Abstract
Objective
Barrett’s oesophagus (BE) is a known precursor to oesophageal adenocarcinoma (OAC) but current clinical data have not been consolidated to address whether BE is the origin of all incident OAC, which would reinforce evidence for BE screening efforts. We aimed to answer whether all expected prevalent BE, diagnosed and undiagnosed, could account for all incident OACs in the US cancer registry data.
Design
We used a multiscale computational model of OAC that includes the evolutionary process from normal oesophagus through BE in individuals from the US population. The model was previously calibrated to fit Surveillance, Epidemiology and End Results cancer incidence curves. Here, we also utilised age-specific and sex-specific US census data for numbers at-risk. The primary outcome for model validation was the expected number of OAC cases for a given calendar year. Secondary outcomes included the comparisons of resulting model-predicted prevalence of BE and BE-to-OAC progression to the observed prevalence and progression rates.
Results
The model estimated the total number of OAC cases from BE in 2010 was 9970 (95% CI: 9140 to 11 980), which recapitulates nearly all OAC cases from population data. The model simultaneously predicted 8%–9% BE prevalence in high-risk males age 45–55, and 0.1%–0.2% non-dysplastic BE-to-OAC annual progression in males, consistent with clinical studies.
Conclusion
There are likely few additional OAC cases arising in the US population outside those expected from individuals with BE. Effective screening of high-risk patients could capture the majority of population destined for OAC progression and potentially decrease mortality through early detection and curative removal of small (pre)cancers during surveillance.
Keywords: Barrett's oesophagus, Barrett's carcinoma, oesophageal cancer, screening, pre-malignancy - gi tract
Significance of this study.
What is already known on this subject?
Barrett’s oesophagus (BE) patients have a 40–50-fold higher risk of developing oesophageal adenocarcinoma (OAC) than the general population yet many remain undiagnosed.
Identified BE patients receiving surveillance can have early cancers discovered endoscopically, which decreases the high overall OAC-associated mortality.
Currently, around 90% of patients who develop OAC were never part of a BE surveillance programme, and those BE patients on surveillance have a low annual progression rate of 0.1%–0.3% to develop OAC.
What are the new findings?
By applying a model that incorporates the evolution from normal cells to BE to OAC in patients, we found that the numbers add up—the expected number of OAC cases in the US population are explained by the published rates of BE described above.
We cohesively examined the published estimates to determine that all OAC likely arises from both identified BE and occult, undiagnosed BE in the population.
How might it impact on clinical practice in the foreseeable future?
Based on current best estimates, our findings suggest that there is no public health need to seek cases of a non-BE alternative pathway to OAC.
Increasing efforts for effective, sensitive screening and surveillance of the true BE population has the potential to decrease OAC mortality in the coming years.
Introduction
Oesophageal adenocarcinoma (OAC) is typically diagnosed when a patient presents with symptoms such as dysphagia. Unfortunately, the majority of these patients do not live past the first year of their diagnosis because by the time dysphagia develops, metastatic cancer is already present. In order to prevent this cancer or detect it at an earlier, more treatable stage, efforts are now made to identify patients with Barrett’s oesophagus (BE), the only known precursor to OAC. Identified BE patients are believed to have a 40–50-fold higher annual incidence of OAC than the general population.1 Metaplastic BE progresses through dysplasia to cancer. Advances in endoscopic eradication therapy for dysplastic BE discovered during surveillance of BE can now prevent cancer.2 However, most cancers arise in patients without previously diagnosed BE suggesting either inadequate screening strategies or, as a recent study proposes, the possible existence of a pathway independent from the BE pathway.3 In this study, we seek to answer a simple question about the unseen origins of OAC: does overall OAC incidence reflect the number of cancers that would be expected to arise only from prevalent BE? In other words, do any OAC cases remain unaccounted for that ergo did not arise from the typical Barrett’s precursor pathway? The answer to this question will importantly guide research and public health efforts. If BE is the major or only precursor of OAC, then investigators should continue to focus on improving BE detection. If BE is not the major precursor of OAC, then research needs to focus on identifying alternative pathways and BE screening programmes will have limited impact on prevention and early detection of OAC.
In reality, very few individuals who have BE are ever offered an upper endoscopy, and therefore most BE remains asymptomatic and undiagnosed.1 Patients with gastro-oesophageal reflux disease (GERD) are technically the only subpopulation of the general public typically recommended BE screening because it is believed they have a 5-fold relative risk (RR) of developing long segment BE,4 yet even so only about 10% of GERD patients will receive an endoscopy.1 This indicates underscreening, likely because patients either do not complain of their GERD symptoms, they respond adequately to medical therapy, or were otherwise not deemed suitably high-risk by their physician to warrant an esophagogastroduodenoscopy. Nonetheless, the prevalence of BE in the general population is 1%–2%, whether diagnosed or not,5 6 and this is likely considerably higher in certain at-risk groups in the USA.7–10 The main concern is that the average rate to develop OAC in these patients is low—around 0.3% per year.11 Therefore, the majority of endoscopies are futile in finding OAC. We aimed to answer whether all prevalent BE expected, diagnosed and undiagnosed in the US population, could account for all the incident OACs expected as progression rates would imply, to fit the national cancer registry data.
Methods
The question above is too complex to answer on the ‘back of an envelope’ because published average rates of progression are dependent on age, birth cohort and calendar year. In particular for OAC, age-specific incidence rates vary drastically between men and women.12 This complexity of timescales involved in normal to premalignant BE to OAC progression has necessitated the creation of quantitative models that analyse cancer incidence rates, and project these trends into the future for public health risk assessments and planning.13 Models also quantify the potential impact of progression rates measured in clinical studies on hypothetical intervention and surveillance scheduling in efficacy and cost-effectiveness studies.14–16 Such models allow us to perform quantitative, comparative analyses on the benefits versus harms of proposed screening and surveillance protocols against watch-and-wait strategies; these simply cannot be done heuristically due to the complex nature of cancer evolution.
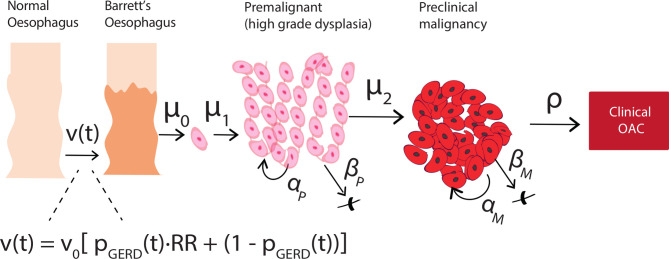
In this study, we model both the onset of BE and the progression of BE to OAC. As a brief background, the multistage clonal expansion model for OAC (herein referred to as the MSCE-OAC model, but also referred to as the MSCE-EAC model elsewhere) is a stochastic model for development of OAC during patient lifetime that includes probabilities of developing BE at various ages, followed by initiation of dysplastic and malignant cell clones in BE with parameters for growth and progression of individual clones to cancer (figure 1). The inputs only include GERD prevalence (calibrated to age-specific and sex-specific estimates)17 18 and OAC age-specific and sex-specific incidence curves provided by Surveillance, Epidemiology and End Results (SEER) registry.12 The BE prevalence and neoplastic progression rates are calibrated to fit those inputs, that is, they are not based on observed BE prevalence nor neoplastic progression rates from empiric studies. Briefly, the model includes a GERD-stratified risk curve to develop BE, which is modelled as an age-dependent rate of exponential BE onset each calendar year with an unknown baseline parameter ν0. The patient-specific BE lengths can vary, derived from a Beta distribution with general population mean length set to 2–3 cm. Beyond ν0, the baseline constant rate for BE onset, the additional model parameters govern the evolutionary dynamics for dysplastic and malignant growth and OAC detection. The model parameters have been previously calibrated such that the resulting hazard functions fit to OAC age-specific and sex-specific incidence curves provided by SEER registry.13 We found during rigorous model selection with likelihood ratio tests that models stratified by birth cohort and sex best fit the incidence data, robust to sensitivity analyses (figure 2). With these fits, the model outputs used for this study include the expected number of OAC cases in an at-risk population at a given year calculated using the hazard function h OAC (see online supplemental material for equation details), along with the BE prevalence and the resulting BE-to-OAC progression rates (predicted as specific to age, sex and birth cohort).
Figure 1.
The stochastic, multiscale model for OAC development (MSCE-OAC) includes conversion from normal squamous epithelium in the oesophagus to BE metaplasia with BE onset rate ν(t), which is a function of a baseline rate ν0 and age-dependent prevalence of GERD pGERD(t) (see Methods for details). Two-hit processes with rates μ0, μ1 can initiate a premalignancy (eg, inactivation of tumour suppressor gene TP53 in non-dysplastic BE due to mutation/copy number alteration in a BE daughter cell creates first cell of a high grade dysplasia lesion). Premalignant cell growth rates are defined as αP = division rate, βP = death/differentiation rate per year. Malignant transformation with rate μ2 creates the first cell of a preclinical clone that can grow with rates αM = division rate, βM = death/differentiation rate per year. Size-based probability ρ for detection of preclinical malignant clone can lead to patient-specific time of incident OAC. BE, Barrett’s oesophagus; OAC, oesophageal adenocarcinoma; GERD, gastro-oesophageal reflux disease; MSCE-OAC, multistage clonal expansion for oesophageal adenocarcinoma.
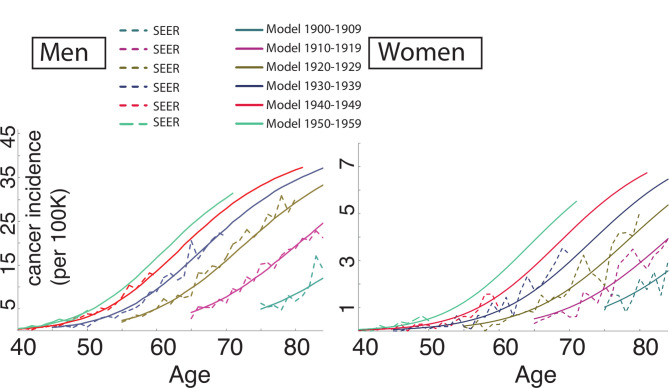
Figure 2.
The MSCE-OAC model was previously calibrated to SEER incidence curve data stratified by sex and 10-year grouped birth cohorts from 1900 - 1909 to 1950–1959.13 The model hazard fits by birth cohort (denoted by colour) represent OAC incidence curves (solid lines) that are consistent with Surveillance, Epidemiology and End Results (SEER) data trends by birth cohort (dashed lines), separately for men (left panel) and women (right panel). MSCE-OAC, multistage clonal expansion for oesophageal adenocarcinoma.
gutjnl-2020-321598supp001.pdf (1.2MB, pdf)
This model has been used and improved in comparative analyses within the NCI Cancer Intervention and Surveillance Modelling Network consortium for the past 9 years, which has enabled numerous studies on sensitivity of biopsy sampling techniques for detection of small dysplastic lesions,14 on influence of patient-specific molecular BE dwell time on future OAC risk,19 and on cost-effectiveness of endoscopic eradication therapy for certain BE risk groups during surveillance.15 In our original study on modelling OAC incidence and mortality rates from 1975 to 2010, we used SEER-specific model fits combined with US census data to estimate past and predict future OAC-related deaths but did not include predicted OAC cases by calendar year when applied to US census data.13
In the Results below, we expand on prior modelling to help elucidate an answer to our general public health question—‘Is BE the precursor of all OAC?’ To do this, we first applied the model to estimate the number of OAC cases using the US age-specific and sex-specific at-risk population estimates from the US census data, to be able to compare with the expected number quoted by Vaughan and Fitzgerald.1 This outcome serves as an independent validation of successful calibration of our model to OAC incidence. Then, we compared the simultaneous predictions of age-specific BE prevalence using the MSCE-OAC model with the published data currently used for screening rationale,20 which included endoscopic reports from the Clinical Outcomes Research Initiative (CORI) for more than 150 000 patients, most of whom were born around 1950. We also compared the mathematical predictions of neoplastic progression rate from non-dysplastic BE to published estimates.
Results
First, Vaughan and Fitzgerald estimated that the newly diagnosed number of cases for ages greater than 40 to be roughly around 10 000 total in the USA every year based on data from 2010 with an average OAC incidence rate across all age groups.1 With the Markov model framework, we can analytically compute the OAC hazard function and estimate the expected number of newly diagnosed OAC cases by age and year separately for men and women when considering also population data. As a starting point using 2010 census person-year data,21 the model predicts that about 2.2 million adults had prevalent BE in 2010, which is around 1.6% of the general US population over age 40. Then, for age groups greater than 40 in both sexes of all races, our single-age calibrated model estimated that the expected number of new OAC cases diagnosed in 2010 was equal to 9970 (95% CI: 9140 to 11 980).
We also computed the analogous estimate for OAC cases using incidence rates quoted directly from the SEER registry for ages 40–90, which was found to be 9400 OAC cases total in 2010.12 Thus, the estimate generated by our computational model of progression from BE to OAC is closely consistent with the total number of OAC cases reported in SEER, which also aligns with the 10K incident cases quoted by Vaughn and Fitzgerald.1 The model therefore suggests that over 90% of OAC cases are attributable to BE.
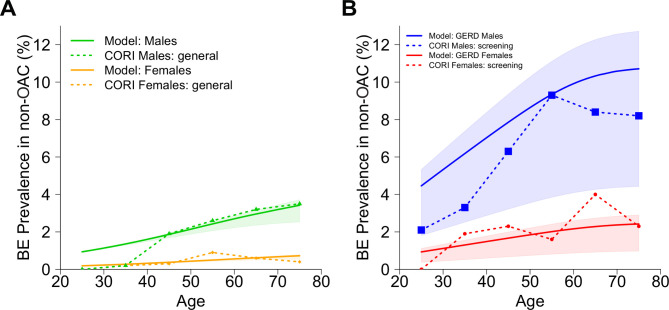
Second, we considered what the model simultaneously predicted for BE prevalence and BE-to-OAC progression rates in order to achieve the expected ~10K cases. Breaking down the contributions of the 2.2 million total BE patients estimated above, the model predicted BE prevalence to be 1.9%–2.4% in men and 0.4%–0.5% in women in the general US population ages 45–55 in 2010 (figure 3A). These predictions concur with best estimates5 6 and influence the total OAC cases predicted by the multistage model. To further explore implications for high-risk patients, we note that the model predicted a BE prevalence of 7.9%–9.3% in US men with symptomatic GERD who are cancer-free ages 45–55 in 2010 when the RR of BE vs non-GERD individuals is assumed to be RR=5 (figure 3B). This is also consistent with the estimate of 8% provided by Vaughan and Fitzgerald1 for prevalence of cancer-free BE diagnoses among GERD patients who undergo an upper endoscopy. Further, the model’s predicted age-specific BE prevalence curves by sex were consistent with previous results on BE prevalence from the CORI study20 (figure 3A). Compared with our model results and 8% quoted above1 for high-risk groups, the CORI study independently found similar BE prevalence in white men with GERD of 6.3% for ages 40–49 and 9.3% for ages 50–59 (figure 3B). To account for likely heterogenous RR of developing BE in GERD populations based on symptom onset age, BE length and other factors,4 22–26 we also considered a range of fixed values (RR=2–6) and found age-specific trends broadly consistent to overall BE prevalence results in CORI. Observed BE prevalence in white women undergoing screening was less precise in the CORI study data yet still coincided with our predictions for women (figure 3B).
Figure 3.
Model predictions for BE-positive yield in a cancer-free population (solid lines) are consistent with observed data (dashed lines) from Clinical Outcomes Research Initiative (CORI).20 (A) Solid lines show model results for the general US population stratified by sex from the 1950 birth cohort, with contributions of relative risk (RR) of BE from the age-specific, prevalent GERD population assumed to be RR=5 (shaded areas, RR=[2,6]). Dashed lines show consistency with observed BE prevalence data for patients without indication for screening in CORI, which are independent of the model. Model BE prevalence estimates are part of the evolutionary multistage process and thus affect predictions of the total OAC cases predicted (see Results). (B) Solid lines show model results for the symptomatic GERD subpopulation stratified by sex from the 1950 birth cohort with RR for BE set to RR=5. The shaded areas are predicted ranges for GERD subpopulations with fixed RR=2–6 to describe a wide range of increased risks of BE in published estimates, based on factors such as onset age of GERD and BE length. The true GERD-specific BE prevalence contributing to mathematical formulation used in (A) is within this region, where individual contributions are based on GERD onset age and underlying distribution of RR. Dashed lines show BE prevalence data for patients with GERD, and/or another indication for screening, in CORI. BE, Barrett’s oesophagus; GERD, gastro-oesophageal reflux disease; OAC, oesophageal adenocarcinoma.
In a sensitivity analysis, we also found these results to be robust to varying GERD prevalence in the model input for men and women in the population (see online supplemental material and online supplemental figure S1). When assuming smaller values of RR that lead to reduced BE prevalence in the GERD subpopulations for both sexes (see online supplemental material for details, online supplemental figure S2), the model still predicts that the majority of expected OACs (over 90%) develop in BE patients.
Finally, we previously found using this model that, for individuals born after 1940, the range of progression rates from BE-to-OAC was 0.10%–0.20% for men, and this was about twice as high as we found for women.13 These are plausibly low rates compared with current best estimates.11 27 Taken together, these secondary outcomes support the plausibility of our model’s predictions for numbers of OAC cases from BE annually.
The modelling results above imply that, even in the most conservative probability estimates, less than 10% of all annual OAC cases are unaccounted for beyond those expected to arise from BE. If there were a more significant alternate non-BE pathway than these numbers imply, then this model (which does not include a non-BE pathway) would have estimated either a much lower predicted population incidence of OAC than what was observed in SEER or shown greater inconsistencies with BE studies. In the latter case, the model would have estimated a greater prevalence of BE than what has been observed, and/or a greater rate of neoplastic progression among non-dysplastic BE than observed.
Discussion
Based on the published epidemiology of BE and OAC, our analysis suggests that a major alternative non-BE pathway to OAC is an unlikely scenario. The existence of such an alternative pathway was suggested by a retrospective analysis of macroscopic reports of OAC specimens diagnosed without BE in two cohorts from the USA and UK by Sawas and colleagues; however, their study conclusions remain speculative due to some important limitations including (1) a lack of longitudinally followed cases to OAC from non-BE patient oesophageal tissue and (2) the plausibility that small BE segments were completely overtaken by malignant expansions and thus were unmeasurable at cancer diagnosis.3 Moreover, our result that BE is the main origin of OAC does not necessarily refute the existence of differing phenotypes for OAC—the finding that the presence of BE was associated with better survival could plausibly be explained by the theory that more aggressive cancers are likely to replace the precursor BE more readily than less aggressive cancers. The stochastic nature of our model allows for variation in progression across a population and we explored a wide range of parameter values for rates defining the stochastic process from birth to clinical OAC and reached similar results, but there is still ultimately some uncertainty.
Indeed, genetic and epigenetic analyses have also consistently shown BE and OAC to be very similar,28–31 and one study that sought genomic differences between adenocarcinomas with and without BE failed to reveal molecular differences between the two.32 Nonetheless, this is fortunate news that, with adequate uptake, screening for BE by upper endoscopy or minimally invasive non-endoscopic technologies16 33 could potentially identify and enrol all patients who are at risk for developing OAC into a surveillance programme.
Although the overall progression to OAC is low in patients diagnosed with BE, for those selected BE patients who have high grade dysplasia and/or early OAC detected during surveillance, effective treatment can save lives. In this way, our analysis reinforces the primary goal in BE screening for OAC prevention—that effective surveillance of the entire BE population could potentially prevent the majority of mortality caused by OAC in the general population. Further, by mathematically analysing the time-dependent nature of cumulative risk of BE in GERD patients, we can also use our multistage model framework to improve identification of at-risk populations by optimising the timing of initial screening recommended for BE in symptomatic GERD.34 Although current intensive ‘one-size-fits-all’ surveillance strategies35–40 would lead to high costs for those over-diagnosed BE screen cases and surveillance strategies clearly need to improve, we conclude that there is a strong rationale for screening for BE to reduce OAC mortality.
Acknowledgments
The authors thank the NIDDK Clinical Outcomes Research Initiative (CORI) for access to endoscopy data.
Footnotes
Twitter: @yosoykit
Contributors: KC: statistical analysis; obtained funding; JHR: acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; KC, AC and JMI: study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; JMI obtained funding.
Funding: This work was supported by funding from NIH grants CISNET U01 CA152926, U01 CA199336 (JMI, KC and JHR), K24 DK080941 (JMI and KC) and a UKRI Rutherford Fund Fellowship (KC). JHR is also supported by NIH grant U54 CA163059 (BETRNET). AC is supported by NIH grants U54 CA163060 and P50 CA150964.
Competing interests: KC, JHR and JMI declare no potential conflicts of interest. AC has founders shares and stock options in LucidDx, serves as a consultant to LucidDx, has sponsored research with LucidDx and has a royalty interest in patents licensed to LucidDx. He is also a consultant for Interpace Diagnostics and receives research support from C2 Therapeutics/Pentax.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as Supplementary material. All data used in our analysis are publicly available. CORI data can be accessed through application with ethical approval to NIDDK (https://niddkrepository.org/studies/cori/). All equations are provided either in Figures and Supplementary material or were previously published along with model parameters. Code to solve equations was developed in R (V.3.6.1). Computational scripts are available at: github.com/yosoykit/BEtoEAC_Results.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Vaughan TL, Fitzgerald RC. Precision prevention of oesophageal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2015;12:243–8. 10.1038/nrgastro.2015.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wani S, Qumseya B, Sultan S, et al. Endoscopic eradication therapy for patients with Barrett's esophagus-associated dysplasia and intramucosal cancer. Gastrointest Endosc 2018;87:907–31. 10.1016/j.gie.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 3. Sawas T, Killcoyne S, Iyer PG, et al. Identification of prognostic phenotypes of esophageal adenocarcinoma in 2 independent cohorts. Gastroenterology 2018;155:1720–8. 10.1053/j.gastro.2018.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor JB, Rubenstein JH. Meta-Analyses of the effect of symptoms of gastroesophageal reflux on the risk of Barrettʼs esophagus. Am J Gastroenterol 2010;105:1730–7. 10.1038/ajg.2010.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zagari RM, Fuccio L, Wallander M-A, et al. Gastro-oesophageal reflux symptoms, oesophagitis and Barrett's oesophagus in the general population: the Loiano-Monghidoro study. Gut 2008;57:1354–9. 10.1136/gut.2007.145177 [DOI] [PubMed] [Google Scholar]
- 6. Ronkainen J, Aro P, Storskrubb T, et al. Prevalence of Barrett's esophagus in the general population: an endoscopic study. Gastroenterology 2005;129:1825–31. 10.1053/j.gastro.2005.08.053 [DOI] [PubMed] [Google Scholar]
- 7. Ward EM, Wolfsen HC, Achem SR, et al. Barrett's esophagus is common in older men and women undergoing screening colonoscopy regardless of reflux symptoms. Am J Gastroenterol 2006;101:12–17. 10.1111/j.1572-0241.2006.00379.x [DOI] [PubMed] [Google Scholar]
- 8. Rex DK, Cummings OW, Shaw M, et al. Screening for Barrett's esophagus in colonoscopy patients with and without heartburn. Gastroenterology 2003;125:1670–7. 10.1053/j.gastro.2003.09.030 [DOI] [PubMed] [Google Scholar]
- 9. Gerson LB, Shetler K, Triadafilopoulos G. Prevalence of Barrett's esophagus in asymptomatic individuals. Gastroenterology 2002;123:461–7. 10.1053/gast.2002.34748 [DOI] [PubMed] [Google Scholar]
- 10. Rubenstein JH, Morgenstern H, Appelman H, et al. Prediction of Barrett's esophagus among men. Am J Gastroenterol 2013;108:353–62. 10.1038/ajg.2012.446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kroep S, Lansdorp-Vogelaar I, Rubenstein JH, et al. An accurate cancer incidence in Barrett's esophagus: a best estimate using published data and modeling. Gastroenterology 2015;149:577–85. 10.1053/j.gastro.2015.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. SEER*Explorer: An interactive website for SEER cancer statistics . Surveillance research program, National cancer Institute. Available: https://seer.cancer.gov/explorer/ [Accessed 15 Apr 2019].
- 13. Kong CY, Kroep S, Curtius K, et al. Exploring the recent trend in esophageal adenocarcinoma incidence and mortality using comparative simulation modeling. Cancer Epidemiol Biomarkers Prev 2014;23:997–1006. 10.1158/1055-9965.EPI-13-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Curtius K, Hazelton WD, Jeon J, et al. A multiscale model evaluates screening for neoplasia in Barrett's esophagus. PLoS Comput Biol 2015;11:e1004272. 10.1371/journal.pcbi.1004272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kroep S, Heberle CR, Curtius K, et al. Radiofrequency ablation of Barrett's esophagus reduces esophageal adenocarcinoma incidence and mortality in a comparative modeling analysis. Clin Gastroenterol Hepatol 2017;15:1471–4. 10.1016/j.cgh.2016.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heberle CR, Omidvari A-H, Ali A, et al. Cost effectiveness of screening patients with gastroesophageal reflux disease for Barrett's esophagus with a minimally invasive cell sampling device. Clin Gastroenterol Hepatol 2017;15:1397–404. 10.1016/j.cgh.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruigómez A, García Rodríguez LA, Wallander M-A, et al. Natural history of gastro-oesophageal reflux disease diagnosed in general practice. Aliment Pharmacol Ther 2004;20:751–60. 10.1111/j.1365-2036.2004.02169.x [DOI] [PubMed] [Google Scholar]
- 18. Ruigómez A, Wallander M-A, Lundborg P, et al. Gastroesophageal reflux disease in children and adolescents in primary care. Scand J Gastroenterol 2010;45:139–46. 10.3109/00365520903428606 [DOI] [PubMed] [Google Scholar]
- 19. Curtius K, Wong C-J, Hazelton WD, et al. A molecular clock infers heterogeneous tissue age among patients with Barrett's esophagus. PLoS Comput Biol 2016;12:e1004919. 10.1371/journal.pcbi.1004919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubenstein JH, Mattek N, Eisen G. Age- and sex-specific yield of Barrett's esophagus by endoscopy indication. Gastrointest Endosc 2010;71:21–7. 10.1016/j.gie.2009.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. United States Census Bureau . Census of population and housing, 2015. Available: http://www.census.gov/prod/www/decennial.html [Accessed Feb 2020].
- 22. Thrift AP, Kramer JR, Qureshi Z, et al. Age at onset of GERD symptoms predicts risk of Barrett's esophagus. Am J Gastroenterol 2013;108:915–22. 10.1038/ajg.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisen GM, Sandler RS, Murray S, et al. The relationship between gastroesophageal reflux disease and its complications with Barrett's esophagus. Am J Gastroenterol 1997;92:27–31. [PubMed] [Google Scholar]
- 24. Anderson LA, Watson RGP, Murphy SJ, et al. Risk factors for Barrett's oesophagus and oesophageal adenocarcinoma: results from the FINBAR study. World J Gastroenterol 2007;13:1585–94. 10.3748/wjg.v13.i10.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edelstein ZR, Bronner MP, Rosen SN, et al. Risk factors for Barrett's esophagus among patients with gastroesophageal reflux disease: a community clinic-based case-control study. Am J Gastroenterol 2009;104:834–42. 10.1038/ajg.2009.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shaheen N, Ransohoff DF. Gastroesophageal reflux, Barrett esophagus, and esophageal cancer: scientific review. JAMA 2002;287:1972–81. 10.1001/jama.287.15.1972 [DOI] [PubMed] [Google Scholar]
- 27. de Jonge PJF, van Blankenstein M, Looman CWN, et al. Risk of malignant progression in patients with Barrett’s oesophagus: a Dutch nationwide cohort study. Gut 2010;59:1030–6. 10.1136/gut.2009.176701 [DOI] [PubMed] [Google Scholar]
- 28. Ross-Innes CS, Becq J, Warren A, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet 2015;47:1038–46. 10.1038/ng.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Secrier M, Li X, de Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 2016;48:1131–41. 10.1038/ng.3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu M, Maden SK, Stachler M, et al. Subtypes of Barrett's oesophagus and oesophageal adenocarcinoma based on genome-wide methylation analysis. Gut 2019;68:389–99. 10.1136/gutjnl-2017-314544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y, Sethi NS, Hinoue T, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell 2018;33:721–35. 10.1016/j.ccell.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferrer-Torres D, Nancarrow DJ, Kuick R, et al. Genomic similarity between gastroesophageal junction and esophageal Barrett's adenocarcinomas. Oncotarget 2016;7:54867–82. 10.18632/oncotarget.10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moinova HR, LaFramboise T, Lutterbaugh JD, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett's esophagus. Sci Transl Med 2018;10:aao5848. 10.1126/scitranslmed.aao5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Curtius K, Dewanji A, Hazelton WH, et al. Optimal timing for cancer screening and adaptive surveillance using mathematical modeling. bioRxiv 2020. 10.1101/2020.02.11.927475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Codipilly DC, Chandar AK, Singh S, et al. The effect of endoscopic surveillance in patients with Barrett's esophagus: a systematic review and meta-analysis. Gastroenterology 2018;154:2068–86. 10.1053/j.gastro.2018.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spechler SJ, Sharma P, Souza RF, et al. American Gastroenterological Association Technical Review on the Management of Barrett's Esophagus. Gastroenterology 2011;140:e18–52. 10.1053/j.gastro.2011.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shaheen NJ, Falk GW, Iyer PG, et al. American College of Gastroenterology. ACG Clinical Guideline: Diagnosis and Management of Barrett’s Esophagus. Am J Gastroenterol 2016;111:30–50. 10.1038/ajg.2015.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fitzgerald RC, di Pietro M, Ragunath K, et al. British Society of Gastroenterology guidelines on the diagnosis and management of Barrett's oesophagus. Gut 2014;63:7–42. 10.1136/gutjnl-2013-305372 [DOI] [PubMed] [Google Scholar]
- 39. di Pietro M, Fitzgerald RC, BSG Barrett's guidelines working group . Revised British Society of Gastroenterology recommendation on the diagnosis and management of Barrett's oesophagus with low-grade dysplasia. Gut 2018;67:392–3. 10.1136/gutjnl-2017-314135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qumseya B, Sultan S, Yang J, et al. ASGE guideline on screening and surveillance of Barrett's esophagus. Gastrointest Endosc 2019;90:335–59. 10.1016/j.gie.2019.05.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-321598supp001.pdf (1.2MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as Supplementary material. All data used in our analysis are publicly available. CORI data can be accessed through application with ethical approval to NIDDK (https://niddkrepository.org/studies/cori/). All equations are provided either in Figures and Supplementary material or were previously published along with model parameters. Code to solve equations was developed in R (V.3.6.1). Computational scripts are available at: github.com/yosoykit/BEtoEAC_Results.