Abstract
Pyruvate kinase M2 (PKM2) catalyzes the conversion of phosphoenolpyruvate (PEP) to pyruvate. It plays a central role in the metabolic reprogramming of cancer cells and is expressed in most human tumors. It is essential in indiscriminate proliferation, survival, and tackling apoptosis in cancer cells. This positions PKM2 as a hot target in cancer therapy. Despite its well-known structure and several reported modulators targeting PKM2 as activators or inhibitors, a comprehensive review focusing on such modulators is lacking. Herein we summarize modulators of PKM2, the assays used to detect their potential, the preferable tense (T) and relaxed (R) states in which the enzyme resides, lacunae in existing modulators, and several strategies that may lead to effective anticancer drug development targeting PKM2.
The review focuses on the tumor pyruvate kinase M2 (PKM2) modulators. Both activators and inhibitors developed against PKM2 are discussed.
1. Introduction
Cancer is a generic term associated with a large group of diseases distinguished by the abnormal growth of cells with inexhaustible replication potential. It is followed by attack and assault on neighboring body parts and spreads to other organs through metastasis.1 The global cancer burden is estimated to have risen to 18.1 million new cases in 2018 with 9.6 million deaths.1 Cancer cells are distinguished from normal cells by relatively unpredictable metabolic characteristics and the consumption of altered amounts of nutrients.2–5 Considering the importance of metabolites generated in cancerous cells, several drug design strategies have been followed, which mainly focus on their metabolic pathways.6,7 In normal cells, oxidative phosphorylation is required for energy production, cell survival, and participation in various metabolic events.8–12 Hypoxic cancerous and normoxic cells generate 2 and 36 ATPs, respectively, per glucose molecule.13 German biochemist and Nobel laureate Prof. Otto Heinrich Warburg hypothesized that the non-oxidative breakdown of glucose generates ATP in tumor cells. He demonstrated that recalcitrant cancer cells are biased towards the generation of lactic acid from glucose even in the presence of oxygen, a phenomenon popularly known as the Warburg effect. Proliferating cancer cells follow the Warburg effect because they require abundant ATP supply to sustain their proliferation and uncontrolled growth.14–18
During glucose metabolism, cancerous cells produce ATP and lactate that is utilized by neighboring cancer cells to produce more ATP via the tricarboxylic acid cycle (TCA) cycle and oxidative phosphorylation (OXPHOS).19 The high bioenergetic demands of cancer cells are fulfilled by alternative metabolic pathways like the pentose phosphate pathway (PPP), the uronic acid pathway (UAP), the polyol pathway (PYP), TCA, and OXPHOS.20 The alternative metabolic pathways help cancer cells in providing nucleic acids to support rapid cell proliferation. The glycolytic pathway plays a vital role in the generation of ATP, where several tightly coupled enzymes are involved.21–25 A number of glycolytic enzymes like hexokinase (HK), phosphoglucose isomerase (PGI), phosphofructokinase (PFK-1), fructose-bisphosphate aldose (ALDO), triosephosphate isomerase (TPI), glyceraldehyde phosphate dehydrogenase (GAPDH), phosphoglycerate kinase (PGK), phosphoglycerate mutase (PGM), enolase, and pyruvate kinase (PK) are involved in glycolysis. The PK enzyme plays a quintessential role in converting phosphoenolpyruvate (PEP) to pyruvate by dephosphorylation.8,26,27 Under normoxic conditions, pyruvate enters the TCA cycle for energy production.
1.1. PKM2 structure, expression, and regulation by endogenous allosteric modulators
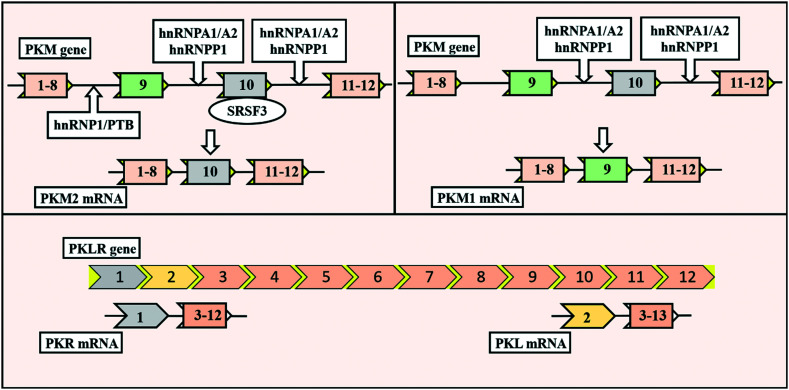
The PK enzyme is coded by two types of genes (namely PK-LR and PK-M), each having two isoforms in mammals.28–30 Each gene expresses a specific isoform in specific tissues; for example, the PK-LR gene expresses the PKR isoform exclusively found in red blood cells. The PKL isoform is expressed primarily in the liver, intestine, and kidney.28–30 The PK-M gene expresses two isoforms: PKM1 and PKM2. The PKM1 isoform is exclusively found in highly catabolic tissues like the heart, brain, and muscles, while PKM2 is generally present in all proliferative and cancer cells. PKM2 is regulatory in nature and can efficiently manipulate the properties of distorted glucose metabolism in cancer.31 PKM gene consists of 12 exons and 11 introns. It has been found that PKM1 contains exclusively exon 9, whereas PKM2 contains exclusively exon 10. PKM1 and PKM2 isoforms have a difference of 22 amino acids due to exon 9 or 10.
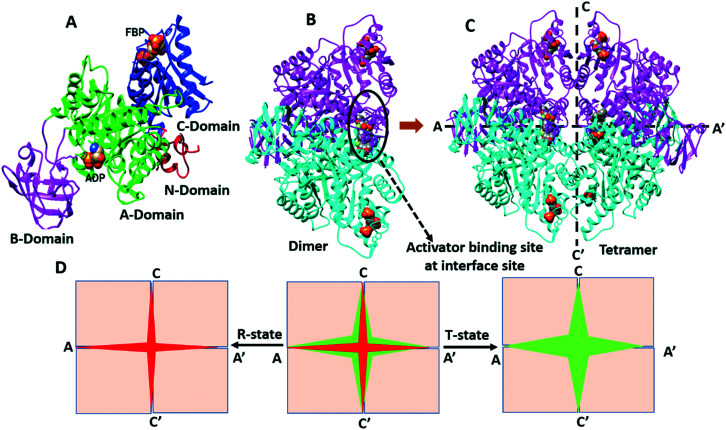
The expression of PKM2 is controlled by oncoprotein c-Myc and three heterogeneous nuclear ribonucleoproteins (hnRNPs) l, A1, and A2, which bind to exon 9. The serine/arginine-rich splicing factor-3 (SRSF-3) is bound to exon 10, which helps in the repression of exon 9 (Fig. 1).32 PKM2 is allosterically regulated by the natural ligand fructose 1,6-bisphosphate (FBP). The FBP binding area is near exon 9 and 10, a distinct region of PKM2. The binding of FBP leads to the altered conformation of the protein.30 The monomeric form of PKM2 consists of 531 amino acids with four different domains (Fig. 2A): A-domain (44–116 and 219–389 amino acids, green color), B-domain (117–218 amino acids, magenta color), C-domain (390–531 amino acids, blue color), and N-domain (1–43 amino acids, red color), respectively (Fig. 2A).28–30
Fig. 1. Structural differences in the four pyruvate kinase (PK) isoforms and splicing variants of pyruvate kinases. The PKR (inclusive of exon 2) isoform chiefly exists in red blood cells. The PKL isoform (inclusive of exon 1) is expressed primarily in the liver, intestine, and kidneys. In alternative splicing of the PKM gene, heterogeneous nuclear ribonucleoproteins (hnRNPs) l, A1, and A2 play an essential role. In the conversion of PKM1 and PKM2, transcription factor serine/arginine-rich splicing factor-3 (SRSF-3) also plays a key role. The PKM1 (exclusively exon 9) isoform is exclusively found in highly catabolic tissues like the heart, brain, and muscles. PKM2 (exclusively exon 10) is generally present in all proliferative and cancer cells. PTB is polypyrimidine tract binding protein.
Fig. 2. A) Monomeric unit of PKM2, B) intermolecular interactions between two monomers along the A–‘A’ axis lead to dimerization of PKM2 and C) two dimers interact at the C–C′ interface which leads to tetramerization of PKM2. D) Tetramer exists in two-states: R-state (active state or tightly bound state) and T-state (inactive state or loosely bound state) of PKM2. In the absence of FBP, PKM2 exists in a mixed population of monomer, dimer, and tetramer that enjoys a T-state conformation (tetramer/inactive). Upon association with FBP, PKM2 changes from the inactive T-state to the active R-state conformation, which favors PEP recognition at the active site and enhances its pyruvate kinase activity.
It hypothesized that the monomer dimerizes at the A–A′ interface (Fig. 2B) followed by tetramerization from the dimer at the C–C′ interface (Fig. 2C). In the absence of FBP, PKM2 exists in the mixed population of monomeric, dimeric, and tetrameric isoforms. Tetrameric PKM2 shows pyruvate kinase activity. Tetrameric PKM2 may adopt the inactive T (tense) state or active R (relaxed) state conformation.33,34 In the case of PKM2, the enzyme achieves stability through a switch between the dimeric and tetrameric states with endogenous ligands' involvement. In cancerous cells, the dimeric form of PKM2 is more abundant than the tetrameric structure. Wang et al. explained the transition of the inactive T-state and active R-state of the tetrameric PKM2 conformer by a seesaw model.31,35 The less active PKM2 is allosterically activated by FBP. It is hypothesized that FBP binds to pyruvate kinase. This induces conformational changes, thereby promoting homotetramer formation. Upon association with FBP, PKM2 undergoes a conformational change from the inactive T-state to the active R-state, which favors PEP recognition at the active site, thereby enhancing the pyruvate kinase activity. The tetrameric PKM2 undergoes significant conformational changes between the active R-state and the inactive T-state by the rotation of five degrees of the α9 helix of each monomer. The tetrameric form represents the most active form of the enzyme and efficiently converts PEP to pyruvate to gain access to the TCA cycle.36 The ratio of dimeric and tetrameric forms of PKM2 decides the fate of the cell. The dimeric state has a role in catabolism and material synthesis in the cells.31 The epigenetic regulation translocates PKM2 into the nucleus by oncogenic stimulation and it acts as a protein kinase.37
1.2. Glycolytic and non-glycolytic functions of PKM2
In the PK family, several oncogenes and tumor suppressors regulate PKM2 through a complex mechanism. The less active form of PKM2 drives glucose through aerobic glycolysis, while active PKM2 directs glucose towards oxidative metabolism.32,38–42 However, the active PKM2 state diverts glucose towards oxidative metabolism to form pyruvate. Instead of using ATP, PKM2 uses the high-energy phosphate from PEP (acts as a phosphate donor) for phosphorylation of its protein substrates to promote tumorigenesis.43 Cytosolic PKM2 regulates the synthesis of amino acids, nucleotides, and NADPH production, whereas nuclear PKM2 subsequently induces epithelial–mesenchymal transition (EMT). After the onset of EMT, the cells lose intercellular adhesion (characteristics of cancerous cells), alter the morphology to a spindle-shaped appearance, and increase the mobility for indiscriminate proliferation and metastasis.38,39,44–46 Cell proliferation and growth is promoted by the up-regulation of PKM2 through hypoxia-inducible factor-1α (HIF-1α) and other transcription factors.47–49 Thus, inhibiting PKM2 or the activating tetramer may eliminate indiscriminate proliferation. PKM2 also localizes to the nucleus, and its levels directly correlate with cellular proliferation. PKM2 gains access to the nucleus as a transcription factor. It further activates specific genes and other transcription factors.43,50 Modulating PKM2 may lead to increased or decreased proliferation of tumors. Collectively, PKM2 seems to be a sensitive prognostic marker in most solid tumors.21
PKM2 regulation is also associated with post-translational modifications (PTMs) and nuclear functions.51 Under cancerous conditions, tetrameric PKM2 favors a dimeric state and subsequently results in the breakdown cascade initiation, thereby generating bulk intermediates. Building blocks for the synthesis of nucleotides, proteins, and membrane components are formed from these intermediates, consequently providing ATP to the proliferative cells. PKM2–HIFα interaction modulates the glycolytic enzyme expression. It results in the diversion of glycolysis towards the biosynthesis of cell division precursors via the pentose phosphate pathway. Cytosolic PKM2 undergoes phosphorylation at tyrosine-105, leading to glycolytic termination and activation of an up-regulated metabolic pathway, providing sufficient fuel for cell proliferation. PTMs in PKM2 include phosphorylation, acetylation, hydroxylation, oxidation, ubiquitination, and sumoylation.52,53 It is established that acetylation of lysine-305 (PKM2K305)54,55 inhibits the pyruvate kinase activity of PKM2 in vitro and affects FBP binding, ultimately preventing its activation. Similarly, phosphorylation of tyrosine-105 (PKM2Y105) inhibits the tetramer formation and pyruvate kinase activity.56 Upon oxidation, cysteine-358 activity is irreversibly inhibited. The metabolic changes for the indiscriminate growth of cancer cells are activated.57–59 It has been observed that in neurogenic tumors, PKM2 undergoes phosphorylation. This is made facile by activating specific transcription factors by fusion of Est variant 6 and neurotrophic tyrosine kinase receptor.60 To meet the escalating biosynthetic demands of cancerous cells, cancer cells carefully rewire their metabolism by increasing the glycolytic rate to support proliferation in multiple ways. Now, the relatively inactive dimeric state terminates the conversion of PEP to pyruvate. The accumulation of glycolytic intermediates allows alternative anabolic pathways to meet the bioenergetic demands of cancer cells.53,61
1.3. Enzymatic assays for measuring the activity of PKM2
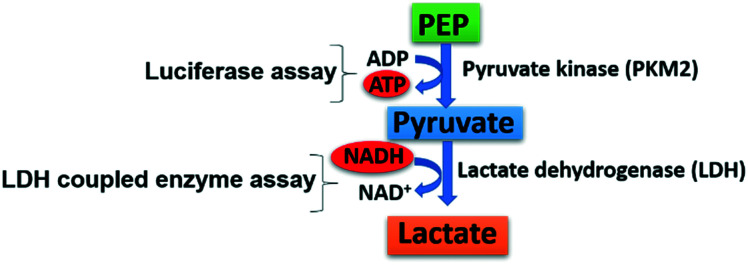
Increased expression of PKM2 in various human cancers is thoroughly established.62 Although several skeletally diverse activators and inhibitors of PKM2 have been screened, none of them has hit the market to date. Bioassay-based screening approaches for PKM2 modulators search for hits at least in the low micromolar range. There are two enzyme assays reported for measuring pyruvate kinase activity: (i) luciferase assay and (ii) lactate dehydrogenase (LDH)-coupled assay (Fig. 3). (i) The luciferase assay has become the assay of choice because of its reliability and flexibility. It is comparatively easier to use, has a sizeable linear signal range, and is compliant to plate reader formats. The pyruvate kinase-luciferase coupled assay is based on the luminescence detection of protein kinases. ATP production is catalyzed by PKM2, which leads to an increasing luminescent signal.63
Fig. 3. Detection of pyruvate kinase activity. The direct method involves the luciferase assay that measures ATP formation during the conversion of PEP to pyruvate. The indirect method involves the LDH-coupled enzyme assay that detects NADH's decreasing absorbance in the second step during the conversion of pyruvate to lactate.
The luciferase assay uses the ATP-dependent reaction catalyzed by the firefly luciferase enzyme. PKM2 catalyzes the ATP production by increasing the luminescent signal. This assay has its advantages like detection of the activation or inhibitory profile of the molecule. It also detects the lower amounts of ATP generated. Yet, some analogs tested in the luciferase assay may activate PKM2 or show fluorescence at the wavelength used for NADH detection, which may not be in line or agreement with the actual profile. (ii) The fluorescence-based PK-lactate dehydrogenase (LDH) coupled assay monitors pyruvate production. In glycolysis, LDH is a cytosolic enzyme that catalyzes pyruvate conversion to lactate with NADH to NAD+. This assay is frequently used to check whether a molecule is an activator or inhibitor of PKM2. Quantification of cellular lactate by the lactate dehydrogenase (LDH) coupled assay is a widely accepted assay for the quantitative determination of PKM2 activity. It is also a sensitive method to ascertain the profile of PKM2 modulators. Another fundamental drawback is interference caused by compounds showing absorbance at 340 nm. Some evidence suggested that phosphate transfer from ATP takes place without involving PKM2. It was also suggested that a lack of PEP mediated phosphorylation in the presence of recombinant PKM2 (rPKM2) takes place.64 However, it isn't easy to distinguish PKM2 catalyzed ATP production or ATP synthesis from other sources.
1.4. Plausible reasons for lack of drug molecules in the market targeting PKM2
Structurally several modulators tested against PKM2 have real structures that define a drug-like moiety, while several are promiscuous compounds, making it difficult for them to move from bench to bedside. Several PKM2 modulators could not reach the market because of their inability for drug development owing to the preponderance of pan assay interference compounds (PAINS).65–70 PAINS may elicit PKM2 modulation and nonspecific bioactivity because of redox cycling, formation of covalent adducts, chelation properties, and aggregation behavior (micelles or vesicles) in aqueous media that directly interferes with the assay outcome. For example, phenolic PKM2 modulators are discussed in detail in this review.71–75
In this review, we have compiled the various categories of compounds that modulate the activity of PKM2. Their mechanism of action, structure–activity relationships (SARs), and interactions of some compounds with PKM2 have been discussed. Also, several strategies that may lead to effective drug development centered on PKM2 are discussed. The article is mainly based on a broad classification according to the mechanism of action serving as PKM2 inhibitors and activators. They are further classified according to their chemical classes like heterocyclics, polyphenolics, amino acids, and miscellaneous PKM2 modulators. The IC50 (inhibitory concentration) and AC50 (activator concentration) values have also been discussed in length. Many different PKM2 modulators have been reported in the last decade; it is worthwhile to have a review article comparing and analyzing them. Discussion in the review is mainly centered on the therapeutic applications of PKM2 modulators as anticancer agents. The study also helps to distinguish between real and unreal PKM2 modulators. The potential uses of PKM2 modulators are much broader, and we hope that this review will inspire their more comprehensive applications in terms of judicious chemical manipulations. To the best of our knowledge, this is the first comprehensive review on PKM2 modulators.
2. PKM2 inhibitors
2.1. Heterocyclic PKM2 inhibitors
The last decade has seen a sharp increase in the research involving the Warburg effect, the typical cancer cell metabolism. PKM2 inhibitors may compete with PEP and inhibit its conversion to pyruvate. Several polyphenolic and heterocyclic molecules have been reported that act as inhibitors. Heterocyclic compounds are the fundamental essence of medicinal chemistry. Heterocyclic compounds exhibit superior biological profiles compared to aromatic ones due to their better interaction with the receptors. Very few molecules have been investigated against PKM2 with their mechanism of action established.
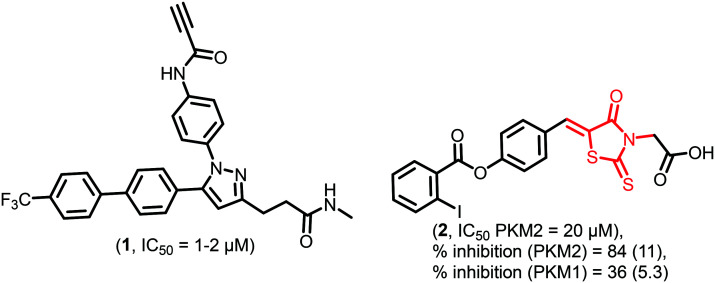
Recently, Chen et al. reported the irreversible PKM2 inhibitor 1 (Fig. 4) having antiproliferative activity against various cell lines like MCF-7, MDA-MB-231, SCC4, SCC2095, PC-3, and LNCaP with IC50 of 1–2 μM. 1 contains a terminal acetylene group, which irreversibly and covalently binds to cysteine-326 (Cys-326) and cysteine-317 (Cys-317) residues of PKM2 and inhibits it. The Cys-326 residue is near the PEP binding site. It is also demonstrated that a 10 mg kg−1 dose of 1 effectively inhibits the growth of tumors in nude mice without any acute toxicity.76
Fig. 4. 1 is a covalent modulator, and 2 is a thiazolidinedione-based PKM2 inhibitor. The red portion in the molecule signifies the part with PAINS propensity.
1 is an example of a covalent inhibitor that modifies the cysteine residue of PKM2 through its terminal alkyne. Although covalent modulators may have some pitfalls, their in vitro and in vivo efficacy makes them a real and developable lead in anticancer drug discovery.77 The inhibitors should also be tested in autochthonous tumor models to check their effectiveness. This inactivation of pyruvate kinase can redirect metabolic flow through the pentose phosphate pathway, as explained in the introduction, making the reduced NADPH required to maintain cellular redox of cancer cells.
Thiazolidinediones (TZDs) provoke apoptosis, cell cycle arrest and also block the differentiation of cells. It is found that TZDs have kinase inhibitory activity for which they are under investigation.78,792 (Fig. 4) belongs to the thiazolidinedione class and shows promising PKM2 inhibitory activity. Vander et al. screened a large number of compounds (107 360) in silico. Among them, 74 compounds showed specific PKM2 inhibition at a concentration of 10 μM.80,812 depicts the selective inhibition of PKM2 over PKM1. 2 binds to the allosteric site of PKM2 and releases FBP from the FBP binding site. Simultaneously, no allosteric site exists on PKM1, which may lead to the selectivity of 2 towards PKM2. Chemically 2 is an alkylidene rhodanine derivative. It is a reactive chelating compound with an exocyclic alkene that may function as a Michael acceptor and yield false results. In 2, there is a 1,3-syn planar relationship between the carbonyl and alkene moiety that may cause transition-state stabilization with an incoming nucleophile. The insidious, photochemically active, and non-selective nature of the rhodanine core to react with multiple proteins may lead to redundant results, which is a typical PAINS feature.82,83 It is also important to mention that the mere presence of PAINS doesn't disqualify them from the drug discovery pipeline. It provides an opportunity for the incorporation of isosteres or bioisosteres for better drug-like characteristics.
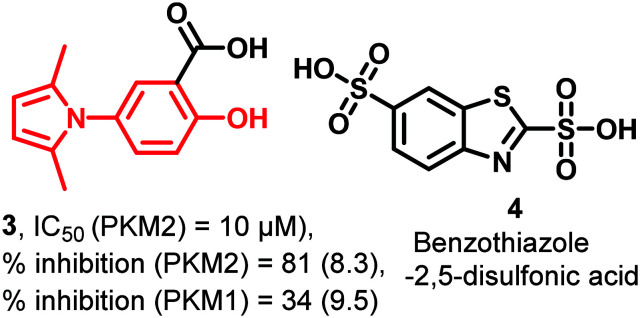
3 (Fig. 5) is a salicylic acid derivative consisting of a five-membered pyrrole ring at position 5 of the aromatic ring. 3 is a more selective PKM2 (81%) inhibitor than PKM1 (34%) at 10 μM concentration. 3 was tested on human lung cancer cells (H1299) and resulted in an 18.5% decrease in the glycolysis rate. Using the lactate dehydrogenase (LDH)-coupled assay, 3 is retested at varying concentrations (0.1–1000 μM), where it showed PKM2 inhibition at 10.00 μM concentration.84 Despite this, it is PAINS positive.85 Another heterocycle, benzothiazole-2,5-disulfonic acid (4) (Fig. 5), binds with pyruvate kinase's active site and inhibits it.86
Fig. 5. 3 is a salicylic acid derivative, and benzothiazole-2,5-disulfonic acid (4) shows PKM2 inhibitory properties. The red portion in the molecule signifies the part with PAINS propensity.
2.2. Indirect PKM2 inhibitors
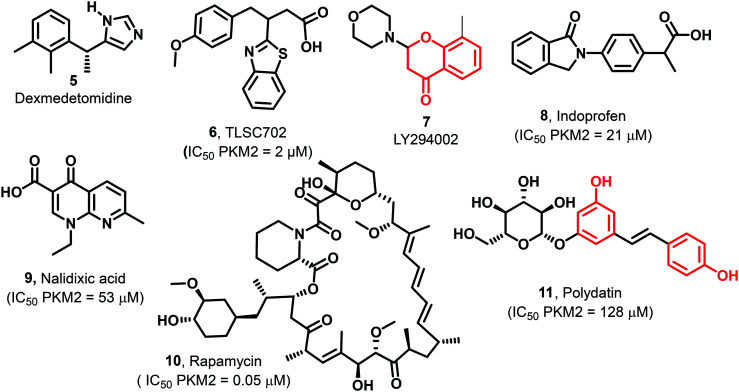
Other heterocyclics established as drugs (5–11, Fig. 6) have also been tested against PKM2. Dexmedetomidine (5) concurrently inhibits the HIF-α/PKM2 axis activating the P13/AKT pathway. It acts as an antiapoptotic agent.87 TLSC702 (6) works against GLO I and exhibits PKM2 inhibitory activity (IC50 2 μM). 6 is a benzothiazole-based molecule, and if used in combination with shikonin (PKM2 inhibitor), reveals a more pronounced anticancer effect than the treatment with either agent.88 LY294002 (7) shows antitumor activity by downregulating the PKM2 expression. 7 also reduces HIF-1α, p-AKT, and p-mTOR protein expression.89 Rapamycin (10) is responsible for abolishing TGF-b1-induced epithelial–mesenchymal transition and inhibits the mTOR pathway, reducing PKM2 expression.90 Indoprofen (8, 21 μM), nalidixic acid (9, 53 μM), and polydatin (11, 128 μM) are identified as inhibitors of PKM2 at micromolar concentrations.91
Fig. 6. Biologically active molecules like dexmedetomidine (5), TLSC702 (6), LY294002 (7), indoprofen (8), nalidixic acid (9), rapamycin (10), and polydatin (11) showing PKM2 inhibitory properties. The red portion in the molecule signifies the part with PAINS propensity.
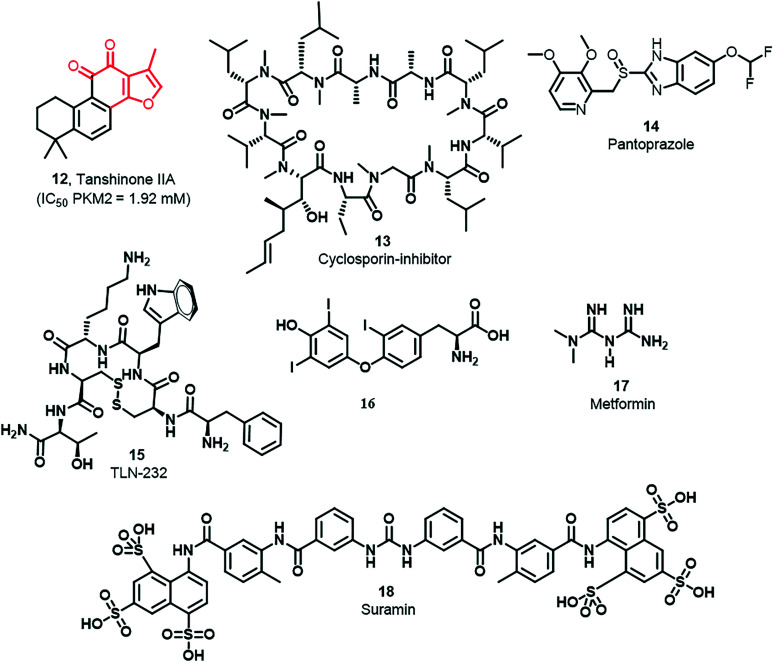
The growth of esophageal cancer cells is inhibited by tanshinone IIA (12) through the miR-122 down-regulating PKM2 pathway, and the reported IC50 of 12 is 1.92 mM (Fig. 7).92 Three breast cancer cell lines possess overexpressed PKM2, namely MDA-MB-435, MCF-7, and MDA-MB-231. Their growth is inhibited by cyclosporine A (CsA) (13) by down-regulation of PKM2 (Fig. 7).93 Pantoprazole (14) suppresses tumor proliferation and induces apoptosis in adenocarcinoma cells (Fig. 7).94 Likewise, 15 is a synthetic cyclic heptapeptide (TLN-232) having potential PKM2 specificity and antineoplastic activity (Fig. 7).95,9616 is a biologically essential hormone known as 3,3,5-triiodothyronine and thyroxine secreted from the thyroid gland (Fig. 7). Treatment with T3 blocks the reactive oxygen species (ROS) and malonic dialdehyde (MDA) expression. PKM2 is inhibited by Si RNA that increases oxidative stress.97 In various cancers, metformin (17) (Fig. 7) is used as an anticancer agent. In cervical carcinoma, 17 abolishes EMT by hindering mTOR signaling to down-regulate the expression of PKM2.98,99 Under nutrient poor conditions, 17 inhibits AMPK, which down regulates PKM2 that initiates apoptosis in breast cancer cells.100 In yet another report by Tang et al., it was confirmed that treatment of tumor tissues with 17 down-regulates PKM2 and enhances the apoptosis rate. 18 (sulfonic acid-based drug suramin)86 has been used to inhibit human pyruvate kinase isoenzymes (M1, M2, and L isoenzymes) (Fig. 7). Its IC50 values against M1, M2, and L isoenzymes were 20 μM, 33 μM, and 2.2 μM, respectively. The Ki of 18 against M1, M2, and L isoenzymes was 10.00 μM, 16.50 μM, and 1.10 μM, respectively.
Fig. 7. Drugs with established PKM2 inhibitory activity. Tanshinone IIA (12), cyclosporin inhibitor (13), pantoprazole (14), TLN-232 (15), 3,3,5-triiodothyronine (16), metformin (17), and suramin (18) show PKM2 inhibitory properties. The red portion in the molecule signifies the part with PAINS propensity.
2.3. Dyes having PKM2 inhibitory properties
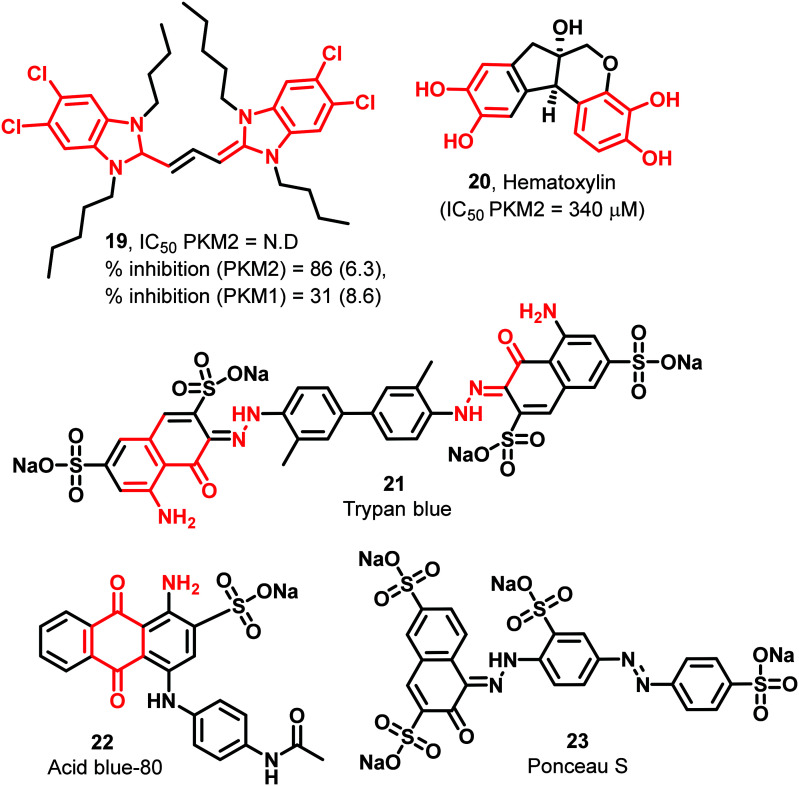
The benzimidazole heterocycle is also a critical pharmacophore. It also shows structural analogy to nucleotides found in the human body. 19 is a benzimidazole derivative with four chlorine atoms in the molecule, increasing the molecule's overall lipophilicity (Fig. 8). 19 is a symmetric molecule having four conformationally flexible alkyl chains of varying carbon numbers. 6.3 μM of 19 exhibits 86% inhibition of PKM2.80 Hematoxylin (20) is also identified as a weak inhibitor of PKM2 (340 μM) (Fig. 8).91 The series of sulfonic acid-based molecules like trypan blue (21), acid blue-80 (22), and Ponceau S (23) (Fig. 8) interact or bind with the active site of pyruvate kinase and show inhibition of PKM2.86 It is also reported that 22 and 23 absorb light at which kinetic studies are difficult to perform, making it impractical to determine their Ki and IC50 values. Although these dyes have strong PKM2 inhibitory properties, their instability problems under physiological conditions still restrict their use.95 Besides, their having PAINS propensity flanked with toxicity threatens their prolonged usage as anticancer agents.
Fig. 8. Dyes have PKM2 inhibitory properties. The red portion in the molecule signifies the part with PAINS propensity.
2.4. Polyphenolic compounds having PKM2 inhibitory properties
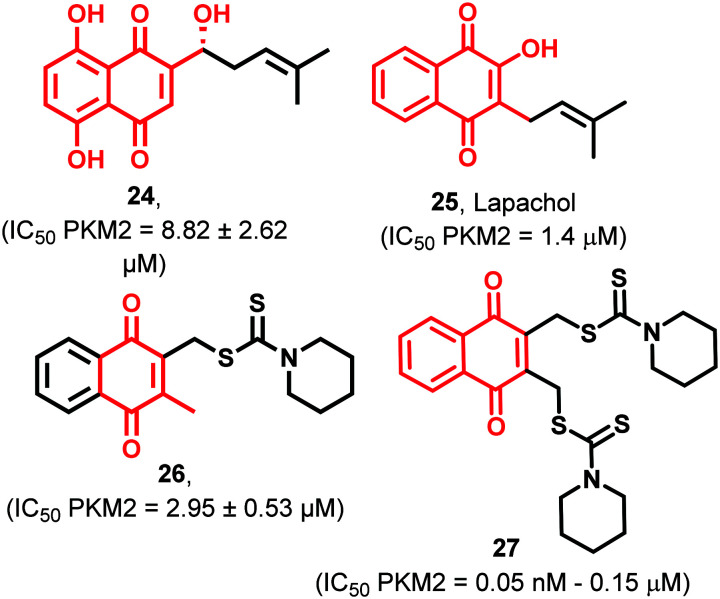
Shikonin (24) selectively inhibits PKM2 without affecting the activities of the PKM1 isoform (Fig. 9). It inhibits glucose consumption and promotes the release of lactate in MCF-7 and A549 tumor cells. This study shows that such specific PKM2 inhibitors have the potential to mature into anticancer hits.101 Although 24 may have metabolically fragile phenolic groups, judicious chemical manipulations may help obtain a potent drug-like molecule. These molecules are also known as oncostatics. These molecules may be used with existing oncology drugs. 1.4 μM of lapachol (25) (Fig. 9) inhibits purified PKM2 enzyme as well as PKM2 in cell extracts of melanoma. Obstruction of glycolysis in melanoma cells by 25 also led to decreased ATP levels. 25 also effectively sensitized cells to the mitochondrial protonophore, promoting apoptosis and antiproliferative effects. Thus 25 acts as an effective PKM2 inhibitor with high affinity towards its binding pocket.102 The estimated free energy and inhibition concentration for 25 are −9.34 kcal mol−1 and 141.8 nM, respectively. Meanwhile the estimated free energy and inhibition concentration for shikonin are −7.98 kcal mol−1 and 1.42 μM, respectively. Henceforth, it becomes apparent that 25 enjoys the distinction of good binding affinity and lower IC50 in comparison to 24.102 Thus 24 and 25 show great promise as selective PKM2 inhibitors though they possess soft metabolic spots. Dithiocarbamates exhibit anticancer and antibacterial activities. Dithiocarbamate is an analog of carbamate where sulfur atoms replace both oxygen atoms.103 The fluorescence cell-based PK-LDH coupled enzyme assay was used to evaluate the PKM2 (cell-free) inhibitory potency of 26 (Fig. 9). 26 is a naphthoquinone-based potent inhibitor of PKM2 (IC50 = 2.95 μM). 26 was tested against various cancer cell lines and exhibited strong antiproliferative potential against several cell lines like HeLa, HCT116, and H1299 with IC50 in the lower micromolar range (2.95 ± 0.53 μM). Despite the proven role of phenols in the treatment of cancer, several identified phenol-based ligands are yet to capture the complexities of this receptor. It is important to mention that phenols may get easily converted to quinones and may lack selectivity.104 This promiscuity can be avoided by rational incorporation of isosteres and bioisosteres of phenols.82,83 Ning et al. designed and synthesized novel naphthoquinone-based derivatives and introduced dithiocarbamate moieties for achieving better PKM2 inhibitory activity and selectivity than shikonin (24).10527 (Fig. 9), a naphthoquinone derivative, shows anticancer potential by inhibiting HeLa, MCF7, B16, MDA-MB231, and HCT116 cells with IC50 ranging from 50–150 nM in a dose-dependent manner.106 Thus, such molecules showing selective antiproliferative potential are established across cell lines, and PKM2 inhibitory activity is ascertained, which can be taken ahead for in vivo and PK/PD studies.
Fig. 9. Polyphenolic compounds have PKM2 inhibitory properties. The red portion in the molecule signifies the part with PAINS propensity.
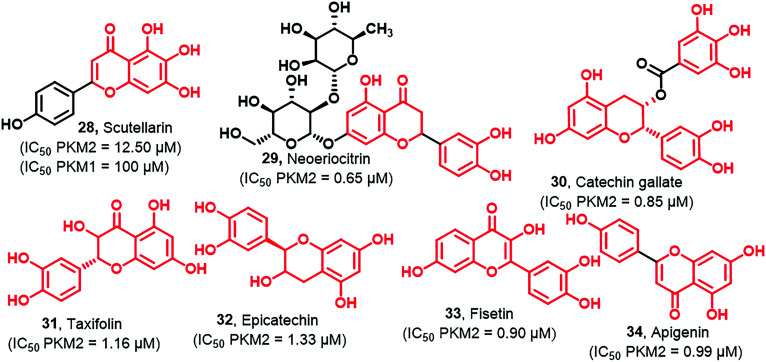
Flavonoids are well known for their anticancer, antibacterial, antifungal, and anti-inflammatory activities. Scutellarin (28) is a natural polyphenol isolated from Erigeron breviscapus (Fig. 10) and is used to treat several diseases in China like cerebral infarction, angina pectoris, and coronary heart diseases. They develop multiple NO-donating scutellarin derivatives as novel, potent anticancer agents. Multiple NO-donating scutellarin derivatives show cytotoxicity against MCF-7, HCT-116, HepG2, and PC-3 cancer cells.107 It decreased glycolytic rates by directly targeting PKM2, and it also triggered an apoptotic pathway. You et al. prepared two probes P4 and P10 of biotinylated scutellarin through the process of bioconjugation.108 One of the biotinylated analogs was the blank probe (P6) used to identify the potential target of 28 (Fig. 10).
Fig. 10. Flavonoids with PKM2 inhibitory properties. The red portion in the molecule signifies the part with PAINS propensity.
Results reflected that P10 and P4 probes exhibited weak antitumor activities on HeLa cells at concentrations of 50–400 μM. Also, it expressed more potent inhibition of PKM2 over PKM1. Again, it is essential to state that considering the selectivity, such molecules should be used as hits to be matured into bona fide PKM2 inhibitors by blocking such molecules' quick metabolism. Other flavanonols like neoeriocitrin (29), catechin gallate (30), taxifolin (31), epicatechin (32), and fisetin (33) have PKM2 inhibitory profiles (Fig. 10). These compounds were found to inhibit the Wnt/β-catenin pathway typically involved in the tumor progression mediated by PKM2.109,11031 exhibits a chemopreventive effect against colon cancer cells. It also exhibits multiple beneficial effects, including antioxidant, antidiabetic, and anti-inflammatory. Its anticancer activity is mediated by inhibiting PKM2 with an IC50 of 1.16 μM. Whether the activity is real or false positive will be clear only after in-depth investigation and in vivo studies. 30 and 32 also act as antitumor agents by inhibiting PKM2 at an IC50 of 0.85 μM and 1.33 μM, respectively. 29, 30, and 32 are noncompetitors, while 31 is a competitor inhibitor of PEP. Apigenin (34, Fig. 10) belongs to the flavone class that blocks cellular glycolysis by inhibiting the expression of PKM2 and producing the anti-colon cancer effect. It was tested against various cancer cell lines such as HCT116, HT29, and DLD1 and exhibited an IC50 of 27.9 ± 2, 2.0 ± 3.01, and 89.5 ± 4.89 μM, respectively.45,48 In the presence of FBP, 34 did not show the reversed inhibitory effect for PKM2; hence it was concluded that apigenin is an allosteric inhibitor of PKM2.111,112 Thus, these flavonoids have strong potential to be developed as PKM2 hits. It may require some semisynthetic manipulations so that the first-pass metabolism can be avoided and bioavailability is not compromised.113–115 Bioavailability of phenolics (flavonoids) is generally compromised in phase II conjugation, facilitating fastidious elimination from the body. It is suggested that the half-lives of flavonoids could be dramatically increased by recycling through phase II metabolic pathways. This may enhance their PKM2 inhibition and anticancer effect for prolonged durations. Moreover, phenolics may act by the cysteine-oxidizing mechanism, which poses limitations that will negatively affect in vivo efficacy or safety. Phenols may also react with thiols (cysteine). Thus phenolic groups present significant challenges in terms of selectivity. Phenolics may be oxidized to reactive quinones. They have attacked as electrophiles the nucleophilic amino acid side chains of PKM2 forming covalent bonds. Thus, reaction mechanisms depend on the phenolic structure and molecular characteristics.
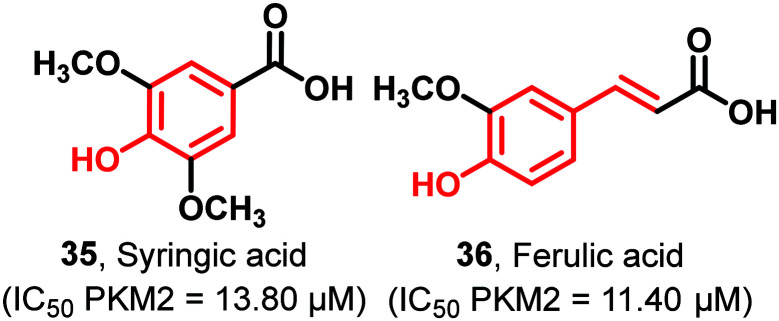
Some phenolic acids, which are biosynthesized in plants from amino acids like l-phenylalanine or l-tyrosine, like syringic acid (35) and ferulic acid (36), exhibit PKM2 inhibition with an IC50 of 13.80 μM and 11.40 μM, respectively (Fig. 11).111 It is worth mentioning that the acid and phenolic moieties are amenable to downstream modifications to achieve better anticancer leads.116
Fig. 11. Phenolic acids with PKM2 inhibitory potential. The red portion in the molecule signifies the part with PAINS propensity.
2.5. Amino acids with PKM2 inhibitory properties
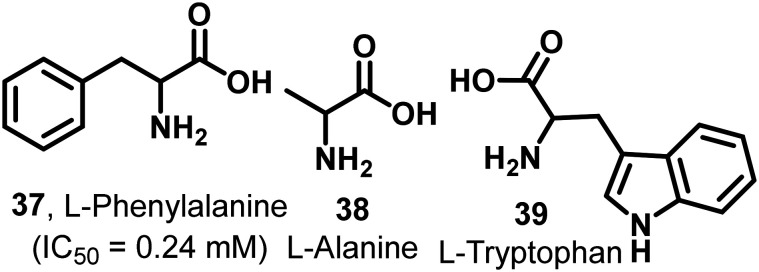
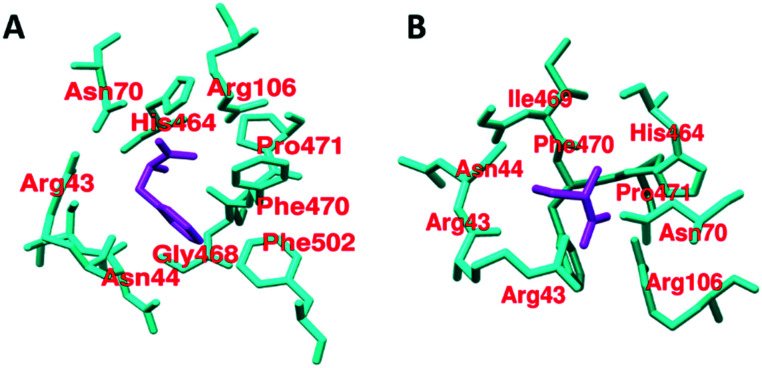
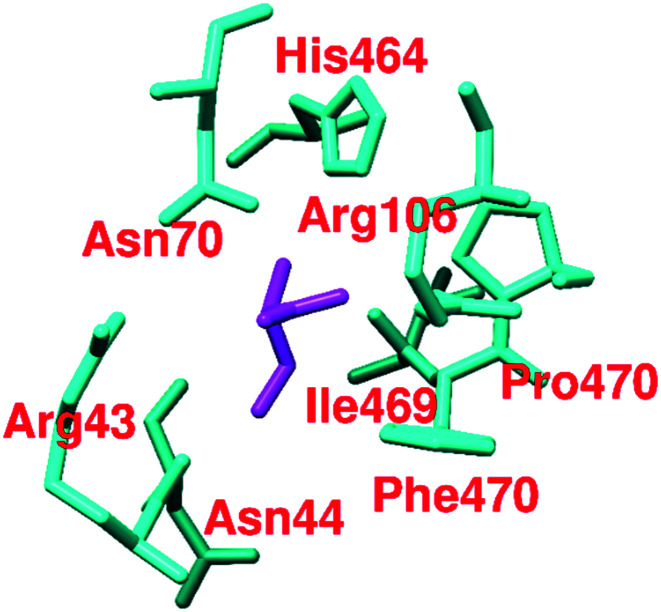
It has been found that out of 20 proteinogenic amino acids, l-phenylalanine (37, IC50 = 0.24 mM), alanine (38), and tryptophan (39) act as inhibitors of PKM2 (Fig. 12). Their inhibitory effect on PKM2 was tested by using size exclusion chromatography. The X-ray structure of the PKM2 complex with phenylalanine (Fig. 13), alanine, and tryptophan showed that PKM2 is locked into the (inactive) T-state conformation. 37, 38, and 39 formed hydrogen bonds (H-bonds) with Asp-70, Arg-106, His-464, and Ile-469.
Fig. 12. Amino acids with PKM2 inhibitory properties.
Fig. 13. A) Cartoon representation of the crystal structure of the PKM2–phenylalanine complex (chain A). The interactive residue is distinct from the FBP binding site. The selected zone is 4 Å around phenylalanine and cysteine. Phenylalanine: magenta color and amino acid: cyan color, PDB ID: 4FXJ. B) Cartoon representation of the crystal structure of the PKM2–cysteine complex (chain A). The interactive residue is distinct from the FBP binding site. The selected zone is 4 Å around cysteine. Cysteine: magenta color and amino acid: cyan color, PDB ID: 6NU1.
These interactions push the N-terminal loop outwards and ultimately stabilize the (inactive) T-state of PKM2 (PDB ID: 4FXJ).117,118 Kinetic studies of l-amino acids (37–39) have been established. They act as uncompetitive allosteric inhibitors of PKM2. Although these l-amino acids have PKM2 inhibitory properties and act as allosteric inhibitors, their chances of being developed as anticancer drugs are very meager. Serine (activator) and cysteine (inhibitor) regulate the PKM2 activity and oligomeric state. Both ligands bind at the amino acid (A–A′)-binding pocket, which is a different binding site than the interface binding site. The PKM2 asparagine (Asn70) and arginine (Arg106) residues directly make contact with the carbonyl group of cysteine (Fig. 13). Due to the interaction, Arg106 flips inward.
Similarly, the amine group of cysteine directly binds with the histidine (His464) and isoleucine (Ile469) residues.59,119 Dynamic population (tetramer and dimer/monomer equilibrium) of wtPKM2 due to cysteine interaction has been observed.59,119 Amino acids like aspartic acid and asparagine act as PKM2 activators, whereas valine acts as a PKM2 inhibitor. These small molecules are similarly bound to Ser/Asn/Asp and act as activators of PKM2.
2.6. Miscellaneous compounds
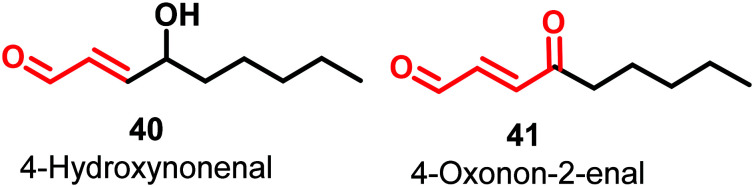
4-Hydroxynonenal (HNE) (40) and 4-oxonon-2-enal (41) (Fig. 14) are lipophilic electrophilic species that covalently modify multiple sites of PKM2 by inhibiting the His-439 and Cys-424 residues of the protein.
Fig. 14. Aliphatic carboxaldehyde as PKM2 inhibitors. The red portion in the molecule signifies the part with PAINS propensity.
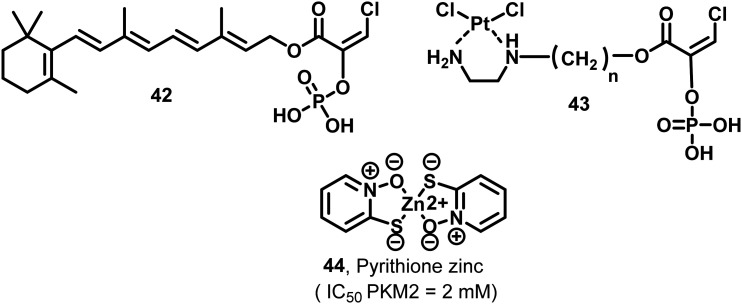
These amino acids play a vital role in protein–protein interactions and binding with allosteric activator FBP.120 This is an example of covalent inhibition of PKM2. The molecules have a Michael acceptor moiety, which helps in kinase inactivation. Michael acceptors are regarded as promiscuous moieties. Given the significance of such covalently acting molecules and terminal carboxaldehyde activity, these molecules have raised the hope that aldehydes can also be used as effective hits. A natural compound, 2′-hydroxycinnamaldehyde (HCA), activates the tetrameric state of PKM2 and inhibits STAT phosphorylation.12142 is a retinoic acid derivative that has a conjugated internal ester group. The cyclohexenyl ring offers some degree of conformational flexibility to the molecule.
43 consists of a platinum complex and phosphoric acid with a halo terminated vinyl chain.12243 and 44 exhibit anticancer activity by inhibiting PKM2 (Fig. 15). In oral squamous cell carcinoma cells (OSCC), pyrithione zinc (PYZ) (44) induced apoptosis in vitro by reducing the expression of PKM2.123 This is one of the examples of drug repurposing where a successful antidiabetic drug may be employed for PKM2 mediated malignancies.
Fig. 15. Miscellaneous compounds with PKM2 inhibitory properties.
3. PKM2 activators
3.1. Natural ligands with PKM2 activator properties
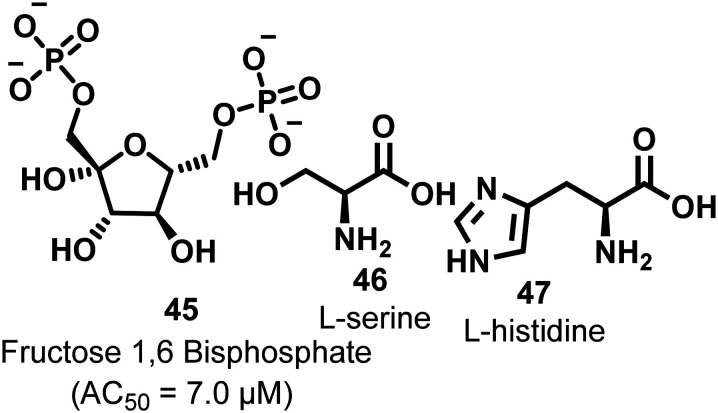
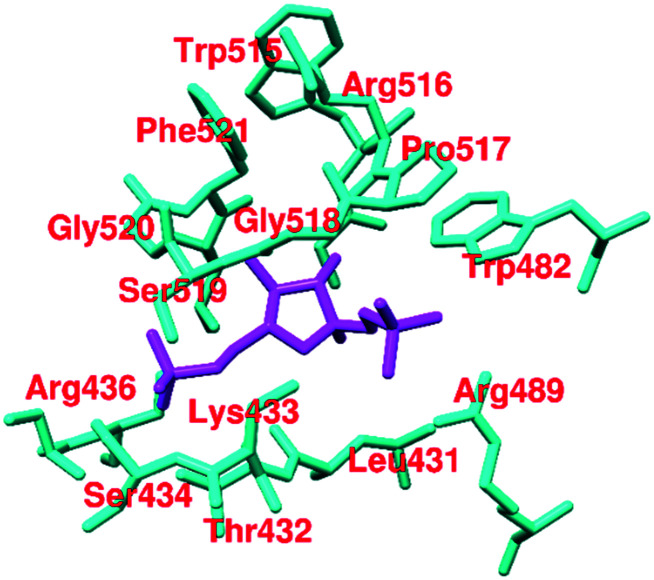
Fructose 1,6-bisphosphate (FBP, 45, Fig. 16) is a natural allosteric activator of PKM2 (AC50 = 7.0 μM). In the absence of FBP, PKM2 is present in monomeric, dimeric, and tetrameric forms. Binding of FBP at the allosteric site of PKM2 results in an increase of its activity by 4–10-fold. The FBP binding site contains Trp-515 and Arg-516 residues, which form the salt bridge, while hydrogen bonding at the C–C′ interface may stabilize the (inactive) T-state of PKM2. Once FBP is overlapped by the amino acid loops (514–524) and forms a hydrogen bond between Arg-516–Asp-487–Trp-515, it typically stabilizes the (active) R-state (Fig. 17). The six phosphate oxygen atoms of FBP form hydrogen bonds with lysine-433, serine-434, serine-437, and glycine-520 (Fig. 17). The different amino acids interact with various oxygen atoms of FBP. These are responsible for stabilizing the active PKM2 state.124,125 Another natural ligand, l-serine (46),118 also acts as an allosteric activator of PKM2. l-Serine binds to PKM2 at the C-terminal site, which is distinct from the FBP binding site. Histidine (47) is also a heterocyclic amino acid that prompts PKM2 activation by stabilizing the active R-state conformation.
Fig. 16. Naturally occurring allosteric activators of PKM2. 45 binds to the allosteric site, whereas 46 and 47 bind at a site other than the FBP binding site.
Fig. 17. Cartoon representation of the crystal structure of the PKM2–FBP complex (chain A). The selected zone is 4 Å around FBP, FBP: magenta color and amino acid: cyan color, PDB ID: 3GR4.
This ultimately thwarts the dissociation of tetramer to dimeric or monomeric forms. As explained in the inhibitor section, l-amino acids like phenylalanine (37), alanine (38), and tryptophan (39) can stabilize the inactive T-state of PKM2. In contrast, serine (46) can efficiently stabilize the active R-state of PKM2 by forming hydrogen bonds (H-bond) with Asp-70, Arg-106, His-464, Iie-469, and Arg-43 (Fig. 18). These extra interactions with Arg-43 push the N-terminal loop containing amino acids inward and stabilize the R-state of PKM2 (Fig. 18).118 In proliferating tumor cells, higher metabolic activity is noticed to gain quick access to nucleic acids for rapid growth and proliferation, demanding fast T to R allosteric switch. Faster T to R switching or slower tetramer–monomer dissociation of PKM2 assists cells in balancing the rate of catabolism and anabolism.126
Fig. 18. Cartoon representation of the crystal structure of the PKM2–serine complex (chain A). The interactive residue is distinct from the FBP binding site. The selected zone is 4 Å around serine. Serine: magenta color, and amino acid: cyan color, PDB ID: 6GG6.
This helps in the quick growth and proliferation of cancer cells. Even in the absence of FBP, serine (46) interactions alone can stabilize the R-state of PKM2, closely resembling the PKM1 isoform. Therefore, the serine-bound structure's enhanced activity may be attributed to the stabilization of the R-state conformation possibly by the hydrogen bond interactions made by serine in the amino-acid binding pocket.118
3.2. Heterocyclics acting as PKM2 activators
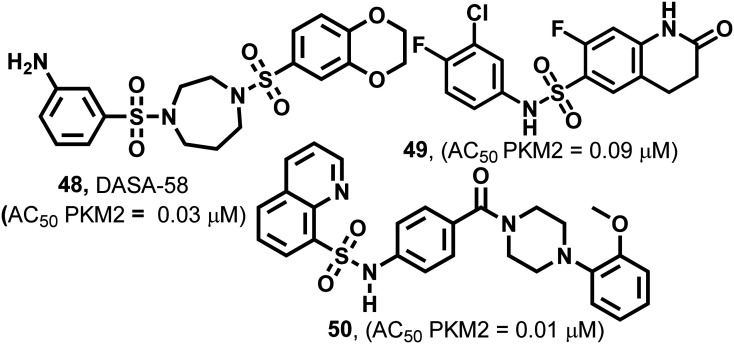
Several synthetic small heterocyclic molecules that activate PKM2 have been reported. N,N′-Diarylsulfonamides (Fig. 19) act in a similar manner to the natural activator FBP. We know that FBP stabilizes the R-state at the C–C′ interface, whereas N,N′-diarylsulfonamides stabilize the R-state of PKM2 by binding at the A–A′ interface (of two monomers of PKM2). These derivatives have considerable potencies, drug-like features, and appreciable aqueous solubilities. They exhibit selective activation of PKM2 and do not interact with PKM1, PKR, or PKL isoforms, which marks their selectivity. DASA-58 (48, Fig. 19) shows half-maximum activation response (AC50) at 0.03 μM. The binding site of DASA-58 at the A–A′ interface site of two monomers is approximately 35 Å away from the FBP binding site. It has been found that the acetylation of lysine-305 (K305) results in the destabilization of PKM2. This is probably because of a salt bridge formation with Glu-384 at the A–A′ interface site (Fig. 20).12748 prevents this interaction between two residues and stabilizes the tetramer of PKM2. Most of the N,N′-diarylsulfonamides possess an AC50 of 0.01–0.09 μM and exhibit a very good response.128 Similar PKM2 activators like 48 which act at a site away from the allosteric or orthosteric sites are developed. However, whether the same set of molecules will be equipotent in autochthonous tumor models is yet to be elucidated.36
Fig. 19. N,N′-Diarylsulfonamides as PKM2 activators bind at the interface of two monomers of PKM2, triggering tetramer formation.
Fig. 20. Cartoon representation of the crystal structure of the PKM2–DASA complex at the interface site between chain A and chain B. The interactive residue is distinct from the FBP binding site. The selected zone is 4 Å around the activator. Activator: magenta color and amino acid: cyan color, PDB ID: 3GR4.
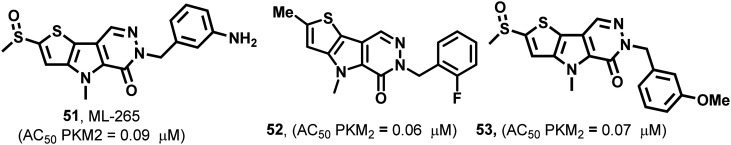
51–53 (Fig. 21) are pyridazinone derivatives and have different substitution patterns that show PKM2 activation at variable concentrations.129,130 TEPP-46 (51), also known as ML-265, acts similarly to DASA-58 (48). It has an AC50 of 0.09 μM in the H1299 cell line.57 PKM2 is closely associated with several biochemical and pharmacological events in the cell.18 The events take place by the impact of modulators on amino acids. TEPP-46 is similarly bound to the DASA binding site. TEPP-46 inhibits the growth of experimental autoimmune encephalomyelitis (EAE) and T helper cells (Th17 and Th1) in vitro. 51 also blocks the nuclear translocation of PKM2, and severely impacts T-cell activation and pathogenicity under biological conditions. This highlights the modulation of amino acid residues using PKM2 modulators like 51. Two equivalents of 51 bind at the dimer–dimer interface and the molecule is entirely buried within this interfacial pocket. 51 is accommodated through van der Waals interactions and water-mediated hydrogen bonds to the pocket lining residues. It binds at the dimer–dimer A–A′ interface. The sulfoxide and carbonyl groups of 51 form water-mediated hydrogen bond networks with PKM2, while the aniline moiety forms a direct hydrogen bond with aspartic acid (D354). Co-crystallization studies of PKM2 with 51 indicated that two small molecules bind per tetramer, each at the dimer–dimer interface. It forms water-mediated and direct hydrogen bonds to the amino acid residues of both monomers in each dimer. Somewhat surprisingly, four equivalents of 1,6-FBP still bind per tetramer, highlighting the distinct location that the endogenous activator binds to compared to the small synthetic molecules.131–133
Fig. 21. ML-265 (51), 52, and 53 show PKM2 activation by binding at the interface site of two monomers of PKM2.
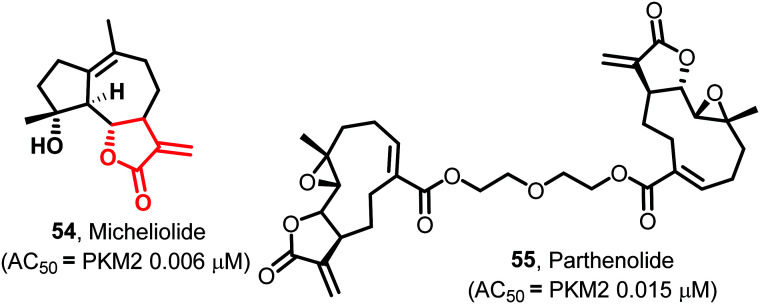
The naturally occurring guaianolide sesquiterpene lactone, micheliolide (54) (MCL, Fig. 22), is found in Michelia compressa and Michelia champaca. 54 is an irreversible, selective, and covalent activator of PKM2. It provides therapeutic efficacy by inhibiting the acetylation of lysine-433 (K-433) of PKM2, preventing its nuclear migration and blocking cancer cells' proliferation. LC-MS/MS data proved that the exocyclic double bond acts as a Michael acceptor, which formed a selectively covalent bond with the cysteine-424 (C-424) residue of PKM2, absent in PKM1. This double bond plays a role in high pyruvate kinase activity by facilitating the irreversible tetramerization of PKM2. The pro-drug dimethylaminomicheliolide (DMAMCL) is in Australia's clinical trials, suppressing leukemia cells' growth in the xenograft model of zebrafish. Molecular dynamics studies suggested that the A–A′ interfacial binding energy of the bonded ligand (MCL–PKM2 complex) is higher (−322.3 kcal mol−1) compared to the non-bonded ligand (PKM2 without complex, −253.8 kcal mol−1). MCL reduces the nuclear translocation of PKM2 by stabilizing the more active tetrameric form. This leads to the prevention of the proliferation of tumor cells.134 Another natural product, parthenolide (55) (PTL, Fig. 22), shows moderate PKM2 activation properties. 55 shows potent PKM2 activation without any impact on the expression of tetramer formation. LC-MS/MS data suggested that similar to micheliolide, 55 covalently binds to a dimer of PKM2 at cysteine-424 and forms a stable tetramer of PKM2. PTL also inhibits the protein kinase activity by preventing the translocation of dimeric PKM2 into the nucleus and promotes pyruvate kinase activity by forming tetramer PKM2 in the cytosol. PTL also inhibits the STAT3 pathway and induces apoptosis in cancerous cells.13554 and 55 may act as bona fide activators of PKM2 (because of their selectivity) with the potential of populating the drug discovery pipeline with their derivatives.
Fig. 22. Micheliolide (54) and parthenolide (55) are PKM2 selective covalent activators. The red portion in the molecule signifies the part with PAINS propensity.
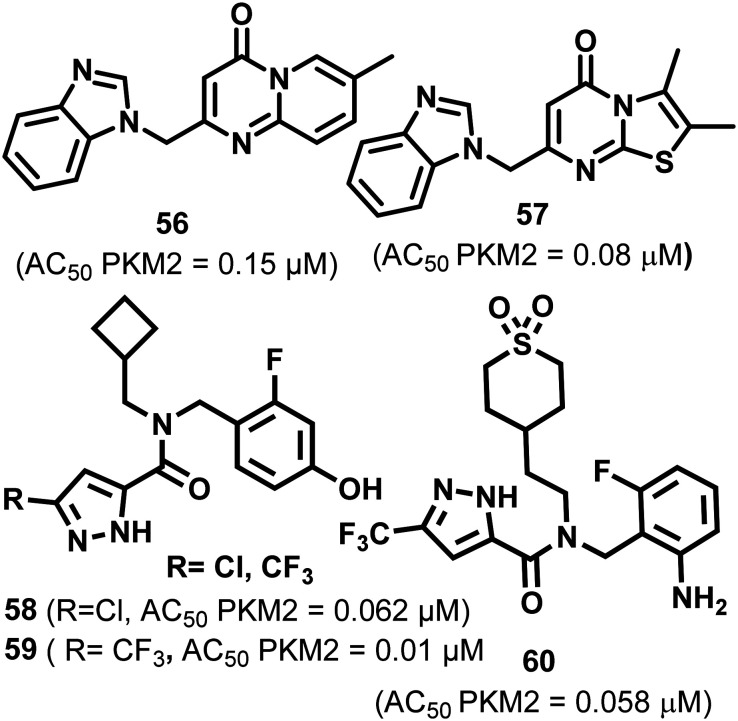
Guo et al. identified pyridopyrimidone derivatives 56–57 as PKM2 activators using high throughput screening which was confirmed later on by the LDH coupled enzyme assay (Fig. 23). 56 and 57 act as potent and selective activators of PKM2.136 They possess good permeability, excellent aqueous solubility, and metabolic stability.136
Fig. 23. Pyridopyrimidone (56–57) and pyrazole (58–60) derivatives that activate PKM2.
These may be utilized in combination with existing oncostatics. Kanner et al. screened pyrazole-5-carboxamide-based derivative 58in vitro and in vivo. Several electron-withdrawing groups tested (R = –Cl and –CF3) showed the highest activation. 58 and 59 exhibited an AC50 of 0.062 and 0.01 μM, respectively.137,138 These are highly cell-permeable, reach the cytosol, and ultimately stabilize the active tetrameric form of PKM2, eventually terminating cell proliferation and metabolism in cancer cells.132,139 Pyrazole-5-carboxamide-based compound 59 (Fig. 23) potentially activates PKM2 in lung cancer cell lines NCI-H1299 and A549. The authors crystallized the PKM2 complex with a Cl derivative. It has a 2.03 Å resolution. They observed that each monomer has one ligand at two dimer interfaces; hence a total of four ligands were found. The binding site is distinct from the allosteric (FBP binding site) and interface binding sites.127 Tolero pharmaceuticals explored the synergistic anticancer effect of 60 with a combination of the heat shock protein 90 (HSP90) and kinase inhibitors. The HSP90 inhibitor used was retaspimycin hydrochloride. The antineoplastic agent and sorafenib were utilized as tyrosine kinase inhibitors. When 60 was used in combination with the HSP90 inhibitor, it decreased ED50 from 59.94 nM to 36.45 nM.
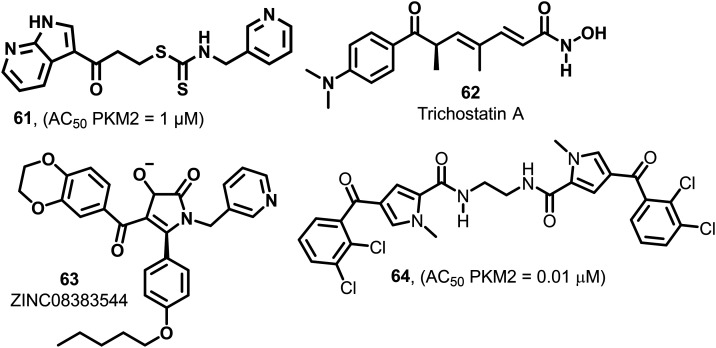
Similarly, when 60 was used in combination with sorafenib, it exhibited a 4-fold decrease in EC50 from 30.126 nM to 7.126 nM. 60 also showed an excellent synergy with bortezomib. In another report, a xenograft study using A549 lung cancer cells was performed to investigate whether there is some in vivo synergy between the representative ROS-producing anticancer drug doxorubicin and PKM2 activator 60. The combination of doxorubicin and 60 showed the best reduction of tumor volume and showed no significant bodyweight reduction.138 Based on the study, it may be concluded that the combination of the PKM2 activator 60 with other anticancer drugs will generate new treatment options. It is found that 61 (Fig. 24) acts as a selective PKM2 activator. The group examined its antiproliferative activity in lung and cervical cancer cell lines (IC50 = 1.12 μM).140In silico studies explored the interaction between PKM2 protein (PDB ID: 4GIN) and 61. The molecular docking studies demonstrated that 61 efficiently fits in the binding pocket with a binding free energy of −9.28 kcal mol−1. Using docking studies, it was found that there is a strong hydrogen bond connection between the hydrogen of seven-azaindole (61), Ala-388 and the carbonyl group of Lys-311 of PKM2. The seventh nitrogen in 61 binds to Tyr-390C and serves as a hydrogen bond acceptor. The activity of azaindole derivatives (1 μM) was much higher than indole [>20 μM] derivatives. Besides this, 61 also showed selective PKM2 activation, which means that 61 does not exhibit any activity on PKM1 or PKR isoforms.14161 prevents the migration of PKM2 to the nucleus and arrests the G2/M phase of the cell cycle.
Fig. 24. Nitrogen heterocycles and hydroxamate-containing compounds that show PKM2 activation.
Trichostatin (TSA, 62, Fig. 24) enhances SRSF3 upregulation that may be responsible for the tetramerization of pyruvate kinase and hypoxia-inducible factor-1α dependent glycolytic gene expression.142 1,5-2H-Pyrrole-dione (63) (Fig. 24), the derivative, also shows a pyruvate kinase activation effect. It stabilizes the tetramer of PKM2, reduces the translocation of PKM2 to the nucleus, and inhibits cellular proliferation and tumorigenesis. 64 (Fig. 24) is 4-(2,3-dichlorobenzoyl)-1-methyl-1H-pyrrole-2-carboxamide that fulfills drug-likeness criteria with low molecular weight, high ligand efficacy, and high PKM2 activation effect with an AC50 of 0.01 μM. The methylene and ethylene bridges in the molecules show a higher impact on PKM2 activation.143 Such selective molecules having an activation effect only on PKM2 may be utilized for therapeutic interventions in cancer.
3.3. Polyphenolic compounds having PKM2 activation properties
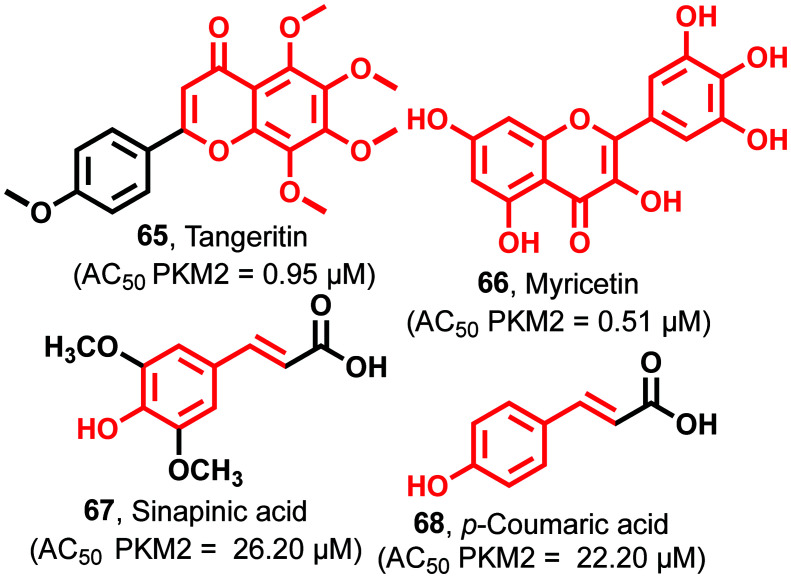
Tangeritin (65)111 contains a chromenone nucleus, and chemically it is known as a 4,′5,6,7,8-pentamethoxyphenyl chromen-4-one derivative with an AC50 of 0.95 μM against PKM2 (Fig. 25). 65 also causes a reduction of intermediate metabolites like lipids, nucleotides, and amino acids, which retard the cell growth in HL-60 cells. 65 inhibits the generation of cyclo-oxygenase throughinterleukin-1-β as an inflammatory mediator. 65 causes the suppression of various genes involved in cancer cell proliferation like P38 MAPK, Ras–Raf–MEK–ERK pathway, C-Jun-N-terminal kinase (JNK), and phosphoinositide3-kinase (PI3K/AKT). Myricetin (66)144 is a trihydroxy phenyl chromanone containing scaffold with an AC50 of 0.51 μM against PKM2 (Fig. 25).
Fig. 25. Polyphenolic compounds as PKM2 activators. The red portion in the molecules signifies the part with PAINS propensity.
It induces apoptosis through the enhancement of the apoptotic/antiapoptotic protein BAX/Bcl-2 ratio. It causes significant changes through the caspase pathway and induces cleavage of caspase-3 and caspase-9, resulting in the release of apoptosis-inducing factor (AIF) from mitochondria to the cytoplasm. This result shows that 66 is a beneficial therapeutic agent for the treatment of colon cancer. Sinapinic acid (67)144–146 also shows PKM2 activation with an AC50 of 26.20 μM (Fig. 25). It shows anticancer activity through inhibition of the dimerization of PKM2. Tyrosine-105 phosphorylation in PKM2 causes the dislodging of binding of FBP. Sinapinic acid forms a tight tetramer junction between PKM2 and PKM1. This provides resistance to inhibition due to the phosphorylation of tyrosine residues. Besides, it prevents the translocation of dimeric PKM2 to the nucleus, avoiding the proliferation of tumor cells. This, in turn, blocks the formation of glycolytic intermediates due to higher pyruvate kinase activity and inhibits Hif-1α. 68 has 4-hydroxy phenyl acrylic acid as the central scaffold with an AC50 of 22.20 μM against PKM2.120,147 It leads to the inhibition of cysteine-358 oxidation and efficiently inhibits tyrosine phosphorylation, together with the diversion of glycolytic intermediates into the auxiliary pathways to sustain the growth and proliferation of tumor cells. The biological and pharmacological potential of these polyphenolics is well established. As mentioned in the inhibitors section, these phenolics may hit the market as anticancer agents after due optimization of the PK/PD properties. They have strong potency and affinity to PKM2, and their drug-likeness can be easily tuned to increase their half-life and prevent their elimination from the body.
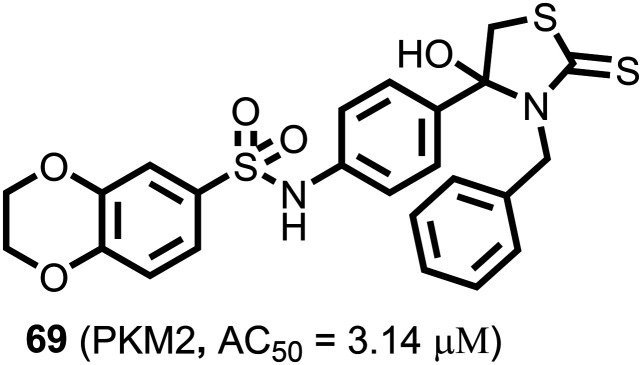
A series of novel 4-hydroxy-thiazolidine-2-thione (69) (Fig. 26) derivatives were identified as the most potent antitumor agents with an AC50 of 3.14 μM against PKM2. 69 showed excellent antiproliferative activity against H1299, HCT116, HeLa, and PC3 cell lines, affecting IC50 values from 0.46 μM to 0.81 μM.
Fig. 26. The thiazolidine-2-thione-based PKM2 activator.
It arrested the cell cycle at the G2/M phase in the HCT116 cell line.148 It is reported that these compounds (65–69) affect metabolic pathways and show anticancer activity with protein kinase activation in cell metabolism, significantly affecting glucose consumption. Therefore, we propose that the activation effect of these compounds on PKM2 may contribute to their anticancer activity.
3.4. Miscellaneous
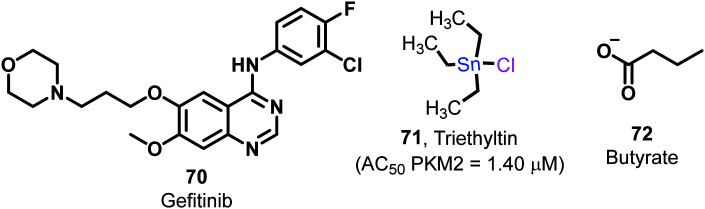
Gefitinib (70, Fig. 27) is a potent tyrosine kinase inhibitory drug, which induces apoptosis in mutant lung cancer cells. It concurrently inhibits glucose utilization and shows the PKM2 activation effect.149 The utilization of this drug as a potent PKM2 activator alone or in combination may help tackle tumors with multiple arms. At low concentrations, triethyltin (71) (Fig. 27) activates PKM2, while at higher concentrations inhibits it.150 Triethyltin (71) shows anticancer activities by various mechanisms like inhibition of macromolecular synthesis, mitochondrial dysfunction, and DNA damage. It may form polymeric coordination complexes with the heterocyclic amino acid histidine.
Fig. 27. Miscellaneous molecules with PKM2 activation properties.
Triethyltin binds selectively and with very high affinity to specific mitochondrial proteins, leading to death of cancer cells. Yet, due to tin metal's presence, the safety profile of the molecule is highly questionable. Compound 72 (Fig. 27), known as butyrate, is the conjugate base of butyric acid used in cancer treatment. It targets HDAC and subsequently regulates the pro-apoptotic system of the cell cycle. Mechanistically butyrate directly inhibits the metabolism of colorectal cancer cells. It directly acts to increase the tetramerization of the dimeric form of PKM2.151 Although the drug-likeness of the molecule again seems low, the development of butyrate salts of PKM2 modulators may be useful to treat cancerous cells and drag the dimer towards tetramer formation.
4. Conclusions
This review was intended to highlight the importance of a structured approach towards drug discovery targeting PKM2 representing a complex target. Here, we highlight the modulators of PKM2 with activities ranging from the micromolar to nanomolar range. These modulators activate or inhibit tumor PKM2. Additionally, developing new and PKM2 specific assays may help achieve consistencies between the IC50 values measured by biochemical versus cellular assays. The assays used for determining the PKM2 modulatory potential must have cost-effectiveness, optimization potential, and scalability and we believe that the customized LDH assay is a better option. However, some studies showed that the depletion of PKM2 did not significantly reduce tumor growth and tumor progression in mice. It is suggested that metabolic plasticity allows tumors to switch to alternate metabolisms, sustaining tumor growth. But importantly, several studies have also demonstrated the significant synergistic effect of PKM2 modulators with reputable oncology drugs present in the market. This also hints that preclinical evaluations check the synergistic impact of PKM2 modulators with other well-established oncology drugs like doxorubicin, vinca alkaloids, and other drugs. In the future, potent and selective PKM2 modulators may be worth exploring in diabetes, inflammation, Bloom syndrome, and other chronic disorders because of the association of PKM2 in several pathways.152,153
Moreover, two sequential conformational states thoroughly control the pyruvate kinase activity and its conformational landscape, one of which is an inactive T state, and the second is an active R-state, indispensable for the conversion of phosphoenolpyruvate to pyruvate. In the case of PKM2 inhibitors, it is challenging to suggest which state is inhibited. There is a need for more studies (crystal structure of a protein with the inhibitor) ascertaining the interactions of PKM2 with the inhibitor.
We also believe that designing drug-like PKM2 activators with adequate pharmacokinetics will be more beneficial than the inhibitors. This is because it will bypass the functional consequences associated with the kinase inhibitors reported to date. Activators like micheliolide, DASA-58, and TEPP-46 act by interacting with sites other than PEP binding sites. Thus, they effectively activate the dimeric PKM2 having a higher affinity towards allosteric sites leading to the Warburg effect's attenuation.60 It is also desirable to study the efficacy of activators like DASA-58 in autochthonous tumors that display overexpression of PKM2. Most of the developed activators possess heterocyclic cores, chemical structures that exhibit superior biological profiles because of the multiple interactions with the kinases, which results in highly efficient binding to the protein. Despite the physical advantages possessed by heterocyclic cores, their low aqueous solubility is a significant limitation. So, efforts must be directed towards making water-soluble analogs.
Depletion of PKM2 by inhibitors may not lead to tumorigenesis in the mouse model, which could be one of the primary reasons for the inhibitors' failure. Although several skeletally diverse activators and inhibitors of PKM2 have been investigated, several of them are not amenable to drug development due to the preponderance of PAINS. Henceforth designing such molecules with adequate drug-like properties with avoidance (as far as possible) of promiscuous (PAINS) moieties may lead to converting the dimeric form to the tetrameric form. We suggest that considering the presence of PAINS154 in several PKM2 selective phenolics, the medicinal chemists should consider their isosteric or bioisosteric replicas155 to avoid attrition once the molecule starts its odyssey from bench to bedside. This will improve the arsenal of PKM2 modulators and provide a smooth sailing of the molecules through the valley of death (drug discovery pathway). Equally exciting is the expectation from potent peptidomimetics targeting PKM2 which may serve with a greater degree of selectivity in the future.
The PKM2 modulators like DASA-58 that stabilize the R-state by binding at the A–A′ interface and afford tetramers should be considered for further development. The derivatives of this class have considerable drug metabolism and pharmacokinetic properties, and appreciable aqueous solubilities. Apart from this, they exhibit selective activation of PKM2 without having any impact on other isoforms. Thus, it is clear from the comprehensive account of activators and inhibitors that they show strong potential to be developed as drugs. It is expected that this and a similar approach will be used by academia and drug companies and may help hit critical metabolic conduit PKM2 in cancer. It is challenging to develop a selective modulator of PKM2 because of PKM1 and PKM2. Notably, designing selective PKM2 modulators, as discussed in the review, would help investigate their role in pathological and physiological processes. It is anticipated that more potent and selective PKM2 modulators acting as drug candidates with concrete mechanisms of action will sizably populate clinical trials. With an activator like 60 entering clinical trials in combination with anticancer therapy, it seems clear that such molecules' future seems promising. It may undoubtedly lead to more effective chemotherapeutic outcomes. To determine the potential of modulators in in vivo models, it is necessary to develop practical, sensitive detection methods or assays that lead to an accurate investigation of the inhibition of PKM2. From the drug development perspective, a difficult challenge ahead will be designing selective PKM2 modulators.
Abbreviations
- PKM2
M2 pyruvate kinase
- WHO
World Health Organization
- RAF
Rapidly accelerated fibrosarcoma
- RAS
Retrovirus-associated DNA sequences
- MEK
Mitogen activated protein kinase
- EGFR
Epidermal growth factor receptor
- Wnt
Wingless-related integration site
- ATP
Adenosine ‘5’-triphosphate
- PKR
Pyruvate kinase R
- PKL
Pyruvate kinase L
- PKM1
Pyruvate kinase M1
- MLC2
Myosin light chain 2
- SAR
Structure activity relationship
- IC50
Inhibitory concentration
- AC50
Activator concentration
- TZDs
Thiazolidinediones
- Bcl-2
B cell lymphoma
- Bcl-X
B cell X
- PPAR-γ
Peroxisome proliferator-activated receptor gamma
- CGZ
Cigilitazone
- RGZ
Rosiglitazone
- μM
Micromolar
- CRC
Colorectal cancer
- DHBA
Dihydroxy benzoic acid
- CDK1
Cyclin-dependent kinase
- LDH assay
Lactate dehydrogenase assays
- MAPKs
Mitogen-activated protein kinase
- SCU-CD
Scutellarin-cyclodextrin
- DNA
Deoxyribonucleic acid
- HDAC
Histone deacetylase
- VK3
Vitamin K3
- VK5
Vitamin K5
- FBP
Fructose 1,6-bisphosphate
- mTOR
Mammalian target of rapamycin
- ELISA
Enzyme-linked immunosorbent assay
- PCAF
P300/CBP-associated factor
- PDGF
Platelet derived growth factor
- FGFR1
Fibroblast growth factor
- PI3K/AKT
Phosphinositide-3-kinase/adenosine tyrosine kinase
- ERK
Extracellular signal-regulated kinase
- MFN1
Mitofusion
- DRP1
Dynamin related protein
- MDM2
Mouse double minute 2
- TCF4
Transcription factor-4
- NPM-ALK
Nucleophosmin-anaplastic lymphoma kinase
- P
Phosphorylation
Author contributions
Bhagyashri Rathod: writing – original draft. Shivam Chak: writing – original draft. Sagarkumar Patel: conceptualization, writing – original draft, software, investigation, data curation, software, visualization. Amit Shard: conceptualization, data curation, writing – review & editing, supervision. All the authors have approved the final version of the manuscript.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgments
BR, SC, and SP are thankful to NIPER-Ahmedabad, Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, and India's Government for their fellowships. The authors gratefully acknowledge Director, NIPER-Ahmedabad for the support and encouragement. The communication number for publication is NIPER-A/218/12/2018.
Biographies
Biography
Bhagyashri Rathod.

Rathod Bhagyashri Ramesh received her B. Pharm degree from Swami Ramanand Teerth Marathwada University, Nanded, in 2017. She obtained her M.S in Medicinal Chemistry from NIPER-A under the guidance of Dr. Amit Shard.
Biography
Shivam Chak.

Shivam Chak obtained his B. Pharm degree from Abdul Kalam Technical University, Uttar Pradesh, in 2017, and received his master's degree (M.S. Pharm) in Medicinal Chemistry from NIPER-Ahmedabad. He worked on the synthesis of the anticancer molecule containing trifluoroethanone substituted thiazoles under the guidance of Dr. Amit Shard.
Biography
Sagarkumar Patel.

Sagarkumar Patel completed his B. Pharm at Ganpat University, Gujarat, in 2014, and obtained his M. Tech (Pharm) degree from NIPER-Mohali in 2016. Now he is pursuing a Ph.D. in Medicinal Chemistry at the National Institute of Pharmaceutical Education and Research (NIPER)-Ahmedabad under the guidance of Dr. Amit Shard. His research area is the design and synthesis of pyruvate kinase M2 (PKM2) modulators.
Biography
Amit Shard.

Dr. Amit Shard received his M.Sc. degree in Pharmaceutical Chemistry and Ph. D from CSIR-Institute of Himalayan Bioresource Technology, Palampur. After completing his doctoral work, he joined as faculty at the National Institute of Pharmaceutical Education and Research-Ahmedabad in 2014. His research interests include the development of novel pyruvate kinase M2 modulators and computer-aided drug design.
References
- www.who.int/news-room/fact-sheets/detail/cancer. www.who.int/news-room/fact-sheets/detail/cancer
- Gatenby R. A. Gillies R. J. Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Kee H. J. Cheong J.-H. BMB Rep. 2014;47:158. doi: 10.5483/BMBRep.2014.47.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-Y. Biomol. Ther. 2015;23:99. doi: 10.4062/biomolther.2015.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H. R. Vander Heiden M. G. Wu N. Asara J. M. Cantley L. C. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Ru P. Williams T. M. Chakravarti A. Guo D. Cancers. 2013;5:1469–1484. doi: 10.3390/cancers5041469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M. Suematsu M. Saya H. Clin. Cancer Res. 2012;18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G. Cantley L. C. Thompson C. B. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrodt E. Reinacher M. Scheefers-Borchel U. Scheefers H. Friis R. Crit. Rev. Oncog. 1992;3:91. [PubMed] [Google Scholar]
- Vander Heiden M. G. Plas D. R. Rathmell J. C. Fox C. J. Harris M. H. Thompson C. B. Mol. Cell. Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M. S. Mesecar A. Heath P. J. Shi W. Nowak T. Stoddard B. L. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- Christofk H. R. Vander Heiden M. G. Harris M. H. Ramanathan A. Gerszten R. E. Wei R. Fleming M. D. Schreiber S. L. Cantley L. C. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Alauddin M. M. Am. J. Nucl. Med. Mol. Imaging. 2012;2:55. [PMC free article] [PubMed] [Google Scholar]
- Garber K. J. Natl. Cancer Inst. 2004;96:1805–1806. doi: 10.1093/jnci/96.24.1805. [DOI] [PubMed] [Google Scholar]
- Bayley J.-P. Devilee P. Curr. Opin. Oncol. 2012;24:62–67. doi: 10.1097/CCO.0b013e32834deb9e. [DOI] [PubMed] [Google Scholar]
- Gupta V. Bamezai R. N. K. Protein Sci. 2010;19:2031–2044. doi: 10.1002/pro.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niekerk G. Engelbrecht A.-M. Cell. Oncol. 2018;41:343–351. doi: 10.1007/s13402-018-0383-7. [DOI] [PubMed] [Google Scholar]
- Liu V. M. Vander Heiden M. G. Brain Pathol. 2015;25:781–783. doi: 10.1111/bpa.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-Y. Biomol. Ther. 2018;26:39. doi: 10.4062/biomolther.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P. Du W. Wu M. Protein Cell. 2014;5:592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.-C. Hung W.-C. Mol. Cancer. 2018;17:1–9. doi: 10.1186/s12943-018-0791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton T. L. Jacks T. Vander Heiden M. G. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori C. F. Mol. Cell. Biochem. 1974;5:47–53. doi: 10.1007/BF01874171. [DOI] [PubMed] [Google Scholar]
- Mazurek S. Int. J. Biochem. Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Alquraishi M. Puckett D. L. Alani D. S. Humidat A. S. Frankel V. D. Donohoe D. R. Whelan J. Bettaieb A. Free Radical Biol. Med. 2019;143:176–192. doi: 10.1016/j.freeradbiomed.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett J. A. Yeast. 2003;20:509–543. doi: 10.1002/yea.986. [DOI] [PubMed] [Google Scholar]
- Stone O. A. El-Brolosy M. Wilhelm K. Liu X. Romão A. M. Grillo E. Lai J. K. H. Günther S. Jeratsch S. Kuenne C. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-06406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T. Inoue H. Tanaka T. J. Biol. Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- Noguchi T. Yamada K. Inoue H. Matsuda T. Tanaka T. J. Biol. Chem. 1987;262:14366–14371. [PubMed] [Google Scholar]
- Gui D. Y. Lewis C. A. Vander Heiden M. G. Sci. Signaling. 2013;6:pe7. doi: 10.1126/scisignal.2003925. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Deng X. Liu Y. Liu Y. Sun L. Chen F. Cell Biosci. 2019;9:52. doi: 10.1186/s13578-019-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. J. Chen M. Assanah M. Canoll P. Manley J. L. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily M. D. Gray J. J. Proteins: Struct., Funct., Bioinf. 2007;67:385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- McEwen B. S. Neurochem. Res. 2000;25:1219–1231. doi: 10.1023/a:1007687911139. [DOI] [PubMed] [Google Scholar]
- Wang P. Sun C. Zhu T. Xu Y. Protein Cell. 2015;6:275–287. doi: 10.1007/s13238-015-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D. Yu Y. Israelsen W. J. Jiang J.-K. Boxer M. B. Hong B. S. Tempel W. Dimov S. Shen M. Jha A. Nat. Chem. Biol. 2012;8:839. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahra K. Dey T. Ashish A. Pandey U. Mishra S. P. Front. Oncol. 2020;10:159. doi: 10.3389/fonc.2020.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N. De Melo J. Tang D. Int. J. Cell Biol. 2013;2013:242513. doi: 10.1155/2013/242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Lu Z. Cell Cycle. 2013;12:3343–3347. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W. Semenza G. L. Trends Endocrinol. Metab. 2012;23:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M. Thomas S. D. Islam A. Muench D. Sedoris K. Clin. Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I. Song M. S. Hobbs R. M. Laurent G. Giorgi C. De Boer V. C. J. Anastasiou D. Ito K. Sasaki A. T. Rameh L. Cell. 2012;149:49–62. doi: 10.1016/j.cell.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. Wang H. Yang J. J. Liu X. Liu Z.-R. Mol. Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Zheng Y. Xia Y. Ji H. Chen X. Guo F. Lyssiotis C. A. Aldape K. Cantley L. C. Lu Z. Nat. Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese E. K. Hitosugi T. Front. Cell Dev. Biol. 2018;6:79. doi: 10.3389/fcell.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka F. Yoshimoto S. Okamura K. Ikebe T. Hashimoto S. Oncotarget. 2018;9:33745. doi: 10.18632/oncotarget.25850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoitei N. Becher A. Steinestel K. Rouhi A. Diepold K. Genze F. Simmet T. Seufferlein T. Mol. Cancer. 2016;15:3. doi: 10.1186/s12943-015-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M. A. Siddiqui F. A. Gupta V. Chattopadhyay S. Gopinath P. Kumar B. Manvati S. Chaman N. Bamezai R. N. K. Mol. Cancer. 2013;12:72. doi: 10.1186/1476-4598-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Zhang D. Chen X. He L. Li T. Xu X. Li M. Sci. Rep. 2015;5:16082. doi: 10.1038/srep16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Hu L. Chen M. Cao W. Chen H. He T. OncoTargets Ther. 2016;9:4277. doi: 10.2147/OTT.S106508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H. P. McNae I. W. Nowicki M. W. Hannaert V. Michels P. A. M. Fothergill-Gilmore L. A. Walkinshaw M. D. J. Biol. Chem. 2010;285:12892–12898. doi: 10.1074/jbc.M109.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakasam G. Iqbal M. A. Bamezai R. N. K. Mazurek S. Front. Oncol. 2018;8:22. doi: 10.3389/fonc.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G. Mao Q. Xia W. Xu Y. Wang J. Xu L. Jiang F. Oncol. Lett. 2016;11:1980–1986. doi: 10.3892/ol.2016.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L. Li D. Zhao D. Lin R. Chu Y. Zhang H. Zha Z. Liu Y. Li Z. Xu Y. Mol. Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L. Xu Y.-P. Zhao D. Li F.-L. Wang W. Sasaki N. Jiang Y. Zhou X. Li T.-T. Guan K.-L. Mol. Cell. 2013;52:340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T. Kang S. Vander Heiden M. G. Chung T.-W. Elf S. Lythgoe K. Dong S. Lonial S. Wang X. Chen G. Z. Sci. Signaling. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Walsh M. J., Brimacombe K. R., Anastasiou D., Yu Y., Israelsen W. J., Hong B.-S., Tempel W., Dimov S. and Veith H., in Probe Reports from the NIH Molecular Libraries Program [Internet], National Center for Biotechnology Information (US), 2013 [PubMed] [Google Scholar]
- Anastasiou D. Poulogiannis G. Asara J. M. Boxer M. B. Jiang J. Shen M. Bellinger G. Sasaki A. T. Locasale J. W. Auld D. S. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Nandi S. Dey M. Biochemistry. 2019;58:3669–3682. doi: 10.1021/acs.biochem.9b00349. [DOI] [PubMed] [Google Scholar]
- Zwerschke W. Mazurek S. Massimi P. Banks L. Eigenbrodt E. Jansen-Dürr P. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G. Locasale J. W. Swanson K. D. Sharfi H. Heffron G. J. Amador-Noguez D. Christofk H. R. Wagner G. Rabinowitz J. D. Asara J. M. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroki T. Yokoyama M. Tanuma N. Maejima R. Tamai K. Yamaguchi K. Oikawa T. Noguchi T. Miura K. Fujiya T. Cancer Sci. 2017;108:931–940. doi: 10.1111/cas.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaja S. Payne A. J. Singh T. Ghuman J. K. Sieck E. G. Koulen P. J. Pharmacol. Toxicol. Methods. 2015;73:1–6. doi: 10.1016/j.vascn.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios A. M. Fiske B. P. Gui D. Y. Vander Heiden M. G. Mol. Cell. 2015;59:850–857. doi: 10.1016/j.molcel.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J. Holzgrabe U. MedChemComm. 2016;7:214–223. [Google Scholar]
- Davis B. J. Erlanson D. A. Bioorg. Med. Chem. Lett. 2013;23:2844–2852. doi: 10.1016/j.bmcl.2013.03.028. [DOI] [PubMed] [Google Scholar]
- Dahlin J. L. Walters M. A. Assay Drug Dev. Technol. 2016;14:168–174. doi: 10.1089/adt.2015.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin J. L. Inglese J. Walters M. A. Nat. Rev. Drug Discovery. 2015;14:279–294. doi: 10.1038/nrd4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin J. L. Walters M. A. Future Med. Chem. 2014;6:1265–1290. doi: 10.4155/fmc.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R. S. Simeonov A. Jadhav A. Eidam O. Mott B. T. Keiser M. J. McKerrow J. H. Maloney D. J. Irwin J. J. Shoichet B. K. J. Med. Chem. 2010;53:4891–4905. doi: 10.1021/jm100488w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N. Auld D. S. Inglese J. Curr. Opin. Chem. Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W. P. Namchuk M. Nat. Rev. Drug Discovery. 2003;2:259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- Baell J. B. Future Med. Chem. 2010;2:1529–1546. doi: 10.4155/fmc.10.237. [DOI] [PubMed] [Google Scholar]
- Czarna A. Beck B. Srivastava S. Popowicz G. M. Wolf S. Huang Y. Bista M. Holak T. A. Dömling A. Angew. Chem., Int. Ed. 2010;49:5352–5356. doi: 10.1002/anie.201001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer J. Schiebel J. Wulsdorf T. Grohe K. Najbauer E. E. Ehrmann F. R. Radeva N. Zitzer N. Linne U. Linser R. Angew. Chem., Int. Ed. 2017;56:1908–1913. doi: 10.1002/anie.201609824. [DOI] [PubMed] [Google Scholar]
- Hsieh I.-S. Gopula B. Chou C.-C. Wu H.-Y. Chang G.-D. Wu W.-J. Chang C.-S. Chu P.-C. Chen C. S. J. Med. Chem. 2019;62:8497–8510. doi: 10.1021/acs.jmedchem.9b00763. [DOI] [PubMed] [Google Scholar]
- Smith A. J. T. Zhang X. Leach A. G. Houk K. N. J. Med. Chem. 2009;52:225–233. doi: 10.1021/jm800498e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquicett C. Roman J. Hart C. M. Cancer Ther. 2008;6:25. [PMC free article] [PubMed] [Google Scholar]
- Clay C. E. Namen A. M. Atsumi G. Willingham M. C. High K. P. Kute T. E. Trimboli A. J. Fonteh A. N. Dawson P. A. Chilton F. H. Carcinogenesis. 1999;20:1905–1911. doi: 10.1093/carcin/20.10.1905. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G. Christofk H. R. Schuman E. Subtelny A. O. Sharfi H. Harlow E. E. Xian J. Cantley L. C. Biochem. Pharmacol. 2010;79:1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen J. C., Richard D. J. and Zask A., in Kinase Drug Discovery, Royal Society of Chemistry, 2011, pp. 161–217 [Google Scholar]
- Baell J. B. Holloway G. A. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Capuzzi S. J. Muratov E. N. Tropsha A. J. Chem. Inf. Model. 2017;57:417–427. doi: 10.1021/acs.jcim.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley L., Vander Heiden M. G. and Christofk H. R., AU Pat., AU2007281911B2, 2007
- Zhu W. Groh M. Haupenthal J. Hartmann R. W. Chem. – Eur. J. 2013;19:8397–8400. doi: 10.1002/chem.201301289. [DOI] [PubMed] [Google Scholar]
- Morgan H. P. McNae I. W. Nowicki M. W. Zhong W. Michels P. A. M. Auld D. S. Fothergill-Gilmore L. A. Walkinshaw M. D. J. Biol. Chem. 2011;286:31232–31240. doi: 10.1074/jbc.M110.212613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F. Kang X. Xie Q. Wu J. Exp. Ther. Med. 2019;17:63–70. doi: 10.3892/etm.2018.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada N. Takasawa R. Tanuma S. Arch. Biochem. Biophys. 2018;638:1–7. doi: 10.1016/j.abb.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Lu J. Chen M. Gao S. Yuan J. Zhu Z. Zou X. Oncol. Lett. 2018;15:4358–4364. doi: 10.3892/ol.2018.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. Hao M. Med. Sci. Monit. 2017;23:2017–2028. doi: 10.12659/MSM.901542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X. Sun Y. Xu Y. Chen Z. Lu S. Med. Chem. 2016;12:613–620. doi: 10.2174/1573406412666160307151535. [DOI] [PubMed] [Google Scholar]
- Zhang H.-S. Zhang F.-J. Li H. Liu Y. Du G.-Y. Huang Y.-H. Arch. Biochem. Biophys. 2016;598:50–56. doi: 10.1016/j.abb.2016.03.031. [DOI] [PubMed] [Google Scholar]
- Jiang K. He B. Lai L. Chen Q. Liu Y. Guo Q. Wang Q. Int. J. Mol. Med. 2012;30:302–308. doi: 10.3892/ijmm.2012.989. [DOI] [PubMed] [Google Scholar]
- Shen Y. Chen M. Huang S. Zou X. Oncol. Lett. 2016;11:717–722. doi: 10.3892/ol.2015.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ibarbia L. Majdanski T. Schubert S. Windhab N. Schubert U. S. Eur. J. Pharm. Sci. 2016;93:264–273. doi: 10.1016/j.ejps.2016.08.026. [DOI] [PubMed] [Google Scholar]
- Crochet R. B., Insights into the Development of Chemotherapeutics targeting PFKFB Enzymes, PhD thesis, Louisiana State university, 2015 [Google Scholar]
- Li Q. Qi X. Jia W. Biochem. Biophys. Res. Commun. 2016;475:51–56. doi: 10.1016/j.bbrc.2016.05.030. [DOI] [PubMed] [Google Scholar]
- Cheng K. Hao M. Int. J. Mol. Sci. 2016;17:2000. doi: 10.3390/ijms17122000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea T. I. Bauman J. E. Bologa C. G. Buranda T. Chigaev A. Edwards B. S. Jarvik J. W. Gresham H. D. Haynes M. K. Hjelle B. Drug Discovery Today: Ther. Strategies. 2011;8:61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri A. Palumbo F. Rasi I. Posca D. Pavlidou T. Paoluzi S. Castagnoli L. Cesareni G. PLoS One. 2015;10:e0136250. doi: 10.1371/journal.pone.0136250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porporato P. E. Dadhich R. K. Dhup S. Copetti T. Sonveaux P. Front. Pharmacol. 2011;2:49. doi: 10.3389/fphar.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar Babu M. Mahanta S. Lakhter A. J. Hato T. Paul S. Naidu S. R. PLoS One. 2018;13:e0191419. doi: 10.1371/journal.pone.0191419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R. Li Y. Tang P. Yuan Q. Eur. J. Med. Chem. 2018;143:320–333. doi: 10.1016/j.ejmech.2017.08.041. [DOI] [PubMed] [Google Scholar]
- Boulos J. C. Rahama M. Hegazy M.-E. F. Efferth T. Cancer Lett. 2019;459:248–267. doi: 10.1016/j.canlet.2019.04.033. [DOI] [PubMed] [Google Scholar]
- Ning X. Qi H. Li R. Li Y. Jin Y. McNutt M. A. Liu J. Yin Y. Eur. J. Med. Chem. 2017;138:343–352. doi: 10.1016/j.ejmech.2017.06.064. [DOI] [PubMed] [Google Scholar]
- Ning X. Li Y. Qi H. Li R. Jin Y. Liu J. Yin Y. MedChemComm. 2018;9:632–638. doi: 10.1039/c8md00062j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. Li J. Xue J. Li H. Xu F. Cheng K. Li D. Li Z. Gao M. Hua H. Eur. J. Med. Chem. 2017;135:270–281. doi: 10.1016/j.ejmech.2017.03.020. [DOI] [PubMed] [Google Scholar]
- You L. Zhu H. Wang C. Wang F. Li Y. Li Y. Wang Y. He B. Bioorg. Med. Chem. Lett. 2017;27:5404–5408. doi: 10.1016/j.bmcl.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Manigandan K. Manimaran D. Jayaraj R. L. Elangovan N. Dhivya V. Kaphle A. Biochimie. 2015;119:103–112. doi: 10.1016/j.biochi.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Rinaldo D. Batista Jr J. M. Rodrigues J. Benfatti A. C. Rodrigues C. M. Dos Santos L. C. Furlan M. Vilegas W. Chirality. 2010;22:726–733. doi: 10.1002/chir.20824. [DOI] [PubMed] [Google Scholar]
- Aslan E. Adem S. J. Biochem. Mol. Toxicol. 2015;29:109–113. doi: 10.1002/jbt.21673. [DOI] [PubMed] [Google Scholar]
- Shan S. Shi J. Yang P. Jia B. Wu H. Zhang X. Li Z. J. Agric. Food Chem. 2017;65:8136–8144. doi: 10.1021/acs.jafc.7b02757. [DOI] [PubMed] [Google Scholar]
- Francioso A. Mastromarino P. Masci A. d'Erme M. Mosca L. Med. Chem. 2014;10:237–245. doi: 10.2174/15734064113096660053. [DOI] [PubMed] [Google Scholar]
- Sęczyk Ł. Świeca M. Kapusta I. Gawlik-Dziki U. Molecules. 2019;24:408. doi: 10.3390/molecules24030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe A. James M. J. Lee V. M.-Y. Smith A. B. Trojanowski J. Q. Ballatore C. Brunden K. R. J. Biol. Chem. 2013;288:11024–11037. doi: 10.1074/jbc.M112.436006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heleno S. A. Martins A. Queiroz M. J. R. P. Ferreira I. C. F. R. Food Chem. 2015;173:501–513. doi: 10.1016/j.foodchem.2014.10.057. [DOI] [PubMed] [Google Scholar]
- Yuan M. McNae I. W. Chen Y. Blackburn E. A. Wear M. A. Michels P. A. M. Fothergill-Gilmore L. A. Hupp T. Walkinshaw M. D. Biochem. J. 2018;475:1821–1837. doi: 10.1042/BCJ20180171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. Holyoak T. McDonald G. Gui C. Fenton A. W. Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- Mitchell A. R. Yuan M. Morgan H. P. McNae I. W. Blackburn E. A. Le Bihan T. Homem R. A. Yu M. Loake G. J. Michels P. A. Biochem. J. 2018;475:3275–3291. doi: 10.1042/BCJ20180556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarillo J. M. Ullery J. C. Rose K. L. Marnett L. J. Chem. Res. Toxicol. 2017;30:635–641. doi: 10.1021/acs.chemrestox.6b00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adem S. Comakli V. Uzun N. Expert Opin. Ther. Pat. 2018;28:61–68. doi: 10.1080/13543776.2018.1391218. [DOI] [PubMed] [Google Scholar]
- Smith B. R., Chan T.-H. and Leyland-Jones B., US Pat., US8026278B2, 2006 [Google Scholar]
- Srivastava G. Matta A. Fu G. Somasundaram R. T. Datti A. Walfish P. G. Ralhan R. Mol. Oncol. 2015;9:1720–1735. doi: 10.1016/j.molonc.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrauckas J. D. Santarsiero B. D. Mesecar A. D. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Zaccaro C., Evaluation of Tumor M2 Pyruvate Kinase and Endocannabinoid System Expression in Colorectal Preneoplastic and Neoplastic Lesions: Possible use of non-invasive Diagnosis, PhD thesis, Alma Mater Studiorum-Universita di Bologna, 2016, 10.6092/unibo/amsdottorato/7357 [DOI] [Google Scholar]
- Morgan H. P. O'Reilly F. J. Wear M. A. O'Neill J. R. Fothergill-Gilmore L. A. Hupp T. Walkinshaw M. D. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5881–5886. doi: 10.1073/pnas.1217157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C. Hixon J. Choe S. Marks K. Gross S. Murphy E. DeLaBarre B. Cianchetta G. Sethumadhavan S. Wang X. Chem. Biol. 2012;19:1187–1198. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer M. B. Jiang J. Vander Heiden M. G. Shen M. Skoumbourdis A. P. Southall N. Veith H. Leister W. Austin C. P. Park H. W. J. Med. Chem. 2010;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Liu X.-H. Saunders M. Pearce S. Foulks J. M. Parnell K. M. Clifford A. Nix R. N. Bullough J. Hendrickson T. F. Bioorg. Med. Chem. Lett. 2014;24:515–519. doi: 10.1016/j.bmcl.2013.12.028. [DOI] [PubMed] [Google Scholar]
- Macpherson J. A. Theisen A. Masino L. Fets L. Driscoll P. C. Encheva V. Snijders A. P. Martin S. R. Kleinjung J. Barran P. E. eLife. 2019;8:e45068. doi: 10.7554/eLife.45068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S. Dey M. J. Biol. Chem. 2020;295:5390–5403. doi: 10.1074/jbc.RA120.013030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiari S. Runtsch M. C. Sutton C. E. Palsson-McDermott E. M. Kelly B. Rana N. Kane H. Papadopoulou G. Pearce E. L. Mills K. H. G. Cell Metab. 2020;31:391–405. doi: 10.1016/j.cmet.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Y. Z. Levy D. J. Barto A. G. Meyer J. S. Front. Mol. Psychiatry. 2013;4:167. doi: 10.3389/fpsyt.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Li S. Guo J. Li Q. Long J. Ma C. Ding Y. Yan C. Li L. Wu Z. J. Med. Chem. 2018;61:4155–4164. doi: 10.1021/acs.jmedchem.8b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. Xue Q. Liu S. Hu K. Wang D. Wang T. Li Y. Guo H. Hao X. Ge W. J. Med. Chem. 2020;63:1597–1611. doi: 10.1021/acs.jmedchem.9b01328. [DOI] [PubMed] [Google Scholar]
- Guo C. Linton A. Jalaie M. Kephart S. Ornelas M. Pairish M. Greasley S. Richardson P. Maegley K. Hickey M. Bioorg. Med. Chem. Lett. 2013;23:3358–3363. doi: 10.1016/j.bmcl.2013.03.090. [DOI] [PubMed] [Google Scholar]
- Granchi C. Minutolo F. ChemMedChem. 2012;7:1318–1350. doi: 10.1002/cmdc.201200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-jain A., Peterson P. W., Whatcott C. J., Bearss D. J. and Warner S. L., US Pat., US20200237766A1, 2018
- Warner S. L. Carpenter K. J. Bearss D. J. Future Med. Chem. 2014;6:1167–1178. doi: 10.4155/fmc.14.70. [DOI] [PubMed] [Google Scholar]
- Liu B. Yuan X. Xu B. Zhang H. Li R. Wang X. Ge Z. Li R. Eur. J. Med. Chem. 2019;170:1–15. doi: 10.1016/j.ejmech.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Liu B. Wu X. Li R. Ning X. Liu Y. Liu Z. Ge Z. Li R. Yin Y. Bioorg. Med. Chem. 2015;23:4815–4823. doi: 10.1016/j.bmc.2015.05.041. [DOI] [PubMed] [Google Scholar]
- Jiang H. Zhang S. Song T. Guan X. Zhang R. Chen X. Front. Pharmacol. 2018;9:612. doi: 10.3389/fphar.2018.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y. Yasumatsu I. Asahi T. Kitamura T. Kanai K. Ubukata O. Hayasaka H. Takaishi S. Hanzawa H. Katakura S. Bioorg. Med. Chem. 2017;25:3540–3546. doi: 10.1016/j.bmc.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Aslan E. Guler C. Adem S. J. Enzyme Inhib. Med. Chem. 2016;31:314–317. doi: 10.3109/14756366.2015.1022173. [DOI] [PubMed] [Google Scholar]
- Chen C. Oxid. Med. Cell. Longevity. 2016;2016:3571614. doi: 10.1155/2016/3571614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott E. M. Curtis A. M. Goel G. Lauterbach M. A. R. Sheedy F. J. Gleeson L. E. van den Bosch M. W. M. Quinn S. R. Domingo-Fernandez R. Johnston D. G. W. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Lu Z. Cancer Lett. 2013;339:153–158. doi: 10.1016/j.canlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Ning X. Zhou S. Lin Z. Wu X. Chen H. Bai X. Wang X. Ge Z. Li R. Eur. J. Med. Chem. 2018;143:48–65. doi: 10.1016/j.ejmech.2017.11.023. [DOI] [PubMed] [Google Scholar]
- Li R. Fan X. Shi D. Zhu G. Wang Y. Luo L. Pan H. Yao X. Leung E. L. Liu L. Chem. Biol. Drug Des. 2018;92:1851–1858. doi: 10.1111/cbdd.13354. [DOI] [PubMed] [Google Scholar]
- Davidoff F. Carr S. Biochemistry. 1973;12:1415–1422. doi: 10.1021/bi00731a023. [DOI] [PubMed] [Google Scholar]
- Li Q. Cao L. Tian Y. Zhang P. Ding C. Lu W. Jia C. Shao C. Liu W. Wang D. Mol. Cell. Proteomics. 2018;17:1531–1545. doi: 10.1074/mcp.RA118.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Filho J. C. Pålsson-McDermott E. M. Front. Immunol. 2016;7:145. doi: 10.3389/fimmu.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W. Keenan H. A. Li Q. Ishikado A. Kannt A. Sadowski T. Yorek M. A. Wu I.-H. Lockhart S. Coppey L. J. Nat. Med. 2017;23:753. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger M. R. Fraga C. A. M. Dantas R. F. Silva Jr F. P. Drug Discovery Today. 2016;21:868–872. doi: 10.1016/j.drudis.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Meanwell N. A. J. Med. Chem. 2011;54:2529–2591. doi: 10.1021/jm1013693. [DOI] [PubMed] [Google Scholar]