Abstract
RASSF1A, one of the eight isoforms of the RASSF1 gene, is a tumor suppressor gene that influences tumor initiation and development. In cancer, RASSF1A is frequently inactivated by mutations, loss of heterozygosity, and, most commonly, by promoter hypermethylation. Epigenetic inactivation of RASSF1A was detected in various cancer types and led to significant interest; current research on RASSF1A promoter methylation focuses on its roles as an epigenetic tumor biomarker. Typically, researchers analyzed genomic DNA (gDNA) to measure the amount of RASSF1A promoter methylation. Cell-free DNA (cfDNA) from liquid biopsies is a recent development showing promise as an early cancer diagnostic tool using biomarkers, such as RASSF1A. This review discusses the evidence on aberrantly methylated RASSF1A in gDNA and cfDNA from different cancer types and its utility for early cancer diagnosis, prognosis, and surveillance. We compared methylation frequencies of RASSF1A in gDNA and cfDNA in various cancer types. The weaknesses and strengths of these analyses are discussed. In conclusion, although the importance of RASSSF1A methylation to cancer has been established and is included in several diagnostic panels, its diagnostic utility is still experimental.
Keywords: RASSF1A, epigenetic alteration, DNA methylation, biomarker
INTRODUCTION
Cancer is a significant cause of mortality worldwide, despite the tremendous progress that has been made in treatments [1]. One constant, however, is that for the successful treatment of cancer, early diagnosis is crucial. Therefore, molecular biomarkers, which can be sensitive enough to detect initial malignant transformations and progressions, are extensively studied. Biomarker candidates are developed from the knowledge of cancer development mechanisms, which involves both genetic and epigenetic abnormalities. Epigenetic abnormalities may allow early tumor detection by noninvasive methods, which would be of significant importance.
In tumor diagnosis and screening, tissue biopsies are still standard in the procedure. They can be invasive, challenging to perform, and generally are not suitable for screening. In contrast, liquid biopsies, or sampling and analysis of non-solid biological tissue such as blood, represents a less invasive diagnostic procedure with the potential to facilitate the early detection of the tumor [2,3]. Therefore, liquid biopsies as cancer biomarker sources are a quickly developing research topic. Most studies regarding liquid biopsies focus on investigating circulating tumor cells and cell-free DNA (cfDNA) released into the bloodstream. CfDNA represents small fragments of DNA released from the normal or tumor tissue as a consequence of cellular apoptosis or necrosis [4,5]. The cancer patients usually have higher cfDNA concentrations in their bloodstream because of increased tumor volume associated with increased cell apoptosis and necrosis [6]. Although blood (plasma and serum) is the most investigated body fluid, the search for specific tumor biomarkers includes other body fluids as well, including urine and saliva [7].
DNA methylation is traditionally the most investigated epigenetic modification [8], and it can be experimentally identified in liquid biopsies. Abnormal DNA methylation may be classified as hyper- or hypomethylation. For the last two decades, DNA methylation patterns of tumor suppressor genes have been investigated as potential biomarkers for many types of cancer. In that time, research has found that global DNA hypomethylation and regional hypermethylation, frequently affecting the promoter region of tumor suppressor genes, are characteristic in cancer [9,10]. One of the highlighted and most-researched biomarkers in liquid biopsies is RASSF1A.
RASSF1A gene is within the region of the 3p21.3 chromosome, which is sensitive to genetic and epigenetic changes in many tumors. Loss of heterozygosity (LOH) often occurs in this region, and promoter hypermethylation represents the mechanisms that lead to loss of RASSF1A gene expression. It is important to note that RASSF1A is more frequently inactivated by promoter hypermethylation than by LOH [11]. Aberrant RASSF1A methylation was detected in various cancers, including breast cancer, lung cancer, gastrointestinal cancer, prostate cancer, and testicular germ cell tumors. In some cancers, RASSF1A is still expressed but in suboptimal or supra-optimal concentration, which leads to disturbed signal transduction and consequent malignant transformation [12].
The Ras association domain family 1 isoform (RASSF1A) is a tumor suppressor gene that belongs to the C-terminal RASSF family. RASSF1 gene has eight isoforms, of which RASSF1A and RASSF1C are the most abundantly expressed [12]. These two isoforms are omnipresent in the normal cells, where they localize microtubules and regulate cell growth. RASSF1A is activated by mitogenic stimuli and KRAS seems to be main RASSF1A activator upon mitogenic stimulation [12]. RASSF1A responds to above described stimuli by regulating the cell cycle progression, apoptosis, and microtubule stability, while RASSF1C is involved in DNA damage response. Normally, RASSF1C is anchored to the the DAXX, a Death domain-associated protein and localized in the nuclei. DNA damage leads to DAXX degradation and RASSF1C releases into the cytoplasm where it activates the c-Jun N-terminal kinases (JNK) pathway [14]. However, in tumorigenesis. RASSF1C acts as an oncogene, which directly contrasts to RASSF1A, a tumor suppressor (Figure 1) [13].
FIGURE 1.
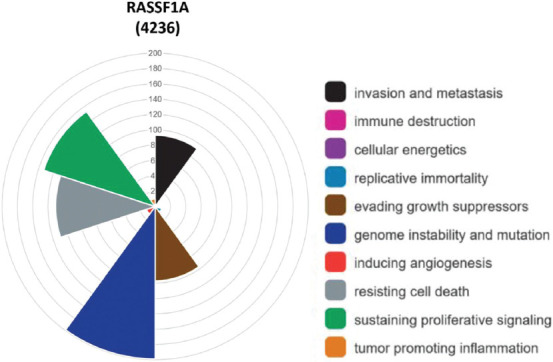
Distribution of hallmarks of cancer for RASSF1A. In 4236 scientific articles was detected that RASSF1A expression contributes the most to genome instability and mutation, and sustains proliferative signaling. Furthermore, it enables the cell to resist death, to evade growth suppressors and favors invasion of cancer and metastasis
Epigenetic inactivation of RASSF1A
The RASSF1 locus is modulated by two CpG islands, A and C; CpG island A is in the regulatory region of RASSF1A. The methylation of CpG islands A has been detected in normal tissue and does not affect gene expression. On the other hand, hypermethylation was associated with loss of RASSF1A expression [13]. DNA methyltransferases (DMNTs) mediate the methylation of these CpG islands. When DMNTs are dysregulated, its actions in cancer cells lead to the epigenetic silencing of RASSF1A. It has been shown that MUC1-C or p53 protein bind to the RASSF1A promoter and consequently activates their corepressors ZEB1 and DAXX. MUC1-C-ZEB1 complex recruits DMNT3B, while p53-DAXX complex recruits DMNT1. This activity causes CpG island’s methylation in the RASSF1A promoter region and the subsequent loss of RASSF1A expression [16]. In cancer cells, loss of RASSF1A because of this cascade leads to the binding of RASSF1C to the RASSF1A effectors, which, in turn, favors tumorigenesis [14,15]. Epigenetic inactivation of RASSF1A causes disturbance in various signaling pathways, as outlined in Figure 2.
FIGURE 2.
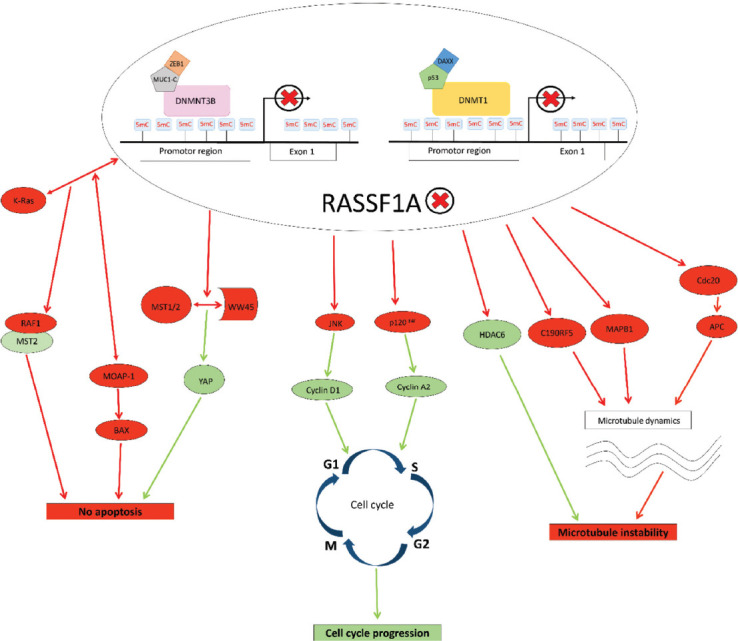
Impact of epigenetic inactivation of RASSF1A to cell signaling pathways. Red arrows represent disturbed or deactivated, while green ones represent activated signaling pathways. When expressed normally, RASSF1A causes repression of cyclin A2 and cyclin D1, which results in cell cycle arrest. RASSF1A also modulates apoptosis. Interactions of RASSF1A with K-Ras activates the MST2-LATS1 apoptotic pathway, i.e., RASSF1A modulates the RAF-1 activity due to competition with MST2 for RAF-1 binding. Also, the interaction of RASSF1A with K-Ras, enhances the interaction of RASSF1A and MOAP-1, promoting RASSF1A’s ability to induce BAX translocation to the mitochondria and cell death. RASSF1A binds to MST1/2 with adaptor protein WW45 that causes phosphorylation, respectively, leading to YAP phosphorylation and inhibition. Additionally, RASSF1A plays an important role in microtubule stability, by inhibiting HDAC6 (histone deacetylase 6), which results in an increase of acetylated microtubules, that are more stable. RASSF1A binds to microtubule-associated proteins (MAPs) which regulate microtubule stability. If RASSF1A is epigenetically inactivated, it causes microtubule instability, repression of apoptosis, and progression of the cell cycle, which favors tumorigenesis
This review summarizes the current knowledge on DNA methylation of RASSF1A in genomic DNA (gDNA) and cfDNA in various tumors. We provide a comprehensive assessment of the diagnostic and prognostic value of RASSF1A in cfDNA and gDNA, highlighting some potential benefits. We will also be discussing some limitations regarding the utility of RASSF1A methylation as a tumor biomarker in clinical practice.
Breast cancer
Breast cancer (BC) is the most frequent malignancy among women worldwide, and early detection of BC broadly defines disease management planning and the course of treatment. When mammography was implemented as a routine diagnostic procedure for BC, there was an overall increase in early detection. However, there were unintended consequences, along with that improvement. According to Nelson et al. [17], the rates of false-positive mammography results were the highest for women between 40 and 49 years old and decreased with increasing age. Also, rate of false-positive mammography results were statistically significantly higher for women with specific risk factors then for those without them [17]. Mammography and ultrasound cannot always differentiate benign and malignant lesions [18,19], so women with false-positive results received biopsies. Therefore, there is significant research interest in RASSF1A methylation in gDNA and cfDNA as a possible BC biomarker.
According to UALCAN, the β value of RASSF1A methylation in BC patients is 0.311, hypermethylated compared to healthy tissue (β value is 0.22). RASSF1A methylation frequency in BC tissue samples in gDNA was detected in around 65.0% of BC tissue samples [20,21]. Regarding cfDNA, the RASSF1A methylation frequency was detected in the range from 63.3% [20,22] to 16.6% [21,23]. All studies that detected such a wide range of RASSF1A methylation in cfDNA used the same method (methyl-specific PCR) for RASSF1A methylation analysis. The only difference was specific primers, which might explain diversity in RASSF1A methylation frequency and show a specific site in the promoter region, which is more methylated than the others and might have a greater impact on BC tumorigenesis. Indeed, two different primer pairs gave different results regarding the methylation frequency of RASSF1A in the same study [20]. Different technologies for methylation detection could give different results than reported by Skalski and Paulszczak [24].
The reported results of variability in the RASSF1A methylation analysis in cfDNA of BC patients show that the RASSF1A methylation pattern in cfDNA requires further investigation before it is considered a possible BC biomarker.
In addition to using different specific primer pairs, the potential challenge could be that the entire first RASSF1A exon contains a CpG island [25]. This CpG island is methylated in normal breast tissue [26], and the methylation of RASSF1A was also detected in the promoter region in some healthy control samples. Methylation of RASSF1A in healthy controls could indicate that methylated RASSF1A would be useless as a breast cancer biomarker because it does not differentiate enough between healthy tissue and tumor tissue. However, RASSF1A methylation was not as frequent as seen in BC patient samples. Based on these results, the authors pointed out that RASSF1A methylation in BC occurs progressively from the first exon to the promoter region and is beginning early in breast tumorigenesis [26]. This finding can help us understand why some studies detected hypermethylated RASSF1A in healthy controls and consequently excluded RASSF1A from further investigation [27].
Strikingly, RASSF1A was more frequently methylated in the serum of healthy controls than in the serum of patients with benign breast disease [22,28], a particularly important finding when considering using these patients for controls. This finding may lead to incorrect associations regarding BC patients’ methylation patterns and healthy, tumor-free women.
Clinicopathological parameters
Hypermethylated RASSF1A was found associated with clinicopathological parameters, meaning that besides diagnostic value, it could have prognostic value as well. A critical clinicopathological parameter for BC diagnosis, treatment, and prognosis is the estrogen receptor status (ER) since ER-positive (ER+) BC tumors are less aggressive. Methylation of RASSF1A in gDNA and cfDNA is associated with ER and progesterone receptor status (PR). Hypermethylated RASSF1A was detected with high frequency in ER-positive/PR-positive (ER+/PR+) (20,29,30). Furthermore, higher methylation levels of RASSF1A were detected in ER+/PR+ BC tissue samples when compared to ER+/PR- tumors. [31]. Hypermethylated RASSF1A from gDNA and cfDNA was strongly associated with tumor size and poor prognosis. However, the literature is not consistent as some authors did not detect any significant association between methylation in cfDNA or gDNA with clinicopathological parameters [21]. Furthermore, DNA methylation of RASSF1A seemed to be associated with lymph node metastasis (LN) [32], showing a similar methylation frequency in both gDNA and cfDNA [20].
Future clinical utility
RASSF1A methylation status as cancer biomarker has high sensitivity and specificity for detecting BC in gDNA, while in cfDNA, RASSF1A biomarker specificity was 100.0%, but sensitivity was low 78.83% [22,31]. However, no single gene was hypermethylated in all BC patients, and a specific gene panel should be favored in the diagnosis [33]. Panel testing for aberrantly methylated genes in BC, which included RASSF1A in gDNA and cfDNA, resulting in a better diagnostic performance compared to RASSF1A methylation analysis exclusively [31]. Furthermore, when analyzing a panel of genes, the panel had a higher diagnostic-sensitivity than mammography for tumors ≤1 cm. For tumors >2 cm, the sensitivity was lower than for mammography [28]. These findings suggest a gene panel has the potential for use in early diagnosis protocols. The same panel could discriminate between BC patients and healthy women with 79.6% sensitivity and a specificity of 72.4%. Importantly, methylation panel testing was able to distinguish BC patients from women with benign breast diseases, with high specificity (78.1%) and even higher sensitivity (82.4%) (28).
Lung cancer
According to GLOBOCAN 2018, lung cancer (LC) has the highest mortality rate of all cancers worldwide. Because of the absence of symptoms in the early stages of the disease, approximately 70% of patients are diagnosed in an advanced stage. Low-dose computed tomography (LD-CT) is currently considered the best LC screening method available [34]. However, LD-CT screening of high-risk smokers had shown 24% positivity among which 96% of the individuals tested positive were false positive [35]. Therefore, there is a need for a new, more accurate screening approach for LC, which can provide early detection.
As with BC, aberrant methylation of RASSF1A was detected in LC. Compared to BC, hypermethylated RASSF1A was detected only in patients with LC, compared to none in healthy control samples [36–39]. This evidence suggests RASSF1A is more discriminative between LC and healthy controls than between BC and healthy controls. Significantly, different RASSF1A methylation frequencies were observed between small-cell (SCLC) and non-small cell lung cancer (NSLC) [40]. RASSF1A methylation frequency in gDNA and cfDNA was higher among SCLC patients than among NSCLC [41,42]. As SCLC represents a more aggressive LC than NSLC, higher RASSF1A methylation frequency could be linked to more aggressive LC [40].
Furthermore, studies with LC patients for cfDNA testing potential detected that RASSF1A methylation in gDNA was not always accompanied by methylation in cfDNA [36]. This relationship should be considered, since methylated RASSF1A in cfDNA could be detectable only in already advanced stages of the disease.
Clinicopathological parameters
Smoking represents a high risk of developing LC, and research has shown that the methylation of RASSF1A was higher in smokers than in non-smokers [43]. Even the duration of the number of years smoking was associated with hypermethylation of RASSF1A, detected with an observational study [39]. Interestingly, LC from men exhibited higher methylation levels of RASSF1A than those from women (7.5% versus 17.9%, P < 0.01) [44]. This finding indicates that RASSF1A methylation could have different efficacies for men than women.
Methylation of RASSF1A was determinative between NSCLC than SCLC [45,46]. This difference was also detected by UALCAN, where β value of adenocarcinomas, one of the main subtypes of NSCLC was slightly higher than of SCLC, i.e., mean of β is 0.265 in adenocarcinomas and 0.243 in SCLC. As adenocarcinoma is the most common type of LC [47], RASSF1A methylation could be used to diagnose most LC, particularly among non-smokers low-risk populations [48]. Considering overall survival, patients with hypermethylated RASSF1A have lower overall survival rates than patients without hypermet0hylated RASSF1A [43,49].
There is also a significant association was detected between RASSF1A and TNM stages. When TNMs were investigated separately, there was no significant difference. However, when TNM I and II were observed as one group vs. TNM III, methylation of RASSF1A was significantly associated with TNM III [40,43]. RASSF1A levels in cfDNA from blood plasma were significantly higher in LC patients with distant metastatic disease [50]. Based on the presented studies, RASSF1A seems like a promising prognostic biomarker for LC. However, the research is inconsistent for associations between clinicopathological parameters and RASSF1A methylation in tissue or blood samples (37,44,45,51). This discrepancy was often explained that the number of samples was significantly lower than in studies in which RASSF1A hypermethylation was associated with the clinicopathological parameters.
Future clinical utility
Specific genes’ hypermethylation can be detected with other noninvasive sampling techniques, including plasma and sputum [52], which is very useful for LC. Sputum contains exfoliated epithelial cells where hypermethylation of RASSF1A was detected. RASSF1A methylation individually showed high specificity (96.5%), while the sensitivity was significantly lower (42.5%) [53]. Regarding cfDNA, RASSF1A, together with p16INK4a, represents the most frequently reported gene in blood-based liquid biopsies displaying 22%–66% sensitivity and 57%–100% specificity for LC detection [54]. However, when incorporated into the methylation gene panel, both the sensitivity and specificity for RASSF1A, together with p16INK4a, were significantly higher in cfDNA [50,55]. Besides, the sensitivity of hypermethylated RASSF1A in gDNA was higher when RASSF1A was incorporated into a methylation gene panel, while specificity was more or less the same [53,56]. Bronchial lavage fluid (BLAF) represents a less invasive method than tissue biopsy and is an excellent candidate for a source of biomarkers for early detection. In gDNA from BLAF and tumor tissue sample, similarly to gDNA from sputum, the methylation gene panel’s sensitivity increased significantly [57,58]. Methylation panel of SHOX2 and RASSF1A in BLAF demonstrated dominantly higher sensitivity than cytological examination or analysis of serum biomarker carcinoembryonic antigen, which is both used for cancer screening [59]. Although BLAF sampling represents an invasive method with potential complications, it represents a promising diagnostic tool for detecting LC’s early detection. Due to LC’s nature, where sputum and BLAF represent a better source of LC biomarkers than blood, methylation of RASSF1A in gDNA showed a better diagnostic value than in cfDNA.
Gastrointestinal cancer
Like LC, gastrointestinal cancer (GC) causes high mortality worldwide since they are diagnosed at an advanced stage when the prognosis is poor, and the treatment options are limited [60]. The diagnosis of GC usually requires endoscopy, followed by a biopsy of tissue from a suspicious area. Therefore, it represents an invasive and unpleasant diagnostic method that many people avoid until the disease’s clinical manifestations. GC is a very heterogeneous and complex disease that involves many genetic and epigenetic alterations. Therefore, biomarkers useful for molecular diagnosis would enable earlier detection and diagnosis. In studies of methylation patterns of different genes in GC such as gastric, liver, and colorectal cancer, aberrant methylation of RASSF1A has emerged as a promising biomarker.
In colorectal cancer, RASSF1A was silenced by hypermethylation in aberrant crypt foci (ACF) [61]. ACF represents the earliest morphologically indeterminate mucosal abnormality in the colon, which can progress to colorectal cancer (CRC), meaning hypermethylated RASSF1A could be used for the detection of possible precancerous subsets [62]. Methylation of RASSF1A in morphologically indeterminate mucosal abnormality in the colon was confirmed in studies where gDNA from adjacent tumor-free mucosal tissue served as a control and hypermethylated RASSF1A was found in these controls [63]. The situation is different in gastric cancer, where hypermethylated RASSF1A was detected in cfDNA of patients with benign gastric disease, such as chronic gastritis, gastric ulcers, and benign polyp, non-malignant adenoma, and ulcerative colitis, but in none of the healthy tumor-free control [64]. Nevertheless, this evidence suggests hypermethylated RASSF1A plays a role in early gastric carcinogenesis, which may be initiated in adjacent tumor-free tissue near the tumor region. In hepatocellular carcinoma (HCC), the most common liver malignancy, hypermethylated RASSF1A, was also detected in cancer patients’ tissue and serum samples, but was not detected in healthy controls [65,66]. In the early stages, a diagnosis of HCC, when the treatment is still favorable, seems complicated because about 75,0% HCCs coexist with liver cirrhosis [67]. However, a comparison of serum samples of patients suffering from liver cirrhosis to those with HCC showed that RASSF1A hypermethylation is relatively specific to HCC [68,69]. A higher RASSF1A methylation frequency was found in cfDNA of patients with chronic hepatitis C infection than in healthy controls, but this increase was lower than in HCC patients [70]. Hepatitis C infection could promote HCC development through disruption of the RASSF1A methylation frequency/pattern.
Clinicopathological parameters
In the context of the possible prognostic biomarker for gastric, colorectal, and liver cancer, RASSF1A showed potential. Hypermethylated RASSF1A in gDNA was associated with TNM stages in gastric cancer, where frequencies of hypermethylated RASSF1A in patients with stages III and IV were significantly higher than in stages I and II [71]. A significant association of RASSF1A promoter hypermethylation and more advanced stages of the disease was also observed in cfDNA from gastric cancer patients [64]. Hypermethylated RASSF1A was associated with advanced stages in liver cancer [72]. A significant association between LN metastasis and RASSF1A methylation was found in cfDNA of gastric cancer patients [64]. Hypermethylated RASSF1A was linked to poor prognosis and low overall survival in liver cancer and CRC patients [73–75], and the same correlation was found in CRC, although in gDNA [63]. On the other hand, in one study on gastric and colorectal adenocarcinoma patients, no significant difference in RASSF1A methylation between preoperative serum samples and four weeks postoperative serum samples was found [42] and implied it not be useful in the surveillance prediction of gastric and colorectal adenocarcinoma patients.
Future clinical utility
An elevation in alfa-fetoprotein (AFP) level is a widely used serum marker for HCC screening. The cut-off value of serum concentration of 20 ng/mL is commonly used to differentiate HCC patients from healthy adults in clinical studies [76]. However, AFP specificity and sensitivity are limited because the AFP level is elevated in non-malignant liver diseases, like inflammation and liver cirrhosis. Hypermethylation of RASSF1A in cfDNA of HCC patients had showed high specificity in discrimination of HCC patients from healthy controls [77]. Moreover, individual RASSF1A methylation status displayed good diagnostic performance regarding discrimination between HCC patients and patients with hepatitis C [70]. Incorporation of RASSF1A into methylation panel with different genes further improved diagnostic performance [77,78]. RASSF1A, APC, GSTP1, and SFRP1 gene panel has successfully discriminated HCC patients from healthy controls with 84,7% sensitivity and 87,8% specificity [79]. Methylation statuses of RASSF1A, BVES, and HOXA9 in serum, when analyzed together, showed 73,5% sensitivity and 91,1% specificity for distinguishing between HCC and chronic hepatitis patients [69]. Additionally, when aberrantly methylated APC, COX2, and RASSF1A, were combined with other epigenetic marker miR-203, 75,0% of HCC cases were underdiagnosed by AFP measurement [80].
Methylation of RASSF1A in other cancers
Besides the association of RASSF1A methylation with BC, LC, and GC, cancers with the highest incidence worldwide, the diagnostic and prognostic value of RASSF1A methylation were also investigated in other malignancies. Patients with renal cell cancer (RCC) and prostate cancer (PC) could be symptom free in the early stage, while diagnosing the disease has already progressed. Also, routine diagnostics methods for RCC and PC involve substantial patient discomfort and have variable sensitivity [81]. Given the shortcomings of current screening methods and predictive biomarkers, the development and implementation of useful biomarkers for early detection are crucial.
For bladder cancer (BCA), a similar RASSF1A methylation level was found in cfDNA from BCA patients and patients with non-malignant bladder disease [82]. These results question the utility of methylation of RASSF1A as a biomarker for BCA because of low specificity [83]. In RCC [84–86] and PC [50], a wide range of RASSF1A methylation frequency in cfDNA was reported. The variable RASSF1A methylation frequency ratio was also detected in gDNA from head and neck squamous cell carcinoma (HNSCC) [87]. A discrepancy regarding HNSCC may arise from the lower importance of RASSF1A hypermethylation in HNSCC tumorigenesis. Indeed, hypermethylation of other genes, such as MGMT, DAPK, and p16, was more frequently detected in HNSCC patients [88]. Methylation of RASSF1A in cfDNA from HNSCC patients has not yet been reported. In testicular germ cell tumor (TGCT), methylation of RASSF1A in gDNA was discriminative between one of the TGCT subgroups, seminomas, and healthy tissue [89]. In contrast, RASSF1A methylation in cfDNA was not investigated to date.
Most cancers are presented with heterogeneous groups of various more or less similar cellular entities, so discrimination between tumor subtypes is crucial. RASSF1A seems to meet this challenge. Combined methylation analysis of two genes, HOXA9/RASSF1A, had successfully distinguished two subgroups, nonseminomas and seminomas in TGCT diagnostics [90,91]. In BCA, significantly higher methylation was detected in muscle-invasive BCA than noninvasive one [66,83]. Regarding RCC, RASSF1A methylation in gDNA had discriminated well clear cell renal carcinoma (ccRCC) from papillary renal carcinoma (pRCC) patients. Higher methylation frequency was detected in pRCC [92,93], but the methylation of RASSF1A in cfDNA was not associated with RCC’s histological subtypes [86].
Besides diagnostic value, the association of RASSF1A methylation and clinicopathological parameters was proven, indicating its prognostic value in several tumors. A strong association of hypermethylated RASSF1A in gDNA and cfDNA, respectively, and aggressive PC was detected, where hypermethylated RASSF1A correlated with Gleason score and serum PSA [94,95]. Hypermethylation of RASSF1A was linked to high grade and advanced-stage tumors of cervical cancer [96], HNSCC [97], RCC [98], BCA (66,81,99), melanoma [100], and brain tumor [101]. Hypermethylated RASSF1A in TGCT was associated with chemotherapy resistance [102] and lower response to treatment [103]. In tumors of the central nervous system (CNS), hypermethylated RASSF1A in cfDNA was found more frequently in glial tumors than in metastatic CNS neoplasms [104]. Hypermethylated RASSF1A was also linked to a lower overall survival rate in HNSCC [71].
Hypermethylated RASSF1A was associated with risk factors for several tumors. Tobacco use and human papillomavirus (HPV) infection are some primary risks for HNSCC [105,106], and it was shown that methylation of RASSF1A can successfully distinguish smokers and non-smokers [107]. HPV positive patients had presented significantly higher methylation of RASSF1A than HPV negative patients [108].
In the investigation of hypermethylated RASSF1A in several cancers, adjacent non-malignant tissue, used as a control group, could present a challenge. As malignant transformation is already present in these patients’ organisms, it could reflect the epigenome in the more or less healthy region near the tumor [109]. The possible reflection of malignant transformation on epigenome in the adjacent non-malignant tissue could explain why several studies had found high RASSF1A methylation frequency in tumor tissue and adjacent non-malignant tissue, e.g., in RCC (85,110,111) and HNSCC [112–114]. The reported level of RASSF1A DNA methylation should be contextualized with lifestyle as well. A lot of HNSCC patients have been smoking, and it was shown that DNA methylation of DAPK1, p16INK4a, and RASSF1A is significantly different in smokers and non-smokers [107], regardless of the presence of malignant tissue. Therefore, DNA methylation patterns of genes in adjacent non-malignant tissue could be altered because of smoking, but not only because of cancer. Adjacent non-malignant tissue does not represent an adequate control group for DNA methylation studies [115].
RASSF1A DNA methylation in gDNA vs. cfDNA
DNA methylation is nowadays widely used to detect cancer, triage, screening, or surveillance. Methylation patterns can be detected in gDNA or cfDNA. Since gDNA can be isolated from circulating tumor cells, its analysis can be used in liquid biopsies samples and cfDNA. Indeed, diagnostic tests detect aberrant methylation in cancer gDNA from circulating tumor cells in liquid biopsies [116]. In the case of RASSF1A methylation, it has been implemented into the diagnostic test for PC detection. Namely, the ConfirmMDx test detects GSTP1, RASSF1A, and APC genes that exhibit increased methylation in prostate tumor tissue [117]. As tumors are very heterogeneous, simple tissue biopsy provides only a fragment of cancer cell subpopulation in the tumor, giving a misleading image of the tumor’s true cellular content. In cfDNA, all tumor cell types contribute to the total cfDNA compartment, providing more accurate tumor composition information. The potential challenge regarding cfDNA could represent rare tumor sub-populations and early stage tumors, in which cfDNA concentration could be too low, making it undetectable by a diagnostic test. Despite all these challenges, various tests for detecting aberrantly methylated genes in cfDNA were developed and integrated into clinical practice. Epi proColon detects methylated SEPT9 in CRC patients, while COLVERA detects methylated BCAT1 and IKZF1 genes, which are associated with colorectal tumor growth. Furthermore, HCCBloodTest detects methylated SEPT9 in plasma samples of cirrhotic patients, which are at high risk for developing HCC. The therascreen MGMT Pyro test detects methylated MGMT, which is associated with the treatment response in the case of glioblastoma, and Epi proLung test detects aberrant methylated PTGER4 and SHOX2 in cfDNA from blood plasma in LC [116].
Challenges in translating RASSF1A DNA methylation as a useful cancer biomarker
As discussed in this review, the expression of RASSF1A is altered by hypermethylation in various tumors. Its methylation was extensively studied as a potential diagnostic and prognostic tumor biomarker. Indeed, based on the evidence, RASSF1A methylation showed an excellent diagnostic performance in distinguishing several tumors from healthy controls. It was linked to various clinicopathological parameters that pointed out its potential value in future clinical use (Figure 3).
FIGURE 3.
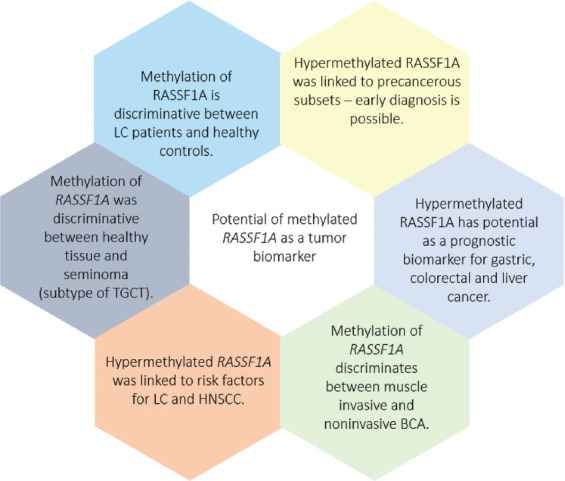
Potential of RASSF1A methylation as a tumor biomarker
However, to validate the hypermethylation of RASSF1A as a useful cancer biomarker, some challenges should be addressed (Figure 4). A contradictory data on RASSF1A methylation frequency was detected in various malignant diseases for cfDNA and gDNA, which could be because different studies analyzed different CpG sites, and not all of them are equally methylated or biologically active in terms of gene expression. Regarding cfDNA, this wide range of contradictory results could be because of studies of cfDNA collected from patients with different cancer stages. The low methylation frequency could reflect early stage tumors, which are not highly vascularized and have a low necrosis rate in the tumor core.
FIGURE 4.
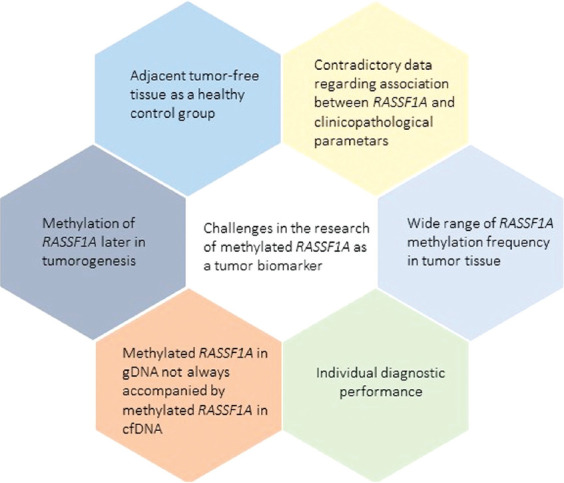
Drawbacks that presented itself in the research of methylated RASSF1A as a tumor biomarker. Highlighted drawbacks should be taken into account and possibly solved, before the translation of methylated RASSF1A as a tumor biomarker in the clinic
Furthermore, there were cases where RASSF1A in gDNA was methylated but not in paired cfDNA samples. This could be explained by the fact that the methylation in cfDNA can be detected after the malignant cells are necrotized. In contrast, gDNA methylation can be detected when the malignant cell is still viable. Thus, the failure of methylated RASSF1A detection in cfDNA does not rule out the possibility of the tumor presence. It can also be that methylation of RASSF1A occurs later in tumorigenesis, so that cfDNA from these tumors carrying aberrantly methylated RASSF1A is not yet present in the blood. This may also suggest that when the methylated RASSF1A is detected in the peripheral blood, the tumor is present in the advanced stage. If this would be the case, detection of aberrant RASSF1A methylation in cfDNA might not be suitable for early diagnosis.
There are additional discrepancies between some studies where various degrees of association of RASSF1A methylation and clinicopathological parameters were analyzed. This inconsistency in RASSF1A methylation could implicate that methylation of RASSF1A does not play such an essential role in disease development as it was presumed. It might be that the aberrant methylation of other genes has a more significant influence on the tumorigenesis of some cancers. Furthermore, from studies of RASSF1A methylation in tumor tissue samples, it must be determined which tissue samples can be used as a control group. The usage of tumor-free benign disease tissue or adjacent tumor-free tissue might lead to wrong conclusions about RASSF1A methylation alterations in tumorigenesis. This suggests that referent healthy tissue should be used more frequently to analyze altered gene methylation in cancers.
Methylation of RASSF1A showed good diagnostic value individually, but in most studies, its sensitivity was low. Therefore, it was considered advantageous to use a panel of genes for cancer screening procedures since it usually provides more accurate diagnostic and prognostic information. Indeed, a gene panel of GASP1/APC/RASSF1A is currently used to diagnose PC. Furthermore, as aberrantly methylated RASSF1A was detected in various cancer types, a panel of aberrantly methylated genes that includes RASSF1A could be more specific for screening various cancer types.
CONCLUSION
A body of evidence shows that epigenetic inactivation of RASSF1A is strongly associated with tumorigenesis and cancer behavior. However, its specificity and sensitivity are increasing when combined with other aberrantly methylated genes. RASSF1A DNA methylation can be a cancer biomarker, although some critical issues must be addressed before translation into routine clinical practice.
ACKNOWLEDGMENTS
This publication was financed by the European Union through the European Regional Development Fund, Operational Programme Competitiveness and Cohesion, under grant agreement No. KK.01.1.1.01.0008, Operational programme competitiveness and cohesion, under grant agreement No. KK.01.1.1.01.0008, Regenerative and Reproductive Medicine - Exploring New Platforms and Potentials. This work has been supported in part by Croatian Science Foundation under the project “Epigenetic biomarkers in blood and ejaculate of patients with testicular seminoma” (IP-2016-06-36-92).
Footnotes
Conflict of interest statement: The authors declare no conflict of interests.
Funding: This publication was financed by the European Union through the European Regional Development Fund and supported in part by Croatian Science Foundation.
REFERENCES
- 1.Mun EJ, Babiker HM, Weinberg U, Kirson ED, Von Hoff DD. Tumor-treating fields:A fourth modality in cancer treatment. Clin Cancer Res. 2018;24(2):266–75. doi: 10.1158/1078-0432.CCR-17-1117. [DOI] [PubMed] [Google Scholar]
- 2.Aghamir SMK, Heshmat R, Ebrahimi M, Khatami F. Liquid Biopsy:The Unique Test for Chasing the Genetics of Solid Tumors. Epigenetics Insights. 2020:13. doi: 10.1177/2516865720904052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krasic J, Sincic N, Samija I. Liquid biopsy for patients with cancer:different approaches and clinical applications. Molecular and Experimental Biology in Medicine. 2018;2:1–8. [Google Scholar]
- 4.Poulet G, Massias J, Taly V. Liquid Biopsy:General Concepts. Acta Cytol. 2019;63(6):449–55. doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 5.Diaz LA, Bardelli A. Liquid biopsies:Genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet) Cancer Cell [Internet] 2017;31(2):172–9. doi: 10.1016/j.ccell.2017.01.002. Available from: http://dx.doi.org/10.1016/j.ccell.2017.01.002 . [DOI] [PubMed] [Google Scholar]
- 7.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–48. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 8.Argadal OG, Mutlu M, Ak Aksoy S, Kocaeli H, Tunca B, Civan MN, Egeli U, Cecener G, Bekar A, Taskapilioglu MO, Tekin C, Tezcan G, Tolunay S. Long noncoding RNA MALAT1 may be a prognostic biomarker in IDH1/2 wild-type primary glioblastomas. Bosn J Basic Med Sci. 2020 Feb 5;20(1):63–69. doi: 10.17305/bjbms.2019.4297. http://dx.doi.org/10.17305/bjbms.2019.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9):1–35. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinčić N, Herceg Z. DNA methylation and cancer:Ghosts and angels above the genes. Curr Opin Oncol. 2011;23(1):69–76. doi: 10.1097/CCO.0b013e3283412eb4. [DOI] [PubMed] [Google Scholar]
- 11.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120(18):3163–72. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 12.García-Gutiérrez L, McKenna S, Kolch W, Matallanas D. RASSF1A tumour suppressor:Target the network for effective cancer therapy. Cancers (Basel) 2020;12(1):1–22. doi: 10.3390/cancers12010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malpeli G, Innamorati G, Decimo I, Bencivenga M, Kamdje AHN, Perris R, et al. Methylation dynamics of RASSF1A and its impact on cancer. Cancers (Basel) 2019;11(7):1–18. doi: 10.3390/cancers11070959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois F, Bergot E, Levallet G. Cancer and RASSF1A/RASSF1C, the Two Faces of Janus. Trends in Cancer. 2019;5(11):662–5. doi: 10.1016/j.trecan.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, He J, Li J, Tian D, Gu L, Zhou M. Methylation of RASSF1A gene promoter is regulated by p53 and DAXX. FASEB J. 2013;27(1):232–42. doi: 10.1096/fj.12-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajabi H, Hata T, Li W, Long MD, Hu Q, Liu S, et al. MUC1-C represses the RASSF1A tumor suppressor in human carcinoma cells. Oncogene [Internet] 2019;38(47):7266–77. doi: 10.1038/s41388-019-0940-1. Available from: http://dx.doi.org/10.1038/s41388-019-0940-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson HD, O'Meara ES, Kerlikowske K, Balch S, Miglioretti D. Factors associated with rates of false-positive and false-negative results from digital mammography screening:An analysis of registry data. Ann Intern Med. 2016;164(4):226–35. doi: 10.7326/M15-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhl CK, Schrading S, Leutner CC, Morakkabati-Spitz N, Wardelmann E, Fimmers R, et al. Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer. J Clin Oncol. 2005;23(33):8469–76. doi: 10.1200/JCO.2004.00.4960. [DOI] [PubMed] [Google Scholar]
- 19.Abudula A, Rouzi N, Xu L, Yang Y, Hasimu A. Tissue-based metabolomics reveals potential biomarkers for cervical carcinoma and HPV infection. Bosn J Basic Med Sci. 2020 Feb 5;20(1):78–87. doi: 10.17305/bjbms.2019.4359. http://dx.doi.org/10.17305/bjbms.2019.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagrass HA, Pasha HF, Shaheen MA, Abdel Bary EH, Kassem R. Methylation status and protein expression of RASSF1A in breast cancer patients. Mol Biol Rep. 2014;41(1):57–65. doi: 10.1007/s11033-013-2837-3. [DOI] [PubMed] [Google Scholar]
- 21.Kajabova V, Smolkova B, Zmetakova I, Sebova K, Krivulcik T, Bella V, et al. RASSF1A promoter methylation levels positively correlate with estrogen receptor expression in breast cancer patients. Transl Oncol. 2013;6(3):297–304. doi: 10.1593/tlo.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloten V, Becker B, Winner K, Schrauder MG, Fasching PA, Anzeneder T, et al. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15(1):1–11. doi: 10.1186/bcr3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadopoulou E, Davilas E, Sotiriou V, Georgakopoulos E, Georgakopoulou S, Koliopanos A, et al. Cell-free DNA and RNA in plasma as a new molecular marker for prostate and breast cancer. Ann N Y Acad Sci. 2006;1075:235–43. doi: 10.1196/annals.1368.032. [DOI] [PubMed] [Google Scholar]
- 24.Skalski M, Paluszczak J. Technological advances in free-circulating tumor-derived DNA methylation analysis. J Med Sci. 2019;88(4):256–60. [Google Scholar]
- 25.Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung breast and ovarian tumours. Oncogene. 2001;20(12):1509–18. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 26.Yan PS, Shi H, Rahmatpanah F, Hsiau THC, Hsiau AHA, Leu YW, et al. Differential distribution of DNA methylation within the RASSF1A CpG island in breast cancer. Cancer Res. 2003;63(19):6178–86. [PubMed] [Google Scholar]
- 27.Braga EA, Burdennyy AM, Pronina I V, Filippova EA, Kazubskaya TP, Fridman M V, et al. System of Markers Based on the Methylation of a Group of Proapoptotic Genes in Combination with MicroRNA in the Diagnosis of Breast Cancer. Bull Exp Biol Med. 2020;168(3):366–70. doi: 10.1007/s10517-020-04710-2. [DOI] [PubMed] [Google Scholar]
- 28.Shan M, Yin H, Li J, Li X, Wang D, Su Y, et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7(14):18485–94. doi: 10.18632/oncotarget.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Shetty PB, Feng W, Chenault C, Bast RC, Issa JPJ, et al. Methylation of HIN-1, RASSF1A, RIL and CDH13 in breast cancer is associated with clinical characteristics, but only RASSF1A methylation is associated with outcome. BMC Cancer. 2012;12:1–7. doi: 10.1186/1471-2407-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tserga A, Michalopoulos N V, Levidou G, Korkolopoulou P, Zografos G, Patsouris E, et al. Association of aberrant DNA methylation with clinicopathological features in breast cancer. Oncol Rep. 2012;27(5):1630–8. doi: 10.3892/or.2011.1576. [DOI] [PubMed] [Google Scholar]
- 31.Salta S P, Nunes S, Fontes-Sousa M, Lopes P, Freitas M, Caldas M, et al. A DNA Methylation-Based Test for Breast Cancer Detection in Circulating Cell-Free DNA. J Clin Med. 2018;7(11):420. doi: 10.3390/jcm7110420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagadi SAR, Prasad CP, Kaur J, Srivastava A, Prashad R, Gupta SD, et al. Clinical significance of promoter hypermethylation of RASSF1A, RARb2, BRCA1 and HOXA5 in breast cancers of Indian patients. Life Sci. 2008;82(25–26):1288–92. doi: 10.1016/j.lfs.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Wittenberger T, Sleigh S, Reisel D, Zikan M, Wahl B, Alunni-Fabbroni M, et al. DNA methylation markers for early detection of women's cancer:Promise and challenges. Epigenomics. 2014;6(3):311–27. doi: 10.2217/epi.14.20. [DOI] [PubMed] [Google Scholar]
- 34.De Koning HJ, Van Der Aalst CM, De Jong PA, Scholten ET, Nackaerts K, Heuvelmans MA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med. 2020;382(6):503–13. doi: 10.1056/NEJMoa1911793. [DOI] [PubMed] [Google Scholar]
- 35.Patz EF, Pinsky P, Gatsonis C, Sicks JRD, Kramer BS, Tammemägi MC, et al. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern Med. 2014;174(2):269–74. doi: 10.1001/jamainternmed.2013.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Begum S, Brait M, Dasgupta S, Ostrow KL, Zahurak M, Carvalho AL, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res. 2011;17(13):4494–503. doi: 10.1158/1078-0432.CCR-10-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Yu Z, Wang T, Zhang J, Hong L, Chen L. Identification of epigenetic aberrant promoter methylation of RASSF1A in serum DNA and its clinicopathological significance in lung cancer. Lung Cancer. 2007;56(2):289–94. doi: 10.1016/j.lungcan.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Dammann R, Takahashi T, Pfeifer GP. The CpG island of the novel tumor suppressor gene RASSF1A is intensely methylated in primary small cell lung carcinomas. Oncogene. 2001;20(27):3563–7. doi: 10.1038/sj.onc.1204469. [DOI] [PubMed] [Google Scholar]
- 39.Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, et al. The tumor suppressor RASSF1A in human carcinogenesis:An update. Histol Histopathol. 2005;20(2):645–63. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- 40.Scott M. Langevin1, Robert A Kratzke2, and Karl T. Kelsey3,4 †. Epigenetics of Lung Cancer. Transl Res [Internet] 2015;165(1):74–90. doi: 10.1016/j.trsl.2014.03.001. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3624763/pdf/nihms412728.pdf . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zöchbauer-Müller S, Minna JD, Gazdar AF. Aberrant DNA Methylation in Lung Cancer:Biological and Clinical Implications. Oncologist. 2002;7(5):451–7. doi: 10.1634/theoncologist.7-5-451. [DOI] [PubMed] [Google Scholar]
- 42.Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol. 2008;14(19):3074–80. doi: 10.3748/wjg.14.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckingham L, Faber LP, Kim A, Liptay M, Barger C, Basu S, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126(7):1630–9. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 44.Vaissière T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69(1):243–52. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niklinska W, Naumnik W, Sulewska A, Kozłowski M, Pankiewicz W, Milewski R. Prognostic significance of DAPK and RASSF1A promoter hypermethylation in Non-Small Cell Lung Cancer (NSCLC) Folia Histochem Cytobiol. 2009;47(2):275–80. doi: 10.2478/v10042-009-0091-2. [DOI] [PubMed] [Google Scholar]
- 46.Tomizawa Y, Iijima H, Nomoto T, Iwasaki Y, Otani Y, Tsuchiya S, et al. Clinicopathological significance of aberrant methylation of RARb2 at 3p24 RASSF1A at 3p21.3 and FHIT at 3p14.2 in patients with non-small cell lung cancer. Lung Cancer. 2004;46(3):305–12. doi: 10.1016/j.lungcan.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Travis WD, Brambilla E, Müller-hermelink HK, Harris CC. WHO Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. World Heal Organ Classif Tumours. 2004;July 2017:125–36. [Google Scholar]
- 48.Subramanian J, Govindan R. Lung cancer in never smokers:A review. J Clin Oncol. 2007;25(5):561–70. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 49.Gao W, Zhou Z, Liu Y, Liu D, Feng Q, Xu Y. Prognostic significance of BRCA1 and RASSF1A promoter hypermethylation in non-small cell lung cancer patients. Int J Clin Exp Pathol. 2016;9(8):8544–9. [Google Scholar]
- 50.Constâncio V, Nunes SP, Moreira-Barbosa C, Freitas R, Oliveira J, Pousa I, et al. Early detection of the major male cancer types in blood-based liquid biopsies using a DNA methylation panel. Clin Epigenetics. 2019;11(1):1–18. doi: 10.1186/s13148-019-0779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu WJ, Tan XH, Guo BP, Ke Q, Sun J, Cen H. Associations between RASSFIA promoter methylation and NSCLC:A meta-analysis of published data. Asian Pacific J Cancer Prev. 2013;14(6):3719–24. doi: 10.7314/apjcp.2013.14.6.3719. [DOI] [PubMed] [Google Scholar]
- 52.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11(18):6505–11. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 53.Hubers AJ, Heideman DAM, Burgers SA, Herder GJM, Sterk PJ, Rhodius RJ, et al. DNA hypermethylation analysis in sputum for the diagnosis of lung cancer:Training validation set approach. Br J Cancer. 2015;112(6):1105–13. doi: 10.1038/bjc.2014.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Constâncio V, Nunes SP, Henrique R, Jerónimo C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells. 2020;9(3):624. doi: 10.3390/cells9030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ponomaryova AA, Rykova EY, Cherdyntseva N V, Skvortsova TE, Dobrodeev AY, Zav'yalov AA, et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer [Internet] 2013;81(3):397–403. doi: 10.1016/j.lungcan.2013.05.016. Available from: http://dx.doi.org/10.1016/j.lungcan.2013.05.016 . [DOI] [PubMed] [Google Scholar]
- 56.Su Y, Fang Bin H, Jiang F. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin Epigenetics [Internet] 2016;8(1):1–9. doi: 10.1186/s13148-016-0275-5. Available from: http://dx.doi.org/10.1186/s13148-016-0275-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grote HJ, Schmiemann V, Geddert H, Bocking A, Kappes R, Gabbert HE, et al. Methylation of RAS association domain family protein 1A as a biomarker of lung cancer. Cancer. 2006;108(2):129–34. doi: 10.1002/cncr.21717. [DOI] [PubMed] [Google Scholar]
- 58.Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, et al. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012;72(22):5692–701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C, Yu W, Wang L, Zhao M, Guo Q, Lv S, et al. DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. J Cancer. 2017;8(17) doi: 10.7150/jca.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25(17):2029–44. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greenspan EJ, Jablonski MA, Rajan T V, Levine J, Belinsky GS, Rosenberg DW. Epigenetic alterations in RASSF1A in human aberrant crypt foci. Carcinogenesis. 2006;27(7):1316–22. doi: 10.1093/carcin/bgi373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matthew P, Hanleya B, Hahnc MA, Arthur X, Lid Xiwei Wue JL, Wangg J, Choic A, Zhengqing Ouyangfh YF, et al. Genome-wide DNA methylation profiling reveals cancerassociated changes within early colonic neoplasia. Oncogene. 2017;36(35):5035–44. doi: 10.1038/onc.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinha R, Hussain S, Mehrotra R, Kumar RS, Kumar K, Pande P, et al. Kras Gene Mutation and RASSF1A, FHIT and MGMT Gene Promoter Hypermethylation:Indicators of Tumor Staging and Metastasis in Adenocarcinomatous Sporadic Colorectal Cancer in Indian Population. PLoS One. 2013;8(4):1–8. doi: 10.1371/journal.pone.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res - Fundam Mol Mech Mutagen [Internet] 2015;778:46–51. doi: 10.1016/j.mrfmmm.2015.05.002. Available from: http://dx.doi.org/10.1016/j.mrfmmm.2015.05.002 . [DOI] [PubMed] [Google Scholar]
- 65.Hu L, Chen G, Yu H, Qiu X. Clinicopathological significance of RASSF1A reduced expression and hypermethylation in hepatocellular carcinoma. Hepatol Int. 2010;4(1):423–32. doi: 10.1007/s12072-010-9164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhan L, Zhang B, Tan Y, Yang C, Huang C, Wu Q, et al. Quantitative assessment of the relationship between RASSF1A gene promoter methylation and bladder cancer (PRISMA) Med (United States) 2017;96(7):1–9. doi: 10.1097/MD.0000000000006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang H, Yi B, Li L, Zhang HY, Sun F, Dong SQ, et al. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol. 2008;85(2):96–100. doi: 10.1016/j.yexmp.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Zhao ZH, Fan YC, Yang Y, Wang K. Association between Ras association domain family 1A promoter methylation and hepatocellular carcinoma:A meta-analysis. World J Gastroenterol. 2013;19(41):7189–96. doi: 10.3748/wjg.v19.i41.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong X, Hou Q, Chen Y, Wang X. Diagnostic Value of the Methylation of Multiple Gene Promoters in Serum in Hepatitis B Virus-Related Hepatocellular Carcinoma. Dis Markers. 2017:2017. doi: 10.1155/2017/2929381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohamed NA, Swify EM, Amin NF, Soliman MM, Tag-Eldin LM, Elsherbiny NM. Is serum level of methylated RASSF1A valuable in diagnosing hepatocellular carcinoma in patients with chronic viral hepatitis C? Arab J Gastroenterol [Internet] 2012;13(3):111–5. doi: 10.1016/j.ajg.2012.06.009. Available from: http://dx.doi.org/10.1016/j.ajg.2012.06.009 . [DOI] [PubMed] [Google Scholar]
- 71.Guo Q, Wang HB, Li YH, Li HF, Li TT, Zhang WX, et al. Correlations of promoter methylation in WIF-1, RASSF1A, and CDH13 genes with the risk and prognosis of esophageal cancer. Med Sci Monit. 2016;22:2816–24. doi: 10.12659/MSM.896877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Honda S, Haruta M, Sugawara W, Sasaki F, Ohira M, Matsunaga T, et al. The methylation status of RASSF1A promoter predicts responsiveness to chemotherapy and eventual cure in hepatoblastoma patients. Int J Cancer. 2008;123(5):1117–25. doi: 10.1002/ijc.23613. [DOI] [PubMed] [Google Scholar]
- 73.Nilsson TK, Löf-Öhlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol. 2013;42(1):127–33. doi: 10.3892/ijo.2012.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saelee P, Wongkham S, Chariyalertsak S, Petmitr S, Chuensumran U. RASSF1A promoter hypermethylation as a prognostic marker for hepatocellular carcinoma. Asian Pacific J Cancer Prev. 2010;11(6):1677–81. [PubMed] [Google Scholar]
- 75.Fodor A, Cozma A, Vulturar R, Tăut AVS, Orăşan OH, Mureşan F, et al. DNA methylation and micro-RNAs:The most recent and relevant biomarkers in the early diagnosis of hepatocellular carcinoma. Med. 2019;55(9) doi: 10.3390/medicina55090607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol. 2006;12(8):1175–81. doi: 10.3748/wjg.v12.i8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma:Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23(31):5650–68. doi: 10.3748/wjg.v23.i31.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su YH, Lin SY, Song W, Jain S. DNA markers in molecular diagnostics for hepatocellular carcinoma. Expert Rev Mol Diagn. 2014;14(7):803–17. doi: 10.1586/14737159.2014.946908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91(3):702–7. doi: 10.1016/j.yexmp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Lu CY, Chen SY, Peng HL, Kan PY, Chang WC, Yen CJ. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget. 2017;8(4):6406–18. doi: 10.18632/oncotarget.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim YJ, Kim WJ. Can we use methylation markers as diagnostic and prognostic indicators for bladder cancer?Investig Clin Urol. 2016;57:S77–88. doi: 10.4111/icu.2016.57.S1.S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hauser S, Kogej M, Fechner G, Von Pezold J, Vorreuther R, Lummen G, et al. Serum DNA hypermethylation in patients with bladder cancer:Results of a prospective multicenter study. Anticancer Res. 2013;33(3):779–84. [PubMed] [Google Scholar]
- 83.Bilgrami SM, Qureshi SA, Pervez S, Abbas F. Promoter hypermethylation of tumor suppressor genes correlates with tumor grade and invasiveness in patients with urothelial bladder cancer. Springerplus. 2014;3(1):1–9. doi: 10.1186/2193-1801-3-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skrypkina I, Tsyba L, Onyshchenko K, Morderer D, Kashparova O, Nikolaienko O, et al. Concentration and Methylation of Cell-Free DNA from Blood Plasma as Diagnostic Markers of Renal Cancer. Dis Markers. 2016:2016. doi: 10.1155/2016/3693096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ellinger J, Holl D, Nuhn P, Kahl P, Haseke N, Staehler M, et al. DNA hypermethylation in papillary renal cell carcinoma. BJU Int. 2011;107(4):664–9. doi: 10.1111/j.1464-410X.2010.09468.x. [DOI] [PubMed] [Google Scholar]
- 86.De Martino M, Klatte T, Haitel A, Marberger M. Serum cell-free DNA in renal cell carcinoma:A diagnostic and prognostic marker. Cancer. 2012;118(1):82–90. doi: 10.1002/cncr.26254. [DOI] [PubMed] [Google Scholar]
- 87.Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics. 2011;2(2):123–50. doi: 10.1007/s13148-011-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Misawa K, Mochizuki D, Imai A, Endo S, Mima M, Misawa Y, et al. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget. 2016;7(18):26087–98. doi: 10.18632/oncotarget.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Honorio S, Agathanggelou A, Wernert N, Rothe M, Maher ER, Latif F. Frequent epigenetic inactivation of the RASSF1A tumour suppressor gene in testicular tumours and distinct methylation profiles of seminoma and nonseminoma testicular germ cell tumours. 2003:461–6. doi: 10.1038/sj.onc.1206119. [DOI] [PubMed] [Google Scholar]
- 90.Costa AL, Moreira-Barbosa C, Lobo J, Vilela-Salgueiro B, Cantante M, Guimarães R, et al. DNA methylation profiling as a tool for testicular germ cell tumors subtyping. Epigenomics. 2018;10(12):1511–23. doi: 10.2217/epi-2018-0034. [DOI] [PubMed] [Google Scholar]
- 91.Hacioglu BM, Kodaz H, Erdogan B, Cinkaya A, Tastekin E, Hacibekiroglu I, et al. K-RAS and N-RAS mutations in testicular germ cell tumors. Bosn J Basic Med Sci. 2017;17(2):159–63. doi: 10.17305/bjbms.2017.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cairns P. Detection of promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Ann N Y Acad Sci. 2004;1022:40–3. doi: 10.1196/annals.1318.007. [DOI] [PubMed] [Google Scholar]
- 93.Costa AL, Lobo J, Jerónimo C, Henrique R. The epigenetics of testicular germ cell tumors:Looking for novel disease biomarkers. Epigenomics. 2017;9(2):155–69. doi: 10.2217/epi-2016-0081. [DOI] [PubMed] [Google Scholar]
- 94.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer [Internet] 2011;11(6):426–37. doi: 10.1038/nrc3066. Available from: http://dx.doi.org/10.1038/nrc3066 . [DOI] [PubMed] [Google Scholar]
- 95.Yang M, Park JY. DNA methylation in promoter region as biomarkers in prostate cancer. Methods in Molecular Biology. 2012;863:67–109. doi: 10.1007/978-1-61779-612-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feng C, Dong J, Chang W, Cui M, Xu T. The Progress of Methylation Regulation in Gene Expression of Cervical Cancer. Int J Genomics. 2018:2018. doi: 10.1155/2018/8260652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li J, El-Naggar A, Mao L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer. 2005;104(4):771–6. doi: 10.1002/cncr.21215. [DOI] [PubMed] [Google Scholar]
- 98.Klacz J, Wierzbicki PM, Wronska A, Rybarczyk A, Stanislawowski M, Slebioda T, et al. Decreased expression of RASSF1A tumor suppressor gene is associated with worse prognosis in clear cell renal cell carcinoma. Int J Oncol. 2016;48(1):55–66. doi: 10.3892/ijo.2015.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Friedrich MG, Chandrasoma S, Siegmund KD, Weisenberger DJ, Cheng JC, Toma MI, et al. Prognostic relevance of methylation markers in patients with non-muscle invasive bladder carcinoma. Eur J Cancer. 2005;41(17):2769–78. doi: 10.1016/j.ejca.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 100.Atsushi Tanemura, Alicia M. Terando1, 3, Myung-Shin Sim2, Anneke Q. van Hoesel1 M, F.G. de Maat1, Donald L, Morton3 and DSBH. CpG Island Methylator Phenotype Predicts Progression of Malignant Melanoma. Clin Cancer Res. 2009;15(5):1801–7. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muñoz J, Inda M, del M, Lázcoz P, Zazpe I, Fan X, Alfaro J, et al. Promoter Methylation of RASSF1A Associates to Adult Secondary Glioblastomas and Pediatric Glioblastomas. ISRN Neurol. 2012;2012:1–10. doi: 10.5402/2012/576578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koul S, McKiernan JM, Narayan G, Houldsworth J, Bacik J, Dobrzynski DL, et al. Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol Cancer. 2004;3:1–12. doi: 10.1186/1476-4598-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Markulin D, Vojta A, Samaržija I, Gamulin M, Bečeheli I, Jukić I, et al. Association between RASSF1A promoter methylation and testicular germ cell tumor:A meta-analysis and a cohort study. Cancer Genomics and Proteomics. 2017;14(5):363–72. doi: 10.21873/cgp.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Majchrzak-Celińska A, Paluszczak J, Kleszcz R, Magiera M, Barciszewska AM, Nowak S, et al. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J Appl Genet. 2013;54(3):335–44. doi: 10.1007/s13353-013-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gandini S, Botteri E, Iodice S, Boniol M, Lowenfels AB, Maisonneuve P, et al. Tobacco smoking and cancer:A meta-analysis. Int J Cancer. 2008;122(1):155–64. doi: 10.1002/ijc.23033. [DOI] [PubMed] [Google Scholar]
- 107.Ovchinnikov DA, Cooper MA, Pandit P, Coman WB, Cooper-White JJ, Keith P, et al. Tumor-suppressor Gene Promoter Hypermethylation in Saliva of Head and Neck Cancer Patients. Transl Oncol. 2012;5(5):321–6. doi: 10.1593/tlo.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Choudhury JH, Ghosh SK. Promoter hypermethylation profiling identifies subtypes of head and neck cancer with distinct viral, environmental, genetic and survival characteristics. PLoS One. 2015;10(6):1–19. doi: 10.1371/journal.pone.0129808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wagner KJ, Cooper WN, Grundy RG, Caldwell G, Jones C, Wadey RB, et al. Frequent RASSF1A tumour suppressor gene promoter methylation in Wilms'tumour and colorectal cancer. Oncogene. 2002;21(47):7277–82. doi: 10.1038/sj.onc.1205922. [DOI] [PubMed] [Google Scholar]
- 110.Costa VL, Henrique R, Ribeiro FR, Pinto M, Oliveira J, Lobo F, et al. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer. 2007;7:1–8. doi: 10.1186/1471-2407-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peters I, Rehmet K, Wilke N, Kuczyk MA, Hennenlotter J, Eilers T, et al. RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis. Mol Cancer. 2007;6:1–9. doi: 10.1186/1476-4598-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Laytragoon-Lewin N, Chen F, Castro J, Elmberger G, Rutqvist LE, Lewin F, et al. DNA content and methylation of p16, DAPK and RASSF1A gene in tumour and distant, normal mucosal tissue of head and neck squamous cell carcinoma patients. Anticancer Res. 2010;30(11):4643–8. [PubMed] [Google Scholar]
- 113.Paluszczak J, Misiak P, Wierzbicka M, Woźniak A, Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol. 2011;47(2):104–7. doi: 10.1016/j.oraloncology.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 114.Righini CA, De Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, et al. Tumor-specific methylation in saliva:A promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13(4):1179–85. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 115.Aran D, Camarda R, Odegaard J, Paik H, Oskotsky B, Krings G, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun [Internet] 2017;8(1):1–13. doi: 10.1038/s41467-017-01027-z. Available from: http://dx.doi.org/10.1038/s41467-017-01027-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KYC, et al. DNA Methylation Cancer Biomarkers:Translation to the Clinic. Front Genet. 2019;10(November):1–22. doi: 10.3389/fgene.2019.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mikeska T, Craig JM. DNA methylation biomarkers:Cancer and beyond. Genes (Basel) 2014;5(3):821–64. doi: 10.3390/genes5030821. [DOI] [PMC free article] [PubMed] [Google Scholar]


