement. In the two habitat networks, the compositional diversity (or quality) and size (or carrying capacity) of habitat components are characterized by node attributes (colours and sizes), the abundance by the number of nodes in the networks, and the spatial arrangement by the patterns of connectivity and the distribution of link weights (both as a function of the Euclidean distances between habitat components)
In habitat networks, nodes represent spatially explicit habitat components (or patches) which can be characterized by attributes (e.g. size, quality, and physical composition). The presence of a link between nodes indicates that animals of a given species can move between patches. In weighted animal habitat networks, link weights can characterize variation in the propensity for individuals to move between patches. Variation can arise from a range of factors, such as the spatial proximity between habitat components or the permeability of the landscape matrix between habitat components. Link weights can also be defined by empirical data (such as the actual rate of movement or gene flow between patches previously observed, which are typically treated as measurements of the extent to which landscapes facilitate/impede movements of individuals of a species, Taylor et al. 1993). Link presences and weights can also be related to properties of habitat patches (i.e. node attributes), such as the extent to which they are similar in their attributes for a species (e.g. types of resources they provide for a species).
The definition of the presences and weights of links in habitat networks highlights a clear distinction between network structures defined as a priori potential connectivity versus those determined by post hoc observational data. The former provides a fundamental template indicating where animals of a species can move, as the outcome of how the species’ intrinsic biological attributes (e.g. locomotion, Hirt et al. 2018) interact with the configurational features of the physical habitat environment. By contrast, the latter reflects realized animal movements (e.g. animal movement networks; see properties of movement networks in Bastille-Rousseau et al. 2018; Jacoby and Freeman 2016) that are typically driven by habitat potential connectivity together with a range of factors. These include the social environment that contribute independently to individuals’ movement decisions (Armansin et al. 2020; Strandburg-Peshkin et al. 2015, 2017), but also methodological factors (e.g. measurement accuracy, effort, decisions about where to collect data). Defining animal habitat networks from the perspective of potential connectivity highlights the fundamental bottom–up role of habitat physical configuration together with focal species’ biology in structuring animal movements and the subsequent processes in populations or communities.
The AHN model
We propose a general and spatially explicit modelling framework for plausible spatial networks (hereafter the ‘AHN’ model) that can depict the diverse configurational features of animal habitats. Although we focus on animals, our framework can equally be applied to characterize the configurational features of habitats for any moving organisms, such as pathogens in moving hosts and dispersing seeds. The model contains eight parameters (Table 1) explicitly encoding five fundamental dimensions characterizing animal habitat physical configuration (Tokeshi and Arakaki 2012) and species-level movement characteristics. We define the model within a 2-dimensional Euclidean space, by conceiving a planar rectangular landscape with an area and a side length . The model can accept any given spatial layouts of habitat components. For example, the coordinates can be determined by the spatial locations of empirically observed natural habitats, approaches developed for simulating point patterns in spatial ecology (Baddeley et al. 2016; see also Baddeley and Turner 2005 for the R package ‘spatstat’), or any spatial distributions relevant to a hypothesis of interest (e.g. the layout simulated using the Gauss-Poisson point process; see also the example in Code Availability). By default, the spatial coordinates and of the habitat components are randomly drawn from the intervals and (e.g. Fig. 3a), respectively. In this way, the AHN can depict landscapes with variable sizes, spatial extents, and aspect ratios, which can be based on the geometric properties of empirical animal habitats. Thus, the model explicitly captures the number (or density) of habitat components at the given spatial scale. Finally, in the model, the compositional diversity (i.e. heterogeneity) and size (or other physical properties) of habitat components can be encapsulated as node attributes, in vectors and respectively, the values of which can be quantitative or qualitative, and can be provided specifically or drawn at random from a given distribution, depending on the hypothesis of interest. By being explicit in spatial scale and node attributes (e.g. heterogeneity), the model can relate spatial scaling (Fletcher et al. 2013) to the functionality of the physical habitat environment for organisms of focal species (Fahrig et al. 2011).
Table 1.
Parameters of the AHN model for depicting habitat physical configuration
| Parameter | Description |
|---|---|
| Area of the conceived landscape | |
| The length of one side of the conceived landscape | |
| Number of habitat components (integer) | |
| Scaler for the weights of rewiring links | |
| Determining the steepness of link filtering-out function | |
| Determining the concave-to-convex transition point of | |
| Heterogeneity (or qualitative properties) of habitat components | |
| Sizes (or quantitative properties) of habitat components |
Fig. 3.
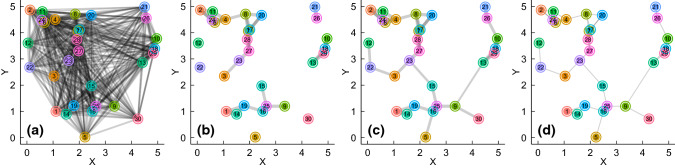
The workflow of the AHN model for generating animal habitat networks. First, (a) the algorithm constructs a fully connected and weighted habitat network. Here, numbered nodes represent 30 habitat components colour-coded by their attributes (such as their sizes, quality or compositions, with continuous or discrete colour palette) and connected by links whose thicknesses indicate the strength of the spatial relationship between the two habitat components, and is determined by the spatial positions of the nodes. The network is defined in a conceived 2-dimensional landscape in which the and axes indicate the spatial extents of the landscape (here the aspect ratio is 1, i.e. , but the model allows any and extents for capturing the diverse landscape geometry), therefore, it inherits spatial properties of the landscape. Next, (b) the algorithm removes the link between node and () from the network with probability ; in this example, it results in a disconnected habitat network. Then, (c) the (disconnected) network components can be rewired with minimal number of links if a connected network is wanted. Finally, (d) the habitat network can be transformed to unweighted, if so desired (e.g. when we are interested only in the patterns of potential connections while their attributes are irrelevant to our hypotheses)
Once the layout of habitat components is defined, the AHN model can then generate links to characterize the patterns of potential connectivity among habitat components for a given species. Links can be weighted (e.g. Fig. 3c, with link weights characterizing the strength of connections) or unweighted (e.g. Fig. 3d, where the strength of connections is not of interest), and are non-directed (the model can easily be extended to have directed links, for example if there is a gradient—such as altitude—a physical features of habitat that might favour animal movements in one direction more than the other).
When generating links, the model starts by allocating a link weight between node and node (). By default, the model uses the power function , where is the Euclidean distance, a primary metric characterizing habitat potential connectivity (Poli et al. 2020), between habitat components that node and node are referring to (Fig. 3a). However, alternative approaches can be used to define link weights, such as relating them to the similarities or differences in the properties of patches themselves (i.e. node attributes characterized by and/or , such as their heterogeneity), using different metrics such as cost-distance (Fletcher et al. 2016), using dispersal kernels (Clobert et al. 2012; Hartfelder et al. 2020; Urban and Keitt 2001; see also examples in Code Availability), or using information on habitat choice of a species, such as evidence that animals can preferentially disperse to habitats with similar properties as their natal habitats (Davis 2008; Hoover et al. 2021)—which would mean that patches with greater similarity would have a stronger link weight.
With our approach, the network starts by being fully connected, with link weights indicating the strength of potential connectivity between pairs of habitat components. In nature, however, animals are often limited by their ability to move between habitat components that could otherwise be connected by movements, for example, due to physical barriers to their movements, and/or spatial distances that they are intrinsically not able to directly cover between habitat components. To capture this realism, we use the sigmoidal function to determine a threshold probability for filtering out the link between node and from the initial complete network, where determines the steepness of the thresholding curve which transits from concave to convex at the species-specific critical distance . We define so consistently increases over (Fig. A1). This function enables us to generate a wide spectrum of curves to cover the diverse and evolving relationships between and by tuning and (Figs. A1–A3). The dependence of the probability on assumes that it is less likely that there exists a direct movement potential between two habitat patches when they are much further away.
The probabilistic nature of the filtering function captures the stochasticity that exists in the relationships between spatial proximity, landscape configurational features and potential connectivity. That is, on one hand, operates on the distances between patches to characterize the baseline stochasticity as the product of the interplay between species-specific characteristics of how individuals typically move (e.g. locomotion mode or capacity) and the given spatial proximity between habitat components, while, on the other hand, is a coefficient which characterizes the stochasticity arising from how the intrinsic biological properties of a species interact with the given configurational features of the physical habitat environment between habitat components with the given spatial proximity, such as the presences/absences of physical barriers, and/or the amount of resistance to movements. With and/or we can generate the patterns of absences/presences of links that are ecologically relevant to our hypotheses or questions (e.g. parameterization with empirical or observational data on focal species’ attributes, spatial proximity between habitat components, and/or the quality of landscape matrix for the focal species).
Our framework provides a starting point for incorporating more complex approaches for simulating spatial networks as habitat networks. It provides a simple and flexible approach to simulating networks that can capture the diverse patterns of potential connectivity of animal habitats as found in nature. For example, with a given set of spatially referenced nodes, allows us to simulate variable patterns of network connectivity, such as node clustering, which can be used as meaningful depictions of the potential connectivity of alternative scenarios of empirically derived habitats or to generate scenarios for habitats based on observed (or hypothesized) locomotion modes of a species. can also determine the presence of links in both probabilistic and deterministic ways, which not only makes the framework general, but also enriches our ability to encode the diverse physical features between habitat components, such as the variation in the quality of habitat matrix (in terms of their effects on animal movements), and species’ intrinsic attributes, such as the ability to exploit the physical landscapes. For instance, in the extreme case when and , the link removal function becomes deterministic (i.e. ), and with the rewiring option in the model (described below), the model can then generate networks that approximate planar networks (McDiarmid et al. 2005), which have previously been used to model landscape functional connectivity (Chubaty et al. 2020).
In some cases, fragments the network into (disconnected) network components (e.g. Fig. 3b). The smaller the gets and the larger the gets, the more links on average will be filtered out, and the more likely it is for the resulting habitat network to be disconnected (Fig. A3). In such cases, a disconnected network would represent habitats containing isolated clusters of habitat components between which animals cannot physically move among them. From a modelling perspective, it is often preferable (at least initially) to consider one habitat as a connected network (i.e. a single network component) which denotes a complete habitat or a section of a larger fragmented habitat in which individuals can theoretically (but not necessarily) move from one patch to any other. This means that the whole of the focal population can be functionally connected as a biologically meaningful unit (e.g. gene flow is possible between any two patches). We, therefore, incorporate the option of using a step-wise approach to rewire network components for connected habitat networks. In the rewiring, the two spatially closest nodes from each of the two network components are wired each time until the network has no disconnected network components, and the algorithm uses the minimum number of rewiring links for doing this (Fig. 3c). While by default the weights of rewiring links (if simulated) are defined in the same way as for the links within connected clusters, the model provides the option to additionally mediate these weights (for example, if we expect lower movement potentials between clusters, given the distance between them). We implement this option with the function , where is the Euclidean distance between node from the network component and node from the network component , and the scaler enables control over the weights of the rewiring links (if any; Fig. A4), and by default (i.e. all the link weights in a network are defined with the mathematical reciprocal of the Euclidean distance between nodes).
We provide the implemented algorithm for simulating habitat networks in the function ahn_gen() in the R package AnimalHabitatNetwork (version 0.1.0, He and Farine 2019; see Code Availability).
Demonstrating the capability of the AHN model in simulating habitat potential connectivity
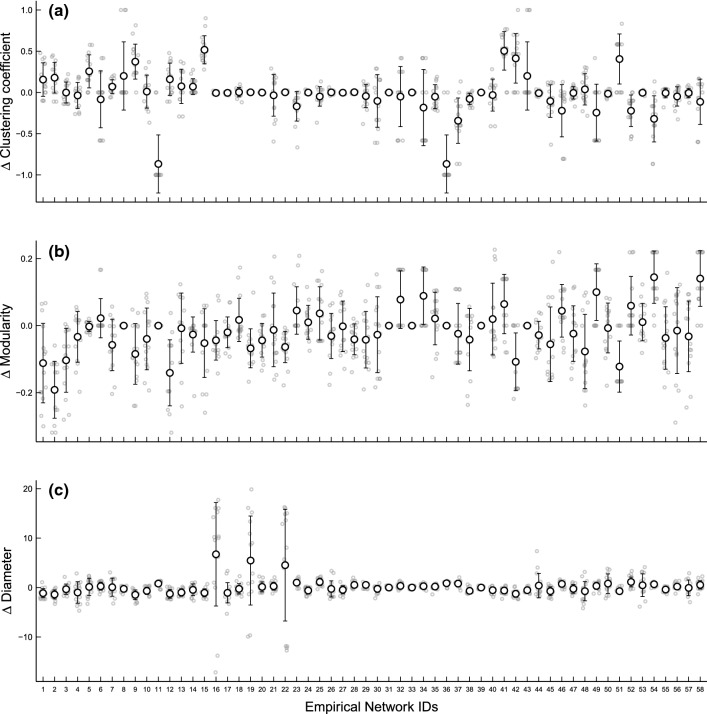
The pattern of connectivity is the key signature of a network (Albert et al. 1999). To test the capability of the AHN model in simulating habitat networks that are similar in terms of their structural properties (i.e. connectivity) to those observed from real habitats, we compared the topological properties of networks generated by the model using a given parameter space with those of empirical habitat networks characterizing habitat potential connectivity by Friesen et al. (2019). Here we consider three network metrics, the (average) clustering coefficient, modularity and diameter. Studies have discussed the relationships between these metrics of (social) networks and population outcomes, such as transmission of pathogens (Sah et al. 2018) and evolutionary dynamics (Marcoux and Lusseau 2013; Raghunandan and Subramanian 2012) in populations. In the context of animal habitat networks, these metrics could explain outcomes where individual movement play a fundamental role. The clustering coefficient (Fagiolo 2007) in animal habitat networks characterize the probability that two patches connected to a third patch are themselves connected, which can capture the extent to which individuals are locally constrained and contained by the physical habitat environments. Modularity (Newman 2006) in animal habitat networks characterizes the extent to which clusters of habitat patches tend to be more densely connected with each other within the cluster than with other clusters, which can capture the extent to which individuals are facilitated in local movements but impeded in movements at larger spatial scales by configurational features of the physical habitat environments (where higher modularity would represent more distinct subpopulations). Diameter (Albert et al. 1999; Jackson 2008) in an animal habitat network captures the length of the longest (yet the most efficient) potential movement path between two patches within a given habitat, which can capture the linearity of habitat potential connectivity.
We extracted the largest network component from each of the 62 empirical habitat networks contained in the Friesen et al. (2019) dataset, and kept 58 of them for benchmarking (the two largest were omitted due to computation limit and the two smallest, each with two nodes, were excluded). Each of these extracted networks is connected, denoting a habitat or a part of a larger habitat where animals can physically move from a given habitat component to any other one (i.e. the habitat can be functionally connected—biological processes such as information or genes flows are possible among habitat components). Next, we simulated random habitat networks with the AHN model and identified those sets of parameters under which the corresponding output habitat networks best approximated the (average) clustering coefficient, modularity, and diameter of each of these empirical networks, respectively. We considered the parameter space , , , across all empirical benchmark networks, while keeping identical to the number of nodes of the corresponding empirical habitat network. This parameter space was determined by considering the effects of each parameter on the resulting network structures (Fig. A1–A7). In total, for each metric of each empirical network, we generated 270 (i.e. size of the parameter space) random habitat networks, and identified the set of parameters from the parameter space that generated the network that most closely approximated the metric of the given empirical network as the ‘best-fitting’ set of parameters for that network. We then simulated 15 habitat networks with each of these sets of parameters as replicates, and evaluated the extent to which each of these metrics of each replicate deviated from that of the corresponding empirical network (see Code Availability). The test of the model with these networks confirmed that our proposed algorithm can generate networks that capture the key structural properties of real habitats (Fig. 4), thereby forming the basis for subsequently exploring on how population outcomes (structures and/or processes) might change under alternative habitat scenarios (e.g. by controlling and/or parameterizing key parameters from empirical and/or observed landscapes, such as increasing or decreasing connectivity by tuning and/or ). For example, if we wanted to test whether a species in a landscape with a given set of configurational features is more prone to infectious pathogens than another species, we can model transmission dynamics with epidemic models on habitat networks defined by species-specific and/or . All network computations were done in R (version 3.6.1, R Development Core Team 2019) with the igraph library (version 1.2.5, Csardi and Nepusz 2006).
Fig. 4.
The AHN model can simulate spatially explicit networks to characterize habitat potential connectivity. Each grey circle denotes the difference in each of the three metrics (y-axes, a, b, c) between each of the 15 replicated random habitat networks generated by the AHN model with each set of best-fitting parameters identified from the given parameter space and the corresponding empirical network; black circles and bars characterize the means and the standard deviations
Key research questions related to the modelling framework
The AHN model can be used to address a range of topics and research questions. Here, identify three key research areas where our framework can be used to address outstanding questions.
What are the consequences of habitat changes for population processes in social animals?
Animal habitats are changing under natural and anthropogenic drivers, typically characterized by changes in the spatial distribution of habitat components (e.g. food and shelter), changes in their potential connectivity (e.g. through fragmentation or reforestation), and/or changes in the physical attributes of the habitat components themselves (e.g. the amount of resources in each patch). These changes can then reshape the movements of animals, which can subsequently affect the patterns of biological or ecological interactions (e.g. inter-individual social structure, predator–prey interactions), or even impose evolutionary pressures on impacted species (Banks et al. 2011; Kokko and Sutherland 2001). The spatial distributions of food resources or habitat fragmentation can shape the spatial organization of individuals (Jacobson et al. 2015; Mourier et al. 2012), with consequences on the evolution of their social or mating systems (Banks et al. 2007; Emlen and Oring 1977; Tuomainen and Candolin 2011; van Schaik 1989). For example, Banks et al. (2011) empirically explored the relationship between the patterns of den-sharing interactions among hollow-dependent Australian mountain brushtail possums and the spatial variation in hollow tree availability, and found a behavioural switch from kin avoidance to kin preference in den sharing when hollow tree availability decreases, highlighting the important role of habitat change in driving individuals’ social behaviours as responses. In another example, Bain et al. (2014) examined the effects of habitat configuration on the frequency of extra-pair paternity (EPP) in cooperatively breeding superb fairy-wren (Malurus cyaneus) by linking spatial arrangements of their territories to the frequency of EPP, and found that the frequency of extra-group paternity (EGP) among groups in linear strips of vegetation was lower than those in more clustered territories in continuous habitats, highlighting the role of habitat spatial configuration in influencing the rates of EGP and the potential consequences of anthropogenic habitat change for mating systems.
Our network-based modelling framework can be used to depict multiple yet diverse configurational properties of animal habitats, thereby providing the starting point for explicitly modelling habitat change and predicting the population outcomes. Moreover, understanding the consequences of habitat fragmentation for populations is one of the central topics in conservation biology (Fischer and Lindenmayer 2007; Haddad et al. 2015), while the impacts on social processes of conservation efforts aimed at reducing habitat fragmentation are almost completely unexplored. With our framework, one can simulate network scenarios that realistically map the potential trajectories of habitat change (e.g. by parameter optimization and/or network manipulation), and generate predictions on the potential consequences for a population.
How is habitat connectivity shaped by landscape and species properties?
The patterns of potential connectivity that form animals’ habitats are shaped by a range of properties. Some of these are biological, such as species attributes. For example, the movement capacity of an animal species can be driven by body mass and locomotion, influencing how they explore their physical habitat environments (Hirt et al. 2018). Many of the properties shaping potential connectivity are abiotic, such as the climatic conditions that determine the composition of habitat patches (e.g. the assemblage of plants, or coral, species in a patch) and geological features that determine the shape of the landscape (e.g. the long and narrow valleys created by a mountain range vs. an open plain). A key question is, therefore, whether certain types of landscapes consistently shape networks with different properties. For example, it is likely that riverine habitats, or habitats in valleys, will have a larger network diameter than habitats that are less restricted by the geometric features of landscapes. Studies have highlighted the importance of linking the configurational features of landscapes and species-level properties to population-level outcomes, and practical guidelines have been proposed for exploring such linkages (e.g. Frank and Wissel 1998). Using our framework and following a fundamental bottom–up approach, it will be possible to develop a mechanistic understanding of the relative roles of the multiple factors underlying population outcomes, such as species-level properties (e.g. body mass and locomotion characteristics, Hirt et al. 2018) and landscape properties (e.g. linearity) in shaping structural properties of habitat networks (e.g. network clustering).
How different do we expect population social structures to be in different landscapes?
Studies have revealed that animal population social structures often exhibit notable variations (Mori and Saito 2005; Nandini et al. 2017; Prehn et al. 2019; Whitehead and Kahn 1992). When habitats vary in their physical configurations, we would expect the social structures of populations in these habitats to exhibit variations (even for the same species), and this is indicated by empirical evidence. For example, Farine and Sheldon (2019) showed that the social network structure (at the network community level) of a woodland bird community observed in the Wytham Woods in the UK, remained consistent across four winters, despite the high turnover rate of individuals within the communities. This study suggests that the predictability of habitat configuration for the emergent social network structures. Our framework can be tuned to depict animal habitats with distinct configurational features, therefore, providing a theoretical tool to examine how much variations in population social structures observed from one habitat to another might be explained by habitat physical configurations (i.e. the extent to which animal habitat networks account for the variations in animal social networks). Figure 5 highlights how different aspects of habitat configuration (such as the aspect ratio and the tendency for distant patches to be connected versus not) can interact with each other to shape the resulting structural features of the habitat networks.
Fig. 5.
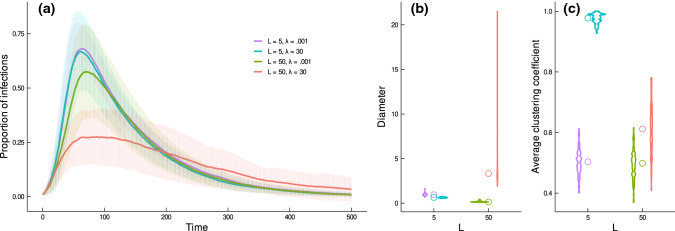
The application of the AHN model for understanding the role of habitat geometry and potential connectivity in mediating pathogen transmission dynamics in habitat-structured animal populations. a The transmission of pathogens in populations is dependent on both the landscape geometry (shape), depicted by where a larger value represents landscapes with a larger aspect ratio, and the extent to which the potential connections between patches to be determined by the habitat features between them, depicted by , where a lower value corresponds to a weaker deterministic effect of how the species’ movement characteristics interact with the environmental features on the potential connectivity between patches with given spatial proximity. Simulations show that the transmission dynamics, when individual mobility is at a medium level () under an infection rate of and a recovery rate of , are impacted by habitat shape and potential connectivity. Specifically, habitats with a larger aspect ratio (a larger value) and with their potential connectivity determined with a stronger deterministic effect of the configurational features on the potential connectivity between patches with given spatial proximity (i.e. a larger value) have the smallest disease outbreaks (each curve indicates the mean percentage of infected individuals in a population of 100 individuals moving on simulated habitat networks comprising 20 nodes, over 500 timesteps, with bars indicating the standard deviations from 100 replications). The interaction between landscape shape and the degree to which between-patch potential connectivity is determined by habitat configurational features affects the structural properties of habitat networks, such as diameter (b) and (average) clustering coefficient (c). Open circles (in b and c) indicate medians
Illustrating the application of the framework: how do habitat networks shape pathogen transmissions?
Our modelling framework can be used for understanding the link between animal habitat network structures and population outcomes. Here, we illustrate a possible application of it in the context of pathogen transmission in a population of 100 individuals (with no birth, death, emigration from and immigration to the landscape) moving among 20 habitat patches, where the patterns of potential connectivity are depicted by (connected) habitat networks simulated using the AHN model. We considered the simplest case where habitat patches are randomly positioned (with their and coordinates drawn from uniform distributions) in landscapes with the same area (i.e. ) but varying aspect ratios, with producing square-shaped landscapes (e.g. resembling forests on open plains) and producing landscapes with a large aspect ratio (e.g. resembling forests in a narrow valley). Within these landscapes, we considered two scenarios of habitat potential connectivity, defined by and . These values depict two distinct outcomes of how the focal species’ intrinsic biological attributes interact with the environmental features between habitat patches, whereby characterizes a weaker deterministic effect of such interactions on the potential connectivity between patches with given spatial proximity, and represents a stronger effect. We maintained other parameters constant ( and ).
We initiated simulations by randomly allocating individuals to the nodes of each habitat networks and modelled individual movements and transmission dynamics for 500 timesteps. When modelling individual movements, we defined the probability for an individual to stay at the current node at each timestep consistently as , and the probability of moving from node to as (where is the weight of the link between node and ). We simulated pathogen transmissions in the population using an SIR epidemic model (Keeling and Eames 2005; Kermack and McKendrick 1927), where each individual is either susceptible (S), infectious (I) or recovered (R). We randomly set one individual (1% of the population) as infectious, and at each timestep, simulated infected individual infecting each of its susceptible neighbours in the same patch (if any) with probability . Individuals recovered and acquires immunity with the probability . We tracked the percentages of infected individuals in the simulated populations over time, and compared the mean percentages of infections observed from 100 replications.
We show that the landscape properties play a role in mediating the pathogen transmission dynamics. Importantly, large- (landscape geometry) and local- (the propensity for patches to be more connected) scale characteristics work together to shape the transmission dynamics of a simulated pathogen. A weaker relationship between inter-patch potential connectivity and inter-patch configurational features (as modelled by a lower value) increases the scale of global disease outbreaks, but this effect is most strongly realized in landscapes with a larger aspect ratio (Fig. 5a), such as a forest habitat in a narrow valley. Our results relate to existing literature linking social (or contact) network structures to patterns of disease transmissions—the stronger tendency for patches to be connected (i.e. ) and squarer landscapes (i.e. ) typically decrease the path length, or diameter (Fig. 5b), and clustering (Fig. 5c) of the habitat networks, and correspondingly, increase the pathogen outbreak size in simulated populations. Our results complement recent work demonstrating that the fragmentation of animal habitats can impact the transmission dynamics of pathogens (Silk et al. 2019), extending it by showing that the shape as well as the internal potential connectivity of a habitat is important.
Discussion
We present a multi-dimensional framework for simulating networks that can realistically capture the diverse physical configurations of animal habitats. Our model provides a tool to develop a more mechanistic understanding of the role of habitat configuration in modulating population processes and outcomes. Such modulating effects are likely to be widespread—for example, we have demonstrated that the structure of the habitat network can have consequences on pathogen transmission, in line with predictions from studies of social networks. Developing such mechanistic knowledge is critical as natural animal populations face increasingly rapid changes in their habitats, which have ecological and evolutionary consequences. For instance, habitat change can affect the magnitude of competition (Calizza et al. 2017) and the spread dynamics of pathogens (Bloomfield et al. 2020), information (Betts et al. 2008), or genes (Keller and Largiader 2003). By taking a bottom–up and spatially explicit approach to capture habitat configurations, our model can be tuned to approximate the potential connectivity of specific habitat configurations for a given species. From these, researchers can produce a range of alternative and realistic scenarios to explore the consequences of different features of the habitat on population processes, such as changes in the spatial arrangements of resource patches and physical barriers to movement.
The fundamental role of the physical environment on animal populations makes the evaluation of the consequences of habitat configuration relevant to both theorists and empiricists. If we do not explicitly consider habitat configuration, we risk missing the importance of its contribution to biological processes. For example, simulating social networks of large populations without considering spatial dependencies could produce networks that are more connected than they should be (i.e. without considering the spatial constraints on social interactions). Doing so can misrepresent the biological processes that network structure shapes, such as the transmissions of pathogens (White et al. 2018; Wilkinson et al. 2018), information (Aplin et al. 2015), genes (Vähä et al. 2007). By contrast, current geometric network models, which are spatially-dependent, may largely overestimate the spatial clustering of habitat components because it does not allow for rare long-distance connections or missing connections among close patches by forcing all closely located components to be connected. The need for models tailored to simulate habitat networks has been highlighted by recent studies that modelled specific habitat scenarios. For example, Carraro et al. (2020) proposed a toolkit for generating networks to capture the topological features of real riverine habitats to understand their role in shaping the key processes in freshwater ecology and evolution. The AHN model herein proposed is a more general and flexible framework for depicting habitat potential connectivity. Notably, when simulating networks, the AHN model allows any spatial distributions of habitat components in any landscape (i.e. by tuning and/or ), and provides a cluster of probability curves (i.e. by tuning the and/or in the ) to model the diverse patterns of potential connectivity among habitat components. In addition, the AHN model can generate alternative representations of the potential connectivity of given habitats, and allows control over the deviations of alternative scenarios from the specific habitats that can be used to generate scenarios. Thus, our proposed model can generate realistic habitat scenarios that are biologically meaningful.
There are many useful applications in generating realistic animal habitat scenarios. For example, there is growing interest in understanding the interplay between habitat physical configurations and individuals’ behaviours to predict the persistence of animal populations (Snijders et al. 2017), and explain the structure and composition of ecological communities (Altermatt and Holyoak 2012; Carraro et al. 2020). Rapid habitat changes are also a major threat to wildlife, as they can alter the movement patterns of individuals which may have consequences for populations (Collingham and Huntley 2000; Todd et al. 2009). As habitat changes, individual animals can experience different spatial distributions of resources and risks, which in turn can alter the patterns of both intraspecific (Banks et al. 2007) and interspecific (Farine et al. 2015; Meise et al. 2019) interactions among individuals, as well as other processes such as dispersal patterns and gene flow (Wey et al. 2015). In population ecology, for example, changes in habitat physical configurations can reduce rates of movements among neighbouring subpopulations, which potentially reduces gene flow at the scale of meta-populations (Keller and Largiader 2003) and impacts the persistence of populations (Frankham 2005). Likewise, changes in habitat physical configurations could alter the transmission of information within social networks (Barkoczi and Galesic 2016; Franz and Nunn 2009; Whitehead and Lusseau 2012) and other complex behavioural traits to specific social groups (Nowak et al. 2010; Ohtsuki et al. 2007; Stilwell et al. 2020). Furthermore, altered habitat physical configurations imply potential changes to the transmission dynamics of pathogens across populations (Green et al. 2006; Keeling et al. 2010; Riley 2007; Silk et al. 2019). Our simulations show that, as animal social networks, animal habitat network structures play an important role in shaping pathogen transmission, thus highlight a fundamental link between the physical habitat environments and emergent biological processes, such as the evolutionary dynamics of cooperation (Stilwell et al. 2020) and animal culture (Gruber et al. 2019; Somveille et al. 2018).
Understanding how habitat physical configuration interacts with behavioural and/or demographic dynamics is crucial to assess how vulnerable wild animal populations—and the ecological communities that they are part of (Ryser et al. 2019)—are to the consequences of habitat changes. Our model provides the necessary first step to integrating animal movement at various spatial scales into existing quantitative frameworks. We have demonstrated that the local properties of connectivity and large-scale properties of the landscape can work together to shape population outcomes, such as the spread of pathogens. Such insights can help us to make better predictions or generate new hypotheses on how population or community structures and dynamics are shaped by the physical configurational features of habitats, and how populations or communities might respond to changing physical habitat environments.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank L. M. Aplin, K. B. Beck, H. B. Brandl, J. Calatayud, M. Chimento, A. A. Maldonado-Chaparro, M. Ogino, D. Papageorgiou, and the four anonymous reviewers for their insightful comments on our manuscript, and the Max Planck Computing and Data Facility for computational support.
Author contribution statement
DRF and PH designed the model. PH conducted the analyses and drafted the manuscript. All the authors contributed to writing and editing.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Max Planck Society, a Deutsche Forschungsgemeinschaft Scientific Network grant (‘The role of interaction structure in eco-evolutionary dynamics (EcoEvoInteract)’, FA 1420/3-1) awarded to DRF and POM, a doctoral scholarship from the China Scholarship Council (No. 201706100183) to PH, a postdoctoral fellowship from CAPES-Brazil (88881.170254/2018–01) to MC, the DFG Centre of Excellence 2117 “Centre for the Advanced Study of Collective Behaviour” under Germany’s Excellence Strategy—EXC 2117—422037984, and a grant from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 850859) awarded to DRF.
Availability of data and material
Empirical data used are available from Friesen et al. (2019).
Code availability
The code for implementing the model is available in the R package AnimalHabitatNetwork on the CRAN and GitHub (https://github.com/ecopeng/AnimalHabitatNetwork, where the supplementary file AnimalHabitatNetwork/Examples/Examples.md for illustrating the use of our model is also deposited). The code for the simulations is available at https://github.com/ecopeng/Simulation_Code_AHN.
Declarations
Conflict of interest
We have no conflicting interest with this paper.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Contributor Information
Peng He, Email: phe@ab.mpg.de.
Damien R. Farine, Email: damien.farine@ieu.uzh.ch
References
- Albert R, Jeong H, Barabasi AL. Internet—diameter of the world-wide web. Nature. 1999;401:130–131. doi: 10.1038/43601. [DOI] [Google Scholar]
- Allen B, et al. Evolutionary dynamics on any population structure. Nature. 2017;544:227–230. doi: 10.1038/nature21723. [DOI] [PubMed] [Google Scholar]
- Altermatt F, Holyoak M. Spatial clustering of habitat structure effects patterns of community composition and diversity. Ecology. 2012;93:1125–1133. doi: 10.1890/11-1190.1. [DOI] [PubMed] [Google Scholar]
- Alther R, Altermatt F. Fluvial network topology shapes communities of native and non-native amphipods. Ecosphere. 2018;9:e02102. doi: 10.1002/ecs2.2102. [DOI] [Google Scholar]
- Altizer S, et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu Rev Ecol Evol S. 2003;34:517–547. doi: 10.1146/annurev.ecolsys.34.030102.151725. [DOI] [Google Scholar]
- Aplin LM, Farine DR, Morand-Ferron J, Sheldon BC. Social networks predict patch discovery in a wild population of songbirds. Proc R Soc B. 2012;279:4199–4205. doi: 10.1098/rspb.2012.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin LM, Farine DR, Morand-Ferron J, Cockburn A, Thornton A, Sheldon BC. Experimentally induced innovations lead to persistent culture via conformity in wild birds. Nature. 2015;518:538–541. doi: 10.1038/nature13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armansin NC, et al. Social barriers in ecological landscapes: the social resistance hypothesis. Trends Ecol Evol. 2020;35:137–148. doi: 10.1016/j.tree.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Baddeley A, Turner R. spatstat: An R package for analyzing spatial point patterns. J Stat Softw. 2005;12:1–42. doi: 10.18637/jss.v012.i06. [DOI] [Google Scholar]
- Baddeley A, Rubak E, Turner R (2015) Spatial point patterns: methodology and applications with R. Chapman and Hall/CRC Press
- Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C. Individual dispersal, landscape connectivity and ecological networks. Biol Rev. 2013;88:310–326. doi: 10.1111/brv.12000. [DOI] [PubMed] [Google Scholar]
- Bain GC, Hall ML, Mulder RA. Territory configuration moderates the frequency of extra-group mating in superb fairy-wrens. Mol Ecol. 2014;23:5619–5627. doi: 10.1111/mec.12959. [DOI] [PubMed] [Google Scholar]
- Banks SC, Piggott MP, Stow AJ, Taylor AC. Sex and sociality in a disconnected world: a review of the impacts of habitat fragmentation on animal social interactions. Can J Zool. 2007;85:1065–1079. doi: 10.1139/Z07-094. [DOI] [Google Scholar]
- Banks SC, Lindenmayer DB, McBurney L, Blair D, Knight EJ, Blyton MDJ. Kin selection in den sharing develops under limited availability of tree hollows for a forest marsupial. Proc R Soc B. 2011;278:2768–2776. doi: 10.1098/rspb.2010.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, Ravasz E, Vicsek T. Deterministic scale-free networks. Phys A. 2001;299:559–564. doi: 10.1016/S0378-4371(01)00369-7. [DOI] [Google Scholar]
- Baranyi G, Saura S, Podani J, Jordan F. Contribution of habitat patches to network connectivity: redundancy and uniqueness of topological indices. Ecol Indic. 2011;11:1301–1310. doi: 10.1016/j.ecolind.2011.02.003. [DOI] [Google Scholar]
- Barkoczi D, Galesic M. Social learning strategies modify the effect of network structure on group performance. Nat Commun. 2016;7:13109. doi: 10.1038/ncomms13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélemy M. Spatial networks. Phys Rep. 2011;499:1–101. doi: 10.1016/j.physrep.2010.11.002. [DOI] [Google Scholar]
- Bastille-Rousseau G, Douglas-Hamilton I, Blake S, Northrup JM, Wittemyer G. Applying network theory to animal movements to identify properties of landscape space use. Ecol Appl. 2018;28:854–864. doi: 10.1002/eap.1697. [DOI] [PubMed] [Google Scholar]
- Beninde J, et al. Cityscape genetics: structural vs. functional connectivity of an urban lizard population. Mol Ecol. 2016;25:4984–5000. doi: 10.1111/mec.13810. [DOI] [PubMed] [Google Scholar]
- Betts MG, Hadley AS, Rodenhouse N, Nocera JJ. Social information trumps vegetation structure in breeding-site selection by a migrant songbird. Proc R Soc B. 2008;275:2257–2263. doi: 10.1098/rspb.2008.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield LSP, McIntosh TL, Lambin EF. Habitat fragmentation, livelihood behaviors, and contact between people and nonhuman primates in Africa. Landscape Ecol. 2020;35:985–1000. doi: 10.1007/s10980-020-00995-w. [DOI] [Google Scholar]
- Bodin O, Norberg J. A network approach for analyzing spatially structured populations in fragmented landscape. Landscape Ecol. 2007;22:31–44. doi: 10.1007/s10980-006-9015-0. [DOI] [Google Scholar]
- Calabrese JM, Fagan WF. A comparison-shopper's guide to connectivity metrics. Front Ecol Environ. 2004;2:529–536. doi: 10.1890/1540-9295(2004)002[0529:Acgtcm]2.0.Co;2. [DOI] [Google Scholar]
- Calizza E, Costantini ML, Careddu G, Rossi L. Effect of habitat degradation on competition, carrying capacity, and species assemblage stability. Ecol Evol. 2017;7:5784–5796. doi: 10.1002/ece3.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor M, Farine DR. Simple foraging rules in competitive environments can generate socially structured populations. Ecol Evol. 2018;8:4978–4991. doi: 10.1002/ece3.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor M, et al. The importance of individual-to-society feedbacks in animal ecology and evolution. J Anim Ecol. 2020 doi: 10.1111/1365-2656.13336. [DOI] [PubMed] [Google Scholar]
- Cantor M, et al. Social network architecture and the tempo of cumulative cultural evolution. Proc R Soc B. 2021 doi: 10.1098/rspb.2020.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro L, et al. Generation and application of river network analogues for use in ecology and evolution. Ecol Evol. 2020;10:7537–7550. doi: 10.1002/ece3.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubaty AM, Galpern P, Doctolero SC. The r toolbox grainscape for modelling and visualizing landscape connectivity using spatially explicit networks. Methods Ecol Evol. 2020;11:591–595. doi: 10.1111/2041-210x.13350. [DOI] [Google Scholar]
- Clobert J, Baguette M, Benton TG, Bullock JM. Dispersal ecology and evolution. Oxford University Press; 2012. [Google Scholar]
- Collingham YC, Huntley B. Impacts of habitat fragmentation and patch size upon migration rates. Ecol Appl. 2000;10:131–144. doi: 10.2307/2640991. [DOI] [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. Int J Complex Syst. 2006;1695:1–9. [Google Scholar]
- Dale MRT, Fortin MJ. From graphs to spatial graphs. Annu Rev Ecol Evol Syst. 2010;41:21–38. doi: 10.1146/annurev-ecolsys-102209-144718. [DOI] [Google Scholar]
- Dall J, Christensen M. Random geometric graphs. Phys Rev E. 2002;66:016121. doi: 10.1103/PhysRevE.66.016121. [DOI] [PubMed] [Google Scholar]
- Davis JM. Patterns of variation in the influence of natal experience on habitat choice. Q Rev Biol. 2008;83:363–380. doi: 10.1086/592851. [DOI] [PubMed] [Google Scholar]
- Doherty TS, Fist CN, Driscoll DA. Animal movement varies with resource availability, landscape configuration and body size: a conceptual model and empirical example. Landscape Ecol. 2019;34:603–614. doi: 10.1007/s10980-019-00795-x. [DOI] [Google Scholar]
- Emlen ST, Oring LW. Ecology, sexual selection, and the evolution of mating systems. Science. 1977;197:215–223. doi: 10.1126/science.327542. [DOI] [PubMed] [Google Scholar]
- Erdős P, Rényi A. On the evolution of random graphs. B Int Statist Inst. 1960;38:343–347. [Google Scholar]
- Fagiolo G. Clustering in complex directed networks. Phys Rev E. 2007 doi: 10.1103/PhysRevE.76.026107. [DOI] [PubMed] [Google Scholar]
- Fahrig L. Non-optimal animal movement in human-altered landscapes. Funct Ecol. 2007;21:1003–1015. doi: 10.1111/j.1365-2435.2007.01326.x. [DOI] [Google Scholar]
- Fahrig L, Merriam G. Habitat patch connectivity and population survival. Ecology. 1985;66:1762–1768. doi: 10.2307/2937372. [DOI] [Google Scholar]
- Fahrig L, et al. Functional landscape heterogeneity and animal biodiversity in agricultural landscapes. Ecol Lett. 2011;14:101–112. doi: 10.1111/j.1461-0248.2010.01559.x. [DOI] [PubMed] [Google Scholar]
- Fall A, Fortin MJ, Manseau M, O'Brien D. Spatial graphs: principles and applications for habitat connectivity. Ecosystems. 2007;10:448–461. doi: 10.1007/s10021-007-9038-7. [DOI] [Google Scholar]
- Farine DR. Structural trade-offs can predict rewiring in shrinking social networks. J Anim Ecol. 2019;00:1–11. doi: 10.1111/1365-2656.13140. [DOI] [PubMed] [Google Scholar]
- Farine DR, Sheldon BC. Stable multi-level social structure is maintained by habitat geometry in a wild bird population. bioRxiv. 2019 doi: 10.1101/085944. [DOI] [Google Scholar]
- Farine DR, Aplin LM, Sheldon BC, Hoppitt W. Interspecific social networks promote information transmission in wild songbirds. Proc R Soc. 2015;282:20142804. doi: 10.1098/rspb.2014.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Lindenmayer DB. Landscape modification and habitat fragmentation: a synthesis. Global Ecol Biogeogr. 2007;16:265–280. doi: 10.1111/j.1466-8238.2007.00287.x. [DOI] [Google Scholar]
- Fletcher RJ, Revell A, Reichert BE, Kitchens WM, Dixon JD, Austin JD. Network modularity reveals critical scales for connectivity in ecology and evolution. Nat Commun. 2013 doi: 10.1038/ncomms3572. [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Burrell NS, Reichert BE, Vasudev D, Austin JD. Divergent perspectives on landscape connectivity reveal consistent effects from genes to communities. Curr Landscape Ecol Rep. 2016;1:67–79. doi: 10.1007/s40823-016-0009-6. [DOI] [Google Scholar]
- Frank K, Wissel C. Spatial aspects of metapopulation survival—from model results to rules of thumb for landscape management. Landscape Ecol. 1998;13:363–379. doi: 10.1023/A:1008054906030. [DOI] [Google Scholar]
- Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. doi: 10.1016/j.biocon.2005.05.002. [DOI] [Google Scholar]
- Franz M, Nunn CL. Network-based diffusion analysis: a new method for detecting social learning. Proc R Soc B. 2009;276:1829–1836. doi: 10.1098/rspb.2008.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen SK, Martone R, Rubidge E, Baggio JA, Ban NC. An approach to incorporating inferred connectivity of adult movement into marine protected area design with limited data. Ecol Appl. 2019;29:e01890. doi: 10.1002/eap.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpern P, Manseau M, Fall A. Patch-based graphs of landscape connectivity: a guide to construction, analysis and application for conservation. Biol Conserv. 2011;144:44–55. doi: 10.1016/j.biocon.2010.09.002. [DOI] [Google Scholar]
- Gilarranz LJ, Rayfield B, Linan-Cembrano G, Bascompte J, Gonzalez A. Effects of network modularity on the spread of perturbation impact in experimental metapopulations. Science. 2017;357:199–201. doi: 10.1126/science.aal4122. [DOI] [PubMed] [Google Scholar]
- Gosling LM. The alternative mating strategies of male topi, Damaliscus lunatus. Appl Anim Behav Sci. 1991;29:107–119. doi: 10.1016/0168-1591(91)90240-X. [DOI] [Google Scholar]
- Granovetter MS. The strength of weak ties. Am J Sociol. 1973;78:1360–1380. doi: 10.1086/225469. [DOI] [Google Scholar]
- Green DM, Kiss IZ, Kao RR. Modelling the initial spread of foot-and-mouth disease through animal movements. Proc R Soc B. 2006;273:2729–2735. doi: 10.1098/rspb.2006.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SJ, Boruff BJ, Grueter CC. From ridge tops to ravines: landscape drivers of chimpanzee ranging patterns. Anim Behav. 2020;163:51–60. doi: 10.1016/j.anbehav.2020.02.016. [DOI] [Google Scholar]
- Gruber T, Luncz L, Morchen J, Schuppli C, Kendal RL, Hockings K. Cultural change in animals: a flexible behavioural adaptation to human disturbance. Palgr Commun. 2019;5:64. doi: 10.1057/s41599-019-0271-4. [DOI] [Google Scholar]
- Haddad NM, et al. Habitat fragmentation and its lasting impact on Earth's ecosystems. Sci Adv. 2015;1:e1500052. doi: 10.1126/sciadv.1500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfelder J, et al. The allometry of movement predicts the connectivity of communities. Proc Natl Acad Sci USA. 2020;117:22274–22280. doi: 10.1073/pnas.2001614117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Farine DR (2019) AnimalHabitatNetwork: networks characterising the physical configurations of animal habitats. R package, version 0.1.0. https://CRAN.R-project.org/package=AnimalHabitatNetwork
- He P, Maldonado-Chaparro AA, Farine DR. The role of habitat configuration in shaping social structure: a gap in studies of animal social complexity. Behav Ecol Sociobiol. 2019;73:9. doi: 10.1007/s00265-018-2602-7. [DOI] [Google Scholar]
- Henriques-Silva R, Lindo Z, Peres-Neto PR. A community of metacommunities: exploring patterns in species distributions across large geographical areas. Ecology. 2013;94:627–639. doi: 10.1890/12-0683.1. [DOI] [PubMed] [Google Scholar]
- Hirt MR, Grimm V, Li YH, Rall BC, Rosenbaum B, Brose U. Bridging scales: allometric random walks link movement and biodiversity research. Trends Ecol Evol. 2018;33:701–712. doi: 10.1016/j.tree.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Hoover BA, Brunk KM, Jukkala G, Banfield N, Rypel AL, Piper WH. Early evidence of natal-habitat preference: Juvenile loons feed on natal-like lakes after fledging. Ecol Evol. 2021;11:1310–1319. doi: 10.1002/ece3.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilany A, Akçay E. Social inheritance can explain the structure of animal social networks. Nat Commun. 2016;7:12084. doi: 10.1038/ncomms12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MO. Average distance, diameter, and clustering in social networks with homophily. Lect Notes Comput Sc. 2008;5385:4–11. doi: 10.1007/978-3-540-92185-1_3. [DOI] [Google Scholar]
- Jacobson B, Grant JWA, Peres-Neto PR. The interaction between the spatial distribution of resource patches and population density: consequences for intraspecific growth and morphology. J Anim Ecol. 2015;84:934–942. doi: 10.1111/1365-2656.12365. [DOI] [PubMed] [Google Scholar]
- Jacoby DMP, Freeman R. Emerging network-based tools in movement ecology. Trends Ecol Evol. 2016;31:301–314. doi: 10.1016/j.tree.2016.01.011. [DOI] [PubMed] [Google Scholar]
- Jordano P. Chasing ecological interactions. Plos Biol. 2016;14:e1002559. doi: 10.1371/journal.pbio.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler PM. Sex roles and adult sex ratios: insights from mammalian biology and consequences for primate behaviour. Philo Trans R Soc B. 2017;372:20160321. doi: 10.1098/rstb.2016.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling MJ. The effects of local spatial structure on epidemiological invasions. Proc Roy Soc B. 1999;266:859–867. doi: 10.1098/rspb.1999.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M. The implications of network structure for epidemic dynamics. Theor Popul Biol. 2005;67:1–8. doi: 10.1016/j.tpb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Keeling MJ, Eames KTD. Networks and epidemic models. J R Soc Interface. 2005;2:295–307. doi: 10.1098/rsif.2005.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling MJ, Danon L, Vernon MC, House TA. Individual identity and movement networks for disease metapopulations. Proc Natl Acad Sci USA. 2010;107:8866–8870. doi: 10.1073/pnas.1000416107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller I, Largiader CR. Recent habitat fragmentation caused by major roads leads to reduction of gene flow and loss of genetic variability in ground beetles. Proc Roy Soc B. 2003;270:417–423. doi: 10.1098/rspb.2002.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermack WO, McKendrick AG. A contribution to the mathematical theory of epidemics. Proc R Soc A. 1927;115:700–721. doi: 10.1098/rspa.1927.0118. [DOI] [Google Scholar]
- Kokko H, Sutherland WJ. Ecological traps in changing environments: ecological and evolutionary consequences of a behaviourally mediated Allee effect. Evol Ecol Res. 2001;3:537–551. [Google Scholar]
- Kovalenko KE, Thomaz SM, Warfe DM. Habitat complexity: approaches and future directions. Hydrobiologia. 2012;685:1–17. doi: 10.1007/s10750-011-0974-z. [DOI] [Google Scholar]
- Laiolo P, Tella JL. Habitat fragmentation affects culture transmission: patterns of song matching in Dupont's lark. J Appl Ecol. 2005;42:1183–1193. doi: 10.1111/j.1365-2664.2005.01093.x. [DOI] [Google Scholar]
- Laiolo P, Tella JL. Landscape bioacoustics allow detection of the effects of habitat patchiness on population structure. Ecology. 2006;87:1203–1214. doi: 10.1890/0012-9658(2006)87[1203:Lbadot]2.0.Co;2. [DOI] [PubMed] [Google Scholar]
- Leu ST, Farine DR, Wey TW, Sih A, Bull CM. Environment modulates population social structure: experimental evidence from replicated social networks of wild lizards. Anim Behav. 2016;111:23–31. doi: 10.1016/j.anbehav.2015.10.001. [DOI] [Google Scholar]
- Loehle C. Social barriers to pathogen transmission in wild animal populations. Ecology. 1995;76:326–335. doi: 10.2307/1941192. [DOI] [Google Scholar]
- Lookingbill TR, Gardner RH, Ferrari JR, Keller CE. Combining a dispersal model with network theory to assess habitat connectivity. Ecol Appl. 2010;20:427–441. doi: 10.1890/09-0073.1. [DOI] [PubMed] [Google Scholar]
- Lovett GM, Jones CG, Turner MG, Weathers KC. Ecosystem function in heterogeneous landscapes. New York: Springer; 2005. [Google Scholar]
- Marcoux M, Lusseau D. Network modularity promotes cooperation. J Theor Biol. 2013;324:103–108. doi: 10.1016/j.jtbi.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Marini L, Bartomeus I, Rader R, Lami F. Species-habitat networks: a tool to improve landscape management for conservation. J Appl Ecol. 2019;56:923–928. doi: 10.1111/1365-2664.13337. [DOI] [Google Scholar]
- McDiarmid C, Steger A, Welsh DJA. Random planar graphs. J Comb Theory B. 2005;93:187–205. doi: 10.1016/j.jctb.2004.09.007. [DOI] [Google Scholar]
- Meise K, Franks DW, Bro-Jorgensen J. Using social network analysis of mixed-species groups in African savannah herbivores to assess how community structure responds to environmental change. Philos Trans R Soc B. 2019 doi: 10.1098/rstb.2019.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliano AB, et al. Hunter-gatherer multilevel sociality accelerates cumulative cultural evolution. Sci Adv. 2020 doi: 10.1126/sciadv.aax5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor ES, Urban DL. A graph-theory framework for evaluating landscape connectivity and conservation planning. Conserv Biol. 2008;22:297–307. doi: 10.1111/j.1523-1739.2007.00871.x. [DOI] [PubMed] [Google Scholar]
- Montiglio PO, McGlothlin JW, Farine DR. Social structure modulates the evolutionary consequences of social plasticity: a social network perspective on interacting phenotypes. Ecol Evol. 2018;8:1451–1464. doi: 10.1002/ece3.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Saito Y. Variation in social behavior within a spider mite genus, Stigmaeopsis (Acari: Tetranychidae) Behav Ecol. 2005;16:232–238. doi: 10.1093/beheco/arh157. [DOI] [Google Scholar]
- Mourier J, Vercelloni J, Planes S. Evidence of social communities in a spatially structured network of a free-ranging shark species. Anim Behav. 2012;83:389–401. doi: 10.1016/j.anbehav.2011.11.008. [DOI] [Google Scholar]
- Naka LN, Brumfield RT. The dual role of Amazonian rivers in the generation and maintenance of avian diversity. Sci Adv. 2018;4:eaar8575. doi: 10.1126/sciadv.aar8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandini S, Keerthipriya P, Vidya TNC. Seasonal variation in female Asian elephant social structure in Nagarahole-Bandipur, southern India. Anim Behav. 2017;134:135–145. doi: 10.1016/j.anbehav.2017.10.012. [DOI] [Google Scholar]
- Nathan R, et al. A movement ecology paradigm for unifying organismal movement research. Proc Natl Acad Sci USA. 2008;105:19052–19059. doi: 10.1073/pnas.0800375105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Tarnita CE, Antal T. Evolutionary dynamics in structured populations. Philos T R Soc B. 2010;365:19–30. doi: 10.1098/rstb.2009.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki H, Pacheco JM, Nowak MA. Evolutionary graph theory: breaking the symmetry between interaction and replacement. J Theor Biol. 2007;246:681–694. doi: 10.1016/j.jtbi.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose M. Random geometric graphs. New York: Oxford University Press; 2003. [Google Scholar]
- Phillipsen IC, Lytle DA. Aquatic insects in a sea of desert: population genetic structure is shaped by limited dispersal in a naturally fragmented landscape. Ecography. 2013;36:731–743. doi: 10.1111/j.1600-0587.2012.00002.x. [DOI] [Google Scholar]
- Plitzko SJ, Drossel B. The effect of dispersal between patches on the stability of large trophic food webs. Theor Ecol. 2015;8:233–244. doi: 10.1007/s12080-014-0247-3. [DOI] [Google Scholar]
- Poli C, Hightower J, Fletcher RJ. Validating network connectivity with observed movement in experimental landscapes undergoing habitat destruction. J Appl Ecol. 2020;57:1426–1437. doi: 10.1111/1365-2664.13624. [DOI] [Google Scholar]
- Prado F, Sheih A, West JD, Kerr B. Coevolutionary cycling of host sociality and pathogen virulence in contact networks. J Theor Biol. 2009;261:561–569. doi: 10.1016/j.jtbi.2009.08.022. [DOI] [PubMed] [Google Scholar]
- Prehn SG, Laesser BE, Clausen CG, Jonck K, Dabelsteen T, Brask JB. Seasonal variation and stability across years in a social network of wild giraffe. Anim Behav. 2019;157:95–104. doi: 10.1016/j.anbehav.2019.08.018. [DOI] [Google Scholar]
- R Development Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
- Raghunandan MA, Subramanian CA. Proceedings of the 11th international conference on autonomous agents and multiagent systems. Richland, SC, USA: International Foundation for Autonomous Agents and Multiagent Systems; 2012. Sustaining cooperation on networks: an analytical study based on evolutionary game theory; pp. 913–920. [Google Scholar]
- Read JM, Keeling MJ. Disease evolution on networks: the role of contact structure. Proc Roy Soc B-Biol Sci. 2003;270:699–708. doi: 10.1098/rspb.2002.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S. Large-scale spatial-transmission models of infectious disease. Science. 2007;316:1298–1301. doi: 10.1126/science.1134695. [DOI] [PubMed] [Google Scholar]
- Robertson EP, et al. Isolating the roles of movement and reproduction on effective connectivity alters conservation priorities for an endangered bird. Proc Natl Acad Sci USA. 2018;115:8591–8596. doi: 10.1073/pnas.1800183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser R, Haussler J, Stark M, Brose U, Rall BC, Guill C. The biggest losers: habitat isolation deconstructs complex food webs from top to bottom. Proc Biol Sci. 2019;286:20191177. doi: 10.1098/rspb.2019.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P, Mann J, Bansal S. Disease implications of animal social network structure: a synthesis across social systems. J Anim Ecol. 2018;87:546–558. doi: 10.1111/1365-2656.12786. [DOI] [PubMed] [Google Scholar]
- Silk MJ, et al. Integrating social behaviour, demography and disease dynamics in network models: applications to disease management in declining wildlife populations. Philos T R Soc B. 2019;374:20180211. doi: 10.1098/rstb.2018.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders L, Blumstein DT, Stanley CR, Franks DW. Animal social network theory can help wildlife conservation. Trends Ecol Evol. 2017;32:567–577. doi: 10.1016/j.tree.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Somveille M, Firth JA, Aplin LM, Farine DR, Sheldon BC, Thompson RN. Movement and conformity interact to establish local behavioural traditions in animal populations. Plos Comput Biol. 2018;14:e1006647. doi: 10.1371/journal.pcbi.1006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel O, Leu ST, Sih A, Bull CM. Socially interacting or indifferent neighbours? Randomization of movement paths to tease apart social preference and spatial constraints. Methods Ecol Evol. 2016;7:971–979. doi: 10.1111/2041-210x.12553. [DOI] [Google Scholar]
- Stilwell P, O'Brien S, Hesse E, Lowe C, Gardner A, Buckling A. Resource heterogeneity and the evolution of public goods cooperation. Evol Lett. 2020;4:155–163. doi: 10.1002/evl3.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandburg-Peshkin A, Farine DR, Couzin ID, Crofoot MC. Shared decision-making drives collective movement in wild baboons. Science. 2015;348:1358–1361. doi: 10.1126/science.aaa5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandburg-Peshkin A, Farine DR, Crofoot MC, Couzin ID. Habitat and social factors shape individual decisions and emergent group structure during baboon collective movement. Elife. 2017 doi: 10.7554/eLife.19505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PD, Fahrig L, Henein K, Merriam G. Connectivity is a vital element of landscape structure. Oikos. 1993;68:571–573. doi: 10.2307/3544927. [DOI] [Google Scholar]
- Tildesley MJ, et al. Impact of spatial clustering on disease transmission and optimal control. Proc Natl Acad Sci USA. 2010;107:1041–1046. doi: 10.1073/pnas.0909047107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischendorf L, Fahring L. On the usage and measurement of landscape connectivity. Oikos. 2000;90:7–19. doi: 10.1034/j.1600-0706.2000.900102.x. [DOI] [Google Scholar]
- Todd BD, Luhring TM, Rothermel BB, Gibbons JW. Effects of forest removal on amphibian migrations: implications for habitat and landscape connectivity. J Appl Ecol. 2009;46:554–561. doi: 10.1111/j.1365-2664.2009.01645.x. [DOI] [Google Scholar]
- Tokeshi M, Arakaki S. Habitat complexity in aquatic systems: fractals and beyond. Hydrobiologia. 2012;685:27–47. doi: 10.1007/s10750-011-0832-z. [DOI] [Google Scholar]
- Tuomainen U, Candolin U. Behavioural responses to human-induced environmental change. Biol Rev. 2011;86:640–657. doi: 10.1111/j.1469-185X.2010.00164.x. [DOI] [PubMed] [Google Scholar]
- Urban D, Keitt T. Landscape connectivity: a graph-theoretic perspective. Ecology. 2001;82:1205–1218. doi: 10.2307/2679983. [DOI] [Google Scholar]
- Urban DL, Minor ES, Treml EA, Schick RS. Graph models of habitat mosaics. Ecol Lett. 2009;12:260–273. doi: 10.1111/j.1461-0248.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- Vähä JP, Erkinaro J, Niemelä E, Primmer CR. Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol. 2007;16:2638–2654. doi: 10.1111/j.1365-294X.2007.03329.x. [DOI] [PubMed] [Google Scholar]
- van Schaik CP. The ecology of social relationships amongst female primates. Comparative socioecology: the behavioural ecology of humans and other mammals. 1. Oxford: Blackwell; 1989. [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of 'small-world' networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wey TW, Spiegel O, Montiglio PO, Mabry KE. Natal dispersal in a social landscape: considering individual behavioral phenotypes and social environment in dispersal ecology. Curr Zool. 2015;61:543–556. doi: 10.1093/czoolo/61.3.543. [DOI] [Google Scholar]
- White LA, Forester JD, Craft ME. Disease outbreak thresholds emerge from interactions between movement behavior, landscape structure, and epidemiology. P Natl Acad Sci USA. 2018;115:7374–7379. doi: 10.1073/pnas.1801383115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead H, Kahn B. Temporal and geographic variation in the social structure of female sperm whales. Can J Zool. 1992;70:2145–2149. doi: 10.1139/z92-289. [DOI] [Google Scholar]
- Whitehead H, Lusseau D. Animal social networks as substrate for cultural behavioural diversity. J Theor Biol. 2012;294:19–28. doi: 10.1016/j.jtbi.2011.10.025. [DOI] [PubMed] [Google Scholar]
- Wilkinson DA, Marshall JC, French NP, Hayman DTS. Habitat fragmentation, biodiversity loss and the risk of novel infectious disease emergence. J R Soc Interface. 2018;15:20180403. doi: 10.1098/rsif.2018.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS. A theory of group selection. Proc Natl Acad Sci USA. 1975;72:143–146. doi: 10.1073/pnas.72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MC, et al. Habitat fragmentation and biodiversity conservation: key findings and future challenges. Landscape Ecol. 2016;31:219–227. doi: 10.1007/s10980-015-0312-3. [DOI] [Google Scholar]
- Ziolkowska E, Ostapowicz K, Radeloff VC, Kuemmerle T. Effects of different matrix representations and connectivity measures on habitat network assessments. Landscape Ecol. 2014;29:1551–1570. doi: 10.1007/s10980-014-0075-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Empirical data used are available from Friesen et al. (2019).
The code for implementing the model is available in the R package AnimalHabitatNetwork on the CRAN and GitHub (https://github.com/ecopeng/AnimalHabitatNetwork, where the supplementary file AnimalHabitatNetwork/Examples/Examples.md for illustrating the use of our model is also deposited). The code for the simulations is available at https://github.com/ecopeng/Simulation_Code_AHN.