Abstract
Background
Histone modifications alter transcriptional gene function and participate in cancer progression. Enhancer-of-Zeste-Homologue-2 (EZH2) and Nuclear-Receptor-Binding-SET-domain2 (NSD2) methylate H3K27 and H3K36, respectively, to regulate transcription. Given the therapeutic interest in these enzymes, we investigated expression and coregulation in hormone-sensitive (HS) and castrate-resistant (CR) prostate cancer (PC).
Methods
EZH2 and NSD2 levels were quantified using VECTRA analysis in HS and CRPC tissue microarrays (n = 105 + 66). Expression data from The Cancer Genome Atlas (n = 498), Memorial Sloan Kettering Cancer Center (n = 240), and Stand Up to Cancer/Prostate Cancer Foundation (n = 444) cBioportal datasets were queried, and associations between EZH2 and NSD2 and clinicopathologic variables determined.
Results
Tumour expression of NSD2, but not EZH2, increased in CRPC (p = 0.05, 0.09). Epithelial nuclei co-expressing NSD2 and EZH2 increased in CRPC compared to HSPC (69 vs 42%, p = 0.02), and in metastatic tissue relative to benign (55 vs 35%, p = 0.02). cBioportal analysis revealed collinear NSD2/EZH2 expression (Spearman = 0.57, 0.58, 0.58, all p < 0.001). NSD2/EZH2 co-expression significantly associates with clinicopathologic characteristics including grade group, stage and seminal vesicle involvement. On univariate and multivariate analysis tumours co-expressing NSD2 and EZH2 conferred increased risk of recurrence (hazard ratio: 2.6, 95% confidence inerval: 1.2–5.4, p = 0.01). Kaplan–Meier analysis revealed reduced progression-free-survival of NSD2 and EZH2 co-expression patients in datasets (p < 0.001, 0.002).
Conclusions
Increased EZH2/NSD2 co-expression is overrepresented in CRPC, metastases and associates with shorter disease-free survival in PC patients. Coregulation of these two histone methyltransferases is a biomarker for aggressive PC and licenses them as therapeutic targets.
Subject terms: Prostate cancer, Prognostic markers, Methylation, Gene expression
Background
Molecular tumour analyses demonstrate that unique subtypes exist within prostate cancer (PC) and interest has arisen in using these findings to drive tumour-specific therapy. However, genetic alterations can represent difficult targets for pharmaceutical agents with the exception of the androgen receptor (AR).1 The overexpression or alteration of enzyme activity responsible for the covalent modification of histones including histone methyltransferases (HMTs) is a developing theme in the pathogenesis of malignancy. Epigenetic control of gene expression plays a critical role in many biological processes and tumour profiling indicates that epigenetic alterations comprise an important component of advanced tumours.2 Recently, HMTs have attracted particular interest due to their potential as therapeutic targets, but our understanding of their alteration in local and advanced PC is underdeveloped.
Methylation of H3 and H4 lysine and arginine residues is a common context-dependent histone modification regulated by HMTs. Methylation of H3K36 is generally associated with transcriptionally active euchromatin, while methylation of H3K27 is associated with transcriptionally repressed, compacted heterochromatin.3 The polycomb-repressive complex 2 (PRC2) contains the H3K27 methyltransferase Enhancer-of-Zeste-Homologue-2 (EZH2), and controls dimethylation and trimethylation of H3K27 (H3K27me2/3). EZH2 demonstrates gain of function and is overexpressed in a number of solid tumours including the prostate.4,5 During development, EZH2 plays an important role in cellular differentiation via transcriptional repression, but as it is upregulated it begins to silence other targets including tumour suppressors, further promoting carcinogenesis.6 EZH2 upregulation in cancer occurs, in part, through the loss of a microRNA (miR-101) that post-transcriptionally represses EZH2.7,8 Direct targeting of EZH2 presents a challenge due to its expression in stem and haematologic cells and putative tumour-suppressive function in some haematologic cancers.9
An alteration in the balance of the antagonistic marks H3K36me2 and H3K27me3 is a hallmark of myeloma and other selected cancers.10,11 Nuclear-Receptor-Binding-SET-domain2 (NSD2) (or MMSET) is an HMT that is responsible for the H3K36 monomethyl and di-methyl mark.12 In wild-type cells, H3K36me2 accumulates on active gene bodies and designates open chromatin and transcriptional activity.13 Reports have suggested that both EZH2 and NSD2 expression are linked to cancer.6 The EZH2–NSD2 HMTase axis is, in part, coordinated by a network of microRNAs and the oncogenic functions of EZH2 appear upstream of NSD2.6
PC is marked by heterogeneous genetic changes in primary and advanced cancers.1,14 In the current study, we examined whether co-expression of EZH2 and NSD2 within individual PC cells occurs in hormone-sensitive and castration-resistant disease. Utilising automated Vectra immunoquantitation allows examination of co-expression at a per-tumour-cell basis. We report that there is tight co-expression of these HMTs with progression to CRPC, and furthermore, in primary cancers, this co-expression predicts worse cancer outcomes.
Methods
Tissue microarrays
To investigate expression across disease progression, a previously described progression tissue microarray (pTMA)15 was utilised consisting of 384 cores (duplicates) from 105 patients. For this analysis, samples included were duplicate cores from 73 primary prostate tumours (54 grade group (GG) 1–2, 19 GG 3–5), 22 metastatic lesions, and 48 benign prostate tissue samples. Patients involved in the study provided written informed consent and the study was approved by our Institutional Review Board.
Exploration of biomarker association with hormone responsiveness was achieved via another TMA (hrTMA) constructed from 264 cores (quadruplicates) from 66 patients as previously described.16 Patients having cores with no epithelial cells identified were excluded from the analysis (n = 12) as well as those representing entirely benign prostate tissue (n = 10). For our analysis, cores from 36 of these patients, for which the hormone status was known (18 hormone refractory, 18 hormone sensitive), were used.
IHC, imaging and image analysis
Slide preparation is as previously described for VECTRA analysis. The TMA slides were taken through routine deparaffinisation and rehydration, pre-treated with an endogenous peroxidase block and retrieval buffer. Slides were then rinsed with dH2O, Tris-buffered saline (TBS), and then TBS with Tween (TBST), followed by protein blocking at room temperature. E-cadherin antibodies (Ventana, 790-4497) was used for epithelial compartmentalisation. Multiple antigen labelling was done with 3,3′-diaminobenzidine for staining of NSD2 (Abcam, Ab106180) and EZH2 (Novus, NBP2-29965), while Harris haematoxylin was used for counterstaining. For image analysis and quantification of the staining intensity, the VECTRA system was used. Positive and negative controls and specific antigen blocking were used for each antibody signal optimisation. Cores with <100 epithelial cells or loss of tissue were excluded from the analysis. Nuance system and INFORM 1.2™ software (Caliper Life Sciences, Hopkinton, MA) were used for building spectral libraries on the basis of target signals of the two stained parameters. This system allows automated quantitation of colorimetric (bright-field) staining on a per-cell basis and selection of cellular subsets (nucleus vs cytoplasmic) for analysis of target signals (Supp. Figure 1). For each tissue core, the total number of objects detected (i.e. cell nuclei), the percent of immunostained objects in each of the four staining intensity categories and the global percentage of immunostained objects were computed. These data were used to calculate a mean H-score for each patient as previously defined.17 This compounded score is obtained when the percentage of cells staining positive for a signal was multiplied by the factor representing the intensity of staining (0, 1, 2 and 3). The results are then aggregated, with a maximum value of 300.
Database analyses
The Cancer Genome Atlas (TCGA, Firehose Legacy), Memorial Sloan Kettering Cancer Center (MSKCC, Cancer Cell 2010) and Stand Up to Cancer/Prostate Cancer Foundation (SU2C/PCF PNAS 2019) PC samples were queried using cBioPortal for Cancer Genomics (www.cbioportal.org).1,18–20 The MSKCC dataset was obtained via Gene Expression Omnibus, which was queried for dataset GSE 21032 using NSD2 and EZH2 probes.18 All prostate adenocarcinoma (TCGA = 499, MSKCC = 218) and metastatic PC (SU2C/PCF = 444) samples were analysed for NSD2 and EZH2 using available messenger RNA (mRNA) expression. There were 498, 140 and 270 tissue samples with available NSD2 and EZH2 mRNA data in the TCGA, MSKCC and SU2C datasets, respectively. Clinical data for all samples were obtained using the TCGAbiolinks (Bioconductor) package in R. Biochemical recurrence indicator was selected as the metric to determine disease progression. This definition of biochemical recurrence was individually determined by each dataset and reported under the biochemical recurrence indicator. Samples with no available recurrence indication were excluded from biochemical recurrence and progression-free time analyses.
Statistical analysis
Analyses were performed using StataSE 16.1 (StataCorp, College Station, TX) and GraphPad Prism 5.04 (GraphPad, San Diego, CA). Staining patterns within the progression TMA of NSD2 and EZH2, and the colocalisation of both signals to individual cells were all compared between benign, cancer and metastatic tissues in the pTMA using a one-way analysis of variance (ANOVA) with Tukey’s post test. Within the cancerous cores, GG 1 and 2 cores were pooled and compared to GG 3–5 cores. In the hormone response tissue microarray (hrTMA), EZH2 and NSD2 protein levels were compared using Student’s t test between cores from hormone-sensitive and hormone-resistant patients.
Association between NSD2 and EZH2 in the TCGA, MSKCC and SU2C/PCF datasets was determined by plotting the mRNA values available for each sample. A D’Agostino–Pearson normality test was performed using GraphPad and revealed the distribution of the mRNA values to be non-gaussian, and therefore a Spearman’s correlation coefficient was calculated.
In the TCGA and MSKCC datasets, NSD2 and EZH2 mRNA expression was compared against selected clinicopathological features (GG, seminal vesicle involvement, pathologic stage, recurrence and overall survival). In this analysis, expression values from all cores belonging to the same patient for a specific lesion type (e.g. benign, primary) were averaged. For each of the above parameters, a Student’s t test or ANOVA was used to determine the relationship between protein level and clinical features.
Samples from the TCGA and MSKCC databases were rank-sorted for each target based on mRNA expression. Patients in the top 25th percentile for NSD2 or EZH2 within each dataset received a designation as a high expressor of that target. Another category of patients who were deemed high expressors for both NSD2 and EZH2 were placed in the top quartile NSD2 and EZH2 subset. These cohorts (high vs low expressor for each target) were compared in regard to clinical and pathologic variables.
Results
NSD2 and EZH2 colocalisation increases with hormone-refractory status and metastasis
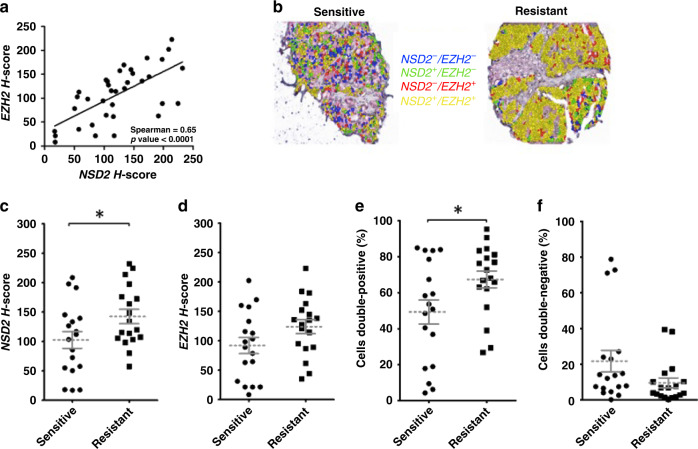
The H3K36 HMT NSD2 has been found to promote the metastatic progression of PC in mouse and human models.10,11,21 Given the putative coregulation with the K27 HMTase EZH2 found in previous work,6 we sought to test this association across the spectrum of hormone-sensitive and advanced PC specimens. Utilising immunohistochemistry and VECTRA™ automated intensity analyses, nuclear staining patterns were determined for NSD2 and EZH2 on a tissue array (hrTMA) consisting of hormone-sensitive and castration-resistant disease. The clinicopathologic features of this androgen-sensitive array have been previously reported.15 A strong positive correlation was seen between NSD2 and EZH2 protein expression across PC specimens (Spearman correlation = 0.65; p < 0.001) (Fig. 1a).
Fig. 1. Concurrent immunostaining demonstrates the percentage of cells with colocalisation of NSD2 and EZH2 significantly increases in castrate-resistant prostate cancer (CRPC) compared to hormone-sensitive disease.
Immunochemistry was quantitated using VECTRA™ and INFORM software using a hormone response tissue microarray (hrTMA) (n = 36) with hormone-sensitive patients (n = 18) and hormone-resistant patients (n = 18). a NSD2 and EZH2 association between core expression in tumour tissue. b Image of VECTRA immunostaining combinations for NSD2 and EZH2 in individual epithelial cells demonstrating overlapping staining. c Mean H-score of NSD2, d mean H-score of EZH2, e percentage of individual epithelial cells positive for both NSD2 and EZH2 signals compared between hormone-sensitive and hormone-resistant PC. f Percentage of individual epithelial cells negative for both NSD2 and EZH2 signals within each core. (p < 0.05 indicated by *, <0.01 indicated by ** and <0.001 indicated by ***).
Quantitation of NSD2 expression demonstrated a strong trend towards increased expression in CRPC compared to hormone-sensitive cancer (p = 0.05) (Fig. 1b, c). There was no increase seen with EZH2 in CRPC (p = 0.09) (Fig. 1d). Investigation of protein co-expression within individual epithelial cells was then examined and the percentage of cells containing the nuclear expression of both NSD2 and EZH2 was determined within each core. The mean percentage of cells that co-stained positive for both proteins increased in CRPC compared to HSPC (p = 0.03) (Fig. 1e). Conversely, the percentage of cells that stained negative for both markers decreased but not to significance (p = 0.09) (Fig. 1f).
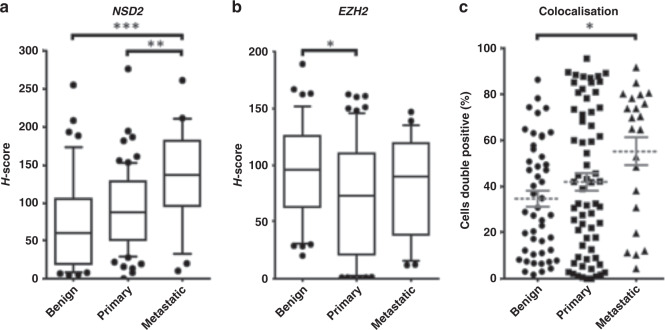
Protein expression was then analysed in benign, primary and metastatic PC tissue within a pTMA consisting of 73 primary PC specimens, of which 54 were low grade (GG 1–2) and 19 were higher grade (GG 3–5). Nuclear NSD2 protein staining was increased in metastases in comparison to benign (p < 0.001) and primary tumour (p < 0.01) (Fig. 2a). In contrast, EZH2 expression in metastatic tissue was not significantly increased vs primary cancer (p = 0.25). Marked heterogeneity in EZH2 staining between tumour samples was noted (Fig. 2b). Analysis of protein colocalisation demonstrates that in comparison to benign and primary cancer tissue, metastatic tissue had a significantly higher percentage of individual cells that co-stained positive for both NSD2 and EZH2 (p = 0.02) (Fig. 2c).
Fig. 2. Core immunostaining of hormone-sensitive prostate cancer (HSPC) demonstrates NSD2 increases during prostate cancer progression and co-expression increases in metastases.
This progression TMA consists of 44 benign, 73 primary PC specimens, of which 54 were low (GG 1–2) and 19 high grade (GG 3–5), and 22 metastases. a, b H-scores of NSD2 and EZH2 compared between benign, primary and metastatic PC radical prostatectomy cores in progression TMA (pTMA). c Percentage of individual cells staining positive for both EZH2 and NSD2 signals in benign, primary and metastatic PC (p < 0.05 indicated by *, <0.01 indicated by ** and <0.001 indicated by ***).
NSD2 strongly correlates with EZH2 in publicly available datasets
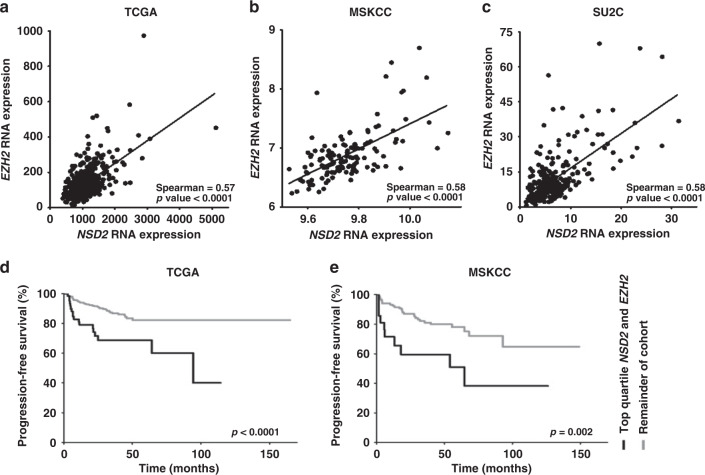
To extend these observations, we examined the correlation of NSD2 and EZH2 expression within the TCGA and MSKCC datasets consisting of primary HSPC from radical prostatectomy specimens, as well as the SU2C dataset that contains samples from patients with metastatic CRPC.1,18–20 Initially, we sorted the expression of all transcripts by the strength of correlation with NSD2 and find that EZH2 was in the top 0.3% of all 20,159 genes in the 333 patient TCGA,19 and the highest gene overall (22,927 transcripts) in the 218 patient MSKCC dataset. Examining the 429 patient SU2C/PCF expression database containing CRPC, EZH2 was in the top 0.4% of 14,265 genes. Using cBioPortal, Spearman correlations of 0.57 (p ≤ 0.001), 0.58 (p ≤ 0.001) and 0.58 (p ≤ 0.001) in the TCGA, MSKCC and SU2C/PCF, respectively, indicating a moderate correlation of expression between the two enzymes in primary and CRPC datasets (Fig. 3a–c). We next examined the NSD2 correlation with other H3K27 and H3K36 HMTases and demethylases (Table 1). Differences in unique HMT expression were noted between primary datasets. NSD1 demonstrates significant but weaker correlations in both TCGA and MSDKCC databases (Spearman correlation = 0.53 and 0.22, respectively, p < 0.001 and p = 0.01). Within the CRPC SU2C dataset, a strong association between NSD2 and K36 histone demethylase KDM2B (Spearman correlation = 0.59, p < 0.001) is noted.
Fig. 3. Prostate cancer database expression data and patient microarray staining demonstrate significant EZH2 and NSD2 correlation within individual tumours and NSD2 and EZH2 expression predicts biochemical recurrence in PC.
a–c For individual tissue samples with expression for both enzymes available, mRNA expression values plotted for each marker. a TCGA (n = 491) consisting of hormone-sensitive primary PC. b MSKCC (n = 128) consisting of primary and metastatic PC samples. c SU2C (n = 270) consists of metastatic CRPC patients. RNA expression data were queried and downloaded using the cBioPortal platform. As sample distribution was found to be non-Gaussian, Spearman correlation coefficients were calculated. d, e Kaplan–Meier analysis performed using clinical outcome data available on cBioPortal for d TCGA (n = 498) and e MSKCC (n = 140). Progression-free time defined as time to biochemical recurrence as determined by PSA cut-offs. In each dataset, samples were assigned individual quartile rank for NSD2 and EZH2. Samples containing top quartile ranks for both NSD2 and EZH2 (n = 70; 21) were compared to all remaining samples in their respective cohorts (n = 428;119). P values are documented.
Table 1.
Association of NSD2 with other H3K27 and H3K36 histone methyltransferases and demethylases in prostate cancer datasets.
| Enzyme | TCGA (n = 491) | MSKCC (n = 128) | SU2C (n = 270) | |||
|---|---|---|---|---|---|---|
| Primary HSPC | Primary + Met HSPC | mCRPC | ||||
| Spearmana | p Value | Spearman | p Value | Spearman | p Value | |
| Histone methylase | ||||||
| H3K27 | ||||||
| EZH1 | −0.08 | 0.072 | −0.09 | 0.297 | 0.29 | <0.001 |
| EZH2 | 0.57 | <0.001 | 0.58 | <0.001 | 0.58 | <0.001 |
| H3K36 | ||||||
| ASH1L | 0.34 | <0.001 | 0.03 | 0.719 | 0.27 | <0.001 |
| NSD1 | 0.53 | <0.001 | 0.22 | 0.012 | ||
| NSD3 | 0.26 | <0.001 | −0.03 | 0.735 | 0.42 | <0.001 |
| SETD2 | 0.483 | <0.001 | 0.01 | 0.896 | ||
| SETD3 | −0.03 | 0.447 | −0.18 | 0.047 | −0.04 | 0.487 |
| SETMAR | −0.17 | <0.001 | 0.01 | 0.908 | ||
| SMYD2 | −0.02 | 0.626 | −0.03 | 0.738 | ||
| Histone demethylase | ||||||
| H3K27 | ||||||
| KDM6A | 0.44 | <0.001 | 0.03 | 0.712 | 0.20 | <0.001 |
| KDM6B | 0.32 | <0.001 | 0.02 | 0.780 | 0.38 | <0.001 |
| H3K36b | ||||||
| KDM2A | 0.44 | <0.001 | −0.07 | 0.439 | 0.41 | <0.001 |
| KDM2B | 0.37 | <0.001 | 0.17 | 0.060 | 0.59 | <0.001 |
Correlations >0.5 are given in bold.
aSpearman correlation coefficient.
bHMTases KDM4A/B/C were not represented in these datasets.
Increased NSD2 and EZH2 co-expression associates with worse outcomes in patients after radical prostatectomy
Within both databases, a series of clinicopathologic patient variables including GG, pathologic stage and seminal vesicle involvement were compared to NSD2 and EZH2 RNA expression values. Using mRNA expression as a continuous variable, only GG was consistently associated with NSD2 or EZH2 in both primary databases (Table 2). Thus, the majority of clinicopathologic features were not consistently associated with NSD2 or EZH2 across multiple databases.
Table 2.
Association of NSD2 and EZH2 with radical prostatectomy pathologic features in the TCGA and MSKCC databases.
| Variable | TCGA | MSKCC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | NSD2 | p Value | EZH2 | p Value | n | NSD2 | p Value | EZH2 | p Value | |
| Grade group | ||||||||||
| 1–2 | 191 | 937 (233) | 100 (42) | 94 | 6.76 (0.28) | 9.72 (0.11) | ||||
| 3–5 | 307 | 1185 (480) | <0.001 | 157 (101) | <0.001 | 44 | 6.95 (0.46) | 0.003 | 9.77 (0.14) | 0.02 |
| Stage | ||||||||||
| 2 | 187 | 969 (255) | 110 (53) | 86 | 6.78 (0.27) | 9.72 (0.10) | ||||
| 3 | 293 | 1137 (401) | 147 (84) | 47 | 6.88 (0.46) | 9.75 (0.14) | ||||
| 4 | 11 | 1796 (1295) | <0.001 | 225 (179) | <0.001 | 7 | 6.94 (0.48) | 0.22 | 9.79 (0.11) | 0.09 |
| SV involved | ||||||||||
| No | 345 | 1018 (331) | 121 (68) | 116 | 6.80 (0.30) | 9.73 (0.11) | ||||
| Yes | 135 | 1208 (395) | <0.001 | 162 (86) | <0.001 | 17 | 6.90 (0.59) | 0.31 | 9.75 (0.17) | 0.50 |
Values are reported as mean (SD).
Biochemical recurrence (e.g. rising PSA) after treatment for PC surgery is an important clinical parameter that associates with patient survival. To analyse the predictive ability of NSD2 and EZH2, individual samples were ranked based on the expression for each enzyme. Samples in the top quartile for each respective HMT were compared to the remainder of patients for the ability to predict biochemical failure. In both the TCGA and MSKCC, the top quartile for each individual HMT as well as the smaller subset containing the top quartile for both transcripts (NSD2 and EZH2) predicted biochemical recurrence on univariate Cox regression analysis in both datasets (Table 3). In this analysis, clinicopathologic variables including GG, stage and seminal vesicle involvement were also predictive of biochemical recurrence in both radical prostatectomy datasets (all p < 0.05).
Table 3.
Univariate cox regression analysis for predicting biochemical recurrence.
| Variable | TCGA | MSKCC | ||||
|---|---|---|---|---|---|---|
| Haz. ratio | 95% Conf. interval | p Value | Haz. ratio | 95% Conf. interval | p Value | |
| Grade group categorya | 6.2 | 2.7–14.5 | <0.001 | 10.6 | 4.9–22.9 | <0.001 |
| Stage | 3.3 | 2.0–5.6 | <0.001 | 3.4 | 2.1–5.4 | <0.001 |
| SV involvement | 3.4 | 2.0–5.7 | <0.001 | 7.0 | 3.4–14.5 | <0.001 |
| Top quartile NSD2 | 2.7 | 1.6–4.5 | <0.001 | 3.5 | 1.8–6.8 | <0.001 |
| Top quartile EZH2 | 2.9 | 1.7-4.9 | <0.001 | 2.4 | 1.2–4.7 | 0.016 |
| Top quartile NSD2 and EZH2 | 2.8 | 1.6–4.9 | 0.001 | 2.9 | 1.4–6.10 | 0.008 |
aGrade group categories: (1–2) vs (3–5).
We then extended this investigation to include a multivariate Cox regression analysis with NSD2 and EZH2 co-expression as a variable. This co-expression was captured by creating a subset of patients that ranked in the top quartile for both NSD2 and EZH2. In both the TCGA and MSKCC datasets, GG and top quartile of NSD2 and EZH2 were independent predictors of biochemical recurrence (Supp. Table S1). This same analysis was performed for the top quartile NSD2 or EZH2 alone and NSD2 was found to be significant in the TCGA only, while EZH2 was predictive in both datasets (Supp. Table A2).
To further examine if a high expression of NSD2 and EZH2 is meaningful in determining biochemical recurrence in PC, we performed a Kaplan–Meier analysis using TCGA and MSKCC outcome data. Being ranked in the top quartile for expression of both NSD2 and EZH2 conferred a significantly worse progression-free survival compared to all other samples (Fig.3d, e). Five-year progression-free survival rates were significantly lower in the top quartile NSD2 and EZH2 cohort when compared to all other patients in both cohorts (TCGA: 68.7 vs 82.1% p < 0.0001, MSKCC: 51 vs 78.1%, p = 0.002). Of note, top quartile expression of NSD2 or EZH2 alone when analysed did also predict worse outcomes in both datasets (Supp. Fig S2). Survival analysis log-rank hazard ratios (HRs) revealed that patients who carry high expressor designation for both NSD2 and EZH2 have a significantly higher probability of progressing compared to all other patients (HR: TCGA 2.8; MSKCC 2.9) (Table 3). Thus, co-expression of both NSD2 and EZH2 in the top quartile consistently predicted worse outcomes in patients with PC across both datasets.
Discussion
Molecular analyses indicate that altered chromatin states generated by histone post-translational (de)methylation impact gene expression and play an important role in the progression to advanced PC. NSD2 has been implicated based on a genetic screen as a driver of metastases in mouse PC models.21 This enzyme binds to transcriptionally active regions of the genome and induces dimethylation of H3K36 that is associated with open chromatin and actively transcribed genes. In the current study, one major finding is the demonstration of a tight positive correlation between NSD2 and EZH2 both in androgen-sensitive and androgen-insensitive, advanced PC. EZH2 is an H3K27 HMT associated with transcriptionally repressed and compacted heterochromatin leading to silencing of PRC2-associated genes. We also demonstrate that protein co-expression occurs at the cellular level more frequently in CRPC than hormone-responsive PC suggesting a role in progression to androgen insensitivity. These observations are further developed in publicly available datasets that confirm this tight correlation between NSD2 and EZH2 in PC and co-expression of these transcripts is associated with worse clinicopathologic outcomes in men undergoing radical prostatectomy. The close correlation between NSD2 and EZH2 suggests that the coregulation may be important in driving a subset of cancers, and thus inform potential therapeutic interventions that target both NSD2 and EZH2.
A unique strength of this study is the ability to analyse protein expression within specific PC cells utilising VECTRA™ imaging technology. This quantitative, automated platform permits the examination of multiple markers simultaneously. We found that NSD2 and EZH2 protein expression are tightly correlated in PC at all stages (Figs. 1a and 3). EZH2 upregulation and its coordination with NSD2 is thought to rely on the expression of several microRNAs including miR-101 that post-transcriptionally repress EZH2.,6–8 Our data demonstrate that co-expression in cells occurs more commonly in CRPC than androgen-sensitive tumours (68 vs. 43%; Fig. 1), as per a new finding. NSD2 specifically interacts with the DNA-binding domain of the AR resulting in increased nuclear translocation that may be, in part, responsible for enhanced AR expression in CRPC.22 In addition, androgen deprivation therapy enhances the activity of EZH2 through the activation of cAMP response element-binding protein leading to angiogenesis and neuroendocrine progression.23 In the current study, NSD2 expression was associated with progression from primary to metastatic cancer, but a significant increase in EZH2 was not noted in contrast to other reports.5,6,24,25 This discrepancy may be, in part, due to the automated, quantitative approach we utilised, which allows cancer cell segmentation and individual quantification in contrast to studies using manual staining assessment or techniques that do not fully capture tumour heterogeneity. In addition, almost 2/3 of the samples in the current array were lower grade tumours representing an earlier stage in the progression of PC. This increase in co-expression in advanced cancer suggests an important dual role for these histone methylases in driving progression to CRPC.
Our analysis of these HMTs in several large cancer databases demonstrates that co-expression is associated with higher grade tumours. However, we note a lack of collinearity between other clinicopathologic features between available datasets, which may be associated with the strength of the observation, the transcript utilised or the size of the observations. Generating a multivariate model indicates combining grade and NSD2/EZH2 in both datasets has a high predictive value for those patients that will have a recurrence. Kaplan–Maier analysis suggests that identifying patients with high levels of both NSD2 and EZH2 further adds to the predictive ability of GG in these datasets. We would anticipate given their tight co-expression that top quartiles of NSD2 and EZH2 alone in multivariate analysis were also predictive of recurrence in combination with grade. However, NSD2 was only significant in TCGA set and NSD2/EZH2 predicted comparatively better than EZH2, which was significant in both. Given the known antagonistic functions of NSD2 and EZH2 in the control of gene expression, the results presented here suggest that the tight positive correlation reveals an epigenetic state in these cancers that favours a dual dependency of at least a subset of genes, which we postulate plays an important role in the pathophysiology of PC.
Conclusions
In this study, we demonstrate that EZH2 and NSD2 display increased co-expression at both protein and mRNA levels in a subset of patients. This group is over-represented in CRPC tissue, in patients with metastases and shorter disease-free survival in those with primary tumours is demonstrated. The co-expression of these targets represents a putative target for therapy via several approaches. One approach would be to target the NSD2/EZH2 HMT regulatory axis network of microRNAs, including miR-203, miR-26a and miR-31.10 Overexpression of miR-203 can suppress tumorigenicity, angiogenesis and metastasis in advanced PC cell lines.26,27 In multiple myeloma, increased levels of NSD2 are associated with a global reduction of H3K27me3, but increased methylation at specific loci.10 Disruption of this balance using existing EZH2 inhibitors, which are currently being evaluated in trials, may be one approach.28 Alternatively, the development of an NSD2 inhibitor might be a therapeutic strategy that could cripple both EZH2 and NSD2 co-regulated pathways. The current work suggests that coregulation of these two HMT enzymes may be used as a biomarker for a more aggressive PC subtype and furthermore identifies them as a putative therapeutic target worthy of further study.
Supplementary information
Acknowledgements
We thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory (TRIP), supported by the UW Department of Pathology and Laboratory Medicine, UWCCC (P30 CA014520) and the Office of the Director-NIH (S10OD023526) for use of its facilities and services.
Author contributions
M.F. organised and interpreted raw data, performed the statistical analysis, designed figures and tables and drafted the manuscript. J.G. was instrumental in designing the experiment and performing data compilation and analysis. A.T. compiled and organised raw data including statistical analysis. G.A. provided statistical analysis and data organisation. W.H. read the H&E slides and was involved in idea development, data development and review, and editing. T.K. provided analysis of raw results and assistance with figures and data presentation. R.M. compiled and analysed raw data involving the tissue staining results. P.L. assisted in idea development, data development and review, and editing of the manuscript. B.Y. assisted in statistical analysis and construction of figures. J.D. was involved in idea development, data development and editing. D.J. designed and supervised the study, interpreted data and edited the manuscript.
Ethics approval and consent to participate
Individual medical centres obtained institutional review board approval exemption or waiver for the use of archived clinical samples for research purposes. This study was performed in accordance with the Declaration of Helsinki. Data and outcomes for cBioportal are made through a data-sharing agreement. Approvals and patient consents were obtained through the University of Wisconsin Carbone Cancer Center Tissue Biobank (IRB #2016-0934) and through the University of Wisconsin Institutional Review Board IRB# XPO5338.
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary information files. RNA expression data from the TCGA, MSKCC and SU2C/PCF is available publicly online from the cBioPortal for cancer genomics.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by the Department of Defense Grant-DODPCRP W81XWH (to D.J.).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01398-7.
References
- 1.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed]
- 2.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. SnapShot: histone-modifying enzymes. Cell. 2007;128:802.e1–02.e2. doi: 10.1016/j.cell.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 6.Asangani IA, Ateeq B, Cao Q, Dodson L, Pandhi M, Kunju LP, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol. Cell. 2013;49:80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman, J. M., Jones, P. A. & Liang, G. The tumor suppressor microRNA-101 becomes an epigenetic player by targeting the polycomb group protein EZH2 in cancer. Cell Cycle 8, 2313–2314 (2009). [DOI] [PMC free article] [PubMed]
- 8.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 10.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Ezponda T, Shah MY, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014;10:e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Ahn JH, Wang GG. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol. Life Sci. 2019;76:2899–2916. doi: 10.1007/s00018-019-03144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 13.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Zhao D, Spring DJ, DePinho RA. Genetics and biology of prostate cancer. Genes Dev. 2018;32:1105–1140. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damodaran S, Damaschke N, Gawdzik J, Yang B, Shi C, Allen GO, et al. Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer. 2017;17:874. doi: 10.1186/s12885-017-3853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JH, Yang B, Lindahl AJ, Damaschke N, Boersma MD, Huang W, et al. Identifying dysregulated epigenetic enzyme activity in castrate-resistant prostate cancer development. ACS Chem. Biol. 2017;12:2804–2814. doi: 10.1021/acschembio.6b01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmeules P, Hovington H, Nguile-Makao M, Leger C, Caron A, Lacombe L, et al. Comparison of digital image analysis and visual scoring of KI-67 in prostate cancer prognosis after prostatectomy. Diagn. Pathol. 2015;10:67. doi: 10.1186/s13000-015-0294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–16.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aytes A, Giacobbe A, Mitrofanova A, Ruggero K, Cyrta J, Arriaga J, et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat. Commun. 2018;9:5201. doi: 10.1038/s41467-018-07511-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang HB, Choi Y, Lee JM, Choi KC, Kim HC, Yoo JY, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009;583:1880–1886. doi: 10.1016/j.febslet.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zheng D, Zhou T, Song H, Hulsurkar M, Su N, et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 2018;9:4080. doi: 10.1038/s41467-018-06177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 25.Saramäki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 26.Tian X, Tao F, Zhang B, Dong JT, Zhang Z. The miR-203/SNAI2 axis regulates prostate tumor growth, migration, angiogenesis and stemness potentially by modulating GSK-3β/β-CATENIN signal pathway. IUBMB Life. 2018;70:224–236. doi: 10.1002/iub.1720. [DOI] [PubMed] [Google Scholar]
- 27.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, et al. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin. Cancer Res. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 28.Taplin M-E, Hussain A, Shah S, Shore ND, Agrawal M, Clark W, et al. ProSTAR: a phase Ib/II study of CPI-1205, a small molecule inhibitor of EZH2, combined with enzalutamide (E) or abiraterone/prednisone (A/P) in patients with metastatic castration-resistant prostate cancer (mCRPC) J. Clin. Oncol. 2019;37(Suppl.):TPS335–TPS335. doi: 10.1200/JCO.2019.37.7_suppl.TPS335. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary information files. RNA expression data from the TCGA, MSKCC and SU2C/PCF is available publicly online from the cBioPortal for cancer genomics.