Abstract
Background
The liver is the most common site for haematogenous metastasis of gastric cancer, and liver metastasis is fatal.
Methods
We conducted a transcriptomic analysis between metastatic foci in the liver, primary tumour and adjacent tissues from gastric cancer patients with metastasis limited to the liver. We determined mRNA expression levels in tumour tissues of 300 patients with gastric cancer via quantitative RT-PCR. The oncogenic phenotypes of GNG4 were determined with knockdown, knockout and forced expression experiments. We established and compared subcutaneous and liver metastatic mouse xenograft models of gastric cancer to reveal the roles of GNG4 in tumorigenesis in the liver.
Results
GNG4 was upregulated substantially in primary gastric cancer tissues as well as liver metastatic lesions. High levels of GNG4 in primary cancer tissues were associated with short overall survival and the likelihood of liver recurrence. Functional assays revealed that GNG4 promoted cancer cell proliferation, the cell cycle and adhesiveness. Tumour formation by GNG4-knockout cells was moderately reduced in the subcutaneous mouse model and strikingly attenuated in the liver metastasis mouse model.
Conclusions
GNG4 expression may provide better disease monitoring for liver metastasis, and GNG4 may be a novel candidate therapeutic target for liver metastasis.
Subject terms: Surgical oncology, Prognostic markers, Translational research, Oncogenes
Background
Gastric cancer represents the third leading cause of death among malignancies.1–3 The 5-year overall survival rate, varying from 20 to 50%, remains dismal, and its trends have improved little or became rather flat in this decade.2 In the early stages, most instances of gastric cancer can be eradicated surgically.4 Distant metastasis will worsen the outcome of advanced gastric cancer, regardless of whether it is synchronous or metachronous. To minimise the risk of metachronous metastases, gastrectomy with perioperative chemotherapy or chemoradiotherapy is the standard therapy. Although several molecular-targeted drugs have also become available, they have only been recently considered indications for metastatic gastric cancer.5–7
The liver is the most common site for haematogenous metastasis of gastric cancer, yet the diagnostic and therapeutic strategies for treating liver metastasis remain to be further investigated.8 Although palliative therapy is standard care for liver metastasis, it is becoming more acceptable to perform single-lobe hepatectomy with R0 margins for a solitary nodule to improve the prognoses for either synchronous or metachronous liver metastases.8–10 Despite this aggressive trend, liver metastasis is still treated by ordinal systemic chemotherapy, unlike intraperitoneal chemotherapies that are being actively developed for peritoneal metastasis.11,12 It is also challenging to diagnose small metastases in the liver accurately prior to surgery. Moreover, early disease recurrence may imply that subclinical dissemination has occurred at the time of surgery.13,14
Gastric cancer cells metastasising to the liver need to acquire a certain gene profile specific to the liver when disseminating. Direct connection of the stomach to the liver via the portal vein might partially explain how liver metastasis of gastric cancer is highly prevalent, but this is yet only part of the ‘seed and soil’ theory. In 1989, Paget proposed that tumour cells have an affinity for certain organs that facilitate their initial survival and outgrowth.15 This theory developed over time, stating that cancer cells acquire diverse malignant phenotypes through various selective pressures, depending on the organs in which cancer cells metastasise during the process of their metastasis.16–18 Disseminated cancer cells in the portal vein circulation are exposed to hypoxia and anoikis, and only a subclone with suitable attributes for a microenvironment of the liver can proliferate to form liver metastasis.19 During recent decades, bioinformatics analyses using next-generation sequencing or microarray techniques have illustrated a genetic profile accounting for metastatic potential, and this has enabled innovative molecular targeting drugs.5–7,20–24 Given this achievement, there must be a unique gene profile that drives cancer cells to form metastases specific to the liver, but this gene profile has yet to be elucidated.
Hence, discovering novel genes that promote liver metastasis by an organ-oriented approach can shed light on strategies for advanced gastric cancer. We hypothesised that primary cancer must have the same gene profile as metastatic liver foci, and that a new driver oncogene could be discovered among these genes. In this study, we used a transcriptomic approach to identify the nucleotide-binding protein (G-protein) subunit gamma-4 gene (GNG4) as a candidate driver gene of liver metastasis. GNG4 is a member of the G-protein γ family, which typically transduces signals from upstream G-protein-coupled receptors (GPCRs).25–28 Some family members of G proteins were previously identified as oncogenes, and a pan-inhibitor of Gβγ reduced cell proliferation and impeded metastasis in prostate and breast cancer cell lines.29,30 GNG4 was, however, reported as a tumour suppressor gene in glioblastoma and renal cell carcinoma.31,32 This study revealed its malignant roles and impact on gastric cancer patient outcomes through expressional and functional studies of GNG4. To the best of our knowledge, this is the first study to reveal malignant roles for GNG4 in metastases and outcomes in gastric cancer.
Methods
Sample collection
For the transcriptome analysis, we used 12 surgically resected specimens consisting of three foci from each of the four patients: primary gastric cancer tissues, normal mucosae and liver metastases. These four gastric cancer patients had synchronous liver metastasis, but were free of other organ metastases (Fig. 1a). To validate the clinical significance of GNG4 expression, we acquired 300 primary gastric cancer specimens of stages I–IV as defined by the Union for International Cancer Control (UICC). These patients underwent gastric resection without preoperative treatment at our institution between 2001 and 2014 and were independent of the four patients for whom we conducted the transcriptomic analysis above. Since 2006, all of those with stage II–III disease underwent adjuvant chemotherapy using oral fluoropyrimidine (S1), unless the patient had a contraindication. Informed consent for the use of clinical samples and data was obtained from all patients in written manners, consistent with the requirement of the Institutional Review Board at Nagoya University, Japan. To validate the results from our institutional data, we employed the GSE62254 dataset and global gene expression experiments.33,34
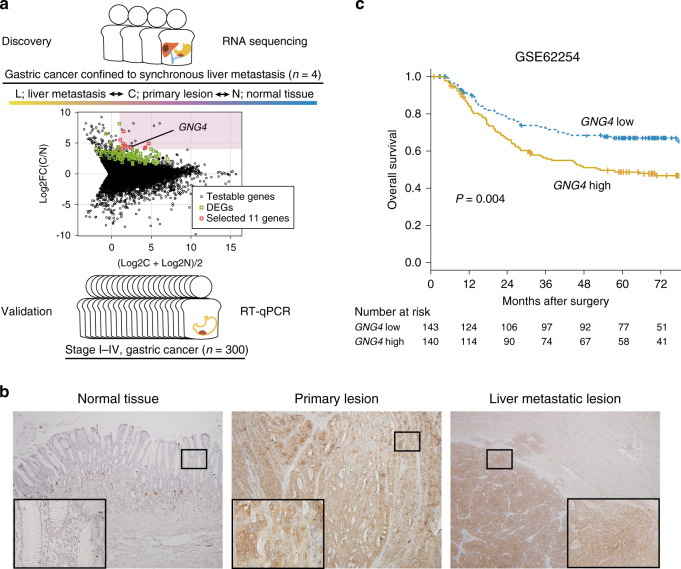
Fig. 1. GNG4 is identified as a promising oncogene to develop liver metastasis of gastric cancer.
The present study’s design is illustrated. In total, 57,749 genes were identified from the RNA-seq analysis of 12 specimens from gastric cancer patients with metastasis confined to the liver. The log-ratio and mean average (MA) plot of the expression levels of C and N was used to select eleven promising genes (a). DEGs indicate differentially expressed genes (upregulated). Immunohistochemistry of gastric cancer samples with liver metastasis of the adjacent gastric mucosa, primary tumour and liver metastatic lesion (b). The black bold square indicates the high-magnification area (×400). Kaplan–Meier plot of overall survival using the GSE62254 dataset stratified by the median value of GNG4 (c).
We acquired 14 human gastric cancer cell lines (AGS, GCIY, KATOIII, IM95, MKN1, MKN7, MKN45, MKN74, N87, NUGC2, NUGC3, NUGC4, OCUM1 and SC-6-JCK) and the non-tumorigenic tubular epithelial cell line FHs74 from the American Type Culture Collection (Manassas, VA, USA), the Japanese Collection of Research BioResources Cell Bank (Osaka, Japan) or our institute’s collection.
Gene expression analyses
For the global gene expression profiling analysis (RNA-seq), we applied the HiSeq Sequencing System (Illumina, San Diego, CA) to compare the expression levels of 57,749 genes as described previously.35 We confirmed GNG4 expression at the protein level by immunohistochemistry as described previously.36 Specifically, we used an anti-GNG4 monoclonal antibody (Product code, 13780-1-AP, Proteintech, Rosemont, IL, USA) diluted 1:50. We used quantitative RT-PCR (qRT-PCR) to determine GNG4 mRNA levels using previously published protocols.37 The primer sequence for qRT-PCR specifically designed for GNG4 is provided in Supplementary Table S1.
Immunoblot assays were performed using a rabbit anti-GNG4 monoclonal antibody diluted 1:500 as previously described.38 For pathway analyses, a capillary-based immunoassay was applied with a Wes and 12-230-kDa Jess or Wes Separation Module (Protein Simple, San Jose, CA, USA) following the manufacturer’s protocol.39 The anti-Akt (Ser473) antibody Duet #8200 and anti-ERK1/2 p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody Duet #8201 were purchased from Cell Signaling Technology (Danvers, MA, USA); ERK1/2 and Akt were diluted 1:50, and p-ERK1/2 and p-AKT were diluted 1:100.
Knockdown, knockout and forced expression of GNG4
For the knockdown assays, four GNG4-specific siRNA sequences (siGNG4s, Supplementary Table S1) were designed using software available online (iScore Designer, https://www.med.nagoya-u.ac.jp/neurogenetics/i_Score/i_score.html). The control nonspecific siRNA (siControl) was purchased from Bioneer (Daejeon, Korea). We transiently transfected 40 nM each siRNA into gastric cancer cells in the presence of LipoTrust EX Oligo (Hokkaido System Science, Sapporo, Japan).
For the forced expression of GNG4, GNG4 cDNA clones ligated as open-reading frame sequences into CMV Flexi Vector pFN21A (GenBank ID: EU621374.1) were purchased (product ID, FHC02107, Promega, Madison, WI, USA). The NEON® (Thermo Fisher) system was used to transfect 0.2 µg of the GNG4 vector into MKN1 cells (1 × 105). Analyses of phenotypes were performed from the second day of transfection.
We performed genome editing by applying the CRISPR/Cas9 system to generate stable GNG4-knockout (dGNG4) gastric cancer cell lines with two different guide RNAs (Supplementary Table S1), as reported previously.40 The GeneArt Genomic Cleavage Detection Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to evaluate the efficiency of genomic cleavage 72 h after the transfection of Cas9. The primer set is described in Supplementary Table S1.
Functional assays and mouse models of gastric cancer
Cell proliferation was evaluated using the Premix WST-8 Cell Proliferation Assay Kit (DOJINDO Inc., Kumamoto, Japan) as previously described.35 Distributions of cells in specific phases of the cell cycle were evaluated by applying the Cell Clock Cell Cycle Assay (Biocolor Ltd., Antrim, UK). To evaluate the apoptotic status, we used the Muse MitoPotential Kit (Merck) by assessing the mitochondrial membrane potential or simply stained cells with annexin-V as described previously.35,41 We tested drug sensitivity to 5FU using a WST-8 kit as described previously.35 Adhesion of gastric cancer cells to a solid matrix was evaluated with the CytoSelect 48 Well Cell Adhesion Assay Kit (Cell Biolabs, San Diego, CA, USA) as described previously.37
To evaluate the influence of GNG4 on tumorigenicity in vivo, we employed a mouse subcutaneous xenograft model as described previously (BALB/c-nu/nu, n = 3 each).40 We evaluated the role of GNG4 in liver metastasis using an already-established mouse model (NOD-SCID, n = 4 each) (CLEA Japan Inc., Tokyo, Japan).40,42 An IVIS Spectrum imaging system (Xenogen, Alameda, CA, USA) and magnetic resonance imaging (MRI) (MRS 3000, MR solutions, Guildford, UK) were employed to observe tumour formation in the livers of engrafted mice. For both mouse models, 4-week-old mice were habituated for 2 weeks before the assays were conducted. The mice were sacrificed by CO2 under inhalational anaesthesia eight or twelve weeks after implantation. All animal experiments were conducted in accordance with the ARRIVE guidelines and were approved by the Animal Research Committee of Nagoya University (no. 30143).
Statistical analysis
The significance of differences between the two variables was assessed using the Student’s t test for normally distributed data and the Wilcoxon test for non-normally distributed data. Fisher’s exact test was used to analyse the significance of categorical data. P values < 0.05 were considered statistically significant. For multiple comparisons, P values were adjusted by the Bonferroni–Hochberg method. For time-survival analyses, survival curves were generated using the Kaplan–Meier method, and Cox proportional hazard models were used to estimate the hazard ratio and P value. For the drug-sensitivity test, curve fitting was performed with four-parameter log-logistic models. The detailed method of transcriptomic analysis was described previously.35 All other statistical analyses were performed using R version 3.6.1 software (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Identification of GNG4 as a candidate driver gene of liver metastasis of gastric cancer
Global expression profiling was conducted to compare the expression levels of 57,749 genes between primary lesion, liver metastasis and adjacent normal tissues. The qualities of the RNA preparations were sufficient for analysis indicated as follows: yield data per sample = 2981 Mb, mean reads per sample = 29,512,162 pairs, mean rate ≥Q30 = 94.40%, mean quality score = 34.05 and mean total mapped-read rate = 89.38%. We filtered 57,749 genes to yield 11 genes according to five criteria: (1) a significant difference (Q < 0.05) between primary cancer lesions (C) and adjacent normal gastric tissues (N), (2) upregulated in C, (3) not significantly different (P ≥ 0.05) between C and liver metastatic lesions (H), (4) the base-2 logarithm of fold change between C and N expression levels [log2FC(C/N)] was >4 and (5) the average expression levels of C and N [(log2C + log2N)/2] were >1 (Fig. 1a and Table 1). Immunohistochemistry also showed that GNG4 was highly expressed in both liver metastatic lesions and primary tumours but not in normal mucosa (Fig. 1b).
Table 1.
Differentially expressed genes associated with liver metastasis.
| Gene symbol | Chromosomal locus | Function | C/N | L/C | ||
|---|---|---|---|---|---|---|
| Log2FC | Q | Log2FC | P | |||
| GNG4 | 1q42.3 | Signal transducer | 4.84 | 0.005 | 0.29 | 0.73 |
| TNFRSF11B | 8q24.12 | Apoptosis modulator | 4.57 | 0.005 | 0.53 | 0.43 |
| VGF | 7q22.1 | Growth factor | 5.46 | 0.005 | −0.81 | 0.34 |
| DNAJC12 | 10q21.3 | Protein modulator | 4.15 | 0.005 | −1.16 | 0.10 |
| RBP4 | 10q23.33 | Retinol carrier | 4.25 | 0.005 | 1.51 | 0.05 |
| SYT7 | 11q12.2 | Signal transducer | 4.29 | 0.005 | 0.30 | 0.63 |
| NPY | 7p15.3 | Signal transducer | 4.86 | 0.005 | 0.09 | 0.90 |
| THBS4 | 5q14.1 | Adhesion modulator | 4.01 | 0.005 | 0.95 | 0.28 |
| KRT6B | 12q13.13 | Structural molecule | 6.83 | 0.034 | −1.53 | 0.08 |
| FNDC1 | 6q25.3 | Signal transducer | 4.50 | 0.005 | −0.89 | 0.16 |
| UTS2R | 17q25.3 | Signal receptor | 4.50 | 0.005 | 0.50 | 0.57 |
C primary cancer tissue, N adjacent normal tissue, L liver metastasis, FC fold change.
We chose GNG4 for further analysis as a candidate driver gene that promotes liver metastasis, as the GSE62254 dataset revealed that high levels of GNG4 expression in gastric cancer tissues were associated with short overall survival (hazard ratio 1.73 (95% confidence interval, 1.20–2.50), P = 0.004) (Fig. 1c). As we obtained little information on GNG4,31 we identified this gene as a potential candidate for a therapeutic target and a biomarker for liver metastasis of gastric cancer and subjected it to further analysis.
Clinical implications of GNG4 expression in tumour tissues
Next, the diagnostic and predictive values of GNG4 were determined using qPCR by expanding the cohort to 300 gastric cancer patients ranging from UICC stage I to IV disease. In addition, we sought to discover the clinical implications of GNG4. By examining the GNG4 mRNA expression levels in cancer tissues among patients with stage I–IV disease, we found that GNG4 expression in patients with stage IV disease was significantly higher than that in patients with stage I and II–III disease, and there was no significant difference between those with stage I and III diseases (Fig. 2a). Among patients with stage IV disease, those with liver metastasis had higher GNG4 levels than those without liver metastasis. However, this was not the case for peritoneal and nodal distal metastases (Fig. 2b).
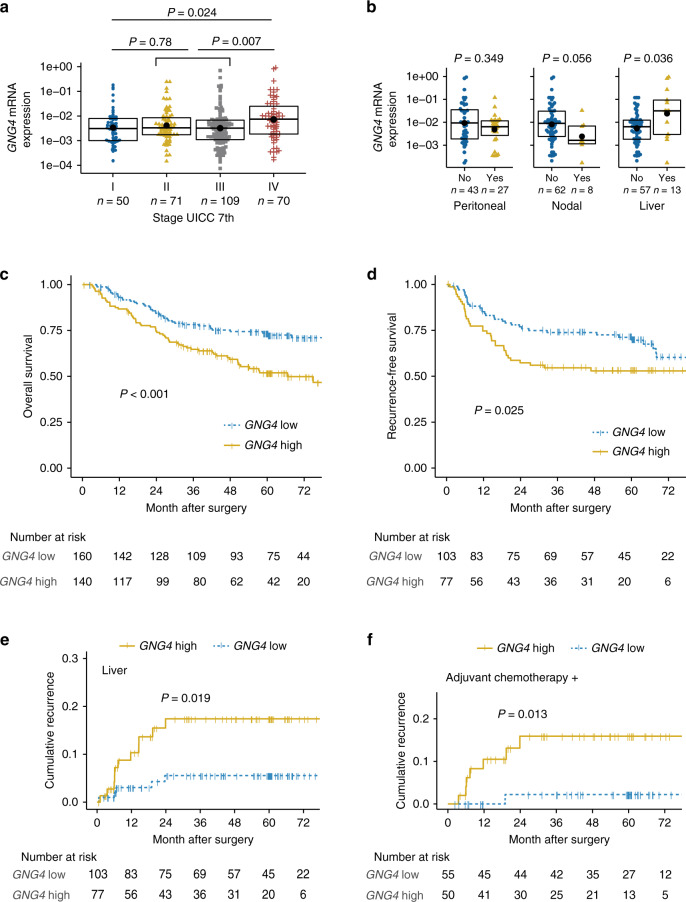
Fig. 2. High GNG4 expression correlated with the poor survival of patients with gastric cancer, especially in terms of liver metastasis.
GNG4 mRNA expression (GNG4/GAPDH) stratified by UICC stage (stage I vs. II+III vs. IV) (a) and the presence (yes/no) of three distant metastatic foci (peritoneal, lymph node and liver) among the UICC stage IV cohort (n = 70) (b). The box, bold bar and black point indicate the interquartile range, median and mean, respectively. Kaplan–Meier plot of overall survival for the whole cohort (n = 300) stratified by GNG4 expression (c) and disease survival analyses for the stage II and III cohorts (n = 180) (d). Liver-specific cumulative recurrence plot (stage II–III, n = 180) (e) and cumulative recurrence plot of the subgroup that received adjuvant chemotherapy (n = 105) (f).
We set the cut-off value to determine the implications of GNG4 expression. A receiver-operating characteristic (ROC) curve was generated to predict liver recurrence within one year postoperatively; the area under the curve was 0.696. By using this ROC curve, we defined high GNG4 expression as GNG4/GAPDH >0.004097 according to the Youden index (Supplementary Fig. S1a). Consistent with the association between UICC stage and GNG4 expression among the whole cohort, the high GNG4 group had significantly shorter overall survival than the low GNG4 group (hazard ratio, 1.94 (1.32–2.85), P < 0.001) (Fig. 2c).
Next, we focused on stage II–III cohort (n = 180) to determine the impact of GNG4 on prognosis, particularly recurrence and its pattern. All GNG4 clinicopathological variables were distributed similarly between the high and low GNG4 expression groups, including stage II or III (Supplementary Table S2). Nevertheless, the high GNG4 group exhibited poorer recurrence-free survival (hazard ratio, 1.73 (1.07–2.79), P = 0.025) (Fig. 2d). The cumulative recurrence rate for liver metastasis was significantly higher in the high GNG4 group than in the low GNG4 group (hazard ratio, 3.34 (1.16–9.63), P = 0.019), but this was not the case for either peritoneal or nodal recurrence (Fig. 2e and Supplementary Fig. S1b). Multivariate analysis demonstrated that high GNG4 expression was the only independent risk factor for liver recurrence (hazard ratio, 3.471 (1.20–10.0), P = 0.022) (Table 2).
Table 2.
Predictive factors of liver recurrence for 180 patients with R0 resection of stage II–III gastric cancer.
| Variables | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI, lower | 95% CI, upper | P | HR | 95% CI, lower | 95% CI, upper |
P | |
| Age >65 years old | 1.59 | 0.551 | 4.57 | 0.380 | ||||
| Male sex | 2.84 | 0.645 | 12.5 | 0.118 | ||||
| Tumour size >50 mm | 0.96 | 0.361 | 2.57 | 0.939 | ||||
| Tumour location (lower third) | 1.62 | 0.605 | 4.36 | 0.343 | ||||
| CA19-9 >37 IU/mL | 2.96 | 1.07 | 8.15 | 0.049* | 2.27 | 0.813 | 6.34 | 0.118 |
| CEA >5 ng/mL | 1.84 | 0.593 | 5.71 | 0.317 | ||||
| Differentiated | 2.59 | 0.943 | 7.14 | 0.059 | ||||
| Invasive growth type | 0.12 | 0.016 | 0.93 | 0.006* | 0.141 | 0.018 | 1.08 | 0.059 |
| Vessel involvement | 5.58 | 0.737 | 42.3 | 0.032* | 3.77 | 0.486 | 29.3 | 0.204 |
| Lymph node metastasis | 0.98 | 0.366 | 2.64 | 0.972 | ||||
| Tumour depth T4 (vs T1-3) | 2.35 | 0.534 | 10.4 | 0.210 | ||||
| UICC stage III | 1.00 | 0.370 | 2.68 | 0.993 | ||||
| High GNG4 | 3.34 | 1.16 | 9.628 | 0.019 | 3.471 | 1.201 | 10.027 | 0.022* |
CI confidence interval, CEA carcinoembryonic antigen, CA19-9 carbohydrate antigen 19-9, UICC Union for International Cancer Control.
*Statistically significant (P < 0.05).
Finally, we further tested whether GNG4 represents a biomarker for resistance to chemotherapy by classifying the stage II–III cohort (n = 180) into two subsets with and without chemotherapy. Among the only subset that received adjuvant chemotherapy, the high GNG4 group showed a significantly higher liver recurrence rate (hazard ratio, 8.32 (1.02–67.7), P = 0.013), but this difference was not significant among the subset that did not receive adjuvant chemotherapy (Fig. 2f and Supplementary Fig. S1c). In summary, we found that GNG4 is a candidate biomarker to diagnose synchronous metastasis and to predict metachronous metastasis, especially of the liver, and possibly to predict chemoresistance.
The oncogenic phenotype of GNG4 in gastric cancer cells
We explored the oncogenic functions of GNG4 on MKN1 and N87 cells because these two cell lines are derived from liver metastatic lesions from gastric cancer patients, and they differentially expressed GNG4 among the 14 gastric cancer cell lines analysed (Supplementary Fig. S2a, b). We observed that siGNG4 decreased MKN1 cell proliferation and deprived the stability of the mitochondrial membrane potential to promote apoptosis (Supplementary Fig. S2c–g). On the other hand, the temporarily forced expression of GNG4 in N87 cells exhibited the opposite results on the proliferation and MitoPotential assays (Fig. 3a, b).
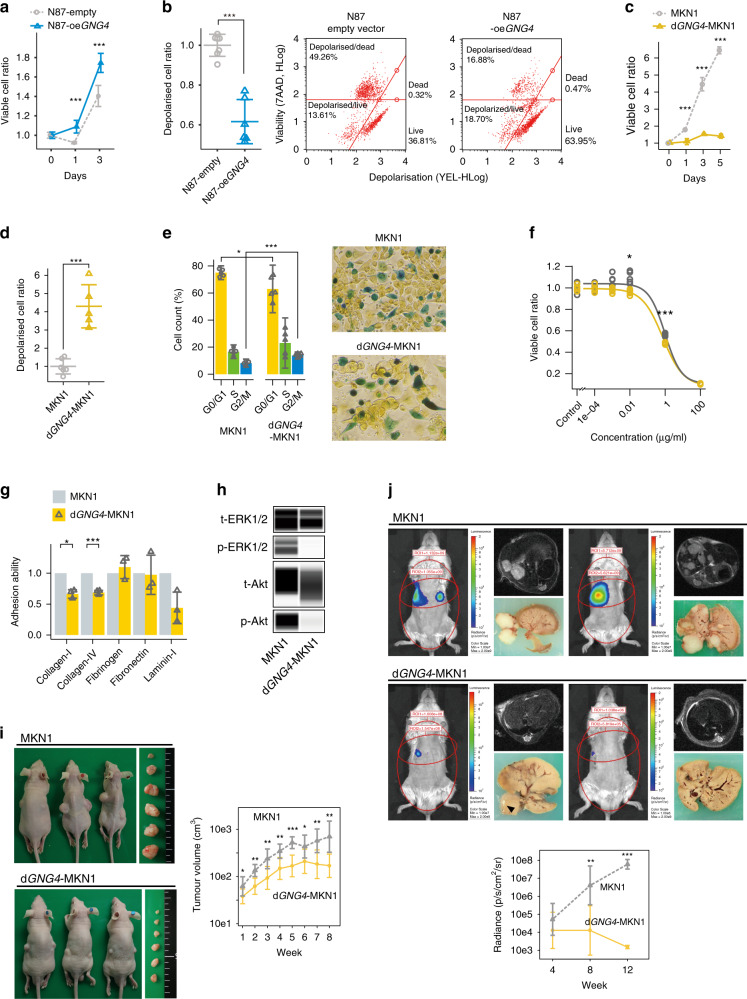
Fig. 3. GNG4 promotes malignant phenotypes of gastric cancer cell lines; in vitro and in vivo assays with transient overexpression of GNG4 (oeGNG4) and stable knockout of GNG4 (dGNG4).
Cell proliferation assay (a) and mitochondrial membrane depolarisation (MitoPotential) assay (b) between N87 cells transfected with an empty vector (N87-empty) and N87-oeGNG4 cells. Cell proliferation assay (c) and MitoPotential assay (d) between parental MKN1 and dGNG4-MKN1 cells. Cell cycle assay comparing MKN1 and dGNG4-MKN1 cells using colorimetric detection of the cell cycle phase (e). Drug-sensitivity assay to 5FU (f) and cell adhesion assay to solid matrices (g) comparing MKN1 and dGNG4-MKN1 cells. Capillary-based immunoassays of phospho-extracellular signal-regulated kinase 1/2 (ERK1/2) (Thr202/Tyr204), total ERK1/2, phospho-Akt (Ser473) and total Akt comparing MKN1 and dGNG4-MKN1 cells (h). p- indicates phosphorylated; t-, total. Mouse subcutaneous xenograft model (i) and liver metastatic model (j) comparing MKN1 and dGNG4-MKN1 cells. Tumour volume was evaluated with an in vivo spectrum imaging system (IVIS) at 4, 8 and 12 weeks after implantation (plotted chronologically in the lower panel) and by MRI at 12 weeks after implantation. Circles on the mice indicate regions of interest. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we used MKN1 cells for genome editing to perform further assays, including in vivo assays, because MKN1 cells had the highest GNG4 expression and were successfully engrafted in nude mice subcutaneously and engrafted in Nod-SCID mice for liver metastasis xenograft models.42 By applying the CRISPR/Cas9 system, we harvested two GNG4-knockout (dGNG4) MKN1 cell lines, one of which was treated with gRNA-GNG4-1 (Supplementary Table S1), resulting in successful stability. GNG4 knockout was confirmed by amplicon cleavage and immunoblot assays (Supplementary Fig. S3a, b, respectively). dGNG4-MKN1 cells demonstrated decreased proliferation and increased proportions of depolarised MitoPotential cells (Fig. 3c, d, and Supplementary Fig. S3c, S3d). dGNG4-MKN1 cells appeared to be arrested in the S/G2 phase and seemed to be apoptotic (Fig. 3e and Supplementary Fig. S3e). In summary, these results indicated that GNG4 promotes the cell cycle to increase proliferation and maintain the polarisation of the mitochondrial membrane to evade apoptosis, and was consistent with the transient knock-in and knockdown assays described above.
We further conducted tests to explore other possible roles of GNG4 (e.g., adherence to the extracellular matrix to form a scaffold for metastasis and avoidance of cell death from a cytotoxic agent). dGNG4-MKN1 cells demonstrated significantly less adherence to collagen I and collagen IV than parental MKN1 cells (Fig. 3f). A drug-sensitivity test demonstrated that dGNG4-MKN1 cells were more sensitive to 5FU than their parental MKN1 cells (Fig. 3g). As we are not aware of any genes with which GNG4 has oncogenic interactions, we conducted pathway analysis. Extracellular signal-regulated kinase 1/2 (ERK1/2) was markedly dephosphorylated, and Akt was moderately dephosphorylated in dGNG4-MKN1 cells compared to parental MKN1 cells (Fig. 3h).
Next, we employed two different xenograft mouse models, subcutaneous and liver metastasis models, to determine how much GNG4 influences tumour formation and growth in the liver. The subcutaneous tumours generated with dGNG4-MKN1 cells did not grow as much as parental MKN1 cells and even stopped growing after six weeks of seeding. The volumes of subcutaneous tumours (= D × d2/2) generated with dGNG4-MKN1 cells were significantly smaller than those generated with parental MKN1 cells (Fig. 3i). Subsequently, we employed a xenograft model of liver metastasis to determine the contribution of GNG4 to the metastatic potential of gastric cancer cells. Metastatic nodules in the liver multiplied 12 weeks after the implantation of parental MKN1 cells. In contrast, those of dGNG4-MKN1 cells were significantly smaller or not detected macroscopically or through IVIS or MRI (Fig. 3j).
Discussion
The outcome of gastric cancer remains dismal, and diagnostic and therapeutic strategies for liver metastasis have not been developed substantially.1–3 The fact that the liver is the most common site for haematogenous metastasis of gastric cancer inspired us to hypothesise that the gene that was overexpressed in the primary gastric tumour was the same as that in the metastatic liver foci. GNG4 was identified as a candidate driver gene for liver metastasis by transcriptome analyses revealing that GNG4 was distinctly overexpressed in both the primary tumour tissues and liver metastatic foci of patients with synchronous liver metastases who were free of other organ metastases. Subsequent qRT-PCR analyses of 300 gastric cancer tissues revealed that higher GNG4 expression correlated with a worse prognosis, particularly regarding the recurrence of liver metastasis. GNG4 expression in a gastric cancer cell line was associated with malignant phenotypes, especially the formation of liver metastases.
GNG4 is a member of the G-protein γ family, which forms heterotrimers with the α- and β-subunits of G proteins. G proteins typically act as switches that transduce signals from upstream GPCRs.25–28 Although GNB1 and GNG2 are well-known oncogenes,25,26,43 we were not aware of the malignant roles of GNG4. Many different mechanisms of Gβγ-mediated ERK1/2 activation have been reported, and each pathway is considered to require a specific cell type, GPCR and G protein,44 that is, the activation of intracellular effectors such as PI3K/Akt,45 the recruitment and formation of a scaffold for MAPK activation46 and the activation of a classic MAPK signalling pathway via the transactivation of an RTK.47 The only studies published implicating GNG4 were those of tumour suppressors in glioblastoma multiform and bladder cancer.31,32 In glioblastoma, GNG4 impedes a signal activating the ERK1/2 axis from CXCR4, a member of the GPCR family.31 Interestingly, although these arguments seemingly opposed our study, CXCR4 was identified as a gene that promotes the peritoneal metastasis of gastric cancer.48 Collectively, these results support the hypothesis that GNG4 promotes liver metastasis instead of peritoneal metastasis.
We are unaware of any oncogenic phenotypes or pathways mediated by GNG4. Several functional assays were performed to reveal the roles of GNG4, such as promoting cell adhesion, cell survival and cell proliferation. To form liver metastasis, disseminated gastric cancer cells need to resist anoikis until attaching to the sinusoidal capillary surface and forming a supportive niche to survive until outgrowth.49 GNB2, which is a member of the G-protein family, reportedly maintains mitochondrial membrane polarity independently.28 Our data showed that GNG4 similarly helps gastric cancer cells evade apoptosis. We characterised the adhesion abilities to several extracellular matrix molecules and revealed that GNG4 promotes adhesion to collagen I and IV. Lung cancer cells with high potential to promote liver metastasis express collagen IV at high levels in the extracellular matrix. Collagen IV activates FAK/PI3K and MEK/ERK signalling through integrin α2/β1.50 We infer that GNG4 may promote adhesion to the sinusoid endothelium cooperatively with collagen IV to form a supportive niche. We indicated that GNG4 promoted gastric cancer cells to specifically form liver metastases through an in vivo study. Notably, these oncogenic phenotypes of GNG4 in vitro were not only reproduced through a subcutaneous model but also much more apparent in a liver metastatic mouse model. This result implies that GNG4 promotes liver metastasis of gastric cancer cells by adapting to the microenvironment of the liver specifically. As we discussed above, every Gβγ-mediated oncogenic pathway requires a specific cell type, GPCR and G protein. We showed that GNG4 mediates the phosphorylation of Akt and the ERK1/2 axis. This observation is similar to that of GNB1 and GNG2, which also activate this well-known oncogenic pathway.25,26,43
The findings of our epidemiological data validated GNG4 to be a biomarker in gastric cancer, especially in the context of liver metastasis. Higher expression of GNG4 in primary gastric cancer tissue was significantly associated with a poorer prognosis caused mainly by liver recurrence. In addition, the high GNG4 group had less benefit from adjuvant chemotherapy aimed at preventing liver recurrence. We chose 5FU as a subject drug for testing because S1 was prescribed routinely to our institutional cohort otherwise contraindicated. The drug-sensitivity test demonstrated that GNG4 is not only a candidate biomarker predicting resistance to chemotherapy, but may also have functional aspects of resistance to 5FU, a representative pyrimidine drug.
The identification of GNG4 as a candidate gene that promotes liver metastasis in gastric cancer can lead to a novel therapeutic or diagnostic strategy. If we recognise the risk of liver metastasis by testing gastric cancer tissues, we may better monitor disease progression by examining liver metastasis using frequent MRI as needed.51 Of note, most cases of liver recurrence in this study occurred within six months to one year after surgery, implying the existence of subclinical dissemination to the liver at the time of surgery. The identification of such a cohort at high risk may enable us to uncover further benefits from perioperative therapy. For these patients, we may selectively apply oxaliplatin-based regimens, which reportedly may control haematogenous metastasis rather than peritoneal metastasis.52–55 Moreover, the discovery of genes specific to liver metastasis, such as GNG4, may also help us to develop molecular-targeted drugs.
This study has certain limitations. First, our GNG4 mRNA expression data were retrospectively acquired, and prospective studies with extensive cohorts are required to validate this gene as a biomarker. Second, we may have to further investigate the mechanisms through which GNG4 promotes liver metastasis. For instance, it may be proven through an organoid model of the sinusoid epithelium. Third, we have not yet shown the specificity of GNG4 for liver metastases in vivo. Further studies using an orthotopic model may be needed to prove whether GNG4 expression affects the metastasising organ directionality of cancer cells.56
Our results showed that higher GNG4 expression correlated with a poorer prognosis, especially in the context of liver metastasis. We also demonstrated that GNG4 likely plays roles in promoting the cell cycle, evading apoptosis and promoting tumour cell adhesion. GNG4 may therefore represent a specific biomarker for detection and a promising therapeutic target for liver metastasis of gastric cancer.
Supplementary information
Acknowledgements
We thank Edanz Group (www.edanzediting.com/ac) and Springer Nature Author Services for editing a draft of this paper. This paper was posted on medRxiv prior to submission. https://medrxiv.org/cgi/content/short/2020.08.14.20175034v1; doi: 2020.08.14.20175034v1.
Author contributions
Study concept and design: M.K. and Y.K. Acquisition of the data: M.K., H.T., T.M., S.U., K.S., C.T. and Y.K. Management of data acquisition: M.K., C.T., D.K., M.H., S.Y., G.N., M.K. and Y.K. Analysis of the present data: H.T., M.K. and Y.K. Statistical analysis: H.T. and M.K. Critical interpretation of the present data: H.T., M.K., T.M., S.U., K.S., C.T., D.K., M.H., S.Y., G.N., M.K. and Y.K. Drafting of the paper: H.T., M.K. and Y.K. Critical revision of the paper for important intellectual content: H.T., M.K., T.M., S.U., K.S., C.T., D.K., M.H., S.Y., G.N., M.K. and Y.K. Obtained funding: H.T., M.K. and Y.K. Technical or material support: H.T., M.K., T.M., S.U., K.S., C.T., D.K. and M.K. Study supervision: Y.K.
Ethics approval and consent to participate
Informed consent for the use of clinical samples and data was obtained from all patients in written manners, consistent with the requirement of the Institutional Review Board at Nagoya University, Japan. All animal experiments were conducted in accordance with the ARRIVE guidelines and were approved by the Animal Research Committee of Nagoya University (no. 30143).
Consent to publish
Not applicable.
Data availability
Data will be available as needed.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by a Grant-in-Aid for the Encouragement of Young Scientists (2017, B, 17K16538), the Japanese Society for Gastroenterological Carcinogenesis (2016), Nakayama Cancer Research Institute (2016) and the Yokoyama Foundation for Clinical Pharmacology (2016).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01366-1.
References
- 1.Tan P, Yeoh KG. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149:1153–1162. doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 4.Hiki N, Katai H, Mizusawa J, Nakamura K, Nakamori M, Yoshikawa T, et al. Long-term outcomes of laparoscopy-assisted distal gastrectomy with suprapancreatic nodal dissection for clinical stage I gastric cancer: a multicenter phase II trial (JCOG0703) Gastric Cancer. 2018;21:155–161. doi: 10.1007/s10120-016-0687-0. [DOI] [PubMed] [Google Scholar]
- 5.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 6.Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 7.Shitara K, Ozguroglu M, Bang YJ, Di Bartolomeo M, Mandala M, Ryu MH, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. 2018;392:123–133. doi: 10.1016/S0140-6736(18)31257-1. [DOI] [PubMed] [Google Scholar]
- 8.Kodera Y, Fujitani K, Fukushima N, Ito S, Muro K, Ohashi N, et al. Surgical resection of hepatic metastasis from gastric cancer: a review and new recommendation in the Japanese gastric cancer treatment guidelines. Gastric Cancer. 2014;17:206–212. doi: 10.1007/s10120-013-0299-x. [DOI] [PubMed] [Google Scholar]
- 9.Markar SR, Mikhail S, Malietzis G, Athanasiou T, Mariette C, Sasako M, et al. Influence of surgical resection of hepatic metastases from gastric adenocarcinoma on long-term survival: systematic review and pooled analysis. Ann. Surg. 2016;263:1092–1101. doi: 10.1097/SLA.0000000000001542. [DOI] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi N, Kanda M, Yoshikawa T, Takiguchi N, Fujitani K, Miyamoto K, et al. A randomized phase II multicenter trial to explore efficacy of weekly intraperitoneal in comparison with intravenous paclitaxel administered immediately after gastrectomy to the patients with high risk of peritoneal recurrence: final results of the INPACT trial. Gastric Cancer. 2018;21:1014–1023. doi: 10.1007/s10120-018-0817-y. [DOI] [PubMed] [Google Scholar]
- 12.Ishigami H, Fujiwara Y, Fukushima R, Nashimoto A, Yabusaki H, Imano M, et al. Phase III trial comparing intraperitoneal and intravenous paclitaxel plus S-1 versus cisplatin plus S-1 in patients with gastric cancer with peritoneal metastasis: PHOENIX-GC trial. J. Clin. Oncol. 2018;36:1922–1929. doi: 10.1200/JCO.2018.77.8613. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto H, Attiyeh MA, Gerold JM, Makohon-Moore AP, Hayashi A, Hong J, et al. The evolutionary origins of recurrent pancreatic cancer. Cancer Discov. 2020;10:792–805. doi: 10.1158/2159-8290.CD-19-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amikura K, Kobari M, Matsuno S. The time of occurrence of liver metastasis in carcinoma of the pancreas. Int. J. Pancreatol. 1995;17:139–146. doi: 10.1007/BF02788531. [DOI] [PubMed] [Google Scholar]
- 15.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 16.Mathot L, Stenninger J. Behavior of seeds and soil in the mechanism of metastasis: a deeper understanding. Cancer Sci. 2012;103:626–631. doi: 10.1111/j.1349-7006.2011.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza M, Khanna C. Revisiting the seed and soil in cancer metastasis. Int. J. Biochem Cell Biol. 2009;41:1452–1462. doi: 10.1016/j.biocel.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Groot AE, Roy S, Brown JS, Pienta KJ, Amend SR. Revisiting seed and soil: examining the primary tumor and cancer cell foraging in metastasis. Mol. Cancer Res. 2017;15:361–370. doi: 10.1158/1541-7786.MCR-16-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge of gastric cancer. Histol. Histopathol. 2018;33:11–26. doi: 10.14670/HH-11-898. [DOI] [PubMed] [Google Scholar]
- 20.Brosnan JA, Iacobuzio-Donahue CA. A new branch on the tree: next-generation sequencing in the study of cancer evolution. Semin Cell Dev. Biol. 2012;23:237–242. doi: 10.1016/j.semcdb.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim R, Schell MJ, Teer JK, Greenawalt DM, Yang M, Yeatman TJ. Co-evolution of somatic variation in primary and metastatic colorectal cancer may expand biopsy indications in the molecular era. PLoS ONE. 2015;10:e0126670. doi: 10.1371/journal.pone.0126670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka H, Kanda M, Shimizu D, Tanaka C, Kobayashi D, Hayashi M, et al. FAM46C serves as a predictor of hepatic recurrence in patients with resectable gastric cancer. Ann. Surg. Oncol. 2016;24:3438–3445. doi: 10.1245/s10434-016-5636-y. [DOI] [PubMed] [Google Scholar]
- 23.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 24.Menard S, Pupa SM, Campiglio M, Tagliabue E. Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003;22:6570–6578. doi: 10.1038/sj.onc.1206779. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Regalado A, Guzman-Hernandez ML, Ramirez-Rangel I, Robles-Molina E, Balla T, Vazquez-Prado J, et al. G protein-coupled receptor-promoted trafficking of Gbeta1gamma2 leads to AKT activation at endosomes via a mechanism mediated by Gbeta1gamma2-Rab11a interaction. Mol. Biol. Cell. 2008;19:4188–4200. doi: 10.1091/mbc.e07-10-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan SM, Sleno R, Gora S, Zylbergold P, Laverdure JP, Labbe JC, et al. The expanding roles of Gbetagamma subunits in G protein-coupled receptor signaling and drug action. Pharm. Rev. 2013;65:545–577. doi: 10.1124/pr.111.005603. [DOI] [PubMed] [Google Scholar]
- 27.Crespo P, Xu N, Simonds WF, Gutkind JS. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Liu W, Liu J, Xiao W, Liu L, Jiang C, et al. G-protein beta2 subunit interacts with mitofusin 1 to regulate mitochondrial fusion. Nat. Commun. 2010;1:101. doi: 10.1038/ncomms1099. [DOI] [PubMed] [Google Scholar]
- 29.Bookout AL, Finney AE, Guo R, Peppel K, Koch WJ, Daaka Y. Targeting Gbetagamma signaling to inhibit prostate tumor formation and growth. J. Biol. Chem. 2003;278:37569–37573. doi: 10.1074/jbc.M306276200. [DOI] [PubMed] [Google Scholar]
- 30.Tang X, Sun Z, Runne C, Madsen J, Domann F, Henry M, et al. A critical role of Gbetagamma in tumorigenesis and metastasis of breast cancer. J. Biol. Chem. 2011;286:13244–13254. doi: 10.1074/jbc.M110.206615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal J, Patil V, Mondal B, Shukla S, Hegde AS, Arivazhagan A, et al. Epigenetically silenced GNG4 inhibits SDF1alpha/CXCR4 signaling in mesenchymal glioblastoma. Genes Cancer. 2016;7:136–147. doi: 10.18632/genesandcancer.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Fang L, Zang Y, Xu Z. Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Med Sci. Monit. 2018;24:3024–3033. doi: 10.12659/MSM.909514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 34.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka H, Kanda M, Miwa T, Tanaka C, Kobayashi D, Umeda S, et al. Pattern-specific transcriptomics identifies ASGR2 as a predictor of hematogenous recurrence of gastric cancer. Mol. Cancer Res. 2018;16:1420–1429. doi: 10.1158/1541-7786.MCR-17-0467. [DOI] [PubMed] [Google Scholar]
- 36.Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, et al. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int. J. Oncol. 2014;44:44–52. doi: 10.3892/ijo.2013.2165. [DOI] [PubMed] [Google Scholar]
- 37.Kanda M, Tanaka C, Kobayashi D, Tanaka H, Shimizu D, Shibata M, et al. Epigenetic suppression of the immunoregulator MZB1 is associated with the malignant phenotype of gastric cancer. Int. J. Cancer. 2016;139:2290–2298. doi: 10.1002/ijc.30286. [DOI] [PubMed] [Google Scholar]
- 38.Oya H, Kanda M, Koike M, Iwata N, Niwa Y, Shimizu D, et al. Detection of serum melanoma-associated antigen D4 in patients with squamous cell carcinoma of the esophagus. Dis. Esophagus. 2016;29:663–669. doi: 10.1111/dote.12373. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki, K., Toshimitsu, K., Matano, M., Fujita, M., Fujii, M., Togasaki, K. et al. An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell183, 1420–1435 (2020). [DOI] [PubMed]
- 40.Kanda M, Tanaka H, Shimizu D, Miwa T, Umeda S, Tanaka C, et al. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018;37:5355–5366. doi: 10.1038/s41388-018-0335-8. [DOI] [PubMed] [Google Scholar]
- 41.Kanda M, Shimizu D, Sawaki K, Nakamura S, Umeda S, Miwa T, et al. Therapeutic monoclonal antibody targeting of neuronal pentraxin receptor to control metastasis in gastric cancer. Mol. Cancer. 2020;19:131. doi: 10.1186/s12943-020-01251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miwa T, Kanda M, Umeda S, Tanaka H, Shimizu D, Tanaka C, et al. Establishment of peritoneal and hepatic metastasis mouse xenograft models using gastric cancer cell lines. Vivo. 2019;33:1785–1792. doi: 10.21873/invivo.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng JQ, Lindsley CW, Cheng GZ, Yang H, Nicosia SV. The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene. 2005;24:7482–7492. doi: 10.1038/sj.onc.1209088. [DOI] [PubMed] [Google Scholar]
- 44.Rozengurt E. Signal transduction pathways in the mitogenic response to G protein-coupled neuropeptide receptor agonists. J. Cell Physiol. 1998;177:507–517. doi: 10.1002/(SICI)1097-4652(199812)177:4<507::AID-JCP2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Hawes BE, Luttrell LM, van Biesen T, Lefkowitz RJ. Phosphatidylinositol 3-kinase is an early intermediate in the G beta gamma-mediated mitogen-activated protein kinase signaling pathway. J. Biol. Chem. 1996;271:12133–12136. doi: 10.1074/jbc.271.21.12133. [DOI] [PubMed] [Google Scholar]
- 46.Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, et al. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 47.Della Rocca GJ, Maudsley S, Daaka Y, Lefkowitz RJ, Luttrell LM. Pleiotropic coupling of G protein-coupled receptors to the mitogen-activated protein kinase cascade. Role of focal adhesions and receptor tyrosine kinases. J. Biol. Chem. 1999;274:13978–13984. doi: 10.1074/jbc.274.20.13978. [DOI] [PubMed] [Google Scholar]
- 48.Yasumoto K, Koizumi K, Kawashima A, Saitoh Y, Arita Y, Shinohara K, et al. Role of the CXCL12/CXCR4 axis in peritoneal carcinomatosis of gastric cancer. Cancer Res. 2006;66:2181–2187. doi: 10.1158/0008-5472.CAN-05-3393. [DOI] [PubMed] [Google Scholar]
- 49.Mielgo A, Schmid MC. Liver tropism in cancer: the hepatic metastatic niche. Cold Spring Harb. Perspect. Med. 2020;10:a037259. doi: 10.1101/cshperspect.a037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaniotis G, Rayes RF, Qi S, Milette S, Wang N, Perrino S, et al. Collagen IV-conveyed signals can regulate chemokine production and promote liver metastasis. Oncogene. 2018;37:3790–3805. doi: 10.1038/s41388-018-0242-z. [DOI] [PubMed] [Google Scholar]
- 51.Borggreve AS, Goense L, Brenkman HJF, Mook S, Meijer GJ, Wessels FJ, et al. Imaging strategies in the management of gastric cancer: current role and future potential of MRI. Br. J. Radiol. 2019;92:20181044. doi: 10.1259/bjr.20181044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M, et al. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol. 2008;9:215–221. doi: 10.1016/S1470-2045(08)70035-4. [DOI] [PubMed] [Google Scholar]
- 53.Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, et al. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N. Engl. J. Med. 2007;357:1810–1820. doi: 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- 54.Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J. Clin. Oncol. 2011;29:4387–4393. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- 55.Noh SH, Park SR, Yang HK, Chung HC, Chung IJ, Kim SW, et al. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-year follow-up of an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:1389–1396. doi: 10.1016/S1470-2045(14)70473-5. [DOI] [PubMed] [Google Scholar]
- 56.Busuttil RA, Liu DS, Di Costanzo N, Schroder J, Mitchell C, Boussioutas A. An orthotopic mouse model of gastric cancer invasion and metastasis. Sci. Rep. 2018;8:825. doi: 10.1038/s41598-017-19025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available as needed.